Abstract
Background:
The apolipoprotein E (APOE) ɛ4 allele is associated with episodic memory and spatial navigation deficits. The brain-derived neurotrophic factor (BDNF) Met allele may further worsen memory impairment in APOE ɛ4 carriers but its role in APOE ɛ4-related spatial navigation deficits has not been established.
Objective:
We examined influence of APOE and BDNF Val66Met polymorphism combination on spatial navigation and volumes of selected navigation-related brain regions in cognitively unimpaired (CU) older adults and those with amnestic mild cognitive impairment (aMCI).
Methods:
187 participants (aMCI [n = 116] and CU [n = 71]) from the Czech Brain Aging Study were stratified based on APOE and BDNF Val66Met polymorphisms into four groups: ɛ4–/BDNFVal/Val, ɛ4–/BDNFMet, ɛ4+/BDNFVal/Val, and ɛ4+/BDNFMet. The participants underwent comprehensive neuropsychological examination, brain MRI, and spatial navigation testing of egocentric, allocentric, and allocentric delayed navigation in a real-space human analogue of the Morris water maze.
Results:
Among the aMCI participants, the ɛ4+/BDNFMet group had the least accurate egocentric navigation performance (p < 0.05) and lower verbal memory performance than the ɛ4–/BDNFVal/Val group (p = 0.007). The ɛ4+/BDNFMet group had smaller hippocampal and entorhinal cortical volumes than the ɛ4–/BDNFVal/Val (p≤0.019) and ɛ4–/BDNFMet (p≤0.020) groups. Among the CU participants, the ɛ4+/BDNFMet group had less accurate allocentric and allocentric delayed navigation performance than the ɛ4–/BDNFVal/Val group (p < 0.05).
Conclusion:
The combination of APOE ɛ4 and BDNF Met polymorphisms is associated with more pronounced egocentric navigation impairment and atrophy of the medial temporal lobe regions in individuals with aMCI and less accurate allocentric navigation in CU older adults.
Keywords: Alzheimer’s disease, apolipoproteins E, brain-derived neurotrophic factor, entorhinal cortex, episodic memory, gene polymorphism, magnetic resonance imaging, mild cognitive impairment, Morris water maze, spatial navigation
INTRODUCTION
Spatial navigation is a complex and multi-modal cognitive process essential for everyday functioning. It encompasses two basic strategies, egocentric (self-centered) and allocentric (world-centered), using different types of spatial reference frames to develop internal representations of surrounding environment. Egocentric navigation is a navigation strategy, where spatial information about locations and objects is encoded from the viewpoint of the navigator to form a self-centered spatial reference frame (self-to-object representations). Allocentric navigation is a navigation strategy, where locations and objects are encoded in relation to one another independently of the position of the navigator to form a world-centered spatial reference frame (object-to-object representations). Previous research has shown that distinct brain regions underlie egocentric and allocentric navigation strategies. Egocentric navigation is associated with the level of function of the posterior parietal cortex [1] including the precuneus [2] and the posterior cingulate cortex [3]. Allocentric navigation is associated with the level of function of the hippocampus [1] and related medial temporal lobe structures including the entorhinal cortex [4]. Recently, more attention has been focused on spatial navigation impairment as a promising early cognitive marker of Alzheimer’s disease (AD) [5, 6]. Allocentric navigation deficits have been observed in individuals with preclinical AD [7], while impairment of both navigation strategies (i.e., egocentric and allocentric) have been observed in individuals with mild cognitive impairment (MCI) [8], especially in those with amnestic MCI (aMCI) [2, 9–11], who are at higher risk of conversion to AD dementia [12]. Spatial navigation impairment observed in the early stages of AD can be explained by the fact that allocentric and egocentric spatial navigation is associated with the level of function of brain regions impaired very early in AD including the hippocampus and entorhinal cortex, and the posterior parietal cortex including precuneus and posterior cingulate cortex, respectively [13–15]. Recently, it has been shown that spatial navigation is a cognitive marker of early AD that shares only limited variance with other cognitive functions and is well distinguishable as a separate cognitive function [16].
Spatial navigation is influenced by genetic background, where the apolipoprotein E gene (APOE) is one of the most important indicators. The APOE ɛ4 allele is the strongest genetic risk factor for sporadic AD dementia [17] that lowers its age at onset [18]. The APOE ɛ4 allele is associated with increased amyloid-β (Aβ) accumulation on positron emission tomography (PET) [19], increased tau load in the entorhinal cortex on PET [20], hippocampal atrophy on MRI [21], posterior cingulate and parietal hypometabolism on fluorodeoxyglucose (FDG) PET [22], and greater cognitive decline in older adults [23]. The APOE ɛ4 allele also increases the risk of progression from MCI to dementia [24] probably due to the fact that individuals with aMCI who are carriers of the APOE ɛ4 allele are more likely to have Aβ pathology [25]. Studies found that the APOE ɛ4 allele is associated with worse allocentric navigation performance in cognitively normal older adults [26] and less accurate egocentric and allocentric navigation in individuals with MCI [27–29]. In addition to the APOE gene, other genetic polymorphisms associated with AD dementia and impairment of cognitive functions including spatial navigation have been identified. One of these polymorphisms is the very long poly-T variant at rs10524523 of the TOMM40 gene that modulates risk and onset age of AD dementia [30]. This polymorphism has been associated with worse memory performance in late middle-aged and older adults [31, 32] and also less accurate allocentric navigation in individuals with aMCI [33]. The other polymorphism is the rs17070145 polymorphism of the KIBRA gene, where non-carriers of the T-allele have had increased risk of AD dementia [34], worse memory performance [35] and less accurate spatial navigation in a study of cognitively normal older adults [36].
Recent studies indicated that among other genetic polymorphisms the brain-derived neurotrophic factor (BDNF) Val66Met polymorphism may also be associated with an increased risk of AD dementia [37] and more pronounced cognitive impairment [38, 39]. BDNF is a critical neurotrophic factor for synaptic plasticity [40], dendritic arborization [41], and facilitation of long-term potentiation [42], especially in the hippocampus and entorhinal cortex where the place cells and grid cells important for spatial navigation are located [43]. BDNF expression is profoundly reduced in the entorhinal cortex, hippocampus, and parietal cortex of individuals with AD dementia [44, 45], that is, in the regions where the earliest pathophysiological changes of AD have been identified [46]. Lower BDNF levels have been found in BDNF Met carriers [47]. The BDNF Met allele has been associated with reduced entorhinal [48] and parietal cortical thickness [49] and smaller hippocampal volume on MRI [50], reduced hippocampal activation on functional MRI [51], lower memory performance [52], and increased reliance on stimulus response rather than allocentric strategy in a human virtual navigation task [53] in cognitively normal younger adults. Next, the BDNF Met allele has been associated with accelerated memory decline and a greater rate of hippocampal volume reduction in cognitively normal older adults but only in those with high Aβ levels on PET [54]. The association between the BDNF Met allele and accelerated memory decline has also been reported in individuals with aMCI and high Aβ levels on PET [55]. The high risk combination of the APOE ɛ4 and BDNF Met alleles has shown the most pronounced negative effect on memory in cognitively normal older adults [56]. The later study has found that the combination of the APOE ɛ4 and BDNF Met alleles is associated with most accelerated memory decline in cognitively normal older adults but only in those with high Aβ levels on PET (i.e., those with preclinical AD) [57]. The combination of the APOE and BDNF Val66Met polymorphisms has also been associated with lowest memory performance in individuals with aMCI [58], prodromal AD and AD dementia [25]. The combined effect of the APOE and BDNF Val66Met polymorphisms on spatial navigation has not been studied.
Our group has demonstrated that egocentric and allocentric navigation is impaired in early clinical AD [11, 27] and can be influenced by genetic risk factors [33], especially by the APOE ɛ4 allele [28, 29]. Next, the BDNF Met allele is associated with structural changes in the navigation-related brain regions (i.e., in the medial temporal lobe structures and parietal cortex [48–50]) and the recent study has indicated that the BDNF Val66Met polymorphism may influence spontaneous navigational strategies in younger adults [53]. Building on this research, we assessed the combined effects of the APOE and BDNF Val66Met polymorphisms on spatial navigation performance and atrophy of brain regions associated with spatial navigation in older adults.
First, we evaluated the effect of combination of the APOE and BDNF Val66Met polymorphisms on two spatial navigation strategies, egocentric and allocentric, in a real-space version of the human analogue of the Morris Water Maze task (hMWM) in cognitively unimpaired (CU) and aMCI individuals. The effect of combination of the polymorphisms on other cognitive functions was also assessed. Second, we assessed the influence of combination of the APOE and BDNF Val66Met polymorphisms on volumes of the selected brain regions associated with spatial navigation (the hippocampus, entorhinal cortex, precuneus, inferior parietal cortex, and posterior cingulate cortex) in CU and aMCI individuals. The association between each spatial navigation strategy and volumes of the relevant brain regions was also tested.
We hypothesized that CU and aMCI participants with the high risk combination of the APOE ɛ4 and BDNF Met alleles would exhibit the least accurate spatial navigation performance in the allocentric and both egocentric and allocentric navigation strategies, respectively. The high risk combination of both alleles would also be associated with the worst episodic memory performance, especially in the participants with aMCI. Further, we hypothesized that the high risk participants, especially those with aMCI, would have the most pronounced atrophy of the brain regions associated with spatial navigation and that less accurate spatial navigation performance would be associated with lower volumes of these brain regions, especially in the participants with aMCI.
METHODS
Participants
A total of 187 participants were recruited from the Czech Brain Aging Study (http://cbas.cz/) cohort at the Memory Clinic of the Charles University, Second Faculty of Medicine and Motol University Hospital in Prague, Czech Republic and signed an informed consent approved by the local ethics committee [59]. The participants were referred to the Memory Clinic by general practitioners, psychiatrists, and neurologists for memory complaints reported by themselves and/or by their informants. All participants underwent clinical and laboratory evaluations, APOE and BDNF Val66Met genotyping, comprehensive neuropsychological assessment, brain magnetic resonance imaging (MRI), and spatial navigation assessment in a real-space version of the hMWM. Data from various modalities were collected within 60 days of each other for every participant.
The participants with aMCI (n = 116) met the clinical criteria for aMCI outlined in recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for AD [60] including memory complaints, evidence of memory impairment (i.e., score lower than 1.5 standard deviations below the age- and education- adjusted norms in any memory test), generally intact activities of daily living (i.e., Clinical Dementia Rating [CDR] global score not greater than 0.5) and absence of dementia. The aMCI group included participants with isolated memory impairment (single domain aMCI [aMCIsd]; n = 31) and those with memory impairment and additional impairment in any other non-memory cognitive domain (multiple domain aMCI [aMCImd]; n = 85). The CU participants (n = 71) had cognitive performance within the normal range (i.e., score higher than 1.5 standard deviations below the age- and education-adjusted norms in any cognitive test). Participants with depressive symptoms (≥6 points on the 15-item Geriatric Depression Scale [GDS-15]) [61], moderate to severe white matter vascular lesions on MRI (Fazekas score > 2 points) [62] or other primary neurological or psychiatric disorders were not included in the study. The CU and aMCI participants were further stratified into 4 groups each based on the APOE and BDNF Val66Met polymorphisms: APOE ɛ4 and BDNF Met noncarriers (ɛ4–/BDNFVal/Val; aMCI [n = 29], CU [n = 27]), APOE ɛ4 noncarriers and BDNF Met carriers (ɛ4–/BDNFMet; aMCI [n = 11], CU [n = 15]), APOE ɛ4 carriers and BDNF Met noncarriers (ɛ4+/BDNFVal/Val; aMCI [n = 52], CU [n = 18]), and APOE ɛ4 and BDNF Met carriers (ɛ4+/BDNFMet; aMCI [n = 24], CU [n = 11]). We did not include the APOE ɛ2 carriers (ɛ2/ɛ2, ɛ2/ɛ3 and ɛ2/ɛ4), as the ɛ2 allele is considered protective and its frequency among our participants was low. Group-wise characteristic are listed in Tables 1 and 2.
