Abstract
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of polycomb repressive complex 2 and contains a SET domain that catalyzes histone H3 trimethylation on lysine 27 (H3K27me3) to generate an epigenetic silencing mark. EZH2 interacts with transcription factors or RNA transcripts to perform its function. In this study, we applied RNA immunoprecipitation sequencing and long intergenic non-coding RNA (lincRNA) sequencing methods to identify EZH2-binding lincRNAs. A total of 356 novel EZH2-binding lincRNAs were identified by bioinformatics analysis and an EZH2-binding lincRNA TCONS-00036665 was characterized. TCONS-00036665 promoted pig skeletal satellite cell proliferation but inhibited cell differentiation, and this function was conserved between pigs and mice. Further mechanistic studies indicated that TCONS-00036665 can bind to EZH2 and recruits EZH2 to the promoters of the target genes p21, MyoG, and Myh4, which leads to the enrichment of H3K27me3 and the repression of target gene expression and pig myogenesis. In conclusion, the lincRNA TCONS-00036665 regulates pig myogenesis through its interaction with EZH2.
Keywords: EZH2, lncRNA, pig, myogenesis, epigenetics
Introduction
Vertebrate skeletal muscle myogenesis is a complex process, which requires a variety of factors to work together. During embryonic development, most skeletal muscle cells originate from dermomyotome. They are termed myogenic progenitor cells and are characterized by high expression of the paired-box transcription factors Pax3 and Pax7 (Hindi et al., 2013), and migrate to the limbs and trunk. Pax3 is necessary for the formation of limb muscles, affecting either the generation of myogenic progenitor cells in the somitic dermomyotome or the migration of these cells into the limb field (Bober et al., 1994). Pax3 and Pax7 can induce expression of myogenic regulatory factors (MRFs) Myf5 and MyoD, thus promoting the specification of muscle progenitor cells to committed myoblasts (Maroto et al., 1997; Buckingham and Relaix, 2007; Lagha et al., 2008). As muscle progenitor cells enter myogenesis, Pax3 and Pax7 are down-regulated, and Myf5, Mrf4, and MyoD are activated; subsequent muscle differentiation depends on MyoD, Mrf4, or MyoG (Buckingham, 2006). In the mouse, Myf5 gene is the first MRF to be expressed in the somite with a role in the early events of myogenic differentiation (Ott et al., 1991). MyoD is also required for the determination of skeletal myoblasts (Rudnicki et al., 1993). Myf5/MyoD double mutant mice are devoid of muscle fiber and myoblasts, while MyoD null or Myf5 null mice develop normal skeletal muscle (Rudnicki et al., 1992; Braun and Arnold, 1996), indicating that Myf5 and MyoD can compensate for each other. MyoG and Myf6 were expressed in a genetic pathway downstream of Myf5 and MyoD. MyoG null mice have virtually no muscle fibers but myoblasts appear normal (Hasty et al., 1993; Nabeshima et al., 1993), suggesting that MyoG is necessary for terminal differentiation. Upon postnatal muscle development, a population of mononucleated precursors, called satellite cells, can generate new myonuclei (Beauchamp et al., 2000) and promote myofiber growth. In mature muscle, satellite cells are mitotically quiescent (Schultz et al., 1978), quiescent satellite cells express Pax7 and Myf5, but not MyoD or MyoG (Yin et al., 2013). Once the muscle is injured, satellite cells can be rapidly activated and give birth to myogenic precursor cells; these cells express Myf5 and MyoD, and later express MyoG for a series of proliferation, differentiation, and fusion to form new myofibers (Kuang et al., 2007; Yin et al., 2013); a part of activated satellite cells also produce offspring that restore quiescent satellite cell pool.
Polycomb group (PcG) proteins are transcriptional inhibitors that remodel chromatin by epigenetic modification and prevent changes of cell characteristics by maintaining transcription states throughout development and into adulthood (Schuettengruber and Cavalli, 2009; Simon and Kingston, 2009; Stojic et al., 2011). In mammals, there are two PcG enhancer of zeste-related genes, enhancer of zeste homologs 1 and 2 (EZH1 and EZH2) (Laible et al., 1997). EZH1 is generally expressed in adults, while EZH2 is expressed during embryonic development (Laible et al., 1997). EZH2 knockout in mouse embryos can lead to embryo post-implantation death (Surface et al., 2010); EZH2 can also recruit transmethylase to its targets in embryonic stem cells (Vire et al., 2006). This indicates that EZH2 is indispensable for mouse embryo development. EZH2, EED, and SUZ12 are the main components of polycomb repressive complex 2 (PRC2), a member of the PcG (Cao et al., 2002). EZH2 is the catalytic subunit of PRC2 and is responsible for the trimethylation of lysine 27 of histone H3 (H3K27me3) (Margueron et al., 2008; Shen et al., 2008) through its SET domain, which is essential for histone lysine methyltransferase activity (Caretti et al., 2004). PRC2 is involved in many biological processes, including cell proliferation, differentiation, stem cell maintenance, embryonic development, and myogenic differentiation (Boyer et al., 2006; Asp et al., 2011; Aloia et al., 2013; Khan et al., 2015). The repression effect of PRC2 on gene expression is mainly through the EZH2 protein (Schuettengruber and Cavalli, 2009; Simon and Kingston, 2009; Margueron and Reinberg, 2011). In skeletal muscle, EZH2 expression is developmentally regulated (Laible et al., 1997); thus, EZH2 may be related to muscle gene expression and differentiation. In proliferating myoblasts, EZH2 and histone deacetylase 1 (HDAC1) are recruited by YY1 to the genomic regions of inactive muscle-specific genes. Upon differentiation, the EZH2-YY1-HDAC1 complex is dissociated, and MyoD and serum response factor are recruited to occupy muscle-specific gene loci (Caretti et al., 2004). Additionally, mice lacking EZH2 in satellite cells will survive with damaged muscle growth (Woodhouse et al., 2013). A previous study showed that EZH2 was required for postnatal muscle growth and adult muscle regeneration (Juan et al., 2011). These observations indicate that EZH2 has important epigenetic roles in skeletal growth and development.
Long non-coding RNAs (lncRNAs) are transcripts greater than 200 nucleotides in length that have no protein coding capacity (Kapranov et al., 2007). lncRNAs have several features including cell type-specific expression patterns (Dinger et al., 2008; Mercer et al., 2008), distinct subcellular localizations (Chen, 2016), regulation of tumorigenesis (Bhan et al., 2017), and evolutionary selection of lncRNA sequences (Ponting et al., 2009), suggesting that lncRNAs may play important roles in various biological processes including skeletal muscle development. For example, lncRNAs such as SYISL (Jin et al., 2018), Malat1 (Chen et al., 2017), Linc-YY1 (Zhou et al., 2015), Linc-RAM (Yu et al., 2017), Myolinc (Militello et al., 2018), MUNC (Mueller et al., 2015), Linc-MD1 (Legnini et al., 2014), Lnc-31 (Dimartino et al., 2018), and Lnc-SMaRT (Martone et al., 2020) regulate myogenesis via multiple mechanisms such as chromosome modification, transcription activation, molecular sponge activity, competitive binding, mRNA translation, and protein stability.
Studies have shown that the majority of lncRNAs are located in the nucleus and bind epigenetic modifiers, particularly PRC2 (Zhao et al., 2008; Redrup et al., 2009; Tsai et al., 2010; Grote et al., 2013; Klattenhoff et al., 2013; Kaneko et al., 2014; Mondal et al., 2015; Li X. et al., 2016; Liu et al., 2017; Wang et al., 2018). RNA immunoprecipitation sequencing (RIP-Seq) in mouse embryonic stem cells identified genome-wide PRC2-interacting RNAs, providing evidence for direct RNA–protein interactions, most likely through EZH2 (Zhao et al., 2010). In fetal porcine skeletal muscle, a catalog of fetal porcine long intergenic non-coding RNAs (lincRNAs) has been identified, some of which might interact with EZH2 (Zhao et al., 2015). However, the global identification of EZH2-binding lncRNAs in porcine skeletal muscle and their roles in pig myogenesis are not yet fully understood. In this study, RIP-Seq combined with a lincRNA sequencing (lincRNAseq) method was used to capture EZH2-binding lincRNA transcripts in skeletal muscle from 1-month-old pigs, and 356 novel lincRNAs were identified. A lincRNA TCONS-00036665 interacting with EZH2 was found to be up-regulated during pig muscle satellite cells (PSCs) and selected for further study. Knockdown and overexpression experiments showed that TCONS-00036665 promotes cell proliferation but inhibits the differentiation of PSCs. Our study provides a genome-wide view of lincRNAs that specifically associate with EZH2 in pig skeletal muscle and clarifies the function of TCONS-00036665 in PSCs.
Materials and Methods
Animal and Tissue Preparation
Pure Large White female pigs were obtained from the Huazhong Agricultural University farm. All longissimus dorsi muscle samples were collected and maintained at −80°C. C57 mice were purchased from the Hubei Center for Disease Control and housed at Huazhong Agricultural University. All animal experiments were conducted based on the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at Huazhong Agricultural University.
