Ito et al. revealed that centrosomal proteins Cep57 and Cep57L1 cooperatively maintain centriole engagement during interphase in human cells. Codepletion of Cep57 and Cep57L1 induces precocious centriole disengagement in interphase, which results in centriole reduplication, centriole amplification, and chromosome segregation errors.
Abstract
Centrioles duplicate in interphase only once per cell cycle. Newly formed centrioles remain associated with their mother centrioles. The two centrioles disengage at the end of mitosis, which licenses centriole duplication in the next cell cycle. Therefore, timely centriole disengagement is critical for the proper centriole duplication cycle. However, the mechanisms underlying centriole engagement during interphase are poorly understood. Here, we show that Cep57 and Cep57L1 cooperatively maintain centriole engagement during interphase. Codepletion of Cep57 and Cep57L1 induces precocious centriole disengagement in interphase without compromising cell cycle progression. The disengaged daughter centrioles convert into centrosomes during interphase in a Plk1-dependent manner. Furthermore, the centrioles reduplicate and the centriole number increases, which results in chromosome segregation errors. Overall, these findings demonstrate that the maintenance of centriole engagement by Cep57 and Cep57L1 during interphase is crucial for the tight control of centriole copy number and thus for proper chromosome segregation.
Introduction
The centrosome is an organelle that serves as a major microtubule-organizing center (MTOC) in animal cells (Conduit et al., 2015). In mitosis, the two centrosomes migrate to opposite sides of the cell and facilitate the formation of a bipolar spindle (Petry, 2016). Therefore, the number of centrosomes must be strictly controlled for proper chromosome segregation (Nigg and Holland, 2018). Abnormalities in centrosome number cause improper spindle formation, chromosome instability, and various disorders, including cancer and congenital abnormalities such as microcephaly (Bettencourt-Dias et al., 2011). After cell division, each daughter cell harbors two centrioles, and a new daughter centriole forms in proximity to the mother centriole during the S phase. As the number of centrioles is halved after cell division, the centrioles are duplicated only once per cell cycle, ensuring that the correct number of centrioles is maintained (Gönczy and Hatzopoulos, 2019). Defects in the centriole duplication cycle can lead to aberrations in centrosome number.
The centrosome is composed of one or two centrioles and the surrounding pericentriolar material (PCM; Conduit et al., 2015). Each newly formed daughter centriole grows at the proximity of the mother centriole during interphase and is orthogonally engaged with the mother centriole until late mitosis (centriole engagement). After mitotic exit, mother and daughter centrioles are dissociated (centriole disengagement), and both centrioles are licensed to duplicate in the next cell cycle (Tsou et al., 2009). When centriole disengagement occurs precociously in interphase, centrioles are reduplicated within the same cell cycle (Lončarek et al., 2010; Martino et al., 2015). Such centriole reduplication results in an increase in the number of centrioles in cycling cells and may lead to chromosomal instability and a failure of cell division (Holmes et al., 2010). Thus, the maintenance of centriole engagement is one of the mechanisms limiting centriole duplication to once per cell cycle and controlling proper centrosome cycle progression. However, the molecular mechanisms underlying centriole engagement and disengagement remain largely unknown.
Recently, it has been suggested that the expanded PCM surrounds the pair of centrioles and maintains centriole engagement during mitosis (Seo et al., 2015). In particular, in human cells, pericentrin (PCNT), a major PCM scaffold protein, has been shown to be a critical factor for centriole engagement during mitosis (Matsuo et al., 2012; Lee and Rhee, 2012). PCNT is an elongated molecule that is radially oriented, with its C-terminus region near the centriole and its N-terminus extending outward to the periphery (Lawo et al., 2012; Mennella et al., 2012). A radial array of PCNT acting as a scaffold for PCM facilitates the recruitment of other PCM proteins during PCM expansion, and depletion of PCNT causes precocious centriole disengagement in early mitosis (Matsuo et al., 2012). Importantly, we recently identified Cep57 (centrosomal protein of 57 kDa) as a binding partner of PCNT (Watanabe et al., 2019). Cep57 localizes at the vicinity of centrioles and binds to the pericentrin-AKAP-450 centrosomal targeting (PACT) domain, a conserved C-terminus domain, of PCNT. Depletion of Cep57 perturbs the Cep57–PCNT interaction and thereby affects PCM organization in early mitosis, leading to precocious centriole disengagement (Watanabe et al., 2019). For the centriole disengagement that normally occurs at the end of mitosis, separase-dependent cleavage of PCNT, which presumably occurs around the metaphase-to-anaphase transition, is required for the disassembly of expanded PCM and subsequent centriole disengagement (Matsuo et al., 2012; Lee and Rhee, 2012). Polo-like kinase 1 (Plk1)-dependent phosphorylation of PCNT has also been reported as a priming step for the separase-dependent cleavage of PCNT (Kim et al., 2015). In the prolonged G2/M phase induced by treatment with a cyclin-dependent kinase 1 (Cdk1) inhibitor or DNA-damaging reagents, Plk1 and separase are aberrantly activated, which subsequently induces precocious centriole disengagement (Prosser et al., 2012; Douthwright and Sluder, 2014; Inanç et al., 2010). However, in contrast to the recent progress in understanding mechanisms of centriole disengagement in mitosis, very little is known about mechanisms maintaining centriole engagement during interphase.
Centrosomal protein 57 kDa-like protein 1 (Cep57L1) is a paralog of the Cep57 gene and is conserved in vertebrates. Cep57L1 was originally named for its homology to Cep57. It has been reported that the reduction of Cep57L1 expression level is responsible for the congenital absence of the anterior cruciate ligament and the posterior cruciate ligament in the knee (Liu et al., 2015). In addition, a recent study showed that double depletion of Cep57 and Cep57L1 suppressed the localization of human spindle assembly abnormal protein 6 homolog (HsSAS-6) and Cep152 at the centrosomes (Zhao et al., 2020). However, whether Cep57 and Cep57L1 are implicated in centriole duplication has not been fully examined, and the exact function of Cep57L1 remains unknown.
In this study, we reveal that Cep57 and Cep57L1 redundantly regulate centriole engagement in interphase. Codepletion of Cep57 and Cep57L1 causes precocious centriole disengagement during interphase in human cells. Such precociously disengaged daughter centrioles acquire PCM and MTOC activity. The precocious centriole disengagement in interphase is accompanied by centriole reduplication, and the number of centrioles per cell gradually increases with each passing cell division because the amplified centrioles are inevitably inherited by the daughter cells. Furthermore, the amplified centrioles cause a higher frequency of chromosome segregation errors. It is therefore most likely that defects in centriole engagement in interphase are more deleterious than those in early mitosis. These findings shed light on the molecules involved in the maintenance of centriole engagement during interphase and clarify the effects of the disruption of centriole engagement on the fidelity of chromosome segregation and cell division.
Results
Codepletion of Cep57 and Cep57L1 causes an increase in the number of centrosomes during interphase
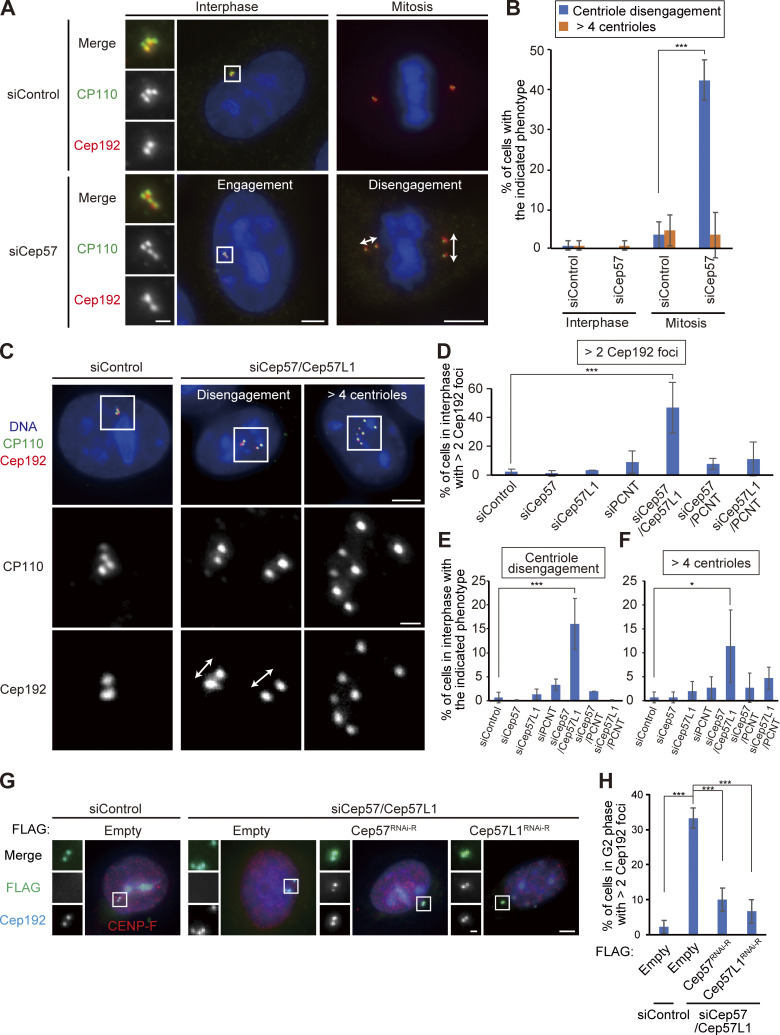
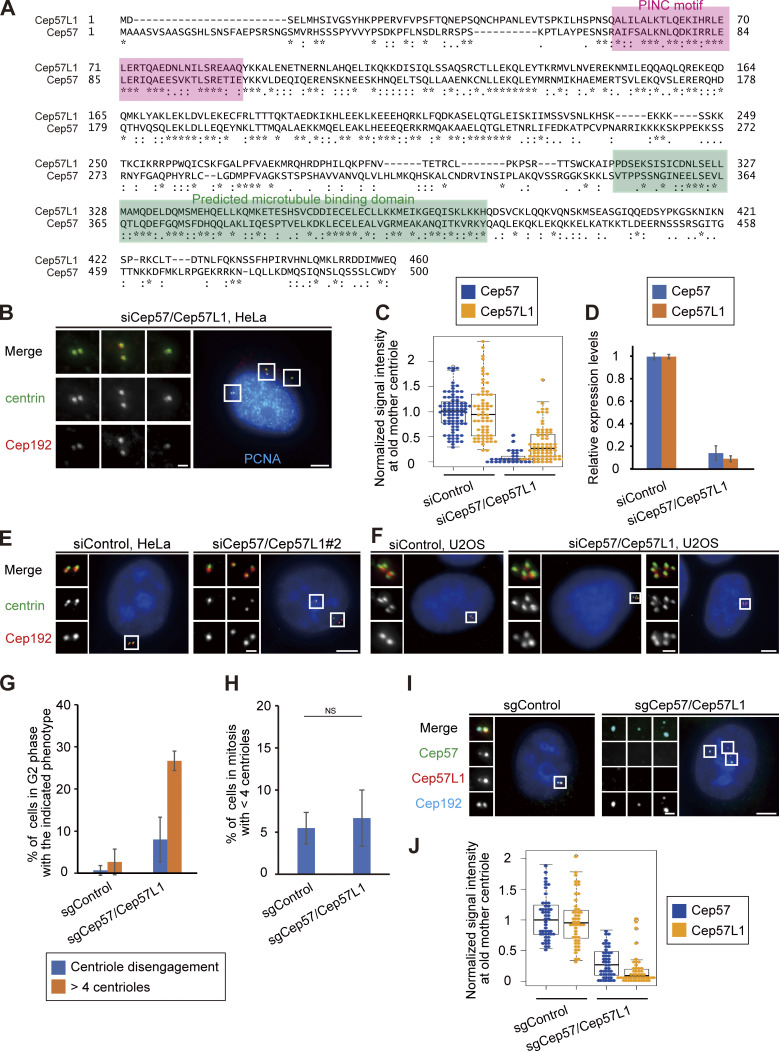
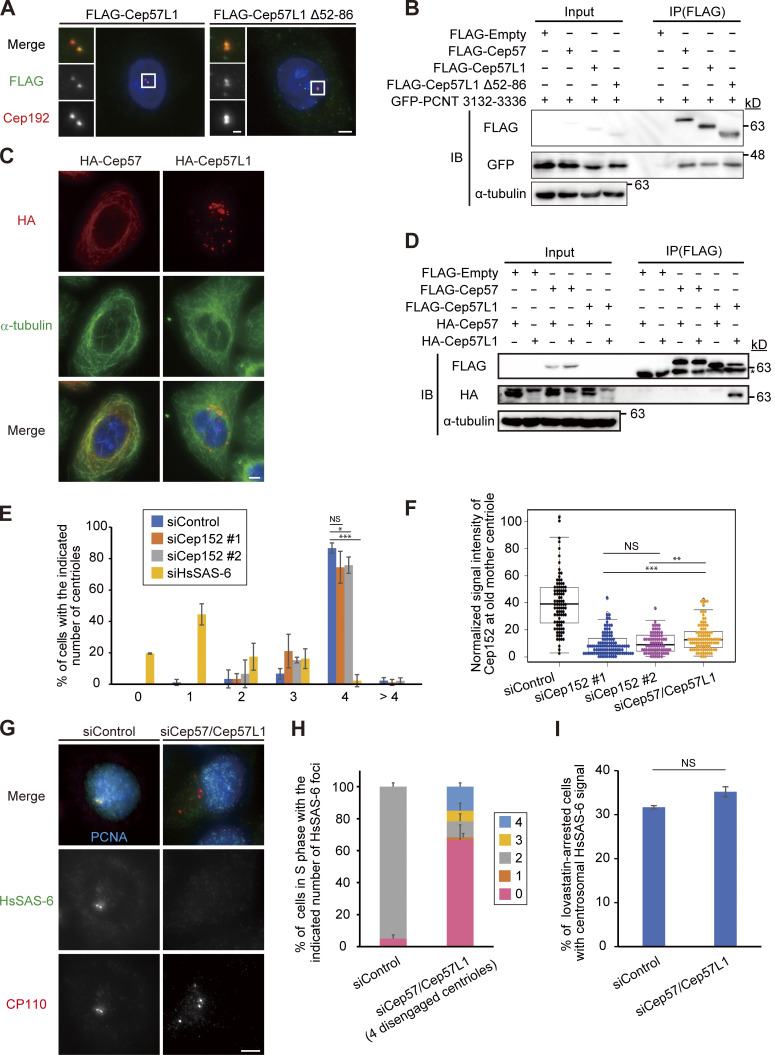
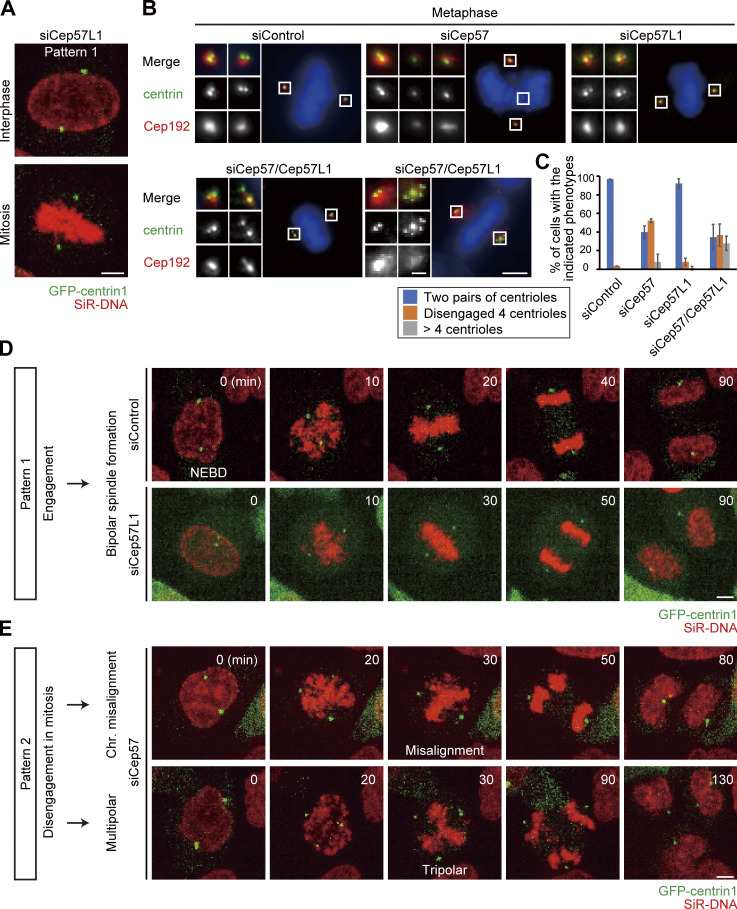
