Abstract
Angiogenesis plays a critical role in skeletal repair and regeneration. Our understanding of the intricate relationship between osteogenesis and angiogenesis at a repair site has been hindered by the lack of an effective approach that allows tracking of bone healing and neovascularization simultaneously at a high spatiotemporal resolution in living animals. To overcome this barrier, we have recently established a cranial bone defect window chamber model in mice that enables high resolution, four-dimensional imaging analyses of bone defect healing using multiphoton laser scanning microscopy (MPLSM). The windowed defect model confers imaging of the defect through both microCT and MPLSM in vivo, facilitating lineage tracing and longitudinal analyses of osteogenesis and angiogenesis in repair. The windowed chamber model further permits insertion of cellular implants or bone graft materials, aiding in spatiotemporal analyses of the interactions between biomaterials and vascular microenvironment in living animals. In this chapter, we will describe the improved technique for establishing the chronic cranial defect window chamber model for long-term imaging as well as the imaging analysis protocols for quantitative analyses of osteogenesis and angiogenesis at the site of bone defect repair.
Keywords: Intravital imaging, osteogenesis, angiogenesis, bone defect repair, lineage tracing, spatiotemporal analyses
1. Introduction
Cranial bone defect healing is driven by coordinated osteogenesis and angiogenesis at the site of repair. Current analyses of bone defect healing heavily rely on histology, which is destructive and challenging to provide a three dimensional perspectives of the defect healing. A microCT-based approach has been used for quantitative and qualitative analyses of osteogenesis and angiogenesis during bone repair and regeneration 1–3. Although successful, microCT-based analyses do not have the spatial and temporal resolution to correlate neovascularization with cellular differentiation during bone defect repair and reconstruction. Furthermore, microCT-based method lacks the resolution to visualize micro-vessels less than 20 μm in diameter. Since bone defect repair is a dynamic process controlled at multiple spatial and temporal scales, establishing the capability to analyze progenitor cell dynamics and their interaction with neovasculature is important for advancing our knowledge of bone defect repair and for further optimizing progenitor cell and material-based approaches for enhanced repair and reconstruction.
Multiphoton laser scanning microscopy (MPLSM) has emerged as a superior in vivo imaging modality for analyses of thick tissue in living animals. MPLSM offers confocal-like imaging quality with enhanced imaging depths and reduced photo damage. Multiphoton microscopy further permits morphological and functional analyses of neovasculature with unprecedented advantages of high spatio-temporal resolution, minimal invasiveness and 3D capability 4, 5. Compared with other existing modalities (e.g. electrodes, MRI, Doppler), multiphoton has the resolution to image structure and function of a single vessel, enabling the evaluation of the impact of local vascular microenvironment on selective donor stem/progenitor cells. In addition to imaging nonlinear fluorescence excitation, multiphoton microscopy can be used for imaging collagen matrix through second harmonic generation (SHG) 6, 7. The unique capability of this technology to allow simultaneously visualizing donor cells, ECM as well as the surrounding vascular network has made it extremely useful in bone tissue repair and engineering applications. Combined with an adequate in vivo bone-healing model, multiphoton microscopy could be especially powerful for in vivo imaging of complex cell-cell and cell-matrix interactions, with added advantages for simultaneously performing functional analyses for cellular motility and vascular ingrowth in a non-destructive, no ninvasive and dynamic fashion 8.
In this chapter, we describe the techniques for the establishment of a chronic window chamber model for visualization and longitudinal analyses of osteogenesis and angiogenesis in bone defect healing. The cranial window chamber model in mice has been previously reported for analyses of brain cell function and tumor-associated neovascularization 9, 10. The model was modified to meet the need for long-term tracking of defect healing via intravital imaging.
2. Materials
2.1. Creation of a cranial defect window chamber model in mice
Col2.3GFP transgenic mice (Jackson Laboratory, Cat No 013134)
Ketamine (Par Pharmaceutical company Inc. Cat No V 42023-113-10) and Xylazine (Akorn Animal Health, Cat No 59399-111-50)
A stereotaxic frame (Stoelting Co. Cat No 51725)
Mouse heating pad (Kent Scientific Co.)
Alpha-MEM medium containing 1% penicillin and streptomycin, 1% glutamine.
Aryl-ether-ether-ketone (PEEK): (McMaster-Carr)
Cyanoacrylate glue (Loctite, Cat #45404 and 49550).