RIP-Seq
Longissimus dorsi muscle tissues from 1-month-old pigs were crushed in liquid nitrogen and lysed in ice-cold lysis buffer [50 mM Tris 7.4, 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.1% sodium dodecyl sulfate (SDS), 0.5% NP-40, and 0.5% deoxycholate] with freshly added 1 mM dithiothreitol, 200 U/ml RNase inhibitor, and protease inhibitor cocktail for 30 min on ice. After centrifugation, 300 μl of supernatant was incubated with 10 μg of anti-EZH2 antibody (Bioss bs-3521R) or IgG control antibody overnight at 4°C. The immunoprecipitates were further incubated with protein A Dynabeads for 3 h at 4°C. After applying a magnet and removing the supernatants, the beads were sequentially washed twice with lysis buffer, high-salt buffer (250 mM Tris 7.4, 750 mM NaCl, 10 mM EDTA, 0.1% SDS, 0.5% NP-40, and 0.5 deoxycholate), and PNK buffer (50 mM Tris, 20 mM ethylene glycol tetraacetic acid, and 0.5% NP-40). The immunoprecipitates were eluted from the beads with elution buffer and the RNAs were purified. Then, purified RNAs were reverse-transcribed into complementary DNA (cDNA). The libraries were prepared according to the manufacturer’s instructions and sequenced on an Illumina HiSeq 2000 platform by ABLife, Inc. (Wuhan, China).
LincRNAseq
Next-generation sequencing was carried out by 1GENE (Hangzhou, China). Briefly, total RNA was extracted from the longissimus dorsi muscle of a 1-month-old pig, ribosomal RNA was removed from total RNA, and the resulting RNA was randomly broken into short segments. Then, the first strand of cDNA was synthesized using random hexamers and the second strand of cDNA was synthesized by adding buffer, dNTPs (except dTTP), dUTP, RNase H, and DNA polymerase I. After purification with a QIAquick PCR kit and elution with EB buffer solution, the ends were repaired, poly(A) tails and sequencing adaptors were added, and the second strand was degraded by uracil-DNA glycosylase enzyme. Then, agarose gel electrophoresis was used to select for fragment size, followed by PCR amplification. Finally, the constructed library was sequenced on the Illumina sequencing platform.
Identification of EZH2-Binding lincRNAs
The raw data from RIP-Seq and lincRNAseq were cleaned through the following quality control steps: trimming of adaptor sequences and removal of reads containing more than 5% Ns, low-quality reads (based on the percentage of bases with quality score < 20), and short reads. Subsequently, clean reads were aligned to the pig reference genome (Sus_scrofa 10.2) using Tophat2 (Kim et al., 2013) and assembled into transcripts using Cufflinks (Trapnell et al., 2012). The assembled transcripts (gene transfer format files) were compared with the reference transcripts annotated in Sus_scrofa 10.2 using Cuffcompare and yielded a cuffcmp.gtf file with which all novel transcripts were selected for lincRNA identification using the following conditions: transcript length ≥200 bp and located in the intergenic region at least 1 kb from neighboring transcripts.
For all selected transcripts, the coding potentials were evaluated using Coding Potential Calculator (CPC) software (Kong et al., 2007) and cross-species conservation was evaluated by BLAST searches against the pig, mouse, and human expressed sequence tag (EST) databases. The expression levels expressed as fragments per kilobase per million mapped fragments (FPKM) for all characterized transcripts were compared using Cuffdiff among different sequencing datasets.
Then, the lincRNAs were identified using the following screening conditions: FPKM value estimated from the lincRNAseq dataset (lincRNA_FPKM) > 1; FPKM value estimated from the EZH2 RIP-Seq dataset (EZH2_FPKM) > 0; and estimated CPC value ≤ −1. Next, the EZH2-enriched lincRNAs were identified using two different strategies. For any specific lincRNA, if the FPKM value estimated from the IgG RIP-Seq dataset (IgG_FPKM) was >0, we could calculate the ratio of EZH2_FPKM relative to IgG_FPKM as log2[(EZH2_FPKM + 2)/(IgG_FPKM + 2)]. If IgG_FPKM was 0, we could calculate the ratio of EZH2_FPKM relative to lincRNA_FPKM as log2[(EZH2_FPKM + 2)/(lincRNA_FPKM + 2)]. For any transcript, if the ratio was ≥1, it could be regarded an EZH2-enriched lincRNA. Finally, Student’s t-test was used to compare the differences in expression levels between different groups.
Isolation and Culture of PSCs
Satellite cells were isolated from hindlimb muscles of Large White piglets within 1 week, as previously described (Wang et al., 2019a). Briefly, skeletal muscles from piglet hindlimbs were cut into pieces and digested with 0.2% collagenase I (Sigma-Aldrich, St. Louis, MO, United States) in a shaking incubator at 37°C for 2 h. The suspension was then added to an equal volume of RPMI 1640 medium (Gibco, Melbourne, Australia) supplemented with 1% penicillin-streptomycin (Gibco) and filtered through 100-, 200-, and 400-mesh sieves to remove tissue debris. The suspension was centrifuged and the precipitated cells were resuspended in RPMI 1640 proliferation medium supplemented with 20% fetal bovine serum (FBS; Gibco), 1% GlutaMAX (Gibco), 1% non-essential amino acids (Gibco), 0.5% chicken embryo extract (Gemini Bio, West Sacramento, CA, United States), 1% penicillin-streptomycin, and 0.25 μg/100 ml basic fibroblast growth factor (Life Technologies, Carlsbad, CA, United States). The cell suspension was cultured in uncoated plates for 2 h to remove fibroblasts by differential adhesion. Then, the purified satellite cells were transferred to a plate coated with Matrigel (BD Biosciences, San Jose, CA, United States) for proliferating culture in humidified air containing 5% CO2 at 37°C in a cell incubator. When the cells reached 60–70% confluence, the RPMI 1640 proliferation medium was replaced with differentiation medium consisting of high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Hyclone Laboratories, San Angelo, TX, United States) supplemented with 2% horse serum (Gibco) and 1% penicillin-streptomycin.
C2C12 Myoblast Culture
C2C12 myoblasts were obtained from the American Type Culture Collection and cultured in 10% (v/v) FBS in DMEM under humidified air containing 5% CO2 at 37°C, and then differentiated at confluence in DMEM with 2% horse serum.
Rapid Amplification of cDNA Ends (RACE)
The Takara SMARTer RACE cDNA amplification Kit (Clontech, Mountain View, CA, United States) was used for 5’ and 3′ RACE according to the manufacturer’s instructions. The gene-specific primers used for RACE PCR were as follows: 5′ RACE (5′-TGCCTTCTTGCTCATGTTCAGGGCATCG-3′); 3′ RACE (5′-TACTCACAGGAAGCCACCTGGCAAACTG-3′). The PCR products were separated by agarose gel electrophoresis on 1.5% agarose gels, and the bands were purified and cloned into the pMD-18T vector. Positive clones were selected and sequenced by Sangon Biological Technology (Shanghai, China).
Small Interfering RNA (siRNA) Synthesis and Transfection
siRNA oligos against TCONS-00036665 were designed using the BLOCK-iT RNAi Designer and synthesized by GenePharma (Shanghai, China). Transfection of siRNA was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States). The TCONS-00036665 siRNA sequences were as follows: sense 5′-GCUCCAUCUUGCCCUAUAUACTT-3′, antisense 5′-GUAUAUAGGGCAAGAUGGAGCTT-3′.
Vector Construction, Virus Production, and Infections
The full-length TCONS-00036665 was obtained by PCR amplification with TCONS-00036665 full-length forward and reverse primers using PSCs cDNA as a template and then introduced into the lentiviral vector pCDH-CMV-MCS-EF1-copGFP. Then, the recombinant virus plasmid was co-transfected with psPAX2, a lentiviral packaging plasmid, and pMD2.G, a lentiviral envelope plasmid, into 293T cells using DNA transfer reagent (Biomed, Beijing, China) to produce the recombinant lentivirus Lenti-TCONS-00036665. At 48 and 72 h after transfection, the virus was harvested and concentrated by ultracentrifugation at 30,000 × g for 2.5 h. The concentrated virus was used to infect cells and animals.
For cell infection, an appropriate amount of Lenti-TCONS-00036665 or Lenti-EV virus was diluted in proliferation medium in the presence of 2 μg/ml of polybrene and the mixture was added to cells.
For animal infection, 6-week-old C57 male mice were intramuscularly injected with an appropriate amount of Lenti-TCONS-00036665 or Lenti-EV virus in the quadriceps (Qu), tibialis anterior (TA), and gastrocnemius (Gas) muscles of the left or right hind legs, respectively. The injection was carried out once a week for 1 month. After 1 month of injections, mice were sacrificed and the Qu, TA, and Gas muscles were collected for further RNA and protein extraction and histological staining.
Quantitative Real-Time PCR (qPCR)
Total RNA from isolated PSC, C2C12, or mouse tissue samples was extracted using TRIzol reagent (Invitrogen). cDNA was synthesized using the RevertAid RT Reverse Transcription Kit (Invitrogen). SYBRGreen Real-time PCR Master Mix (Toyobo, Osaka, Japan) was used for qPCR, which was performed in a CFX96/384 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). The 2–Δ Δ Ct method was used to analyze the relative gene expression data, as described previously (Livak and Schmittgen, 2001). The primers used for qPCR are listed in Supplementary Table S1.