We recently reported that the Cep57–PCNT interaction is crucial for the maintenance of centriole engagement during mitosis and that depletion of Cep57 causes precocious centriole disengagement in mitosis, but not in interphase (mitosis, 42.2 ± 5.1%; interphase, 0%; n = 30 and 50, respectively, from three independent experiments; Fig. 1, A and B; Watanabe et al., 2019). Although the mechanism of centriole engagement in mitosis is gradually being elucidated, that of interphase remains completely unknown. To address this, we sought to identify the molecules required for centriole engagement during interphase. Given that comprehensive RNAi-based screens have failed to identify such molecules thus far, we assumed that two or more molecules have redundant functions in the maintenance of centriole engagement during interphase. Taking this possibility into account, we considered Cep57 and PCNT as potential targets. We also focused on an uncharacterized paralog of Cep57 in addition to Cep57 and PCNT: Cep57L1. Cep57L1 is a conserved protein in vertebrates and consists of 460 amino acid residues with 39% sequence homology to Cep57 in Homo sapiens (Fig. S1 A). We found that depletion of Cep57L1 did not affect centriole engagement in either interphase or mitosis (Fig. 1 E). We accordingly tested codepletion of two of the three proteins Cep57, Cep57L1, and PCNT by treating human cells with two distinct siRNAs, and we observed the centrosome and centriole behaviors. Among these combinations, we first found that codepletion of Cep57 and Cep57L1 (Cep57/Cep57L1) caused an increase in the number of centrosomes, marked by Cep192, in interphase (46.6 ± 17.7%; n = 50; from three independent experiments; Fig. 1, C and D). Interestingly, we next found by using a centriole maker, coiled-coil protein 110 (CP110), that ∼30% of the Cep57/Cep57L1-depleted cells exhibited an abnormal centriole pattern, including precocious centriole disengagement in interphase (16.0 ± 5.3%; n = 50; from three independent experiments; we defined precocious centriole disengagement as being when at least one centriole is more than 0.75 µm away from the others; Fig. 1, C and E; and more than four centrioles [11.3 ± 7.6%; Fig. 1, C and F]). We also noticed that such Cep57/Cep57L1-depleted cells could possess more than two Cep192 foci even in the S phase (Fig. S1 B). The efficiency of the siRNAs was validated by quantifying mRNAs using quantitative RT-PCR (RT-qPCR) and also by measuring the signal intensities of Cep57 and Cep57L1 at old mother centrioles (Fig. S1, C and D). The phenotypes provoked by codepletion of Cep57 and Cep57L1 were also confirmed by using a different siRNA and other human cell lines (Fig. S1, E and F). As expected, the phenotypes were rescued by expressing a synthetic RNAi-resistant Cep57 or Cep57L1 construct (siControl, 2.2 ± 1.9%; siCep57/Cep57L1, 33.2 ± 2.9%; siCep57/Cep57L1 + Cep57 expression, 10.0 ± 3.3%; siCep57/Cep57L1 + Cep57L1 expression, 6.7 ± 3.3%; n = 30; from three independent experiments; Fig. 1, G and H). Furthermore, the phenotypes were also confirmed by CRISPR/CRISPR-associated protein 9 (Cas9)-mediated codepletion of Cep57 and Cep57L1 (8.0 ± 5.3% [precocious centriole disengagement]; 26.7 ± 2.3% [more than four centrioles]; n = 50; from three independent experiments; Fig. S1 G). The efficiency of the CRISPR/Cas9-mediated depletion was validated by measuring the signal intensities of Cep57 and Cep57L1 at the centrosomes (Fig. S1, I and J). Taking these data together, we conclude that codepletion of Cep57 and Cep57L1 increases the number of centrosomes and centrioles during interphase.
Figure 1.
Codepletion of Cep57 and Cep57L1 causes the increase of centrosome number during interphase. (A) Depletion of Cep57 induced precocious centriole disengagement in mitosis, but not in interphase. HeLa cells were treated with siControl or siCep57 and immunostained with antibodies against CP110 (green) and Cep192 (red). White arrows indicate the distance between the disengaged centrioles. (B) Histograms represent the frequency of cells in interphase and mitosis with the indicated phenotypes observed in A. Values are mean percentages ± SD from three independent experiments (n = 50 for interphase and n = 30 for mitosis for each experiment). (C) Codepletion of Cep57 and Cep57L1 (Cep57/Cep57L1) induced precocious centriole disengagement and increase of centrosome number in interphase. HeLa cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against CP110 (green) and Cep192 (red). White arrows indicate the distance between the disengaged centrioles. Note that because the centrosomal linker does not function well in HeLa cells, the disengaged centrioles were sometimes well separated in Cep57/Cep57L1-depleted cells. (D–F) Histograms represent the frequency of the interphase cells with more than two Cep192 foci (D), centriole disengagement (E), or more than four centrioles (F), respectively. The quantification was performed on all interphase cells at random. Values are the mean percentages ± SD from three independent experiments (n = 50 for each experiment). (G) Precocious centriole disengagement in interphase induced by Cep57/Cep57L1 codepletion was rescued by exogenous expression of Cep57 or Cep57L1. HeLa cells were treated with siControl or siCep57/Cep57L1, followed by the transfection with FLAG empty (control), FLAG-Cep57 (RNAi resistant [RNAi-R]), or FLAG-Cep57L1 (RNAi-R). The cells were immunostained with antibodies against FLAG (green), CENP-F (red), and Cep192 (cyan). (H) Histograms represent the frequency of cells in the G2 phase with more than two Cep192 foci in each condition observed in G. Values are percentages from three independent experiments (n = 30 for each experiment). All scale bars, 5 µm in the low-magnification view, 1 µm in the inset. A two-tailed, unpaired Student’s t test was used in B to obtain the P value. Dunnett’s multiple comparisons test was used in D, E, F, and H to obtain P values. ***, P < 0.001; *, P < 0.05.
Figure S1.
Double depletion of Cep57 and Cep57L1 causes precocious centriole disengagement in interphase. (A) Alignments of full-length H. sapiens Cep57L1 and Cep57. Asterisks indicate the residues identical in aligned sequence; colons indicate conserved substitutions; periods indicate semiconserved substitutions. The positions of the PINC motif and the predicted microtubule-binding domain are indicated in pink and green boxes, respectively. (B) Cep57/Cep57L1-depleted cells with disengaged or more than four centrioles possessed more than two Cep192 foci even in the S phase. HeLa cells were treated with siCep57/Cep57L1 and immunostained with antibodies against centrin (green), Cep192 (red), and PCNA (cyan). (C) Beeswarm plots piled on boxplots represent the normalized signal intensity of Cep57 and Cep57L1 at the old mother centrioles upon siControl and siCep57/Cep57L1 (n = 50). (D) Expression levels of Cep57 and Cep57L1 were quantified by RT-qPCR in control and Cep57/Cep57L1-depleted cells. Values are normalized expression levels ± SD from triplicated experiments. (E) The phenotype induced by Cep57/Cep57L1 codepletion was confirmed by using another siRNA. HeLa cells were treated with siCep57 and siCep57L1 no. 2 and immunostained with antibodies against centrin (green) and Cep192 (red). siCep57L1 no. 2 targets a different sequence in ORF from siCep57L1 no. 1, which is used in the main figures. (F) U2OS cells also exhibited precocious centriole disengagement and amplified centrioles upon Cep57/Cep57L1 codepletion. U2OS cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against centrin (green) and Cep192 (red). (G) CRISPR/Cas9-mediated codepletion of Cep57 and Cep57L1 induces precocious centriole disengagement and centriole amplification. HeLa cells stably expressing Cas9 were treated with control sgRNA (sgControl) or sgRNAs targeting Cep57 and Cep57L1 (sgCep57/Cep57L1). Values are mean percentages ± SD from three independent experiments (n = 50 for each experiment). (H) CRISPR/Cas9-mediated codepletion of Cep57 and Cep57L1 did not induce a reduction in the centriole number in mitosis. HeLa cells stably expressing Cas9 were treated with control sgRNA (sgControl) or sgRNAs targeting Cep57 and Cep57L1 (sgCep57/Cep57L1). Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (I) CRISPR/Cas9-mediated codepletion of Cep57 and Cep57L1 was validated. HeLa cells stably expressing Cas9 were treated with control sgRNA (sgControl) or sgRNAs targeting Cep57 and Cep57L1 (sgCep57/Cep57L1) and immunostained with antibodies against Cep57 (green), Cep57L1 (red), and Cep192 (cyan). (J) Beeswarm plots piled on boxplots represent the normalized signal intensity of Cep57 and Cep57L1 at the old mother centrioles upon sgControl and sgCep57/Cep57L1 (n = 50). All scale bars, 5 µm in the low-magnification view, 1 µm in the inset. A two-tailed, unpaired Student’s t test was used in H to obtain P value. NS, P > 0.05.