Tungsten vanadium inverted cone burs, various size (Armstrong Tool & Supply Company)
High-speed micro-drill (Stoelting Co. Cat No 58600).
Eight mm round cover-glass slip in #1 thickness (Warner Instruments. Cat No 64–0730)
Bone Cement (A-M Systems, Cat No 525000).
2.2. Imaging and image analyses
Olympus FV1000-AOM multiphoton imaging system (Olympus), equipped with a Titanium:Sapphire laser (Spectra-Physics), a C-Apochromat 10X/0.45 (Zeiss), and a 25X/1.05 (Olympus) water immersion objective.
Q-tracker 655 nanocrystals (ThermoFisher Scientific, Cat No Q21021MP).
Amira software developed by Visualization Sciences Group (VSG, Burlington MA).
Image J with plugins for simple image processing.
3. Methods
3.1. Surgical procedures for establishing a chronic window chamber model for cranial defect repair.
Anesthetize Col2.3GFP mice via intraperitoneal injection of 0.2 ml/20 g body weight of a mix of 20 mg/ml Ketamine and 3 mg/ml Xylazine in 0.9% NaCl. (See note 1) The Col2.3GFP mice express Enhanced Green Fluorescent Protein (GFP) gene under the control of the 2.3 kb rat procollagen, type 1, alpha 1(Col1a1) promoter, which allows visualization and tracking of the proliferation, differentiation and migration of osteoblasts in living animals.
To control any acute pain induced by the surgery, pre-emptive analgesia, buprenorphine (0.5mg/kg SQ) should be given prior to surgery and after surgery every 12 hours for 3 days.
The hair should be thoroughly shaved over most of the scalp using a conventional electric razor.
Set a heating pad on low (~35°C) and place it under the mouse on the stereotaxic frame to maintain body temperature.
To stabilize the mouse head for surgery, the animal should be raised to the level of the ear bars mounted on the platform of stereotaxic frame. First lock the nose clamp and incision bar to mount the head on the stereotaxic device and then lock the two ear bars into the external auditory meatus. Tighten both sides of the bars to make sure the mouse head is not movable and stably fixed onto the stereotaxic platform. Optionally apply ophthalmic ointment or mineral oil to the eyes to protect them from accidental spills.
Following stabilization of the mice with iodine surgical scrub, make a midline scalp incision using microsurgical tools. Clear the soft tissue and excessive fluid on the skull with Q-tips.
A customized Polyetheretherketone (PEEK) spacer with a circular opening of 6 mm (diameter) in its center is glued on the skull with a thin layer of glue (Loctite, Cat #45404). Press it for a few minutes to ensure the plate is tightly glued to the skull and secured it with a small amount of glue around the edges.
Seal the custom-made plate on the skull with a mix of bone cement and glue (Loctite, Cat No 49550) at about 1:1 ratio.
Wait 10–15 minutes until the metal plate is firmly attached to the skull.
Wash the open chamber with extensive sterile PBS with antibiotics (see Note 2).
Use a tungsten vanadium inverted cone bur attached to a high-speed micro-drill to create a full thickness defect in the parietal bone of mouse calvarium. The size of the defect can be controlled using a 0.9 mm or 1.8 mm size Busch inverted cone bur which generates a 1 mm or 2 mm full thickness defect, respectively, in the parietal bone. Avoid suture and regions with visible vasculatures. (see Note 3)
Carefully remove the debris of bone and clear the area with an air blower, avoiding damaging the dura and the vessels on the dura.
Thoroughly rinse the opening with room temperature alpha-MEM medium containing 1% penicillin and streptomycin, 1% glutamine.
Cover the clear dura with a thin layer of a-MEM medium containing 1% penicillin and streptomycin, 1% glutamine and keep the wound wet.
Carefully place a custom-made circular glass cover-glass slips on top of the custom-made plate to cover the open-skull region.
Seal the edge of the circular cover slip onto the custom-made plate with a thin layer of glue (Loctite, Cat No 45404).
Allow the mouse to wake up and recover on a warm pad.
Return the mouse to vivarium and keep each mouse separately in a cage to avoid fight during the first few weeks after surgery.
Once established, the mouse with a window chamber can survive over a 6-month period with no visible signs of distress (Fig. 1) (see Note 4 and 5).
Figure 1.