Western Blotting
Cells and muscle tissue were lysed in RIPA buffer (Beyotime Biotechnology, Jiangsu, China) according to the manufacturer’s instructions. Protein lysates were heated at 95°C for 5 min in 5 × SDS sample buffer, then separated by 10% SDS polyacrylamide gel electrophoresis (30 μg each lane), followed by transfer to a polyvinylidene fluoride membrane (Millipore, Burlington, MA, United States) using a Mini Trans-Blot Cell system (Bio-Rad). The membrane was blocked with 5% non-fat milk for 3 h. The primary antibodies were incubated overnight at 4°C. The membranes were washed and incubated with secondary antibodies for 1 h at 37°C followed by visualization by enhanced chemiluminescence (Bio-Rad). Primary antibodies specific for EZH2 (ab3748, 1:1,000; Abcam, Cambridge, United Kingdom), MyoD (sc-760, 1:1,000; Santa Cruz Biotechnology, Dallas, TX, United States), MyoG (sc-12732, 1:200; Santa Cruz Biotechnology), MyHC (sc-376157, 1:3,00; Santa Cruz Biotechnology), Ki67 (ab16667, 1:1,000; Abcam), p21 (sc-6246, 1:1,000; Santa Cruz Biotechnology), and β-actin (sc-4777, 1:1,000; Santa Cruz Biotechnology), along with goat anti-mouse IgG-HRP (sc-2005, 1:3,000; Santa Cruz Biotechnology) and goat anti-rabbit IgG-HRP (sc-2004, 1:3,000; Santa Cruz Biotechnology) secondary antibodies were used to detect protein expression.
Cell Proliferation Assays
For flow cytometry, isolated PSCs were immobilized in 70% (v/v) ethanol overnight at −20°C and incubated in 50 mg/ml propidium iodide for 30 min at 4°C. Cell cycles were detected using a FACSCalibur Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ, United States) and the data were analyzed using ModFit software (Verity Software House, Topsham, ME, United States).
For 5-ethynyl-2′-deoxyuridine (EdU) staining, isolated PSCs or C2C12 cells were fixed in 4% paraformaldehyde and permeated in 0.5% Triton X-100. The cells were then stained with EdU using an EDU kit (RiboBio, Guangdong, China) according to the manufacturer’s instructions. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) reagent. Images were captured using an Olympus IX51-A21PH fluorescence microscope (Olympus, Tokyo, Japan) and the percentage of EdU+ cells was further analyzed using ImageJ.
Immunofluorescence Staining of Cells
Cells were washed twice with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 30 min, and permeated in 0.5% Triton X-100 for 30 min. Cells were then blocked with 3% bovine serum albumin for 2 h. Next, cells were incubated with MyHC (1:200) antibody overnight at 4°C. The next day, cells were washed three times with PBS and incubated with a secondary antibody (anti-mouse CY3; Beyotime Biotechnology) for 1 h at 37°C. Then, cell nuclei were stained with DAPI reagent. Images were visualized using an Olympus IX51-A21PH fluorescence microscope. To calculate the cell differentiation index, total and MyHC+ nuclei were quantified using ImageJ; the cell differentiation index is represented as the proportion of MyHC+ nuclei relative to total nuclei in one field of view.
Immunofluorescence Staining of Tissue Sections
The Qu, TA, and Gas muscles were fixed in 4% paraformaldehyde and embedded in paraffin, and 4 μm-thick serial sections were obtained. The sections were subjected to dewaxing and antigen repair processes and then immunofluorescence staining was performed as described above for immunofluorescence staining of cells. The antibodies were myosin (M4276, 1:1,000; Sigma-Aldrich) and dystropin (ab15277, 1:200; Abcam). The images were captured using an Olympus DP80 upright Metallurgical Microscope (Olympus), and the cross-section areas of individual myofibers were qualified using ImageJ.
RNA Pulldown Assay
The full-length TCONS-00036665 was cloned to the pGEM-3z vector, and the recombinant vector was linearized by single-enzyme digestion. The linearized DNA was labeled with biotin and in vitro transcribed using Biotin RNA Labeling Mix and T7 RNA polymerase (Roche, Basel, Switzerland). The obtained RNA was purified with an RNeasy Mini Kit (Qiagen, Hilden, Germany). Total protein was extracted from PSCs using RNA immunoprecipitation (RIP) buffer, then 1 mg of protein was incubated with 3 μg of biotinylated RNA at room temperature for 1 h. Next, each RNA–protein complex was incubated with 40 μl of streptavidin-coupled Dynabeads (Invitrogen) at room temperature for 1 h. Finally, the beads were washed five times with RIP buffer and the pulled down proteins were used for Western blotting.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin Immunoprecipitation assays were conducted using a ChIP Assay Kit (P2078; Beyotime) according to the manufacturer’s instructions. Briefly, the cells were fixed with 37% formaldehyde, neutralized with glycine solution, and then washed three times with PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF). Next, the cells were centrifuged and resuspended in SDS lysis buffer containing 1 mM PMSF. After ultrasonic crushing treatment, the samples were resuspended in ChIP dilution buffer containing 1 mM PMSF. Each ChIP reaction was incubated with 1 μg of antibody against EZH2 (ab3748; Abcam), H3K27me3 (ab6002; Abcam), or IgG (negative control) overnight at 4°C. The next day, 60 μl of protein A + G agarose/salmon sperm DNA was added and the mixture was incubated for 1 h at 4°C. Then, the pulled down DNA was eluted and purified. Fold enrichment was quantified by qPCR. All promoter primers are listed in Supplementary Table S2.
Data Availability
The raw data of RIP-Seq and lincRNAseq generated in this study have been deposited to the Gene Expression Omnibus (GEO) database with the accession number GSE155260.
Statistical Analysis
Data are presented as means ± standard deviation. Statistical analyses between different groups were determined by Student’s t-test. For all analyses, a P < 0.05 was considered to indicate statistical significance.
Results
RIP-Seq Identified EZH2-Binding lincRNAs
We performed RIP assays with skeletal muscle of a 1-month-old pig. After incubation with EZH2 or IgG antibody, the samples were centrifuged, and supernatant and precipitate were collected separately for Western blotting. Specific bands could be detected for precipitates of the EZH2 antibody but not the IgG antibody (Supplementary Figure S1), indicating that EZH2 antibody enrichment was successful and its precipitates could be used for RIP-Seq. The pulled down RNAs were purified for RIP sequencing on an Illumina HiSeq 2000 system. To provide a potential control, we also performed lincRNAseq with the same sample. To identify EZH2-enriched lincRNAs, we conducted several filtering steps (Supplementary Figure S2). First, we set the lincRNA_FPKM value to >0.1 and obtained 21,455 novel expressed transcripts that displayed significantly lower expression levels than the mRNA annotated in the reference genome (Figure 1A). Second, we added the condition of an EZH2_FPKM value >0 and obtained 9,709 transcripts, of which 5,963 had a CPC value ≤ -1. The transcripts with a CPC value > −1 displayed significantly higher expression levels than those of transcripts with a CPC < -1 (Figure 1B), whereas their enrichment levels in precipitates of the EZH2 antibody showed no significant difference (Figure 1C). Third, based on the above screening, we further set the lincRNA_FPKM value to >1 to remove some lowly expressed lincRNAs. Furthermore, we performed enrichment analysis to further identify EZH2-enriched lincRNAs. Based on the ratio of EZH2_FPKM to lincRNA_FPKM, we identified a total of 496 EZH2-binding lincRNAs and 598 non-EZH2-binding lincRNAs (Figure 1D). The EZH2-binding lincRNAs displayed significantly higher EZH2_FPKM values (Figure 1E) but lower expression levels (Figure 1F) than the non-EZH2-binding lincRNAs. Based on the ratio of EZH2_FPKM to IgG_FPKM, we identified a total of 58 EZH2-binding lincRNAs and 116 non-EZH2-binding lincRNAs (Figure 1G). The EZH2-binding lincRNAs displayed significantly higher EZH2_FPKM values (Figure 1H) and expression levels (Figure 1I) than the non-EZH2-binding lincRNAs. After manually removing the repetitive lincRNAs, we finally obtained 356 EZH2-binding lincRNAs (Supplementary Table S3), of which 24 have ESTs in pigs, mice, and humans, 138 have no ESTs in the three species, and 160 lincRNAs have ESTs only in pigs (Figure 2A). Not surprisingly, almost all EZH2-binding lincRNAs were in intergenic regions (96.75%) (Figure 2B).
FIGURE 1.
