Abstract
Background
Scaling SARS-CoV-2 testing to meet demands of safe reopenings continues to be plagued by assay costs and supply chain shortages. In response, we developed SalivaDirect, which received Emergency Use Authorization (EUA) from the U.S. Food and Drug Administration (FDA).
Methods
We simplified our saliva-based diagnostic test by (1) not requiring collection tubes with preservatives, (2) replacing nucleic acid extraction with a simple enzymatic and heating step, and (3) testing specimens with a dualplex qRT-PCR assay. Moreover, we validated SalivaDirect with reagents and instruments from multiple vendors to minimize supply chain issues.
Findings
From our hospital cohort, we show a high positive agreement (94%) between saliva tested with SalivaDirect and nasopharyngeal swabs tested with a commercial qRT-PCR kit. In partnership with the National Basketball Association (NBA) and National Basketball Players Association (NBPA), we tested 3,779 saliva specimens from healthy individuals and detected low rates of invalid (0.3%) and false-positive (<0.05%) results.
Conclusions
We demonstrate that saliva is a valid alternative to swabs for SARS-CoV-2 screening and that SalivaDirect can make large-scale testing more accessible and affordable. Uniquely, we can designate other laboratories to use our sensitive, flexible, and simplified platform under our EUA (https://publichealth.yale.edu/salivadirect/).
Funding
This study was funded by the NBA and NBPA (N.D.G.), the Huffman Family Donor Advised Fund (N.D.G.), a Fast Grant from Emergent Ventures at the Mercatus Center at George Mason University (N.D.G.), the Yale Institute for Global Health (N.D.G.), and the Beatrice Kleinberg Neuwirth Fund (A.I.K.). C.B.F.V. is supported by NWO Rubicon 019.181EN.004.
Keywords: COVID-19, SARS-CoV-2, molecular testing, saliva, population screening
Context and significance
Frequent testing is critical to limit SARS-CoV-2 transmission. In response to this need, we developed SalivaDirect, a sensitive, simplified, and flexible testing framework, which received Emergency Use Authorization (EUA) from the U.S. Food and Drug Administration (FDA). We tested saliva collected from a hospital cohort and showed a high positive agreement (94%) as compared to paired nasopharyngeal swabs tested with a commercial diagnostic kit. Then, we partnered with the National Basketball Association (NBA) to test a large cohort of mostly healthy individuals, and we detected low rates of invalid (0.3%) and false-positive (0.03%–0.05%) results. Our study shows that SalivaDirect can help to increase testing capacity by providing access to an affordable framework that is less prone to supply chain shortages.
Graphical abstract
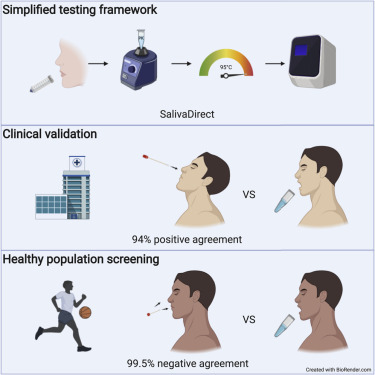
SalivaDirect is a sensitive saliva-based COVID-19 diagnostic test, which received Emergency Use Authorization from the U.S. FDA. With the ability to designate other laboratories, the flexible and simplified framework helps to increase the capacity of existing laboratory infrastructure for SARS-CoV-2 testing.
Introduction
SARS-CoV-2, a novel beta-coronavirus, emerged in late 2019 in Wuhan, China, and the subsequent coronavirus disease 2019 (COVID-19) pandemic rapidly followed.1 , 2 In many parts of the world, including the United States, COVID-19 cases continue to rise.3 , 4 The implementation of mass testing efforts followed by contact tracing will be necessary to quell the pandemic. Routine state-level screening and surveillance of healthy individuals is particularly important for safe reopening of the economy and schools and can minimize the risk of relapsing local outbreaks. However, the scalability and availability of currently authorized assays for SARS-CoV-2 diagnostic testing are still limited, and large-scale application is hampered by worldwide supply chain issues.5 To overcome these challenges, mass testing efforts must be (1) safe, both at the point of specimen collection and specimen processing; (2) affordable; (3) flexible, without the need for specific reagents or instrumentation from specific vendors; (4) adaptable to high-throughput workflows; and (5) amenable to quick turnaround times. While several different types of diagnostic assays have been recently authorized for emergency use by the U.S. Food and Drug Administration (FDA) such as qRT-PCR, LAMP, CRISPR, and sequencing-based assays, alternatives are still needed for large-scale testing efforts.6
Based on established diagnostic practices for other respiratory infections, the nasopharyngeal swab was initially adopted as the preferred sampling technique for SARS-CoV-2. However, we and others have shown that saliva can serve as an alternative upper respiratory tract specimen type for SARS-CoV-2 detection.7, 8, 9, 10, 11, 12, 13, 14 This is significant as saliva offers a number of advantages over nasopharyngeal swabs when considering the aforementioned criteria for mass testing efforts. Specifically, saliva does not require a certified swab and collection receptacle and does not necessarily have to be obtained by a skilled healthcare provider, both of which increase diagnostic-associated costs. Nasopharyngeal sampling requires a swab being inserted into the back of the nares, which can cause irritation that could promote sneezing and coughing. Thus, the noninvasive collection of saliva is safer, as it protects healthcare workers from being inadvertently exposed to potentially infectious droplets. In addition to being more affordable and safer, collection of nasopharyngeal swabs has been associated with variable, inconsistent, and false-negative test results due to the technical difficulties of taking a proper swab.10 , 15, 16, 17, 18, 19
To increase testing capacity for large-scale screening efforts, we developed SalivaDirect, a saliva-based, nucleic-acid-extraction-free, dualplex qRT-PCR method for SARS-CoV-2 detection. Our approach can be broadly implemented, as it does not require expensive saliva collection tubes containing preservatives20 and does not require specialized reagents or equipment for nucleic acid extraction. We validated SalivaDirect for use with products from multiple vendors. Thus, the simplicity and flexibility of SalivaDirect mean that it will not be as affected by supply chain bottlenecks as some other assays that rely on swabs and/or nucleic acid extraction. We show that SalivaDirect has a low limit of detection (6–12 copies/μL) and yields highly concordant results as compared to currently validated qRT-PCR assays. The unique features of SalivaDirect is that it is noninvasive, less expensive ($1.21–$4.39/sample in reagents), and validated for use with reagents and instruments from multiple vendors. Through our partnerships with the National Basketball Association (NBA) and the National Basketball Players Association (NBPA), we conducted a large usability study of SalivaDirect and comparison to standard qRT-PCR testing of paired anterior nares/oropharyngeal (AN/OP) swabs for asymptomatic/presymptomatic detection of SARS-CoV-2. Our results demonstrate how our specialized protocols for saliva collection produce mostly valid specimens for testing (99.7%), and the specificity of SalivaDirect leads to very few false-positive results (0.03%–0.05%), showcasing that SalivaDirect, and saliva testing in general, can be used to enhance large-scale testing of symptomatic and asymptomatic individuals. The data presented here were used to support our Emergency Use Authorization (EUA) for SalivaDirect granted by the FDA on August 15th, 2020.21
Results
Development of a simplified SARS-CoV-2 molecular diagnostic framework
To reduce the cost, time, and effort for SARS-CoV-2 detection, we developed SalivaDirect (https://publichealth.yale.edu/salivadirect/), a simplified and flexible saliva-based platform. SalivaDirect consists of three steps: (1) collecting saliva without preservative buffers, (2) proteinase K treatment and heat inactivation in place of nucleic acid extraction, and (3) dualplex qRT-PCR SARS-CoV-2 detection (Figure 1 A).
Figure 1.
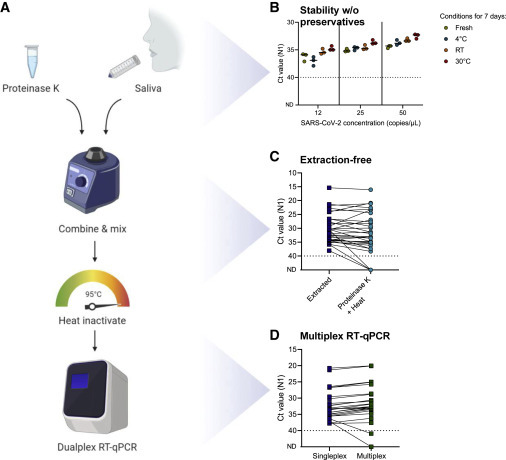
SalivaDirect is a simplified method for SARS-CoV-2 detection
(A) Schematic overview of SalivaDirect workflow depicting the main steps of mixing saliva with proteinase K, heat inactivation, and dualplex qRT-PCR testing. Figure created with Biorender.com.
(B) SARS-CoV-2 is stable in saliva for at least 7 days at 4°C, room temperature (RT; ∼19°C), and 30°C without addition of stabilizing buffers. Spiked-in saliva samples of low virus concentrations (12, 25, and 50 SARS-CoV-2 copies/μL) were kept at the indicated temperature for 7 days and then tested with SalivaDirect. N1 cycle threshold (Ct) values were lower when kept for 7 days at 30°C as compared to fresh specimens (Kruskal-Wallis; p = 0.03). Horizontal bars indicate the median.
(C) Comparing Ct values for saliva treated with proteinase K and heat as compared to nucleic extraction yields higher N1 Ct values without extraction (Wilcoxon; p < 0.01).
(D) Testing extracted nucleic acid from saliva with the N1 primer-probe set (singleplex) as compared to a multiplex assay showed stronger N1 detection in multiplex (Wilcoxon; p < 0.01). The dotted line in (B)–(D) indicates the limit of detection.
Data used to make this figure can be found in Data S1.
