Abstract
Aims
MicroRNAs (miRNAs) might be used as prospective biomarkers for the identification of unexplained heart failure caused by a viral and/or inflammatory process. The aim of this study was to identify and to evaluate prognostic miRNAs in serum of patients with inflammatory heart diseases diagnosed by endomyocardial biopsies.
Methods and results
After TaqMan® OpenArray® screening of 754 unique circulating miRNAs in serum of biopsy‐proven patients [184 patients with inflammatory and/or virally induced myocardial diseases (DCMi), 25 patients with dilated cardiomyopathy (DCM), and 25 healthy donors], we identified seven miRNAs of interest (P < 0.05). These data have been verified by single qRT–PCR assays in other biopsy‐proven patients (159 patients with viral and/or inflammatory myocardial diseases, 46 patients with DCM, and 60 healthy donors). The expression of let‐7f, miR‐197, miR‐223, miR‐93, and miR‐379 allowed us to differentiate between patients with a virus and/or inflammation and healthy donors (P < 0.05) with the specificity over 93%. Based on the expression of miR‐21 and miR‐30a‐5p, we could sort out patients with DCM from all other study groups (P < 0.05) with the specificity over 95%.
Conclusions
This miRNA profile provides for the first time a new non‐invasive diagnostic perspective to identify patients with intramyocardial inflammation and/or viral persistence only from single serum sample, independently of prescribed therapy and time of symptoms onset. It allows the early finding of those patients relevant for myocardial biopsy for exact diagnosis and further proscription of causal aetiology‐driven specific treatment.
Keywords: MiRNA, Virus, Inflammation, Myocarditis, DCM
Introduction
Endomyocardial biopsies (EMBs) are the gold standard 1 for identification of causative factors in patients with unexplained heart failure (HF) as a prerequisite of an aetiology‐driven treatment. 2 , 3 , 4 , 5 Nevertheless, EMB is used infrequently because of missing non‐invasive, clear defined diagnostic criteria. 6
MicroRNAs (miRNA) are small, 19–23 nucleotides long non‐coding RNAs that bind to complementary sequences on the 3′ untranslated region of target messenger RNAs (mRNA) post‐transcriptionally regulating mRNA expression. 7 It is already known that miRNAs are involved in cardiac differentiation, 8 proliferation, 9 apoptosis 8 as well as in myocardial injury, 10 and inflammation. 11 Because of their stability in the circulation, miRNA profile has emerged as prospective biomarker for many human diseases, in particular in cardiovascular diseases, 12 , 13 , 14 , 15 providing novel molecular insight and new therapeutic strategies to treat diseases.
Cardio‐enriched miRNAs play a crucial role in cardiac development 16 and have been associated with the development of dilated cardiomyopathy (DCM). 17
Despite their promise, miRNAs still have not entered the clinical routine scenario, mainly because of a lack of large cohort studies, 15 or several challenges related to technical aspects, miRNAs normalization, drugs interaction, and quality reporting of statistical multivariable analysis of the miRNAs observational studies. 18
Several data indicate that miRNA profiling in serum samples might be a diagnostic marker in specific inflammatory heart disease, beyond being a biomarker of disease course and survival. 19 Therefore, in this retrospective study that was designed to ascertain differential miRNA patterns in human serum samples, miRNA expression levels of biopsy‐proven patients with viral/inflammatory myocardial diseases and DCM were compared with healthy controls.
The aim of the study was to identify clinically relevant novel diagnostic markers of HF in form of miRNAs for the selection of patients who require EMBs as a prerequisite of exact diagnosis and causal specific treatment.
Methods
Study patients
This retrospective study composed of two parts (screening and validation) evaluated blood serum samples (500 μL) of 414 patients in total with clinically unexplained HF (343 patients with inflammatory and/or virally induced heart diseases diagnosed by EMB and 71 patients with DCM diagnosed by EMB) based on different disease entities sent to the CAP‐accredited laboratory IKDT (Institute for Cardiac Diagnostic and Therapy Berlin, Germany). The patients complained about symptoms of HF with fatigue, reduced physical capacity or dyspnoea on exertion, and cardiac dysfunction. Patients with coronary artery disease (CAD), other possible causes of myocardial dysfunction (e.g. valvular heart disease, hypertension, restrictive, or constrictive heart disease) diagnosed by angiography and echocardiography, and concomitant chronic inflammatory disease (e.g. rheumatological disorders) were excluded from this study.
Left ventricular ejection fraction was determined by echocardiography. Therapy at the time of serum samples taking was not known as we received only blood or serum samples and EMBs for diagnostic evaluation.
Blood serum samples (500 μL) of 85 healthy donors have been used as a control group.
Endomyocardial biopsy analysis
Histological and immunohistochemical staining for assessment of inflammation
We diagnosed patients based on EMB as follows:
An active myocarditis was diagnosed according to the Dallas criteria 20 as a histological evidence of inflammatory infiltrates of >14.0 lymphocytes/mm2, including >7.0 CD3 + lymphocytes/mm2 according to the European Society of Cardiology guidelines 21 within the myocardium associated with myocyte degeneration and necrosis.
A borderline myocarditis was diagnosed with histological evidence of inflammatory infiltrates within the myocardium without myocyte degeneration and necrosis.
Inflammatory cardiomyopathy (DCMi) was diagnosed by evidence of intramyocardial inflammation association with cardiac dysfunction.
The diagnosis DCM was made on morphological and functional characterization with significantly impaired left ventricular ejection fraction and/or dilated left ventricle (Table 1 ). EMBs from DCM patients were negative for histologically or immunohistologically detected inflammation and for the detection of cardiotropic viral genomes.
TABLE 1.
Clinical data, ejection fraction, and histological findings of all patients
| Characteristic | Inflammatory and/or virally induced cardiomyopathy patients | DCM patients | Healthy controls | All patients |
|---|---|---|---|---|
| Number | 343 | 71 | 85 | 499 |
| Age, mean (years) | 49 ± 29 | 55 ± 11 | 44.5 ± 16.5 | 53 ± 25 |
| Male sex (%) | 268 (78) | 38 (53) | 27 (31) | 333 (67) |
| LVEF (%), mean ± SD | 48 ± 19 | 28 ± 9 | — | 46 ± 19 |
| EF < 55% (%) | 60 | 100 | 0 | — |
| LVEDD (mm), mean ± SD | 55.7 ± 21.3 | 66.3 ± 15.7 | — | 58.3 ± 23.7 |
| Fibrosis (%) | No fibrosis a —83% | No fibrosis—86% | — | — |
| Low fibrosis b —17% | Low fibrosis—14% |
DCM, dilated cardiomyopathy; EF, ejection fraction; LVEDD, left ventricular end‐diastolic diameter; LVEF, left ventricular ejection fraction; SD, standard deviation.
Up to 3% of connective tissue.
5‐15% of connective tissue.
Patients with a genetic form of cardiomyopathy were excluded from the study due to missing data.
