Abstract
Aims
This study aimed to review the performance of machine learning (ML) methods compared with conventional statistical models (CSMs) for predicting readmission and mortality in patients with heart failure (HF) and to present an approach to formally evaluate the quality of studies using ML algorithms for prediction modelling.
Methods and results
Following Preferred Reporting Items for Systematic Reviews and Meta‐Analyses guidelines, we performed a systematic literature search using MEDLINE, EPUB, Cochrane CENTRAL, EMBASE, INSPEC, ACM Library, and Web of Science. Eligible studies included primary research articles published between January 2000 and July 2020 comparing ML and CSMs in mortality and readmission prognosis of initially hospitalized HF patients. Data were extracted and analysed by two independent reviewers. A modified CHARMS checklist was developed in consultation with ML and biostatistics experts for quality assessment and was utilized to evaluate studies for risk of bias. Of 4322 articles identified and screened by two independent reviewers, 172 were deemed eligible for a full‐text review. The final set comprised 20 articles and 686 842 patients. ML methods included random forests (n = 11), decision trees (n = 5), regression trees (n = 3), support vector machines (n = 9), neural networks (n = 12), and Bayesian techniques (n = 3). CSMs included logistic regression (n = 16), Cox regression (n = 3), or Poisson regression (n = 3). In 15 studies, readmission was examined at multiple time points ranging from 30 to 180 day readmission, with the majority of studies (n = 12) presenting prediction models for 30 day readmission outcomes. Of a total of 21 time‐point comparisons, ML‐derived c‐indices were higher than CSM‐derived c‐indices in 16 of the 21 comparisons. In seven studies, mortality was examined at 9 time points ranging from in‐hospital mortality to 1 year survival; of these nine, seven reported higher c‐indices using ML. Two of these seven studies reported survival analyses utilizing random survival forests in their ML prediction models. Both reported higher c‐indices when using ML compared with CSMs. A limitation of studies using ML techniques was that the majority were not externally validated, and calibration was rarely assessed. In the only study that was externally validated in a separate dataset, ML was superior to CSMs (c‐indices 0.913 vs. 0.835).
Conclusions
ML algorithms had better discrimination than CSMs in most studies aiming to predict risk of readmission and mortality in HF patients. Based on our review, there is a need for external validation of ML‐based studies of prediction modelling. We suggest that ML‐based studies should also be evaluated using clinical quality standards for prognosis research. Registration: PROSPERO CRD42020134867
Keywords: Machine learning, Heart failure, Readmission, Mortality, Hospitalization, Death, Statistical models, Prognosis
Introduction
Despite major technological advances in the diagnosis, assessment, and management of cardiovascular disease, heart failure (HF) remains a major global public health concern, with an estimated prevalence of 64 million individuals around the world. 1 HF hospitalizations have more than tripled in the last 30 years and are associated with high mortality. 2 HF results in a substantial financial strain on the public health‐care system and critically impairs the quality of life of those afflicted with it. 3
Patients with HF present with diverse clinical profiles such that physicians must assess a wide range of data to make fulsome assessments of their patients and to appropriately manage and predict prognosis. Artificial intelligence is a rapidly growing field in cardiovascular medicine that may aid in organizing past, current, and incoming data. 4 Machine learning (ML) is a sub‐field of artificial intelligence that comprises several algorithms such as artificial neural networks, random forests, decision trees, and other supervised or unsupervised models. 5 These algorithms utilize existing and incoming data to identify patterns and predict future clinical events. 6 Many studies have investigated the role of ML in the diagnosis of HF from electronic health records. 7 However, we are still nascent in our understanding of the potential applications of ML in other aspects of patient management.
While there is interest in whether ML could improve our ability to predict outcomes, 5 there have been few studies that have systematically reviewed the current literature comparing it with conventional statistical models (CSMs). 8 , 9 To date, there is no systematic review that quantitatively compares ML with CSMs in HF prognosis. This systematic review presents an approach to summarize results graphically and a novel approach to formally evaluating the quality of these studies. The objective of this study was to review the performance of ML methods compared with conventional statistical models in the prognosis of hospitalized HF patients.
Methods
Literature search
This systematic review complies with Preferred Reporting Items for Systematic Reviews and Meta‐Analyses (PRISMA) guidelines (Table S1). 10 A comprehensive systematic literature search was performed in MEDLINE, EPUB, Cochrane CENTRAL, EMBASE, INSPEC, ACM, and Web of Science electronic databases for articles published between 1 January 2000 and 26 July 2020. The search terms included synonyms of HF such as cardiac failure, myocardial edema, and cardiac insufficiency, combined with synonyms or subcategories of ML terms such as neural networks, expert systems, and support vector machines. Search terms and keywords used in our search strategy are provided in Table S2. All titles and abstracts were manually filtered for outcomes of hospitalizations, readmissions, mortality, and related terms for inclusion in the study. If required, for clarity, we conducted a full‐text review of the document to determine if relevant outcomes were examined. No restrictions were applied on language or sex.
Study selection
Two reviewers (S. S. and M. M.) independently screened all titles and abstracts. Only primary articles that compared ML with CSMs in the prognosis of initially hospitalized HF patients were considered for inclusion. This criterion was instituted to allow for the evaluation of hospital readmission outcomes and for a clear inception point for the assessment of mortality. Among studies deemed potentially relevant for a full‐text review, articles were excluded if (i) the full‐text manuscript could not be accessed or they were conference or symposium abstracts, (ii) the paper did not assess hospitalized HF patients, (iii) there was no comparison between ML and CSMs, or (iv) the outcomes examined did not include mortality or readmission. Discrepancies were resolved by the consensus of a group of reviewers (C. F., D. C., G. T., H. A. Q., and D. S. L.).