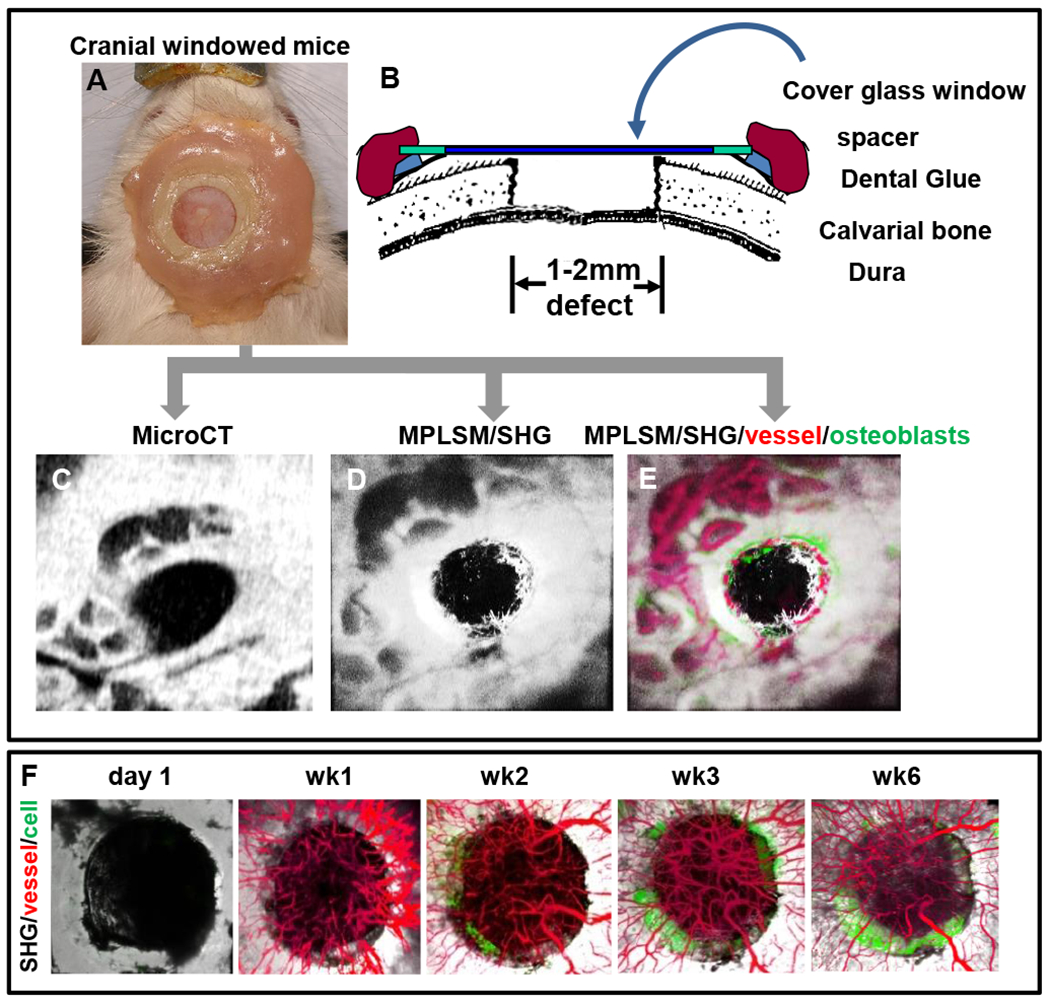
Superior view photograph of a Col 2.3 GFP mouse with the instrumented head piece exposing a cranial defect is shown (A). Schematic illustration of the model (B). Windowed mouse is subjected to both micro-CT scanning (C) and MPLSM (D&E). Three-dimension reconstructed images of the healing defect derived from microCT scanning (C) and SHG microscopy (D) are presented to illustrate the remarkable agreement between the current standard and our new MPLSM approach. Longitudinal tracking of bone defect healing in Col2.3GFP transgenic mice over 6 week period to illustrate osteogenesis and angiogenesis (F). SHG, white; vessel, red; osteoblasts, green.