Used antibodies and immunohistological staining procedure and evaluation of EMBs can be found elsewhere. 3 , 22
Detection of viral genomes
DNA and RNA were extracted from frozen heart muscle tissue probes. RT–PCR was performed for the detection of enteroviruses (including Coxsackievirus), adenovirus, parvovirus B19 (B19V), and human herpesvirus type 6 (HHV6) (Table 2 ) using methods published previously. 23 , 24 In addition, DNA was extracted from peripheral blood cells to exclude a systemic infection with B19V and HHV6. As a control for successful extraction of DNA and RNA from heart muscle tissue, oligonucleotide sequences were chosen from the DNA sequence of the GAPDH gene.
TABLE 2.
Immunohistological analysis of endomyocardial biopsy (median) of all patients with unexplained heart failure
| Diagnosis | CD3 + cells/mm2 | CD45RO + cells/mm2 | LFA‐1 + cells/mm2 | Mac‐1 + cells/mm2 | HLA‐1% area fraction | |
|---|---|---|---|---|---|---|
| Inflammatory myocardial disease patients | Active myocarditis (MCA) | 58.78 | 104.18 | 138.27 | 219.23 | 9.43 |
| Borderline myocarditis | 20.65 | 73.92 | 29.92 | 48.09 | 10.59 | |
| DCM with inflammation (DCMi) | 16.08 | 44.18 | 39.08 | 53.50 | 7.40 | |
| Virally and/or inflammatory myocardial disease patients | Adenovirus (ADV) | 0.00 | 0.00 | — | — | 8.05 |
| Enterovirus (Coxsackie virus) | 2.12 | 16.19 | 11.04 | 20.82 | 6.83 | |
| Human herpesvirus 6 (HHV6) | 2.80 | 19.13 | 8.65 | 25.35 | 6.72 | |
| Parvovirus B19 (B19V) | 3.86 | 29.31 | 10.15 | 23.69 | 6.80 | |
| Dilated cardiomyopathy (DCM) | 2.85 | 10.69 | 7.85 | 18.05 | 5.37 | |
The calculated objects were related to the unit heart area (mm2) or % area fraction.
The clinical and EMB‐based molecular virological and immunohistochemical data of all study patients are summarized in Tables 1 and 2 .
MicroRNAs isolation
Ten millilitres of patients' blood have been collected using BD Vacutainer and centrifuged for 10 min by 10 000–2000 g by 4°C. Serum (approximately 4 mL serum) was collected in tubes and immediately frozen by −80°C for further miRNA analyses.
MicroRNAs were obtained from 500 μL of frozen (−80°C) patients' serum using mirVANA™ PARIS™ RNA and Native Protein Purification Kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions.
MicroRNA reverse transcription, pre‐amplification, and expression analysis using TaqMan® OpenArray and qRT–PCR
Total RNA including miRNA fraction was initially reversely transcribed to cDNA using Megaplex stem‐loop RT primer (Thermo Fisher Scientific, Waltham, MA, USA) for Human Pool A and B in combination with the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) for low content samples. The entire procedure for quantification of miRNAs is described elsewhere. 25 Hsa‐miR‐30a‐3p was used as reference miRNA used for data normalization.
Screening of samples for microRNA profile identification
MicroRNAs from serum samples of 184 patients with biopsy‐proven inflammatory and/or virally induced heart muscle diseases, 25 patients with DCM, and 25 healthy subjects (in.vent Diagnostica GmbH, Hennigsdorf) were measured using TaqMan® OpenArray® (Table 3 ). This allowed us to measure the expression of 754 unique circulating miRNAs (Human MicroRNA Panels A and B; https://www.thermofisher.com/order/catalog/product/4470187#/4470187) in each sample to find out the miRNAs of interest.
TABLE 3.
Number of screened samples for microRNA analysis sorted by diagnosis
| Diagnosis by endomyocardial biopsy | Number of serum samples | |||||
|---|---|---|---|---|---|---|
| With inflammation | Without inflammation | With virus | Without virus | Total | ||
| Inflammatory myocardial disease patients | Myocarditis | 29 | 0 | 5 | 24 | 29 |
| DCM with inflammation (DCMi) | 22 | 0 | 0 | 22 | 22 | |
| Borderline myocarditis | 8 | 0 | 4 | 4 | 8 | |
| Virally and/or inflammatory myocardial disease patients | Adenovirus (ADV) | 2 | 7 | 9 | 0 | 9 |
| Enterovirus (Coxsackie virus) | 21 | 60 | 81 | 0 | 81 | |
| Human herpesvirus 6 (HHV6) | 8 | 4 | 12 | 0 | 12 | |
| Parvovirus B19 (B19V) | 10 | 13 | 23 | 0 | 23 | |
| Total | 184 | |||||
| Dilated cardiomyopathy (DCM) patients | 0 | 25 | 0 | 25 | 25 | |
| Healthy blood donors | 0 | 25 | 0 | 25 | 25 | |
Validation experiments
After identification of miRNA profile consisting of eight miRNAs with TaqMan® OpenArray®, the results have been confirmed in single assays of qRT–PCR in serum of another 159 patients with biopsy‐proven inflammatory and/or virally induced diseases, 46 DCM patients, and 60 healthy donors (Table 4 ).
TABLE 4.
Number of validated samples for microRNA analysis sorted by diagnosis
| Diagnosis by endomyocardial biopsy | Number of serum samples | |||||
|---|---|---|---|---|---|---|
| With inflammation | Without inflammation | With virus | Without virus | Total | ||
| Inflammatory myocardial disease patients | Myocarditis | 35 | 0 | 0 | 35 | 35 |
| DCM with inflammation (DCMi) | 32 | 0 | 0 | 32 | 32 | |
| Borderline myocarditis | 8 | 7 | 0 | 15 | 15 | |
| Virally and/or inflammatory myocardial disease patients | Adenovirus (ADV) | 2 | 16 | 18 | 0 | 18 |
| Enterovirus (Coxsackie virus) | 3 | 21 | 24 | 0 | 24 | |
| Human herpesvirus 6 (HHV6) | 2 | 11 | 13 | 0 | 13 | |
| Parvovirus B19 (B19V) | 2 | 20 | 22 | 0 | 22 | |
| Total | 159 | |||||
| Dilated cardiomyopathy (DCM) patients | 0 | 46 | 0 | 46 | 46 | |
| Healthy blood donors | 0 | 60 | 0 | 60 | 60 | |
Ethical approval
The study was performed within the CRC Transregio 19 and was approved by the local ethics committees of the participating clinical centres as well as by the committees of the respective federal states. The study complies with the Declaration of Helsinki. An informed written consent was obtained from each study patient.
Statistical analysis
To normalize the data, we first performed the normalizationo using geometrical mean of all measured miRNAs. 26 However, we have not detected any significant differences between the groups by any expressed miRNA (Figure 1 ).
FIGURE 1.
Normalization of data using geometrical mean of all measured microRNAs (miRNAs). (A–E) MiRNAs of patients with inflammatory and/or viral cardiomyopathies (IMD); (F, G) miRNAs of dilated cardiomyopathy (DCM) patients. IMD, inflammatory myocardial disease.