Data extraction
Data extraction included (i) author name and year of publication, (ii) country of data origin, (iii) specific patient population, (iv) distribution of age and sex in cohort, (v) sample size of developmental/derivation/training cohorts and validation/testing cohorts, (vi) internal and external validation methods, (vii) outcome of interest, (viii) ML algorithm, and (ix) classification and performance statistics (e.g. hazard ratio value, odds ratio, p‐value, c‐statistics, and calibration). Where the outcome was a composite of death or readmission, the study was included in analyses for both mortality and readmission. As model performance is often over‐optimistic in the dataset in which it was derived, we used the c‐indices from the validation set for our analyses. We also extracted the type of CSM methods employed, defining these approaches as logistic regression, Poisson regression, or Cox proportional hazards regression models.
Quality assessment
Two reviewers independently assessed the quality of each included study using a modified version of the CHARMS checklist, which is a validated review tool for quality evaluation. 11 The CHARMS checklist was selected because it is applicable to both clinical studies and those published in the ML literature. 12 Modifications to the CHARMS tool were made in consultation with experts in ML, epidemiology, and biostatistics, using frameworks developed in previously published studies. 13 Studies received an overall score of low, moderate, or high risk of bias based on seven domains: (i) source of data, (ii) outcomes, (iii) candidate predictors, (iv) sample size/missing data, (v) attrition, (vi) model development, and (vii) model performance/evaluation. For the criterion of model evaluation, external validation was defined in the narrow sense, as previously described by Reilly and Evans, 14 and defined by the Evidence‐Based Medicine Working Group. 15 A detailed description of the modified CHARMS checklist is available in Tables S3 and S4.
Statistical analysis
Continuous variables were reported as mean (standard deviation) or median (inter‐quartile range) as published in the original reports. Categorical variables were reported as proportions. Extracted c‐indices were not combined or pooled across studies because standard errors of the individual c‐indices were not reported consistently in the original publications. Therefore, we constructed scatterplots with the c‐indices for the CSMs on the x‐axis and ML on the y‐axis to show graphically the distributions between studies.We also determined the difference, Δc‐index = c‐indexML − c‐indexCSM, and classified studies into three groups on the basis of Δc‐index ≤ 0 or > 0 but ≤ 0.05, or > 0.05.
Results
Study characteristics
The initial literature search yielded 4322 articles, of which 3309 remained after exclusion of duplicates, and these articles underwent title and abstract screening. Full‐text screening was performed for 172 prognostic studies with 20 articles included in the final set. The PRISMA flow chart is shown in Figure 1 . The characteristics of each included study are shown in Table S5. In the final inclusion set, most studies were published from 2015 onward [n = 18 (90%)], and more than half of the studies were from the USA [n = 11 (55%)]. Two studies utilized data from registry datasets, while the remainder used multicentre clinical datasets. The total sample size of the 20 studies was 686 842 patients. The weighted average age of reported means or medians of patients across all studies was 74 years, and the weighted proportion of women was 49%. Fifteen articles reported readmission outcomes, seven articles reported mortality outcomes, and two articles reported composite outcomes.
Figure 1.
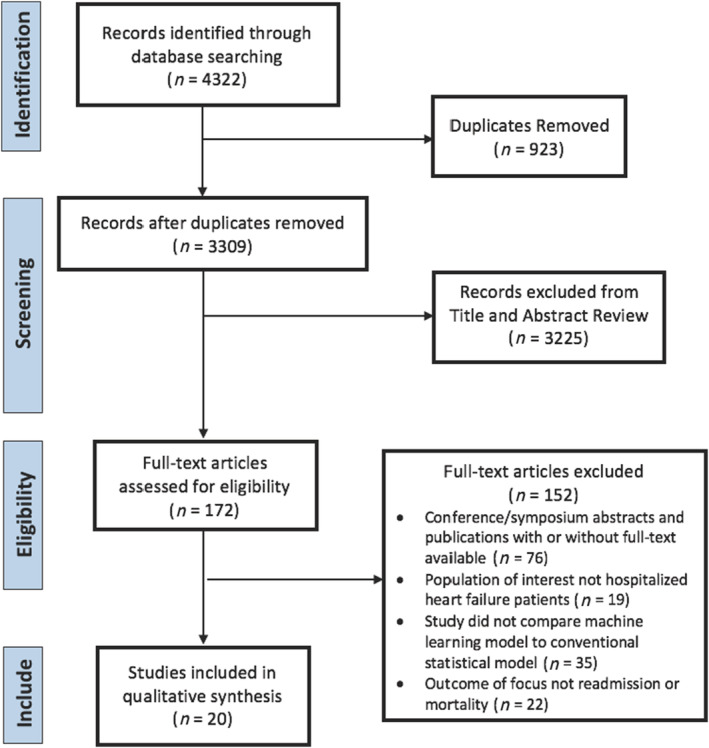
Preferred Reporting Items for Systematic Reviews and Meta‐Analyses (PRISMA) flow chart.