Table 1.
Characteristics of study participants with amnestic mild cognitive impairment
| Variables | ɛ4–/BDNFVal/Val (n = 29) | ɛ4–/BDNFMet (n = 11) | ɛ4+/BDNFVal/Val (n = 52) | ɛ4+/BDNFMet (n = 24) | p | Effect size |
| Demographic characteristics | ||||||
| Women/Men | 10/19 | 5/6 | 24/28 | 16/8 | 0.13 | 0.22d |
| aMCIsd/aMCImd | 5/24 | 4/7 | 16/36 | 6/18 | 0.51 | 0.14d |
| Age in years; range | 72.14 (5.23); 59–85 | 71.91 (8.87); 51–86 | 70.10 (6.97); 52–84 | 72.67 (6.29); 62–85 | 0.356 | 0.03e |
| Education in years | 15.76 (3.80) | 14.18 (2.96) | 15.02 (3.09) | 13.54 (3.36) | 0.098 | 0.05e |
| MMSE score | 26.90 (2.30) | 27.91 (1.22) | 27.12 (2.24) | 26.08 (2.04) | 0.099 | 0.05e |
| GDS-15 score | 2.52 (1.95) | 2.80 (1.55) | 2.60 (1.94) | 2.38 (1.41) | 0.928 | 0.00e |
| Cognitive characteristics | ||||||
| Memory verbala | 0.20 (0.84)* | 0.31 (0.55) | –0.01 (0.79) | –0.35 (0.52) | 0.026 | 0.08e |
| Memory nonverbala | 0.21 (1.11) | 0.04 (0.90) | –0.03 (1.01) | –0.21 (0.89) | 0.472 | 0.01e |
| Executive functiona | –0.22 (1.03) | 0.07 (0.51) | 0.04 (0.70) | 0.16 (0.61) | 0.300 | 0.05e |
| Attention and working memorya | –0.19 (0.80) | 0.20 (0.49) | –0.01 (0.75) | 0.15 (0.70) | 0.268 | 0.00e |
| Languagea | –0.12 (1.03) | –0.12 (0.43) | 0.06 (0.77) | 0.07 (0.62) | 0.674 | 0.02e |
| Visuospatiala | –0.23 (1.06) | 0.25 (0.58) | 0.06 (0.69) | 0.03 (0.62) | 0.270 | 0.04e |
| Navigational characteristics | ||||||
| Egocentric navigation | 28.45 (17.41)* | 24.65 (13.50)* | 34.49 (26.54)* | 56.63 (40.04) | 0.037 | 0.13f |
| Allocentric navigation | 65.69 (27.43) | 69.44 (34.56) | 73.98 (36.65) | 80.49 (40.51) | 0.918 | 0.02f |
| Delayed navigation | 76.08 (42.97) | 71.41 (43.66) | 77.65 (52.14) | 100.73 (60.35) | 0.692 | 0.04f |
| MRI characteristicsb | ||||||
| Left hippocampal volumec (mm3) | 3504.42 (474.37)* | 3755.13 (429.41)** | 3224.87 (647.65) | 2877.19 (421.63) | <0.001 | 0.24g |
| Right hippocampal volumec (mm3) | 3419.31 (762.38) | 3915.11 (592.66)* | 3318.58 (635.68) | 3151.07 (595.28) | 0.020 | 0.14g |
| Left entorhinal volumec (mm3) | 1401.27 (232.61)** | 1365.48 (383.70)* | 1044.38 (277.14) | 1004.16 (158.99) | <0.001 | 0.29g |
| Right entorhinal volumec (mm3) | 1053.15 (194.65) | 1333.24 (392.48)* | 1042.99 (266.38) | 1036.85 (139.04) | 0.002 | 0.19g |
| Left inferior parietal volumec (mm3) | 9020.13 (1242.56) | 10047.24 (1879.06) | 9252.97 (1163.27) | 8991.20 (873.12) | 0.210 | 0.07g |
| Right inferior parietal volumec (mm3) | 11550.19 (1380.26) | 11297.88 (2921.69) | 11226.68 (1295.92) | 10337.68 (1198.49) | 0.333 | 0.05g |
| Left posterior cingulate volumec (mm3) | 2532.60 (498.63) | 2399.50 (594.13) | 2698.89 (348.26) | 2500.84 (491.74) | 0.167 | 0.07g |
| Right posterior cingulate volumec (mm3) | 2646.09 (307.10) | 2460.46 (642.13) | 2739.80 (289.77) | 2639.67 (398.46) | 0.226 | 0.06g |
| Left precuneus volumec (mm3) | 7067.74 (913.77) | 7119.46 (1337.66) | 7232.81 (750.66) | 6852.11 (507.84) | 0.684 | 0.02g |
| Right precuneus volumec (mm3) | 7352.30 (846.09) | 7455.15 (1501.38) | 7695.30 (863.16) | 7351.45 (669.35) | 0.679 | 0.02g |
Values are mean (SD) except for gender and age range. For p indicating the level of significance compared with the ɛ4+/BDNFMet group are *p < 0.05 and **p < 0.01. aValues are presented in z-scores (SD). bBased on a sample restricted to those with brain imaging data (n = 85; ɛ4–/BDNFVal/Val [n = 23], ɛ4–/BDNFMet [n = 11], ɛ4+/BDNFVal/Val [n = 38] and ɛ4+/BDNFMet [n = 13]). cAdjusted for estimated total intracranial volume. dEffect size reported using Cramér’s V (the χ2 test). eEffect size reported using partial eta-squared (one-way ANOVA). fEffect size reported using partial eta-squared (linear mixed effects regression analyses). gEffect size reported using partial eta-squared (MANCOVA controlled for age, gender and education). ɛ4–/BDNFVal/Val, APOE ɛ4 and BDNF Met noncarriers’ group; ɛ4–/BDNFMet, APOE ɛ4 noncarriers and BDNF Met carriers’ group; ɛ4+/BDNFVal/Val, APOE ɛ4 carriers and BDNF Met noncarriers’ group; ɛ4+/BDNFMet, APOE ɛ4 and BDNF Met carriers’ group; aMCIsd, single domain amnestic mild cognitive impairment; aMCImd, multiple domain amnestic mild cognitive impairment; MMSE, Mini-Mental State Examination; GDS-15; 15-item Geriatric Depression Scale; MRI, magnetic resonance imaging.
Table 2.
Characteristics of cognitively unimpaired study participants
| Variables | ɛ4–/BDNFVal/Val (n = 27) | ɛ4–/BDNFMet (n = 15) | ɛ4+/BDNFVal/Val (n = 18) | ɛ4+/BDNFMet (n = 11) | p | Effect size |
| Demographic characteristics | ||||||
| Women/Men | 16/11 | 13/2 | 9/9 | 10/1 | 0.035 | 0.35d |
| Age in years; range | 68.44 (6.39); 54–86 | 64.87 (8.08); 52–82 | 65.61 (7.30); 52–82 | 66.55 (5.56); 55–74 | 0.364 | 0.05e |
| Education in years | 16.93 (2.62) | 17.07 (2.87) | 16.17 (2.26) | 15.00 (2.83) | 0.162 | 0.07e |
| MMSE score | 29.04 (1.09) | 28.80 (1.37) | 29.06 (0.97) | 28.91 (1.38) | 0.911 | 0.01e |
| GDS-15 score | 2.67 (3.28) | 3.27 (3.61) | 2.53 (2.32) | 2.10 (1.51) | 0.782 | 0.02e |
| Cognitive characteristics | ||||||
| Memory verbala | 0.08 (0.82) | 0.01 (0.82) | –0.05 (0.80) | 0.06 (0.83) | 0.944 | 0.00e |
| Memory nonverbala | –0.07 (1.03) | –0.02 (1.18) | 0.19 (0.85) | 0.05 (1.03) | 0.865 | 0.01e |
| Executive functiona | 0.00 (0.78) | –0.11 (0.69) | 0.06 (0.48) | 0.22 (0.99) | 0.693 | 0.02e |
| Attention and working memorya | 0.23 (0.77) | 0.08 (0.57) | –0.25 (0.79) | –0.22 (0.57) | 0.122 | 0.08e |
| Languagea | –0.10 (0.86) | –0.22 (0.85) | 0.23 (0.58) | 0.24 (0.88) | 0.252 | 0.06e |
| Visuospatiala | –0.08 (0.92) | –0.15 (0.61) | 0.31 (0.60) | –0.10 (0.83) | 0.279 | 0.06e |
| Navigational characteristics | ||||||
| Egocentric navigation | 19.99 (6.57) | 21.09 (6.33) | 21.71 (7.40) | 22.24 (10.10) | 0.732 | 0.02f |
| Allocentric navigation | 19.00 (3.88)* | 23.82 (5.50) | 27.38 (8.22) | 35.28 (16.67) | 0.017 | 0.22f |
| Delayed navigation | 15.72 (6.04)* | 21.25 (5.32) | 21.77 (8.89) | 25.99 (12.78) | 0.028 | 0.19f |
| MRI characteristicsb | ||||||
| Left hippocampal volumec (mm3) | 3733.92 (657.66) | 3941.28 (504.83) | 3906.37 (339.6) | 3862.18 (273.81) | 0.206 | 0.09g |
| Right hippocampal volumec (mm3) | 3697.89 (646.54) | 4124.32 (418.13) | 3888.16 (517.43) | 4006.76 (378.62) | 0.126 | 0.11g |
| Left entorhinal volumec (mm3) | 1217.14 (195.58) | 1375.90 (230.86) | 1306.00 (275.89) | 1404.36 (303.92) | 0.112 | 0.12g |
| Right entorhinal volumec (mm3) | 1139.21 (242.38) | 1159.44 (179.72) | 1193.61 (263.86) | 1207.06 (344.16) | 0.395 | 0.06g |
| Left inferior parietal volumec (mm3) | 10193.91 (957.03) | 10002.32 (1616.12) | 9789.21 (1183.13) | 10134.89 (1096.83) | 0.702 | 0.03g |
| Right inferior parietal volumec (mm3) | 11903.11 (1197.12) | 11780.80 (1386.19) | 11734.89 (995.51) | 12038.19 (1533.04) | 0.758 | 0.02g |
| Left posterior cingulate volumec (mm3) | 2755.31 (320.93) | 2798.20 (382.01) | 2767.77 (215.05) | 2767.62 (420.88) | 0.544 | 0.04g |
| Right posterior cingulate volumec (mm3) | 2729.43 (329.39) | 2789.67 (383.44) | 2849.55 (407.77) | 2786.89 (293.95) | 0.170 | 0.10g |
| Left precuneus volumec (mm3) | 7733.91 (595.22) | 8350.33 (895.09) | 7673.38 (793.67) | 7870.21 (576.64) | 0.245 | 0.08g |
| Right precuneus volumec (mm3) | 8225.50 (682.58) | 8563.94 (816.38) | 8068.20 (786.82) | 8344.45 (477.81) | 0.733 | 0.03g |
Values are mean (SD) except for gender and age range. For p indicating the level of significance compared with the ɛ4+/BDNFMet group is *p < 0.05. aValues are presented in z-scores (SD). bBased on a sample restricted to those with brain imaging data (n = 85; ɛ4–/BDNFVal/Val [n = 18], ɛ4–/BDNFMet [n = 12], ɛ4+/BDNFVal/Val [n = 18] and ɛ4+/BDNFMet [n = 9]). cAdjusted for estimated total intracranial volume. dEffect size reported using Cramér’s V (the χ2 test). eEffect size reported using partial eta-squared (one-way ANOVA). fEffect size reported using partial eta-squared (linear mixed effects regression analyses). gEffect size reported using partial eta-squared (MANCOVA controlled for age, gender, and education). ɛ4–/BDNFVal/Val, APOE ɛ4 and BDNF Met noncarriers’ group; ɛ4–/BDNFMet, APOE ɛ4 noncarriers and BDNF Met carriers’ group; ɛ4+/BDNFVal/Val, APOE ɛ4 carriers and BDNF Met noncarriers’ group; ɛ4+/BDNFMet, APOE ɛ4 and BDNF Met carriers’ group; MMSE, Mini-Mental State Examination; GDS-15; 15-item Geriatric Depression Scale; MRI, magnetic resonance imaging.