Thus, it has been detected that Ct values of U6 (Ct = 16.86 ± 2.75) and hsa‐30a‐3p (Ct = 23.83 ± 1.17) have not changed in all study groups, so hsa‐30a‐3p has been used as reference miRNA for data normalization. 27 , 28
All log‐transformed expression data were analysed and represented with GraphPad Prism 7 (GraphPad Software, La Jolla, USA) log‐transformed to create a normal distribution. The Student's t‐test and one‐way ANOVA were used to compare miRNA expression levels between all study groups. Multivariate regression analysis has been used to exclude the influence of gender, age, and ejection fraction (EF) on the expression levels of each miRNA.
The receiver operating characteristic curves were plotted for every single miRNA, and the areas under the curve were calculated to prove their value and diagnostic accuracy.
All data were presented as single values with mean, with a significance level of *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001.
Results
Serum samples were obtained from patients with biopsy‐proven inflammatory and/or virally induced cardiomyopathies (n = 343), DCM (n = 71), and corresponding healthy controls (n = 85) (Tables 1 and 2 ). All data were generated in the same laboratory to facilitate comparative data analysis.
Identification of microRNAs differentially expressed in patients with unexplained heart failure using TaqMan® OpenArray® analysis
First, we performed miRNA expression studies (screening) with TaqMan® OpenArray® (Table 3 ). Missing miRNA expression data were excluded from the study. 29 Based on the expression levels of 323 differently expressed miRNAs measured in duplicate, only 12 miRNAs (Figure 2 and Table 5 ) were significantly deregulated [P (−log 10) > 1.3] in all groups of patients with heart diseases (n = 184) including DCM (n = 25) compared with healthy donors (n = 25) (Table 3 and Figure 2 ).
FIGURE 2.
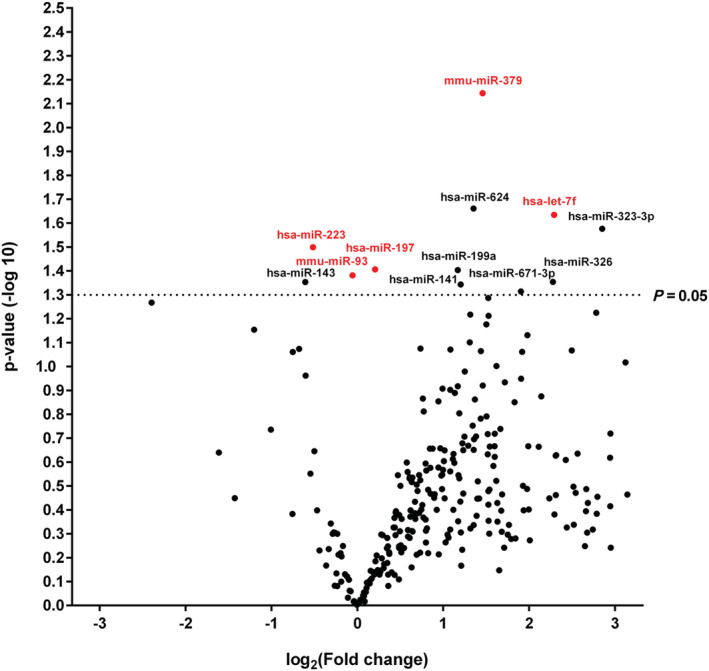
Volcano plot: patients with inflammatory and/or virally induced myocardial disease (including dilated cardiomyopathy) vs. healthy donors. Comparisons of expression of 323 microRNAs (miRNAs) assessed in OpenArray analysis isolated from serum of patients with inflammatory and/or virally induced myocardial diseases (n = 184) or dilated cardiomyopathy (n = 25) and healthy controls (n = 25). The volcano plot displays the relationship between fold change and significance between the two groups, applying a Student's t‐test. The y‐axis depicts the negative log 10 of P‐values of the t‐tests (the horizontal slider at 1.3 corresponds to a P‐value of 0.05; a higher value indicates greater significance), and the x‐axis is the difference in expression between the two experimental groups as log2 fold. Highlighted are five abundant miRNAs (let‐7f, miR‐197, miR‐223, miR‐379, and miR‐93) significantly expressed in all study groups (Tables 3 and 5 ).
TABLE 5.
Significantly deregulated microRNAs in serum samples of patients with inflammatory and/or virally induced myocardial disease detected by the OpenArray® screening
| MiRNA | Fold change | P‐value (−log 10) |
|---|---|---|
| mmu‐miR‐379 | 2.75 | 2.14 |
| hsa‐miR‐624 | 2.55 | 1.66 |
| hsa‐let‐7f | 4.90 | 1.63 |
| hsa‐miR‐323‐3p | 7.22 | 1.58 |
| hsa‐miR‐223 | 0.70 | 1.50 |
| hsa‐miR‐197 | 1.15 | 1.41 |
| hsa‐miR‐199a | 2.25 | 1.40 |
| mmu‐miR‐93 | 0.96 | 1.38 |
| hsa‐miR‐326 | 4.84 | 1.35 |
| hsa‐miR‐143 | 0.66 | 1.35 |
| hsa‐miR‐141 | 2.30 | 1.34 |
| hsa‐miR‐671‐3p | 3.74 | 1.31 |
MiRNA, microRNA.
As indicated in Figure 3 and Table 6 , only two miRNAs (has‐miR‐21 and has‐miR‐30a‐5p) allowed us to distinguish DCM patients from all other study groups.
FIGURE 3.
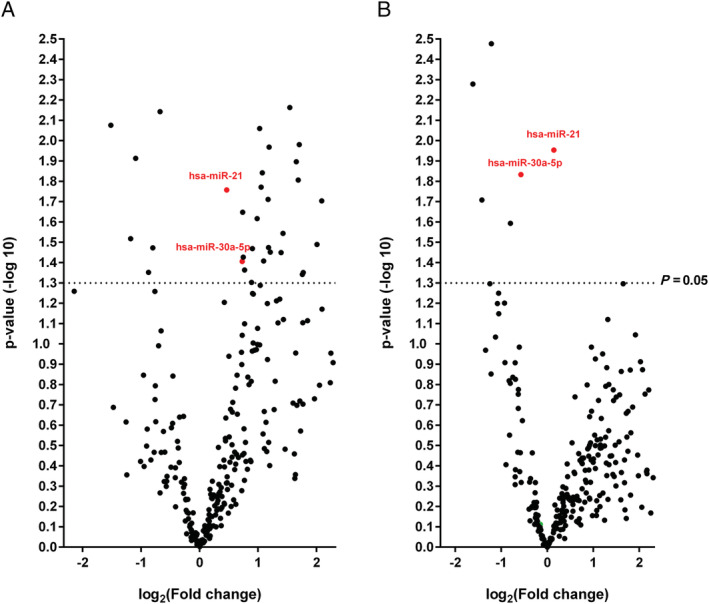
Volcano plot: patients with dilated cardiomyopathy (DCM) vs. healthy donors (A) and vs. patients with inflammatory and/or virally induced myocardial disease (B). Comparison of 323 microRNAs (miRNAs) assessed in OpenArray analysis isolated from serum of DCM patients (n = 25), patients with inflammatory and/or virally induced (n = 184) myocardial disease, and controls (n = 25). Highlighted in green are two abundant miRNAs (miR‐21 and miR‐30a‐5p) significantly expressed in DCM group (Tables 3 and 6 ).
