Abstract
Introduction
MTHFR methylation status is associated with microvascular complications in diabetes, but the factors influencing this profile remain unknown.
Objective
The aim of this study was to evaluate the influence of physical activity level and nutritional status on the methylation profile of the MTHFR gene in patients with type 2 diabetes mellitus (T2DM).
Methods
A total of 111 patients, 43 men and 68 women diagnosed with DM (7.0 ± 2.3 years), answered the International Physical Activity Questionnaire (IPAQ) and underwent blood collection for biochemical analysis, DNA extraction, and MTHFR gene methylation profile determination.
Result
The comparison of the methylation pattern showed that the partially methylated profile predominates in the insufficiently active group (85%), which does not occur in the sufficiently active group (54%) (p = 0.012). No differences were found in the nutritional status comparison. Logistic regression including overweight, waist circumference, gender, age, time of DM, hypertension, dyslipidemia, smoking, alcoholism, and family DM revealed that the association of the level of physical activity with methylation profile proved to be independent of these confounding variables. Considering the partially methylated profile as a result, being physically inactive favors the partially methylated MTHFR pattern in patients with DM.
Conclusion
We concluded that insufficient physical activity is associated with partially methylated pattern of MTHFR promoter.
Keywords: diabetes, MTHFR, methylation, physical activity (exercise), diabetes complication
Introduction
Research in the area of physical activity epidemiology in recent decades has revealed that there is a clear negative association between physical activity level and the emergence of various chronic diseases including diabetes mellitus (DM) (Lackland and Voeks, 2014; Aune et al., 2015; Zhao et al., 2019). These associations need to be studied in clinical, physiological, and molecular aspects. As an example, the level of physical activity (LPA) has been reported as an epigenetic modulator, affecting the methylation pattern of specific genes and global methylation, generally favoring hypomethylation (Voisin et al., 2015). However, data of this nature are still very scarce.
The MTHFR gene encodes an enzyme responsible for converting 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate (MTHF), donating in this way methyl groups that are used in the synthesis of S-adenosyl methionine, which is the substrate for methylation reactions, including DNA. MTHFR activity may be influenced by single nucleotide polymorphisms that are related to the risk of individuals developing diabetes and its major chronic complications (Fekih-Mrissa et al., 2017; Chen et al., 2018; Ramanathan et al., 2019). There are evidences that MTHFR gene expression is influenced by its promoter’s methylation (Wang et al., 2007). Thus, detection of the methylation pattern of this gene may indicate metabolic aspects of patients. In fact, MTHFR methylation profile has been associated with microvascular diabetes complications diabetic retinopathy (DR) and nephropathy (DN) (Ghattas et al., 2014; Yang et al., 2016; Nunes et al., 2017).
Knowing that the environment regulates gene expression, and that people with higher levels of physical activity are better protected against both diabetes and its complications (Sailani et al., 2019), in this study, we hypothesized that the LPA of diabetics is involved in modulating the methylation profile of the MTHFR gene. Thus, the objective of this study was to evaluate whether the LPA and nutritional status influence the methylation profile of the MTHFR gene in patients with type 2 diabetes mellitus (T2DM).
Methodology
Participants
The study consisted of 111 individuals found in the Reference Services for the diabetic patient of the Lauro Wanderley University Hospital of the Federal University of Paraiba (HULW/UFPB) from December 2013 to November 2016. Patients with T2DM of both sexes aged more than 40 years old with at least 5 and at most 10 years of disease evolution with (n = 43) or without diabetic (n = 68) complications were included. Considering the universe of diabetics of João Pessoa (Paraíba State, Brazil), this sample size was representative of this population for a confidence level of 95% and a margin of error of 9.3%.
Clinical Characterization
The diagnosis of DR was based on ophthalmoscopy after pupil dilation with tropicamide 0.5%. Images of the retina (macula and central disk) were captured at an angle of 45° by a background camera. The photographs were analyzed according to the ACCORD (Action to Control Cardiovascular Risk in Diabetes) standards and recommendations (Chew et al., 2007) and the Early Treatment Diabetic Retinopathy Study (ETDRS) (Gangnon et al., 2008). DN was found by determining urinary albumin excretion in 24 h and by the patient’s clinical condition through the nephrologist physician’s anamnesis. Samples with values below 30 mg/L in 24 h of microalbuminuria were considered normal and absence of DN; samples with values equal to or above 30 mg/L in 24 h were considered indicative of DN (American Diabetes Association, 2018).
This research was approved by the Ethical Committee of the Lauro Wanderley University Hospital (João Pessoa, Paraíba State, Brazil). All ethical procedures followed the National Health Council Resolution 466/2012.
Sociodemographic Profile
Gender, age, risk factors (smoking and drinking), and disease history (time of diagnosis of DM, family history of DM, and complications of DM) were collected.
Anthropometric Profile
To measure weight, an electronic scale with capacity up to 150 kg and sensitivity of 100 g (Filizola®) was used. Subjects were weighed in lightweight, barefoot, upright posture, feet parallel and fully supported on the scale platform and with arms across their body (Ministério da Saúde, 2018). Height was measured using a stadiometer attached to the scale, which consists of a steel tube with anodized aluminum ruler, measuring from 97 to 192 cm with 0.5 cm divisions. The subjects were in an upright posture, feet together and heels against the wall. The apex of the ear and the outer corner of the eye were in a line parallel to the floor, forming a 90° angle with the stadiometer bar; thus, the horizontal bar of the stadiometer was lowered and resting on the head, allowing reading in centimeters (Ministério da Saúde, 2018). To measure abdominal circumference (AC), patients should have their legs slightly apart and the inelastic tape measure pass the umbilical scar line for the diagnosis of the result. The reference value for men is 102 cm and for Caucasian women 88 cm (National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults, 2002).