3.2. Imaging of osteogenesis and angiogenesis
3.2.1. MPLSM.
An Olympus FV1000-AOM multiphoton imaging system, equipped with a Titanium:Sapphire laser, a C-Apochromat 10X/0.45, and a 25X/1.05 (Olympus) water immersion objective, is used for live imaging of the cranial defect healing. Images are acquired at 512x512 pixels, 0.2 ms pixel dwell with the laser tuned to 780nm. The fluorescence of GFP and second harmonic signals (SHG) is collected with a 517/23-nm and a 390/20- nm bandpass filters (Semrock), respectively. To visualize the blood vessel network, Q-tracker 655 nanocrystals are dissolved in sterilized water according to the instruction from the manufacturer and injected intraveneously via retro-orbital venous sinus into mouse circulation. Far red fluorescence from nanocrystals is detected using a far red bandpass emission filter (Semrock, 655/40 nm). Using the 10x water immersion objective lens (Zeiss), a 1.3 x1.3 mm multichannel z-series stack with 5mm steps is obtained. The z-series stack allowed 3D reconstruction of the defect up to a depth of 300mm (see Note 6). To characterize collagen matrix propagation and Col2.3GFP (+) cell dynamics, the defect can be repeatedly imaged over 3 month period by MPLSM. To ensure accuracy of the analyses, all imaging parameters, including laser power, PMT voltages and compensation should be maintained constant throughout the entire experiment.
3.3. MPLSM Image analyses
3.3.1. Quantification of SHG propagation and Col2.3GFP cell dynamics in cranial defect repair.
The multichannel 2D slice viewing and 3D reconstruction of the defect are conducted using Amira software. Based on the SHG microscopy, which provided the contour of the defect region, a region of interest (ROI) can be established in a time-series, referencing back to the circular defect region at the day 1 following surgery.
To quantify SHG propagation within the ROI, a global thresholding method is used to quantify the total number of SHG (+) voxels above an appropriate threshold in time-series images. The percentage volume of SHG occupying the total volume of the defect can be used to depict the propagation of collagen bone matrix within the defect. Similarly, the volume of Col2.3GFP (+) cells occupying the defect regions within defined ROI can be obtained by quantifying the total number of green fluorescent voxels above an appropriate threshold across the time-series images. SHG signal from bone matrix can be easily distinguished from soft tissue by its distinct morphology and its association with Col2.3GFP cells during healing. A schematic illustrating the measurements of SHG and GFP (+) cells can be found in Fig. 2. All measurements can be conducted using a combination of Image J and Amira.