Stability of SARS-CoV-2 detection in saliva without preservatives
Several protocols imply that stabilizing buffers (e.g., Tris-borate-EDTA [TBE], Tris-EDTA [TE], or PBS) and additives (e.g., Triton X-100, Tween 20, or NP-40) are required to preserve the detection of SARS-CoV-2 RNA in saliva specimens, while other studies suggest that these buffers are not required and may even inhibit qRT-PCR.20 , 22 , 23 To determine the stability of SARS-CoV-2 RNA detection using SalivaDirect, we stored saliva specimens for 7 days at 4°C, room temperature, or 30°C without the addition of preservatives. We quantified the virus copies from a positive saliva specimen and spiked-in different concentrations of the positive sample to achieve concentrations of 12, 25, and 50 SARS-CoV-2 copies/μL into negative saliva collected from healthcare workers.10 After 7 days, we tested the spiked-in saliva specimens with SalivaDirect and compared results to “fresh” samples. We found that SARS-CoV-2 detection was stable in saliva for at least 7 days at each of the three thermal conditions (Figure 1B). Surprisingly, we even detected significantly lower N1 cycle threshold (Ct) values (e.g., better detection) when saliva was kept for 7 days at 30°C as compared to fresh specimens (median difference across concentrations of 1.4 Ct, p = 0.03; Figure 1B). In contrast, we found that Ct values for human RNase P (RP) were significantly higher after 7 days at room temperature (median difference of 3.8 Ct, p < 0.01) or 30°C (median difference of 5.0 Ct, p < 0.001) as compared to fresh specimens, which suggests that the human RNA degraded over time (Figure S1). We observed similar patterns when testing additional temperature profiles that the samples may encounter during transport, with no significant differences between N1 Ct values for fresh samples as compared to samples kept at 40°C for 72 h (p > 0.99) or samples kept under summer (alternating 28°C–40°C; p = 0.54) or winter profiles (alternating −20°C to room temperature; p > 0.99; Figure S2). Moreover, in a parallel clinical study, we showed that SARS-CoV-2 RNA from saliva collected from COVID-19 patients without preservatives is stable at 30°C for at least 3 days (n = 20) and at room temperature for up to 25 days (n = 20), though the samples were tested by a standard PCR test and not SalivaDirect.20 Thus, our data suggest that SARS-CoV-2 RNA, or at least the targeted nucleocapsid RNA, is stable in saliva without preservatives for at least 3 days, and likely longer, when stored at temperatures of up to 40°C.
Nucleic-acid-extraction-free PCR detection of SARS-CoV-2
Nucleic acid extraction is included in most authorized PCR diagnostic assays to detect SARS-CoV-2 RNA by qRT-PCR. However, nucleic acid extraction is relatively expensive, time-consuming, and subject to supply chain bottlenecks, which limit the scalability of testing that is critical for safe reopenings. Previous studies have shown that the nucleic acid extraction step can be omitted with a relatively small impact on analytical sensitivity.23, 24, 25, 26, 27, 28 Therefore, we explored the potential of proteinase K and heat as an affordable, fast, and easy alternative to nucleic acid extraction. We used the modified Centers for Disease Control and Prevention (CDC) assay29 to compare qRT-PCR detection of SARS-CoV-2 in saliva specimens processed with nucleic acid extraction or by simply mixing the specimen with proteinase K followed by heat inactivation (Figure 1C). As compared to nucleic acid extraction, our data show that our extraction-free approach minimally decreases detection (median N1 Ct increase = 1.8 Ct; p < 0.01). The reduction in detection that we observed is equivalent to what we would expect from omitting the ∼4-fold concentration step that occurs during nucleic acid extraction. Our findings demonstrate that proteinase K and heat can be used as an alternative to nucleic acid extraction with only a minor loss in sensitivity.
Dualplex PCR detection of SARS-CoV-2 and a human control gene
Our final modification to improve the scalability of SARS-CoV-2 diagnostic assays was to increase the high-throughput testing potential of the qRT-PCR step. We previously found that the U.S. CDC primer-probe sets are among the most sensitive and reliable for SARS-CoV-2 detection.29 The CDC assay consists of three separate reactions targeting two regions of the SARS-CoV-2 nucleocapsid (N1 and N2) and a human RP control.30 We previously modified the CDC assay by multiplexing the three primer-probe sets, thereby reducing the number of tests from three to one, without a significant impact on its sensitivity.31 When testing the multiplex qRT-PCR assay on saliva treated with proteinase K and heat, however, we were not able to detect consistent results for the N2 primer-probe set or the Sarbeco-E (E) or HKU-ORF1 (ORF1) primer-probe sets with HEX fluorophores (Table S1). We previously showed that SARS-CoV-2 detection by the N1 primer-probe set is more consistent and stronger as compared to N2.29 Therefore, to further simplify the qRT-PCR assay, we developed a dualplex qRT-PCR assay based on N1 and RP and modified the fluorophore (Cy5, ATTO647, or Quasar670 instead of FAM) on the RP probe. When comparing the modified singleplex CDC assay with the dualplex assay on extracted nucleic acid, median N1 Ct values were 0.9 Ct lower when tested in multiplex (p < 0.01; Figure 1D). Thus, SalivaDirect allows for a reduction in the number of qRT-PCR reactions to one reaction per sample.
Lower limit of detection using reagents and equipment from multiple vendors
Tests that depend on specific reagents from single vendors leave them vulnerable to supply chain shortages, as happened throughout the COVID-19 pandemic. Thus our approach was to validate SalivaDirect using reagents and instruments from multiple vendors to avoid dependence on a single vendor for each step (Table 3). In addition to what is shown here, we will continue to amend our SalivaDirect EUA through the use of bridging studies to ensure that it remains free of supply chain issues and to provide cheaper alternatives. A current list of validated products can be found in our updated EUA summary.21
Table 3.
SalivaDirect is a relatively inexpensive method for SARS-CoV-2 diagnostic testing
| Vendor | Item | Catalog Number | Price/Sample |
|---|---|---|---|
| Sample Collection (Pick one of the Listed Options) | |||
| Thomas Scientific | screw-cap tube, 5 mL, sterile | 1188R46 | $0.22 |
| VWR | 5 mL screw-cap centrifuge tubes, sterile | 10002-738 | $0.25 |
| Eppendorf | Eppendorf tubes 5.0 mL with screw cap, sterile | 0030122321 | $0.41 |
| Salimetrics | saliva collection aid | 5016.02 | $1.40 |
| Sample Processing (Pick one of the Listed Options) | |||
| AmericanBio | proteinase K | AB00925-00100 | $0.13 |
| ThermoFisher Scientific | MagMAX viral/pathogen proteinase K | A42363 | $0.16 |
| New England Biolabs | proteinase K, molecular biology grade | P8107S | $0.26 |
| qRT-PCR Primers and Probes (Pick one of the Listed Sets) | |||
| Eurofins | SalivaDirect primer probe set, 50–100 nmol | 12YS-010YST | $0.12 |
| Integrated DNA technologies | nCOV_N1 forward primer aliquot, 50 nmol | 10006821 | $0.02 |
| nCOV_N1 forward primer aliquot, 100 nmol | 10006830 | $0.02 | |
| nCOV_N1 reverse primer aliquot, 50 nmol | 10006822 | $0.02 | |
| nCOV_N1 reverse primer aliquot, 100 nmol | 10006831 | $0.02 | |
| nCOV_N1 probe aliquot, 25 nmol | 10006823 | $0.04 | |
| nCOV_N1 probe aliquot, 50 nmol | 10006832 | $0.03 | |
| RNase P forward primer aliquot, 50 nmol | 10006827 | $0.01 | |
| RNase P forward primer aliquot, 100 nmol | 10006836 | $0.01 | |
| RNase P reverse primer aliquot, 50 nmol | 10006828 | $0.01 | |
| RNase P reverse primer aliquot, 100 nmol | 10006837 | $0.01 | |
| RP probe (Cy5-IBRQ) | custom | $0.10 | |
| RP probe (ATTO657-IBRQ), 25 nmol | 10007061 | $0.09 | |
| RP probe (ATTO657-IBRQ), 50 nmol | 10007062 | $0.06 | |
| LGC Biosearch Technologies | nCOV_N1 forward primer, 100 nmol | nCoV-N1-F-100 | $0.01 |
| nCOV_N1 forward primer, 1000 nmol | nCoV-N1-F-1000 | $0.01 | |
| nCOV_N1 reverse primer, 100 nmol | nCoV-N1-R-100 | $0.01 | |
| nCOV_N1 reverse primer, 1000 nmol | nCoV-N1-R-1000 | $0.01 | |
| nCOV_N1 probe, 25 nmol | nCoV-N1-P-25 | $0.04 | |
| nCOV_N1 probe, 250 nmol | nCoV-N1-P-250 | $0.03 | |
| RNase P forward primer, 20 nmol | RNP-F-20 | <$0.01 | |
| RNase P forward primer, 100 nmol | RNP-F-100 | <$0.01 | |
| RNase P forward primer, 1000 nmol | RNP-F-1000 | <$0.01 | |
| RNase P reverse primer, 20 nmol | RNP-R-20 | <$0.01 | |
| RNase P reverse primer, 100 nmol | RNP-R-100 | <$0.01 | |
| RNase P reverse primer, 1000 nmol | RNP-R-1000 | <$0.01 | |
| RP probe, 25 nmol | RNP-PQ670-25 | $0.06 | |
| RP probe, 250 nmol | RNP-PQ670-250 | $0.03 | |
| RT-qPCR Kits (Pick One of the Listed Options) | |||
| New England Biolabs | Luna Universal Probe One-Step RT-qPCR Kit | E3006S | $0.75–$1.08 |
| E3006L | |||
| E3006X | |||
| E3006E | |||
| Bio-Rad | Reliance One-Step Multiplex RT-qPCR Supermix | 12010176 | $1.84–$2.11 |
| 12010220 | |||
| 12010221 | |||
| ThermoFisher Scientific |
TaqPath 1-Step RT-qPCR Master Mix, GC |
A15299 | $1.94–$2.06 |
| A15300 | |||
| Controls | |||
| Twist Bioscience | synthetic SARS-CoV-2 RNA control 2 | 102024 | <$0.01 |
The price per sample is calculated based on prices listed on the vendor websites and does not include additional costs for general laboratory consumables (e.g., pipette tips) or required equipment and instruments (e.g., pipette and qRT-PCR instruments). Total minimum reagent cost per sample is $1.21–$4.39
To validate reagents and instruments, we spiked a known concentration of SARS-CoV-2-positive saliva into negative saliva from healthcare workers to prepare a 2-fold dilution series of 400, 200, 100, 50, 25, 12, and 6 virus copies/μL. By testing each concentration in triplicate, we determined the preliminary limit of detection, which was then confirmed by testing another 20 replicates (Figure 2 ). Treating saliva with proteinase K from three different vendors resulted in a limit of detection of 6 SARS-CoV-2 copies/μL, suggesting that SalivaDirect is not dependent on proteinase K from a specific vendor (Figures 2A–2C).