Body mass index (BMI) was calculated by dividing weight (in kg) by height (in meters squared). The values obtained will be categorized into low weight, normal weight, overweight, or obesity for adult individuals and thinness, normal weight, and overweight for elderly individuals according to the World Health Organization cutoff points (WHO, 2018).
Physical Activity Level
To quantify the LPA of patients, the long version International Physical Activity Questionnaire (IPAQ) was used (Pardini et al., 2001), allowing to generate an estimate of the weekly time patients spent in physical activity at different intensities (vigorous or moderate). In different daily activities, such as work, transportation, domestic tasks, and leisure, as well as the estimated time spent in activities in the sitting position, the IPAQ was applied individually by a properly trained researcher.
According to WHO (2018) to be classified as active, at least 150 min of mild or moderate physical activity per week or at least 75 min of intense physical activity per week is required.
The results divided the patients into two groups, the active and the insufficiently active according to the IPAQ nomenclature.
Biochemical Determinations
For the biochemical determinations, blood was collected after 12 h fasting. Enzymatic commercial tests were employed in glycemia, total cholesterol, high-density lipoprotein (HDL), and triglyceride determinations. Glycated hemoglobin was determined by immunoturbidimetry technique. All tests were performed in an automated analyzer (LabMax 240; Labtest, Lagoa Santa, Brazil) using a standardized kit and following the manufacturer’s recommended guidelines (Labtest, Lagoa Santa, Brazil).
Low-density lipoprotein (LDL) concentration was determined by Friedewald formula, where: (LDL) = (total cholesterol) − (HDL) − (triglycerides ÷ 5) (Ohkawa et al., 1979).
The cutoffs applied were: total cholesterol < 200 mg/dl, LDL < 100 mg/dl, HDL > 60 mg/dl, triglycerides < 150 mg/dl (Brazilian Society of Cardiology, 2007), blood glucose < 126 mg/dl, and glycated hemoglobin 7% (American Diabetes Association (ADA), 2010).
Dyslipidemia was defined as increased levels of total cholesterol, LDL, or triglycerides or decreased levels of HDL (Brazilian Society of Cardiology, 2007).
Methylation Profile Analysis
Venous blood (4 ml) was collected by venipuncture into sterile tubes containing 7.2 mg K3 EDTA and stored in a freezer at −20°C for 15 days until extraction.
Blood samples were diluted in a lysis solution (10 mM Tris–HCl pH 8.5, 5 mM EDTA, 0.3 M sucrose, and 1% Triton-X-100). After centrifugation was followed at 3,200 rpm, the supernatant was discharged. This process was repeated three times. The precipitate was then resuspended in a second lysis solution [10 mM Tris–HCl pH 8, 0.5% sodium dodecyl sulfate (SDS), 5 mM EDTA] and 0.2 μg proteinase K (Invitrogen, Carlsbad, CA, United States) and incubated at 55°C in a water bath. After 7 h of incubation 500 μl of a 1 mM EDTA 7.5 M ammonium acetate solution was added followed by vigorous mixing. The mixture was centrifuged for 10 min at 14,000 × g at 4°C, and 700 μl of the supernatant was transferred to a new tube where DNA precipitation with 540 μl of isopropanol was performed. The DNA precipitate was washed with 70% ethanol, centrifuged (12,000 × g for 5 min), dried, and resuspended in Tris–EDTA pH 8.0 buffer (Miller et al., 1988).
The extracted DNA was converted (500 ng) by sodium bisulfite using the EZ DNA MethylationTM Kit (Zymo Research) according to the manufacturer’s instructions (conversion efficiency ≥ 99%). The principle of the technique is to transform unmethylated cytosine into uracil without causing alteration in methylated cytosine from the DNA sample. Different methylation patterns will be recognized through PCRs that employ specific pairs of primers for each methylation condition (methylated and unmethylated). Methylated and unmethylated DNA (Cells-to-CpGTM Methylated and Unmethylated DNA Control Kit; Life Technologies) were used as controls, which were modified, as previously mentioned, and amplified by PCR as control of the reactions with primers for the methylated and unmethylated conditions, respectively.
For each methylation-specific PCR, a mixture containing 100 ng bisulfite-transformed DNA (both controls and samples), 0.7 μl (7 μM) from each methylated (sense: 5′-tagatttaggtacgtgaagtagggtagac-3′ and antisense: 5′-gaaaaactaataaaaaaaccgaca-3′) and unmethylated target-specific primer (sense: 5′-tttaggtatgtgaagtagggtagatgt-3′ and antisense: 5′-caaaaaactaataaaaaaccaacaaa-3′) as previously described (Khazamipour et al., 2009) and 1 × Go Taq Hot Start Green Master Mix (Promega Corporations, Madison, WI, United States) in a 25 μl final reaction was performed. The annealing temperature was 58°C for 40 s and 40 cycles. Amplified PCR samples were loaded (7 μl) on 3% red gel agarose gels and electrophoresed. The DNA bands were visualized in ultraviolet light transluminator.
