Abstract
Background
Colorectal cancer (CRC) is one of the most frequently diagnosed tumors worldwide with high mortality and morbidity. There is an urgent need for biomarkers to improve the outcomes and early detection of CRC. The sensitivity of traditional CRC tumor markers (carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9)) is not ideal. The levels of leucine-rich-alpha-2-glycoprotein 1 (LRG1) and stem cell factor (SCF) were evaluated, but the combined value of both markers is unclear. This case-control study included four groups: CRC patients before treatments (n = 22), CRC patients after treatments (n = 26), 20 patients with benign tumor, and 20 healthy subjects. Levels of routine biochemical and hematological markers, traditional tumor markers (CA19.9 and CEA), and candidate markers (LRG1 and SCF) were determined. Univariate and multivariate logistic regression analysis and area receiver-operating characteristic analysis (ROC) were used for evaluation the diagnostic performances of single and combined markers.
Results
No significance difference in traditional tumor markers CEA, CA 19.9, and neutrophil–lymphocyte ratio (NLR) were found among study groups. SCF, LRG1, and platelet–lymphocyte ratio (PLR) were significantly decreased (p < 0.05) in non-treated CRC patients than after treated CRC. The combination between SCF and LRG1 showed highly significant difference in CRC patients compared with benign, healthy subjects, and among CRC groups (treated and non-treated) (p < 0.0001). The highest areas under curve (AUCs) were observed when LRG1 was used as a single predictor for discriminating CRC from healthy (0.87), benign (0.84), and non-treated CRC vs treated CRC (0.82). AUCs were jumped to 0.90, 0.84, and 0.84 when LRG1 and SCF were combined.
Conclusion
Our study revealed that LRG1 and SCF were potential diagnostic and follow-up markers for CRC.
Impact statement
Colorectal cancer is a major health problem worldwide. The current work aimed to evaluate the diagnostic performances of CEA, CA19-9, LRG1, and SCF as single and combined markers.
Background
Worldwide, CRC is the third most commonly detected cancer, and being generally symptomatic, it is often detected in advanced stage of growth [1]. The most reliable method for the CRC screening is endoscope, but it physically burdens and has high cost. Thus, alternative, noninvasive markers have been evaluated and validated to improve early detection and treatment outcome of CRC [2]. In addition, the blood examinations are accessible and easily performed, but the precision to detect the CRC in early stages is restricted. The routine tumor markers (CEA and CA 19–9) are raised in malignancies and benign diseases and have unsatisfactory sensitivity and specificity [3]. LRG is a plasma glycoprotein elevated in the acute-phase response to bacterial or viral infection [4, 5]. LRG1 has several biological functions including cell migration, cell proliferation, cell apoptosis, immune response and neovascularization, and malignancy [5, 6]. Stem cell factor is one of the hematopoietic cytokines (HC) which induce hematopoietic progenitor cell proliferation. Receptor of HC has been established on non-hematopoietic cancer cell lines including CRC [7]. Clinical prospective studies open the door for non-invasive biomarkers which assist in monitoring therapy for CRC patients. CEA levels should be detected before starting in chemotherapy and every 2 to 3 months [8]. In our study, we aimed to evaluate diagnostic performances of traditional CRC tumor markers (CEA, CA19.9) vs LRG1 and SCF for early CRC diagnosis and assess its potential usefulness for monitoring CRC patient’s treatment.
Methods
Patients
Sixty-eight patients were recruited from Egyptian National Cancer institute. Patients were divided into four groups: CRC patients before treatments (n = 22), CRC patients after treatments (n = 26) with histologically affirmed adenocarcinoma of colon or rectum cancer, 20 patients with benign tumor (includes hyperplastic or adenomatous polyps), and 20 healthy subjects. Treatment strategies included chemotherapy, radiotherapy, concurrent chemo and/or radiotherapy, or surgery. Clinical data of patients including age, sex, tumor histology, stage, and treatment strategies were collected from medical records. Pretreatment staging included the following: physical and blood examinations (CEA, CA 19.9, NLR and PLR), computed tomography (CT), chest roentgenogram, abdominal ultrasound scanning, and colonoscopy. TNM (tumor, node, and metastasis) classification system for colorectal cancer was utilized to play out the pathological staging of the study [9].