Machine learning methods
Seventeen of 20 studies incorporated a tree‐type ML algorithm ( Table S6). Two studies assessed survival time using random survival forests, which is an extension of the random forest ML approach. Support vector machines were the second most utilized ML algorithm (n = 9), followed by neural networks (n = 7). The remaining studies presented combinations of these with other ML algorithms such as deep learning, 16 , 17 Bayesian techniques (including naïve Bayes classifiers and Bayesian networks), 18 , 19 and K‐nearest neighbour. 20 Several studies employed multiple ML algorithms and compared them with one or more CSMs. Many studies took advantage of ensemble learning algorithms, which are ML techniques that aggregate the outcomes of multiple‐based trained models, producing a unified general result for each data sample (e.g. random forests, gradient boosting machine, and boosted classification tree; Table S6 ). 21 Ensemble learning techniques can be very powerful but lack interpretability, which is crucial in biomedical studies. 22
Conventional statistical model approaches
Logistic regression models were employed in 16 studies, three studies employed Cox regression models, and three studies employed a Poisson regression model. Some studies compared ML with previously derived, clinically validated models, including the Poisson‐based Meta‐Analysis Global Group in Chronic (MAGGIC) HF model, 23 the Cox regression‐based Get With The Guidelines HF (GWTG‐HF) model, 24 Seattle Heart Failure Model, 25 MUerte Subita en Insuficiencia Cardiaca (MUSIC) risk score, 26 SENIORS model, 27 and the logistic regression‐based LACE index. 28 Nearly all studies that compared ML with one of the previously developed validated models also conducted comparisons with re‐fit statistical models using one of the three CSM approaches described earlier. Only one study compared ML exclusively with one of the aforementioned clinical prediction models without developing de novo CSMs or re‐fitting the model covariates in the new dataset. 29
Readmission
For the outcome of readmission, most studies reported superior performance using ML compared with CSMs. Of 15 studies examining readmission outcomes, 11 reported higher c‐indices using ML, and one study reported higher c‐indices at some (but not all time points). When each time point was counted separately, there were 21 comparisons, of which higher c‐indices were reported using ML in 16. These outcome studies are depicted in Figure 2 , where the dashed line (blue diagonal) indicates equivalence between ML and CSMs. Figure 3 shows the magnitude of the differences (delta) between ML and CSMs. Four studies comprising comparisons at eight different time points reported a Δc‐index > 0.05.
Figure 2.
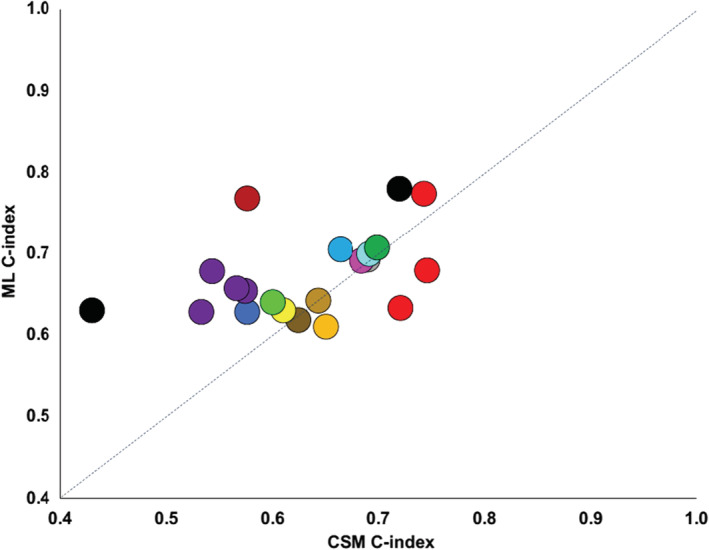
Scatterplot of the highest reported c‐index for machine learning and conventional statistical approaches for readmission studies. Circles of the same colour indicate different time points in the same study publication. CSM, conventional statistical model; ML, machine learning.
Figure 3.
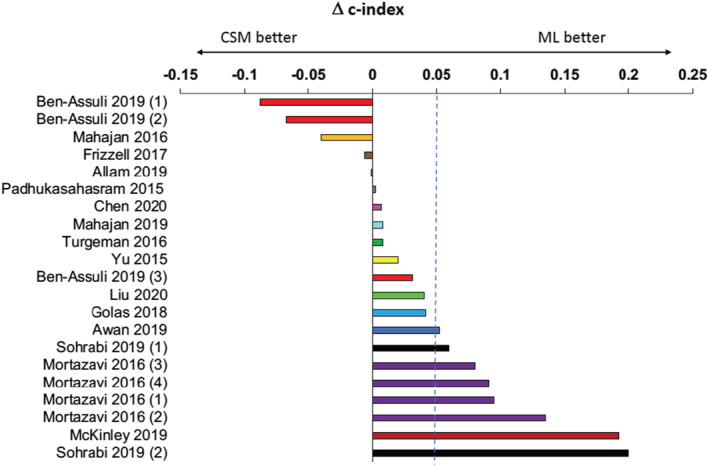
Cluster‐bar plot of the difference in the highest reported c‐index for machine learning and conventional statistical approaches for readmission studies. Bars of the same colour indicate different time points in the same study publication. Bars on the right side of the zero line indicate that c‐indices were higher with ML; bars on the left of the zero line indicate c‐indices were higher with CSMs. Outcomes: Ben‐Assuli 2019 (1) = 90 day readmission; Ben‐Assuli 2019 (2) = 60 day readmission; Ben‐Assuli 2019 (3) = 30 day readmission; Mortazavi 2016 (1) = 30 day all‐cause readmission, Mortazavi 2016 (2) = 30 day HF readmission, Mortazavi 2016 (3) = 180 day all‐cause readmission, Mortazavi 2016 (4) = 180 day HF readmission, Sohrabi 2019 (1) = 1 month HF readmission, Sohrabi 2019 (2) = 3 month readmission. CSM, conventional statistical model; ML, machine learning.