TABLE 6.
Significantly deregulated microRNAs in serum samples of dilated cardiomyopathy patients detected by the OpenArray® screening compared with healthy donors (left side) and patients with inflammatory and/or virally induced cardiomyopathies (right side)
| MiRNA | Fold change | P‐value (−log 10) | MiRNA | Fold change | P‐value (−log 10) |
|---|---|---|---|---|---|
| hsa‐miR‐101 | 3.22 | 1.81 | hsa‐let‐7f | 0.33 | 2.28 |
| hsa‐miR‐106b | 2.31 | 2.73 | hsa‐miR‐195 | 0.41 | 2.94 |
| HSA‐MIR‐1180 | 3.38 | 1.34 | hsa‐miR‐21 | 1.10 | 1.95 |
| hsa‐miR‐125b | 0.57 | 1.47 | hsa‐miR‐30a‐5p | 0.67 | 1.83 |
| hsa‐miR‐130a | 2.26 | 1.47 | hsa‐miR‐422a | 0.37 | 1.71 |
| hsa‐miR‐130b | 1.87 | 1.47 | hsa‐miR‐590‐5p | 0.43 | 2.48 |
| hsa‐miR‐132 | 1.66 | 1.65 | |||
| hsa‐miR‐140‐3p | 3.26 | 1.98 | |||
| hsa‐miR‐141 | 2.75 | 2.60 | |||
| hsa‐miR‐146b‐3p | 4.25 | 1.70 | |||
| hsa‐miR‐155 | 2.69 | 1.54 | |||
| hsa‐miR‐185 | 1.98 | 1.62 | |||
| hsa‐miR‐18a | 1.97 | 2.81 | |||
| hsa‐miR‐21 | 1.38 | 1.76 | |||
| hsa‐miR‐223 | 0.29 | 2.51 | |||
| hsa‐miR‐23a | 2.25 | 1.71 | |||
| hsa‐miR‐296 | 2.10 | 1.84 | |||
| hsa‐miR‐30a‐5p | 1.66 | 1.41 | |||
| hsa‐miR‐30d | 2.04 | 2.06 | |||
| hsa‐miR‐320 | 4.13 | 5.62 | |||
| HSA‐MIR‐320B | 7.64 | 4.01 | |||
| hsa‐miR‐323‐3p | 5.69 | 1.70 | |||
| hsa‐miR‐335 | 2.14 | 1.41 | |||
| hsa‐miR‐363 | 2.31 | 1.45 | |||
| hsa‐miR‐365 | 0.47 | 1.91 | |||
| hsa‐miR‐455 | 4.02 | 1.49 | |||
| hsa‐miR‐486‐3p | 10.16 | 1.86 | |||
| hsa‐miR‐502 | 3.42 | 4.30 | |||
| hsa‐miR‐502‐3p | 3.42 | 1.35 | |||
| hsa‐miR‐574‐3p | 0.63 | 2.14 | |||
| hsa‐miR‐579 | 2.92 | 2.16 | |||
| hsa‐miR‐624 | 2.28 | 1.97 | |||
| hsa‐miR‐885‐5p | 0.35 | 2.08 | |||
| hsa‐miR‐92a | 2.07 | 1.77 | |||
| hsa‐miR‐942 | 1.68 | 1.43 | |||
| hsa‐miR‐99a | 0.44 | 1.52 | |||
| mmu‐miR‐134 | 2.63 | 1.45 | |||
| mmu‐miR‐140 | 0.55 | 1.35 |
Only two microRNAs (miRNAs) (hsa‐miR‐21 and hsa‐miR‐30a‐5p) have been significantly different in dilated cardiomyopathy patients compared with healthy or inflammatory and/or virally induced subjects.
Confirmation of differentially expressed microRNAs using qRT–PCR
To confirm the significant changes in miRNA expression detected by TaqMan® OpenArray®, 15 miRNAs (12 miRNAs from Table 5 , 2 miRNAs from Table 6 , and hsa‐miR‐30a‐3p) were assessed by qRT–PCR using independent sets of another patients with inflammatory and/or virally induced cardiomyopathies (n = 159), DCM (n = 46), and control healthy subjects (n = 60) (Table 4 ). Only five miRNAs (let‐7f, miR‐197, miR‐223, miR‐379, and miR‐93) showed statistically significant deregulation in patients with inflammatory and/or virally induced cardiomyopathies, and two miRNAs (miR‐21 and miR‐30a‐5p) were significantly deregulated in patients with DCM compared with healthy controls and patients with inflammatory and/or virally induced cardiomyopathies (Figure 4 ).
FIGURE 4.
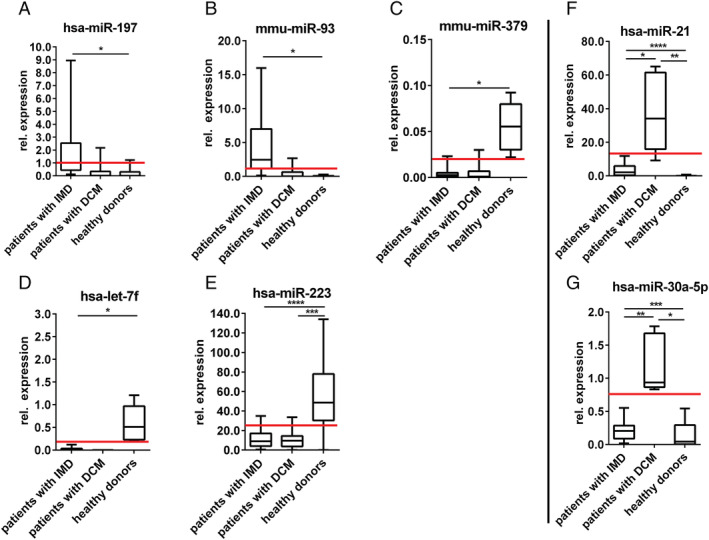
Confirmation of differentially expressed microRNAs (miRNAs) in patients with inflammatory myocardial disease (IMD), dilated cardiomyopathy (DCM), and healthy subjects using qRT–PCR. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001. According to single measurements of relative expression referred to hsa‐miR‐30a‐3p of each patient, it was possible to plot expression level limits for each miRNA (red lines). These boundaries can be used further in the diagnostics to distinguish between healthy donors and patients with IMD (A–E) as between healthy donors, patients with IMD, and patients with DCM (F, G).
Expression level
As the achieved results have to be used further in the diagnostic, it was necessary to set the expression level for each miRNA in three study groups (Table 7 ). Given their significantly different levels in patient serum, these seven miRNAs (referred to hsa‐miR‐30a‐3p) may have potential as diagnostic biomarkers (Figure 4 ).
TABLE 7.