Neuropsychological assessment
The neuropsychological battery comprised the following tests for each cognitive domain: 1) verbal memory measured with the Logical Memory I – Immediate and 20-minute Delayed Recall conditions and the Rey Auditory Verbal Learning Test – trials 1–5 and 30-minute Delayed Recall trial; 2) non-verbal memory measured with the Rey-Osterrieth Complex Figure Test (ROCFT) – the Recall condition after 3 minutes; 3) executive function measured with the Trail Making Test B, Prague Stroop Test – colors and Controlled Oral Word Association Test – Czech version with letters N, K, and P; 4) attention and working memory measured with the Forward and Backward Digit Spans and Trail Making Test A; 5) language measured with the Boston Naming Test – 60-item version and Category Fluency Test – animals and vegetables; and 6) visuospatial function measured with the ROCFT – the Copy condition and the Clock Drawing Test. The Mini-Mental State Examination (MMSE) was administered to measure global cognitive function. The GDS-15 was used to assess depressive symptoms among participants. Group-wise neuropsychological characteristics are listed in Tables 1 and 2.
APOE and BDNF Val66Met genotyping
To determine the APOE genotype, DNA was isolated from blood samples (ethylenediaminetetraacetic acid; Qiagen extraction) and genotyping was performed according to Idaho-tech protocol (LunaProbes Genotyping Apolipoprotein [ApoE] Multiplexed Assay) for high-resolution melting (HRM) analysis [28, 63].
The BDNF rs6265 (Val66Met) polymorphism was analyzed with a novel HRM method using polymerase chain reaction (PCR). The PCR product of 59bp was amplified with primers. Subsequent HRM analysis of PCR product was performed on Light Scanner (IdahoTech, USA) [58].
MRI acquisition and analysis
Brain scans were performed on a 1.5 T scanner (Siemens AG, Erlangen, Germany) using the T1-weighted 3-dimensional high resolution magnetization-prepared rapid gradient-echo (MP-RAGE) sequence with the following parameters: TR/TE/TI = 2000/3.08/1100 ms, flip angle 15°, 192 continuous partitions, slice thickness = 1.0 mm, and in-plane resolution = 1 mm. Scans were visually inspected by a neuroradiologist to ensure appropriate data quality and to exclude patients with a major brain pathology that could interfere with cognitive functioning such as cortical infarctions, tumor, subdural hematoma, and hydrocephalus.
FreeSurfer image analysis suite (version 5.3; http://surfer.nmr.mgh.harvard.edu) was used to compute regional brain volumes and estimated total intracranial volume (eTIV), the internal reference. The procedure was described in detail elsewhere [64–66]. We selected the regions that play a key role in spatial navigation [1, 4, 67] and are affected early in AD [68, 69] to limit the number of analyses to those that were aligned with our hypothesis. The selected regions included the hippocampus, entorhinal cortex, precuneus, inferior parietal cortex, and posterior cingulate cortex. Volumes were reported separately for the left and right hemisphere, since spatial navigation is thought to be lateralized to the right hemisphere [70]. Volumes were normalized to eTIV using the previously published regression formula [71]. MRI data were available for 85 participants with aMCI (ɛ4–/BDNFVal/Val [n = 23], ɛ4–/BDNFMet [n = 11], ɛ4+/BDNFVal/Val [n = 38], and ɛ4+/BDNFMet [n = 13]) and 57 CU participants (ɛ4–/BDNFVal/Val [n = 18], ɛ4–/BDNFMet [n = 12], ɛ4+/BDNFVal/Val [n = 18], and ɛ4+/BDNFMet [n = 9]). Group-wise MRI characteristics are listed in Tables 1 and 2.
Spatial navigation assessment
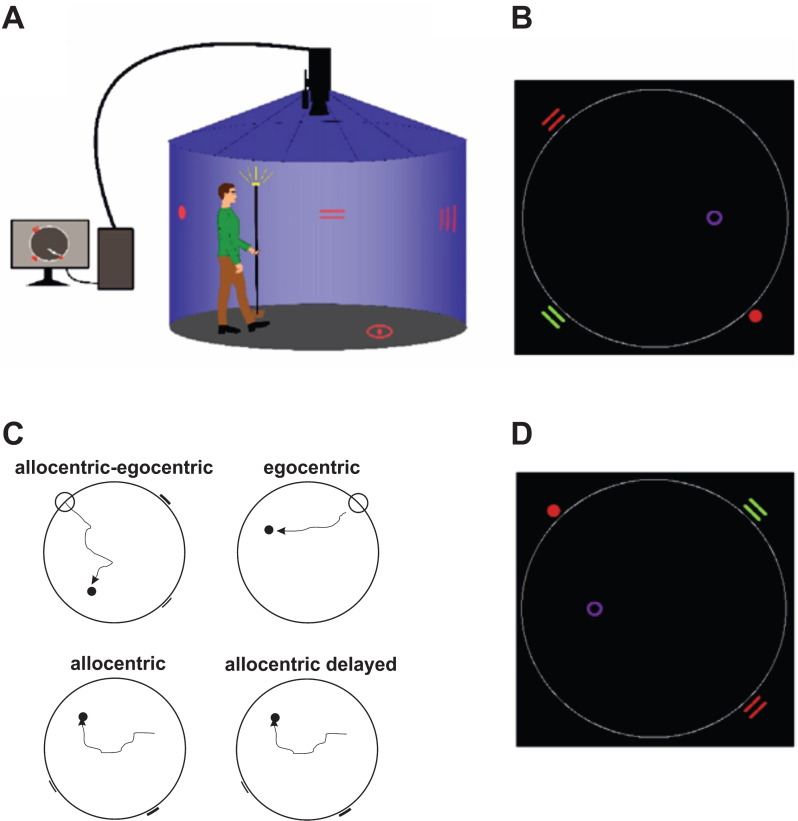
For spatial navigation assessment, we used the real-space version of the hMWM that was performed in a real-space navigational setting, a fully enclosed cylindrical arena 2.8 m in diameter surrounded by a 2.9 m high dark blue velvet curtain (Fig. 1A). The task and the real-space navigational setting were described in detail elsewhere [9, 10]. The participants located an invisible goal on the arena floor in 4 different tasks using a start position (egocentric) or 2 distal orientation cues on the wall (allocentric), respectively (Fig. 1B, 1C). The first, allocentric–egocentric, task was a training task designed to familiarize participants with the testing procedure and involved locating the goal using both the start position and 2 distal orientation cues on the arena wall. The second, egocentric, task involved using the start position to locate the goal with no distal orientation cues displayed. The third, allocentric, task involved using 2 distal orientation cues at the arena wall for navigation to the goal from the start position that was unrelated to the goal position. The fourth, allocentric delayed, task was identical to the allocentric task and was administered 30 minutes after its completion. The training (egocentric-allocentric), egocentric, and allocentric tasks had 8 trials and the correct position of the goal was shown after each trial to provide the feedback. The delayed task had 2 trials and no feedback through showing the position of the goal was provided. The relative positions (distances and directions) of the goal to the start position or to both orientation cues were constant across all trials in all tasks. After each trial, the goal position along with the start position and the positions of 2 distal orientation cues were rotated in a pseudorandom sequence and the participants were instructed to go to the new start position at each consecutive trial in all tasks (Fig. 1D). The tasks had no time limit. Spatial navigation performance derived as distance error in centimeters from the correct goal position was automatically recorded by in-house developed software. A TV camera with a sampling frequency of 25 frames per second located above the center of the arena was used to capture the position of an infrared light-emitting diode placed on top of a hat that was worn by the participants during the testing and indicated their location.
Fig.1.
Human analogue of the Morris Water Maze task. A) The real-space navigation setting. B) The scheme of the task showing an aerial view of the arena (large white circle) with starting point (red filled circle), orientation cues (red and green lines), and goal (purple circle). C) The scheme of 4 individual tasks: allocentric-egocentric (i.e., training; designed to use the starting point and 2 orientation cues for navigation), egocentric (designed to use the starting point for navigation), allocentric (designed to use 2 orientation cues for navigation) and allocentric delayed (identical to the allocentric task administered 30 minutes after its completion). D) An aerial view of the arena rotated 180 degrees from the previous trial shown in B.
Statistical analyses
Data standardization and analyses were performed separately for the CU and aMCI participants. Scores for neuropsychological and spatial navigation tests and MRI data were standardized to z-scores calculated from the overall mean and SD of the sample to enable comparison across the measurements scored on different scales. The values from the Prague Stroop Test, Trail Making Tests A and B (seconds to completion), and the Boston Naming Test (number of errors) were reversed before the z-scores were generated. The scores for each cognitive domain were expressed as a unit-weighted composite z-score from the relevant neuropsychological tests. Higher z-scores reflected better neuropsychological performance, but less accurate spatial navigation performance (distance error in centimeters). All data were found to be adequate for parametric analysis. Spatial navigation data were log transformed prior to the standardization and analyses because of their right-skewed distribution. The transformation satisfactorily solved the skewness.
A one-way analysis of variance (ANOVA) with post hoc Sidak’s test evaluated between-group differences in age, years of education, GDS and MMSE scores, and cognitive domains’ composite scores. The χ2 test evaluated differences in gender proportions. Differences in proportions of aMCI subtypes (aMCIsd versus aMCImd) were evaluated in the participants with aMCI.
In the spatial navigation analyses, scores from all trials in each spatial navigation task were entered into linear mixed effects regression models [72] that were used to properly account for the repeated measures structure of the spatial navigation data. The distance error in each spatial navigation task was the outcome and the polymorphism group was the independent variable. As in the one-way ANOVA, our primary interest was in the main effect for group and differences in spatial navigation performance across the individual polymorphism groups (ɛ4–/BDNFVal/Val versus ɛ4–/BDNFMet versus ɛ4+/BDNFVal/Val versus ɛ4+/BDNFMet) that were evaluated by post hoc Sidak’s test. We also report the main effect for trial (trials 1–8 for the egocentric and allocentric tasks and trials 1–2 for the allocentric delayed task) and the group-by-trial interaction, which reflect learning and differential learning by group, respectively. The intercept and trial were specified as random effects. Based on model fit, the final models used the unstructured covariance matrix. Gender distribution was not equal across the groups, so gender was added as a covariate to the analyses. This decreased the model fit and the main effects for gender and group-by-gender interactions were not significant. Therefore, gender was not used in the final analyses.
Next, we estimated a multivariate analysis of covariance (MANCOVA) to assess the between-group differences in volumes of the brain regions relevant for spatial navigation. Age, gender, and education were controlled in the analysis. Initially, we performed multivariate tests and when the Wilks’ Lambda indicating overall differences across the four groups in volumes of the brain regions was significant, we subsequently performed univariate tests with post hoc Sidak’s test, where the polymorphism group was the independent variable and volumes of the relevant brain regions were separately entered as the outcome.
Finally, we calculated Pearson correlation coefficients to explore relationships between spatial navigation performance in the egocentric, allocentric, and allocentric delayed tasks and volumes of the selected brain regions. A Bonferroni correction for multiple comparisons was used, resulting in a p value of 0.005 (0.05/10 regions). The subsequent multivariate linear regression analysis was used to evaluate the associations between spatial navigation performance in each task and selected regional brain volumes controlling for age, gender, education, and polymorphism group membership. Volumes of the navigation-related brain regions that correlated with spatial navigation performance were separately entered as the independent variables. Spatial navigation performance was the dependent variable and was calculated as the distance error averaged across the trials for each spatial navigation task.