Mortality
For the outcome of mortality, five of seven studies reported higher c‐indices with ML than CSMs (Figure S1). When each time point was counted separately, there were nine comparisons, of which higher c‐indices were reported using ML in seven. Two studies comprising three different time point comparisons reported minimally improved differences in c‐indices with ML [(Δc‐index < 0.05), Figure S2]. Four studies reported a Δc‐index > 0.05.
Quality assessment
Upon assessment using the modified CHARMS checklist ( Tables S3 and S4), all studies demonstrated moderate to high risk of bias in at least three major categories of quality assessment. Low risk of bias was demonstrated in two major categories for 90% of the studies: ‘source of data’ and ‘outcomes’ (Table 1 ). However, 95% of studies demonstrated high risk of bias in the ‘sample size and missing data’ and ‘attrition’ (i.e. completeness of follow‐up) domains. All studies demonstrated moderate risk of bias in the ‘model performance and evaluation’ domain. Critically, only one study performed a comparison of ML and CSMs in an entirely different external validation dataset. 17 This study reported c‐indices of 0.913 for ML, 0.835 for logistic regression, and 0.806 for MAGGIC. 17 All other studies performed internal validation in the same dataset where the ML models were derived by cross‐validation (n = 11), random split sample (n = 8), chronological split sample (n = 2), or bootstrap resampling (n = 3) techniques.
Table 1.
CHARMS checklist evaluations for each included study
| Study ID | Source of data | Outcomes | Candidate predictors | Sample size/missing data | Attrition | Model development | Model performance/evaluation |
|---|---|---|---|---|---|---|---|
| Allam 2019 | L | L | M | H | H | L | M |
| Austin 2010 | L | L | M | H | L | L | M |
| Austin 2012 | L | L | L | H | H | L | M |
| Awan 2019 | L | L | M | H | H | M | M |
| Ben‐Assuli 2019 | L | M | M | H | H | M | M |
| Chen 2020 | L | L | H | H | H | H | M |
| Frizzell 2017 | L | L | L | H | L | H | M |
| Golas 2018 | L | L | M | H | H | L | M |
| Kwon 2019 | L | L | M | H | H | H | M |
| Liu 2020 | L | L | H | H | H | L | M |
| Mahajan 2016 | L | L | H | H | H | M | M |
| Mahajan 2020 | L | L | H | H | H | H | H |
| McKinley 2019 | L | M | H | H | H | H | M |
| Miao 2018 | L | L | M | H | H | L | M |
| Mortazavi 2016 | L | L | M | L | H | L | M |
| Padhukasahasram 2015 | L | M | H | H | H | M | M |
| Sohrabi 2019 | L | M | H | H | H | H | H |
| Turgeman 2016 | L | L | H | H | H | L | M |
| Wang 2020 | L | L | M | H | H | M | M |
| Yu 2015 | L | L | H | H | H | L | M |
H, high risk of bias; L, low risk of bias; M, moderate risk of bias.
Calibration
Notably, although calibration is a component of model evaluation that is mentioned in the CHARMS checklist, only two studies reported ML and CSM calibration results. 18 , 30 Austin et al. found that neither CSMs nor ML had uniformly superior calibration; however, both logistic regression and random forests resulted in good calibration among subjects with a lower predicted probability of death. 30 Frizzell et al. found that while logistic regression was well calibrated, some ML methods (e.g. tree‐augmented Bayesian network and gradient boosting model) were poorly calibrated when predicted readmissions were higher, and the miscalibration was observable in the validation set. 18
Discussion
In this systematic review, we found that ML methods had better performance than CSMs for prediction of readmission and mortality among patients with HF. All studies applied supervised learning algorithms predicting readmission and/or mortality. The most used method of supervised ML was tree‐type ML algorithms, and logistic regression modelling was the most frequent conventional statistical approach. Unsupervised ML algorithms, which provide inputs with no pre‐specified outcomes, were not utilized in any of the identified studies reviewed. Of the comparative studies, 90% demonstrated high risk of bias in at least two major domains of the modified CHARMS checklist, with >60% demonstrating high risk in at least three major domains. Importantly, most studies showing higher c‐indices performed internal validation but lacked external validation (in even a narrow sense) in an independent dataset. Additionally, only a small minority of studies reported on calibration, which is an important component of predictive model development.