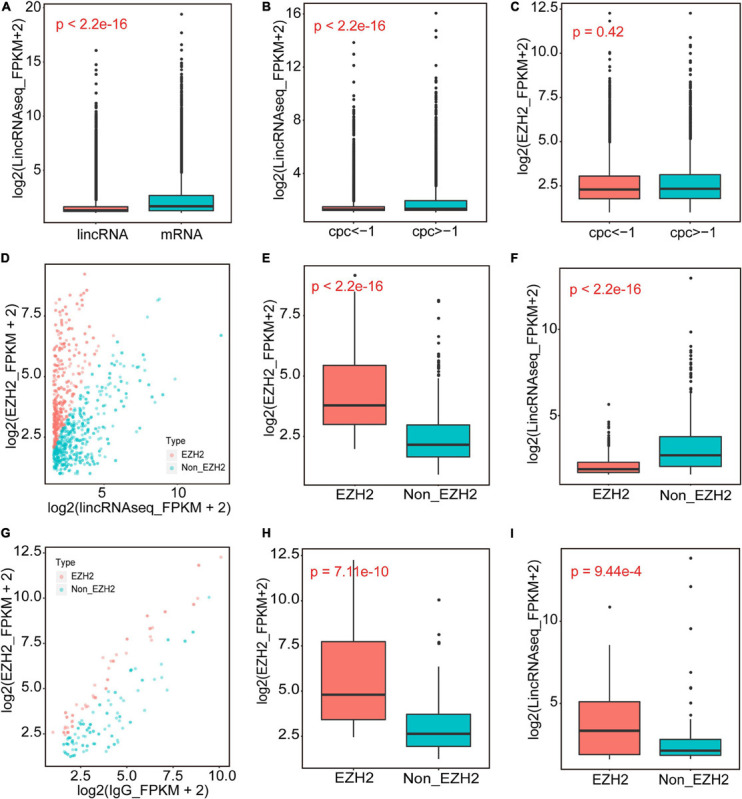
The identification of EZH2-binding novel lincRNAs. (A) The expression levels of novel lincRNAs and mRNAs annotated in Ensembl. (B,C) The expression levels of novel lincRNAs with different CPC groups. (D–F) The EZH2-enriched lincRNAs identified when IgG_FPKM was 0. Volcano plots representing the distribution of log2(lincRNAseq_FPKM + 2) (X-axis) and log2(EZH2_FPKM + 2) (Y-axis); the red dots represent EZH2-enriched lincRNAs, and the blue dots represent Non-EZH2-enriched lincRNAs (D). The EZH2_FPKM values of EZH2-enriched lincRNAs group and Non-EZH2-enriched lincRNAs group (E). The lincRNA expression levels of EZH2-enriched lincRNAs group and Non-EZH2-enriched lincRNAs group (F). (G–I) The EZH2-enriched lincRNAs identified when IgG_FPKM was >0. Volcano plots representing the distribution of log2(IgG_FPKM + 2) (X-axis) and log2(EZH2_FPKM + 2) (Y-axis) (G). The red dots represent EZH2-enriched lincRNAs, and the blue dots represent Non-EZH2-enriched lincRNAs. The EZH2_FPKM values of EZH2-enriched lincRNAs group and Non-EZH2-enriched lincRNAs group (H). The lincRNA expression levels of EZH2-enriched lincRNAs group and Non-EZH2-enriched lincRNAs group (I).
FIGURE 2.
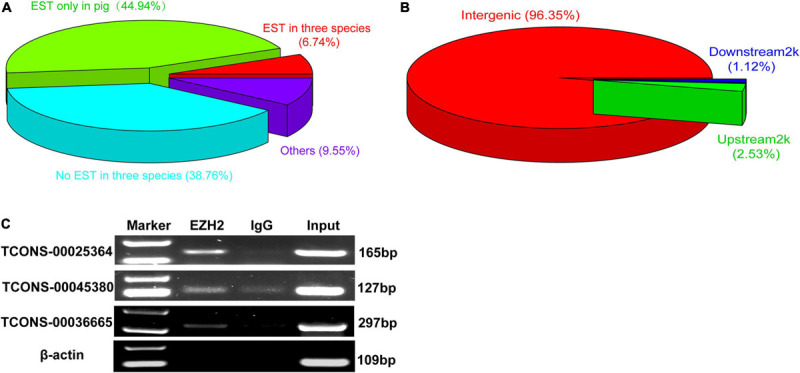
The verification of EZH2-binding novel lincRNAs. (A) The EST information of those novel lincRNA by EZH2 RIP. (B) The location of those novel lincRNA by EZH2 RIP. (C) RIP-PCR results showed that three lincRNAs, TCONS-00025364, TCONS-00045380, and TCONS-00036665, could bind to EZH2.
Verification of EZH2-Binding lincRNAs and Molecular Characterization of TCONS-00036665
Three lincRNAs, TCONS-00025364, TCONS-00045380, and TCONS-00036665, were selected and verified to bind EZH2 by RIP-PCR experiments (Figure 2C). Among the three lincRNAs, the sequences of TCONS-00036665 aligned with the pig EF397601 transcript, which shows similarity to human and mouse NEAT1 gene when aligned it to the National Center for Biotechnology Information (NCBI) database. EF397601 was first identified in Tongcheng pig with a full length of 3409 bp and up-regulated in prenatal skeletal muscle (Ren et al., 2009), indicating that it may be involved in skeletal muscle development. Therefore, we selected TCONS-00036665 for further study. To examine the function of TCONS-00036665, the full-length sequence of TCONS-00036665 was obtained using rapid amplification of cDNA ends (RACE). RACE results showed that TCONS-00036665 was a 3,450 bp polyadenylated transcript (Figure 3A and Supplementary Table S4); it has been deposited in GenBank (accession number MT248265). Next, cell-fractionation assays were performed in PSCs at the proliferation and differentiation stages for 3 days. The results demonstrated that TCONS-00036665 was distributed in the nuclei of proliferating and differentiated PSCs (Figures 3B,C). Finally, we detected the expression of TCONS-00036665 during PSC differentiation and found that the expression of TCONS-00036665 increased during PCS differentiation at 0, 2, and 4 days (Figure 3D). All of these characteristics of TCONS-00036665 imply that it may play a role in PSC proliferation and differentiation.
FIGURE 3.
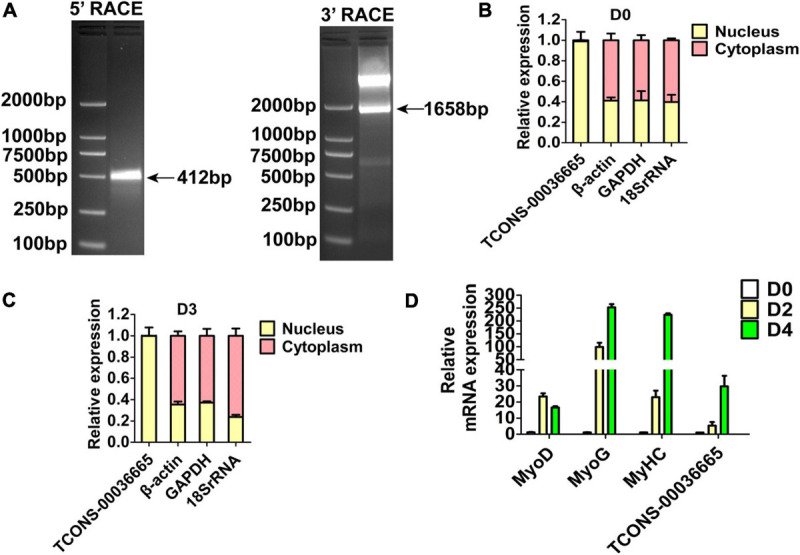
The molecular characterization of TCONS-00036665. (A) Detection of full-length TCONS-00036665 by 5′ RACE and 3′ RACE. (B,C) Detection of the distribution of TCONS-00036665 in the cytoplasm and nucleus of proliferating pig primary myoblasts (B) and differentiated 3 days (C). (D) qPCR results showed that the expression of TCONS-00036665 was induced during PSC differentiation. The relative RNA levels were normalized to those of the control β-actin. Error bars represent the mean ± SD of three biological replicates.
TCONS-00036665 Promotes PSC Proliferation and Inhibits PSC Differentiation
To investigate the role of TCONS-00036665 in PSC proliferation, we knocked down TCONS-00036665 expression with siRNA (Figure 4A). TCONS-00036665 knockdown significantly decreased protein expression of the key proliferation marker Ki67 (Figure 4B). Then, EdU staining and flow cytometry were used to detect the role of TCONS-00036665 in PSC proliferation. EdU staining revealed that knockdown of TCONS-00036665 significantly reduced the proportion of EdU+ cells compared to control (Figure 4C). Flow cytometry showed that TCONS-00036665 knockdown significantly decreased the percentage of S-phase cells (Figure 4D). These observations demonstrated that TCONS-00036665 knockdown repressed PSC proliferation. To verify the effect of TCONS-00036665 knockdown on PSC differentiation, Western blotting and immunofluorescence staining were used to detect expression of myogenic marker genes. Western blotting revealed that the protein expression of MyoD, MyoG, and MyHC was significantly increased by TCONS-00036665 knockdown (Figure 4E). Immunofluorescence staining of MyHC showed that TCONS-00036665 knockdown remarkably enhanced PSC differentiation (Figure 4F). The above results indicated that TCONS-00036665 knockdown inhibits PSC proliferation and accelerates PSC differentiation.
FIGURE 4.