Relative expression level limits in three study groups based on Figure 3
| Diagnosis | Relative expression levels | ||||||
|---|---|---|---|---|---|---|---|
| hsa‐let7f | hsa‐miR‐197 | mmu‐miR‐93 | mmu‐miR‐379 | hsa‐miR‐223 | hsa‐miR‐21 | hsa‐miR‐30a‐5p | |
| Patients with inflammatory/virally induced cardiomyopathies | 0–0.2 | >1 | 1–10 | >0.005 | 0–30 | 0–12 | 0–7 |
| Patients with DCM | 0–0.2 | 0–1 | 0–3 | 0–0.005 | 0–20 | >12 | >7 |
| Healthy donors | >0.2 | 0–1 | 0–1 | 0–0.005 | >30 | 0–1 | 0–1 |
DCM, dilated cardiomyopathy.
The relative expression of each microRNA is referred to hsa‐miR‐30a‐3p used as housekeeping microRNA.
Diagnostic value of microRNAs for dilated cardiomyopathy and cardiac diseases
To discriminate between the patients with inflammatory and/or virally induced cardiomyopathies, DCM patients, and healthy controls (diagnostic value), the receiver operating characteristic curves were plotted for every single miRNA in confirmation of qRT–PCR that was expressed at a significantly higher level in patient serum (Figure 5 ). Despite the overlaps in the expression levels (Figure 4 ), the areas under the curve ranged from 0.904 to 1 in all of miRNAs, proving that these circulating miRNAs are of particular interest for cardiac disease, DCM detection, and diagnosis.
FIGURE 5.
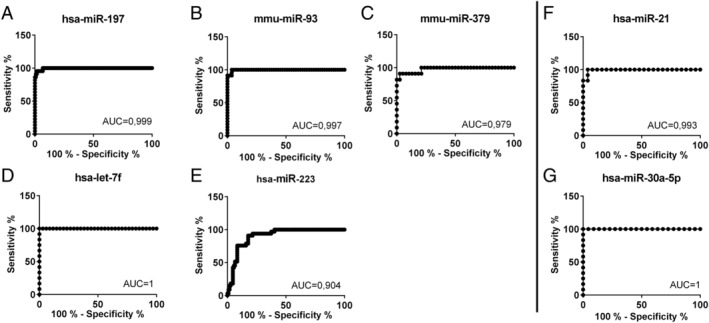
Diagnostic value of microRNAs (miRNAs) for dilated cardiomyopathy and unexplained heart failure presented in receiver operating characteristic curves with area under the curve (AUC) compared with each other. As receiver operating characteristic curve illustrates the diagnostic ability of a binary classifier system as its discrimination threshold is varied, it is created by plotting the true positive rate (sensitivity) against the false positive rate (1‐specificity). AUC is equal to the probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one. (A–E) Heart failure patients against healthy donors; (F, G) dilated cardiomyopathy patients against the rest.
Influence of ejection fraction, gender, and age on the microRNA expression
We evaluate the role of miRNAs in the classification of patients with inflammatory myocardial diseases and DCM.
The relationship between the expression levels of circulating miRNAs and cardiac function, gender, and age was further studied. In seven selected miRNAs, we have not detected any significant dependence of EF (Figure 6 ), gender (Figure 7 ), and age (Figure 8 ) on the miRNA expression level.
FIGURE 6.
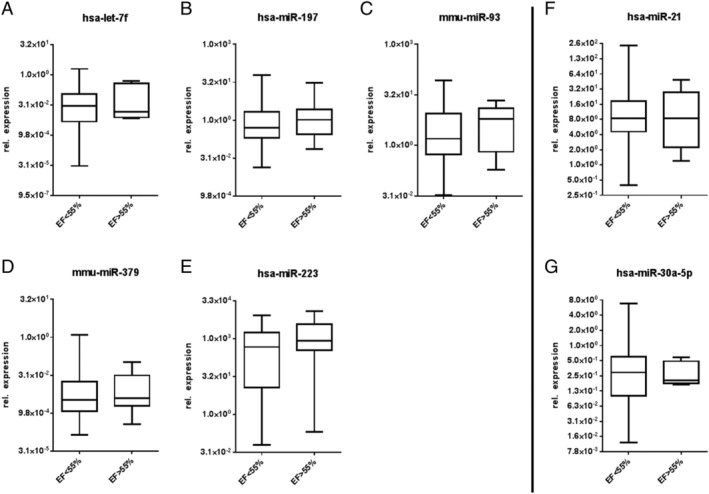
Influence of ejection fraction (EF) (<55%, >55%) on the miRNA expression. (A–E) MiRNAs of patients with inflammatory and/or viral cardiomyopathies; (F, G) miRNAs of DCM patients.
FIGURE 7.
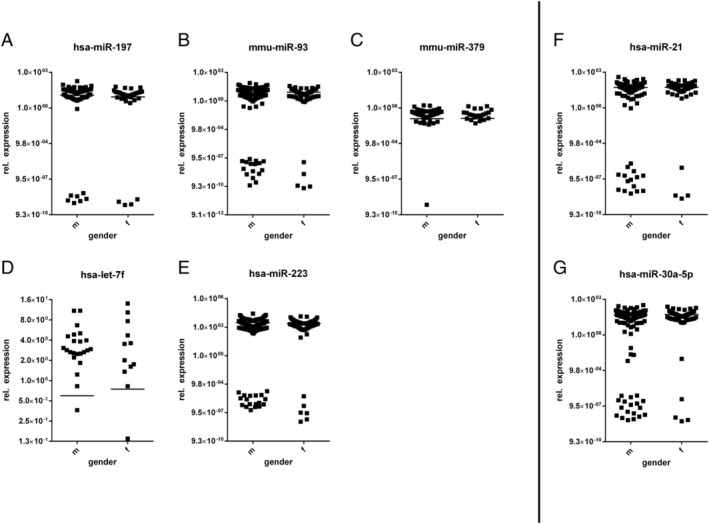
Influence of gender on the microRNA (miRNA) expression. (A–E) MiRNAs of patients with inflammatory and/or viral cardiomyopathies; (F, G) miRNAs of dilated cardiomyopathy patients.
FIGURE 8.
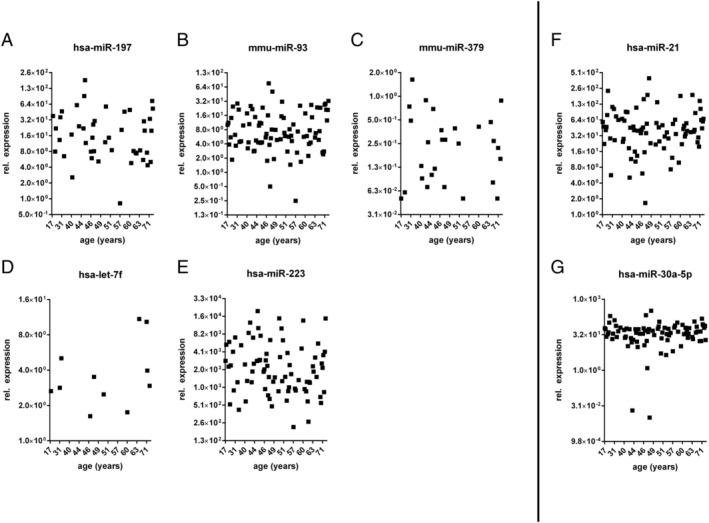
Influence of age on the microRNA (miRNA) expression. (A–E) MiRNAs of patients with inflammatory and/or viral cardiomyopathies; (F, G) miRNAs of dilated cardiomyopathy patients.