Figure 2. Quantitative and volumetric analyses of GFP and SHG signals in cranial defect healing.
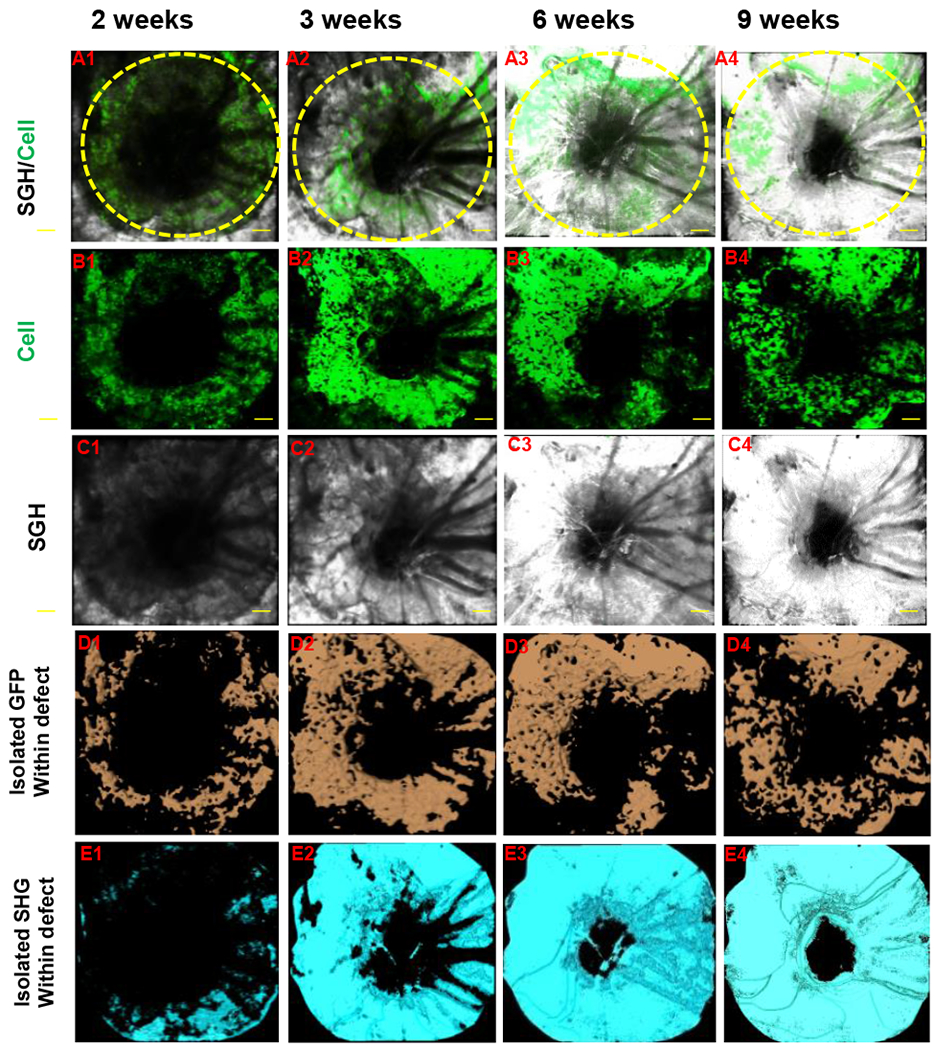
A bony defect was created in Col2.3GFP transgenic mouse as previously described. The defect healing was imaged over a period of 9 weeks using MPLSM. Propagation of SHG and differentiation of Col2.3GFP (+) osteoblasts were illustrated in panel A (SHG and GFP + cells), B (GFP + cells) and C (SHG, white). Signals of GFP and SHG were isolated within the defect region by proper thresholding across all samples. Volume of GFP (+) cells (D, orange) and SHG (E, cyan) were further calculated based on isolated signals.
To examine the extent of bone defect healing, the area of the defect is measured in ImageJ by tracing the circular defect region in the 2D images obtained from MPLSM. Since we are using a Busch inverted cone bur to create the defect, the shape of the defect is circular or nearly circular. Based on mathematical calculations, the radius of a circle can be approximated using the area of the circular region, i.e., . The mean of the cell advancing distance from the edges of the bone is determined by calculating the difference of the outer radius (corresponding to the original edge of the defect) and the inner radius (corresponding to the leading edge of the expanding cells). The original radius of the defect at day 1 is calculated as , where A_0 is the area of the defect at day 1. The area of the defect region that is not occupied by the cells at each indicated time point can be used to approximately calculate the radius of the inner circle as: , where A_inner is the area of the void in the defect. The advancing distance of osteoblasts at the indicated time points is then given by . The difference is further normalized by the radius of the original defect and plotted as a function of time.
3.3.2. Quantitative and histomorphometric analyses of neovascularization at the site of cranial bone defect repair.
Quantitative and histomorphometric analyses of neovasculature can be performed simultaneously with volumetric quantification of Col2.3GFP cells and SHG. A schematic for quantitative analyses of blood vessels is illustrated in Fig. 3. All analyses including segmentation, skeletonization and filament editing are performed using available modules in Amira.
Figure 3.
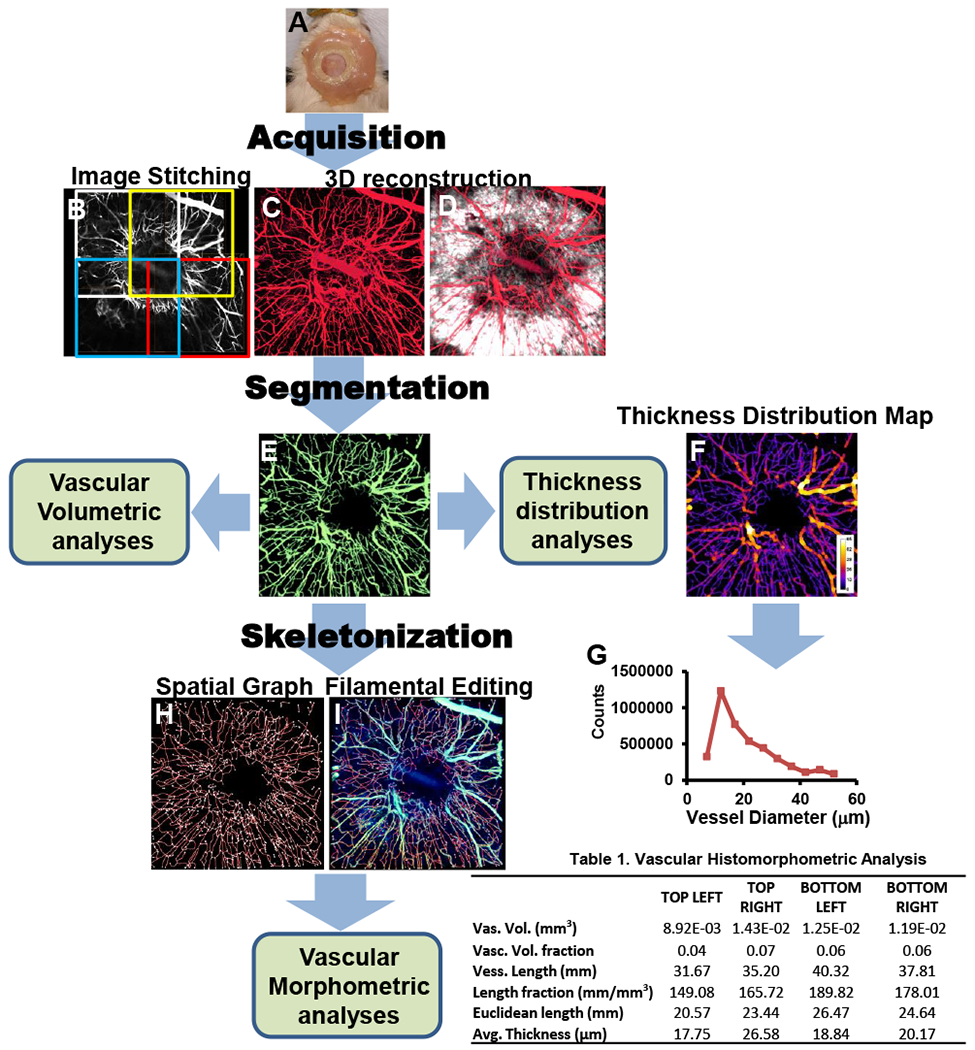