Figure 2.
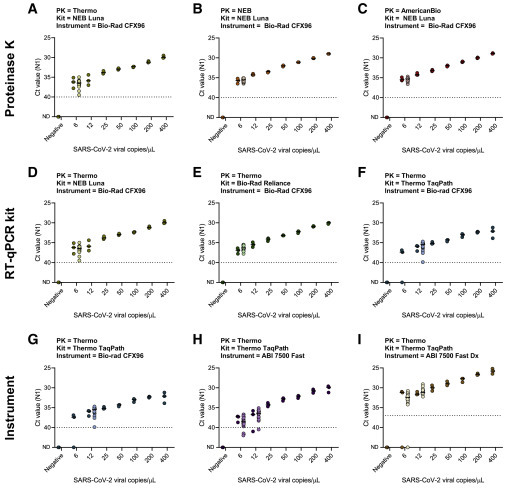
SalivaDirect is validated for use with reagents and instruments from multiple vendors
We determined the lower limit of detection of SalivaDirect with a 2-fold dilution series (400, 200, 100, 50, 25, 12, and 6 copies/μL) of positive saliva spiked-in negative saliva. Initially, each concentration and negative saliva were tested in triplicate to determine the preliminary limit of detection (dark-colored dots). The limit of detection was confirmed with 20 additional replicates (light-colored dots) for which 19 out of 20 needed to be detected. Limit of detection when tested with (A–C) proteinase K, (D–F) RT-qPCR kits, and (G–I) qRT-PCR instruments from different vendors, while keeping the other conditions constant. (A) and (D), as well as (F) and (G), are duplicates to enable comparisons between the different combinations of reagents or instruments within a single row. Shown are the Ct values for the N1 primer-probe set. The horizontal bars indicate the median and the dotted line indicates the limit of detection. Data used to make this figure can be found in Data S1.
Next, we determined the limit of detection by comparing three different qRT-PCR kits obtained from New England Biolabs, Bio-Rad, and ThermoFisher Scientific (Table 3). As each kit specifies the use of slightly different PCR cycle times and temperatures, we first sought to standardize these into a “universal” thermocycler program to make it easier to switch between products when needed. Comparing the results from each kit using the manufacturer’s protocol and the universal qRT-PCR program, we found no significant differences in Ct values (Luna: p = 0.69, Reliance: p = 0.06, TaqPath: p = 0.44; Figure S3). One additional qRT-PCR kit, Invitrogen EXPRESS One-Step SuperScript qRT-PCR kit, which we tested under their recommended protocol as well as our universal program, did not seem compatible with SalivaDirect and was therefore excluded from our limit-of-detection experiment. Using the universal thermocycler program with the Bio-Rad CFX96 instrument, New England Biolabs (NEB) Luna Universal Probe One-Step kit and Bio-Rad Reliance One-Step Multiplex RT-qPCR Supermix had a lower limit of detection of 6 SARS-CoV-2 copies/μL, whereas the ThermoFisher Scientific TaqPath 1-Step RT-qPCR Master Mix resulted in a slightly higher limit of detection of 12 SARS-CoV-2 copies/μL (Figures 2D–2F). Importantly, this indicates that the specific qRT-PCR kit can influence the lower limit of virus detection and that not all kits may be suitable for use with SalivaDirect.
Using the qRT-PCR kit with the highest limit of detection, TaqPath 1-Step RT-qPCR Master Mix, we compared the detection across three commonly used qRT-PCR thermocycler instruments: Bio-Rad CFX96, Applied Biosystems (ABI) 7500 Fast, and ABI 7500 Fast Dx. We found that the Bio-Rad CFX96 and ABI 7500 Fast had similar lower limits of detection at 12 SARS-CoV-2 copies/μL, whereas the ABI 7500 Fast Dx had a slightly lower limit of detection of 6 SARS-CoV-2 copies/μL (Figures 2G–2I). Interestingly, when determining the preliminary limit of detection for the ABI 7500 Fast Dx, we found that Ct values were on average 4.7 lower than Ct values generated on the ABI 7500 Fast. This suggests a difference in the auto-threshold that the machine sets, and therefore, we have increased the positive threshold to 37 Ct for the ABI 7500 Fast Dx to correspond to the positive threshold for the ThermoFisher Scientific TaqPath COVID-19 combo kit using the ABI 7500 Fast Dx. Changing the threshold did not affect the confirmed lower limit of detection of 6 copies/μL for the ABI 7500 Fast Dx. Overall, we found that SalivaDirect has a low limit of detection (6–12 SARS-CoV-2 copies/μL) using reagents and instruments from multiple vendors.
Sensitivity of SalivaDirect compared to saliva tested using a standard qRT-PCR assay
After determining the lower limit of detection of SalivaDirect, we compared the sensitivity of SARS-CoV-2 detection from saliva using a standard approach, a modified CDC assay with nucleic acid extraction and singleplex qRT-PCR.29 We found that the median N1 Ct values were 1.2 higher (e.g., weaker detection) for SalivaDirect as compared to the modified CDC assay (p < 0.001; Figure 3 ). Overall, the reduction in analytical sensitivity contributed to a 7.3% (3/41) false-negative rate for SalivaDirect, but only for weakly positive samples (all three false-negative specimens had N1 Ct values of 35–40 when using the modified CDC assay; Figure 3). Our findings show ∼93% positive agreement of SalivaDirect compared to a standard testing approach.
Figure 3.
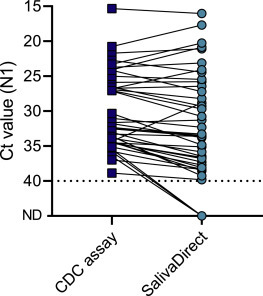
Sensitivity of SalivaDirect is comparable to a standard approach for SARS-CoV-2 detection in saliva
We compared Ct values for N1 between the modified CDC assay (nucleic acid extraction and singleplex qRT-PCR) and SalivaDirect for 41 saliva specimens tested with both methods. Overall, detection of SARS-CoV-2 with SalivaDirect is weaker (median 1.2 Ct, Wilcoxon; p < 0.001) than the modified CDC assay, but with a high agreement in outcomes of both tests of (93%). Shown are the Ct values for the N1 primer-probe set and the dotted line indicates the limit of detection. Data used to make this figure can be found in Data S1.
Clinical validation with paired nasopharyngeal swabs and saliva
For consideration of an EUA, the FDA requires clinical validation of any new laboratory developed test by comparing to currently authorized tests. For our validation study, we compared both across tests (SalivaDirect to the authorized ThermoFisher Scientific TaqPath COVID-19 combo kit)32 and across sample types (saliva to nasopharyngeal swabs) (Figure 4 ; Tables 2 and 3). We collected 37 paired positive and 30 paired negative nasopharyngeal swabs and saliva specimens from inpatients and healthcare workers at the Yale-New Haven Hospital. The ThermoFisher Scientific TaqPath COVID-19 combo kit combines nucleic acid extraction using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit with a multiplex qRT-PCR diagnostic assay targeting three regions of the SARS-CoV-2 genome on the ABI 7500 Fast Dx instrument. For SalivaDirect, we used the ThermoFisher Scientific proteinase K, ThermoFisher Scientific TaqPath RT-PCR kit, and Bio-Rad CFX96 instrument. We selected the positive and negative pairs based on preliminary results of our modified CDC assay.
Figure 4.
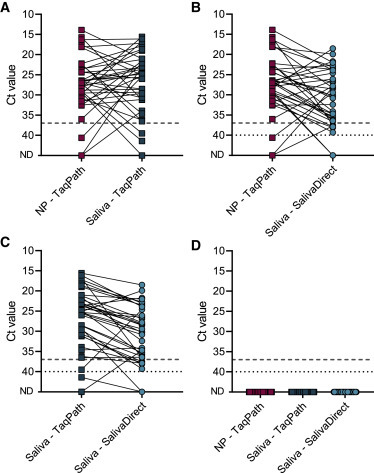
SalivaDirect is highly comparable to standard qRT-PCR tests with nucleic acid extraction from nasopharyngeal swabs and saliva
We selected 37 paired positive and 30 paired negative nasopharyngeal swabs and saliva specimens. Paired samples were collected a maximum 4 days apart. Nasopharyngeal swabs and saliva specimens were tested with the ThermoFisher Scientific TaqPath COVID-19 combo kit, and average Ct values for N, S, and ORF1ab were compared to N1 Ct values for saliva specimens tested with SalivaDirect.
(A) Comparison of 37 paired nasopharyngeal swabs and saliva tested with the TaqPath COVID-19 combo kit showed 84% positive agreement and no significant differences in each of the three virus targets (Wilcoxon; N: p = 0.51, S: p = 0.72, ORF1ab: p = 0.39).
(B) Comparison of nasopharyngeal swabs tested with the TaqPatch COVID-19 combo kit and saliva tested with SalivaDirect showed 94% positive agreement. Median N1 Ct values were 3.3 Ct higher for SalivaDirect (Wilcoxon; p < 0.01).
(C) Comparison of saliva tested with TaqPath COVID-19 combo kit and SalivaDirect again shows that SalivaDirect showed 97% positive agreement. Median N1 Ct values were 5.0 Ct higher for SalivaDirect (Wilcoxon; p < 0.001).
(D) 30 paired nasopharyngeal swabs and saliva specimens tested negative with both the TaqPath COVID-19 combo kit and SalivaDirect. Shown are average Ct values for N, S, and ORF1ab for the TaqPath combo kit and N1 Ct values for SalivaDirect. The dashed line indicates the limit of detection for the TaqPath combo kit (37 Ct) and the dotted line indicates the limit of detection for SalivaDirect (40 Ct).
Data used to make this figure can be found in Data S1.
Table 2.
Parallel testing of anterior nares/oropharyngeal swabs and saliva from NBA players, staff, and contractors
| Quest/BioReference |
|||
|---|---|---|---|
| AN/OP Swab |
|||
| Positive | Negative | ||
| SalivaDirect Saliva | positive | 17 | 2 |
| negative | 2 | 3,746 | |
| invalid | 0 | 12 | |
| Total | 19 | 3,760 | |
Invalid samples = 0.3% (12/3,779). Positive agreement = 89.5% (17/19). Negative agreement = 99.9% (3,746/3,748 valid samples). Overall agreement = 99.9% (3,763/3,767 valid samples).