Statistical Analysis
Frequency distribution was used for categorical variables, as well as descriptive statistics (mean, standard deviation) for continuous variables. Chi-square test was used to test the relationship between methylation profile and physical activity level, along with a binary logistic regression. Statistical analyses were performed using SPSS version 24.0 (SPSS, Inc., Chicago, IL, United States) and p < 0.05 as statistical significance.
Results
Population Characteristics
Participants were 43 males and 68 females, mean age 58.2 ± 9.4 years with a diagnosis of DM at about 7.0 ± 2.3 years, with or without microvascular complications. The epidemiological, clinical, anthropometric, and metabolic characteristics of the studied groups are shown in Table 1. It was observed that most individuals were hypertensive, dyslipidemic, and overweight or obese, and this occurred in both groups (active and insufficiently active). The group classified as active had a lower percentage of males (p = 0.01), dyslipidemics (p = 0.01), smokers (p = 0.02), and hypertension carriers (0.01). Active and insufficiently active groups were similar in the distribution of elderly people percentual, complications (DR and DN), overweight/obesity, DM duration, and treatment in course.
TABLE 1.
Characteristics of diabetic patients as a function of levels of physical activity.
| Active n = 91 | Insufficiently active n = 20 | ||||
| n | % | N | % | P | |
| <60 years | 50 | 54.9 | 8 | 40 | 0.317 |
| Male | 32 | 35.2 | 11 | 55 | 0.01* |
| Dyslipidemia | 69 | 75.8 | 18 | 90 | 0.01* |
| Smoking habit | 7 | 7.7 | 2 | 10 | 0.02* |
| Alcoholism | 13 | 14.3 | 1 | 5 | 0.01* |
| Overweight/obesity | 77 | 84.6 | 15 | 75 | 0.070 |
| Hypertension | 62 | 68.1 | 17 | 85 | 0.01* |
| DM duration | 87 | 95.6 | 20 | 100 | 0.09 |
| Insulin use | 21 | 23.1 | 1 | 5 | 0.187 |
| Diabetes in the family | 61 | 67 | 14 | 70 | 0.022* |
| DM complications | 43 | 47.2 | 10 | 50 | 0.09 |
| Diabetic retinopathy | 20 | 22 | 5 | 25 | 0.07 |
| Diabetic nephropathy | 30 | 33 | 8 | 40 | 0.26 |
*DM diagnosis time ≥ 5 years. Test x2, p < 0.05. João Pessoa, 2016 (n = 111).
In the present study, patients were grouped according to the IPAQ: active (n = 91) and the insufficiently active (n = 20) with a mean time of physical activity of 949.3 ± 850.7 and 33.2 ± 51.7 min, respectively.
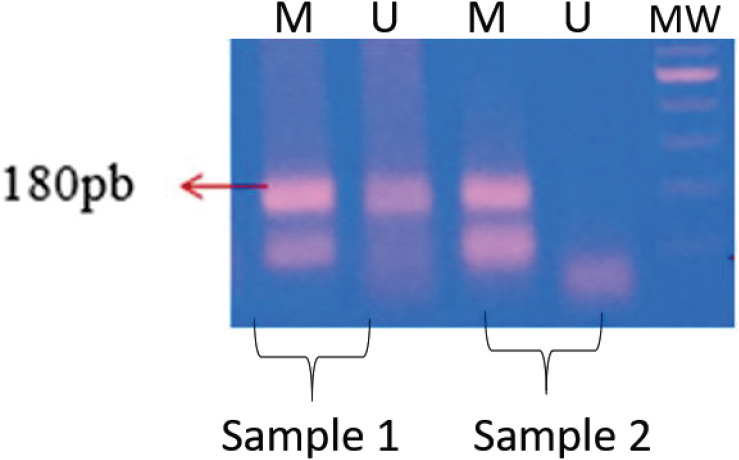
The methylation profile was identified by agarose gel electrophoresis as shown in Figure 1. The presence of the band with only the pair of primers for the methylated condition results in a methylated profile. The presence of bands resulting from the amplification reactions with the two pairs of primers (methylated and non-methylated) simultaneously is related to the partially methylated profile. We did not observe the amplification of any sample only with the unmethylated condition, so we do not have this category of methylation profile for this gene in the population studied.
FIGURE 1.
Methylation profile of the MTHFR gene. Methylation patterns of MTHFR gene samples. Sample 1, partially methylated pattern; sample 2, fully methylated pattern; MW, 100 bp pattern. M = methylated/U = unmethylated.
As the primer pairs are specific to each tested condition (methylated and non-methylated), only samples after transformation by the bisulfite technique were tested. Commercial methylated and non-methylated controls were used to make sure that the efficiency of the transformation process and the amplification reaction worked properly.