Statistical significance was set at 2-tailed (alpha) of 0.05. Effect sizes are reported using Cramér’s V for the χ2 test and partial eta-squared (ηp2) for one-way ANOVA, MANCOVA, and linear mixed effects regression analyses [73]. Cramér’s V of about 0.47 and partial eta-squared of 0.12 correspond to Cohen’s d of about 1.0. All analyses were conducted with IBM SPSS 25.0 software.
RESULTS
Demographic and neuropsychological characteristics
The participants with aMCI
There were no significant differences in age, gender, years of education, aMCI subtypes, depressive symptoms (assessed by GDS-15), and global cognitive function (assessed by MMSE) between the polymorphism groups. The main effect of group on verbal memory performance was significant (F[3,112] = 5.18, p = 0.026, ηp2 = 0.08). The post hoc analysis revealed that the ɛ4+/BDNFMet group had lower verbal memory performance than the ɛ4–/BDNFVal/Val (p = 0.007) and similar performance to the ɛ4–/BDNFMet (p = 0.071) and ɛ4+/BDNFVal/Val (p = 0.240) groups. There were no significant differences in other cognitive domains including attention and working memory, nonverbal memory, executive function, language and visuospatial function between the polymorphism groups. The results are presented in detail in Table 1.
The CU participants
There were no significant differences in age, years of education, depressive symptoms, and global cognitive function between the polymorphism groups but there were more women in the ɛ4–/BDNFMet and ɛ4+/BDNFMet groups than in the ɛ4–/BDNFVal/Val and ɛ4+/BDNFVal/Val groups (13/2 and 10/1 versus 16/11 and 9/9; χ2[3] = 3.09, p = 0.035, Cramér’s V2 = 0.35). There were no significant differences in cognitive performance in any cognitive domain including attention and working memory, verbal memory, nonverbal memory, executive function, language and visuospatial function between the polymorphism groups. The results are presented in detail in Table 2.
Spatial navigation performance
The participants with aMCI
Using the linear mixed models we found main effects for group on egocentric navigation performance (F[3,110] = 2.93; p = 0.037; ηp2 = 0.13), where the ɛ4+/BDNFMet group had less accurate egocentric navigation performance than the ɛ4–/BDNFVal/Val (p = 0.028), ɛ4–/BDNFMet (p = 0.013) and ɛ4+/BDNFVal/Val (p = 0.045) groups. The estimated pairwise differences were around 0.5 SD (ɛ4+/BDNFMet versus ɛ4–/BDNFVal/Val and ɛ4+/BDNFMet versus ɛ4+/BDNFVal/Val) and approached 1 SD (ɛ4+/BDNFMet versus ɛ4–/BDNFMet) for these comparisons. There were no significant differences in egocentric navigation performance between the ɛ4–/BDNFVal/Val, ɛ4–/BDNFMet and ɛ4+/BDNFVal/Val groups (ps≥0.654). There were no significant differences in allocentric (F[3,107] = 0.17; p = 0.918; ηp2 = 0.02) and allocentric delayed (F[3,105] = 0.49; p = 0.692; ηp2 = 0.04) navigation performance between the polymorphism groups. The between-group comparisons of spatial navigation performance in each task are listed in Tables 1 and 3. The main effects for trial were not significant in the egocentric (F[1,110] = 0.24; p = 0.625; ηp2 = 0.00), allocentric (F[1,106] = 0.61; p = 0.438; ηp2 = 0.03), and allocentric delayed (F[1,105] = 0.14; p = 0.707; ηp2 = 0.00) tasks, indicating no significant learning effect across consecutive trials in the sample overall. Finally, there were no significant group-by-trial interactions, suggesting no significant differences in learning between the polymorphism groups in the egocentric (F[3,110] = 0.38; p = 0.767; ηp2 = 0.05), allocentric (F[3,106] = 1.48; p = 0.225; ηp2 = 0.02), and allocentric delayed (F[3,105] = 0.09; p = 0.964; ηp2 = 0.00) tasks.
Table 3.
Group-wise comparisons of adjusted mean error distances from the goal in spatial navigation tasks in the participants with amnestic mild cognitive impairment
| (I) Groupcode | (J) Groupcode | Mean difference (I-J) | p | 95% Confidence interval for difference | |
| Lower bound | Upper bound | ||||
| Egocentric task | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | 0.485 | 0.045 | 0.006 | 0.963 |
| ɛ4–/BDNFMet | 0.812 | 0.013 | 0.120 | 1.505 | |
| ɛ4–/BDNFVal/Val | 0.586 | 0.028 | 0.043 | 1.129 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.328 | 0.654 | –0.296 | 0.951 |
| ɛ4–/BDNFVal/Val | 0.101 | 0.992 | –0.351 | 0.553 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | –0.226 | 0.938 | –0.901 | 0.448 |
| Allocentric task | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | 0.084 | 0.998 | –0.415 | 0.582 |
| ɛ4–/BDNFMet | 0.240 | 0.935 | –0.470 | 0.951 | |
| ɛ4–/BDNFVal/Val | 0.161 | 0.970 | –0.401 | 0.723 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.157 | 0.986 | –0.480 | 0.794 |
| ɛ4–/BDNFVal/Val | 0.078 | 0.998 | –0.388 | 0.543 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | –0.079 | 1.000 | –0.768 | 0.609 |
| Delayed task | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | 0.264 | 0.808 | –0.335 | 0.864 |
| ɛ4–/BDNFMet | 0.450 | 0.651 | –0.405 | 1.306 | |
| ɛ4–/BDNFVal/Val | 0.246 | 0.917 | –0.441 | 0.933 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.186 | 0.987 | –0.581 | 0.953 |
| ɛ4–/BDNFVal/Val | –0.018 | 1.000 | –0.591 | 0.555 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | –0.204 | 0.987 | –1.041 | 0.632 |
Linear mixed models. Mean differences are in standard deviation units. Values in bold indicate significant between-group differences (p < 0.05). ɛ4+/BDNFMet, APOE ɛ4 and BDNF Met carriers’ group; ɛ4+/BDNFVal/Val, APOE ɛ4 carriers and BDNF Met noncarriers’ group; ɛ4–/BDNFMet, APOE ɛ4 noncarriers and BDNF Met carriers’ group; ɛ4–/BDNFVal/Val, APOE ɛ4 and BDNF Met noncarriers’ group.
The CU participants
We found main effects for group on allocentric (F[3,127] = 3.83; p = 0.017; ηp2 = 0.22) and allocentric delayed (F[3,69] = 3.35; p = 0.028; ηp2 = 0.19) navigation performance where the ɛ4+/BDNFMet group had less accurate performance than the ɛ4–/BDNFVal/Val group (p = 0.047 and p = 0.048, respectively). The estimated pairwise differences were around 0.5 and 0.7 SD, respectively, for these comparisons. There were no significant differences in egocentric navigation performance between the polymorphism groups (F[3,125] = 0.43; p = 0.732; ηp2 = 0.02). The between-group comparisons of spatial navigation performance in each task are listed in Tables 2 and 4. The main effects for trial were not significant in the egocentric (F[1,125] = 0.03; p = 0.999; ηp2 = 0.00), allocentric (F[1,127] = 0.87; p = 0.534; ηp2 = 0.02), and allocentric delayed (F[1,69] = 0.05; p = 0.826; ηp2 = 0.00) tasks. There were no significant group-by-trial interactions in the egocentric (F[3,125] = 1.03; p = 0.423; ηp2 = 0.05), allocentric (F[3,127] = 1.30; p = 0.173; ηp2 = 0.09), and allocentric delayed (F[3,69] = 0.09; p = 0.968; ηp2 = 0.01) tasks.
Table 4.
Group-wise comparisons of adjusted mean error distances from the goal in spatial navigation tasks in the cognitively unimpaired participants
| (I) Groupcode | (J) Groupcode | Mean difference (I-J) | p | 95% Confidence interval for difference | |
| Lower bound | Upper bound | ||||
| Egocentric task | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.014 | 1.000 | –0.477 | 0.449 |
| ɛ4–/BDNFMet | 0.002 | 1.000 | –0.478 | 0.482 | |
| ɛ4–/BDNFVal/Val | 0.120 | 0.975 | –0.315 | 0.555 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.016 | 1.000 | –0.407 | 0.439 |
| ɛ4–/BDNFVal/Val | 0.134 | 0.911 | –0.237 | 0.505 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.118 | 0.961 | –0.274 | 0.510 |
| Allocentric task | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | 0.091 | 0.997 | –0.412 | 0.594 |
| ɛ4–/BDNFMet | 0.080 | 0.999 | –0.451 | 0.611 | |
| ɛ4–/BDNFVal/Val | 0.501 | 0.047 | 0.005 | 0.997 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –0.011 | 1.000 | –0.482 | 0.460 |
| ɛ4–/BDNFVal/Val | 0.410 | 0.070 | –0.021 | 0.841 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.421 | 0.093 | –0.043 | 0.884 |
| Delayed task | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | 0.223 | 0.949 | –0.488 | 0.934 |
| ɛ4–/BDNFMet | 0.108 | 0.999 | –0.645 | 0.862 | |
| ɛ4–/BDNFVal/Val | 0.705 | 0.048 | 0.004 | 1.406 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –0.115 | 0.998 | –0.805 | 0.575 |
| ɛ4–/BDNFVal/Val | 0.482 | 0.224 | –0.150 | 1.114 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.596 | 0.113 | –0.083 | 1.276 |
Linear mixed models. Mean differences are in standard deviation units. Values in bold indicate significant between-group differences (p < 0.05). ɛ4+/BDNFMet, APOE ɛ4 and BDNF Met carriers’ group; ɛ4+/BDNFVal/Val, APOE ɛ4 carriers and BDNF Met noncarriers’ group; ɛ4–/BDNFMet, APOE ɛ4 noncarriers and BDNF Met carriers’ group; ɛ4–/BDNFVal/Val, APOE ɛ4 and BDNF Met noncarriers’ group.
Brain MRI characteristics and their association with spatial navigation
The participants with aMCI
Using the MANCOVA adjusted for age, gender, and education, the multivariate tests showed the between-group differences in volumes of the navigation-related brain regions (Wilks’ Lambda = 2.33; p≤0.001; ηp2 = 0.29). In the subsequent univariate tests analyzing each brain region separately, the between-group differences were found for left and right hippocampal volumes (F[3,85] = 7.13; p < 0.001; ηp2 = 0.24 and F[3,85] = 3.50; p = 0.020; ηp2 = 0.14, respectively) and left and right entorhinal cortical volumes (F[3,85] = 9.29; p < 0.001; ηp2 = 0.29 and F[3,85] = 5.34; p = 0.002; ηp2 = 0.19, respectively). The between-group differences in volumes of other navigation-related brain regions were not significant (F[3,85]≤1.74; p≥0.167; ηp2≤0.07). The post hoc tests showed that the ɛ4+/BDNFMet group had smaller left hippocampal and left entorhinal cortical volumes compared to the ɛ4–/BDNFVal/Val (p = 0.019 and p = 0.004) and ɛ4–/BDNFMet (p = 0.001 and p = 0.020) groups and smaller right hippocampal and right entorhinal cortical volumes compared to the ɛ4–/BDNFMet group (p = 0.038 and p = 0.030). The ɛ4+/BDNFVal/Val group had smaller left and right hippocampal and right entorhinal cortical volumes compared to the ɛ4–/BDNFMet group (p = 0.006, p = 0.020 and p = 0.001) and left entorhinal cortical volume compared to the ɛ4–/BDNFVal/Val (p < 0.001) and ɛ4–/BDNFMet (p = 0.007) groups. The between-group comparisons of volumes of the navigation-related brain regions are listed in Tables 1 and 5.