Cep57 and Cep57L1 redundantly regulate centriole engagement during interphase
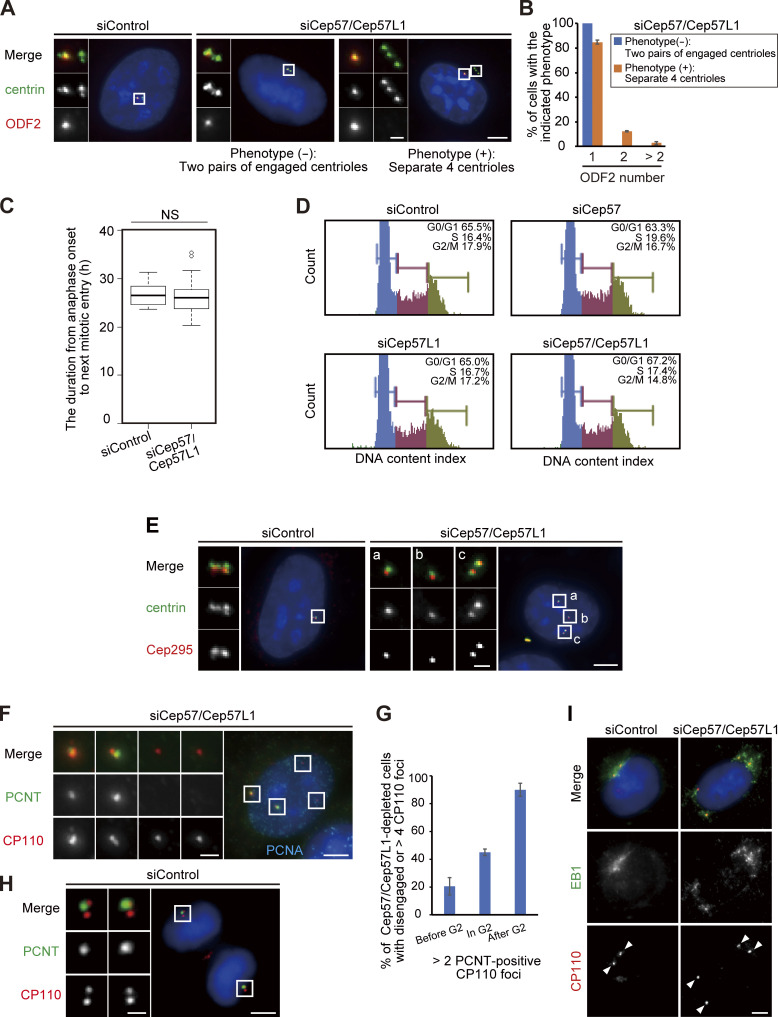
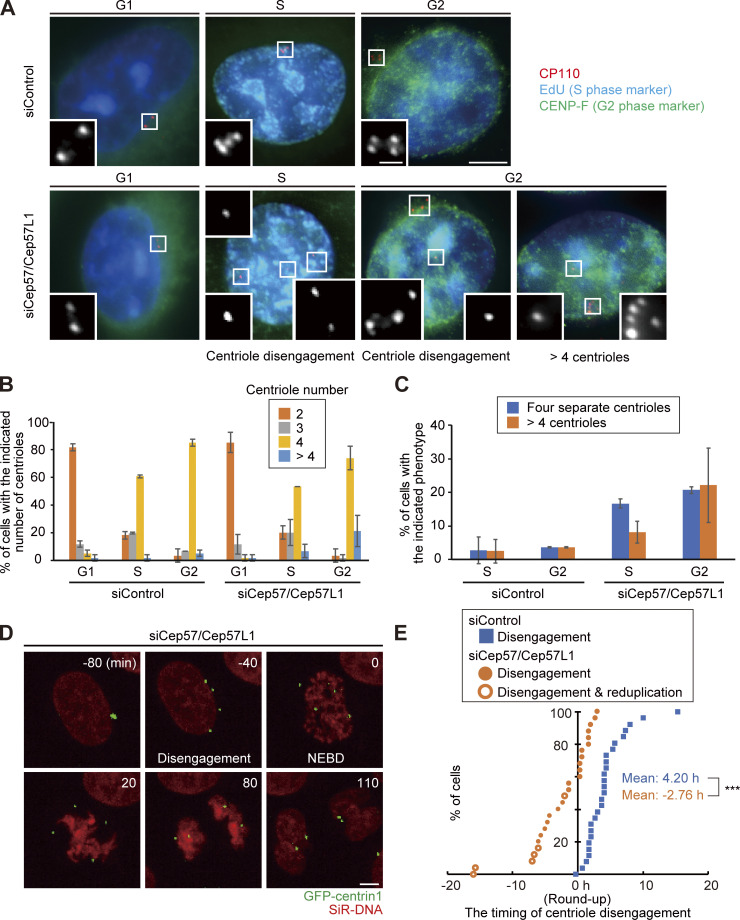
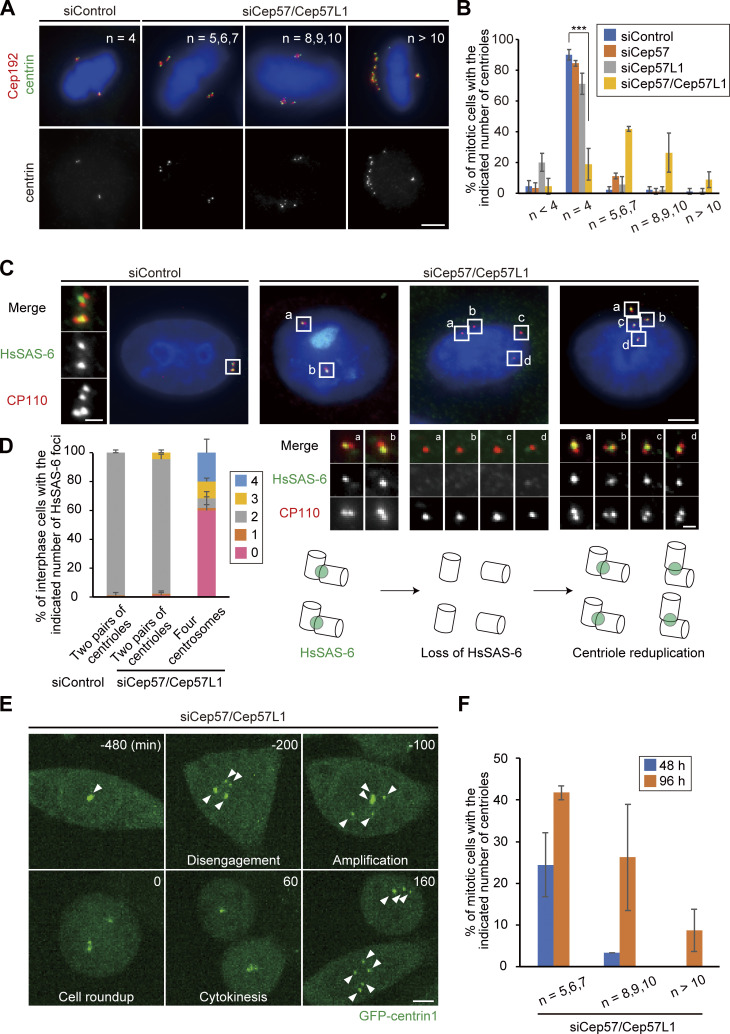
In general, the presence of four separate centrioles in interphase can stem from a failure of cytokinesis. To investigate whether the phenotype seen with codepletion of Cep57 and Cep57L1 is due to a failure of cytokinesis or to precocious centriole disengagement during interphase, we immunostained HeLa cells with an antibody against outer dense fiber protein 2 (ODF2), a marker of old mother centrioles (Fig. S2, A and B). If the four separate centrioles were the consequence of a failure of cytokinesis, the number of old mother centrioles in an interphase cell should be two. However, >80% of the HeLa cells with four separate centrioles or amplified centrioles possessed only one ODF2 focus (84.8 ± 1.7%; n > 50; from two independent experiments), as was the case in control cells (100%; Fig. S2, A and B). We therefore reasoned that the separate centrioles likely resulted from precocious centriole disengagement in interphase rather than from cytokinesis failure. To confirm this idea, we determined when in the cell cycle the phenotype could be observed in Cep57/Cep57L1-depleted cells using 5-ethynyl-2′-deoxyuridine (EdU; an S phase marker) and an antibody against centromere protein F (CENP-F; a G2 phase marker). Immunofluorescence (IF) with the cell cycle markers indicated that, as in control cells, there were two centrioles in the G1 phase in Cep57/Cep57L1-depleted cells (EdU-negative, CENP-F–negative cells), but ∼25% of S phase cells (EdU-positive, CENP-F–negative cells) and 43% of G2 phase cells (CENP-F–positive cells) exhibited four or more separate centrioles (S phase, 24.9 ± 2.0%; G2 phase, 42.9 ± 10.1%; n = 30; from three independent experiments; Fig. 2, A–C). These data strongly suggest that the presence of the four separate centrioles observed during interphase in Cep57/Cep57L1-depleted cells stem from precocious centriole disengagement in the S and G2 phases. Furthermore, live-cell imaging analysis using HeLa cells stably expressing GFP-centrin1 (HeLa-GFP-centrin1) confirmed that precocious centriole disengagement occurred during interphase in Cep57/Cep57L1-depleted cells (Fig. 2 D and Video 1). We also measured cumulative percentages of the disengagement phenotype on the basis of live-cell imaging data and found that ∼60% of Cep57/Cep57L1-depleted cells (16 of 26 cells) exhibited a disengagement phenotype before cell roundup; that is, in interphase (Fig. 2 E; mean time, siControl t = 4.20 h; siCep57/Cep57L1 t = −2.76 h; time 0 corresponds to the cell roundup). The precociously disengaged centrioles were repeatedly assembled and dispersed during interphase (Video 2). Importantly, we also found that centriole disengagement was occasionally followed by centriole reduplication (Fig. 2 E) and that around one-fifth of Cep57/Cep57L1-depleted cells possessed more than four centrioles in the G2 phase (22.1 ± 11.1%; n = 30; from three independent experiments; Fig. 2 C). Taken together, these data suggest that Cep57 and Cep57L1 cooperatively regulate the maintenance of centriole engagement during interphase and thus suppress centriole reduplication within the same cell cycle.
Figure S2.
Phenotypic analyses about the cell cycle in Cep57/Cep57L1-depleted cells. (A) The number of old mother centriole was one, regardless of the phenotypes in Cep57/Cep57L1-depleted cells. HeLa cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against centrin (green) and ODF2 (red). (B) Histograms represent the frequency of the interphase cells with the indicated number of ODF2 foci observed in A. Values are mean percentages ± SD from two independent experiments (n = 50 for each experiment). (C) Quantification of the duration from anaphase onset to next mitotic entry. HeLa-GFP-centrin1 cells were treated with siControl or siCep57/Cep57L1 and visualized for live-cell imaging for 48 h (n = 25). (D) Cep57/Cep57L1 codepletion did not affect cell cycle progression. HeLa cells were treated with the indicated siRNAs and followed by flow cytometric analysis. (E) Cep295 was recruited to the disengaged centrioles. HeLa cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against centrin (green) and Cep295 (red). The insets in a–c are the magnified views of the corresponding regions in the low-magnification view. (F) PCNT was not recruited to disengaged daughter centrioles in the S phase. HeLa cells were treated with siCep57/Cep57L1 and immunostained with antibodies against PCNT (green), CP110 (red), and PCNA (cyan). (G) Histograms represent the frequency of cells with more than two PCNT-positive centrioles among cells with separate or more than four CP110 foci in S phase, G2 phase, and mitosis. Values are mean percentages ± SD from two independent experiments (n = 50 for each experiment). (H) PCNT was not recruited to disengaged daughter centrioles immediately after centriole disengagement in control cells as in Cep57/Cep57L1-depleted cells. HeLa cells were immunostained with antibodies against PCNT (green) and CP110 (red). (I) Ectopic MTOC activity of precociously disengaged daughter centrioles in Cep57/Cep57L1-depleted cells. HeLa cells were treated with siControl or siCep57/Cep57L1 and followed by nocodazole treatment (10 µM) for 3 h. After nocodazole treatment, the cells were cold treated for 1 h, followed by 1-min incubation at 37°C and immunostaining with antibodies against EB1 (green) and CP110 (red). White arrowheads indicate centrosomes. All scale bars, 5 µm in the low-magnification view, 1 µm in the inset. The normal distribution of datasets was confirmed using the Kolmogorov–Smirnov test in C. A two-tailed, unpaired Student’s t test was used in C to obtain P value. NS, P > 0.05.
Figure 2.
Cep57 and Cep57L1 redundantly regulate centriole engagement during interphase. (A) Cep57/Cep57L1 codepletion induced four separated centrioles and more than four centrioles in the G2 phase. HeLa cells were treated with siControl or siCep57/Cep57L1 for 36 h in the presence of EdU (S phase marker; cyan) for the last 30 min before fixation and immunostained with antibodies against CP110 (red) and CENP-F (green). Scale bars, 5 µm in the low-magnification view, 1 µm in the inset. (B) Histograms represent the number of centrioles in the G1, S, and G2 phases treated with the indicated siRNAs in A. (C) Histograms represent the frequency of cells in the S and G2 phases treated with the indicated siRNAs exhibiting the indicated phenotype. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment) in B and C. (D) Time-lapse observation of cells upon Cep57/Cep57L1 codepletion. NEBD indicates nuclear envelope breakdown. HeLa cells stably expressing GFP-centrin1 (HeLa-GFP-centrin1, green) were treated with siControl or siCep57/Cep57L1 in the presence of SiR-DNA (200 nM, red). Scale bar, 5 µm. (E) Cumulative scatterplot indicates the duration from cell roundup to centriole disengagement observed in D. Orange open circles indicate precocious centriole disengagement accompanied by centriole reduplication. The normal distribution of datasets was confirmed using the Kolmogorov–Smirnov test in E. A two-tailed, unpaired Student’s t test was used in E to obtain the P value. ***, P < 0.001.
Video 1.
Precocious centriole disengagement in Cep57/Cep57L1-depleted cells. HeLa-GFP-centrin1 (green) cells were treated with siCep57 and siCep57L1 in the presence of SiR-DNA (200 nM, red). 25 × 1.2–µm z-stacks were acquired every 10 min at 40× magnification. Image frames shown are maximum-intensity projections at each time point.
Video 2.
The precociously disengaged centrioles are repeatedly assembled and dispersed during interphase. HeLa-GFP-centrin1 (green) cells were treated with siCep57 and siCep57L1 in the presence of SiR-DNA (200 nM, red). 25 × 1.2–µm z-stacks were acquired every 10 min at 40× magnification. Image frames shown are maximum-intensity projections at each time point.
Previous studies have reported that the G2 phase cell cycle arrest induced by treatment with a Cdk1 inhibitor or DNA-damaging reagents caused precocious centriole disengagement (Prosser et al., 2012; Douthwright and Sluder, 2014). To examine whether codepletion of Cep57 and Cep57L1 induces cell cycle arrest in interphase, we performed a live-cell imaging analysis and measured the duration of the period from anaphase onset to cell roundup in the next mitosis. In Cep57/Cep57L1-depleted cells, the duration was not significantly altered compared with control cells (siControl, 26.8 ± 2.2 h; siCep57/Cep57L1, 26.2 ± 3.6 h; n = 30; Fig. S2 C). In addition, FACS profiling analysis revealed that the distribution of cell cycle phases was not affected by Cep57/Cep57L1 codepletion (Fig. S2 D). Overall, these findings reveal that codepletion of Cep57 and Cep57L1 causes precocious centriole disengagement starting in the S phase without affecting the cell cycle progression.