In the past decade, the incorporation of ML algorithms into prognostic models has increased. For example, multiple studies discuss the utility of ML in prognostic models for mortality following myocardial infarction. 30 , 31 , 32 ML has also been applied in cardiac diagnostics, to predict the occurrence of atrial fibrillation. 33 Recent reviews emphasize the tremendous interest in combining these techniques for clinical guidance and the need for additional prognostic studies. 34 The emergence of promising studies that leverage natural language processing or ML is illustrative of this burgeoning interest, but these early studies including patients with HF did not directly compare artificial intelligence algorithms with CSMs. 35 , 36
While the interest in using ML in health care is growing exponentially, few studies have evaluated if it can potentially surpass CSMs in predictive performance. Christodoulou et al. compared ML algorithms with logistic regression in primarily non‐cardiac disease conditions, and they found that ML did not perform better than logistic regression. 37 However, this study explored health‐care outcomes other than mortality or readmission, included diagnostic studies, did not include studies that utilized Cox regression or Poisson regression, and did not capture studies published in the computer science literature. 37 In the specific clinical context of predicting mortality from gastrointestinal bleeding, a systematic review demonstrated higher c‐indices and predictive capacity with ML compared with clinical risk scores. 38 Another specific study aiming to predict bleeding risk following percutaneous coronary intervention reported that ML better characterized bleeding risk than a standard registry model. 39 An important distinction of our review was that we included disease‐specific studies that were published in the computer science literature, which were underrepresented in the aforementioned earlier comparisons of ML and CSMs. Finally, a recent comparison of ML vs. CSMs using the TOPCAT trial dataset found that ML methods had higher c‐indices than CSMs for both readmission (0.76 vs. 0.73) and mortality (0.72 vs. 0.66). 40 However, the study was restricted to heart failure with preserved ejection fraction in the setting of an ambulatory clinical trial and did not examine hospitalized HF cohorts and readmission outcomes, which were the focus of our report. 40 Additionally, although a separate external validation was not performed, overall, the higher c‐indices with ML were consistent with our reported findings.
CSMs have been used successfully in the clinical setting, 41 and ML offers promise for clinical use. ML allows rapid examination of constantly expanding datasets and allows identification of patterns and trends not readily visible to clinicians. 42 The advantages provided by ML are that it is flexible, is nonparametric, does not require a data model for the probability distribution of the outcome variable, does not require pre‐specification of covariates, and can handle a large number of input variables simultaneously. 43 , 44 Indeed, in our review, the maximum number of variables (or features) that could be input simultaneously was >3500. 16 In the context of cardiology, this may allow clinicians to utilize these algorithms for high‐performing prognostic models to enhance care of HF patients. There has been an extensive amount of research suggesting that improved ability to predict risk and implement transitional care interventions may improve HF patient outcomes and reduce readmissions. 41 , 45 , 46 With improved accuracy of predictive modelling, clinicians and other health‐care providers may be better equipped to offer the best care for each patient using individualized predictive data. Clinical decisions and discharge care for HF patients may be guided more effectively to reduce adverse events and improve quality of life.
Our study highlights the heterogeneity that currently exists in the literature for prognostic studies using ML, where many studies often did not report confidence intervals or standard deviations for their performance measures. The heterogeneity in the rigour of evaluation also extended to the lack of a priori cut points when validating prediction algorithms using ML. Importantly, we found that the term ‘external validation’ in the ML literature often referred to methods that would be considered ‘internal validation’ methods (e.g. split sample, cross‐validation, or bootstrap resampling) using CSMs and accepted epidemiological standards. 14 , 47 Thus, there is a strong need for future studies of prognostication using ML to use standardized reporting protocols, where the definitions of risk strata and a clear distinction between the derivation/training and validation/testing sets are explicitly stated.
Recommendations
To improve the quality of reporting of future comparisons of ML and CSMs, we recommend that standard errors of the differences in the c‐statistics between the two analytical approaches should be reported, to allow pooling of multiple studies meta‐analytically. Second, the calibration of ML algorithms was not provided in the majority of studies, but it should be routinely reported. As future studies expand to include high‐dimensional and ‐omics data sources, the potential applications for ML, predictive analytics, and advanced statistical learning techniques are likely to grow. 48 However, our study suggests that the same rigorous principles of model development, internal validation, considering potential sources of bias, and external validation should apply. Furthermore, collaborations between computer scientists, biostatisticians, and clinicians are necessary to ensure that the high quality and standards applicable to clinical prediction rules are also applied to ML methods. 4
Limitations must be acknowledged. Owing to the heterogeneity of reported performance and descriptive statistics, only a narrative synthesis was possible for this study. In addition, we used a modified version of the CHARMS checklist to conduct quality assessment, a tool that was not originally constructed to assess the quality of ML studies or for comparison between ML and CSMs. We modified the CHARMS in consultation with ML and biostatistical experts in order to best suit the objectives of the study and incorporated the framework from a previously published modification of the CHARMS as well. 11 , 12 Other available approaches to assessing quality of prognostic studies, such as the TRIPOD, were considered, but its aim is to increase transparency of reporting, and from an operational standpoint, it was not ideally suited to evaluate prognostic studies using ML. 49 The modified CHARMS checklist does set a high bar for predictive models because it was originally developed for assessing applications for use in clinical practice. However, there is often little distinction between a pre‐clinical and clinical predictive model from a mathematical standpoint. Rather, the major distinction is whether the model has undergone validation—initially in a narrow sense, followed by broad validation, and then in an impact analysis. Some of the CSMs were previously developed models, whose performance may be inferior to purpose‐build CSMs tested in the derivation sample. However, in these studies, we also examined the performance of purpose‐build CSMs derived in the study dataset, which was available in all but one study reviewed. Finally, we cannot exclude the possibility of publication bias, whereby studies showing an advantage of ML could be more likely to be published. However, given the novelty of ML in the health science literature, we would anticipate that studies showing either better performance with CSMs or ML would merit publication irrespective of the directionality of effects.