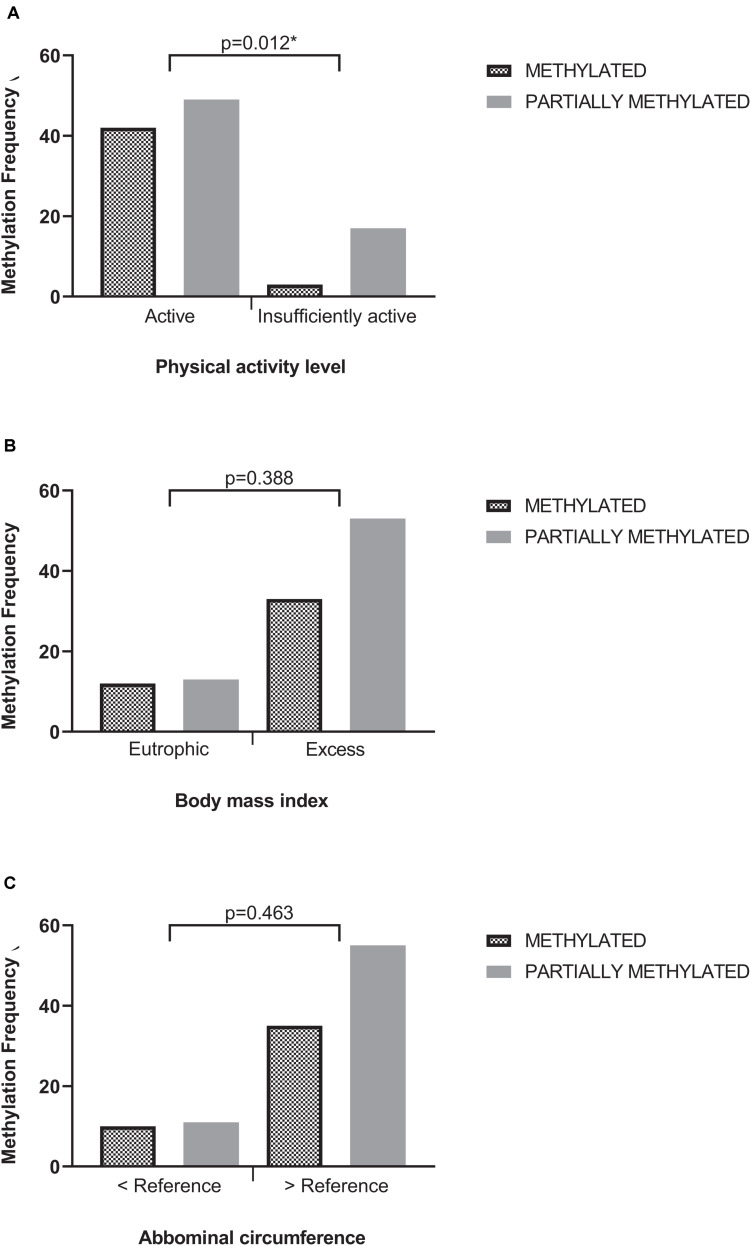
Among the samples, 66 (59.5%) presented the methylated profile, and 45 (45.5%) presented the partially methylated profile. Samples with the unmethylated profile of the MTHFR gene were not present (Figure 2A).
FIGURE 2.
Methylation profile according to physical activity level (A), body mass index (B), and abdominal circumference (C). Chi-square test; p = value of significance. AC, Abdominal circumference.
Comparing the methylation profile in relation to the LPA, it was possible to observe a balance between the methylated and partially methylated states among those classified as active (respectively, 46 and 54%). On the other hand, among the insufficiently active, there was an imbalance in distribution, with 85% of this group with partially methylated profile to only 15% with methylated profile. The chi-square test showed that this difference in distribution was statistically significant (Figure 2A). According to odds ratio (OR), DM patients with inactive profile have a partially methylated MTHFR gene pattern.
Figures 2B,C shows that the methylation profile of the MTHFR was not influenced by the anthropometric parameters BMI (p = 0.388) and waist circumference (p = 0.463) once methylated and partially methylated samples were equally distributed in the BMI and AC categories in the population studied.
Table 2 presents the result of logistic regression, in which the methylation profile was considered the dependent variable and as independent variables gender, age, time of DM, hypertension, dyslipidemia, smoking, alcoholism, family DM, LPA, presence of microvascular complications (DR and/or DN), overweight, and AC. This test revealed that none of the independent variables besides LPA was found to influence the methylation profile, suggesting that LPA is an independent factor influencing the methylation profile. The OR test revealed that being physically active represents a protection factor against the partially methylated profile outcome in patients with DM.
TABLE 2.
Logistic regression model verifying the influence of LPA and confounding variables on the occurrence of MTHFR gene methylation profile.
| OR | Confidence interval | p-value | |
| Age | 0.874 | 0.330–2.310 | 0.785 |
| Male | 0.767 | 0.302–1.946 | 0.576 |
| Hypertension | 0.668 | 0.254–1.754 | 0.413 |
| Hb1AC | 0.652 | 0.206–2.061 | 0.466 |
| Blood glucose | 1.413 | 0.496–4.023 | 0.517 |
| Total cholesterol | 1.081 | 0.328–3.564 | 0.898 |
| HDL | 0.939 | 0.196–4.489 | 0.937 |
| LDL | 1.352 | 0.445–4.110 | 0.595 |
| Triglycerides | 1.436 | 0.500–4.130 | 0.502 |
| LPA | 6.185 | 1.476–25.911 | 0.013* |
| Sex | 0.352 | 0.300–1.906 | 0.553 |
| Smoking | 1.661 | 0.407–6.776 | 0.479 |
| Alcoholism | 0.365 | 0.075–1.897 | 0.227 |
| Diabetes in the family | 1.688 | 0.664–4.291 | 0.271 |
| DM time | 0.918 | 0.353–2.383 | 0.860 |
| Insulin use | 0.604 | 0.224–1.630 | 0.319 |
| Overweight | 0.643 | 0.198–2.083 | 0.462 |
| AC | 1.292 | 0.338–4.560 | 0.707 |
LPA, Physical activity level; AC, Abdominal circumference. *p < 0.05.