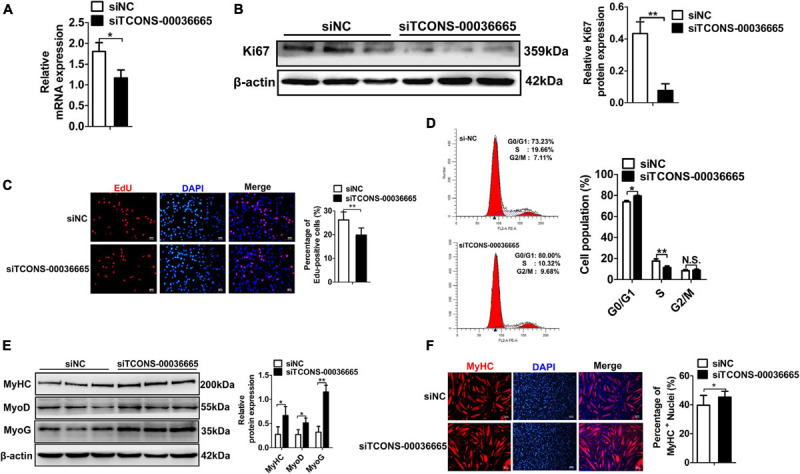
Knockdown of TCONS-00036665 inhibits PSC proliferation but promotes PSC differentiation. (A) qPCR results showed that TCONS-00036665 expression was significantly reduced. (B) Western blotting results showed that the proliferating marker gene Ki67 protein expression was significantly reduced after TCONS-00036665 knockdown, and relative Ki67 protein expression was quantified by ImageJ software. (C) EdU staining results showing that cell proliferation ability was remarkably inhibited by TCONS-00036665 knockdown. (D) Flow cytometry analysis results showing that knockdown of TCONS-00036665 remarkably decreased the proportion of S phase, but increased the proportion of G1 phase. (E) Western blotting results showing that the protein expression of myogenic marker genes MyoD, MyoG, and MyHC was significantly increased by TCONS-00036665 knockdown, and the relative MyoD, MyoG, and MyHC protein expression was quantified by ImageJ software. (F) MyHC immunofluorescence staining results showing that the proportion of MyHC+ was significantly enhanced by TCONS-00036665 knockdown. The relative RNA and protein levels were normalized to those of the control β-actin. Error bars represent the mean ± SD of three biological replicates. *p < 0.05, **p < 0.01, N.S. indicates not significant.
To further verify the role of TCONS-00036665 in the proliferation and differentiation of PSCs, TCONS-00036665 was overexpressed in PSCs (Figure 5A). Overexpression of TCONS-00036665 significantly increased the protein expression of Ki67 (Figure 5B) and the percentage of EdU+ cells (Figure 5C), indicating that overexpression of TCONS-00036665 promoted PSC proliferation. TCONS-00036665 overexpression remarkably reduced protein expression of MyoD, MyoG, and MyHC (Figure 5D) as well as the proportion of MyHC+ cells (Figure 5E), indicating that TCONS-00036665 overexpression inhibited the differentiation of PSCs. These results all demonstrate that TCONS-00036665 can promote the proliferation and inhibit the differentiation of PSCs.
FIGURE 5.
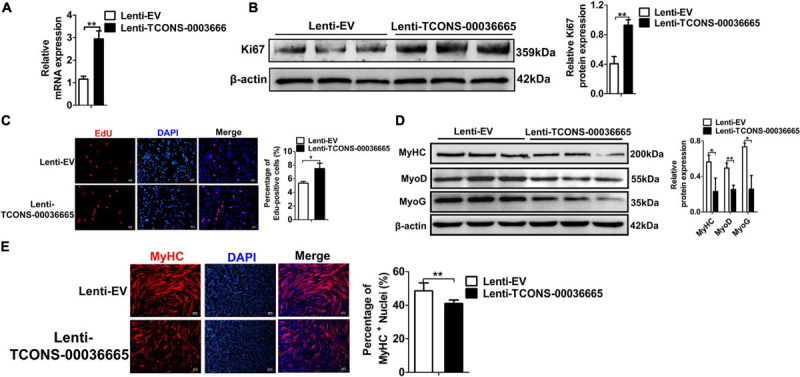
TCONS-00036665 overexpression promotes PSC proliferation but inhibits PSC differentiation. (A) qPCR results showing that TCONS-00036665 expression was significantly increased. (B) Western blotting results showed that Ki67 protein expression was significantly enhanced by TCONS-00036665 overexpression, and relative Ki67 protein expression was quantified by ImageJ software. (C) EdU staining results showing that TCONS-00036665 overexpression remarkably enhanced cell proliferation ability. (D) Western blotting results showing that the protein expression of MyoD, MyoG, and MyHC was significantly inhibited by TCONS-00036665 overexpression, and relative MyoD, MyoG, and MyHC protein expression was quantified by ImageJ software. (E) MyHC immunofluorescence staining results showing that the proportion of MyHC+ was significantly decreased by TCONS-00036665 overexpression. The relative RNA and protein levels were normalized to those of the control β-actin. Error bars represent the mean ± SD of three biological replicates. *p < 0.05, **p < 0.01.
Overexpression of TCONS-00036665 Promotes Proliferation but Inhibits Differentiation of Mouse C2C12 Myoblasts
To verify whether TCONS-00036665 function is conserved between pigs and mice, we overexpressed TCONS-00036665 in mouse C2C12 myoblasts (Figure 6A) and examined its roles in C2C12 proliferation and differentiation. Western blot analysis of proliferating C2C12 cells showed that overexpression of TCONS-00036665 increased the protein expression of proliferation-related genes Ki67 and PCNA (Figure 6B). EdU staining revealed that the percentage of EdU+ cells was significantly increased after TCONS-00036665 overexpression (Figure 6C). These results indicated that TCONS-00036665 overexpression enhanced the proliferation ability of C2C12 cells. To investigate the effect of TCONS-00036665 on C2C12 differentiation, the protein expression of differentiation marker genes was analyzed by Western blotting and the expression of MyoD, MyoG, and MyHC proteins was decreased by TCONS-00036665 overexpression (Figure 6D). Immunofluorescence staining of MyHC revealed that TCONS-00036665 overexpression remarkably inhibited C2C12 differentiation (Figure 6E), suggesting that TCONS-00036665 overexpression suppressed C2C12 differentiation. These observations indicate that TCONS-00036665 overexpression promotes the proliferation but inhibits the differentiation of mouse C2C12 myoblasts, indicating the conserved function of TCONS-00036665 during pig and mouse myogenesis.
FIGURE 6.
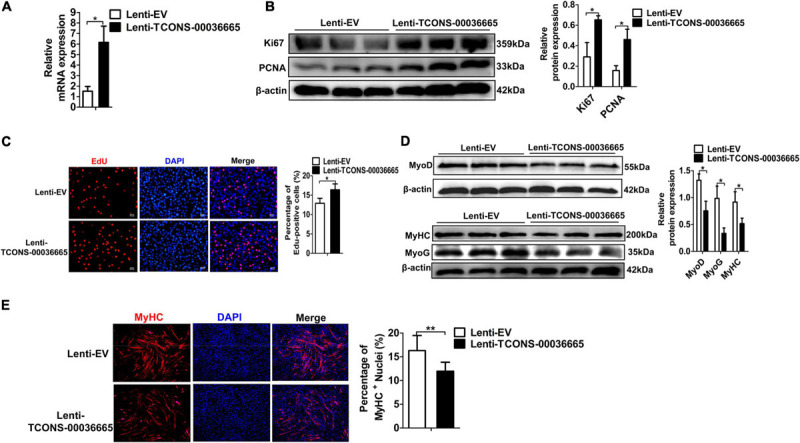
TCONS-00036665 overexpression promotes the proliferation but inhibits the differentiation of mouse C2C12 myoblasts. (A) qPCR results showed that TCONS-00036665 expression was significantly increased in C2C12 cells. (B) Western blotting results showed that TCONS-00036665 overexpression significantly increased Ki67 and PCNA protein expression in C2C12 cells, and the relative Ki67 and PCNA protein expression was quantified by ImageJ software. (C) EdU staining results showed that TCONS-00036665 overexpression remarkably promoted C2C12 cell proliferation ability. (D) Western blotting results showed that MyoD, MyoG, and MyHC protein expression was significantly inhibited by TCONS-00036665 overexpression in C2C12 cells, and the relative MyoD, MyoG, and MyHC protein expression was quantified by ImageJ software. (E) MyHC immunofluorescence staining results showing that TCONS-00036665 overexpression significantly decreased MyHC+ proportion of C2C12 myotubes. The relative RNA and protein levels were normalized to those of the control β-actin. Error bars represent the mean ± SD of three biological replicates. *p < 0.05, **p < 0.01.