Biochemical analyses
Blood samples were assembled from all individuals after clinical determination and blood samples were inspected for routine laboratory examinations, including biochemical profile [liver function tests] and [kidney function tests] using an automated biochemistry analyzer (Beckman Coulter AU680 chemistry analyzer, USA). Complete blood count was estimated using an automated hematology analyzer (Sysmex XN 1000, Japan). Serum CA19.9 and CEA were estimated by ELISA assay according to the manufacturer’s protocol (ARCHITECT i 1000 SR immunoassay analyzer, Abbott USA). LRG1 (Assay Pro/Human LRG1, MO, USA) and SCF Glory Science Co. Ltd/Human SCF/Del Rio, TX, USA) were determined by immunoassay according to the manufacturer’s protocol.
Statistical analyses
Statistical Package for the Social Sciences (SPSS) operating system, version 22.0 (SPSS Inc., Chicago, IL), was utilized to operate the statistical analysis information. Continuous data was presented as mean ± standard deviation (SD). Statistically significant differences were determined using Kruskal-Wallis, ANOVA, Mann-Whitney U test, and Student’s t test. Univariate and multivariate logistic regression analysis and AUC were used for evaluation the diagnostic performances of routine and candidate tumor markers. Platelet count-to-lymphocyte ratio (PLR) was calculated as the platelet count divided by the lymphocyte count. Neutrophil-to-lymphocyte ratio (NLR) was calculated as the neutrophil count divided by the lymphocyte count. The best cutoff points and diagnostic performances were determined based on receiver-operating characteristic analysis (ROC). The best combination was developed using the logistic regression function that combined the most discriminatory independent factors. The combination can be represented as = (1.445 + LRG (μg/ml) × 0.038 + SCF (ng/ml) × 0.001).
Results
Baseline clinical features of study groups
Baseline clinical features are listed in Table 1; 48 patients (24 men and 24 women) with diagnosis of CRC including 22 non-treated CRC and 26 treated CRC, 20 with benign (13 men and 7 women), and 20 healthy subjects (14 men and 6 women) were enrolled into this study. Levels of routine laboratory parameters show non- significant difference between CRC and benign compared with healthy subjects.
Table 1.
Clinicopathologic parameters of patients with colorectal cancer and control characteristics
| Parameters | Value (%) |
|---|---|
| Cancer patients no. | |
| No. of patients | 48 |
| Treated CRC | 26 (54.2%) |
| Non-treated CRC | 22 (45.8%) |
| Mean age, years (range) | 48 (28-81) |
| Cancer position, no. | |
| Colon | 28 (58.3%) |
| Rectal | 20 (41.7%) |
| Tumor size | |
| T1 | 7 (14.6%) |
| T2 | 19 (39.6%) |
| T3 | 10 (20.8%) |
| Unknown | 12 (25%) |
| Tumor grade, no. | |
| G1 | 2 (4.2%) |
| G2 | 26 (54.2%) |
| G3 | 9 (18.7%) |
| Unknown | 11 (22.9%) |
| Lymph node status, no. | |
| Positive | 6 (12.5%) |
| Negative | 30 (62.5%) |
| Unknown | 12 (25%) |
| Metastasis | |
| Positive | 3 (6.25%) |
| Negative | 33 (68.75%) |
| Unknown | 12 (25%) |
| Stage, no. | |
| I | 23 (47.9%) |
| II | 7 (14.6%) |
| III | 3 (6.25%) |
| IV | 3 (6.25%) |
| Unknown | 12 (25%) |
| Benign cancer diseases | |
| No. of patients | 20 |
| Mean age, years (range) | 51 (27-73) |
| Healthy controls | |
| No. of cases | 20 |
| Mean age, years | 34 (23-61) |
Levels of biomarkers in study groups
The levels of CEA, CA19.9, PLR, and SCF and LRG1 in benign patients, CRC (treated and non-treated), and healthy subjects are shown in Table 2. The levels of SCF and LRG1 were significantly increased in patients with CRC than in benign (p = 0.001) and healthy subjects (p = 0.001). No significant difference in traditional markers CEA, CA19.9, and PLR among study groups. SCF, LRG1, and PLR were significantly decreased (p values raged from 0.014 to 0.001) in non-treated CRC patients than treated CRC. The combination between SCF and LRG1showed highly significant difference in CRC compared with benign and healthy subjects and among CRC groups (treated and non-treated) (p value ranged from 0.001 to < 0.0001).
Table 2.