Discussion
In the present study, we identified for the first time a miRNA profile from patient's serum samples, which matches the criteria for different inflammatory and/or virally induced cardiomyopathies. The expression of let‐7f, miR‐197, miR‐223, miR‐93, and miR‐379 allowed us to differentiate between patients with a virus or/and inflammation and healthy donors with average specificity of about 93%, while based on the expression of miR‐21 and miR‐30a‐5p, we could distinguish between patients with DCM, inflammatory, and/or virally induced cardiomyopathies and healthy controls with specificity of 95%. This new approach is of high clinical relevance to clarify the indication of taking EMB from patients with suspected inflammation and/or viral heart muscle disease for identification of causative factors (inflammation and/or virus), getting a clear diagnosis and biopsy‐proven causal treatment. The main message of this study is that accurate molecular signatures in serum essentially improve the clinical detection of patients with inflammatory/viral diseases as a prerequisite for the targeted diagnostics by taking EMBs, which are the gold standard for correct diagnosis.
Moreover, in this study, we showed that EF, gender, and age have no significant influence on the expression level of miRNAs in this setting, meaning that the miRNA profile can identify patients with inflammatory and/or virally induced cardiomyopathies by the expression level of particular miRNAs regardless of other factors.
Investigations of other microRNAs in cardiovascular diseases
As miRNAs are highly investigated, their dysregulation has been widely reported in cancer and different cardiac and vascular diseases. 30 , 31 However, all investigations have been performed on the research level so far. MiR‐208 production was detected exclusively in the heart being a candidate biomarker of myocardial injury. 32 Plasma concentrations of miR‐208 increased significantly (P < 0.0001) after isoproterenol‐induced myocardial injury and showed a similar time course to the concentration of cTnI, a classic biomarker of myocardial injury. 32
MicroRNA‐155 plays a crucial role in haematopoietic lineage differentiation, immunity, inflammation, viral infections, and vascular remodelling, which is linked to cardiovascular diseases such as CAD, abdominal aortic aneurysm, HF, and diabetic heart disease. 33 The results, however, are not consistent.
Declining levels of circulating miRNAs, including miR‐18a, miR‐27a, miR‐30e, miR‐26b, miR‐199a, miR‐106a, and miR‐652, are found in patients with HF. Reductions in circulating miRNAs let‐7i, miR‐18b, miR‐18a, miR‐223, miR‐301a, miR‐652, and miR‐423 have been reported within 48 h after acute HF admission and are associated with an increased risk of 180 days of mortality. MiR‐21 is up‐regulated and miR‐1 is down‐regulated in patients with symptomatic HF. 34
MicroRNAs profile in cardiovascular diseases
Let‐7 was subsequently found as the first known human miRNA. The human let‐7 family of miRNAs contains now 12 members that become the most studied miRNAs in development, stem cell biology, aging, and metabolism. 35 The aberrant expression of let‐7 members has been revealed in cardiovascular diseases, such as heart hypertrophy, cardiac fibrosis, DCM, myocardial infarction, arrhythmia, angiogenesis, atherosclerosis, and hypertension. 36 The circulating let‐7b is suspected to be the biomarker of acute myocardial infarction and let‐7i, the biomarker of DCM, 36 and, as stated during our investigations, let‐7f is suspected to be the biomarker of inflammatory and/or virally induced heart diseases.
MiR‐197 represents a molecule that is associated with a wide range of pathologic conditions—from non‐neoplastic diseases (e.g. myocardial infarction) to major human malignancies (e.g. cancers). 37 The dysregulation of miR‐197 has been already reported in human heart disease 38 that confirms our results achieved in patients with unexplained HF.
MiR‐223 expression is a biomarker and therapeutic target in cancer and inflammation. 39 MiR‐223 is deregulated in damaged smooth, skeletal, and cardiac muscles. Many studies have demonstrated a possible role of miR‐223 in vascular smooth muscle cells and in cardiomyocytes: miR‐223 is proposed to play a protective and anti‐inflammatory role, and its expression is dysregulated in cardiovascular diseases such as vascular calcification, acute myocardial infarction, and diabetic and non‐diabetic HF. 40 Thus, from our experiments, we can confirm a crucial role of miR‐223 in patients with unexplained HF.
MiR‐197 and miR‐223 predict cardiovascular death in patients with symptomatic CAD 41 and are involved in endovascular inflammation and platelet activation and have been described as biomarkers in the diagnosis of CAD. 42
MiR‐93 is elevated in the blood of CAD patients. 43 Similar to human CAD, miR‐93 is elevated in both ventricle tissue and blood in mice and is secreted from cardiomyocytes cultured under hypoxia. Interestingly, miR‐93 inhibits apoptosis and protects cardiomyocytes from ischaemia/reperfusion injury. In another type of ischaemic disease like stoke and peripheral arterial disease, miR‐93 shows long‐term protective effects via enhancing angiogenesis. 43 In our research, we confirmed the influence of pathological cardiac conditions on the miRNA expression.
MiR‐379 may be a novel biomarker for the diagnosis of acute myocardial infarction, as plasma miR‐379 level was significantly decreased in patients with acute myocardial infarction compared with healthy donors 44 and increased in patients with unexplained HF that has been detected during our experiments.
MiR‐21 is one of the most intensively studied miRNAs in recent years. 45 Due to the critical functions of its target proteins in various signalling pathways, miR‐21 has become an attractive target for genetic and pharmacological modulation in various disease conditions. MiR‐21 has been found to be up‐regulated in many pathological conditions including cancer and cardiovascular diseases 46 that has been confirmed in our experiments by DCM patients.
MiR‐30a‐5p was significantly elevated on admission in those patients who developed left ventricular dysfunction and HF symptoms 6 months after acute myocardial infarction: a bioinformatics analysis indicated that miR‐30a‐5p may regulate genes involved in cardiovascular pathogenesis 47 and up‐regulated in patients with DCM as stated by our experiments.
We think that miRNA profile consisted of let‐7f, miR‐197, miR‐223, miR‐93, miR‐379, miR‐21, and miR‐30a‐5p normalized by miR‐30a‐3p has great clinical potential in the future. These accurate molecular signatures in serum essentially improve the clinical detection of patients with inflammatory diseases of the heart.
Study limitations
Further in‐depth studies have to be conducted to demonstrate the relevance of miRNA profiling in clinical routine practice and to investigate the more value of differential miRNAs.