Daughter centrioles can disengage from the mother centrioles before structural maturation
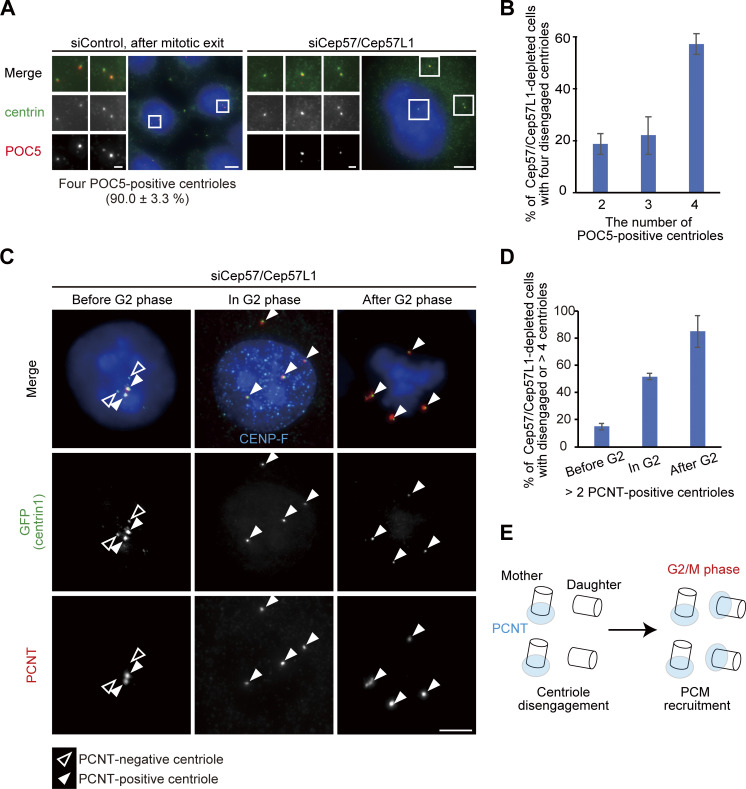
Given that, after codepletion of Cep57 and Cep57L1, precocious centriole disengagement was apparent starting in the S phase and that centriole formation proceeds during the S phase, we next examined whether the precocious centriole disengagement occurs before full elongation of daughter centrioles. To address this, we used proteome of centriole 5 (POC5), which is known to be incorporated at the final stage of daughter centriole formation (Chang et al., 2016; Azimzadeh et al., 2009). In control cells, all of the disengaged centrioles were POC5 positive after mitotic exit (90.0 ± 3.3%; Fig. 3 A). In contrast, in Cep57/Cep57L1-depleted cells, POC5 signal was not detected at approximately half of the disengaged centrioles (42.9% ± 4.0%; n = 30; from three independent experiments), suggesting that upon codepletion of Cep57 and Cep57L1, a daughter centriole can disengage from the mother centriole during the process of its formation (Fig. 3, A and B).
Figure 3.
Precociously disengaged centrioles convert into centrosomes during the G2 phase and mitosis. (A) A portion of disengaged daughter centrioles lack POC5. HeLa cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against centrin (green) and POC5 (red). Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (B) Histograms represent the frequency of cells with the indicated number of POC5-positive centrioles among cells with two pairs of centrioles (siControl) or disengaged four centrioles (siCep57/Cep57L1) observed in A. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (C) PCNT was recruited to disengaged daughter centrioles mainly in the G2 phase and mitosis. HeLa-GFP-centrin1 cells were treated with siCep57/Cep57L1 and immunostained with antibodies against GFP (green), PCNT (red), and CENP-F (cyan). White and black arrowheads indicate PCNT-positive and PCNT-negative centrioles, respectively. (D) Histograms represent the frequency of cells with more than two PCNT-positive centrioles among cells with disengaged four centrioles before, during, and after the G2 phase observed in C. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (E) Schematic illustration of the results in C and D. PCNT was recruited into disengaged daughter centrioles mainly in the G2 phase and mitosis. All scale bars, 5 µm in the low-magnification view, 1 µm in the inset.
Precociously disengaged daughter centrioles are converted to centrosomes in the G2 phase and mitosis
Because disengaged daughter centrioles are converted into centrosomes after mitosis in normal cells (a process called “centriole-to-centrosome conversion”; Wang et al., 2011), we then asked if the precociously disengaged daughter centrioles accomplish the centriole-to-centrosome conversion in interphase. In normal cells, around mitotic exit, a disengaged daughter centriole becomes a functional centrosome and recruits PCM components and centriole duplication factors, such as PCNT and Cep152, respectively. For this conversion, procentriole formation requires the centriolar recruitment of Cep295 and Cep192 (Tsuchiya et al., 2016; Fu et al., 2016). Similar to Cep192 (Fig. S1 B), Cep295 was localized at almost all of the precociously disengaged daughter centrioles (Fig. S2 E). In contrast, the disengaged daughter centrioles did not always acquire PCM components (PCNT), suggesting that PCM proteins were gradually recruited to the disengaged daughter centrioles. Indeed, Cep57/Cep57L1-depleted cells with more than two PCNT-positive centrioles were relatively rare before the G2 phase (15.0 ± 2.4%; n = 30 from three independent experiments) but gradually increased during the G2 phase (51.7 ± 2.4%) and were most frequently observed after the G2 phase was completed (mitosis, 85.0 ± 11.8%; Fig. 3, C and D; Fig. S2, F and G). These observations show that the precociously disengaged daughter centrioles first lacked PCM and then gradually recruited PCM components to their surroundings mainly during the G2 phase and mitosis (Figs. 3 E and S2 H). Furthermore, such disengaged daughter centrioles nucleated microtubules after depolymerization of microtubules by cold treatment, indicating that these centrioles had acquired MTOC activity during interphase (Fig. S2 I). Taken together, these results indicate that upon Cep57/Cep57L1 codepletion, the precociously disengaged centrioles can be converted to centrosomes starting in interphase.
Codepletion of Cep57 and Cep57L1 induces centriole disengagement in interphase even without Plk1 kinase activity
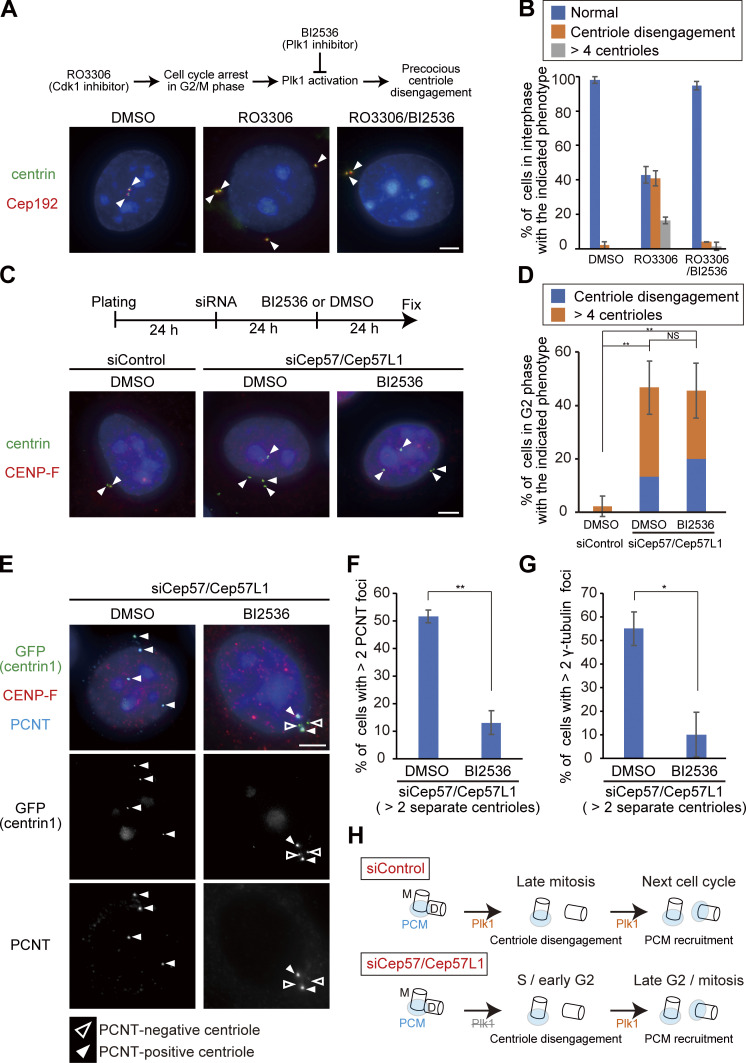
To gain insight into the mechanisms by which Cep57/Cep57L1 codepletion causes precocious centriole disengagement in interphase, we tested a requirement of the known factors involved in centriole disengagement at the end of mitosis. Previous studies have reported that canonical centriole disengagement requires Plk1 activity and that the inhibition of Plk1 perturbs centriole disengagement at the end of mitosis (Tsou et al., 2009). Precocious centriole disengagement in G2/M phase-arrested cells was also suppressed by Plk1 inhibition (Prosser et al., 2012). To determine whether the precocious centriole disengagement in Cep57/Cep57L1-depleted cells also requires Plk1 activity, we treated G2 phase-arrested cells or Cep57/Cep57L1-depleted cells with a small-molecule inhibitor of Plk1 (BI2536). As expected, the precocious centriole disengagement or the increase in the number of centrioles in G2/M-arrested cells was suppressed by BI2536 treatment (RO3306, 57.2 ± 4.8%; RO3306/BI2536, 5.3 ± 2.4%; n = 50 from three independent experiments; Fig. 4, A and B). In contrast, BI2536 treatment did not suppress the precocious centriole disengagement or the increase in the number of centrioles in Cep57/Cep57L1-depleted cells (DMSO, 46.7 ± 10.0%; BI2536, 45.6 ± 10.1%; n = 30 from three independent experiments; Fig. 4, C and D). These data indicate that the precocious centriole disengagement that occurs in Cep57/Cep57L1-depleted cells does not depend on Plk1 activity, unlike canonical centriole disengagement or cell cycle arrest–induced precocious centriole disengagement.
Figure 4.
Inhibition of Plk1 prevents disengaged daughter centrioles from acquiring PCM but does not suppress precocious centriole disengagement. (A) Precocious centriole disengagement caused by G2/M phase arrest was suppressed by BI2536 (Plk1 inhibitor; 100 nM). HeLa cells were synchronized in the G1/S phase by aphidicolin (1.2 µg/ml) for 17 h, then released into fresh medium for 4 h. Next, the cells were treated with DMSO (control), RO3306 (Cdk1 inhibitor; 10 µM), or both of RO3306 and BI2536 (100 nM) for 24 h. HeLa cells were immunostained with antibodies against centrin (green) and Cep192 (red). White arrowheads indicate centrioles. Scale bar, 5 µm. (B) Histograms represent the frequency of the interphase cells with the indicated phenotype observed in A. Values are mean percentages ± SD from three experiments (n = 50 for each experiment). (C) Precocious centriole disengagement in Cep57/Cep57L1-depleted cells was not suppressed by BI2536. HeLa cells were treated with siControl or siCep57/Cep57L1 for 24 h, followed by treatment of DMSO (control) or BI2536 (100 nM) for an additional 24 h. The cells were immunostained with antibodies against centrin (green) and CENP-F (red). White arrowheads indicate centrioles. Scale bar, 5 µm. (D) Histograms represent the frequency of cells in the G2 phase with the indicated phenotype observed in C. Values are mean percentages ± SD from three experiments (n = 30 for each experiment). (E) BI2536 suppressed recruitment of PCNT at disengaged daughter centrioles. HeLa-GFP-centrin1 cells were treated as in C and immunostained with antibodies against GFP (green), CENP-F (red), and PCNT (cyan). White and black arrowheads indicate PCNT-positive and PCNT-negative centrioles, respectively. Scale bars, 5 µm. (F and G) Histograms represent the frequency of cells with more than two PCNT or γ-tubulin foci among the G2 phase cells with disengaged or more than four centrioles. Values are mean percentages ± SD from three experiments (n = 30 for each experiment). (H) Schematic illustration of the results in Fig. 4, C–G. The activity of Plk1 is required for the recruitment of PCM components at the disengaged daughter centrioles, but not for precocious centriole disengagement itself. M and D indicate mother and daughter centrioles, respectively. Tukey’s multiple comparisons test was used in D against the total value of centriole disengagement and more than four centrioles to obtain the P value. A two-tailed, unpaired Student’s t test was used in F and G to obtain P values. **, P < 0.01; *, P < 0.05; NS, P > 0.05.
Plk1 activity is required for PCM recruitment to disengaged daughter centrioles, presumably in the G2 phase
Because Plk1 is necessary not only for centriole disengagement but also for daughter centrioles to acquire PCM in normal cells (Wang et al., 2011), we next asked if Plk1 activity is also required for PCM recruitment at precociously disengaged daughter centrioles in Cep57/Cep57L1-depleted cells. To address this, we treated Cep57/Cep57L1-depleted cells with BI2536 and counted the number of PCM-positive centrioles. Interestingly, in Cep57/Cep57L1-depleted cells, BI2536 treatment significantly reduced the number of cells with disengaged centrioles that have acquired PCNT (13.1 ± 4.3%; n = 30; from three independent experiments) compared with the DMSO-treated cells (51.6 ± 2.4%; Fig. 4, E and F), indicating that Plk1 activity is required for PCNT recruitment to the disengaged daughter centrioles. Similarly, γ-tubulin recruitment to the disengaged daughter centrioles was also suppressed by BI2536 treatment (DMSO, 55.1 ± 7.1%; BI2536, 10.0 ± 9.4%; n = 30; from three independent experiments; Fig. 4 G). Overall, these findings demonstrate that the precociously disengaged daughter centrioles acquire PCM during the interphase in a Plk1-dependent manner (Fig. 4 H). Given that Plk1 is activated starting from the late G2 stage and preceding mitosis (Schmucker and Sumara, 2014) and is necessary for PCM recruitment to the disengaged daughter centrioles, it is reasonable that precociously disengaged daughter centrioles gradually acquire PCNT in the G2 phase (Fig. 3, C and D).