No results were significant when comparing the methylation profile with the lipid and glycemic profile of the patients.
Discussion
According to this study, the lack of physical activity in diabetics with a diagnosis time of 5–10 years is associated with the MTHFR promoter partially methylated profile, and this association was independent of the nutritional status.
It is well established in the literature that exercise is an important protective factor for diabetes (Esteghamati et al., 2014; Li et al., 2015; Roya et al., 2019). There is a direct beneficial effect of exercise on β cell function (Li et al., 2015). It is also consensus that increased homocysteine (Hcy) is an independent risk factor for cardiovascular disease and is modifiable by nutrition and exercise (Bostom et al., 1999; Ganguly and Alam, 2015).
Deminice et al. (2016) provided evidence that acute exercise increases plasma Hcy concentration regardless of the duration or intensity of exercise performed. An analysis of physical intervention in groups of pre-diabetic and normoglycemic individuals also found a relationship between high intensity physical activity and increased Hcy levels and impact on other transulfurization pathway metabolites impacting elevated glutathione levels possibly due to a metabolic response to increased production of reactive oxygen species under conditions of intense physical activity (Lee et al., 2019). Similar results were found in experimental studies using acute swimming exercise in rats that showed acute exercise increased the flow of transmethylation reactions that elevate Hcy formation (Riberio et al., 2018). Evidence of the relationship between the effects of physical activity and the metabolism of methyl groups was also demonstrated in the analysis of gene polymorphisms related to the transfer of methyl groups in elite athletes (Terruzzi et al., 2011). A predominance of genotypes that form less functional versions of enzymes involved in DNA methylation, including MTHFR, has been demonstrated in elite athletes (Terruzzi et al., 2011). Similar results were found in strength and speed athletes (Zarebska et al., 2014). Thus, studies that analyzed polymorphism found a relationship between less functional versions of MTHFR and differential physical performance.
In this work, we evaluated the methylation profile of the MTHFR gene, which plays a central role in the methionine Hcy remethylation pathway. We identified predominance of partially methylated profile, which possibly leads to higher levels of expression than the fully methylated profile, in individuals with insufficient levels of physical activity. Knowing that MTHFR is involved in remethylation (and consequent removal) of Hcy, we can speculate that there will be less expression of the enzyme (due to promoter hypermethylation) and consequently higher levels of Hcy in the most active individuals. Assuming that (1) there are evidences that methylation impacts MTHFR gene expression (Wang et al., 2007), (2) in general, DNA promoter methylation is a silencing expression mechanism, (3) the methylated profile predominated in the physically active, (4) there are relations described between high physical activity level and Hcy, and (5) the involvement of MTHFR on the Hcy metabolism, it is possible to suggest that methylation is a mechanism of MTHFR expression control mediated by physical activity.
In the context of diabetes complications, while the hypermethylated MTHFR profile has been associated with DR (Nunes et al., 2017), in the DN, the results are contradictory, because while hypermethylation predominated in patients with end-stage renal disease (Ghattas et al., 2014), diabetics with initial DN or in less advanced stages had a higher occurrence of the non-methylated profile (Yang et al., 2016; Nunes et al., 2017). Hypermethylation of the MTHFR promoter in blood samples has been linked to other relevant clinical conditions, such as women who have had recurrent miscarriages (Mishra et al., 2019) and mothers of Down syndrome patients (Coppedè et al., 2016). In both cases, the method of analyzing methylation employed quantitative techniques, making it difficult to compare with the qualitative data presented here.
It is important to understand the factors that modify the MTHFR methylation pattern, especially those that can be modified (e.g., physical activity), taking into account that DNA methylation is a potentially reversible epigenetic mechanism. Therefore, it is possible from these relationships to identify strategies for preventing or monitoring chronic complications in diabetes and other clinical conditions related to specific profiles.
Some aspects of this work need to be considered. First, the group of patients we studied is made up of type 2 diabetics, and comparisons are made with studies that analyzed athletes. A second aspect is that, in this work, the LPA was accessed through a structured questionnaire, and there was no physical intervention; in addition, there are other ways to measure this variable, such as the use of accelerometers. Third, we analyzed the methylation profile and not the quantification, which would offer more accurate results.
In this study, we found a higher prevalence of hypertensive patients in the inactive group, which corroborates with other studies (Jurik and Stastny, 2019; Sharman et al., 2019; Alpsoy, 2020) that demonstrated the association between aerobic and resistance exercises and reduced blood pressure (BP). The guidelines of the Canadian Journal of Cardiology (Rabi et al., 2020) strongly recommend aerobic exercises of moderate intensity for at least 30 min on at least 3 days of the week or resistance exercises on 2–3 days of the week, aiming at BP control.
Dyslipidemia in this study was more frequent among the inactive, but a high percentage (75.8%) was also found in the active group. The effect of physical activity on the control of lipid metabolism and consequently of circulating levels has a wide repercussion in the literature (Gordon et al., 2014; Mann et al., 2014; Fikenzer et al., 2018).