Simultaneous imaging of SHG (white) and vessels (red) allows 3D reconstruction of the healing defect and vasculature at 8 weeks post-surgery (A). The total vasculature within the window (B) was segmented using the SHG volume as region of interest (ROI) to isolate the vessels in the new bone (C). These vessels were skeletonized (D), and superimposed on the original image of the vessels (E). From binarized 3D images, a vessel thickness distribution map was obtained of the ROI (F), and used for morphometric analyses (G). Table lists quantification data from 4 quadrants of the defect.
A multichannel imaging stack that includes the entire defect as illustrated by SHG and the surrounding vascular network is reconstructed in 3D format using the Votex Function available in Amira.
To analyze the vascular network, a region of interest (ROI) is defined to include all the vessels associated with bone defect as determined by SHG microscopy.
The vascular network is isolated using Amira Segmentation Editor to obtain a segmented vascular image, excluding non-specific signals as well as noises generated during imaging.
The segmented vascular image stack is used to perform volumetric analyses available from the MaterialStatistics function in Amira: vascular volume (Vasc. Vol.), total volume (T. Vol.), and vascular volume fraction (Vol. Fract.) (i.e., the ratio of Vasc. Vol. over T. Vol.).
Using Amira’s AutoSkeleton module, which implements a distance-ordered homotopic thinning operation, the segmented 3D vascular network is further skeletonized to generate a line-based network that is topologically equivalent to the original network 11, 12. The skeleton can be superimposed on the original image to assess the relative accuracy of this method.
The final skeletonized vessel network can be obtained by manually re-tracing of the skeletons using Amira’s Filamental Editor to remove false or redundant segments.
Based on the skeletonized network, the number of vascular segments (NV), total vessel length (T. Length), and vessel length fraction (L.Fract.) (i.e., ratio of vessel length to total volume) can be read from SpatialGraphStatistics in Amira.
To analyze the average vessel thickness (Avg. Vess. Th.) and the associated vessel thickness distribution, the segmented vascular image stack is imported into the Image J. Using an ImageJ plugin developed by Robert Dougherty 13, vascular thickness and thickness distribution can be output in excel spread sheet. The complete process for quantification of vascularity is illustrated in Fig. 3.