First, when we compared nasopharyngeal swabs and saliva specimens when tested with the TaqPath COVID-19 combo kit, we found a positive agreement of 91.2% (Figure 4A). For both sample types, there were three specimens that tested negative, invalid, or inconclusive, while the other sample type tested positive. However, we did not find significant differences in Ct values for the three virus targets between both sample types (p = 0.39–0.72), with the median difference for each of the virus targets <2 Ct. This again confirms that some variation exists between sample types but that saliva is a valuable alternative.7, 8, 9, 10
Next, we found a 94% positive agreement with SalivaDirect compared to nasopharyngeal swabs tested with the TaqPath COVID-19 combo kit (Table 1 ). The N1 Ct values were higher using SalivaDirect (median difference of 3.3 Ct; p < 0.01; Figure 4B), and the increased Ct values are likely due to a combination of removing the nucleic acid step (Figures 1C and 3) and using different thermocycler instruments (Figure 2). Out of the 37 nasopharyngeal swabs that were tested with the TaqPath COVID-19 combo kit, three specimens tested negative (Table 1 and Figure 4B). However, earlier results with the modified CDC assay indicated a (weakly) positive signal, and the paired saliva specimen tested positive with both SalivaDirect and the TaqPath COVID-19 kit. While this is not captured in the percentage of positive agreement, SalivaDirect was able to detect SARS-CoV-2 in saliva of three individuals for which the nasopharyngeal swab tested negative.
Table 1.
Parallel testing of nasopharyngeal swabs and saliva from inpatients and healthcare workers with SalivaDirect and a commercial qRT-PCR kit
| TaqPath COVID-19 |
|||||||
|---|---|---|---|---|---|---|---|
| Nasopharyngeal Swab |
Saliva |
||||||
| Positive | Negative | Positive | Inconclusive | Invalid | Negative | ||
| SalivaDirect | positive | 32 | 3a | 33 | 0 | 2 | 0 |
| Saliva | negative | 2 | 30 | 1 | 1 | 0 | 30 |
| Total | 34 | 33 | 34 | 1 | 2 | 30 | |
NP-Saliva: positive agreement = 94.1% (32/34) and negative agreement = 90.9% (30/33). Saliva-Saliva: positive agreement = 97.1% (33/34) and negative agreement = 100% (30/30).
Three nasopharyngeal swabs tested negative, while previous outcomes of the modified CDC assay indicated that they were weakly positive.
When we directly compared the results of SARS-CoV-2 detection from saliva using SalivaDirect and the TaqPath COVID-19 combo kit, we found a high positive (97.1%) as well as negative agreement (100%; Table 1). Ct values for N1 were higher when comparing SalivaDirect with the TaqPath COVID-19 combo kit (median difference of 5.0 Ct, p < 0.001; Figure 4C), likely for the reasons described above. We intentionally included this comparison to enable a direct comparison of test results based on the same input specimen.
Finally, we compared results of negative paired nasopharyngeal swabs and saliva specimens tested with both the TaqPath COVID-19 combo kit and SalivaDirect (Figure 4D). No SARS-CoV-2 was detected in any of the specimens, while we did detect the internal controls. Thus, we did not detect any false-positive results with any of the assays.
Evaluation of off-target amplification
Background amplification or cross-reactivity of primer-probe sets with related human respiratory pathogens can cause false-positive results. Previous in vitro evaluations by the CDC showed no cross-reactivity with other human coronaviruses (229E, OC43, NL63, and HKU1), MERS-coronavirus, SARS-coronavirus, and 14 additional human respiratory viruses.30 These findings are in accordance with our previous investigation of nine primer-probe sets, including the N1 set, which did not detect any background amplification.29 To test for possible cross-reactivity of the dualplex qRT-PCR assay, we tested 52 saliva specimens collected from adults in the 2018/2019 and 2019/2020 fall and winter (pre-COVID-19; Figure S4). We did not detect off-target amplification or false positives, which is in agreement with previous findings from the CDC.30
Asymptomatic validation with paired AN/OP swabs and saliva
To conduct effective SARS-CoV-2 screening programs to allow populations to return to school and work, the test must be able to detect asymptomatic and/or presymptomatic cases and have a low false-positive rate. To evaluate SalivaDirect for these uses, we compared 3,779 saliva samples to paired combined AN/OP swabs from healthy NBA players, staff, and contractors (Table 2 ). The saliva samples were sent to the Yale School of Public Health for testing by SalivaDirect, and the AN/OP swabs were tested by commercial clinical diagnostic laboratories (Quest Diagnostics or BioReference Laboratories) using standard qRT-PCR assays.
There are concerns with using saliva as a testing specimen, as accidental collection of sputum and/or remnants from food or drinks can interfere with sample processing or inhibit PCR. For example, in a study with 124 symptomatic outpatients without optimized saliva collection protocols, approximately one-third of the samples were difficult to pipet.33 In our NBA cohort study, we developed explicit instructions on how to collect true saliva,34 , 35 used a saliva collection aid (a straw that fits into the collection tube) to promote providing true saliva and minimize potential aerosolization, and added proteinase K as the first laboratory step to help degrade any mucus. As a result, all 3,779 saliva samples collected from the cohort could be tested by SalivaDirect. Furthermore, the use of the RP human control gene in the dualplex PCR helps to determine if there are inhibitors in the specimens. We found that 12 of the 3,779 saliva samples had RP values above a Ct of 35, the threshold for an invalid sample (Table 2; Figure S5A). Thus 0.3% of the saliva samples collected from our cohort were invalid by PCR. Overall, we had a high rate of success for testing saliva by SalivaDirect.
During the study, 19 AN/OP swabs were positive for SARS-CoV-2, and 17 of those were also positive from saliva tested by SalivaDirect (89.5% positive agreement; Table 2). Out of the 19 AN/OP swabs that tested positive by commercial clinical diagnostic laboratories, we received 10 for comparative testing in our lab with the modified multiplex CDC assay (Figure S5B). When comparing the Ct values for the paired AN/OP swabs and saliva, we found no significant differences between both sample types (p = 0.91; Figure S5B). Upon retesting with the modified multiplex CDC assay, we found that two AN/OP swabs tested below our positive threshold, indicating that these samples may have been weakly positive or false positive from the commercial labs. Paired saliva of one of these two swab specimens also tested negative with SalivaDirect. Thus, the true sensitivity of SalivaDirect to AN/OP swabs by standard qRT-PCR for asymptomatic/presymptomatic detection may be higher than 90%; however, a larger positive sample size is needed to further evaluate.
Out of the 3,748 valid samples that tested negative by AN/OP swabs, 3,746 were also identified as negative by SalivaDirect, resulting in a negative agreement of 99.9%. For one of the samples that was negative by AN/OP swabs but positive by SalivaDirect, the subsequent saliva and AN/OP swabs from the same individual tested positive, suggesting that the previous SalivaDirect result was a true positive. For the other incongruent result (AN/OP negative, SalivaDirect positive), however, subsequent saliva and AN/OP swabs tested negative, suggesting that the previous SalivaDirect result was a false positive. Thus, our data indicate that the false-positive rate for SalivaDirect is between 1 and 2 per 3,748 (0.03%–0.05%) samples tested.
Supply costs for SalivaDirect testing
We aimed to develop a simplified testing method that is not dependent on commercialized kits, which may be subject to supply chain issues. Therefore, we reduced the number of steps and initially validated SalivaDirect with reagents and instruments from three different vendors. By doing so, we have reduced the cost per sample to a minimum of $1.21, if saliva is collected without a saliva collection aid, and a maximum of $4.39 when using a saliva collection aid (Table 3 ). These cost estimates are based on list prices; therefore, the actual costs may be lower. Additional reagents and instruments can be validated by performing a bridging study to show an equal limit of detection and can be submitted to the FDA as an amendment to the authorized EUA. Thus, the supply costs for SalivaDirect are relatively inexpensive, though these prices do not include labor or other laboratory expenses.
Discussion
SalivaDirect is a simplified and flexible platform
We developed SalivaDirect to adapt to the needs and budgets of heterogeneous SARS-CoV-2 surveillance systems. Testing saliva as an alternative to invasive swabs allows for safe and easy specimen collection. Furthermore, high-throughput testing can be maximized without the need for expensive saliva collection tubes with stabilizing reagents and nucleic acid extraction kits and a reduction in qRT-PCR reagents needed per specimen. We validated SalivaDirect with multiple reagents and instruments from different vendors to provide alternative options to minimize bottlenecks associated with supply chain issues. Furthermore, we demonstrated its effectiveness using multiple cohorts. (1) From our hospital cohort consisting of inpatients and healthcare workers with known SARS-CoV-2 infections, we found a positive agreement (94%) between SalivaDirect and NP swabs tested using a commercial qRT-PCR kit. (2) From our large NBA cohort consisting of mostly healthy individuals, our results showed that SalivaDirect had low rates of invalid (0.3%) and false-positive (0.03%–0.05%) samples when compared to AN/OP swabs tested by commercial clinical diagnostic laboratories. Together, our clinical studies showcase how our saliva collection and testing protocols are conducive for large-scale and repeated SARS-CoV-2 screening in mostly healthy individuals.
Uniquely, the U.S. FDA authorized us to designate other high-complexity Clinical Laboratory Improvement Amendments (CLIA) certified laboratories to use SalivaDirect, allowing our flexible and simplified framework to help increase the capacity of existing laboratory infrastructure for SARS-CoV-2 testing. As of December 7, 2020, 52 high-complexity CLIA certified laboratories from 25 U.S. states have been designated to use SalivaDirect, and another 265 laboratories have initiated the designation process. An outline of the designation process and a list of the designated labs can be found on our website (https://publichealth.yale.edu/salivadirect/).
To adapt to interested laboratories, avoid supply chain interruptions, or expand product availability, additional reagents and instruments can be added to our SalivaDirect FDA EUA. This can be done by performing bridging studies to establish equivalent performance between parallel testing of saliva specimens with new and previously validated components.36 The FDA recommends testing 2- to 3-fold serial dilutions of SARS-CoV-2-spiked saliva specimens in a pooled negative saliva matrix in triplicate until a hit rate of <100% is reached. Both tests can be considered to have equivalent performance if the resultant limit of detection is the same (e.g., ≤2–3 times the limit of detection) as the unmodified authorized test. Thus, our SalivaDirect EUA can continue to be modified to fill future needs.