Table 5.
Group-wise comparisons of hippocampal and cortical volumes in the participants with amnestic mild cognitive impairment
| (I) Groupcode | (J) Groupcode | Mean difference (I-J) | p | 95% Confidence interval for difference | |
| Lower bound | Upper bound | ||||
| Left hippocampal volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.376 | 0.655 | –1.099 | 0.346 |
| ɛ4–/BDNFMet | –1.394 | 0.001 | –2.345 | –0.443 | |
| ɛ4–/BDNFVal/Val | –0.967 | 0.019 | –1.823 | –0.111 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –1.018 | 0.006 | –1.825 | –0.211 |
| ɛ4–/BDNFVal/Val | –0.590 | 0.123 | –1.270 | 0.090 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.428 | 0.767 | –0.500 | 1.355 |
| Right hippocampal volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.093 | 1.000 | –0.926 | 0.739 |
| ɛ4–/BDNFMet | –1.136 | 0.038 | –2.232 | –0.041 | |
| ɛ4–/BDNFVal/Val | –0.348 | 0.919 | –1.334 | 0.638 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –1.043 | 0.020 | –1.973 | –0.113 |
| ɛ4–/BDNFVal/Val | –0.255 | 0.944 | –1.038 | 0.528 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.788 | 0.263 | –0.280 | 1.856 |
| Left entorhinal volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.055 | 1.000 | –0.825 | 0.715 |
| ɛ4–/BDNFMet | –1.135 | 0.020 | –2.148 | –0.122 | |
| ɛ4–/BDNFVal/Val | –1.202 | 0.004 | –2.114 | –0.290 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –1.079 | 0.007 | –1.939 | –0.219 |
| ɛ4–/BDNFVal/Val | –1.147 | 0.000 | –1.871 | –0.422 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | –0.067 | 1.000 | –1.055 | 0.921 |
| Right entorhinal volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | 0.166 | 0.989 | –0.546 | 0.879 |
| ɛ4–/BDNFMet | –1.003 | 0.030 | –1.940 | –0.065 | |
| ɛ4–/BDNFVal/Val | 0.017 | 1.000 | –0.827 | 0.861 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –1.169 | 0.001 | –1.965 | –0.373 |
| ɛ4–/BDNFVal/Val | –0.149 | 0.991 | –0.820 | 0.521 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 1.020 | 0.021 | 0.105 | 1.934 |
| Left inferior parietal volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.153 | 0.998 | –1.041 | 0.735 |
| ɛ4–/BDNFMet | –0.818 | 0.319 | –1.987 | 0.351 | |
| ɛ4–/BDNFVal/Val | –0.014 | 1.000 | –1.066 | 1.038 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –0.665 | 0.368 | –1.657 | 0.327 |
| ɛ4–/BDNFVal/Val | 0.139 | 0.998 | –0.697 | 0.974 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.804 | 0.311 | –0.336 | 1.943 |
| Right inferior parietal volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.459 | 0.655 | –1.339 | 0.422 |
| ɛ4–/BDNFMet | –0.600 | 0.661 | –1.759 | 0.559 | |
| ɛ4–/BDNFVal/Val | –0.669 | 0.420 | –1.713 | 0.374 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | –0.141 | 0.999 | –1.125 | 0.843 |
| ɛ4–/BDNFVal/Val | –0.211 | 0.983 | –1.039 | 0.618 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.669 | 0.420 | –0.374 | 1.713 |
| Left posterior cingulate volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.499 | 0.619 | –1.424 | 0.426 |
| ɛ4–/BDNFMet | 0.240 | 0.996 | –0.977 | 1.458 | |
| ɛ4–/BDNFVal/Val | –0.096 | 1.000 | –1.192 | 1.000 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.739 | 0.296 | –0.295 | 1.772 |
| ɛ4–/BDNFVal/Val | 0.403 | 0.764 | –0.468 | 1.274 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | –0.336 | 0.971 | –1.523 | 0.851 |
| Right posterior cingulate volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.293 | 0.950 | –1.215 | 0.629 |
| ɛ4–/BDNFMet | 0.487 | 0.861 | –0.726 | 1.701 | |
| ɛ4–/BDNFVal/Val | –0.025 | 1.000 | –1.117 | 1.068 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.780 | 0.236 | –0.250 | 1.811 |
| ɛ4–/BDNFVal/Val | 0.268 | 0.956 | –0.600 | 1.136 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | –0.512 | 0.815 | –1.696 | 0.671 |
| Left precuneus volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.400 | 0.809 | –1.316 | 0.517 |
| ɛ4–/BDNFMet | –0.310 | 0.982 | –1.517 | 0.896 | |
| ɛ4–/BDNFVal/Val | –0.211 | 0.996 | –1.297 | 0.875 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.089 | 1.000 | –0.935 | 1.113 |
| ɛ4–/BDNFVal/Val | 0.189 | 0.992 | –0.674 | 1.052 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.100 | 1.000 | –1.077 | 1.276 |
| Right precuneus volumea | |||||
| ɛ4+/BDNFMet | ɛ4+/BDNFVal/Val | –0.296 | 0.941 | –1.199 | 0.606 |
| ɛ4–/BDNFMet | –0.120 | 1.000 | –1.308 | 1.068 | |
| ɛ4–/BDNFVal/Val | 0.031 | 1.000 | –1.038 | 1.101 | |
| ɛ4+/BDNFVal/Val | ɛ4–/BDNFMet | 0.176 | 0.998 | –0.832 | 1.185 |
| ɛ4–/BDNFVal/Val | 0.328 | 0.882 | –0.522 | 1.177 | |
| ɛ4–/BDNFMet | ɛ4–/BDNFVal/Val | 0.151 | 1.000 | –1.007 | 1.310 |
Analysis of covariance adjusted for age, gender and education. Mean differences are in standard deviation units. Values in bold indicate significant between-group differences (p < 0.05). aAdjusted for estimated total intracranial volume. ɛ4+/BDNFMet, APOE ɛ4 and BDNF Met carriers’ group; ɛ4+/BDNFVal/Val, APOE ɛ4 carriers and BDNF Met noncarriers’ group; ɛ4–/BDNFMet, APOE ɛ4 noncarriers and BDNF Met carriers’ group; ɛ4–/BDNFVal/Val, APOE ɛ4 and BDNF Met noncarriers’ group.
In the correlational analyses (Supplementary Table 1), right hippocampal volume correlated with allocentric navigation performance (r = –0.39; p = 0.001) indicating that smaller volume was associated with greater distance error in the navigational task. Right hippocampal volume correlated with allocentric delayed (r = –0.33; p = 0.006) and egocentric (r = –0.32; p = 0.006) navigation performance, left hippocampal volume correlated with allocentric (r = –0.28; p = 0.020), allocentric delayed (r = –0.30; p = 0.014), and egocentric (r = –0.28; p = 0.017) navigation performance, right entorhinal cortical volume correlated with allocentric delayed navigation performance (r = –0.27; p = 0.028) and left posterior cingulate volume correlated with allocentric delayed navigation performance (r = –0.27; p = 0.028); however, these relationships did not surpass the threshold of Bonferroni-corrected p-value.
In the multivariate linear regression analyses, right hippocampal volume was associated with allocentric navigation performance (β= –0.41; p = 0.001) indicating that smaller volume was associated with greater distance error in the navigational task above and beyond demographic characteristics. Right hippocampal volume was associated with allocentric delayed (β= –0.30; p = 0.014) and egocentric (β= –0.27; p = 0.022) navigation performance, left hippocampal volume was associated with allocentric (β= –0.37; p = 0.007), allocentric delayed (β= –0.32; p = 0.020), and egocentric (β= –0.26; p = 0.048) navigation performance and left posterior cingulate volume was associated with allocentric delayed navigation performance (β= –0.23; p = 0.042); however, these associations did not surpass the threshold of Bonferroni-corrected p-value.
The CU participants
The multivariate tests did not show significant differences between the polymorphism groups in volumes of the navigation-related brain regions (Wilks’ Lambda = 0.96; p = 0.532; ηp2 = 0.20). There was no significant correlation between spatial navigation performance in the egocentric, allocentric, and allocentric delayed tasks and volumes of the selected brain regions. The results are presented in detail in Table 2 and Supplementary Tables 2 and 3.
DISCUSSION
In this study, we examined for the first time the combined effect of APOE and BDNF Val66Met polymorphisms on spatial navigation and volume of brain regions associated with spatial navigation in CU and aMCI individuals. Our findings indicate that the combination of the APOE ɛ4 and BDNF Met alleles is associated with least accurate spatial navigation performance in individuals with aMCI. Specifically, we found that those with aMCI who carry both APOE ɛ4 and BDNF Met alleles, although similar to other polymorphism groups in demographic characteristics, aMCI subtypes, global cognitive function, or depressive symptoms, have the least accurate egocentric spatial navigation performance. Our findings further indicate that the combination of the APOE ɛ4 and BDNF Met alleles may influence spatial navigation in CU individuals. Specifically, we found that those CU individuals with the high risk combination of the APOE ɛ4 and BDNF Met alleles compared to those with the low risk combination of the APOE ɛ3 and BDNF Val/Val alleles have less accurate allocentric spatial navigation performance. Egocentric (self-centered) navigation, where spatial information about locations and objects is encoded from the viewpoint of the navigator, is associated with the level of function of the posterior parietal cortex including the precuneus [2]. The function of the parietal cortex is adversely affected by the APOE ɛ4 allele [74, 75], which is associated with less accurate egocentric navigation in individuals with aMCI [27–29]. In addition, levels of BDNF protein in this region are prominently decreased [45, 76] due to the presence of the BDNF Met allele [52]. This may explain why the combination of the APOE ɛ4 and BDNF Met alleles was negatively associated with egocentric navigation in our participants with aMCI. Allocentric (world-centered) navigation, where locations and objects are encoded in relation to one another and are independent of the position of the navigator, is associated with the level of function of the medial temporal lobe structures, especially of the hippocampus [1]. BDNF expression is profoundly reduced in the hippocampus and adjacent entorhinal cortex [44] that are affected by APOE ɛ4-related changes [20, 21]. Next, the previous studies showed that the APOE ɛ4 allele is associated with less accurate allocentric navigation performance in cognitively normal older adults [26] and that the BDNF Met allele is associated with decreased use of allocentric spatial strategy in cognitively normal younger adults [53]. This may explain why the combination of the APOE ɛ4 and BDNF Met alleles was negatively associated with allocentric navigation in our CU participants. One would expect that the combination of the APOE ɛ4 and BDNF Met alleles could also negatively influence allocentric navigation in individuals with aMCI. However, in the current study we did not find the differences in allocentric spatial navigation performance between the polymorphism groups of participants with aMCI. This finding may be explained by the severity of cognitive impairment in our participants who were in the late stage of aMCI (with a mean MMSE score of 26.9 and predominant multiple domain cognitive impairment) and whose results could thus be affected by the floor effect. Similar findings were reported in our previous study, where the results for allocentric unlike egocentric navigation as a function of APOE ɛ4 categorization have not been significant in aMCI participants with a mean MMSE score of 26.4 [29]. Allocentric navigation is impaired in the course of aging [77, 78] and especially in preclinical AD and early aMCI [9, 79]. Impairment of egocentric navigation becomes increasingly more prominent later in the course of aMCI and in AD dementia [9, 10] and unlike impairment of allocentric navigation more accurately discriminates AD dementia from other types of dementia [80]. Therefore, aMCI participants with both APOE ɛ4 and BDNF Met alleles who have more pronounced egocentric navigation impairment may represent the more advanced stage of the disease compared to other polymorphism groups and may be more likely to have steeper cognitive decline and progress to AD dementia. The CU participants with both APOE ɛ4 and BDNF Met alleles who have less accurate allocentric navigation performance may also be more prone to cognitive decline and progression to MCI. Since individuals with aMCI who are carriers of the APOE ɛ4 allele are more likely to have Aβ pathology (i.e., prodromal AD) [81], it is plausible that the negative effect of the BDNF Met allele on spatial navigation has been influenced by abnormally high levels of Aβ in our aMCI cohort. Indeed, the BDNF Met allele enhances the brain vulnerability to Aβ toxicity [82] and therefore it may further worsen Aβ-related egocentric navigation impairment in individuals with aMCI and APOE ɛ4 allele [27, 28]. In the current study, there was no evidence of a learning effect across the trials in individual spatial navigation tasks in the participants with aMCI, which is in line with previous studies showing impairment of spatial navigation learning in individuals with aMCI and early AD [9, 28, 29].