The contribution of this work is to demonstrate that there is a relationship between MTHFR methylation, a molecular marker previously related to microvascular complications, and activity levels in diabetic individuals. The data presented here aggregate molecular information at the epigenetic level on the well-known relationship between physical activity and diabetes prevention.
Studies of exercise physiology and biochemistry have previously demonstrated the relationship between physical activity and Hcy levels. They also demonstrated an association between physical exercise and global hypomethylation and also a relationship between polymorphisms that impact MTHFR activity and outstanding physical performance. Our study brings the unprecedented approach of evaluating the methylation profile of the MTHFR gene in the context of physical activity, showing that physical inactivity favors the occurrence of the partially methylated profile.
This is a first observation that opens the way for more detailed investigations, exploring the biological and physiological aspects of the association. The association found in the present study of descriptive and cross-sectional characteristics encourages the development of an intervention study to test the effect of a physical training program on the methylation profile of previously sedentary diabetics.
Conclusion
We concluded that insufficient physical activity is associated with partially methylated pattern of MTHFR promoter in patients with diabetes. Our results encourage further studies focused on determining if this result is affected by the metabolic profile of participants with diabetes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Comitê de Ética em Pesquisa do Hospital Universitário Lauro Wanderley. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
TD performed statistical analysis, interpretation of results, and writing of the manuscript. MR performed statistical analysis. AS analyzed data from the manuscript. MS participated in data collection, biochemical analysis, and molecular biology experiments. JF participated in data collection and performed endocrinological consultation in individuals. RN participated in data collection and biochemical analysis. CG participated in data collection and carried out individual consultations. IQ participated in data collection and performed ophthalmological consultation in individuals. NO participated in experiments in molecular biology. DP participated in all stages of the research (study design and logistics, data collection, molecular biology experiments, interpretation of results, statistical analysis, and writing of the manuscript). All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The results presented in this article are product of master’s thesis of Diniz (2020). We thank the Lauro Wanderley University Hospital of the Federal University of Paraíba and all study participants for accepting and allowing the collection of blood samples and financial support institutions, the National Council for Scientific and Technological Development (CNPq, Brasilia, Brazil). Coordination for the Improvement of Higher Education Personnel (CAPES, Brasília, Brazil) and the Paraíba State Research Support Foundation (FAPESQ, Paraíba, Brazil).
Footnotes
Funding. This study was funded by grants from the Paraíba State University Research Support Foundation (FAPESQ, Paraíba, Brazil) the project of the National Council for Scientific and Technological Development (CNPq, Brasília, Brazil) 057/13, public notice 001/2013 FAPESQ/Ministry of Health/CNPq under the agreement SINCOV 774379/2012 FAPESQ/Ministry of Health/CNPq in the PPSUS program for the acquisition of inputs, reagents, and materials necessary for the development of research. This study was funded in part by the Higher Education Personnel Improvement Coordination (CAPES) – Financial Code 001.
References
- Alpsoy S. (2020). Exercise and hypertension. Adv. Exp. Med. Biol. 1228 153–167. 10.1007/978-981-15-1792-1_10 [DOI] [PubMed] [Google Scholar]
- American Diabetes Association (2018). Microvascular Complications and foot care: standards of medical care in diabetesd-2018. Diabetes Care 41 105–118. 10.2337/dc18-S010 [DOI] [PubMed] [Google Scholar]
- American Diabetes Association (ADA) (2010). Standards of medical care in diabetes-2010. Diabetes Care 33 11–61. 10.2337/dc10-S011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune D., Norat T., Leitzmann M., Tonstad S., Vatten L. J. (2015). Physical activity and the risk of type 2 diabetes: a systematic review and dose-response meta-analysis. Eur. J. Epidemiol. 30 529–542. 10.1007/s10654-015-0056-z [DOI] [PubMed] [Google Scholar]
- Bostom A. G., Silbershatz H., Rosenberg I. H., Selhub J., D’Agostino R. B., Wolf P. A., et al. (1999). Nonfasting plasma total homocysteine levels and all-cause and cardiovascular disease mortality in elderly Framingham men and women. Arch. Intern. Med. 159 1077–1080. 10.1001/archinte.159.10.1077 [DOI] [PubMed] [Google Scholar]
- Brazilian Society of Cardiology (2007). IV brazilian guideline on dyslipidemias and prevention of atherosclerosis. Arq. Bras. Cardiol. 88 2–19. [DOI] [PubMed] [Google Scholar]