Sensitivity and cost comparison to other authorized tests
In the rush to develop and authorize SARS-CoV-2 diagnostic tests,37 sparse and unstandardized data made it difficult to compare the performance. This led to several independent evaluations of SARS-CoV-2 diagnostic tests or test components,29 , 38, 39, 40, 41, 42, 43 though these were not always comprehensive enough to help clinicians or public health officials to determine which to use in the sea of growing options. To better establish true performance and directly compare among assays, the FDA developed reference material to establish an absolute limit of detection for each assay.37 We participated in this post-authorization validation using our least sensitive combination of reagents and equipment: ThermoFisher proteinase K, ThermoFisher TaqPath RT-qPCR Master Mix, and the Bio-Rad CFX96 thermocycler (detailed in our EUA summary21). Our measured limit of detection with the FDA reference material was 18 detectable units/μL (18,000 units/mL), similar to what we measured in this study (12 copies/μL). Compared to other FDA EUA authorized SARS-CoV-2 PCR tests, SalivaDirect has a limit of detection similar to or better than most manufactured PCR assays for swabs with RNA extraction (range = 540–540,000 units/mL), including the CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel (18,000 units/mL).44 Furthermore, SalivaDirect is one of only seven PCR tests that completed the reference panel for saliva, and based on the detection limit ranges (600–180,000 units/mL), SalivaDirect has intermediate overall performance and is approximately three times more sensitive than the Fluidigm Advanta Dx SARS-CoV-2 RT-PCR Assay (54,000 units/mL).44 Finally, as expected for a PCR assay, the limit of detection for SalivaDirect is substantially better than antigen based tests, including the Abbott ID NOW COVID-19 test (300,000 units/mL) and the Quidel Lyra Direct SARS-CoV-2 Assay (540,000 units/mL).44 Thus the analytical sensitivity of SARS-CoV-2 detection for SalivaDirect is similar to many other PCR tests with RNA extraction using swabs or saliva, and it is more sensitive than the rapid antigen tests.
One of our motivating factors behind developing SalivaDirect was to help reduce the costs of PCR testing. Most manufacturers and laboratories do not make their SARS-CoV-2 testing prices easily available to the public. However, those that do show that the cost to the individual being tested is typically between $100 and $250, sometimes with additional expenses for sample collection and/or shipping.45, 46, 47, 48, 49 While these prices may be justifiable for diagnostic testing covered by health insurance, they are not conducive for large-scale screening. By removing the need for swabs, expensive saliva collection devices,20 RNA extraction reagents, and kits manufactured specifically for SARS-CoV-2, we reduced the raw supply costs to $1.21–4.39 per sample. Even with that, some have argued that the labor of processing saliva samples ends up making SalivaDirect more expensive than swab-based tests.50 In reality, most of our more than 50 designated laboratories are offering SalivaDirect to the public for $25 or less, citing the inexpensive reagents and simplified workflow as the driving forces for the reduced costs. While these prices are similar to isothermal methods (e.g., LAMP assays),51 SalivaDirect will be more expensive than some of the less sensitive rapid antigen tests, such as the Abbott BinaxNOW being offered at a subsidized cost of $5 per test.52 Our hope is that other government subsidies and/or pooling approaches will further reduce the costs associated with testing saliva for SARS-CoV-2 by PCR to aid large-scale screening programs.53 , 54
Target populations and testing approaches
There is an increasing debate over the appropriate uses of high-sensitivity diagnostic tests (primarily PCR) and rapid lower sensitivity screening tests (primarily antigen).55 This revolves around the need to shift from clinical diagnostics, in which the demand has mostly been met, to population-level routine screening for safe reopenings.56 There is a reason why most schools, for example, do not have testing programs in place, despite the obvious need for them: most available local testing options are from low-capacity clinical labs offering primarily expensive swab-based tests, sometimes with long (>48 hour) turnaround times. Until rapid and inexpensive tests become widely available, there is a critical need to increase the capacities of existing labs and expand access to saliva-based tests.
To date, there are only a few SARS-CoV-2 laboratory diagnostic tests authorized by the FDA for asymptomatic screening.57, 58, 59, 60 While we validated SalivaDirect using our hospital cohort and our EUA is in the category of a high-sensitivity diagnostic test for suspected COVID-19 cases, the simplification, reduced costs, and flexibility of the platform was designed to facilitate routine screening of mostly healthy populations. Through our partnerships with the NBA and NBPA, we conducted the largest evaluation to date of saliva to swabs (AN/OP combined) for screening of healthy individuals. Importantly, the saliva collection for this study was observed by individuals without healthcare training, and still 99.7% of the samples were valid for PCR testing (i.e., samples could be pipetted and human control values were acceptable), showcasing how saliva could be used in non-healthcare settings. We will use this study to apply for an EUA for asymptomatic screening, and we are already working with organizations to use SalivaDirect in school settings.
Safety precautions for saliva collection and testing
Like with all clinical samples, we strongly recommend that anyone handling saliva for SARS-CoV-2 testing to follow “universal precautions”: treat all samples as infectious, wear proper personal protective equipment, and decontaminate any spills or exposed areas. Additional precautions should be taken during saliva collection where the individual may contaminate the sample collection tube through their breath or by directly spilling saliva on the outside surfaces. Because of this, we outline in our protocol (which was reviewed by the FDA) to disinfect the outside of the tube with 70% ethanol and for the clinical and laboratory staff to always wear gloves when handling the tubes. We do not believe that our method of collecting saliva without preservatives, as some other methods use,32 , 61, 62, 63 makes the samples any more hazardous to handle. First, anything added to the inside of the tubes would not help with saliva spilled on the outside. Second, preservatives were included in some tests to stabilize the genetic material, and to our knowledge, none of the commercial preservatives have been evaluated for inactivating SARS-CoV-2. Finally, SARS-CoV-2 RNA is still considered as a biosafety level 2 risk, and thus, even inactivated samples should be handled with caution. Thus, as similar to nasal swabs, collecting saliva contains risks to the collector and laboratory staff that can be minimized through following proper safety protocols.
Limitations of study
Our intended use of SalivaDirect is for the clear and liquid saliva that naturally pools in the mouth. The protocol as currently written is not intended for use with individuals who are unable to produce “true” saliva, whether this is due to illness or other reasons. While our previous analysis indicates that saliva is more sensitive for SARS-CoV-2 detection than nasopharyngeal swabs in COVID-19 inpatients,10 saliva can contain blood, mucus, or foreign substances (e.g., food, drink, or tobacco), which can interfere with PCR or make it difficult to pipet.33 We can overcome many of these issues by having specific collection procedures for saliva collection and using proteinase K to make samples easier to pipet, but some samples can still be invalid if they are not collected properly. While we show that invalid samples are rare from our NBA cohort, our study was biased toward adult men. Thus we were not able to test our approach in two key demographics: elderly individuals (>65 years of age) who are the most at risk for disease and school-aged children (<18 years of age) who critically need testing to help support in-person learning, though others have recently demonstrated that saliva is a suitable specimen for pediatric testing.64 Moreover, including professional athletes in our trial of repeat SARS-CoV-2 screening may not be representative of the general population, and we will therefore continue to monitor any laboratories using SalivaDirect for asymptomatic screening.
Conclusions
We designed SalivaDirect to be a simplified approach for sample collection and PCR testing for SARS-CoV-2. By using many different vendors, not seeking commercialization, and making the protocol completely open, our goal is to make SalivaDirect as accessible as possible. We encourage other groups to work with us or make their own adjustments to fit their specific needs. In particular, development and validation of automated liquid handling platforms could significantly enhance testing throughput, or replacing PCR with isothermal amplification techniques could make SalivaDirect more accessible for lower complexity laboratory settings. Furthermore, our ability to designate laboratories to use SalivaDirect under our EUA application provides a direct pathway for organizations looking to use noninvasive sampling coupled with a simplified molecular testing scheme without having to submit their own application to the FDA. Thus, SalivaDirect is not only a unique assay but also a unique way to approach testing during a pandemic.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Clinical samples | Yale New Haven Health | N/A |
| Clinical samples | National Basketball Association | N/A |
| Critical Commercial Assays | ||
| Proteinase K | American Bio | AB00925-00100 |
| MagMAX Viral/Pathogen Proteinase K | ThermoFisher Scientific | A42363 |
| Proteinase K, Molecular Biology Grade | New England Biolabs | P8107S |
| Luna Universal Probe One-Step RT-qPCR Kit | New England Biolabs | E3006S; E2006L; E3006X; E3006E |
| Reliance One-Step Multiplex RT-qPCR Supermix | Bio-Rad | 12010176; 12010220; 12010221 |
| TaqPath 1-Step RT-qPCR Master Mix, GC | ThermoFisher Scientific | A15299; A15300 |
| Synthetic SARS-CoV-2 RNA Control 2 | Twist Bioscience | 102024 |
| Deposited Data | ||
| Raw and analyzed data | This paper; and GitHub | Table S1; Data S1; https://github.com/grubaughlab/paper_2021_salivadirect |
| Oligonucleotides | ||
| Forward primer SARS-CoV-2 Nucleocapsid 1: N1-F: GACCCCAAAATCAGCGAAAT | Eurofins; IDT; LGC Biosearch Technologies | See Table 3 |
| Reverse primer SARS-CoV-2 Nucleocapsid 1: N1-R: TCTGGTTACTGCCAGTTGAATCTG | Eurofins; IDT; LGC Biosearch Technologies | See Table 3 |
| Probe SARS-CoV-2 Nucleocapsid 1: N1-P: FAM-ACCCCGCATTACGTTTGGTGGACC-IBFQ | Eurofins; IDT; LGC Biosearch Technologies | See Table 3 |
| Forward primer human RNase P: RP-F: AGATTTGGACCTGCGAGCG | Eurofins; IDT; LGC Biosearch Technologies | See Table 3 |
| Reverse primer human RNase P: RP-R: GAGCGGCTGTCTCCACAAGT | Eurofins; IDT; LGC Biosearch Technologies | See Table 3 |
| Probe human RNase P: Cy5-TTCTGACCTGAAGGCTCTGCGCG-IBRQ | Eurofins; IDT; LGC Biosearch Technologies | See Table 3 |
| Forward primer SARS-CoV-2 Nucleocapsid 2: N2-F: TTACAAACATTGGCCGCAAA | IDT | https://www.idtdna.com/pages |
| Reverse primer SARS-CoV-2 Nucleocapsid 2: N2-R: GCGCGACATTCCGAAGAA | IDT | https://www.idtdna.com/pages |
| Probe SARS-CoV-2 Nucleocapsid 2: N2-P: HEX-ACAATTTGCCCCCAGCGCTTCAG-IBFQ | IDT | https://www.idtdna.com/pages |
| Forward primer SARS-CoV-2 Envelope: E-F: ACAGGTACGTTAATAGTTAATAGCGT | IDT | https://www.idtdna.com/pages |
| Reverse primer SARS-CoV-2 Envelope: E-R: ATATTGCAGCAGTACGCACACA | IDT | https://www.idtdna.com/pages |
| Probe SARS-CoV-2 Envelope: E-P: HEX-ACACTAGCCATCCTTACTGCGCTTCG-IBFQ | IDT | https://www.idtdna.com/pages |
| Forward primer SARS-CoV-2 ORF1: ORF1-F: TGGGGYTTTACRGGTAACCT | IDT | https://www.idtdna.com/pages |
| Reverse primer SARS-CoV-2 ORF1: ORF1-R: AACRCGCTTAACAAAGCACTC | IDT | https://www.idtdna.com/pages |
| Probe SARS-CoV-2 ORF1: ORF1-P: HEX-TAGTTGTGATGCWATCATGACTAG-IBFQ | IDT | https://www.idtdna.com/pages |
| Software and Algorithms | ||
| GraphPad Prism 8.3.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Bio-Rad CFX Maestro 1.1 V4.1.2435.1219 | Bio-Rad | https://www.bio-rad.com/en-us/category/qpcr-analysis-software?ID=42a6560b-3ad7-43e9-bb8d-6027371de67a |
| ABI 7500 Software v2.3 | ThermoFisher Scientific | https://www.thermofisher.com/us/en/home/technical-resources/software-downloads/applied-biosystems-7500-real-time-pcr-system.html |
| ABI 7500 Fast System SDS software v1.4.1 | ThermoFisher Scientific | https://www.thermofisher.com/us/en/home/technical-resources/software-downloads/applied-biosystems-7500-real-time-pcr-system.html |
| Other | ||
| SalivaDirect: RNA extraction-free SARS-CoV-2 diagnostics V.5 | Protocols.io | https://www.protocols.io/view/salivadirect-rna-extraction-free-sars-cov-2-diagno-bkjgkujw |
| CFX96 Touch Real-Time PCR Detection System | Bio-Rad | https://www.bio-rad.com/en-us/product/cfx96-touch-deep-well-real-time-pcr-detection-system?ID=LZJTUJ15 |
| Applied Biosystems 7500 Fast Real-Time PCR System | ThermoFisher Scientific | https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/7500-fast-real-time-pcr-system.html |
| Applied Biosystems 7500 Fast Dx Real-Time PCR System | ThermoFisher Scientific | https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/7500-fast-real-time-pcr-system.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Nathan Grubaugh (nathan.grubaugh@yale.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Additional Supplementary Items are available from GitHub at https://github.com/grubaughlab/paper_2021_salivadirect.