As expected, the combination of the APOE ɛ4 and BDNF Met alleles was associated with more pronounced memory dysfunction in participants with aMCI. Specifically, the carriers of both APOE ɛ4 and BDNF Met alleles had lower verbal memory performance compared to the non-carriers. These results are in agreement with previous findings in individuals with preclinical AD [57], aMCI [58], prodromal AD and AD dementia [25] and support the hypothesis that combination of the APOE ɛ4 and BDNF Met alleles may specifically interfere with memory function. Again, as suggested previously [55, 57, 83], the negative effect of the BDNF Met allele on memory in our aMCI cohort may be influenced by abnormally high levels of Aβ that are more frequent in APOE ɛ4 carriers [81]. However, neither the current nor the previous studies [25, 58] have found the differences in memory performance between the BDNF Met carriers and non-carriers within a group of individuals with aMCI and the APOE ɛ4 allele, that is, within the group at increased risk of progressing to AD dementia [84]. The noteworthy finding in the current study was that the APOE ɛ4/BDNF Met carriers with aMCI compared to all other polymorphism groups including those with APOE ɛ4/BDNF Val/Val alleles have less accurate egocentric spatial navigation performance. This result may indicate that spatial navigation testing could more reliably reflect the deleterious effect of the BDNF Met allele on cognition than commonly used episodic memory tests. Given that spatial navigation is distinguishable from other cognitive functions [16] strongly influenced by genetic polymorphisms [29, 33] and impaired very early in AD [5, 6], its assessment along with examination of other cognitive functions could be highly beneficial when characterizing the cognitive profile of individuals with aMCI and their risk of progression to AD dementia.
The combination of the APOE ɛ4 and BDNF Met alleles was also associated with smaller volumes of the navigation-related brain regions in the participants with aMCI. Specifically, the carriers of both APOE ɛ4 and BDNF Met alleles had lower volumes of the hippocampus and the entorhinal cortex, that is, of the brain regions affected very early in AD and known to be important for allocentric navigation [1, 4]. The results are consistent with previous research demonstrating more pronounced hippocampal and entorhinal cortical atrophy in APOE ɛ4 [21, 85] and BDNF Met [48, 50] carriers and findings of the previous study showing lower right hippocampal volume in individuals with aMCI and combination of the APOE ɛ4 and BDNF Met alleles [58]. These APOE and BDNF Val66Met polymorphism-related structural changes in participants with aMCI could be explained by increased vulnerability of APOE ɛ4 carriers to AD-related pathological changes in this region [20] and by the fact that levels of BDNF protein that are decreased in BDNF Met carriers [47] are prominently reduced in the medial temporal lobe of individuals with AD [44]. This may negatively influence BDNF-induced neurogenesis [86] and dendritic arborization [41] and consequently lead to volumetric changes in this region. In our study, the APOE ɛ4/BDNF Met carriers with aMCI had lower volumes of the hippocampus and the entorhinal cortex compared to the APOE ɛ4 non-carriers. However, lower volumes of these regions were also found in the APOE ɛ4/BDNF Val/Val carriers. Therefore, it seems that structural differences in the medial temporal lobe between our aMCI polymorphism groups may be mainly driven by the presence of the APOE ɛ4 allele [21, 29]. Another noteworthy result was that right hippocampal volume was associated with allocentric navigation performance in participants with aMCI. This is in line with our previous findings in individuals with aMCI and AD dementia [16, 29, 70, 87] and underlines the important role of the right hippocampus in allocentric navigation. The combination of the APOE ɛ4 and BDNF Met alleles in individuals with aMCI was not associated with volume reduction of the inferior parietal cortex, precuneus, and posterior cingulate cortex, that is, of the brain regions known to be important for egocentric navigation [1, 2]. These brain regions have been associated with lower BDNF expression in AD dementia [45] and hypometabolism on FDG-PET in APOE ɛ4 carriers [22, 88] but their structural changes related to the APOE and BDNF Val66Met polymorphisms have not been reported. This may be explained by the fact that AD-related pathological changes associated with tissue loss occur later in the parietal cortex than in the medial temporal lobe during the disease [89] and that functional changes in these areas associated with cognitive impairment precede structural abnormalities [90]. Therefore, more pronounced egocentric navigation impairment in individuals with aMCI and combination of the APOE ɛ4 and BDNF Met alleles may not be reflected in volumetric changes of these navigation-related brain regions.
One of the strengths of the current study is the fact that this is the first study to date to focus on the influence of combination of the APOE and BDNF Val66Met polymorphisms on spatial navigation in a well-defined cohort of CU and aMCI participants. In addition, we used the real-space version of the hMWM, a well-established method mimicking navigation in the real world, to examine spatial navigation, a specific and neglected cognitive function, whose impairment is observed and frequently reported by patients in the early stages of AD [5, 91]. The real-space version allows the use of vestibular and proprioceptive information that is missing in virtual reality and therefore may better reflect real-world navigation [15]. Finally, we assessed the influence of these polymorphisms on volumes of specific brain regions relevant for spatial navigation and being affected very early in the course of AD [13, 14].
This study also has some limitations. First, the number of participants was relatively small, which may increase the chances of bias towards the Type II error. Next, our hypotheses required many statistical comparisons, which may increase the chances of bias towards the Type I error, although the post hoc tests and corrections for multiple comparisons were used in all analyzes. Because of these limitations our results should be interpreted with caution. Further studies with larger study cohorts are required to validate our findings. In addition, specific AD biomarkers (Aβ1–42 and p-tau181 in cerebrospinal fluid and amyloid PET imaging) were assessed only in a subset of the participants and therefore we could not evaluate the presence of AD pathology in all CU and aMCI participants. Because of the relatively small number of participants in some groups we could not evaluate the differences between the polymorphism groups in associations between spatial navigation performance and volumes of the brain regions, which should be a focus of future studies. Spatial navigation tasks were always performed in the same order and therefore we were not able to control for changes in participants’ performance across the tasks. Future studies randomly varying order of egocentric and allocentric tasks between participants may be needed to control for the practice effect. Finally, the cross-sectional design did not allow evaluating the combined effect of APOE and BDNF Val66Met polymorphisms on spatial navigation changes over time but longitudinal follow-up is ongoing.
Conclusion
In conclusion, the current study demonstrated that the high risk combination of the APOE ɛ4 and BDNF Met alleles is associated with more pronounced egocentric spatial navigation impairment and smaller volumes of the medial temporal lobe structures in individuals with aMCI and allocentric spatial navigation deficits in CU individuals. This finding may indicate that aMCI individuals with both APOE ɛ4 and BDNF Met alleles may have more advanced AD pathology and higher risk of progression to AD dementia and CU individuals may be at greater risk of cognitive decline and progression to MCI. Further, our findings, in line with previous research [56–58], suggest that the combination of the APOE ɛ4 and BDNF Met alleles may interfere with memory function in individuals with aMCI. However, memory testing, unlike spatial navigation testing, did not distinguish BDNF Met carriers from non-carriers among the individuals with the APOE ɛ4 allele. These findings may indicate that spatial navigation testing could more reliably assess the deleterious effect of the BDNF Met allele on cognition than traditionally used episodic memory tests. The focus of future studies should be to evaluate the effect of combination of APOE and BDNF Val66Met polymorphisms on longitudinal spatial navigation changes in individuals with aMCI and those in the earlier stage of the disease, such as individuals with subjective cognitive decline.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. J. Kalinova, Ms. K. Kopecka, Mr. J. Korb, Dr. T. Nikolai, Dr. H. Markova, Ms. M. Dokoupilova and Ms. S. Krejcova for help with data collection, Dr. I. Wolfova, Dr. I. Trubacik Mokrisova, and Dr. J. Cerman for help with participant recruitment and Mr. M. Uller for help with MRI data processing.
This study was supported by the Ministry of Health of the Czech Republic grants no. 19-04-00560, 18-04-00346 and 18-04-00455; the project no. LQ1605 from the National Program of Sustainability II (MEYS CR); the Ministry of Health, Czech Republic—conceptual development of research organization, University Hospital Motol, Prague, Czech Republic grant no. 00064203; the Institutional Support of Excellence 2. LF UK grant no. 6990332; and the Grant Agency of Charles University grants no. 176317, 693018 and 654217. Dr. R. Andel acknowledges support through a University of South Florida Nexus Initiative (UNI) Award.
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/20-0615r2).
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JAD-200615.