It has to be mentioned that results might vary across the source material (i.e. serum, plasma, and tissue), the disease process including other forms of cardiomyopathies being investigated, and platforms (PCR devices, software, and thresholds). There is a huge number of factors that may alter the miRNAs level between cohorts and studies including methodology and therapy of the patients, requiring careful and critical evaluation when interpreting findings and comparing result from different groups.
Conclusions
Application of miRNA profiling in blood samples is a clinical option to early identify patients who have unexplained HF based on virus positive/negative inflammatory myocardial disease. This blood‐based miRNA screening will reduce the disproportion of underdiagnosed cardiac patients by strong indication for an EMB to diagnose the origin of heart muscle disease.
This is the first study investigating the miRNA expression based only on EMB diagnostics.
Clinical perspectives
This study provides miRNA expression profiling in patient serum in which we could find a distinct pattern of differentially expressed miRNAs for identification of different inflammatory and/or virally induced heart muscle diseases. The analysis of miRNA expression profile is of high clinical relevance to clarify the indication for EMB in unexplained HF to get a reliable diagnosis and improve the prognosis by direct therapeutic implications for the patients. Thus, according to our data, the cost‐effective expression analysis of miRNA profile in the serum at the time of diagnosis can be used as a novel molecular tool that will contribute to a more optimized clinical management for patients with unexplained HF. However, it has to be mentioned that it is not replacing EMB, as EMB is the only diagnostic tool for establishing aetiological diagnosis (viral or immune‐mediated) in myocarditis and DCMi. 3 An incomplete diagnosis may provide an incomplete picture of the disease, leading to misinterpretation and possibly incorrect treatment decisions. MiRNA profile just determines which patient requires an EMB, as a prerequisite for an aetiology‐driven diagnosis and specific, causal, personalized therapy.
Conflict of interest
None declared.
Acknowledgements
This work was supported by grants of ERA‐Net on Cardiovascular Diseases (ERA‐CVD; JTC2016‐40‐158; Berlin, Germany; Rome, Italy; Madrid, Spain), a ProFIT grant of the Investitionsbank Berlin/EFRE (No. 10169028, Berlin, Germany), a grant of the German Research Foundation (DFG)’, Transregional Collaborative Research Center ‘Inflammatory Cardiomyopathy – Molecular Pathogenesis and Therapy’ (CRC TR19), and grants from the Federal Ministry of Education and Research (BMBF, 03WKP55D). For their excellent technical assistance, we thank K. Winter, S. Ochmann, C. Seifert, P. Liebig, J. Klostermann, and K. Errami.
Aleshcheva, G. , Pietsch, H. , Escher, F. , and Schultheiss, H.‐P. (2021) MicroRNA profiling as a novel diagnostic tool for identification of patients with inflammatory and/or virally induced cardiomyopathies. ESC Heart Failure, 8: 408–422. 10.1002/ehf2.13090.
References
- 1. Schultheiss H‐P, Kühl U, Cooper LT. The management of myocarditis. Eur Heart J 2011; 32: 2616–2625. [DOI] [PubMed] [Google Scholar]
- 2. Escher F, Lassner D, Kühl U, Gross U, Westermann D, Poller W, Skurk C, Weitmann K, Hoffmann W, Tschöpe C, Schultheiss H‐P. Analysis of endomyocardial biopsies in suspected myocarditis—diagnostic value of left versus right ventricular biopsy. Int J Cardiol 2014; 177: 76–78. [DOI] [PubMed] [Google Scholar]
- 3. Escher F, Tschöpe C, Lassner D, Schultheiss H‐P. Myocarditis and inflammatory cardiomyopathy: from diagnosis to treatment. Turk Kardiyol Dern Ars 2015; 43: 739–748. [DOI] [PubMed] [Google Scholar]
- 4. Schultheiss H‐P, Piper C, Sowade O, Waagstein F, Kapp J‐F, Wegscheider K, Groetzbach G, Pauschinger M, Escher F, Arbustini E, Siedentop H, Kuehl U. Betaferon in chronic viral cardiomyopathy (BICC) trial: effects of interferon‐β treatment in patients with chronic viral cardiomyopathy. Clin Res Cardiol 2016; 105: 763–773. [DOI] [PubMed] [Google Scholar]
- 5. Escher F, Kühl U, Lassner D, Poller W, Westermann D, Pieske B, Tschöpe C, Schultheiss H‐P. Long‐term outcome of patients with virus‐negative chronic myocarditis or inflammatory cardiomyopathy after immunosuppressive therapy. Clin Res Cardiol 2016; 105: 1011–1020. [DOI] [PubMed] [Google Scholar]
- 6. Kühl U, Schultheiss H‐P. Myocarditis: early biopsy allows for tailored regenerative treatment. Dtsch Arztebl Int 2012; 109: 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ling H, Fabbri M, Calin GA. MicroRNAs and other non‐coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013; 12: 847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hodgkinson CP, Kang MH, Dal‐Pra S, Mirotsou M, Dzau VJ. MicroRNAs and cardiac regeneration. Circ Res 2015; 116: 1700–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res 2008; 103: 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang E, Nie Y, Zhao Q, Wang W, Huang J, Liao Z, Zhang H, Hu S, Zheng Z. Circulating miRNAs reflect early myocardial injury and recovery after heart transplantation. J Cardiothorac Surg 2013; 8: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang L, Wang B, Zhou Q, Wang Y, Liu X, Liu Z, Zhan Z. MicroRNA‐21 prevents excessive inflammation and cardiac dysfunction after myocardial infarction through targeting KBTBD7. Cell Death Dis 2018; 9: 769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schulte C, Zeller T. microRNA‐based diagnostics and therapy in cardiovascular disease—summing up the facts. Cardiovasc Diagn Ther 2015; 5: 17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou S, Jin J, Wang J, Zhang Z, Freedman JH, Zheng Y, Cai L. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin 2018; 39: 1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dimmeler S, Zeiher AM. Circulating microRNAs: novel biomarkers for cardiovascular diseases? Eur Heart J 2010; 31: 2705–2707. [DOI] [PubMed] [Google Scholar]
- 15. Condorelli G, Latronico MVG, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol 2014; 63: 2177–2187. [DOI] [PubMed] [Google Scholar]
- 16. Gidlöf O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, Erlinge D. Circulating cardio‐enriched microRNAs are associated with long‐term prognosis following myocardial infarction. BMC Cardiovasc Disord 2013; 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schultheiss H‐P, Fairweather D, Caforio ALP, Escher F, Hershberger RE, Lipshultz SE, Liu PP, Matsumori A, Mazzanti A, McMurray J, Priori SG. Dilated cardiomyopathy. Nat Rev Dis Primers 2019; 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cavarretta E, Frati G. MicroRNAs in coronary heart disease: ready to enter the clinical arena? Biomed Res Int 2016: 2016, 2150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM, Voinea SC. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cell 2020; 9: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aretz HT. Myocarditis: the Dallas criteria. Hum Pathol 1987; 18: 619–624. [DOI] [PubMed] [Google Scholar]