Overexpression of TCONS-00036665 Inhibits Mouse Skeletal Muscle Development in vivo
To study the function of TCONS-00036665 in muscle development, TCONS-00036665 was packed with lentivirus to produce Lenti-TCONS-00036665 and injected into the muscles of mice. Lenti-TCONS-00036665 and Lenti-EV were injected into the right and left hindlimbs of 6-week-old C57 mice, respectively (Figure 7A). Reverse transcription qPCR showed that TCONS-00036665 expression was significantly increased by Lenti-TCONS-00036665 injection (Figure 7B). The injection of Lenti-TCONS-00036665 led to a significant reduction in the protein expression of MyoD, MyoG, and MyHC compared to Lenti-EV injection (Figure 7C). The volume and weight of Qu, Gas, and TA muscles were reduced by TCONS-00036665 overexpression (Figures 7D,E). Immunofluorescence staining with myosin and dystropin revealed that the cross-sectional areas of Qu, Gas, and TA muscles were significantly decreased after TCONS-00036665 overexpression (Figures 7F,G). These results demonstrate that TCONS-00036665 overexpression can repress mouse skeletal muscle development.
FIGURE 7.
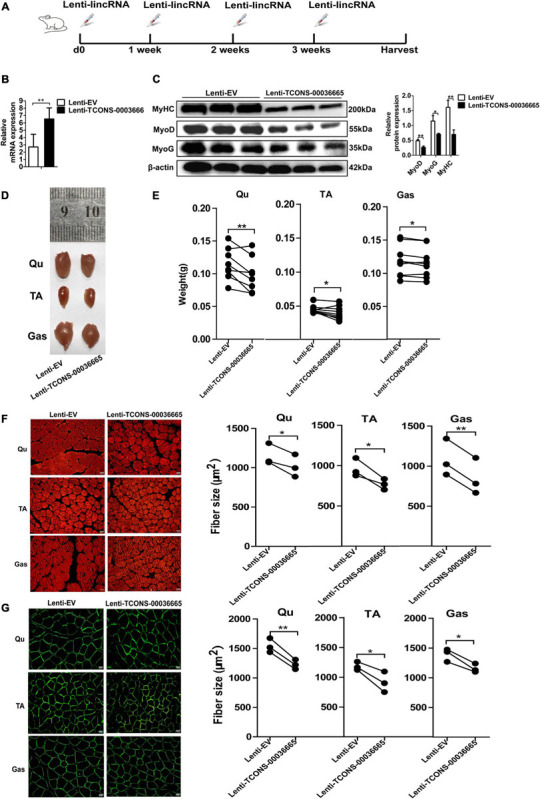
TCONS-00036665 overexpression inhibits mouse skeletal muscle development in vivo. (A) The injection scheme of lenti-TCONS-00036665 or lenti-EV particles into the hindlimb muscles of C57 mice. The injection was performed every 1 week, and the mice were sacrificed after a month injection. (B) qPCR results showing that TCONS-00036665 expression was significantly increased. (C) Western blotting results showing that the protein expression of MyoD, MyoG, and MyHC was significantly inhibited by TCONS-00036665 overexpression, and the relative MyoD, MyoG, and MyHC protein expression was quantified by ImageJ software. (D) Representative pictures of quadriceps (Qu), tibialis anterior (TA), and gastrocnemius (Gas) muscles from the right or left hindlimbs of mice. (E) The statistical analysis of the weight of Qu, TA, and Gas muscles from the right or left hindlimbs of nine injected mice; the results showed that overexpression of TCONS-00036665 decreased the weight of each muscle. (F,G) Representative pictures of myosin (F) and dystrophin (G) immunofluorescence staining of the cross section of Qu, TA, and Gas muscles in mice showing that TCONS-00036665 overexpression remarkably reduced the cross-sectional area of each muscle. The cross-sectional area of each muscle was quantified by ImageJ. The relative RNA and protein levels were normalized to those of the control β-actin. Error bars represent the mean ± SD of three biological replicates. *p < 0.05, **p < 0.01.
TCONS-00036665 Regulates Myogenesis Through EZH2
To verify whether TCONS-00036665 regulates myogenesis by interacting with EZH2, we first confirmed the interaction between TCONS-00036665 and EZH2 by RNA pulldown assay (Figure 8A). Then, Western blotting revealed that TCONS-00036665 knockdown significantly enhanced p21 protein expression (Figure 8B) while TCONS-00036665 overexpression repressed p21 protein expression (Figure 8C), indicating that TCONS-00036665 can inhibit p21 protein expression. Furthermore, ChIP assay revealed that knockdown of TCONS-00036665 significantly decreased the enrichment of EZH2 and H3K27me3 on p21, MyoG, and Myh4 promoters (Figures 8D,E), while EZH2 and H3K27me3 enrichment on the promoters of p21, MyoG, and Myh4 genes was significantly increased by TCONS-00036665 overexpression (Figures 8F,G). These results indicate that TCONS-00036665 can recruit EZH2 to the promoters of p21, MyoG, and Myh4, inhibiting their gene expression and thus promoting PSC proliferation and repressing PSC differentiation.
FIGURE 8.
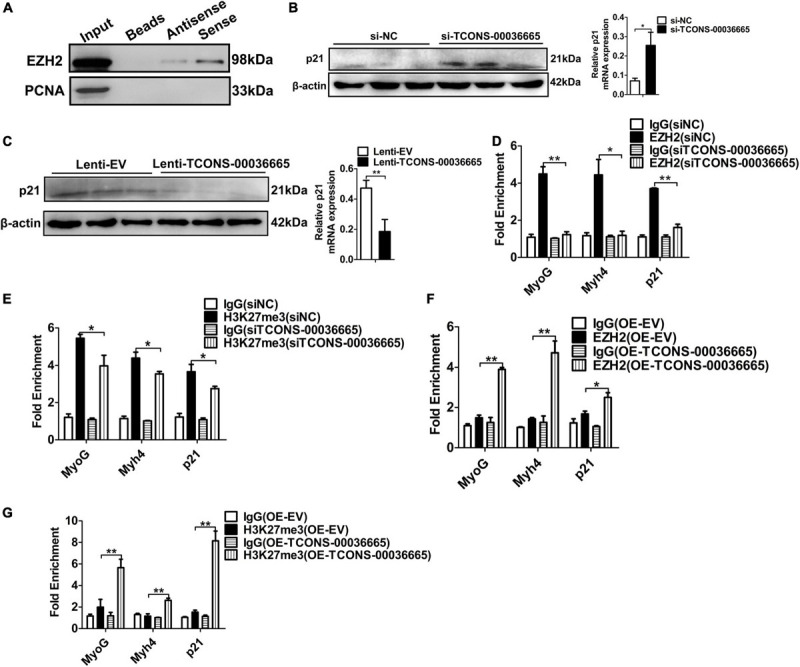
TCONS-00036665 regulates myogenesis through EZH2. (A) Biotin-labeled full-length TCONS-00036665 was used to pull down EZH2. Western blotting result showed that TCONS-00036665 could interact with EZH2. PCNA is a nucleoprotein used as negative controls. (B,C) Western blotting results showed that p21 protein expression was increased by TCONS-00036665 knockdown (B) but decreased by TCONS-00036665 overexpression (C). Relative p21 protein expression was quantified by ImageJ software. (D,E) ChIP-qPCR results showed that the enrichment of EZH2 (D) and H3K27me3 (E) proteins on the promoters of p21, MyoG, and Myh4 were significantly reduced after TCONS-00036665 knockdown in PSCs. (F,G) ChIP-qPCR results showed that the enrichment of EZH2 (F) and H3K27me3 (G) proteins on the promoters of p21, MyoG, and Myh4 were significantly enhanced after TCONS-00036665 overexpression in PSCs. The relative protein levels were normalized to those of the control β-actin. Error bars represent the mean ± SD of three biological replicates. *p < 0.05, **p < 0.01.
Discussion
EZH2-mediated epigenetic repression plays an important role in skeletal muscle specification (Stojic et al., 2011; Dudakovic et al., 2015; Lui et al., 2016; Adhikari and Davie, 2018). EZH2 represses gene expression by catalyzing H3K27me3 on target gene promoters, which serves as an epigenetic signal for chromatin condensation and transcriptional repression (Lui et al., 2016). Recent studies have reported that EZH2 function may be mediated by other factors, such as lncRNAs. For example, lncRNA HERES regulates canonical and non-canonical Wnt signaling pathways through interacting with EZH2 in esophageal squamous cell carcinoma (You et al., 2019). lncRNA ANCR regulates breast cancer progression and metastasis by reducing the stability of EZH2 (Li et al., 2017). Our previous study also found that lncRNA SYISL and Neat1 promote myogenic proliferation but repress myogenic differentiation through interaction with PRC2 (Jin et al., 2018; Wang et al., 2019b). Despite these reports, little is known about EZH2-interacting lincRNAs in pig skeletal muscle. Using unbiased RIP-seq combined with lincRNAseq screening for EZH2-interacting lincRNAs in pig skeletal muscle tissue, we identified 356 new lincRNA transcripts binding to EZH2, which provides a database for the study of the EZH2 regulatory network in pigs.