REFERENCES
- [1]. Maguire EA, Burgess N, Donnett JG, Frackowiak RSJ, Frith CD, O’Keefe J (1998) Knowing where and getting there: A human navigation network. Science 280, 921–924. [DOI] [PubMed] [Google Scholar]
- [2]. Weniger G, Ruhleder M, Lange C, Wolf S, Irle E (2011) Egocentric and allocentric memory as assessed by virtual reality in individuals with amnestic mild cognitive impairment. Neuropsychologia 49, 518–527. [DOI] [PubMed] [Google Scholar]
- [3]. Shine JP, Valdés-Herrera JP, Hegarty M, Wolbers T (2016) The human retrosplenial cortex and thalamus code head direction in a global reference frame. J Neurosci 36, 6371–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Ekstrom AD, Kahana MJ, Caplan JB, Fields TA, Isham EA, Newman EL, Fried I (2003) Cellular networks underlying human spatial navigation. Nature 425, 184–188. [DOI] [PubMed] [Google Scholar]
- [5]. Coughlan G, Laczó J, Hort J, Minihane A-M, Hornberger M (2018) Spatial navigation deficits — overlooked cognitive marker for preclinical Alzheimer disease? Nat Rev Neurol 14, 496–506. [DOI] [PubMed] [Google Scholar]
- [6]. Vlček K, Laczó J (2014) Neural correlates of spatial navigation changes in mild cognitive impairment and Alzheimer’s disease. Front Behav Neurosci 8, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Allison S, Babulal GM, Stout SH, Barco PP, Carr DB, Fagan AM, Morris JC, Roe CM, Head D (2018) Alzheimer disease biomarkers and driving in clinically normal older adults: Role of spatial navigation abilities. Alzheimer Dis Assoc Disord 32, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Rusconi ML, Suardi A, Zanetti M, Rozzini L (2015) Spatial navigation in elderly healthy subjects, amnestic and non amnestic MCI patients. J Neurol Sci 359, 430–437. [DOI] [PubMed] [Google Scholar]
- [9]. Hort J, Laczo J, Vyhnalek M, Bojar M, Bures J, Vlcek K (2007) Spatial navigation deficit in amnestic mild cognitive impairment. Proc Natl Acad Sci U S A 104, 4042–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Laczó J, Vlček K, Vyhnálek M, Vajnerová O, Ort M, Holmerová I, Tolar M, Andel R, Bojar M, Hort J (2009) Spatial navigation testing discriminates two types of amnestic mild cognitive impairment. Behav Brain Res 202, 252–259. [DOI] [PubMed] [Google Scholar]
- [11]. Laczó J, Andel R, Vyhnalek M, Vlcek K, Magerova H, Varjassyova A, Nedelska Z, Gazova I, Bojar M, Sheardova K, Hort J (2012) From Morris water maze to computer tests in the prediction of Alzheimer’s disease. Neurodegener Dis 10, 153–157. [DOI] [PubMed] [Google Scholar]
- [12]. Petersen RC (2004) Mild cognitive impairment as a diagnostic entity. J Intern Med 256, 183–194. [DOI] [PubMed] [Google Scholar]
- [13]. Killiany RJ, Gomez-Isla T, Moss M, Kikinis R, Sandor T, Jolesz F, Tanzi R, Jones K, Hyman BT, Albert MS (2000) Use of structural magnetic resonance imaging to predict who will get Alzheimer’s disease. Ann Neurol 47, 430–439. [PubMed] [Google Scholar]
- [14]. Kogure D, Matsuda H, Ohnishi T, Asada T, Uno M, Kunihiro T, Nakano S, Takasaki M (2000) Longitudinal evaluation of early Alzheimer’s disease using brain perfusion SPECT. J Nucl Med 41, 1155–1162. [PubMed] [Google Scholar]
- [15]. Mokrisova I, Laczo J, Andel R, Gazova I, Vyhnalek M, Nedelska Z, Levcik D, Cerman J, Vlcek K, Hort J (2016) Real-space path integration is impaired in Alzheimer’s disease and mild cognitive impairment. Behav Brain Res 307, 150–158. [DOI] [PubMed] [Google Scholar]
- [16]. Laczó J, Andel R, Nedelska Z, Vyhnalek M, Vlcek K, Crutch S, Harrison J, Hort J (2017) Exploring the contribution of spatial navigation to cognitive functioning in older adults. Neurobiol Aging 51, 67–70. [DOI] [PubMed] [Google Scholar]
- [17]. Saunders A, Hulette C, Welsh-Bohmer K, Schmechel D, Crain B, Burke J, Alberts M, Strittmatter W, Breitner J, Rosenberg C, Scott S, Gaskell P, Pericak-Vance M, Roses A (1996) Specificity, sensitivity, and predictive value of apolipoprotein-E genotyping for sporadic Alzheimer’s disease. Lancet 348, 90–93. [DOI] [PubMed] [Google Scholar]
- [18]. Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923. [DOI] [PubMed] [Google Scholar]
- [19]. Lim YY, Mormino EC, Alzheimer’s Disease Neuroimaging Initiative (2017) APOE genotype and early β-amyloid accumulation in older adults without dementia. Neurology 89, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Mattson MP, Moehl K, Ghena N, Schmaedick M, Cheng A (2018) Intermittent metabolic switching, neuroplasticity and brain health. Nat Rev Neurosci 19, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Li B, Shi J, Gutman BA, Baxter LC, Thompson PM, Caselli RJ, Wang Y, Alzheimer’s Disease Neuroimaging Initiative (2016) Influence of APOE genotype on hippocampal atrophy over time - an N=1925 surface-based ADNI study. PLoS One 11, e0152901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Small GW, Ercoli LM, Silverman DH, Huang SC, Komo S, Bookheimer SY, Lavretsky H, Miller K, Siddarth P, Rasgon NL, Mazziotta JC, Saxena S, Wu HM, Mega MS, Cummings JL, Saunders AM, Pericak-Vance MA, Roses AD, Barrio JR, Phelps ME (2000) Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer’s disease. Proc Natl Acad Sci U S A 97, 6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, Baxter LC, Rapcsak SZ, Shi J, Woodruff BK, Locke DEC, Snyder CH, Alexander GE, Rademakers R, Reiman EM (2009) Longitudinal modeling of age-related memory decline and the APOE ɛ4 effect. N Engl J Med 361, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Xu W-L, Caracciolo B, Wang H-X, Santoni G, Winblad B, Fratiglioni L (2013) Accelerated progression from mild cognitive impairment to dementia among APOE ɛ4ɛ4 carriers. J Alzheimers Dis 33, 507–515. [DOI] [PubMed] [Google Scholar]
- [25]. Gomar JJ, Conejero-Goldberg C, Huey ED, Davies P, Goldberg TE (2016) Lack of neural compensatory mechanisms of BDNF val66met met carriers and APOE E4 carriers in healthy aging, mild cognitive impairment, and Alzheimer’s disease. Neurobiol Aging 39, 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Berteau-Pavy F, Park B, Raber J (2007) Effects of sex and APOE ɛ4 on object recognition and spatial navigation in the elderly. Neuroscience 147, 6–17. [DOI] [PubMed] [Google Scholar]
- [27]. Laczó J, Andel R, Vyhnalek M, Vlcek K, Magerova H, Varjassyova A, Tolar M, Hort J (2010) Human analogue of the morris water maze for testing subjects at risk of Alzheimer’s disease. Neurodegener Dis 7, 148–152. [DOI] [PubMed] [Google Scholar]
- [28]. Laczó J, Andel R, Vlček K, Macoška V, Vyhnálek M, Tolar M, Bojar M, Hort J (2011) Spatial navigation and APOE in amnestic mild cognitive impairment. Neurodegener Dis 8, 169–177. [DOI] [PubMed] [Google Scholar]
- [29]. Laczó J, Andel R, Vyhnalek M, Vlcek K, Nedelska Z, Matoska V, Gazova I, Mokrisova I, Sheardova K, Hort J (2014) APOE and spatial navigation in amnestic MCI: Results from a computer-based test. Neuropsychology 28, 676–684. [DOI] [PubMed] [Google Scholar]
- [30]. Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, Reiman EM (2010) A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J 10, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, Saunders AM, Lutz MW, Welsh-Bohmer KA, Roses AD, Chiba-Falek O (2012) A homopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimers Dement 8, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Johnson SC, La Rue A, Hermann BP, Xu G, Koscik RL, Jonaitis EM, Bendlin BB, Hogan KJ, Roses AD, Saunders AM, Lutz MW, Asthana S, Green RC, Sager MA (2011) The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE ɛ3/ɛ3 genotype. Alzheimers Dement 7, 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Laczó J, Andel R, Vyhnalek M, Matoska V, Kaplan V, Nedelska Z, Lerch O, Gazova I, Moffat SD, Hort J (2015) The effect of TOMM40 on spatial navigation in amnestic mild cognitive impairment. Neurobiol Aging 36, 2024–2033. [DOI] [PubMed] [Google Scholar]
- [34]. Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, Joshipura KD, Pearson J V., Hu-Lince D, Craig DW, Coon KD, Dunckley T, Bandy D, Lee W, Chen K, Beach TG, Mastroeni D, Grover A, Ravid R, Sando SB, Aasly JO, Heun R, Jessen F, Kölsch H, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Papassotiropoulos A, Stephan DA, Huentelman MJ (2010) Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiol Aging 31, 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Milnik A, Heck A, Vogler C, Heinze H-J, de Quervain DJ-F, Papassotiropoulos A (2012) Association of KIBRA with episodic and working memory: A meta-analysis. Am J Med Genet B Neuropsychiatr Genet 159B, 958–969. [DOI] [PubMed] [Google Scholar]
- [36]. Schuck NW, Doeller CF, Schjeide B-MM, Schröder J, Frensch PA, Bertram L, Li S-C (2013) Aging and KIBRA/WWC1 genotype affect spatial memory processes in a virtual navigation task. Hippocampus 23, 919–930. [DOI] [PubMed] [Google Scholar]
- [37]. Budni J, Bellettini-Santos T, Mina F, Garcez ML, Zugno AI (2015) The involvement of BDNF, NGF and GDNF in aging and Alzheimer’s disease. Aging Dis 6, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Kambeitz JP, Bhattacharyya S, Kambeitz-Ilankovic LM, Valli I, Collier DA, McGuire P (2012) Effect of BDNF val66met polymorphism on declarative memory and its neural substrate: A meta-analysis. Neurosci Biobehav Rev 36, 2165–2177. [DOI] [PubMed] [Google Scholar]
- [39]. Brown DT, Vickers JC, Stuart KE, Cechova K, Ward DD (2020) The BDNF Val66Met polymorphism modulates resilience of neurological functioning to brain ageing and dementia: A narrative review. Brain Sci 10, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Deinhardt K, Chao MV (2014) Shaping neurons: Long and short range effects of mature and proBDNF signalling upon neuronal structure. Neuropharmacology 76, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Moya-Alvarado G, Gonzalez A, Stuardo N, Bronfman FC (2018) Brain-derived neurotrophic factor (BDNF) regulates Rab5-positive early endosomes in hippocampal neurons to induce dendritic branching. Front Cell Neurosci 12, 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Panja D, Bramham CR (2014) BDNF mechanisms in late LTP formation: A synthesis and breakdown. Neuropharmacology 76, 664–676. [DOI] [PubMed] [Google Scholar]
- [43]. Moser M-B, Rowland DC, Moser EI (2015) Place cells, grid cells, and memory. Cold Spring Harb Perspect Biol 7, a021808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Narisawa-Saito M, Wakabayashi K, Tsuji S, Takahashi H, Nawa H (1996) Regional specificity of alterations in NGF, BDNF and NT-3 levels in Alzheimer’s disease. Neuroreport 7, 2925–2928. [DOI] [PubMed] [Google Scholar]
- [45]. Hock C, Heese K, Hulette C, Rosenberg C, Otten U (2000) Region-specific neurotrophin imbalances in Alzheimer disease. Arch Neurol 57, 846–851. [DOI] [PubMed] [Google Scholar]
- [46]. Price JL, Ko AI, Wade MJ, Tsou SK, McKeel DW, Morris JC (2001) Neuron number in the entorhinal cortex and CA1 in preclinical Alzheimer disease. Arch Neurol 58, 1395–1402. [DOI] [PubMed] [Google Scholar]
- [47]. Ozan E, Okur H, Eker Ç, Eker ÖD, Gönül AS, Akarsu N (2010) The effect of depression, BDNF gene val66met polymorphism and gender on serum BDNF levels. Brain Res Bull 81, 61–65. [DOI] [PubMed] [Google Scholar]
- [48]. Voineskos AN, Lerch JP, Felsky D, Shaikh S, Rajji TK, Miranda D, Lobaugh NJ, Mulsant BH, Pollock BG, Kennedy JL (2011) The brain-derived neurotrophic factor Val66Met polymorphism and prediction of neural risk for Alzheimer disease. Arch Gen Psychiatry 68, 198–206. [DOI] [PubMed] [Google Scholar]