Cep57 and Cep57L1 have distinct properties in spite of their relatively high amino acid sequence homology
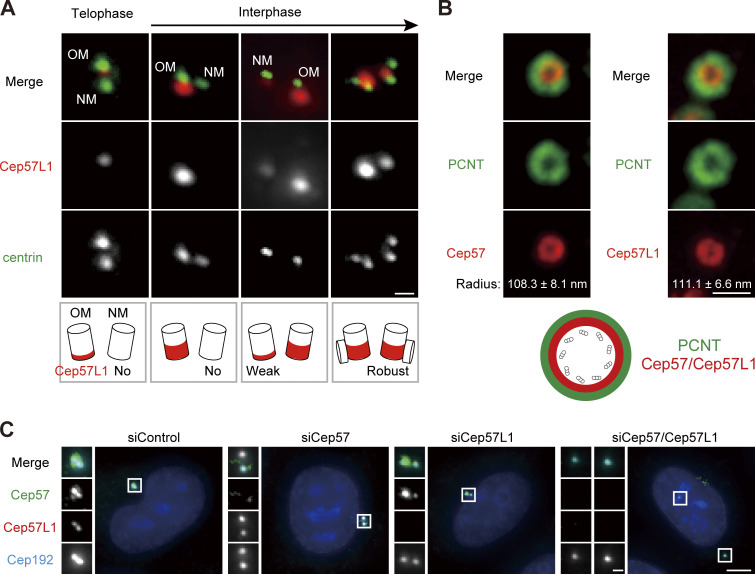
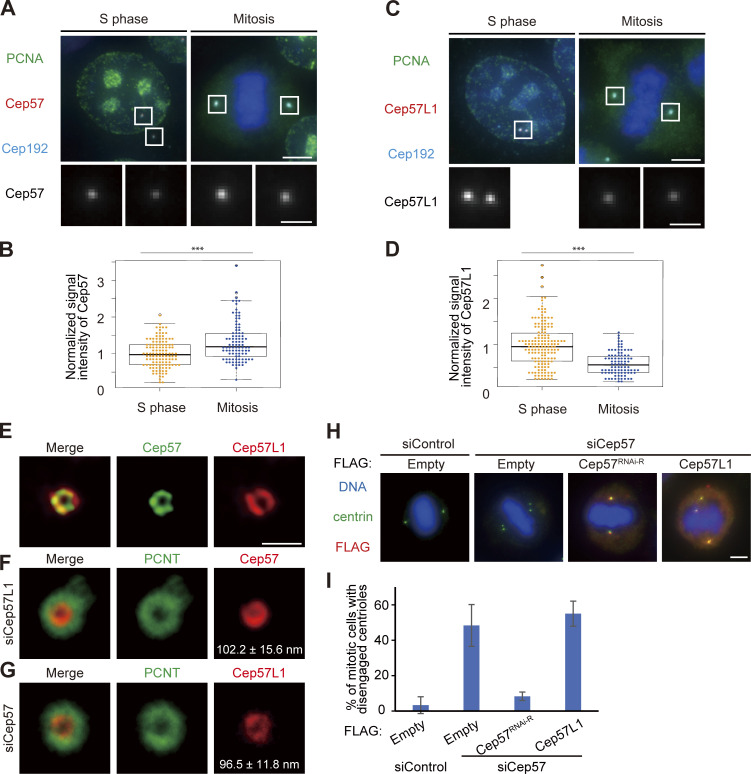
Although Cep57 and Cep57L1 cooperatively regulate centriole engagement in interphase, centriole engagement in mitosis is maintained by Cep57 but not by Cep57L1. This suggests that Cep57 and Cep57L1 have both redundant and distinct functions in centriole engagement. To investigate the similarities and differences between the functions of Cep57 and Cep57L1 in the regulation of centrioles, we performed localization and domain analysis of Cep57 and Cep57L1. We first sought to compare the detailed localization pattern of Cep57 and Cep57L1 at the centrosomes. To do this, we immunostained HeLa cells with an antibody against Cep57L1 and found that endogenous Cep57L1 localized at mother centrioles (Fig. 5 A). Similarly to Cep57, the signal intensity of Cep57L1 at new mother centrioles gradually increased in interphase (Fig. 5 A; Watanabe et al., 2019). We also noticed that the signal intensity of Cep57 further increased toward mitosis, whereas that of Cep57L1 decreased (Fig. S3, A–D). Moreover, stimulated emission depletion (STED) analysis showed that Cep57L1 formed a ring-like structure around the mother centriole (Fig. 5 B). The radius of the Cep57L1 ring was 111.1 ± 6.6 nm, which was close to that of the Cep57 ring (108.3 ± 8.1 nm; Fig. 5 B). STED microscopy also revealed the colocalization of Cep57 and Cep57L1 around the mother centriole (Fig. S3 E). The specificity of the antibody was validated using siRNA against Cep57L1 (Fig. 5 C). To examine the mutual dependency on centriole localization, using STED microscopy, we observed the localization pattern of Cep57 and Cep57L1 at centrioles upon reciprocal siRNA treatment. Depletion of Cep57 or Cep57L1 did not significantly affect each other’s localization pattern at centrioles (Fig. 5 C; Fig. S3, F and G). Overall, these results indicate that Cep57 and Cep57L1 show a similar centriolar distribution but independently localize to the centrosome.
Figure 5.
Cep57L1, a paralog of Cep57 conserved in vertebrates, shows a centriolar distribution similar to that of Cep57. (A) Centriolar distribution of Cep57L1 at different cell cycle stages. HeLa cells were immunostained with antibodies against Cep57L1 (red) and centrin (green). Scale bar, 500 nm. OM and NM indicate an old mother centriole and a new mother centriole, respectively. (B) STED images representing top view of Cep57/Cep57L1 (red) and PCNT (green) at mother centrioles. Scale bar, 500 nm. Values are the mean radius ± SD (n = 5). (C) The signals of Cep57L1 at the centrosomes were attenuated by siCep57L1 or siCep57/Cep57L1, but not by siCep57. HeLa cells were treated with siControl, siCep57, siCep57L1, or siCep57/Cep57L1 and immunostained with antibodies against Cep57 (green), Cep57L1 (red), and Cep192 (cyan). Scale bars, 5 µm in the low-magnification view, 1 µm in the inset.
Figure S3.
Cep57 and Cep57L1 independently localize around mother centrioles. (A) Cep57 is more associated with mitotic centrosomes than those of interphase. HeLa cells were immunostained with antibodies against PCNA (green), Cep57 (red), and Cep192 (cyan). Scale bars, 5 µm in the low-magnification view, 1 µm in the inset. (B) Beeswarm plots piled on boxplots represent the normalized signal intensity of Cep57 at the centrosomes in the S phase and mitosis (n = 50). (C) Cep57L1 is more associated with interphase centrosomes than those of mitosis. HeLa cells were immunostained with antibodies against PCNA (green), Cep57L1 (red), and Cep192 (cyan). Scale bars, 5 µm in the low-magnification view, 1 µm in the inset. (D) Beeswarm plots piled on boxplots represent the normalized signal intensity of Cep57L1 at the centrosomes in the S phase and mitosis (n = 50). (E) STED images representing top view of Cep57L1 (red) and Cep57 (green) at mother centrioles. (F) STED images representing top view of Cep57 (red) and PCNT (green) at mother centrioles upon siRNA treatment against Cep57L1. (G) STED images representing top view of Cep57L1 (red) and PCNT (green) at mother centrioles upon siRNA treatment against Cep57. Note that the radius of the Cep57L1 ring was significantly smaller in Cep57-depleted cells than in control cells (P < 0.05). Scale bar, 500 nm in E–G. (H) Precocious centriole disengagement in mitosis induced by Cep57 depletion was not rescued by exogenous expression of Cep57L1. HeLa cells were treated with siControl or siCep57/Cep57L1, followed by transfection with FLAG empty (control), FLAG-Cep57 (RNAi resistant [RNAi-R]), or FLAG-Cep57L1. The cells were immunostained with antibodies against FLAG (red) and centrin (green). (I) Histograms represent the frequency of cells in mitosis with precocious centriole disengagement in each condition observed in H. Values are percentages from two independent experiments (n = 30 for each experiment). Scale bars, 5 µm in the low-magnification view, 1 µm in the inset, except STED images in E–G. The normal distribution of datasets was confirmed using the Kolmogorov–Smirnov test in B and D. A two-tailed, unpaired Student’s t test was used in B and D to obtain the P value. ***, P < 0.001.
To further examine the similarities and differences between Cep57 and Cep57L1 function, we next performed domain analysis of Cep57 and Cep57L1. We previously reported that the conserved PINC (present in N-terminus of Cep57) domain in the N-terminus of Cep57 is required for its centriolar localization and the interaction with PCNT, a functional binding partner (Watanabe et al., 2019). Because Cep57L1 also has a PINC domain, we asked if the role of the PINC motif in Cep57L1 was similar to its role in Cep57. To address this, we constructed plasmids expressing the full-length Cep57L1 or a mutant lacking the PINC motif (52–86 aa). Unexpectedly, IF and coimmunoprecipitation (co-IP) analyses with the mutant revealed that the PINC motif of Cep57L1 was dispensable for the centrosomal localization (Fig. S4 A) and for the binding to the PCNT C-terminal region, including the PACT domain (Fig. S4 B). These data suggest that the PINC motif has different functions in Cep57 and Cep57L1. In addition to the PINC motif, the Pfam protein family database predicted the presence of microtubule-binding domains in both Cep57 and Cep57L1 (Fig. S1 A). As previously reported, overexpressed Cep57 was occasionally localized on the microtubule network via its microtubule-binding domain (Fig. S4 C; Momotani et al., 2008). In contrast, overexpressed Cep57L1 did not show such a localization pattern, but instead aggregated in the cytoplasm (Fig. S4 C). This result suggests that the putative microtubule-binding domain of Cep57L1 does not possess a binding affinity for microtubules. Furthermore, co-IP assays of Cep57 and Cep57L1 proteins in HEK293T cells detected self-interaction of Cep57L1 but not of Cep57 (Fig. S4 D). Overall, these findings show that Cep57 and Cep57L1 have distinct properties in spite of their relatively high amino acid sequence homology.
Figure S4.
Cep57 and Cep57L1 show distinct molecular properties. (A) The Cep57L1 PINC motif was not required for the centrosomal localization. HeLa cells expressing FLAG-Cep57L1 or FLAG-Cep57L1 mutant lacking the PINC motif (Δ52–86 aa) were immunostained with antibodies against FLAG (green) and Cep192 (red). (B) The Cep57L1 PINC motif was not required for binding to the C-terminal region of PCNT containing the conserved PACT domain. HEK293T cells coexpressing FLAG empty (control), FLAG-Cep57, FLAG-Cep57L1, or FLAG-Cep57L1 mutant lacking the PINC motif and GFP-PCNT (3132–3336 aa) were immunoprecipitated with FLAG antibodies and immunoblotted with the indicated antibodies. (C) Overexpressed Cep57 was accumulated on the microtubules, but overexpressed Cep57L1 aggregated in the cytoplasm. HeLa cells expressing HA-Cep57 or HA-Cep57L1 were immunostained with antibodies against HA (red) and α-tubulin (green). (D) Cep57L1 formed a homodimer. HEK293T cells coexpressing FLAG empty (control), FLAG-Cep57 or FLAG-Cep57L1, and HA-Cep57 or HA-Cep57L1 were immunoprecipitated with FLAG antibodies and immunoblotted with the indicated antibodies. (E) Depletion of Cep152 has a modest effect on centriole duplication. Histograms represent the number of centrioles in mitosis treated with the indicated siRNAs. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (F) Beeswarm plots piled on boxplots represent the normalized signal intensity of Cep152 at the old mother centrioles upon the indicated siRNAs (n = 50). (G) HsSAS-6 was frequently absent from disengaged centrioles in the S phase. HeLa cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against HsSAS-6 (green), CP110 (red), and PCNA (cyan). (H) Histograms represent the frequency of the number of HsSAS-6 foci on the two pairs of centrioles (siControl) or four disengaged centrioles (siCep57/Cep57L1) in G. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (I) HsSAS-6 was normally recruited to the centrosomes at the onset of centriole formation in Cep57/Cep57L1-depleted cells. Histograms represent frequency of the lovastatin-arrested G1 phase cells with two HsSAS-6 foci. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). The normal distribution of datasets was confirmed using the Kolmogorov–Smirnov test in F. Tukey’s multiple comparisons test was used in E and F to obtain the P values. A two-tailed, unpaired Student’s t test was used in I to obtain the P values. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, P > 0.05. All scale bars, 5 µm in the low-magnification view, 1 µm in the inset.