Skeletal muscle growth and development depend on myoblast proliferation and differentiation and determine the quality and quantity of meat production. Elucidating the regulatory network of skeletal muscle growth and development will be helpful for the improvement of agricultural animal meat traits. In the recent years, owing to the development of RNA-Seq and other high-throughput technologies, tens of thousands of lncRNAs have been identified in the skeletal muscle of agricultural animal. However, only a very small part of lncRNAs have been functionally studied. In particular, lncRNAs, like lncRNA-Six1 (Ma et al., 2018) and lncIRS1 (Li et al., 2019), were found to regulate chicken skeletal muscle development. LncRNAs, like lncMD (Sun et al., 2016), Lnc133b (Jin et al., 2017), and MDNCR (Li et al., 2018), were found to regulate bovine skeletal muscle development. Lnc-SEMT has a role in regulating sheep skeletal muscle development (Wei et al., 2018). In this study, a lincRNA TCONS-00036665 was functionally characterized. We found that TCONS-00036665 was specifically localized in the nucleus and induced during PSCs differentiation. Follow-up studies demonstrated that TCONS-00036665 can promote PSC proliferation but repress differentiation. Furthermore, overexpression of TCONS-00036665 in mouse C2C12 myoblasts and mouse leg muscle also inhibited C2C12 differentiation and muscle growth. Therefore, we concluded that TCONS-00036665 plays an important role in pig skeletal muscle development.
The function of lncRNAs is associated with their subcellular localization; lncRNAs that exhibit nuclear localization patterns are important for nuclear function regulation (Chen, 2016). These lncRNAs execute their regulatory roles by multiple mechanisms; for example, they can accumulate to their transcriptional sites and influence the expression of neighboring genes or recruit transcription factors or chromatin-modifying complexes to target gene promoter (Chen, 2016). The subcellular localization results of TCONS-00036665 showed that it is specifically located in the nucleus, indicating that it may play a role at the transcriptional level. So, we firstly consider that TCONS-00036665 regulates myogenesis through the epigenetic modifier EZH2. As expected, the interaction between TCONS-00036665 and EZH2 was confirmed by our RIP and RNA pulldown assays. Our ChIP assays also demonstrated that TCONS-00036665 can increase the enrichment of EZH2 and H3K27me3 on corresponding target gene promoters. These results confirmed our conjecture that TCONS-00036665 promotes PSC proliferation but inhibits PSC differentiation by recruiting EZH2 to target gene promoters. LncRNAs may bind to RNA binding proteins (RNPs) through a core binding region; for example, Linc-YY1 regulates myogenesis by binding to YY1 with its middle part (386-851nt) (Zhou et al., 2015). LncRNA HOXC-AS3 interacts with YBX1 to mediate gastric cancer tumorigenesis, and a 115 nucleotide region of HOXC-AS3 are responsible for its binding ability to YBX1 (Zhang et al., 2018). LncRNA CPR recruits DNMT3A to Mcm3 promoter and represses cardiomyocyte cell proliferation by binding to DNMT3A with a 244 nucleotide region at the 3′ end (Ponnusamy et al., 2019). Whether TCONS-00036665 interacts with EZH2 through a core binding region is unknown, which needs to be further study.
In this study, we found that TCONS-00036665 serves as a scaffold to link proteins to target gene promotes. However, the mechanism of TCONS-00036665 recruitment to gene promoter remains unclear. Previous studies have proposed two opposing types of RNA–DNA interaction, one of which described that the RNA molecule interacts with one of the two DNA strands by base-pairing to substitute the second strand to form an “R loop” (Skourti-Stathaki and Proudfoot, 2014). The other described that the DNA remains paired and the RNA is wound within the major groove of DNA and forms triplex base pairing between RNA and DNA bases (Li Y. et al., 2016). Furthermore, other studies also reported that proteins may be involved in the interaction and maintenance of RNA–DNA hybridization. For example, the ubiquitous dsRNA-binding protein ILF2 and ILF3 were found to be associated with RNA–DNA hybrids (Nadel et al., 2015), suggesting that it may play a role in RNA–DNA interaction. Thus, we speculated that the recruitment of TCONS-00036665 to different promoters may be caused by forming an “R loop” or triplex base pairing, and RNA-binding proteins or other factors may also affect the recruitment of TCONS-00036665 to different promoters.
In summary, using RIP-Seq and RNA-Seq, we identified a total of 356 novel EZH2-binding lincRNAs in pig skeletal muscle. A lincRNA, TCONS-00036665, was found to regulate myogenesis and muscle development by increasing the enrichment of EZH2 and H3K27me3 on p21, MyoG, and Myh4 promoters.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE155260.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee at Huazhong Agricultural University.
Author Contributions
SW performed all experiments, interpreted results, and wrote the manuscript. XX conducted the bioinformatics analysis of RIP-seq and revised the primary version of manuscript. YL performed the data analysis of RIP-seq. JJ performed nuclear and cytoplasmic RNA fractionation and isolated pig satellite cells. FZ collected tissue samples for RIP-seq. WB, YG, and JZ isolated pig satellite cells. HZ provided support and suggestions for histology staining. BZ and ZX conceived the project, revised the manuscript, and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was financially supported by the National Key Project of Transgenic Research (Grant No. 2016ZX08006-002), Key R & D projects of Hubei Province (2020BBB069), the Agricultural Innovation Fund of Hubei Province (2016-620-000-001-043), the Fundamental Research Funds for the Central Universities (Program No. 2662018PY045), and the China Postdoctoral Science Foundation (Program 590319103).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.618617/full#supplementary-material
References
- Adhikari A., Davie J. (2018). JARID2 and the PRC2 complex regulate skeletal muscle differentiation through regulation of canonical Wnt signaling. Epigenet. Chrom. 11:46. 10.1186/s13072-018-0217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L., Di Stefano B., Di Croce L. (2013). Polycomb complexes in stem cells and embryonic development. Development 140 2525–2534. 10.1242/dev.091553 [DOI] [PubMed] [Google Scholar]
- Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., et al. (2011). Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. U.S.A. 108 E149–E158. 10.1073/pnas.1102223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp J. R., Heslop L., Yu D. S., Tajbakhsh S., Kelly R. G., Wernig A., et al. (2000). Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 151 1221–1234. 10.1083/jcb.151.6.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A., Soleimani M., Mandal S. S. (2017). Long noncoding RNA and cancer: a new paradigm. Cancer Res. 77 3965–3981. 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bober E., Franz T., Arnold H. H., Gruss P., Tremblay P. (1994). Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120 603–612. [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441 349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Braun T., Arnold H. H. (1996). Myf-5 and myoD genes are activated in distinct mesenchymal stem cells and determine different skeletal muscle cell lineages. EMBO J. 15 310–318. 10.1002/j.1460-2075.1996.tb00361.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M. (2006). Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 16 525–532. 10.1016/j.gde.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Buckingham M., Relaix F. (2007). The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. 23 645–673. 10.1146/annurev.cellbio.23.090506.123438 [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., et al. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 1039–1043. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- Caretti G., Di Padova M., Micales B., Lyons G. E., Sartorelli V. (2004). The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18 2627–2638. 10.1101/gad.1241904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41 761–772. 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Chen X., He L., Zhao Y., Li Y., Zhang S., Sun K., et al. (2017). Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 3:17002. 10.1038/celldisc.2017.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimartino D., Colantoni A., Ballarino M., Martone J., Mariani D., Danner J., et al. (2018). The long non-coding RNA lnc-31 Interacts with Rock1 mRNA and mediates its YB-1-dependent translation. Cell Rep. 23 733–740. 10.1016/j.celrep.2018.03.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger M. E., Amaral P. P., Mercer T. R., Pang K. C., Bruce S. J., Gardiner B. B., et al. (2008). Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18 1433–1445. 10.1101/gr.078378.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A., Camilleri E. T., Xu F., Riester S. M., Mcgee-Lawrence M. E., Bradley E. W., et al. (2015). Epigenetic control of skeletal development by the histone Methyltransferase Ezh2. J. Biol. Chem. 290 27604–27617. 10.1074/jbc.M115.672345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24 206–214. 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., et al. (1993). Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364 501–506. 10.1038/364501a0 [DOI] [PubMed] [Google Scholar]
- Hindi S. M., Tajrishi M. M., Kumar A. (2013). Signaling mechanisms in mammalian myoblast fusion. Sci. Signal. 6:re2. 10.1126/scisignal.2003832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C. F., Li Y., Ding X. B., Li X., Zhang L. L., Liu X. F., et al. (2017). lnc133b, a novel, long non-coding RNA, regulates bovine skeletal muscle satellite cell proliferation and differentiation by mediating miR-133b. Gene 630 35–43. 10.1016/j.gene.2017.07.066 [DOI] [PubMed] [Google Scholar]