- [49]. Yang X, Liu P, Sun J, Wang G, Zeng F, Yuan K, Liu J, Dong M, von Deneen KM, Qin W, Tian J (2012) Impact of brain-derived neurotrophic factor Val66Met polymorphism on cortical thickness and voxel-based morphometry in healthy Chinese young adults. PLoS One 7, e37777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR (2004) The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci 24, 10099–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR (2003) Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci 23, 6690–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR (2003) The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112, 257–269. [DOI] [PubMed] [Google Scholar]
- [53]. Banner H, Bhat V, Etchamendy N, Joober R, Bohbot VD (2011) The brain-derived neurotrophic factor Val66Met polymorphism is associated with reduced functional magnetic resonance imaging activity in the hippocampus and increased use of caudate nucleus-dependent strategies in a human virtual navigation task. Eur J Neurosci 33, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Lim YY, Villemagne VL, Laws SM, Ames D, Pietrzak RH, Ellis KA, Harrington KD, Bourgeat P, Salvado O, Darby D, Snyder PJ, Bush AI, Martins RN, Masters CL, Rowe CC, Nathan PJ, Maruff P (2013) BDNF Val66Met, Aβ amyloid, and cognitive decline in preclinical Alzheimer’s disease. Neurobiol Aging 34, 2457–2464. [DOI] [PubMed] [Google Scholar]
- [55]. Lim YY, Villemagne VL, Laws SM, Ames D, Pietrzak RH, Ellis KA, Harrington K, Bourgeat P, Bush AI, Martins RN, Masters CL, Rowe CC, Maruff P, Lim YY, Villemagne VL, Laws SM, Ames D, Pietrzak RH, Ellis KA, Harrington K (2014) Effect of BDNF Val66Met on memory decline and hippocampal atrophy in prodromal Alzheimer’s disease: A preliminary study. PLoS One 9, e86498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Ward D, Summers MJ, Saunders NL, Janssen P, Stuart KE, Vickers JC (2014) APOE and BDNF Val66Met polymorphisms combine to influence episodic memory function in older adults. Behav Brain Res 271, 309–315. [DOI] [PubMed] [Google Scholar]
- [57]. Lim YY, Villemagne VL, Laws SM, Pietrzak RH, Snyder PJ, Ames D, Ellis KA, Harrington K, Rembach A, Martins RN, Rowe CC, Masters CL, Maruff P (2015) APOE and BDNF polymorphisms moderate amyloid β-related cognitive decline in preclinical Alzheimer’s disease. Mol Psychiatry 20, 1322–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Cechova K, Andel R, Angelucci F, Chmatalova Z, Markova H, Laczó J, Vyhnalek M, Matoska V, Kaplan V, Nedelska Z, Ward DD, Hort J (2020) Impact of APOE and BDNF Val66Met gene polymorphisms on cognitive functions in patients with amnestic mild cognitive impairment. J Alzheimers Dis 73, 247–257. [DOI] [PubMed] [Google Scholar]
- [59]. Sheardova K, Vyhnalek M, Nedelska Z, Laczo J, Andel R, Marciniak R, Cerman J, Lerch O, Hort J (2019) Czech Brain Aging Study (CBAS): Prospective multicentre cohort study on risk and protective factors for dementia in the Czech Republic. BMJ Open 9, e030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60]. Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Petersen RC, Snyder PJ, Carrillo MC, Thies B, Phelps CH (2011) The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Yesavage JA (1988) Geriatric Depression Scale. Psychopharmacol Bull 24, 709–711. [PubMed] [Google Scholar]
- [62]. Fazekas F, Chawluk J, Alavi A, Hurtig H, Zimmerman R (1987) MR signal abnormalities at 1.5 T in Alzheimer’s dementia and normal aging. Am J Roentgenol 149, 351–356. [DOI] [PubMed] [Google Scholar]
- [63]. Hixson JE, Vernier DT (1990) Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res 31, 545–548. [PubMed] [Google Scholar]
- [64]. Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM (2004) Automatically parcellating the human cerebral cortex. Cereb Cortex 14, 11–22. [DOI] [PubMed] [Google Scholar]
- [65]. Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002) Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355. [DOI] [PubMed] [Google Scholar]
- [66]. Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ (2006) An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. [DOI] [PubMed] [Google Scholar]
- [67]. Ohnishi T, Matsuda H, Hirakata M, Ugawa Y (2006) Navigation ability dependent neural activation in the human brain: An fMRI study. Neurosci Res 55, 361–369. [DOI] [PubMed] [Google Scholar]
- [68]. Mosconi L, Brys M, Switalski R, Mistur R, Glodzik L, Pirraglia E, Tsui W, De Santi S, de Leon MJ (2007) Maternal family history of Alzheimer’s disease predisposes to reduced brain glucose metabolism. Proc Natl Acad Sci U S A 104, 19067–19072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69]. Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D (1996) Preclinical evidence of Alzheimer’s disease in persons homozygous for the ɛ4 allele for apolipoprotein E. N Engl J Med 334, 752–758. [DOI] [PubMed] [Google Scholar]
- [70]. Nedelska Z, Andel R, Laczó J, Vlcek K, Horinek D, Lisy J, Sheardova K, Bures J, Hort J (2012) Spatial navigation impairment is proportional to right hippocampal volume. Proc Natl Acad Sci U S A 109, 2590–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71]. Jack CR, Petersen RC, O’Brien PC, Tangalos EG (1992) MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology 42, 183–188. [DOI] [PubMed] [Google Scholar]
- [72]. Singer JD, Willett JB (2009), Applied Longitudinal Data Analysis: Modeling Change and Event Occurrence, Oxford University Press. [Google Scholar]
- [73]. Tabachnick BG, Fidell LS (2007) Using Multivariate Statistics, Allyn & Bacon, Boston, MA. [Google Scholar]
- [74]. Koelewijn L, Lancaster TM, Linden D, Dima DC, Routley BC, Magazzini L, Barawi K, Brindley L, Adams R, Tansey KE, Bompas A, Tales A, Bayer A, Singh K (2019) Oscillatory hyperactivity and hyperconnectivity in young APOE-ɛ4 carriers and hypoconnectivity in Alzheimer’s disease. Elife 8, e36011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75]. Chen Y, Liu Z, Zhang J, Chen K, Yao L, Li X, Gong G, Wang J, Zhang Z (2017) Precuneus degeneration in nondemented elderly individuals with APOE ɛ4: Evidence from structural and functional MRI analyses. Hum Brain Mapp 38, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76]. Angelucci F, Gelfo F, De Bartolo P, Caltagirone C, Petrosini L (2011) BDNF concentrations are decreased in serum and parietal cortex in immunotoxin 192 IgG-Saporin rat model of cholinergic degeneration. Neurochem Int 59, 1–4. [DOI] [PubMed] [Google Scholar]
- [77]. Gazova I, Laczó J, Rubinova E, Mokrisova I, Hyncicova E, Andel R, Vyhnalek M, Sheardova K, Coulson EJ, Hort J (2013) Spatial navigation in young versus older adults. Front Aging Neurosci 5, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78]. Rodgers MK, Sindone JA, Moffat SD (2012) Effects of age on navigation strategy. Neurobiol Aging 33, 202.e15–202.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Allison SL, Fagan AM, Morris JC, Head D (2016) Spatial navigation in preclinical Alzheimer’s disease. J Alzheimers Dis 52, 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Tu S, Spiers HJ, Hodges JR, Piguet O, Hornberger M (2017) Egocentric versus allocentric spatial memory in behavioral variant frontotemporal dementia and Alzheimer’s disease. J Alzheimers Dis 59, 883–892. [DOI] [PubMed] [Google Scholar]
- [81]. Mattsson N, Groot C, Jansen WJ, Landau SM, Villemagne VL, Engelborghs S, Mintun MM, Lleo A, Molinuevo JL, Jagust WJ, Frisoni GB, Ivanoiu A, Chételat G, Resende de Oliveira C, Rodrigue KM, Kornhuber J, Wallin A, Klimkowicz-Mrowiec A, Kandimalla R, Popp J, Aalten PP, Aarsland D, Alcolea D, Almdahl IS, Baldeiras I, van Buchem MA, Cavedo E, Chen K, Cohen AD, Förster S, Fortea J, Frederiksen KS, Freund-Levi Y, Gill KD, Gkatzima O, Grimmer T, Hampel H, Herukka S-K, Johannsen P, van Laere K, de Leon MJ, Maier W, Marcusson J, Meulenbroek O, Møllergård HM, Morris JC, Mroczko B, Nordlund A, Prabhakar S, Peters O, Rami L, Rodríguez-Rodríguez E, Roe CM, Rüther E, Santana I, Schröder J, Seo SW, Soininen H, Spiru L, Stomrud E, Struyfs H, Teunissen CE, Verhey FRJ, Vos SJB, van Waalwijk van Doorn LJC, Waldemar G, Wallin ÅK, Wiltfang J, Vandenberghe R, Brooks DJ, Fladby T, Rowe CC, Drzezga A, Verbeek MM, Sarazin M, Wolk DA, Fleisher AS, Klunk WE, Na DL, Sánchez-Juan P, Lee DY, Nordberg A, Tsolaki M, Camus V, Rinne JO, Fagan AM, Zetterberg H, Blennow K, Rabinovici GD, Hansson O, van Berckel BNM, van der Flier WM, Scheltens P, Visser PJ, Ossenkoppele R (2018) Prevalence of the apolipoprotein E ɛ4 allele in amyloid β positive subjects across the spectrum of Alzheimer’s disease. Alzheimers Dement 14, 913–924. [DOI] [PubMed] [Google Scholar]
- [82]. Franzmeier N, Ren J, Damm A, Monté-Rubio G, Boada M, Ruiz A, Ramirez A, Jessen F, Düzel E, Rodríguez Gómez O, Benzinger T, Goate A, Karch CM, Fagan AM, McDade E, Buerger K, Levin J, Duering M, Dichgans M, Suárez-Calvet M, Haass C, Gordon BA, Lim YY, Masters CL, Janowitz D, Catak C, Wolfsgruber S, Wagner M, Milz E, Moreno-Grau S, Teipel S, Grothe MJ, Kilimann I, Rossor M, Fox N, Laske C, Chhatwal J, Falkai P, Perneczky R, Lee J-H, Spottke A, Boecker H, Brosseron F, Fliessbach K, Heneka MT, Nestor P, Peters O, Fuentes M, Menne F, Priller J, Spruth EJ, Franke C, Schneider A, Westerteicher C, Speck O, Wiltfang J, Bartels C, Araque Caballero MÁ, Metzger C, Bittner D, Salloway S, Danek A, Hassenstab J, Yakushev I, Schofield PR, Morris JC, Bateman RJ, Ewers M (2019) The BDNFVal66Met SNP modulates the association between beta-amyloid and hippocampal disconnection in Alzheimer’s disease. Mol Psychiatry, doi: 10.1038/s41380-019-0404-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83]. Boots EA, Schultz SA, Clark LR, Racine AM, Darst BF, Koscik RL, Carlsson CM, Gallagher CL, Hogan KJ, Bendlin BB, Asthana S, Sager MA, Hermann BP, Christian BT, Dubal DB, Engelman CD, Johnson SC, Okonkwo OC (2017) BDNF Val66Met predicts cognitive decline in the Wisconsin Registry for Alzheimer’s Prevention. Neurology 88, 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84]. Petersen RC, Smith GE, Ivnik RJ, Tangalos EG, Schaid DJ, Thibodeau SN, Kokmen E, Waring SC, Kurland LT (1995) Apolipoprotein E status as a predictor of the development of Alzheimer’s disease in memory-impaired individuals. JAMA 273, 1274–1278. [PubMed] [Google Scholar]
- [85]. Bunce D, Anstey KJ, Cherbuin N, Gautam P, Sachdev P, Easteal S (2012) APOE genotype and entorhinal cortex volume in non-demented community-dwelling adults in midlife and early old age. J Alzheimers Dis 30, 935–942. [DOI] [PubMed] [Google Scholar]
- [86]. Hsiao Y-H, Hung H-C, Chen S-H, Gean P-W (2014) Social interaction rescues memory deficit in an animal model of Alzheimer’s disease by increasing BDNF-dependent hippocampal neurogenesis. J Neurosci 34, 16207–16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87]. Parizkova M, Lerch O, Moffat SD, Andel R, Mazancova AF, Nedelska Z, Vyhnalek M, Hort J, Laczó J (2018) The effect of Alzheimer’s disease on spatial navigation strategies. Neurobiol Aging 64, 107–115. [DOI] [PubMed] [Google Scholar]
- [88]. Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J (2005) From The Cover: Correlations between apolipoprotein E 4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci U S A 102, 8299–8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89]. Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82, 239–259. [DOI] [PubMed] [Google Scholar]
- [90]. Chetelat G, Desgranges B, de la Sayette V, Viader F, Berkouk K, Landeau B, Lalevée C, Le Doze F, Dupuy B, Hannequin D, Baron J-C, Eustache F (2003) Dissociating atrophy and hypometabolism impact on episodic memory in mild cognitive impairment. Brain 126, 1955–1967. [DOI] [PubMed] [Google Scholar]
- [91]. Cerman J, Andel R, Laczo J, Vyhnalek M, Nedelska Z, Mokrisova I, Sheardova K, Hort J (2018) Subjective spatial navigation complaints - A frequent symptom reported by patients with subjective cognitive decline, mild cognitive impairment and Alzheimer’s disease. Curr Alzheimer Res 15, 219–228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.