Levels of traditional and candidate markers in studied groups
| Study groups | CEA (ng/ml) | CA19.9 (μ/ml) | PLR | SCF (ng/ml) | LRG1 (μg/ml) | SCF-LRG1 (μg/ml) |
|---|---|---|---|---|---|---|
| Healthy subjects | 43.1 ± 14.0 | 98.1 ± 32.2 | 4.8 ± 2.2 | 231.9 ± 33.2 | 39.6 ± 16.3 | 3.1 ± 0.64 |
| Benign | 88.8 ± 21.8 | 118.7 ± 37.5 | 5.2 ± 2.3 | 440.1 ± 35.4 | 44.6 ± 8.7 | 3.6 ± 0.38 |
| CRC | 47.7 ± 13.5 | 108.5 ± 34.2 | 5.0 ± 2.2 | 605.7 ± 38.5 | 52.5 ± 16.9 | 4.6 ± 1.3 |
| p value a | 0.349 | 0.216 | 0.025 | 0.001 | 0.0001 | 0.0001 |
| Non-treated CRC | 40.1 ± 33.7 | 155.9 ± 49.3 | 6.2 ± 2.1 | 1185.3 ± 39.5 | 69.3 ± 12.0 | 5.3 ± 1.6 |
| Treated CRC | 26.0 ± 22.2 | 68.6 ± 48.3 | 4.2 ± 1.9 | 530.2 ± 39.1 | 54.2 ± 13.2 | 4.0 ± 0.59 |
| p valueb | 0.978 | 0.167 | 0.014 | 0.019 | 0.001 | 0.001 |
aFor discriminating among studied groups
bFor discriminating between non-treated and treated CRC groups
Colon cancer patients had significant high level of SCF compared with rectum cancer (p = 0.039) whereas LRG1 levels were significantly elevated in high grade G3 than low grade of CRC (p = 0.05). Level of SCF was highly significantly higher in high depth T3 than T2 and T1 (p = 0.001), while LRG1 level was significantly higher in T1 than high depth (p = 0.032). Levels of CEA, CA19.9, PLR, SCF, and LRG1 had non-significant difference in lymph nodes and presence or absence of metastasis in CRC patients and early or late tumor stage (p ≥ 0.05).
The diagnostic power of single and combined markers
ROC analysis was performed to evaluate the discriminatory power of single or combined markers to differentiate patients with CRC from benign and healthy individuals (Table 3). The highest discriminatory AUC was observed when LRG1was used as a single marker with AUC value of 0.87 which jumped to 0.90 when LRG1 and SCF were combined in discriminating CRC from healthy subjects (Fig. 1a). LRG1 recorded the highest efficiency, specificity, positive prediction, and AUC value (66%, 95% and 96%, 0.84, respectively) as an individual marker for discriminating CRC from benign tumors, while AUC was not improved when LRG1 combined with SCF (Fig. 1b). LRG1 had an excellent ability to monitor the treatment (AUC = 0.82) with highest efficiency of 73%. Combined LRG1 and SCF increased AUC to 0.84 for discriminating treated CRC patients from non-treated (Fig. 1c).
Table 3.
Diagnostic performances of single and combined markers
| Variables | AUC | Cut-off | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Efficiency (%) |
|---|---|---|---|---|---|---|---|
| Healthy vs CRC | |||||||
| CEA ng/ml | 0.37 | 2.8 | 79 | 10 | 68 | 17 | 59 |
| CA 19.9 μ/ml | 0.41 | 23.4 | 77 | 10 | 67 | 15 | 57 |
| PLR | 0.73 | 4.9 | 47 | 65 | 76 | 34 | 52 |
| SCF ng/ml | 0.80 | 539 | 58 | 90 | 93 | 47% | 63 |
| LRG1 μg/ml | 0.87 | 60 | 52 | 90 | 93 | 44 | 68 |
| SCF-LRG1 | 0.9 | 3.6 | 85 | 80 | 91 | 70 | 84 |
| CRC vs Benign | |||||||
| CEA ng/ml | 0.44 | 2.8 | 79 | 20 | 70 | 29 | 62 |
| CA 19.9 μ/ml | 0.50 | 23.4 | 77 | 30 | 73 | 35 | 63 |
| PLR | 0.48 | 4.9 | 45 | 60 | 72 | 32 | 49 |
| SCF ng/ml | 0.72 | 539 | 58 | 70 | 82 | 41 | 61 |
| LRG1 μg/ml | 0.84 | 60 | 54 | 95 | 96 | 46 | 66 |
| SCF-LRG1 | 0.84 | 3.6 | 79 | 70 | 86 | 58 | 77 |
| Treated CRC vs non-treated CRC | |||||||
| CEA ng/ml | 0.59 | 2.8 | 82 | 23 | 47 | 60 | 50 |
| CA 19.9 μ/ml | 0.64 | 23.4 | 82 | 27 | 49 | 64 | 52 |
| PLR | 0.76 | 4.55 | 71 | 69 | 65 | 75 | 70 |
| SCF ng/ml | 0.68 | 539 | 73 | 54 | 57 | 70 | 63 |
| LRG1 μg/ml | 0.82 | 61.5 | 82 | 73 | 72 | 83 | 73 |
| SCF-LRG1 | 0.84 | 3.6 | 77 | 70 | 68 | 78 | 77 |
Fig. 1.