- Chen D., Wang J., Dan Z., Shen X., Ci D. (2018). The relationship between methylenetetrahydrofolate reductase C677T polymorphism and diabetic retinopathy: A meta-analysis in multiethnic groups. Ophthalmic Genet. 39 200–207. 10.1080/13816810.2017.1401087 [DOI] [PubMed] [Google Scholar]
- Chew E. Y., Ambrosius W. T., Howard L. T., Greven C. M., Johnson S., Danis R. P., et al. (2007). Rationale, design, and methods of the action to control cardiovascular risk in diabetes eye study (ACCORD-EYE). Am. J. Cardiol. 99 103i–111i. 10.1016/j.amjcard.2007.03.028 [DOI] [PubMed] [Google Scholar]
- Coppedè F., Stoccoro A., Tannorella P., Gallo R., Nicolì V., Migliore L. (2016). Increased MTHFR promoter methylation in mothers od down syndrome individuals. Mutat. Res. Fundament. Mol. Mech. Mutag. 787 1–6. 10.1016/j.mrfmmm.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Deminice R., Ribeiro D. F., Frajacomo F. T. T. (2016). The effects of acute exercise and exercise training on plasma homocysteine: a meta-analysis. PLoS One 11:e0151653. 10.1371/journal.pone.0151653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz T. G. (2020). Influência do nível de atividade física e estado nutricional no perfil de metilação do gene MTHFR em pacientes diabéticos. Dissertation/Master’s Thesis, Universidade Federal da Paraíba, João Pessoa, Paraíba, Brazil: Available online at: https://repositorio.ufpb.br/jspui/handle/123456789/18344 [Google Scholar]
- Esteghamati A., Etemad K., Koohpayehzadeh J., Abbasi M., Meysamie A., Noshad S., et al. (2014). Trends in the prevalence of diabetes and impaired fasting glucose in association with obesity in Iran: 2005–2011. Res. Clin. Pract. Diab. 103 319–327. 10.1016/j.diabres.2013.12.034 [DOI] [PubMed] [Google Scholar]
- Fekih-Mrissa N., Mrad M., Ibrahim H., Akremi I., Sayeh A., Jaidane A., et al. (2017). Methylenetetrahydrofolate reductase (MTHFR) (C677T and A1298C) polymorphisms and vascular complications in patients with type 2 diabetes. Can. J. Diabetes. 41 366–371. 10.1016/j.jcjd.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Fikenzer K., Fikenzer S., Laufs U., Werner C. (2018). Effects of endurance training on serum lipids. Vas. Pharmacol. 101 9–20. 10.1016/j.vph.2017.11.005 [DOI] [PubMed] [Google Scholar]
- Gangnon R. E., Davis M. D., Hubbard L. D., Aiello L. M., Chew E. Y., Ferris F. L., III, et al. (2008). A severity scale for diabetic macular edema developed from ETDRS data. Investigat. Ophthalmol. Vis. Sci. 49 5041–5047. 10.1167/iovs.08-2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly P., Alam S. F. (2015). Role of homocysteine in the development of cardiovascular disease. Nutr. J. 14:6. 10.1186/1475-2891-14-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghattas M., El-Shaarawy E., Mesbah N., Abo-Elmatty D. (2014). DNA methylation status of the methylenetetrahydrofolate reductase gene promoter in peripheral blood of end-stage renal disease patients. Mol. Biol. Rep. 41 683–688. 10.1007/s11033-013-2906-7 [DOI] [PubMed] [Google Scholar]
- Gordon B., Chen S., Durstine J. L. (2014). The effects of exercise training on the traditional lipid profile and beyond. Curr. Sports Med. Rep. 13 253–259. 10.1249/JSR.0000000000000073 [DOI] [PubMed] [Google Scholar]
- Jurik R., Stastny P. (2019). Role of nutrition and exercise programs in reducing blood pressure: a systematic review. J. Clin. Med. 8:1393. 10.3390/jcm8091393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazamipour N., Noruzinia M., Fatehmanesh P., Keyhanee M., Pujol P. (2009). MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: the role of epigenetics in male infertility. Hum. Reprod. 24 2361–2364. 10.1093/humrep/dep194 [DOI] [PubMed] [Google Scholar]
- Lackland D. T., Voeks J. h. (2014). Metabolic syndrome and hypertension: regular exercise as part of lifestyle management. Curr. Hypertens Rep. 16:492. 10.1007/s11906-014-0492-2 [DOI] [PubMed] [Google Scholar]
- Lee S., Olsen T., Vinknes K. J., Refsum H., Gulseth H. L., Birkeland K. I., et al. (2019). Plasma sulphur-containing amino acids, physical exercise and insulin sensitivity in overweight dysglycemic and normal weight normoglycemic men. Nutrients 11:10. 10.3390/nu11010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., He S., Liu S., Kong Z., Wang J., Zhang Y. (2015). Effects of different exercise durations on Keap1-Nrf2-ARE pathway activation in mouse skeletal muscle. Free Radic. Res. 49 1269–1274. 10.3109/10715762.2015.1066784 [DOI] [PubMed] [Google Scholar]
- Mann S., Beedie C., Jimenez A. (2014). Differential effects of aerobic exercise, resistance training and combined exercise modalities on cholesterol and the lipid profile: review, synthesis and recommendations. Sports Med. 44 211–221. 10.1007/s40279-013-0110-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. A. W., Dykes D. D., Polesky H. F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16 1215. 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministério da Saúde (2018). SISVAN – FOOD AND NUTRITIONAL SURVEILLANCE SYSTEM: Basic Guidelines for the Collection, Processing, Data Analysis and Information in Health Services. Ministry of Health, Secretariat of Health Care, Department of Primary Care. – Brasilia: Ministry of Health, 2011. 76. Available online at: http://bvsms.saude.gov.br/bvs/publicacoes/orientacoes_coleta_analise_dados_antropometricos.pdf (acessed November 25, 2018). [Google Scholar]