In conclusion, our study has shown that ML methods demonstrated an overall stronger predictive performance over CSMs for HF prognosis. The heterogeneity in reported outcomes and descriptive statistics warrants the need for established standards of reporting for ML studies. In particular, it is important to externally validate ML models and demonstrate that performance is preserved in new cohorts if the intention is to utilize them clinically.
Conflict of interest
The authors have no relevant disclosures.
Funding
ICES (formerly, the Institute for Clinical Evaluative Sciences) is supported in part by a grant from the Ontario Ministry of Health and Long‐Term Care. The opinions, results, and conclusions are those of the authors and no endorsement by the Ministry of Health and Long‐Term Care or by ICES is intended or should be inferred. This study was supported by a Foundation Grant from the Canadian Institutes of Health Research (grant no. FDN 148446). Dr. D. Lee is supported by a mid‐career investigator award from the Heart and Stroke Foundation and the Ted Rogers Chair in Heart Function Outcomes, a Hospital‐University Chair of the University Health Network at the University of Toronto. Dr. Austin is supported by Mid‐Career investigator award from the Heart and Stroke Foundation.
Supporting information
Figure S1. Scatterplot of the highest reported c‐index for machine learning and conventional statistical approaches for mortality studies.
Figure S2. Cluster‐bar plot of the difference in highest reported c‐index for machine learning and conventional statistical approaches for mortality studies. Bars on the right side of the zero‐line indicate that c‐indices were higher with ML; Bars on the left of the zero‐line indicate c‐indices were higher with CSM.
Table S1. PRISMA guidelines.
Table S2. Literature search strategy.
Table S3. Modified CHARMS checklist.
Table S4. Additional CHARMS criteria.
Table S5. Characteristics of included studies.
Table S6. Machine learning algorithms.
Shin, S. , Austin, P. C. , Ross, H. J. , Abdel‐Qadir, H. , Freitas, C. , Tomlinson, G. , Chicco, D. , Mahendiran, M. , Lawler, P. R. , Billia, F. , Gramolini, A. , Epelman, S. , Wang, B. , and Lee, D. S. (2021) Machine learning vs. conventional statistical models for predicting heart failure readmission and mortality. ESC Heart Failure, 8: 106–115. 10.1002/ehf2.13073.
From the Ted Rogers Centre for Heart Research (SS, CF, MM, HAQ, PRL, AG, SE, HJR, and DSL), Peter Munk Cardiac Centre of University Health Network (CF, HAQ, BW, PRL, FB, SE, HJR, and DSL), ICES (HAQ, PCA, and DSL), Institute for Health Policy, Management and Evaluation (HAQ, PCA, GT, and DSL), Women's College Hospital (HAQ), Biostatistics Research Unit of University Health Network (GT), Krembil Research Institute (DC), Toronto General Hospital Research Institute (PRL, FB, SE, and DSL), University of Toronto (CF, MM, HAQ, PCA, GT, BW, PRL, FB, AG, SE, HJR, and DSL).
Contributor Information
Sheojung Shin, Email: sshin037@uottawa.ca.
Douglas S. Lee, Email: dlee@ices.on.ca.
References
- 1. G. B. D. Causes of Death Collaborators . Global, regional, and national age‐sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017; 390: 1151–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fang J, Mensah GA, Croft JB, Keenan NL. Heart failure‐related hospitalization in the U.S., 1979 to 2004. J Am Coll Cardiol 2008; 52: 428–434. [DOI] [PubMed] [Google Scholar]
- 3. Ziaeian B, Fonarow GC. Epidemiology and aetiology of heart failure. Nat Rev Cardiol 2016; 13: 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shameer K, Johnson KW, Glicksberg BS, Dudley JT, Sengupta PP. Machine learning in cardiovascular medicine: are we there yet? Heart 2018; 104: 1156–1,164. [DOI] [PubMed] [Google Scholar]
- 5. Shameer K, Johnson KW, Yahi A, Miotto R, Li LI, Ricks D, Jebakaran J, Kovatch P, Sengupta PP, Gelijns S, Moskovitz A, Darrow B, David DL, Kasarskis A, Tatonetti NP, Pinney S, Dudley JT. Predictive modeling of hospital readmission rates using electronic medical record‐wide machine learning: a case‐study using Mount Sinai Heart Failure Cohort. Pac Symp Biocomput 2017; 22: 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen JH, Asch SM. Machine learning and prediction in medicine—beyond the peak of inflated expectations. N Engl J Med 2017; 376: 2507–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blecker S, Katz SD, Horwitz LI, Kuperman G, Park H, Gold A, Sontag D. Comparison of approaches for heart failure case identification from electronic health record data. JAMA Cardiol 2016; 1: 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Tanna GL, Wirtz H, Burrows KL, Globe G. Evaluating risk prediction models for adults with heart failure: a systematic literature review. PLoS ONE 2020; 15: e0224135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patel B, Sengupta P. Machine learning for predicting cardiac events: what does the future hold? Expert Rev Cardiovasc Ther 2020; 18: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred Reporting Items for Systematic Reviews and Meta‐Analyses: the PRISMA statement. PLoS Med 2009; 6: e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moons KG, de Groot JA, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, Reitsma JB, Collins GS. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med 2014; 11: e1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Damen JA, Hooft L, Schuit E, Debray TP, Collins GS, Tzoulaki I, Lassale CM, Siontis GC, Chiocchia V, Roberts C, Schlüssel MM. Prediction models for cardiovascular disease risk in the general population: systematic review. BMJ 2016; 353: i2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smit HA, Pinart M, Anto JM, Keil T, Bousquet J, Carlsen KH, Moons KG, Hooft L, Carlsen KC. Childhood asthma prediction models: a systematic review. Lancet Respir Med 2015; 3: 973–984. [DOI] [PubMed] [Google Scholar]