Experimental model and subject details
Ethics
The collection of clinical samples from COVID-19 inpatients and healthcare workers at the Yale-New Haven Hospital was approved by the Institutional Review Board of the Yale Human Research Protection Program (Protocol ID. 2000027690). Informed consent was obtained from all patients and healthcare workers prior to sample collection. We used deidentified saliva specimens collected pre-COVID-19 to test for possible cross-reactivity of SalivaDirect. The collection of these saliva specimens was approved by the Institutional Review Board of the Yale Human Research Protection Program (Protocol ID. 0409027018). The collection of deidentified specimens from healthy or asymptomatic individuals from the NBA was approved by the Institutional Review Board of the Yale Human Research Protection Program (Protocol ID. 2000028394). Study participants were informed in writing about the purpose and procedure of the study, and consented to study participation through the act of providing the saliva sample; the requirement for written informed consent was waived by the Institutional Review Board.
Sample size and replication
We followed guidelines from the U.S. FDA to determine sample size and replication for each of the components of the performance evaluation. To determine the lower limit of detection, we initially tested each concentration in triplicate, and confirmed the lowest concentration for which all 3 replicates were detected with an additional 20 replicates for confirmation. For the clinical evaluation, the FDA recommends to test a minimum of 30 paired nasopharyngeal and saliva specimens to compare SalivaDirect to a test that previously received Emergency Use Authorization. For the asymptomatic validation we tested all samples we received from the NBA, and we will continue testing to meet the requirements of the FDA for a minimum of 20 positive specimens.
Method details
Clinical specimens
Clinical samples were collected from COVID-19 diagnosed patients and healthcare workers at the Yale-New Haven Hospital as described earlier.10 , 29 Briefly, nasopharyngeal swabs were collected in viral transport medium, and saliva was collected in containers without the addition of stabilizing reagents. All specimens were aliquoted upon arrival in the laboratory, with nucleic acid extracted from one aliquot,35 tested using a modified CDC RT-qPCR assay,29 and the remainder stored at −80°C. We modified the CDC assay by using the 2019-nCoV_N1 (N1), 2019-nCoV-N2 (N2), and human RNase P (RP) primer-probe sets (500 nM of forward and reverse primer and 250 nM of probe per reaction; Integrated DNA Technologies, Coralville, IA, US) with the Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs, Ipswich, MA, US). Thermocycler conditions were reverse transcription for 10 minutes at 55°C, initial denaturation for 1 min at 95°C, followed by 45 cycles of 10 s at 95°C and 30 s at 55°C on the CFX96 qPCR machine (Bio-Rad, Hercules, CA, US).
SalivaDirect protocol
A detailed step-by-step SalivaDirect protocol has been published.34 SalivaDirect has been validated with proteinase K (American Bio, ThermoFisher Scientific, New England Biolabs) and RT-qPCR kits (New England Biolabs, Bio-Rad, and ThermoFisher Scientific) from three vendors, as well as three RT-qPCR instruments (Table 3; Key Resources Table). At least 500 μL of saliva that naturally pools in the mouth was collected in tubes without preservatives. The sample provider was observed by a trained healthcare professional, and was instructed not to eat, drink, smoke, or conduct dental hygiene for 30 minutes prior to the saliva collection. Saliva collection tubes were decontaminated with 70% ethanol or a disinfecting wipe. Saliva specimens were then transferred to the laboratory for testing. Processing of saliva specimens which could potentially be positive for SARS-CoV-2 should be conducted in BSL2+ settings. A total of 2.5 μL (50 mg/mL) or 6.5 μL (20 mg/mL) of Proteinase K was added to 50 μL of saliva in 8-strip tubes. The tubes were placed in a rack and vortexed for 1 minute at 3200 RPM. Samples were heated for 5 minutes at 95°C on a thermocycler, and then 5 μL of processed saliva was used as input for the dualplex RT-qPCR assay. The dualplex RT-qPCR assay includes the 2019-nCoV_N1 (N1) primer-probe set that targets the nucleocapsid (N1-F: GACCCCAAAATCAGCGAAAT, N1-R: TCTGGTTACTGCCAGTTGAATCTG, N1-P: FAM-ACCCCGCATTACGTTTGGTGGACC-IBFQ) and the human RNase P control (RP) primer-probe set (RP-F: AGATTTGGACCTGCGAGCG, RP-R: GAGCGGCTGTCTCCACAAGT, RP-P: Cy5-TTCTGACCTGAAGGCTCTGCGCG-IBRQ) developed by the CDC (Integrated DNA Technologies, Coralville, IA, US), which are highly specific for SARS-CoV-2 detection (Figure S4). The fluorophore on the human RNase P probe was modified to combine both primer-probe sets in a dualplex assay, reducing the number of tests to a single assay. Additional fluorophores on the human RNase P probe have been validated and include ATTO647 and Quasar670. Each of these fluorophores are detected by the Cy5 channel and have equal performance (Table 3). We will continue to monitor the performance of our primer-probe sets and make adjustments if mismatches are identified with newly emerging SARS-CoV genotypes.
In the initial development, we included N2 (Fwd: TTACAAACATTGGCCGCAAA, Rev: GCGCGACATTCCGAAGAA, Probe: HEX-ACAATTTGCCCCCAGCGCTTCAG-IBFQ),30 E (Fwd: ACAGGTACGTTAATAGTTAATAGCGT, Rev: ATATTGCAGCAGTACGCACACA, Probe: HEX-ACACTAGCCATCCTTACTGCGCTTCG-IBFQ),65 or ORF1 (Fwd: TGGGGYTTTACRGGTAACCT, Rev: AACRCGCTTAACAAAGCACTC, Probe: HEX-TAGTTGTGATGCWATCATGACTAG-IBFQ)66 as a second virus target with HEX-fluorophore. However, this second virus target was removed from the final assay, because unlike the promising results with extracted nucleic acid,31 we were not able to consistently detect SARS-CoV-2 in saliva treated with proteinase K and heat (Table S1). Thus, the final SalivaDirect dualplex RT-qPCR assay consisted of the N1 and RP primer-probe sets.
The RT-qPCR master mix was prepared following the vendor’s recommended instructions, with 400 nM of N1 forward and reverse primer, 200 nM of N1 probe, 150 nM of RP forward and reverse primer, and 200 nM of RP probe per reaction. Thermocycler conditions were unified for all three RT-qPCR kits (universal protocol) with 10 minutes at 52°C, 2 minutes at 95°C, and 45 cycles of 10 s at 95°C and 30 s at 55°C. Specimens were considered positive if N1 Ct < 40 (or < 37 on the ABI 7500 Fast Dx) and any value for RP, negative if N1 Ct ≥ 40 (or ≥ 37 on the ABI 7500 Fast Dx) and RP < 35, and invalid if N1 Ct ≥ 40 and RP ≥ 35. Thus, RP is considered as a sample quality control and invalid samples should be retested on a new aliquot of saliva re-run through the entire SalivaDirect protocol.