Interdependency of centrosomal localization between Cep57, Cep57L1, Cep63, and Cep152
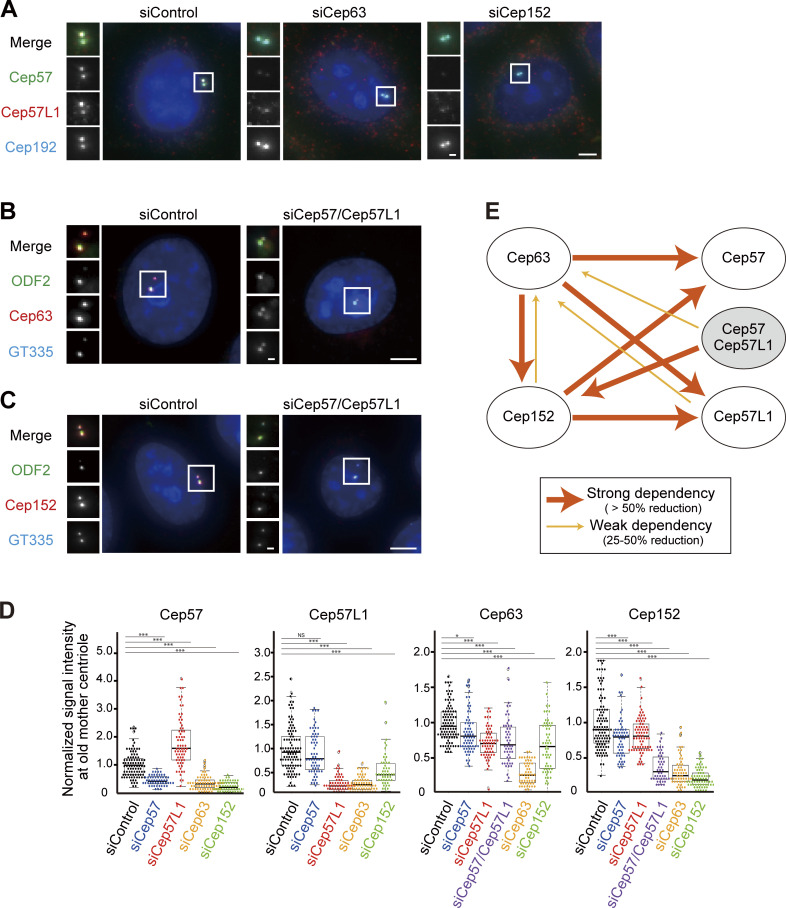
We next searched for the proteins recruiting Cep57L1 to the centrosome. Considering the similar distribution patterns of Cep57 and Cep57L1 at the centrosomes, we postulated that the localization of Cep57 and Cep57L1 around the centriole wall is regulated by common proteins. Cep57 is recruited to the centrosomes dependent on Cep63, Cep152, and NEDD1 (Aziz et al., 2018; Wu et al., 2012; Lukinavičius et al., 2013). Of these three proteins, Cep63 and Cep152 were demonstrated to form a trimeric complex with Cep57 at the centrosomes (Lukinavičius et al., 2013). Consistent with previous studies (Aziz et al., 2018; Lukinavičius et al., 2013), we confirmed that Cep63 (median, 30.6% compared with siControl; n > 50) or Cep152 depletion (19.3%) significantly decreased the signal intensity of Cep57 at the centrosomes (Fig. 6, A and D). We then assumed that Cep63 and Cep152 were also responsible for centrosomal localization of Cep57L1. As expected, depletion of Cep63 (24.8%) or Cep152 (45.8%) reduced the signal intensity of Cep57L1 at the centrosomes, indicating that both Cep57 and Cep57L1 localization at the centrosomes was partially dependent on Cep63 and Cep152 (Fig. 6, A and D). On the other hand, the signal intensity of Cep63 and Cep152 at the centrosomes was only slightly affected by depletion of Cep57 (Cep63, 81.0%; Cep152, 80.2%) or Cep57L1 (Cep63, 70.6%; Cep152, 81.0%), whereas codepletion of Cep57 and Cep57L1 reduced the signal intensity of Cep152 at the centrosomes more drastically (31.1%; Fig. 6, B–D). Moreover, as reported in previous studies (Lukinavičius et al., 2013; Kim et al., 2019; Brown et al., 2013), the signal intensity of Cep152 was attenuated by Cep63 depletion (25.8%) and vice versa (66.4%; Fig. 6 D). Hence, we propose that Cep57, Cep57L1, Cep63, and Cep152 mutually influence centrosomal localization (Fig. 6 E).
Figure 6.
Interdependency of centrosomal localization of Cep57, Cep57L1, Cep63, and Cep152. (A) The centrosomal localization of Cep57 and Cep57L1 was dependent on Cep63 and Cep152. HeLa cells were treated with siControl, siCep63, or siCep152 and immunostained with antibodies against Cep57 (green), Cep57L1 (red), and Cep192 (cyan). (B) The centrosomal localization of Cep63 was partially dependent on Cep57 and Cep57L1. HeLa cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against ODF2 (green), Cep63 (red), and GT335 (cyan). (C) The centrosomal localization of Cep152 was partially dependent on Cep57 and Cep57L1. HeLa cells were treated with siControl or siCep57/Cep5L1 and immunostained with antibodies against ODF2 (green), Cep152 (red), and GT335 (cyan). (D) Beeswarm plots piled on boxplots represent the normalized signal intensity of Cep57, Cep57L1, Cep63, and Cep152 at the old mother centrioles upon the indicated siRNAs (n = 50). (E) Schematic of the dependency of the centrosomallocalization among Cep57, Cep57L1, Cep63, and Cep152. The wide arrows indicate strong dependency, defined as >50% reduction of the signal intensity, and narrow arrows indicate weak dependency, defined as 25–50% reduction of the signal intensity. All scale bars, 5 µm in the low-magnification view, 1 µm in the inset. The normal distribution of datasets was confirmed using the Kolmogorov–Smirnov test in D. Tukey’s multiple comparisons test was used in D to obtain P value. *, P < 0.05; ***, P < 0.001; NS, P > 0.05.
Precocious centriole disengagement in interphase results in centriole reduplication and thereby accelerates numerical centrosome abnormalities
We next examined the long-term consequence of precocious centriole disengagement in Cep57/Cep57L1-depleted cells. To this end, we codepleted Cep57 and Cep57L1 in HeLa cells for 96 h and observed the centriole number during mitosis. IF analysis revealed that ∼80% of the Cep57/Cep57L1-depleted cells had more than four centrioles during mitosis (80.1 ± 9.0%; n = 50; from three independent experiments). In contrast, this was not the case for Cep57-depleted cells, in which centriole disengagement precociously occurred only in mitosis (siControl, 4.7 ± 3.6%; siCep57, 10.7% ± 1.3%; Fig. 7, A and B). This result suggests that the precociously disengaged centrioles, upon codepletion of Cep57 and Cep57L1, were already licensed to reduplicate during the interphase within the same cell cycle. This may have thereby led to the increase in the number of centrioles. To test this idea, we monitored the number of cartwheels using an HsSAS-6 marker. The Cep57/Cep57L1-depleted cells with two pairs of engaged centrioles possessed two HsSAS-6 foci, as in control cells (siControl, 98.9 ± 1.9%; siCep57/Cep57L1, 93.3 ± 3.3%; n = 30; from three independent experiments; Fig. 7, C and D). On the other hand, more than half of the Cep57/Cep57L1-depleted cells with four separate centrosomes had no HsSAS-6 foci in interphase (60.0 ± 6.7%; Fig. 7, C and D), suggesting that HsSAS-6 disappeared from the centrosomes, presumably concomitant with centriole disengagement. To further confirm this observation, we counted the number of HsSAS-6 foci in the S phase and in lovastatin-mediated G1 arrest. In the S phase, the Cep57/Cep57L1-depleted cells with four disengaged centrioles lack HsSAS-6 foci, in contrast to control cells, most of which possessed two HsSAS-6 foci (Fig. S4, G and H). However, in lovastatin-mediated G1 arrest, HsSAS-6 was loaded onto centrioles with similar frequency in the control and Cep57/Cep57L1-depleted cells (siControl, 31.7 ± 0.4%; siCep57/Cep57L1, 35.2% ± 1.1%; n = 30; from three independent experiments; Fig. S4 I). This result suggests that codepletion of Cep57 and Cep57L1 did not suppress the initial recruitment of HsSAS-6 to the centriole. Considering these results, we assume that in the Cep57/Cep57L1-depleted cells, mother centrioles normally recruit HsSAS-6 to initiate daughter centriole formation, whereas HsSAS-6 is lost from centrioles, concomitantly with precocious centriole disengagement in the S phase. The previous studies also showed the absence of HsSAS-6 foci in the S phase upon Cep57 or Cep57/Cep57L1 codepletion (Zhao et al., 2020; Wei et al., 2020), which is consistent with our observation. However, we speculate that those phenotypes might also be coupled with the precocious centriole disengagement rather than a defect in centriole duplication. Future investigations will be needed to clarify this point.
Figure 7.
Precocious centriole disengagement results in an increase of the number of centrioles. (A) Long-term Cep57/Cep57L1 codepletion increased the number of centrioles. HeLa cells were treated with siCep57/Cep57L1 for 96 h and immunostained with antibodies against centrin (green) and Cep192 (red). (B) Histograms represent the frequency of mitotic cells with the indicated number of centrioles observed in A. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). Note that depletion of Cep57L1 for 96 h slightly reduced the number of centrioles. (C) HsSAS-6 was absent from disengaged centrioles and was reacquired. HeLa cells were treated with siControl or siCep57/Cep57L1 and immunostained with antibodies against HsSAS-6 (green) and CP110 (red). The insets in a–d are the magnified views of the corresponding regions in the low-magnification view. (D) Histograms represent the frequency of the number of HsSAS-6 foci on the two pairs of centrioles or four centrosomes in C. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (E) Precocious centriole disengagement and centriole reduplication in interphase and inevitable inheritance of amplified centrioles to daughter cells observed in Cep57/Cep57L1-depleted cells. HeLa-GFP-centrin1 (green) cells were treated with siCep57/Cep57L1. White arrowheads indicate centrosomes. (F) Histograms represent the frequency of mitotic cells with the indicated number of centrioles 48 h or 96 h after Cep57/Cep57L1 codepletion observed in A and in Fig. S5 B. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). All scale bars, 5 µm in the low-magnification view, 1 µm in the inset. Dunnett’s multiple comparisons test was used in B to obtain the P value. ***, P < 0.001.
Interestingly, in Cep57/Cep57L1-depleted cells with four HsSAS-6 foci, each HsSAS-6 focus was associated with a pair of centrioles (Fig. 7 C), which leads us to reason that the precociously disengaged centrioles newly acquired HsSAS-6 and reduplicated within the same cell cycle. To address this further, we performed live-cell imaging using the HeLa-GFP-centrin1 cell line and revealed that centriole reduplication occurred after the precocious centriole disengagement in interphase (Fig. 7 E). Intriguingly, we noticed that the earlier the precocious centriole disengagement occurred, the more frequently centriole reduplication could be observed (Fig. 2 E). In addition, live-cell imaging data revealed that the centrioles amplified by centriole reduplication could be inherited by the two daughter cells (Fig. 7 E and Video 3). The inherited amplified centrioles could normally duplicate in the subsequent cell cycle, which further led to centrosome numerical abnormalities. Indeed, comparing the number of centrioles between 48 h and 96 h after the siRNA treatment, the numerical abnormalities were more significant at 96 h post-treatment (Fig. 7 F). Overall, these findings demonstrate that precocious centriole disengagement in interphase results in centrosome numerical abnormalities with each passing cell division due to continuous centriole reduplication and the inevitable inheritance of amplified centrioles by daughter cells.
Video 3.
Precocious centriole disengagement results in an increase in the number of centrioles. HeLa-GFP-centrin1 (green) cells were treated with siCep57 and siCep57L1. 25 × 1.2–µm z-stacks were acquired every 10 min at 40× magnification. Image frames shown are maximum-intensity projections at each time point.