- Jin J. J., Lv W., Xia P., Xu Z. Y., Zheng A. D., Wang X. J., et al. (2018). Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. U.S.A. 115 E9802–E9811. 10.1073/pnas.1801471115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan A. H., Derfoul A., Feng X., Ryall J. G., Dell’orso S., Pasut A., et al. (2011). Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 25 789–794. 10.1101/gad.2027911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Bonasio R., Saldana-Meyer R., Yoshida T., Son J., Nishino K., et al. (2014). Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 53 290–300. 10.1016/j.molcel.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D. A., Duttagupta R., Willingham A. T., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316 1484–1488. 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- Khan A. A., Lee A. J., Roh T. Y. (2015). Polycomb group protein-mediated histone modifications during cell differentiation. Epigenomics 7 75–84. 10.2217/epi.14.61 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C. A., Scheuermann J. C., Surface L. E., Bradley R. K., Fields P. A., Steinhauser M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152 570–583. 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z. Q., Liu X. Q., Zhao S. Q., Wei L., et al. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35 W345–W349. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M. A. (2007). Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129 999–1010. 10.1016/j.cell.2007.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M., Kormish J. D., Rocancourt D., Manceau M., Epstein J. A., Zaret K. S., et al. (2008). Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev. 22 1828–1837. 10.1101/gad.477908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G., Wolf A., Dorn R., Reuter G., Nislow C., Lebersorger A., et al. (1997). Mammalian homologues of the polycomb-group gene enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J 16 3219–3232. 10.1093/emboj/16.11.3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. (2014). A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell. 53 506–514. 10.1016/j.molcel.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yang J., Jiang R., Wei X., Song C., Huang Y., et al. (2018). Long non-coding RNA profiling reveals an abundant MDNCR that promotes differentiation of myoblasts by sponging miR-133a. Mol. Ther. Nucleic Acids 12 610–625. 10.1016/j.omtn.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lin Y., Yang X., Wu X., He X. (2016). Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 473 913–919. 10.1016/j.bbrc.2016.03.150 [DOI] [PubMed] [Google Scholar]
- Li Y., Syed J., Sugiyama H. (2016). RNA-DNA triplex formation by long Noncoding RNAs. Cell Chem. Biol. 23 1325–1333. 10.1016/j.chembiol.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Li Z., Cai B., Abdalla B. A., Zhu X., Zheng M., Han P., et al. (2019). LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachex. Sarcopen. Muscle 10 391–410. 10.1002/jcsm.12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. W., Hou P. F., Fan D. M., Dong M. C., Ma M. S., Li H. Y., et al. (2017). The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 24 59–71. 10.1038/cdd.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Peng B., Wu S., Xu N. (2017). Epigenetic regulation of regulatory T cells in kidney disease and transplantation. Curr. Gene Ther. 17 461–468. 10.2174/1566523218666180214093813 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lui J. C., Garrison P., Nguyen Q., Ad M., Keembiyehetty C., Chen W., et al. (2016). EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat. Commun. 7:13685. 10.1038/ncomms13685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Cai B., Jiang L., Abdalla B. A., Li Z., Nie Q., et al. (2018). lncRNA-Six1 is a target of miR-1611 that functions as a ceRNA to regulate Six1 Protein expression and fiber type switching in chicken myogenesis. Cells 7:243. 10.3390/cells7120243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C. L., et al. (2008). Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 32 503–518. 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. (2011). The polycomb complex PRC2 and its mark in life. Nature 469 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M., Reshef R., Munsterberg A. E., Koester S., Goulding M., Lassar A. B. (1997). Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell 89 139–148. 10.1016/s0092-8674(00)80190-7 [DOI] [PubMed] [Google Scholar]
- Martone J., Mariani D., Santini T., Setti A., Shamloo S., Colantoni A., et al. (2020). SMaRT lncRNA controls translation of a G-quadruplex-containing mRNA antagonizing the DHX36 helicase. EMBO Rep. 21:e49942. 10.15252/embr.201949942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Sunkin S. M., Mehler M. F., Mattick J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105 716–721. 10.1073/pnas.0706729105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Militello G., Hosen M. R., Ponomareva Y., Gellert P., Weirick T., John D., et al. (2018). A novel long non-coding RNA Myolinc regulates myogenesis through TDP-43 and Filip1. J. Mol. Cell Biol. 10 102–117. 10.1093/jmcb/mjy025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., et al. (2015). MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6:7743. 10.1038/ncomms8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A. C., Cichewicz M. A., Dey B. K., Layer R., Reon B. J., Gagan J. R., et al. (2015). MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell. Biol. 35 498–513. 10.1128/MCB.01079-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., et al. (1993). Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364 532–535. 10.1038/364532a0 [DOI] [PubMed] [Google Scholar]
- Nadel J., Athanasiadou R., Lemetre C., Wijetunga N. A., Broin P., Sato H., et al. (2015). RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenet. Chrom. 8:46. 10.1186/s13072-015-0040-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M. O., Bober E., Lyons G., Arnold H., Buckingham M. (1991). Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development 111 1097–1107. [DOI] [PubMed] [Google Scholar]
- Ponnusamy M., Liu F., Zhang Y. H., Li R. B., Zhai M., Liu F., et al. (2019). Long Noncoding RNA CPR (Cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation 139 2668–2684. 10.1161/Circulationaha.118.035832 [DOI] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W. (2009). Evolution and functions of long noncoding RNAs. Cell 136 629–641. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Redrup L., Branco M. R., Perdeaux E. R., Krueger C., Lewis A., Santos F., et al. (2009). The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136 525–530. 10.1242/dev.031328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Li Y., Tang Z., Yang S., Mu Y., Cui W., et al. (2009). Genomic structure, chromosomal localization and expression profile of a porcine long non-coding RNA isolated from long SAGE libraries. Anim. Genet. 40 499–508. 10.1111/j.1365-2052.2009.01868.x [DOI] [PubMed] [Google Scholar]
- Rudnicki M. A., Braun T., Hinuma S., Jaenisch R. (1992). Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparentlynormal muscle development. Cell 71 383–390. 10.1016/0092-8674(92)90508-a [DOI] [PubMed] [Google Scholar]
- Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993). MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75 1351–1359. 10.1016/0092-8674(93)90621-v [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Cavalli G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136 3531–3542. 10.1242/dev.033902 [DOI] [PubMed] [Google Scholar]
- Schultz E., Gibson M. C., Champion T. (1978). Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J. Exp. Zool. 206 451–456. 10.1002/jez.1402060314 [DOI] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y. J., Fujiwara Y., Kim J., Mao X., et al. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 32 491–502. 10.1016/j.molcel.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E. (2009). Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10 697–708. 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Proudfoot N. J. (2014). A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28 1384–1396. 10.1101/gad.242990.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L., Jasencakova Z., Prezioso C., Stutzer A., Bodega B., Pasini D., et al. (2011). Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenet. Chrom. 4:16. 10.1186/1756-8935-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Li M., Sun Y., Cai H., Lan X., Huang Y., et al. (2016). The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. Biochim. Biophys. Acta 1863 2835–2845. 10.1016/j.bbamcr.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Surface L. E., Thornton S. R., Boyer L. A. (2010). Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7 288–298. 10.1016/j.stem.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., Lan F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., et al. (2006). The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439 871–874. 10.1038/nature04431 [DOI] [PubMed] [Google Scholar]
- Wang S., Sun Y., Ren R., Xie J., Tian X., Zhao S., et al. (2019a). H3K27me3 depletion during differentiation promotes myogenic transcription in porcine satellite cells. Genes 10:231. 10.3390/genes10030231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zuo H., Jin J., Lv W., Xu Z., Fan Y., et al. (2019b). Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 10:505. 10.1038/s41419-019-1742-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xie Y., Li L., He Y., Zheng D., Yu P., et al. (2018). EZH2 RIP-seq identifies tissue-specific long Non-coding RNAs. Curr. Gene Ther. 18 275–285. 10.2174/1566523218666181008125010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Wu M., Wang C., Liu R., Zhao H., Yang L., et al. (2018). Long Noncoding RNA Lnc-SEMT modulates IGF2 expression by sponging miR-125b to promote sheep muscle development and growth. Cell Physiol. Biochem. 49 447–462. 10.1159/000492979 [DOI] [PubMed] [Google Scholar]
- Woodhouse S., Pugazhendhi D., Brien P., Pell J. M. (2013). Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J. Cell Sci. 126 565–579. 10.1242/jcs.114843 [DOI] [PubMed] [Google Scholar]
- Yin H., Price F., Rudnicki M. A. (2013). Satellite cells and the muscle stem cell niche. Physiol. Rev. 93 23–67. 10.1152/physrev.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You B. H., Yoon J. H., Kang H., Lee E. K., Lee S. K., Nam J. W. (2019). HERES, a lncRNA that regulates canonical and noncanonical Wnt signaling pathways via interaction with EZH2. Proc. Natl. Acad. Sci. U.S.A. 116 24620–24629. 10.1073/pnas.1912126116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhang Y., Li T., Ma Z., Jia H., Chen Q., et al. (2017). Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 8:14016. 10.1038/ncomms14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E., He X., Zhang C., Su J., Lu X., Si X., et al. (2018). A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 19:154. 10.1186/s13059-018-1523-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Ohsumi T. K., Kung J. T., Ogawa Y., Grau D. J., Sarma K., et al. (2010). Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 40 939–953. 10.1016/j.molcel.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sun B. K., Erwin J. A., Song J. J., Lee J. T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322 750–756. 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Mu Y., Ma L., Wang C., Tang Z., Yang S., et al. (2015). Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 5:8957. 10.1038/srep08957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Sun K., Zhao Y., Zhang S., Wang X., Li Y., et al. (2015). Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 6:10026. 10.1038/ncomms10026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of RIP-Seq and lincRNAseq generated in this study have been deposited to the Gene Expression Omnibus (GEO) database with the accession number GSE155260.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE155260.