Limit of detection
We spiked a positive saliva specimen from a confirmed COVID-19 patient with a known virus concentration (3.7 × 104 copies/μL) into saliva collected from 25 healthcare workers who tested negative for SARS-CoV-2 using the modified CDC assay.29 We tested a 2-fold dilution series of 400, 200, 100, 50, 25, 12, and 6 SARS-CoV-2 copies/μL in triplicate to determine the preliminary limit of detections, and confirmed the final limit of detection with 20 additional replicates. We used this approach to determine the lower limit of detection of different proteinases K, RT-qPCR kits, and RT-qPCR instruments from multiple vendors (Key Resources Table), by using the same input volumes, matrices and RT-qPCR programs for each combination of reagents and instruments. We found no differences in the limit of detection between proteinase K from three vendors and therefore selected one (ThermoFisher Scientific MagMAX proteinase K) to validate the three RT-qPCR kits. The RT-qPCR kit (ThermoFisher TaqPath) with the weakest limit of detection was then used to validate additional RT-qPCR instruments.
Stability
We determined the stability of SARS-CoV-2 RNA detection in spiked-in saliva samples (positive saliva spiked into a pool of negative saliva from 25 individuals to achieve concentrations of 12, 25, and 50 copies/μL; as prepared for the limit of detection experiment) by placing them for 7 days at 4°C, room temperature (RT, ∼19°C), or 30°C. In addition, we tested stability at 40°C for 72 hours, and under summer and winter profiles - conditions which could be encountered during sample transport. The summer profile consisted of 40°C for 8 hours, room temperature for 4 hours, 40°C for 2 hours, 28°C for 36 hours, and 40°C for 6 hours. The winter profile consisted of −20°C for 8 hours, room temperature for 4 hours, −20°C for 2 hours, 4°C for 36 hours, and −20°C for 6 hours. Results were compared to results obtained in the limit of detection experiment (fresh). Saliva specimens were tested in triplicate and were treated with ThermoFisher Scientific proteinase K and tested with the ThermoFisher TaqPath RT-qPCR kit on the Bio-Rad CFX96.
Cross-reactivity
We tested 52 saliva specimens, collected from adults during the 2018/2019 and 2019/2020 (pre-COVID19) autumn/winter influenza seasons in New Haven, CT to test for possible cross-reactivity of SalivaDirect with other human respiratory pathogens. Saliva specimens were treated with ThermoFisher Scientific proteinase K and tested with the NEB Luna Universal Probe One-Step RT-qPCR kit on the Bio-Rad CFX96.
Clinical validation
Paired nasopharyngeal swabs and saliva specimens (collected maximum 4 days apart) were selected from the Yale IMPACT biorepository. In total 67 paired nasopharyngeal swabs and saliva specimens were tested with the US FDA EUA ThermoFisher Scientific TaqPath COVID-19 combo kit following the vendor’s protocol. Briefly, nucleic acid was extracted using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit on the KingFisher Flex Magnetic Particle Processor. In total 200 μL of specimen was used as input and eluted in 50 μL. For each reaction, 5 μL of extracted nucleic acid was used as input and tested with the ThermoFisher Scientific TaqPath RT-qPCR reaction on the ABI 7500 Fast Dx. Ct values were exported through the 7500 Fast System SDS software v1.4.1. For saliva specimens that were too thick to pipette, 100 μL sample was mixed with 100 μL PBS, and 10 μL was used in the RT-qPCR reaction. For the clinical validation of SalivaDirect, saliva samples were treated with ThermoFisher Scientific proteinase K and tested with the ThermoFisher Scientific TaqPath RT-qPCR kit on the Bio-Rad CFX96.
Asymptomatic testing
Paired AN/OP swabs and saliva specimens (collected maximum 4 days apart) were collected from NBA players, staff, and other vendors. Combined AN/OP swabs were collected by Quest and followed their EUA specimen collection guidelines.67 , 68 Saliva was collected, under observation of Drug Free Sport International collectors, who are not trained healthcare workers, using a Salimetrics Saliva Collection Aid (Salimetrics, State College, PA, US) into a 2 mL screw-top tube with O-ring cap (Millipore Sigma, Burlingston, MA, US). Saliva was shipped overnight in NanoCool cooling system boxes to our laboratory at the Yale School of Public Health for testing.
A total of 3,779 paired samples were tested by SalivaDirect for saliva specimens at Yale and with Quest’s protocol for swabs, in their labs during the in-market phase of the study while swab testing was completed by BioReference while the teams were in Orlando, FL. Additional swab testing was performed in our lab using the modified CDC multiplex assay protocol for positive swabs sent on for sequencing.31 In total 10 matched samples, reported positive by the NBA, were tested in-house.
Quantification and statistical analysis
We used the Bio-Rad CFX Maestro 1.1 V4.1.2435.1219, ABI 7500 Software v2.3, and ABI 7500 Fast System SDS Software v1.4.1 to analyze and export Ct values. GraphPad Prism 8.3.0 was used to make the figures and perform all statistical analyses. Kruskal-Wallis tests were used to test for statistical differences in SARS-CoV-2 RNA stability kept at different temperatures and multiple comparisons were corrected with Dunn’s test. The Wilcoxon matched pairs test was used to test for statistical differences between paired samples. If a virus target was not detected, the Ct value was set to 45 Ct. In all statistical tests, p ≤ 0.05 was considered significant.
Consortia
The members of the Yale IMPACT Research Team (in alphabetical order) are Kelly Anastasio, Michael H. Askenase, Maria Batsu, Sean Bickerton, Kristina Brower, Molly L. Bucklin, Staci Cahill, Yiyun Cao, Edward Courchaine, Giuseppe DeIuliis, Rebecca Earnest, Bertie Geng, Benjamin Goldman-Israelow, Ryan Handoko, William Khoury-Hanold, Daniel Kim, Lynda Knaggs, Maxine Kuang, Sarah Lapidus, Joseph Lim, Melissa Linehan, Alice Lu-Culligan, Anjelica Martin, Irene Matos, David McDonald, Maksym Minasyan, Maura Nakahata, Nida Naushad, Jessica Nouws, Abeer Obaid, Camila Odio, Ji Eun Oh, Saad Omer, Annsea Park, Hong-Jai Park, Xiaohua Peng, Mary Petrone, Sarah Prophet, Tyler Rice, Kadi-Ann Rose, Lorenzo Sewanan, Lokesh Sharma, Albert C. Shaw, Denise Shepard, Mikhail Smolgovsky, Nicole Sonnert, Yvette Strong, Codruta Todeasa, Jordan Valdez, Sofia Velazquez, Pavithra Vijayakumar, Elizabeth B. White, and Yexin Yang.
Acknowledgments
We are honored to have been supported by the NBA, NBPA, and the Yale community, who shared our vision to democratize SARS-CoV-2 testing. Among them, we gratefully acknowledge David Weiss, Anjali Salooja, Peter Meisel, and Wesley Harris from the NBA, who helped design and coordinate the study, and the study participants from the NBA and the Yale-New Haven Hospital for their time and commitment to the efforts. We also thank all members of the collection teams from Drug Free Sport International and the clinical teams at Yale-New Haven Hospital for their dedication and work, which made this study possible, as well as the FDA and many staff and faculty at Yale University for helping bring SalivaDirect to life. We also thank the NBA testing partners, Quest and BioReference, for their continued willingness to support our research. Finally, we are appreciative of the advice and support from the COVID-19 Sports and Society Working Group and for the moral support from our friends and family (particularly V. Parsons, S. Taylor, and P. Jack).
This study was funded by a clinical research agreement with the National Basketball Association, US and National Basketball Players Association, US (N.D.G.), the Huffman Family Donor Advised Fund (N.D.G.), Fast Grant funding support from the Emergent Ventures at the Mercatus Center, George Mason University (N.D.G.), the Yale Institute for Global Health (N.D.G.), and the Beatrice Kleinberg Neuwirth Fund (A.I.K.). C.B.F.V. is supported by NWO Rubicon 019.181EN.004.
Author contributions
C.B.F.V., D.E.B., A.L.W., and N.D.G. designed the laboratory experiments. C.B.F.V., A.E.W., C.A.H., D.E.B., J.W., C.C.K., J.R.F., and I.M.O. performed the experiments. J.S., E.K., P.L., A.V., M.T., A.J.M., M.C.M., A.C.-M., J.F., S.B., M.C., R.D., and A.N. provided clinical samples. C.S.D.C., A.I.K., A.I., J.M., S.F.F., and R.S. are members of the Yale IMPACT Research Team. J.S., C.C., H.M.K., J.M., S.F.F., R.S., A.L.W., and N.D.G. designed the NBA study. C.B.F.V., A.E.W., C.A.H., J.W., and J.R.F. analyzed the data. C.S.D.C., A.I.K., A.I., J.M., P.H., C.L., S.F.F., R.S., A.L.W., and N.D.G. supervised the project. C.B.F.V., A.E.W., C.A.H., D.E.B., A.L.W., and N.D.G. wrote and edited the manuscript. All authors read and approved the final manuscript.
Declaration of interests
A.L.W. has received research funding through grants from Pfizer to Yale and has received consulting fees for participation in advisory boards for Pfizer. N.D.G. and A.L.W. have received research funding from Tempus to Yale to develop future versions of SalivaDirect. The remaining authors declare no competing interests.
Published: December 26, 2020
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2020.12.010.