- 14. Reilly BM, Evans AT. Translating clinical research into clinical practice: impact of using prediction rules to make decisions. Ann Intern Med 2006; 144: 201–209. [DOI] [PubMed] [Google Scholar]
- 15. McGinn TG, Guyatt GH, Wyer PC, Naylor CD, Stiell IG, Richardson WS. Users' guides to the medical literature: XXII: how to use articles about clinical decision rules. Evidence‐Based Medicine Working Group. JAMA 2000; 284: 79–84. [DOI] [PubMed] [Google Scholar]
- 16. Golas SB, Shibahara T, Agboola S, Otaki H, Sato J, Nakae T, Hisamitsu T, Kojima G, Felsted J, Kakarmath S, Kvedar J, Jethwani K. A machine learning model to predict the risk of 30‐day readmissions in patients with heart failure: a retrospective analysis of electronic medical records data. BMC Med Inform Decis Mak 2018; 18: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwon JM, Kim KH, Jeon KH, Park J. Deep learning for predicting in‐hospital mortality among heart disease patients based on echocardiography. Echocardiography 2019; 36: 213–218. [DOI] [PubMed] [Google Scholar]
- 18. Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, Bhatt DL, Fonarow GC, Laskey WK. Prediction of 30‐day all‐cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol 2017; 2: 204–209. [DOI] [PubMed] [Google Scholar]
- 19. Turgeman L, May JH. A mixed‐ensemble model for hospital readmission. Artif Intell Med 2016; 72: 72–82. [DOI] [PubMed] [Google Scholar]
- 20. McKinley D, Moye‐Dickerson P, Davis S, Akil A. Impact of a pharmacist‐led intervention on 30‐day readmission and assessment of factors predictive of readmission in African American men with heart failure. Am J Mens Health 2019; 13: 1557988318814295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang C, Ma Y. Ensemble Machine Learning, First ed. New York: Springer‐Verlag; 2012. [Google Scholar]
- 22. Dietterich TG. Ensemble methods in machine learning In Multiple Classifier Systems: MCS 2000 Lecture Notes in Computer Science, Vol. 1857 Heidelberg: Springer; 2000. [Google Scholar]
- 23. Rich JD, Burns J, Freed BH, Maurer MS, Burkhoff D, Shah SJ. Meta‐Analysis Global Group in Chronic (MAGGIC) heart failure risk score: validation of a simple tool for the prediction of morbidity and mortality in heart failure with preserved ejection fraction. J Am Heart Assoc 2018; 7: e009594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peterson PN, Rumsfeld JS, Liang L, Albert NM, Hernandez AF, Peterson ED, Fonarow GC, Masoudi FA, American Heart Association Get With the Guidelines‐Heart Failure Program . A validated risk score for in‐hospital mortality in patients with heart failure from the American Heart Association get with the guidelines program. Circ Cardiovasc Qual Outcomes 2010; 3: 25–32. [DOI] [PubMed] [Google Scholar]
- 25. Levy WC, Mozaffarian D, Linker DT, Sutradhar SC, Anker SD, Cropp AB, Anand I, Maggioni A, Burton P, Sullivan MD, Pitt B, Poole‐Wilson PA, Mann DL, Packer M. The Seattle Heart Failure Model: prediction of survival in heart failure. Circulation 2006; 113: 1424–1433. [DOI] [PubMed] [Google Scholar]
- 26. Vazquez R, Bayes‐Genis A, Cygankiewicz I, Pascual‐Figal D, Grigorian‐Shamagian L, Pavon R, Gonzalez‐Juanatey JR, Cubero JM, Pastor L, Ordonez‐Llanos J, Cinca J, de Luna AB, MUSIC Investigators . The MUSIC Risk score: a simple method for predicting mortality in ambulatory patients with chronic heart failure. Eur Heart J 2009; 30: 1088–1096. [DOI] [PubMed] [Google Scholar]
- 27. Manzano L, Babalis D, Roughton M, Shibata M, Anker SD, Ghio S, van Veldhuisen DJ, Cohen‐Solal A, Coats AJ, Poole‐Wilson PPA, Flather MD, SENIORS Investigators . Predictors of clinical outcomes in elderly patients with heart failure. Eur J Heart Fail 2011; 13: 528–536. [DOI] [PubMed] [Google Scholar]
- 28. van Walraven C, Dhalla IA, Bell C, Etchells E, Stiell IG, Zarnke K, Austin PC, Forster AJ. Derivation and validation of an index to predict early death or unplanned readmission after discharge from hospital to the community. CMAJ 2010; 182: 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miao F, Cai Y, Zhang Y, Fan X, Li Y. Predictive modeling of hospital mortality for patients with heart failure by using an improved random survival forest. IEEE Access 2018; 6: 7244–7,253. [Google Scholar]