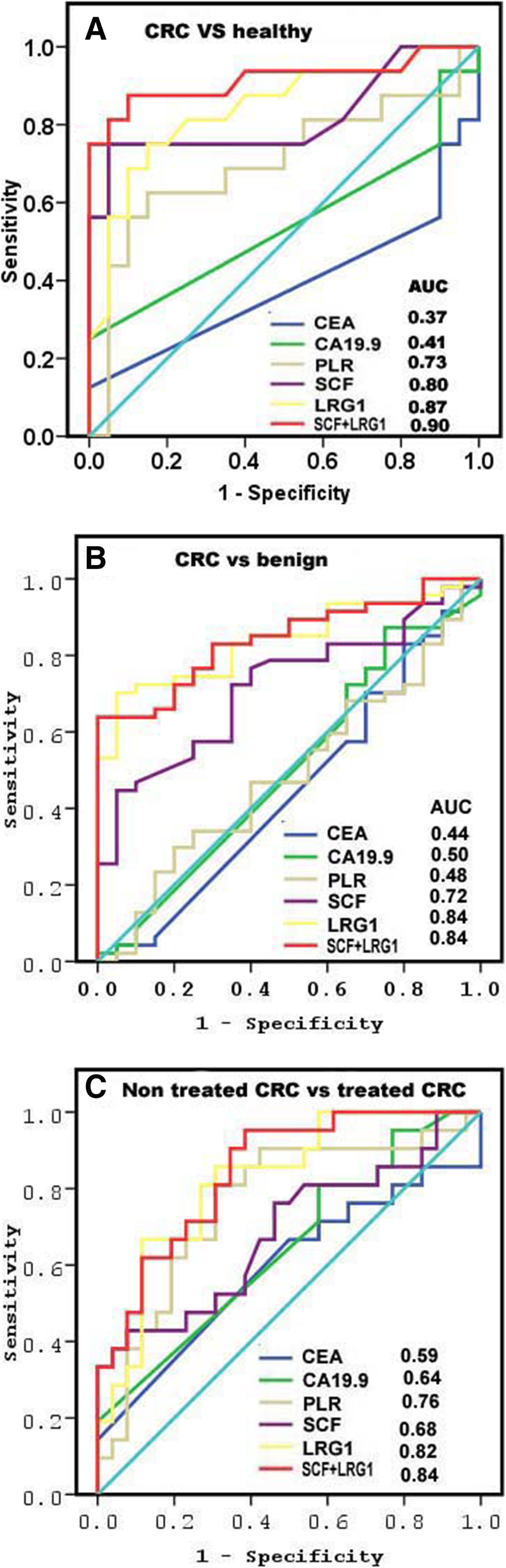
Diagnostic power of single and combined markers. a CRC from healthy individuals. b CRC from benign. c Treated CRC from non-treated CRC patients
Discussion
CRC is one of the main causes of death worldwide, so it is very imperative to early detect CRC. Levels of CEA and CA 19–9 were raised in malignancies and benign diseases and are not specific to malignancies only [10]. The sensitivity and specificity of traditional markers (CEA and CA 19–9) were limited in differentiation malignant from benign disease. We evaluated the diagnostic power of LRG1, SCF, PRL, CEA, and CA 19–9 for early detection of CRC. Our study was the first one that estimated the levels of SCF and LRG1 in CRC patients. Our results demonstrated that SCF and LRG1 levels were significantly increased in patients with CRC compared with benign tumors and healthy subjects. High level of LRG has been attended related to various diseases, such as inflammation and cancer [11, 12]. LRG was informed to show critical roles in CRC tumorigenesis [13] and could be considered as a diagnostic tumor marker [14, 15]. Inflammation response is associated not only to tumorigenicity but also to tumor progression. Inflammatory response plays a dual role in tumorigenicity due to its induced local accumulation of different types of white blood cells that secrete cytokines to stimulate tumor angiogenesis and metastasis, and the rise of monocytes and lymphocytes counts creates a resistance to tumor invasion. Tumor progression is induced by the production of inflammatory cytokines and chemokines by white blood cells, which are themselves stimulated by the tumors [16]. Elevated level of several circulating cytokines was detected in patients with CRC and other malignancies diseases [17, 18]. In the present study, the SCF and LRG1 levels were significantly decreased in treated CRC patients than non-treated CRC patients, while the level of CEA and CA 19–9 had no significant difference in treated CRC compared with non-treated CRC individuals. During treatment of CRC, CEA and CA19-9 traditional tumor markers were undergoing dynamic changes [19, 20]. Our results disagreed with the results of Perez et al. [21]. The levels of CEA were decreased after CRC therapy. Our levels of LRG1 and SCF were correlated with clinicopathological characters and agreed with Kaminska et al. [22]. Increased hematopoietic cytokines correlated with clinicopathological features, and it may have diagnostic value [23].