- Mishra J., Talwar S., Kaur L., Chandiok K., Yadav S., Puri M., et al. (2019). Differential global and MTHFR gene specific methylation patterns in preeclampsia and recurrent miscarriages: a case-control study from North India. Gene 704 68–73. 10.1016/j.gene.2019.04.036 [DOI] [PubMed] [Google Scholar]
- National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults (2002). Third report of cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) 2000. NIH publication n. 01-3670. Circulation 106 3143–3421. 10.1161/circ.106.25.3143 [DOI] [PubMed] [Google Scholar]
- Nunes M. K. S., Silva A. S., Evangelista I. W. Q., Filho J. M., Gomes C. N. A. P., do Nascimento R. A. F., et al. (2017). Hypermethylation in the promoter of the MTHFR gene is associated with diabetic complications and biochemical indicators. Diabetol. Metab Syndr. 9:84. 10.1186/s13098-017-0284-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa H., Ohishi N., Yagi K. (1979). Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 95 351–358. 10.1016/0003-2697(79)90738-3 [DOI] [PubMed] [Google Scholar]
- Pardini R., Matsudo S., Araújo T., Matsudo V., Andrade E., Braggio G., et al. (2001). Validation of the international physicalactivity questionaire (IPAQ version 6): pilot study in Brasilian Young adults. Ver. Bras. Ciên. E Mov. 9 45–51. [Google Scholar]
- Rabi D. M., McBrien K. A., Sapir-Pichhadze R., Nakhla M., Ahmed S. B., Dumanski S. M., et al. (2020). Hypertension Canada’s 2020 comprehensive guidelines for the prevention, diagnosis, risk assessment, and treatment of hypertension in adults and children. Can. J. Cardiol. 36 596–624. 10.1016/j.cjca.2020.02.086 [DOI] [PubMed] [Google Scholar]
- Ramanathan G., Harichandana B., Kannan S., Elumalai R., Sfd P. (2019). Association between end-stage diabetic nephropathy and MTHFR (C677T and A1298C) gene polymorphisms. Nephrology (Carlton) 24 155–159. 10.1111/nep.13208 [DOI] [PubMed] [Google Scholar]
- Riberio D. F., Cella P. S., Silva L. E. C. M., Jordao A. A., Deminice R. (2018). Acute exercise alters homocysteine plasma concentration in an intensity-dependent manner due increased methyl flux in liver of rats. Life Sci. 196 63–68. 10.1016/j.lfs.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Roya S. F., Fatemeh R., Kamran T., Behrooz H., Yahya P., Mehdi M., et al. (2019). Prevalence, awareness, treatment, control, and the associated factors of diabetes in an iranian kurdish population. J Diabetes Res. 2019:5869206. 10.1155/2019/5869206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailani R., Halling J. F., Møller D. H., Lee H., Plomgaard P., Pilegaard H., et al. (2019). Lifelong physical activity is associated with promoter hypomethylation of genes involved in metabolism, myogenesis, contractile properties and oxidative stress resistance in aged human skeletal muscle. Sci. Rep. 9:3272. 10.1038/s41598-018-37895-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman J. E., Smart N. A., Coombes J. S., Stowasser M. (2019). Exercise and sport science australia position stand update on exercise and hypertension. J. Hum. Hypertens 33 837–843. 10.1038/s41371-019-0266-z [DOI] [PubMed] [Google Scholar]
- Terruzzi I., Senesi P., Montesano A., La Torre A., Alberti G., Benedini S., et al. (2011). Genetic polymorphisms of the enzymes involved in DNA methylation and synthesis in elite athletes. Physiol. Genomics 43 965–973. 10.1152/physiolgenomics.00040.2010 [DOI] [PubMed] [Google Scholar]
- Voisin S., Eynon N., Yan X., Bispo D. J. (2015). Exercise training and DNA methylation in humans. Acta Physiol. (Oxf.) 213 39–59. 10.1111/apha.12414 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang J., Wang S. (2007). Demethylation in the promoter region of MTHFR gene and its mRNA expression in cultured human vascular smooth muscle cells induced by homocysteine. Wei Sheng Yan Jiu 36 291–294. Chinese. [PubMed] [Google Scholar]
- WHO (2018). Obesity: Preventing and Managing the Global Epidemic, 2000. Available online at: https://www.who.int/nutrition/publications/obesity/WHO_TRS_894/en/ (acessed June 11, 2018). [Google Scholar]
- Yang X. H., Cao R. F., Yu Y., Sui M., Zhang T., Xu J. Y., et al. (2016). A study on the correlation between MTHFR promoter methylation and diabetic nephropathy. Am. J. Transl. Res. 8 4960–4967. [PMC free article] [PubMed] [Google Scholar]
- Zarebska A., Ahmetov I. I., Sawczyn S., Weiner A. S., Kaczmarczyk M., Ficek K., et al. (2014). Association of the MTHFR 1298A>C (rs1801131) polymorphism with speed and strength sports in Russian and Polish athletes. J. Sports Sci. 32 375–382. 10.1080/02640414.2013.825731 [DOI] [PubMed] [Google Scholar]
- Zhao M., Veeranki S. P., Li S., Steffen L. M., Xi B. (2019). Beneficial associations of low and large doses of leisure time physical activity with all-cause, cardiovascular disease and cancer mortality: a national cohort study of 88,140 US adults. Br. J. Sports Med. 53 1405–1411. 10.1136/bjsports-2018-099254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.