- 30. Austin PC, Lee DS, Steyerberg EW, Tu JV. Regression trees for predicting mortality in patients with cardiovascular disease: what improvement is achieved by using ensemble‐based methods? Biom J 2012; 54: 657–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shouval R, Hadanny A, Shlomo N, Iakobishvili Z, Unger R, Zahger D, Alcalai R, Atar S, Gottlieb S, Matetzky S, Goldenberg I, Beigel R. Machine learning for prediction of 30‐day mortality after ST elevation myocardial infraction: an Acute Coronary Syndrome Israeli Survey data mining study. Int J Cardiol 2017; 246: 7–13. [DOI] [PubMed] [Google Scholar]
- 32. Pieszko K, Hiczkiewicz J, Budzianowski P, Budzianowski J, Rzeźniczak J, Pieszko K, Burchardt P. Predicting long‐term mortality after acute coronary syndrome using machine learning techniques and hematological markers. Dis Markers 2019; 2019: 9056402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Attia ZI, Noseworthy PA, Lopez‐Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, Kapa S, Friedman PA. An artificial intelligence‐enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019; 394: 861–867. [DOI] [PubMed] [Google Scholar]
- 34. Topol EJ. High‐performance medicine: the convergence of human and artificial intelligence. Nat Med 2019; 25: 44–56. [DOI] [PubMed] [Google Scholar]
- 35. Press MJ, Gerber LM, Peng TR, Pesko MF, Feldman PH, Ouchida K, Sridharan S, Bao Y, Barron Y, Casalino LP. Postdischarge communication between home health nurses and physicians: measurement, quality, and outcomes. J Am Geriatr Soc 2015; 63: 1299–1305. [DOI] [PubMed] [Google Scholar]
- 36. Ahmad T, Lund LH, Rao P, Ghosh R, Warier P, Vaccaro B, Dahlström U, O'Connor CM, Felker GM, Desai NR. Machine learning methods improve prognostication, identify clinically distinct phenotypes, and detect heterogeneity in response to therapy in a large cohort of heart failure patients. J Am Heart Assoc 2018; 7: e008081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol 2019; 110: 12–22. [DOI] [PubMed] [Google Scholar]
- 38. Shung D, Simonov M, Gentry M, Au B, Laine L. Machine learning to predict outcomes in patients with acute gastrointestinal bleeding: a systematic review. Dig Dis Sci 2019; 64: 2078–2087. [DOI] [PubMed] [Google Scholar]
- 39. Mortazavi BJ, Bucholz EM, Desai NR, Huang C, Curtis JP, Masoudi FA, Shaw RE, Negahban SN, Krumholz HM. Comparison of machine learning methods with national cardiovascular data registry models for prediction of risk of bleeding after percutaneous coronary intervention. JAMA Netw Open 2019; 2: e196835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Angraal S, Mortazavi BJ, Gupta A, Khera R, Ahmad T, Desai NR, Jacoby DL, Masoudi FA, Spertus JA, Krumholz HM. Machine learning prediction of mortality and hospitalization in heart failure with preserved ejection fraction. JACC Heart Fail 2020; 8: 12–21. [DOI] [PubMed] [Google Scholar]
- 41. Lee DS, Lee JS, Schull MJ, Borgundvaag B, Edmonds ML, Ivankovic M, McLeod SL, Dreyer JF, Sabbah S, Levy PD, O'Neill T, Chong A, Stukel TA, Austin PC, Tu JV. Prospective validation of the emergency heart failure mortality risk grade for acute heart failure. Circulation 2019; 139: 1146–1156. [DOI] [PubMed] [Google Scholar]
- 42. Deo RC. Machine learning in medicine. Circulation 2015; 132: 1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harrell FE, Jr . Glossary of statistical terms. Vanderbilt University School of Medicine, 2019. http://hbiostat.org/doc/glossary.pdf. (11 August 2019).
- 44. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist‐level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Phillips CO, Wright SM, Kern DE, Singa RM, Shepperd S, Rubin HR. Comprehensive discharge planning with postdischarge support for older patients with congestive heart failure: a meta‐analysis. JAMA 2004; 291: 1358–1,367. [DOI] [PubMed] [Google Scholar]
- 46. van Walraven C, Bennett C, Jennings A, Austin PC, Forster AJ. Proportion of hospital readmissions deemed avoidable: a systematic review. CMAJ 2011; 183: E391–E402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee DS, Stitt A, Austin PC, Stukel TA, Schull MJ, Chong A, Newton GE, Lee JS, Tu JV. Prediction of heart failure mortality in emergent care: a cohort study. Ann Intern Med 2012; 156: 767–775. [DOI] [PubMed] [Google Scholar]
- 48. Afshar M, Lee DS, Epelman S, Gramolini AO, Ross HJ, Lawler PR. Next‐generation approaches to predicting the need for heart failure hospitalization. Can J Cardiol 2019; 35: 379–381. [DOI] [PubMed] [Google Scholar]
- 49. Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015; 162: W1–W73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Scatterplot of the highest reported c‐index for machine learning and conventional statistical approaches for mortality studies.
Figure S2. Cluster‐bar plot of the difference in highest reported c‐index for machine learning and conventional statistical approaches for mortality studies. Bars on the right side of the zero‐line indicate that c‐indices were higher with ML; Bars on the left of the zero‐line indicate c‐indices were higher with CSM.
Table S1. PRISMA guidelines.
Table S2. Literature search strategy.
Table S3. Modified CHARMS checklist.
Table S4. Additional CHARMS criteria.
Table S5. Characteristics of included studies.
Table S6. Machine learning algorithms.