In the present study, LRG1 has the highest discriminatory power as a single predictor for discriminating CRC from healthy subjects with an AUC value of 0.87, which jumped to 0.90 when SCF and LRG1 were combined. For discriminating patients with CRC from benign, LRG1 has the highest AUC of 0.84 which improved to 0.89 when LRG1 and SCF combined. LRG1 had a great ability to monitor the treatment of CRC with an AUC value = 0.82. Combination of different biomarkers may enhance the diagnostic power and would be used for diagnosis of cancers. The specificity, sensitivity, and AUC of combined CEA and LRG-FT were higher than each marker alone [22]. The AUC of SCF was higher than the IL-3 but less than the AUCs of CEA and CA 19–9 [24]. Mroczko et al. [18] reported that combination of SCF with granulocyte-macrophage colony-stimulating factor (GM-CSF) had greatest diagnostic sensitivity, but GM-CSF and CEA combination had the highest specificity and positive and the negative predictive values. The AUC for osteopontin (OPN), B7-H4, tissue polypeptide-specific antigen, and TPS were 0.81, 0.86, 0.83, and 0.81, respectively. The sensitivity and specificity of B7-H4 for discriminating CRC patients from healthy controls were 88.2% and 86.7%, respectively. However, combination of CEA and B7-H4 had AUC with 0.93, 80% specificity, and 99% sensitivity to differentiate between CRP patients and healthy controls [25]. AUC of miR-125a-3p was 0.69 and 0.84 for CEA for differentiating CRC patients from healthy. Combined CEA and miR-125a-3P enhanced the AUC to 0.86% [26]. AUC of CEA was 0.86 followed by CA 19–9, CYFRA 21-1, IL-8, CA 125, and OPN reaching AUCs between 0.70 and 0.74. AUC of CEA and CA 19–9 increased to 0.89 [27]. This study had two limits. First, it included a small population size and the second limit was including cancer rectum with colon cancer. In the coming research papers, we will increase the population size and include cancer rectum as a separate disease group.
Conclusions
LRG1 had the highest AUC for discriminating patients with CRC from healthy and benign individuals and treated CRC from non-treated CRC. LRG1 combined with SCF could be considered a promising candidate marker for CRC diagnosis and follow-up.
Acknowledgements
Not applicable
Abbreviations
- AUC
Area under the curve
- CA19-9
Carbohydrate antigen 19-9
- CEA
Carcinoembryonic antigen
- CT
Computed tomography
- CRC
Colorectal cancer
- HC
Hematopoietic cytokines
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- LRG1
Leucine-rich-alpha-2-glycoprotein 1
- OPN
Osteopontin
- NLR
Neutrophil–lymphocyte ratio
- NPV
Negative predictive value
- PPV
Positive predictive value
- PLR
Platelet–lymphocyte ratio
- ROC
Receiver-operating characteristic analysis
- SCF
Stem cell factor
- SD
Standard deviation
- SPSS
Statistical Package for the Social Sciences
- TNM
Tumor, node, and metastasis
Authors’ contributions
OM, MF, and AK conceived and designed the experiments. OM, MF, and AM contributed to the analysis and/or interpretation of data. OM, MF, and AM drafted the manuscript. OM and MF revised it critically for important intellectual content. All authors have read the manuscript and approved the submission.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The authors declare that all generated and analyzed data are included in the article.