Centriole reduplication in Cep57/Cep57L1-depleted cells leads to the high frequency of chromosome segregation errors
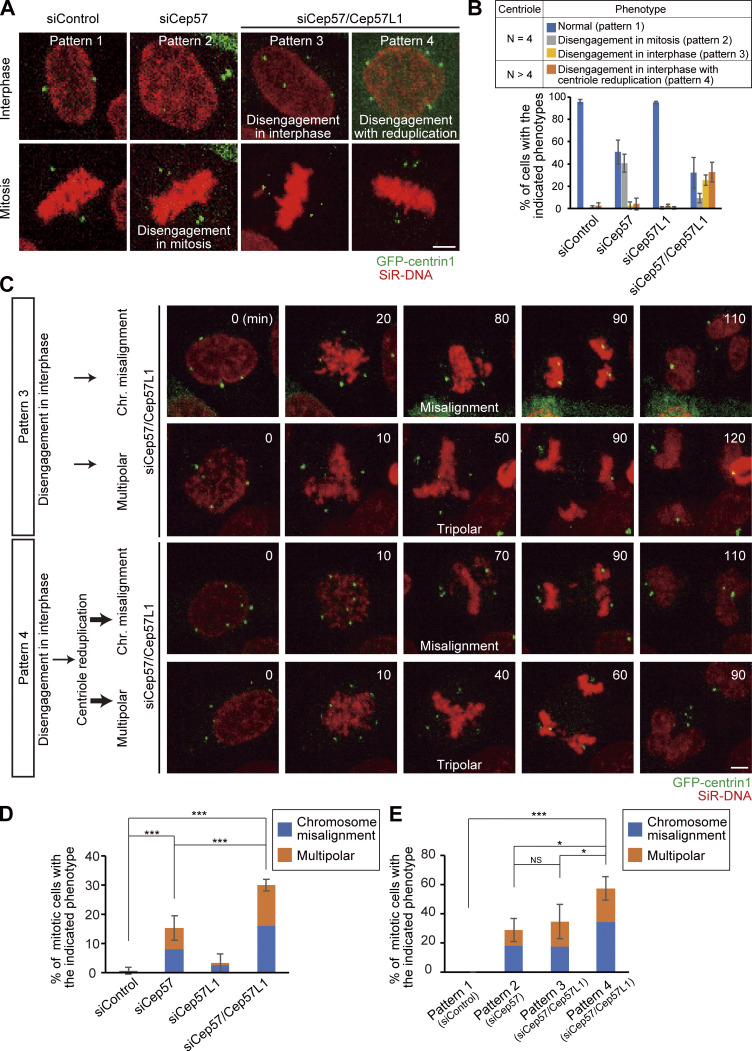
Because precocious centriole disengagement causes defects in chromosome segregation (Watanabe et al., 2019), we then explored the fate of Cep57/Cep57L1-depleted cells in mitosis. We first grouped cells into four categories based on the timing of centriole disengagement and whether centriole reduplication occurred: normal (pattern 1), precocious centriole disengagement in mitosis (pattern 2), precocious centriole disengagement in interphase without centriole reduplication (pattern 3), and precocious centriole disengagement in interphase with centriole reduplication (pattern 4). To track centrioles and chromosomes in the mitotic cells, we performed live-cell imaging using the HeLa-GFP-centrin1 cell line and SiR-DNA, and we quantified the frequency of the phenotypes in each siRNA condition. In control cells or Cep57L1-depleted cells, two pairs of centrioles were engaged throughout the interphase and until the end of mitosis (pattern 1; Fig. 8, A and B; Fig. S5 A). Single depletion of Cep57 caused precocious centriole disengagement only in mitosis, but not in interphase (pattern 2; Fig. 8, A and B). As we mentioned above, most Cep57/Cep57L1-depleted cells exhibited precocious centriole disengagement in interphase (pattern 3), which was frequently followed by centriole reduplication (pattern 4; Fig. 8, A and B). These phenotypic patterns were consistent with what we had observed in the fixed cells (Fig. S5, B and C). We next quantified the frequency of chromosome segregation errors in each siRNA condition. Cep57- or Cep57/Cep57L1-depleted cells exhibited chromosome segregation errors, which were associated with chromosome misalignment and multipolar spindle formation (siControl, 0.7% ± 1.2%; siCep57, 15.3 ± 4.2%; siCep57/Cep57L1, 30.0 ± 2.0%; n = 30; from three independent experiments; Fig. 8, C and D; and Video 4, Video 5, Video 6, and Video 7). The frequency of chromosome segregation errors in Cep57/Cep57L1-depleted cells was about twice that in Cep57-depleted cells. To further investigate what factors most effectively led to the increase of the chromosome segregation errors in Cep57/Cep57L1-depleted cells, we next compared the frequency of chromosome segregation errors according to the phenotypic patterns. We revealed that Cep57/Cep57L1-depleted cells with precocious centriole disengagement and centriole reduplication (pattern 4) showed the highest frequency of chromosome segregation errors (57.5 ± 8.0%; Fig. 8, C and E; Fig. S5, D and E). On the other hand, the frequency of chromosome segregation errors between the cells with precocious centriole disengagement in mitosis (pattern 2, in Cep57-depleted cells, 29.0 ± 8.0%) and interphase (pattern 3, in Cep57/Cep57L1-depleted cells, 34.8 ± 11.6%) were not significantly different (Fig. 8 E). These results suggest that, upon Cep57/Cep57L1 codepletion, the centriole reduplication induced by precocious centriole disengagement in interphase is a more direct cause of chromosome segregation errors than the precocious centriole disengagement itself.
Figure 8.
Amplified centrioles caused by codepletion of Cep57 and Cep57L1 result in high frequency of chromosome segregation errors. (A) Phenotypic patterns were categorized into four groups: normal (pattern 1), precocious centriole disengagement in mitosis (pattern 2), precocious centriole disengagement in interphase without (pattern 3) and with centriole reduplication (pattern 4). HeLa-GFP-centrin1 (green) cells were treated with siControl, siCep57, or siCep57/Cep57L1 in the presence of SiR-DNA (200 nM, red). (B) Histograms represent the frequency of mitotic cells with the indicated phenotypes in A. Values are mean percentages ± SD from three independent experiments (n = 50 for each experiment). (C) Chromosome segregation errors observed in Cep57/Cep57L1-depleted cells with the indicated phenotype. HeLa-GFP-centrin1 (green) cells were treated with siCep57/Cep57L1 in the presence of SiR-DNA (200 nM, red). (D and E) Histograms represent the frequency of the mitotic cells with the indicated chromosome segregation errors observed in C. Values are mean percentages ± SD from three independent experiments (n = 50 for each experiment in D; normal [pattern 1, siControl], n = 48, 47, 49; disengagement in mitosis [Pattern 2, siCep57], n = 25, 18, 18; disengagement in interphase [pattern 3, siCep57/Cep57L1], n = 14, 14, 10; disengagement in interphase with reduplication [pattern 4, siCep57/Cep57L1], n = 21, 16, 12 in E). All scale bars, 5 µm. Tukey’s multiple comparisons test was used in D and E against the total value of chromosome misalignment and multipolar to obtain P values. ***, P < 0.001; *, P < 0.05; NS, P > 0.05.
Figure S5.
Phenotypic analyses of the control and Cep57-, Cep57L1-, and Cep57/Cep57L1-depleted cells in mitosis. (A) Normal bipolar spindle formation (pattern 1) was observed in Cep57L1-depleted cells. HeLa-GFP-centrin1 (green) cells were treated with siCep57L1 in the presence of SiR-DNA (200 nM, red). (B) Phenotypic patterns observed in fixed cells are consistent with live-cell imaging analysis. HeLa cells were treated with siControl, siCep57, siCep57L1, or siCep57/Cep57L1 and immunostained with antibodies against centrin (green) and Cep192 (red). (C) Histograms represent the frequency of mitotic cells with the indicated phenotypes observed in B. Values are mean percentages ± SD from three independent experiments (n = 30 for each experiment). (D) Normal bipolar spindle formation observed in control and Cep57L1-depleted cells. HeLa-GFP-centrin1 (green) cells were treated with siControl or siCep57L1 in the presence of SiR-DNA (200 nM, red). NEBD indicates nuclear envelope breakdown. (E) Chromosome segregation errors observed in Cep57-depleted cells. HeLa-GFP-centrin1 (green) cells were treated with siCep57 in the presence of SiR-DNA (200 nM, red). All scale bars, 5 µm in the low-magnification view, 1 µm in the inset.
Video 4.
Chromosome misalignment in Cep57/Cep57L1-depleted cells with precociously disengaged centrioles. HeLa-GFP-centrin1 (green) cells were treated with siCep57 and siCep57L1 in the presence of SiR-DNA (200 nM, red). 25 × 1.2–µm z-stacks were acquired every 10 min at 40× magnification. Image frames shown are maximum-intensity projections at each time point.
Video 5.
Multipolar spindle formation in Cep57/Cep57L1-depleted cells with precociously disengaged centrioles. HeLa-GFP-centrin1 (green) cells were treated with siCep57 and siCep57L1 in the presence of SiR-DNA (200 nM, red). 25 × 1.2–µm z-stacks were acquired every 10 min at 40× magnification. Image frames shown are maximum-intensity projections at each time point.
Video 6.
Chromosome misalignment in Cep57/Cep57L1-depleted cells with amplified centrioles. HeLa-GFP-centrin1 (green) cells were treated with siCep57 and siCep57L1 in the presence of SiR-DNA (200 nM, red). 25 × 1.2–µm z-stacks were acquired every 10 min at 40× magnification. Image frames shown are maximum-intensity projections at each time point.
Video 7.
Multipolar spindle formation in Cep57/Cep57L1-depleted cells with amplified centrioles. HeLa-GFP-centrin1 (green) cells were treated with siCep57 and siCep57L1 in the presence of SiR-DNA (200 nM, red). 25 × 1.2–µm z-stacks were acquired every 10 min at 40× magnification. Image frames shown are maximum-intensity projections at each time point.
Discussion
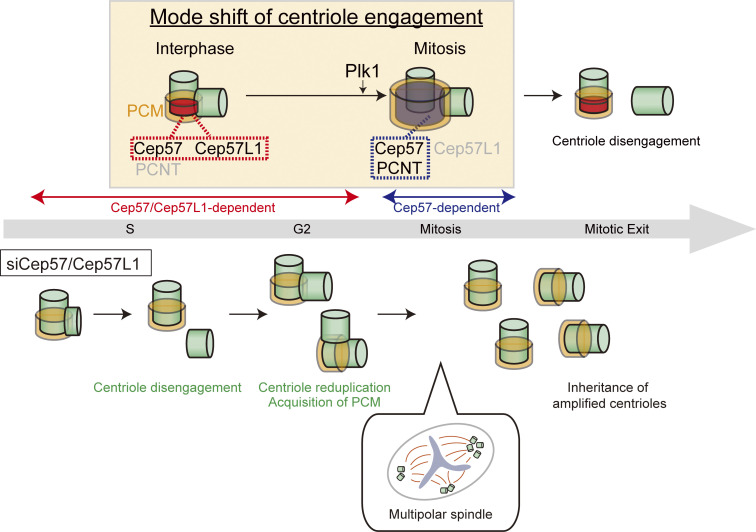
Our work is the first, to our knowledge, to identify Cep57 and Cep57L1 as essential factors maintaining centriole engagement in interphase. In this study, we also show that this tight regulation of centriole engagement is critical for a proper centriole duplication cycle and chromosome segregation (Fig. 9). Surprisingly, codepletion of Cep57 and Cep57L1 induced precocious centriole disengagement in interphase independent of Plk1 activity or cell cycle arrest. Consistent with previous studies (Lončarek et al., 2010), precocious centriole disengagement in interphase released mother centrioles from a block to reduplication and thereby resulted in an increase in the number of centrioles. Furthermore, the number of centrioles per cell increased with each cell division because of the continuous centriole reduplication and inevitable inheritance of the amplified centrioles by daughter cells. In addition, Cep57/Cep57L1-depleted cells exhibited a higher frequency of multipolar spindle formation and chromosome instability mainly because of the amplified centrioles.
Figure 9.
A speculative model for the mode shift of centriole engagement. Centriole engagement in interphase is maintained cooperatively by Cep57 and Cep57L1, and Plk1 changes the engagement mode to a Cep57- and PCNT-dependent one at the mitotic entry. Cep57/Cep57L1 codepletion induces precocious centriole disengagement during interphase, and such disengaged daughter centrioles are converted into centrosomes and can reduplicate before entering mitosis. Consequently, the centriole number increases, which results in the high frequency of chromosome segregation errors.
One interesting issue raised in this study is the difference in the timing of precocious centriole disengagement: Codepletion of Cep57 and Cep57L1 causes the phenotype in interphase, whereas a single depletion of Cep57 causes the phenotype only in mitosis (Fig. 1; Watanabe et al., 2019). These results imply that the mode of centriole engagement somehow shifts from interphase to mitosis. Given that the centriolar localization of Cep57L1 in mitosis was approximately half of that in interphase (Fig. S3, A–D), this might explain why a single depletion of Cep57 is sufficient for inducing the precocious centriole disengagement in mitosis. In this scenario, overexpression of Cep57L1 should rescue the phenotype induced by Cep57 depletion; however, that was not the case (Fig. S3, H and I). Therefore, we assume that the difference in the mode of centriole engagement between interphase and mitosis cannot be explained simply by the change of the expression level of Cep57L1 at centrioles in the cell cycle.
Similar to Cep57, the PCM surrounding centrioles is reported to be important for the maintenance of centriole engagement in mitosis (Seo et al., 2015), and depletion of PCNT, a major component of PCM, disrupts centriole engagement only in mitosis (Fig. 1). Importantly, the PCM expands dynamically and functionally matures toward mitosis (Khodjakov and Rieder, 1999), which is likely a critical event for switching the status of centriole engagement. This PCM maturation at the G2/M transition is known to be a Plk1-dependent process, which includes dynamic reorganization of the PCM from interphase (highly ordered state) to mitosis (amorphous state; Lawo et al., 2012). In addition, a recent study showed that the activity of Plk1 is needed for extending the distance between mother and daughter centrioles from 50 nm to 80 nm in early mitosis (Shukla et al., 2015). Given these observations, we speculate that the Plk1-dependent dynamic rearrangement of PCM components couples the PCM expansion with changes in the status of centriole engagement. In line with this idea, the precocious centriole disengagement was induced only in mitosis by Cep57 depletion, and this could be suppressed by treatment with a Plk1 inhibitor (BI2536; Watanabe et al., 2019). Meanwhile, the precocious centriole disengagement that occurred in the interphase after codepletion of Cep57 and Cep57L1 was not suppressed by Plk1 inhibition. Considering these results, we suggest that Plk1 induces the mode shift of centriole engagement from the Cep57/Cep57L1-dependent mode to Cep57-PCNT–dependent mode at the mitotic entry. However, the critical substrates of Plk1 in this process have not been determined. Given that the functional partner of Cep57 in centriole engagement changes from Cep57L1 to PCNT, we speculate that Cep57, Cep57L1, and PCNT may be critical Plk1 substrates and that phosphorylation of these proteins by Plk1 may alter the organization of these proteins to increase the dependency of the Cep57-PCNT module in mitosis. Unraveling the critical target of Plk1 in triggering this mode shift of centriole engagement will be a fascinating topic for future study.