Contributor Information
Yale IMPACT Research Team:
Kelly Anastasio, Michael H. Askenase, Maria Batsu, Sean Bickerton, Kristina Brower, Molly L. Bucklin, Staci Cahill, Yiyun Cao, Edward Courchaine, Giuseppe DeIuliis, Rebecca Earnest, Bertie Geng, Benjamin Goldman-Israelow, Ryan Handoko, William Khoury-Hanold, Daniel Kim, Lynda Knaggs, Maxine Kuang, Sarah Lapidus, Joseph Lim, Melissa Linehan, Alice Lu-Culligan, Anjelica Martin, Irene Matos, David McDonald, Maksym Minasyan, Maura Nakahata, Nida Naushad, Jessica Nouws, Abeer Obaid, Camila Odio, Ji Eun Oh, Saad Omer, Annsea Park, Hong-Jai Park, Xiaohua Peng, Mary Petrone, Sarah Prophet, Tyler Rice, Kadi-Ann Rose, Lorenzo Sewanan, Lokesh Sharma, Albert C. Shaw, Denise Shepard, Mikhail Smolgovsky, Nicole Sonnert, Yvette Strong, Codruta Todeasa, Jordan Valdez, Sofia Velazquez, Pavithra Vijayakumar, Elizabeth B. White, and Yexin Yang
Supplemental information
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Coronavirus Disease (COVID-19) Situation Reports. World Health Organization. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 5.Patel R., Babady E., Theel E.S., Storch G.A., Pinsky B.A., St George K., Smith T.C., Bertuzzi S. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: value of diagnostic testing for SARS-CoV-2/COVID-19. MBio. 2020;11 doi: 10.1128/mBio.00722-20. e00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.U.S. FDA In vitro diagnostics EUAs. 2020. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas
- 7.Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T., Sato K., Oguri S., Taki K., Senjo H., et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 2020;81:e145–e147. doi: 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.To K.K.-W., Tsang O.T.-Y., Yip C.C., Chan K.-H., Wu T.-C., Chan J.M.C., Leung W.-S., Chik T.S.-H., Choi C.Y.-C., Kandamby D.H., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., Fasano M., Sessa F., Tettamanti L., Carinci F., et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan J., Yu F., Wang X., Zou Q., Lou B., Xie G., Yang X., Chen W., Wang Q., Zhang D., et al. Hock-a-loogie saliva as a diagnostic specimen for SARS-CoV-2 by a PCR-based assay: a diagnostic validity study. Clin. Chim. Acta. 2020;511:177–180. doi: 10.1016/j.cca.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z., Shen Z., guo f., zhang q., jin y., et al. Detection of 2019-ncov in saliva and characterization of oral symptoms in COVID-19 patients. Cell Prolif. 2020;53:e12923. doi: 10.1111/cpr.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00776-20. e00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 16.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winichakoon P., Chaiwarith R., Liwsrisakun C., Salee P., Goonna A., Limsukon A., Kaewpoowat Q. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00297-20. e00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinloch N.N., Ritchie G., Brumme C.J., Dong W., Dong W., Lawson T., Jones R.B., Montaner J.S.G., Leung V., Romney M.G., et al. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J. Infect. Dis. 2020;222:899–902. doi: 10.1093/infdis/jiaa370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott I.M., Strine M.S., Watkins A.E., Boot M., Kalinich C.C., Harden C.A., Vogels C.B.F., Casanovas-Massana A., Moore A.J., Muenker M.C., et al. Simply saliva: stability of SARS-CoV-2 detection negates the need for expensive collection devices. medRxiv. 2020 doi: 10.1101/2020.06.16.20133041. [DOI] [Google Scholar]
- 21.U.S. FDA EUA summary SalivaDirect. 2020. https://www.fda.gov/media/141192/download
- 22.Griesemer S.B., Van Slyke G., Ehrbar D., Strle K., Yildirim T., Centurioni D.A., Walsh A.C., Chang A.K., Waxman M.J., St. George K. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. medRxiv. 2020 doi: 10.1101/2020.06.16.20133041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranoa D.R.E., Holland R.L., Alnaji F.G., Green K.J., Wang L., Brooke C.B., Burke M.D., Fan T.M., Hergenrother P.J. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. bioRxiv. 2020 210.1101/020.06.18.159434. [Google Scholar]
- 24.Smyrlaki I., Ekman M., Lentini A., Vondracek M., Papanicoloau N., Aarum J., Safari H., Muradrasoli S., Albert J., Högberg B., et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-qPCR. Nat Commun. 2020;11:4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marzinotto S., Mio C., Cifu’ A., Verardo R., Pipan C., Schneider C., Curcio F. A streamlined approach to rapidly detect SARS-CoV-2 infection, avoiding RNA extraction. medRxiv. 2020 doi: 10.1101/2020.04.06.20054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallmann L., Schallenberger K., Demolliner M., Eisen A.K.A., Hermann B.S., Heldt F.H., Hansen A.W., Spilki F.R., Fleck J.D. Pre-treatment of the clinical sample with proteinase K allows detection of SARS-CoV-2 in the absence of RNA extraction. bioRxiv. 2020 doi: 10.1101/2020.05.07.083139. [DOI] [Google Scholar]
- 27.Arizti-Sanz J., Freije C.A., Stanton A.C., Boehm C.K., Petros B.A., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.-S.F., Kemball M.E., et al. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.05.28.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalli M.A., Langmade S.J., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C., Wilkinson M.N., Fulton R.S., Heinz M., Buchser W.J., et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 2020:hvaa267. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020;5:1299. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:26. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudo E., Israelow B., Vogels C.B.F., Lu P., Wyllie A.L., Tokuyama M., Venkataraman A., Brackney D.E., Ott I.M., Petrone M.E., et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. bioRxiv. 2020 doi: 10.1371/journal.pbio.3000867. 2020.06.16.155887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Applied Biosystems TaqPath COVID-19 Combo Kit instructions for use. 2020. https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0019181_TaqPath_COVID-19_IFU_EUA.pdf
- 33.Landry M.L., Criscuolo J., Peaper D.R. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J. Clin. Virol. 2020;130:104567. doi: 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogels, C., Doug, E., Kalinich, C., Ott, I., Grubaugh, N., and Wyllie, A. (2020). SalivaDirect: RNA extraction-free SARS-CoV-2 diagnostics v5. https://doi.org/10.17504/protocols.io.bkjgkujw.
- 35.Ott I., Vogels C., Grubaugh N., Wyllie A. Saliva collection and RNA extraction for SARS-CoV-2 detection v2. J. Dent. Res. 2020 doi: 10.17504/protocols.io.bh6mj9c6. Published online November 2, 2020. [DOI] [Google Scholar]
- 36.U.S. FDA (2020). Policy for coronavirus disease-2019 tests during the public health emergency (revised). https://www.fda.gov/media/135659/download.
- 37.Shuren J., Stenzel T. Covid-19 molecular diagnostic testing: lessons learned. N. Engl. J. Med. 2020;383:e97. doi: 10.1056/NEJMp2023830. [DOI] [PubMed] [Google Scholar]
- 38.Fung B., Gopez A., Servellita V., Arevalo S., Ho C., Deucher A., Thornborrow E., Chiu C., Miller S. Direct comparison of SARS-CoV-2 analytical limits of detection across seven molecular assays. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01535-20. e01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00783-20. e00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craney A.R., Velu P.D., Satlin M.J., Fauntleroy K.A., Callan K., Robertson A., La Spina M., Lei B., Chen A., Alston T., et al. Comparison of two high-throughput reverse transcription-PCR systems for the detection of severe acute respiratory syndrome coronavirus 2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00890-20. e00890-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman J.A., Pepper G., Naccache S.N., Huang M.-L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00821-20. e00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00743-20. e00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrington A., Cox B., Snowdon J., Bakst J., Ley E., Grajales P., Maggiore J., Kahn S. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00798-20. e00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.U.S. FDA SARS-CoV-2 reference panel comparative data. 2020. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data
- 45.Avellino 2020. https://www.avellino.com/en/products/avellinocov2/
- 46.Boston Heart 2020. https://bostonheartdiagnostics.com/
- 47.Curative 2020. https://static.curative.com/legal/billing.html
- 48.Dxterity 2020. https://dxterity.com/covid-19/
- 49.Healthquest Esoterics 2020. https://www.hqesoterics.com/covid-19
- 50.ASM What is the COVID-19 SalivaDirect test? 2020. https://asm.org/Articles/2020/August/What-is-the-COVID-19-SalivaDirect-Test
- 51.RAPS COVID rapid diagnostic options expand with at-home LAMP test. 2020. https://www.raps.org/news-and-articles/news-articles/2020/11/covid-rapid-diagnostic-options-expand-with-at-home
- 52.HHS Trump administration deploys Abbott BinaxNOW tests to states. 2020. https://www.hhs.gov/about/news/2020/09/28/trump-administration-deploys-abbott-binaxnow-tests-to-states.html
- 53.Watkins A.E., Fenichel E.P., Weinberger D.M., Vogels C.B.F., Brackney D.E., Casanovas-Massana A., Campbell M., Fournier J., Bermejo S., Datta R., et al. Pooling saliva to increase SARS-CoV-2 testing capacity. medRxiv. 2020 doi: 10.1101/2020.09.02.20183830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mirimus SalivaClear COVID-19 pool testing from Mirimus. 2020. https://www.salivaclear.com/
- 55.Mina M.J., Parker R., Larremore D.B. Rethinking COVID-19 test sensitivity: a strategy for containment. N. Engl. J. Med. 2020;383:e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 56.Paltiel A.D., Zheng A., Walensky R.P. Assessment of SARS-CoV-2 screening strategies to permit the safe reopening of college campuses in the United States. JAMA Netw. Open. 2020;3:e2016818. doi: 10.1001/jamanetworkopen.2020.16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.U.S. FDA EUA summary COVID-19 RT-PCR test (Laboratory Corporation of America) 2020. https://www.fda.gov/media/136151/download
- 58.U.S. FDA Aptima SARS-CoV-2 assay: instructions for use. 2020. https://www.fda.gov/media/138096/
- 59.U.S. FDA Panther Fusion SARS-CoV-2 (Hologic, Inc.) 2020. https://www.fda.gov/media/136153/
- 60.U.S. FDA Helix COVID-19 test: EUA summary. 2020. https://www.fda.gov/media/140420/
- 61.U.S. FDA Wren Laboratories COVID-19 PCR test: EUA summary. 2020. https://www.fda.gov/media/140776/
- 62.U.S. FDA Advanta Dx SARS-CoV-2 RT-PCR assay: instructions for use. 2020. https://www.fda.gov/media/141541/
- 63.U.S. FDA Infinity BiologiX TaqPath SARS-CoV-2 assay: letter of authorization. 2020. https://www.fda.gov/media/137773/
- 64.Delaney M., Simpson J., Thomas B., Ralph C., Evangalista M., Moshgriz M., Granados J., McGuire M., DeBiasi R., Campos J. The use of saliva as a diagnostic specimen for SARS CoV-2 molecular diagnostic testing for pediatric patients. medRxiv. 2020 2020.11.11.20223800. [Google Scholar]
- 65.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.U.S. FDA EUA summary Quest Diagnostics. 2020. https://www.fda.gov/media/136228/download
- 68.Quest Diagnostics Quest Diagnostics COVID-19 specimen collection guidelines. 2020. https://www.questdiagnostics.com/dms/Documents/covid-19/COVID19_Specimen_Collection_Device_Guidelines/COVID19%20Specimen%20Acceptability_LDT%20Roche%20Revised%20Version%20v4%2032020.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional Supplementary Items are available from GitHub at https://github.com/grubaughlab/paper_2021_salivadirect.