3.3.3. Longtudinal live microCT scanning.
Mice are sedated with isoflurane and restrained in a custom-made chamber containing isoflurane. The mouse skull is scanned by a Scanco Viva CT40 system (Scanco Medical) at a voxel size of 17.5 μm. Imaging is performed on the same groups of mice repeated over three months. From the 2D images generated, the defect is reconstructed in 3D and analyzed using Amira software combined with Image J. The rate of the defect healing was evaluated by calculating the area of the defect closure over a period of 3 months. We recommend performing microCT and MPLSM on alternating days/ weeks.
4. Notes:
The mix of Ketamine and Xylazine will ensure deep anesthesia for about 1 hour. We suggest weighting animals before surgery in order to provide an adequate amount of anesthetic agents to the surgical animals.
Washing the wound with PBS containing antibiotics such as penicillin and streptomycin will reduce infection and improve the success of establishing a clear window for imaging.
Damaging suture or vasculature on the dura could lead to excessive bleeding, which could result in the failure of constructing a clear window for imaging. If bleeding occurs, press the area with sterilized Q-tip for several minutes until bleeding stops.
During the first week, some window chambers could become a bit cloudy due to the inflammatory infiltration from the wound. Most of the cloudy windows will become clear in week 2 or 3.
The improved window system provides a stable and clear window for long term imaging. Compared with published cranial window models, the current model allows repeated imaging over months with about 80% successful rate.
The maximum penetration of laser scanning in bone tissue is ~250 μm, which is close to the full thickness of calvaria bone. By adjusting the laser power, the imaging resolution and depth can be further improved.
Acknowledgements:
This study is supported by grants from the Musculoskeletal Transplant Foundation, NYSTEM N08G-495 and N09G346, and the National Institutes of Health (R21 DE021513, RC1 AR058435, AR067859, R01DE019902, R21DE026256, P50 AR054041, and P30AR061307).
References:
- 1.Duvall CL, Robert Taylor W, Weiss D, et al. Quantitative microcomputed tomography analysis of collateral vessel development after ischemic injury. Am J Physiol Heart Circ Physiol. 2004;287:H302–310. [DOI] [PubMed] [Google Scholar]
- 2.Duvall CL, Taylor WR, Weiss D, et al. Impaired angiogenesis, early callus formation, and late stage remodeling in fracture healing of osteopontin-deficient mice. J Bone Miner Res. 2007;22:286–297. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Xie C, Lin AS, et al. Periosteal progenitor cell fate in segmental cortical bone graft transplantations: implications for functional tissue engineering. J Bone Miner Res. 2005;20:2124–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lecoq J, Parpaleix A, Roussakis E, et al. Simultaneous two-photon imaging of oxygen and blood flow in deep cerebral vessels. Nat Med. 2011;17:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoover EE, Squier JA. Advances in multiphoton microscopy technology. Nature photonics. 2013;7:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown E, McKee T, diTomaso E, et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med. 2003;9:796–800. [DOI] [PubMed] [Google Scholar]
- 7.Osswald M, Winkler F. Insights into cell-to-cell and cell-to-blood-vessel communications in the brain: in vivo multiphoton microscopy. Cell Tissue Res. 2013;352:149–159. [DOI] [PubMed] [Google Scholar]
- 8.Sidani M, Wyckoff J, Xue C, et al. Probing the microenvironment of mammary tumors using multiphoton microscopy. J Mammary Gland Biol Neoplasia. 2006;11:151–163. [DOI] [PubMed] [Google Scholar]
- 9.Holtmaat A, Bonhoeffer T, Chow DK, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madden KS, Zettel ML, Majewska AK, et al. Brain tumor imaging: live imaging of glioma by two-photon microscopy. Cold Spring Harbor protocols. 2013;2013. [DOI] [PubMed] [Google Scholar]
- 11.Cassot F, Lauwers F, Fouard C, et al. A novel three-dimensional computer-assisted method for a quantitative study of microvascular networks of the human cerebral cortex. Microcirculation. 2006;13:1–18. [DOI] [PubMed] [Google Scholar]
- 12.Pudney C Distance-Ordered Homotopic Thinning: A Skeletonization Algorithm for 3D Digital Images. Computer Vision and Image Understanding. 1998;72:404–413. [Google Scholar]
- 13.Dougherty R, Kunzelmann KH Computing Local Thickness of 3D Structures with ImageJ. Microscopy & Microanalysis Meeting , Ft. Lauderdale, Florida 2007 [Google Scholar]