Ethics approval and consent to participate
The Ethics and Scientific Committees of the National Cancer Institute approved this study; the research approval code is 2018190314. Biochemical studies on biomarkers for colorectal cancer. Informed written consent was signed by all patients in compliance with the ethical guidelines of the 1975 Helsinki Declaration.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Manar S. Fouda, Email: m.fouda6980@gmail.com
Rokaia M. Aljarwani, Email: rokaia.aljarwani@science.helwan.edu.eg
Khaled Aboul-Enein, Email: kenein@nci.cu.edu.eg.
Mohamed M. Omran, Email: drmmomran@science.helwan.edu.eg, Email: drmmomran@yahoo.com
References
- 1.Araghi M, Soerjomataram I, Jenkins M, Brierley J, Morris E. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer. 2019;144:2992–3000. doi: 10.1002/ijc.32055. [DOI] [PubMed] [Google Scholar]
- 2.Attallah AM, El-Far M, Ibrahim AR, El-Desouky MA. Clinical value of a diagnostic score for colon cancer based on serum CEA, CA19-9, cytokeratin-1 and mucin-1. Br J Biomed Sci. 2018;75:122–7. doi: 10.1080/09674845.2018.1456309. [DOI] [PubMed] [Google Scholar]
- 3.Yiu AJ, Yiu CY. Biomarkers in colorectal cancer. Anticancer Res. 2016;36:1093–1102. [PubMed] [Google Scholar]
- 4.Haupt H, Baudner S. Isolation and characterization of an unknown, leucine-rich 3.1-S-alpha2-glycoprotein from human serum. Hoppe-Seyler’s Zeitschrift fur physiologische Chemie. 1977;358:639–646. doi: 10.1515/bchm2.1977.358.1.639. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Abraham S, McKenzie JAG, Jeffs N, Swire M, Tripathi VB, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature. 2013;499:306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong D, Zhao S, He G, Li J, Lang Y, Ye W, et al. Stable knockdown of LRG1 by RNA interference inhibits growth and promotes apoptosis of glioblastoma cells in vitro and in vivo. Tumour Biol. 2015;36:4271–4278. doi: 10.1007/s13277-015-3065-3. [DOI] [PubMed] [Google Scholar]
- 7.Smith MA, Court EL, Smith JG. Stem cell factor: laboratory and clinical aspects. Blood Rev. 2001;15:191–197. doi: 10.1054/blre.2001.0167. [DOI] [PubMed] [Google Scholar]
- 8.Sturgeon CM, Duffy MJ, Stenman UH, Lilja H, Brünner N, Chan DW, et al. National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers. Clin Chem. 2008;54:e11–e79. doi: 10.1373/clinchem.2008.105601. [DOI] [PubMed] [Google Scholar]
- 9.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 10.Tong Y, Song Z, Zhu W. Study of an elevated carbohydrate antigen 19-9 concentration in a large health check-up cohort in China. Clin Chem Lab Med. 2013;51:1459–1466. doi: 10.1515/cclm-2012-0458. [DOI] [PubMed] [Google Scholar]
- 11.Weivoda S, Andersen JD, Skogen A, Schlievert PM, Fontana D, Schacker T, et al. ELISA for human serum leucine-rich alpha-2-glycoprotein-1 employing cytochrome c as the capturing ligand. J Immunol Methods. 2008;336:22–29. doi: 10.1016/j.jim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen JD, Boylan KL, Jemmerson R, Geller MA, Misemer B, Harrington KM, et al. Leucine-rich alpha-2-glycoprotein-1 is upregulated in sera and tumors of ovarian cancer patients. J Ovarian Res. 2010;3:21. doi: 10.1186/1757-2215-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JW, Liu H, Shin DH, Yu GI, Hwang JS, Kim ES, et al. Proteomic and cytokine plasma biomarkers for predicting progression from colorectal adenoma to carcinoma in human patients. Proteomics. 2013;13:2361–2374. doi: 10.1002/pmic.201200550. [DOI] [PubMed] [Google Scholar]