- 21. Caforio ALP, Pankuweit S, Arbustini E, Basso C, Gimeno‐Blanes J, Felix SB, Fu M, Heliö T, Heymans S, Jahns R, Klingel K, Linhart A, Maisch B, McKenna W, Mogensen J, Pinto YM, Ristic A, Schultheiss H‐P, Seggewiss H, Tavazzi L, Thiene G, Yilmaz A, Charron P, Elliott PM, European Society of Cardiology Working Group on Myocardial and Pericardial Diseases . Current state of knowledge on aetiology, diagnosis, management, and therapy of myocarditis: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2013; 34: 2636–2648. [DOI] [PubMed] [Google Scholar]
- 22. Kühl U, Lassner D, Dorner A, Rohde M, Escher F, Seeberg B, Hertel E, Tschope C, Skurk C, Gross UM, Schultheiss H‐P, Poller W. A distinct subgroup of cardiomyopathy patients characterized by transcriptionally active cardiotropic erythrovirus and altered cardiac gene expression. Basic Res Cardiol 2013; 108: 372. [DOI] [PubMed] [Google Scholar]
- 23. Kühl U, Pauschinger M, Noutsias M, Seeberg B, Bock T, Lassner D, Poller W, Kandolf R, Schultheiss H‐P. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation 2005; 111: 887–893. [DOI] [PubMed] [Google Scholar]
- 24. Kühl U, Schultheiss H. Viral myocarditis. Swiss Med Wkly 2014; 144: w14010. [DOI] [PubMed] [Google Scholar]
- 25. Denk J, Boelmans K, Siegismund C, Lassner D, Arlt S, Jahn H. MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer's disease. PLoS ONE 2015; 10: e0126423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kok MGM, Halliani A, Moerland PD, Meijers JCM, Creemers EE, Pinto‐Sietsma S‐J. Normalization panels for the reliable quantification of circulating microRNAs by RT‐qPCR. FASEB J 2015; 29: 3853–3862. [DOI] [PubMed] [Google Scholar]
- 27. Benz F, Roderburg C, Vargas Cardenas D, Vucur M, Gautheron J, Koch A, Zimmermann H, Janssen J, Nieuwenhuijsen L, Luedde M, Frey N, Tacke F, Trautwein C, Luedde T. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp Mol Med 2013; e42: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Masè M, Grasso M, Avogaro L, D'Amato E, Tessarolo F, Graffigna A, Denti MA, Ravelli F. Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Sci Rep 2017; 7: 41127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ronde MWJ, de Ruijter JM, Lanfear D, Bayes‐Genis A, Kok MGM, Creemers EE, Pinto YM, Pinto‐Sietsma S‐J. Practical data handling pipeline improves performance of qPCR‐based circulating miRNA measurements. RNA 2017; 23: 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang Y, Wang H, Li Y, Guo S, Zhang L, Cai J. Peripheral blood miRNAs as a biomarker for chronic cardiovascular diseases. Sci Rep 2014; 4: 5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duchnowski P, Hryniewiecki T, Kuśmierczyk MSP. The usefulness of selected biomarkers in patients with valve disease. Biomark Med 2018; 12: 1341–1346. [DOI] [PubMed] [Google Scholar]
- 32. Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR‐208 as a biomarker of myocardial injury. Clin Chem 2009; 55: 1944–1949. [DOI] [PubMed] [Google Scholar]
- 33. Cao RY, Li Q, Miao Y, Zhang Y, Yuan W, Fan L, Liu G, Mi Q, Yang J. The emerging role of microRNA‐155 in cardiovascular diseases. Biomed Res Int 2016; 2016: 9869208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ovchinnikova ES, Schmitter D, Vegter EL, Maaten JM, ter Valente MAE, Liu LCY, Harst P, van der Pinto YM, Boer RA, de Meyer S, Teerlink JR, O'Connor CM, Metra M, Davison BA, Bloomfield DM, Cotter G, Cleland JG, Mebazaa A, Laribi S, Givertz MM, Ponikowski P, van der Meer P, van Veldhuisen DJ, Voors AA, Berezikov E. Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail 2016; 18: 414–423. [DOI] [PubMed] [Google Scholar]
- 35. Roush S, Slack FJ. The let‐7 family of microRNAs. Trends Cell Biol 2008; 18: 505–516. [DOI] [PubMed] [Google Scholar]
- 36. Bao M‐H, Feng X, Zhang Y‐W, Lou X‐Y, Cheng Y, Zhou H‐H. Let‐7 in cardiovascular diseases, heart development and cardiovascular differentiation from stem cells. Int J Mol Sci 2013; 14: 23086–23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mavridis K, Gueugnon F, Petit‐Courty A, Courty Y, Barascu A, Guyetant S, Scorilas A. The oncomiR miR‐197 is a novel prognostic indicator for non‐small cell lung cancer patients. Br J Cancer 2015; 112: 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Condorelli G, Latronico MVG, Dorn Gerald WII. microRNAs in heart disease: putative novel therapeutic targets? Eur Heart J 2010; 31: 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taïbi F, Metzinger‐Le Meuth V, Massy ZA, Metzinger L. miR‐223: an inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta 2014; 1842: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 40. Rangrez A, Kumari M, Frey N.An emerging role of microRNA miR‐223 in cardiovascular pathophysiology. microRNAs Cardiovasc Res 2013: 23. [Google Scholar]
- 41. Schulte C, Molz S, Appelbaum S, Karakas M, Ojeda F, Lau DM, Hartmann T, Lackner KJ, Westermann D, Schnabel RB, Blankenberg S, Zeller T. miRNA‐197 and miRNA‐223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLoS ONE 2015; 10: e0145930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orenes‐Piñero E, Marín F, Lip GYH. miRNA‐197 and miRNA‐223 and cardiovascular death in coronary artery disease patients. Ann Transl Med 2016; 4: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li K, Lin T, Chen L, Wang N. MicroRNA‐93 elevation after myocardial infarction is cardiac protective. Med Hypotheses 2017; 106: 23–25. [DOI] [PubMed] [Google Scholar]
- 44. Sanson M, Mournetas V, Massourides E, Barthélémy I, Blot S, Pinset C, Richard I, Israeli D. Expression pattern and biological function of miR‐379 in muscular dystrophy. Neuromuscul Disord 2017; 27: S166. [Google Scholar]
- 45. Li X, Wei Y, Wang Z. microRNA‐21 and hypertension. Hypertens Res 2018; 41: 649–661. [DOI] [PubMed] [Google Scholar]
- 46. Cheng Y, Zhang C. MicroRNA‐21 in cardiovascular disease. J Cardiovasc Transl Res 2010; 3: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maciejak A, Kostarska‐Srokosz E, Gierlak W, Dluzniewski M, Kuch M, Marchel M, Opolski G, Kiliszek M, Matlak K, Dobrzycki S, Lukasik A, Segiet A, Sygitowicz G, Sitkiewicz D, Gora M, Burzynska B. Circulating miR‐30a‐5p as a prognostic biomarker of left ventricular dysfunction after acute myocardial infarction. Sci Rep 2018; 8: 9883. [DOI] [PMC free article] [PubMed] [Google Scholar]