- 14.Wang CH, Li M, Liu LL, Zhou RY, Fu J, Zhang CZ, et al. LRG1 expression indicates unfavorable clinical outcome in hepatocellular carcinoma. Oncotarget. 2015;6:42118–42129. doi: 10.18632/oncotarget.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Luo Q, Wang N, Hu F, Jin H, Ge T, et al. LRG1 suppresses the migration and invasion of hepatocellular carcinoma cells. Med Oncol. 2015;32:146. doi: 10.1007/s12032-015-0598-7. [DOI] [PubMed] [Google Scholar]
- 16.Cao Y, Ke S, Gu J, Mao F, Yao S, Deng S, et al (2020) The Value of Haematological Parameters and Tumour Markers in the Prediction of Intestinal Obstruction in 1474 Chinese Colorectal Cancer Patients. Dis markers 2020:8860328 [DOI] [PMC free article] [PubMed]
- 17.Lahm H, Amstad P, Yilmaz A, Borbenyi Z, Wyniger J, Fischer JR, et al. Interleukin 4 down-regulates expression of c-kit and autocrine stem cell factor in human colorectal carcinoma cells. Cell Growth Differentiation. 1995;6:1111–1118. [PubMed] [Google Scholar]
- 18.Mroczko B, Szmitkowski M, Okulczyk B. Hematopoietic growth factors in colorectal cancer patients. Clin Chem Lab Med. 2003;41:646–651. doi: 10.1515/CCLM.2003.098. [DOI] [PubMed] [Google Scholar]
- 19.Gronlund B, Høgdall C, Hilden J, Engelholm SA, Høgdall EV, Hansen HH. Should CA-125 response criteria be preferred to response evaluation criteria in solid tumors (RECIST) for prognostication during second-line chemotherapy of ovarian carcinoma? J Clin Oncol. 2004;22:4051–4058. doi: 10.1200/JCO.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Petrioli R, Licchetta A, Roviello G, Pascucci A, Francini E, Bargagli G, et al. CEA and CA19.9 as early predictors of progression in advanced/metastatic colorectal cancer patients receiving oxaliplatin-based chemotherapy and bevacizumab. Cancer Invest. 2012;30:65–71. doi: 10.3109/07357907.2011.629380. [DOI] [PubMed] [Google Scholar]
- 21.Perez RO, São Julião GP, Habr-Gama A, Kiss D, Proscurshim I, Campos FG, et al. The role of carcinoembriogenic antigen in predicting response and survival to neoadjuvant chemoradiotherapy for distal rectal cancer. Dis Colon Rectum. 2009;52:1137–1143. doi: 10.1007/DCR.0b013e31819ef76b. [DOI] [PubMed] [Google Scholar]
- 22.Shinozaki E, Tanabe K, Akiyoshi T, Tsuchida T, Miyazaki Y, Kojima N, et al. Serum leucine-rich alpha-2-glycoprotein-1 with fucosylated triantennary N-glycan: a novel colorectal cancer marker. BMC Cancer. 2018;18:406. doi: 10.1186/s12885-018-4252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaminska J, Nowacki MP, Kowalska M, Rysinska A, Chwalinski M, Fuksiewicz M, et al. Clinical significance of serum cytokine measurements in untreated colorectal cancer patients: soluble tumor necrosis factor receptor type I--an independent prognostic factor. Tumour Biol. 2005;26:186–194. doi: 10.1159/000086951. [DOI] [PubMed] [Google Scholar]
- 24.Mroczko B, Szmitkowski M, Wereszczyńska-Siemiatkowska U, Okulczyk B. Stem cell factor (SCF) and interleukin 3 (IL-3) in the sera of patients with colorectal cancer. Dig Dis Sci. 2005;50:1019–1024. doi: 10.1007/s10620-005-2697-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Li C, Zhang F, Ma X, Gai X (2018) Clinical Value of Combined Determination of Serum B7-H4 with Carcinoembryonic Antigen, Osteopontin, or Tissue Polypeptide-Specific Antigen for the Diagnosis of Colorectal Cancer. Dis Markers 2018:4310790 [DOI] [PMC free article] [PubMed]
- 26.Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, et al. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7:4150. doi: 10.1038/s41598-017-04386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dressen K, Hermann N, Manekeller S, Walgenbach-Bruenagel G, Schildberg FA, Hettwer K, et al. Diagnostic performance of a novel multiplex immunoassay in colorectal cancer. Anticancer Res. 2017;37:2477–2486. doi: 10.21873/anticanres.11588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that all generated and analyzed data are included in the article.