In Cep57/Cep57L1-depleted cells, we observed the reduction of Cep152 at the centrosomes (Fig. 6). Consistently, recent studies indicated that depletion of Cep57 or codepletion of Cep57 and Cep57L1 decreases the centrosomal localization of Cep152 and therefore affects centriole duplication (Zhao et al., 2020; Wei et al., 2020). However, although previous studies showed that Cep152 serves as a scaffold for Plk4 and is required for centriole duplication (Hatch et al., 2010), we did not detect a significant reduction of the centriole number in Cep57/Cep57L1-depleted cells (Fig. 2 B and Fig. S1 H), which led us to assume that the reduction of Cep152 at the centrosomes was not sufficient to prevent centriole duplication in our experimental condition. To test the effect of the reduction of centrosomal Cep152 on centriole duplication, we depleted Cep152 for 48 h using two siRNAs targeting distinct Cep152 sequences and counted the centriole number in mitosis. As expected, >70% of the Cep152-depleted cells possessed four centrioles, in contrast to the HsSAS-6–depleted cells (siControl, 86.7 ± 3.3%; siCep152 no. 1, 74.4 ± 10.2%; siCep152 no. 2, 75.8 ± 5.2%; siHsSAS-6, 2.2 ± 3.8%; n = 30 from three independent experiments; Fig. S4 E). Similar results were reported by Hatch et al. and Sonnen et al. Hatch et al. showed that depletion of Cep152 for 48 h caused a reduction in centriole number only in 20% of the mitotic cells (Hatch et al., 2010). Sonnen et al. indicated that ∼70% of the interphase cells treated with siCep152 for 72 h possessed more than two centrioles as normal cells and that codepletion of Cep152 and Cep192 caused a more drastic reduction of centriole number (Sonnen et al., 2013). Considering that the centrosomal signal of Cep152 was more effectively reduced by Cep152 depletion than by Cep57/Cep57L1 codepletion (Fig. S4 F), we suggest that the reduction of Cep152 at centrioles in Cep57/Cep57L1-depleted cells was not sufficient to prevent centriole duplication in our experimental condition.
According to the findings in this study, the timing of centriole disengagement in the cell cycle is critical for the occurrence of centriole reduplication. Given that precocious centriole disengagement in Cep57-depleted cells was not accompanied by centriole reduplication, the reloading of centriole duplication factors must be tightly restricted in mitosis so as not to increase the centrosome number. In contrast, in Cep57/Cep57L1-depleted cells, precocious centriole disengagement occurs in interphase, which secures sufficient centriole components and enough time for centriole duplication to occur. Such centriole reduplication thereby results in the drastic increase in centrosome numbers, which then more frequently leads to chromosome segregation errors. Thus, on the basis of these observations, it is conceivable that the tight control of maintenance of centriole engagement is more important in interphase than in mitosis and also that this might be a reason for the redundancy of Cep57 and Cep57L1 in the interphase centriole engagement.
Materials and methods
Cell culture and transfection
HeLa, U2OS, and HEK293T cells were obtained from the European Collection of Authenticated Cell Cultures. All cells were cultured in DMEM supplemented 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a 5% CO2 atmosphere. Transfection of siRNA or DNA constructs into HeLa, U2OS, and HEK293T cells was conducted using Lipofectamine RNAiMAX (Life Technologies) or Lipofectamine 2000 (Life Technologies), respectively. Unless otherwise noted, the transfected cells were analyzed 48 h after transfection with siRNA and 24 h after transfection with DNA constructs. When the cells were analyzed 96 h after transfection with siRNA, additional siRNA was transfected 48 h after the first transfection.
RNAi
The following siRNAs were used: Silencer Select siRNA (Life Technologies) against Cep57 (s18692), Cep57L1 no. 1 (s226224), Cep57L1 no. 2 (s226223), Cep63 (s37123), Cep152 no. 1 (s225921), Cep152 no. 2 (s225922), PCNT (s10138), HsSAS-6 (s45487), and negative control (4390843). Unless otherwise noted, Cep57L1 no. 1 and Cep152 no. 1 were used in this study.
Plasmids
cDNA encoding Cep57L1 isoform 1 (National Center for Biotechnology Information Protein database identifier NP_001258781.1) was amplified from a cDNA library of A549 cells. The Cep57L1 cDNA was subcloned into pCMV5-HA (Addgene) and pCMV5-FLAG (Addgene). pCMV5 constructs encoding full-length Cep57 and pTB701 constructs encoding PCNT that were generated in the previous studies were used in this study (Watanabe et al., 2019; Takahashi et al., 2002). The Cep57L1 deletion mutant constructs were created using the PrimeSTAR mutagenesis basal kit (TaKaRa) and the In-Fusion HD cloning kit (Clontech) according to the manufacturers’ protocols.
The sequences of the primers (sense and antisense, respectively) were 5′-TTCCTGCAGGTCGACGATTCTGAATTAATGCATAGTATAGTAGGAAGCTATCA-3′ and 5′-CCGGGATCAGGATCCTTACTGTTCCCACATGATATCATCTCTTCTC-3′ for subcloning human Cep57L1, 5′-CAATTGGAGTACACAAAGAGAATGGTTCTC-3′ and 5′-TGTGTACTCCAATTGCTTCTCTAGAAGAGT-3′ for constructing RNAi-resistant Cep57L1 mutant, and 5′-GCCCAAATGCAGCACAGTATAAGAAG-3′ and 5′-GTGCTGCATTTGGGCTATGAAGTAT-3′ for constructing Cep57L1 mutant lacking the PINC motif (Δ52–86 aa).
Antibodies
The following primary antibodies were used in this study: rabbit antibodies against Cep57 (GeneTex; GTX115931; IF, 1:1,000; STED, 1:500), Cep57L1 (Proteintech; 24957-1-AP; IF, 1:500; STED, 1:200), Cep63 (Proteintech; 16268-1-AP; IF, 1:1,000), PCNT (Abcam; ab4448; IF, 1:2,000), Cep192 (Bethyl Laboratories; A302-324A; IF, 1:1,000), Cep152 (Bethyl Laboratories; A302-480A; IF, 1:1,000), CP110 (Proteintech; 12780-1-AP; IF, 1:500), ODF2 (Abcam; ab43840; IF, 1:1,000), GFP (MBL; 598; WB, 1:1,000), Cep295 (Merck; HPA038596; IF, 1:500), CENP-F (Abcam; ab108483; IF, 1:500), POC5 (Bethyl Laboratories; A303-341A; IF, 1:1,000), FLAG-tag (Merck; F7425; IF, 1:1,000; WB, 1:1,000), and HA-tag (Abcam; ab9110; IF, 1:1,000; WB, 1:1,000); mouse antibodies against Cep57 (Abcam; ab169301; IF, 1:1,000), PCNT (Abcam; ab28144; IF, 1:1,000; STED, 1:500), centrin (Merck; 20H5; IF, 1:500), EB1 (BD Transduction Laboratories; 610534; IF, 1:1,000), HsSAS-6 (Santa Cruz Biotechnology; sc-81431; IF, 1:300), polyglutamylation modification (GT335; AdipoGen; AG-20-B0020-C100; IF, 1:2,000), γ-tubulin (GTU88; Merck; T5192; IF, 1:1,000), mitosin (BD Transduction Laboratories; 610768; IF, 1:1,000), FLAG-tag (Merck; F3165; IF, 1:1,000; WB, 1:1,000), and α-tubulin (Merck; DM1A; IF, 1:1,000; WB, 1:1,000); goat antibody against GFP (Abcam; ab6662; IF, 1:500; conjugated to FITC); and rat antibody against proliferating cell nuclear antigen (PCNA; Chromotek; 16d10-25; IF, 1:500). Alexa Fluor 488–labeled ODF2 (Abcam; ab43840; IF, 1:500) and Alexa Fluor 647–labeled Cep192 (Bethyl Laboratories; A302–324A; IF, 1:500) were generated with Alexa Fluor labeling kits (Life Technologies). The following secondary antibodies were used: Alexa Fluor 488 goat antimouse IgG (H+L; Molecular Probes; A-11001; 1:1,000), Alexa Fluor 647 goat antimouse IgG (H+L; Abcam; ab150115; 1:1,000), Alexa Fluor 568 goat antirabbit IgG (H+L; Molecular Probes; A-11011; 1:1,000), Alexa Fluor 647 goat antirat IgG (H+L; Invitrogen; A21247; 1:1,000) for IF; goat polyclonal antibodies HRP against mouse IgG (Promega; W402B; 1:10,000) and rabbit IgG (Promega; W401B; 1:10,000) for WB.
Single-guide RNA (sgRNA)
HeLa cells stably expressing Cas9 (HeLa-Cas9) that were generated in the previous study were used in this study (Watanabe et al., 2019). sgRNA oligos targeting Cep57 (forward, 5′-AGCGTCGTAGATCACTATTA-3′; reverse, 5′-TAATAGTGATCTACGACGCT-3′) and Cep57L1 (forward, 5′-GGAACAAATACTCTTTCTGG-3′; reverse, 5′-CCAGAAAGAGTATTTGTTCC-3′) were transcribed in vitro with the HiScribe T7 transcription kit (New England Biolabs) and purified using the RNA Clean and Concentrator kit (Zymo Research). The purified sgRNAs were introduced into HeLa-Cas9 cells using Lipofectamine RNAiMAX (Life Technologies).
RT-qPCR
cDNA was synthesized with the QuantiTect Reverse Transcription Kit (Qiagen) from RNAs extracted from cells 48 h after siRNA treatment using TRIzol reagent (Thermo Fisher Scientific). RT-qPCR was performed using TB Green Premix Ex Taq Tli RNaseH Plus (TaKaRa). All reactions were triplicated, and GAPDH was used as an internal control. The sequences of the primers (sense and antisense, respectively) were 5′-AAGCCTACACTTGCCTATCCAG-3′ and 5′-TTCCAAGCGTCGAATCTTATC-3′ for human Cep57, 5′-GCCTTAGAGAATGAAACAAATGAGAG-3′ and 5′-GAAGAGTACAACGAGACTGGG-3′ for human Cep57L1, and 5′-TCCACTGGCGTCTTCACC-3′ and 5′-GGCAGAGATGATGACCCTTTT-3′ for human GAPDH.
Chemicals
The following chemicals were used in this study: nocodazole (Wako; 31430-18-9), RO3306 (Merck; SML0569), BI2536 (AdooQ Bioscience; A10134), aphidicolin (Merck; A0781), lovastatin (Merck; 438185), and SiR-DNA (Spirochrome; CY-SC007).
Microscopy
For IF analysis, the cells cultured on coverslips (Matsunami; 18 mm, thickness no. 1_0.12-0.17 mm for widefield microscope) were fixed using −20°C methanol for 7 min and washed with PBS. The cells were permeabilized after fixation with PBS/0.05% Triton X-100 (PBSX) for 5 min twice and incubated for blocking in 1% BSA in PBSX for 30 min at RT. The cells were incubated with primary antibodies for 24 h at 4°C or 2 h at RT, washed with PBSX three times, and incubated with secondary antibodies for 1 h at RT. The cells were thereafter washed with PBSX twice, stained with 0.2 µg/ml Hoechst 33258 (Dojindo, shown in blue in all IF figures) in PBS for 5 min at RT, washed again with PBSX, and mounted onto glass slides with ProLong Gold (P36930; Thermo Fisher Scientific).
Acquiring the images and counting the number of IF signals were preformed using an Axioplan2 fluorescence microscope (Carl Zeiss) with 63× or 100×/1.4 NA plan apochromatic lens objective.
STED images were acquired by using a Leica TCS SP8 STED 3X system with a Leica HC PL APO 100×/1.40 NA OIL STED WHITE objective and 660-nm gated STED. Scan speed was set to 400 Hz in combination with threefold line average in a 1,024 × 1,024 format (pixel size, 14.2 nm). The resolution of green signal (−80 nm; Alexa Fluor 488) is generally lower than that of red signal (−50 nm; Alexa Fluor 555) in this system. In this study, we mainly looked at the proteins of interest at centrioles in red, whereas the other protein was visualized in green.
Live-cell imaging
A Cell Voyager CQ1 system (Yokogawa Electric Corp.) equipped with a 40× dry objective lens was used for live-cell imaging. HeLa cells stably expressing GFP-centrin1 generated in the previous study(Tsuchiya et al., 2016) were cultured on 24-well glass-bottomed plate (Greiner Bio-One; 662892) at 37°C in a 5% CO2 atmosphere. Before imaging, cells were treated with siRNAs for 24 h or 72 h and with 200 nM of SiR-DNA for 6 h. Images were obtained by using a scientific complementary metal-oxide semiconductor camera. After 24 or 72 h from transfection, the cells were visualized every 10 min over 24 h or 48 h. The images were collected at 1.2-µm z steps. Maximum projections were generated using ImageJ (National Institutes of Health).
IP and Western blotting (WB)
For preparation of human cell lysates for WB, HEK293T cells were collected 24 h after transfection, lysed on ice in lysis buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1% Triton X-100, 5 mM EGTA, 1 mM DTT, 2 mM MgCl2, and 1:1,000 protease inhibitor cocktail [Nakarai Tesque]). Insoluble material was removed after centrifugation for 10 min. For IP of FLAG-tagged proteins, whole-cell lysates were incubated with FLAG antibody–conjugated M2 agarose gel (Merck) for 2 h at 4°C. The beads were washed at least three times with lysis buffer and resuspended in SDS sample buffer before being loaded onto 8% polyacrylamide gels, followed by transfer onto Immobilon-P membrane (Merck). The membrane was probed with the primary antibodies, followed by incubation with their respective HRP-conjugated secondary antibodies (Promega). The membrane was soaked with Amersham ECL Prime (GE Healthcare) or Chemi-Lumi One Ultra (Nakarai Tesque). Washes were performed in PBS containing 0.02% Tween-20. The signal was detected with ChemiDoc XRS+ (Bio-Rad Laboratories). Unless otherwise specified, the WB experiments were repeated at least three times. The antibody against α-tubulin was used as a loading control.
Statistical analysis
The normal distribution of datasets was confirmed using the Kolmogorov-Smirnov test (Fig. 2 E; Fig. 6 D; Fig. S2 C; Fig. S3, B and D; and Fig. S4 F). Homoscedasticity of the data was assumed, but this was not formally tested. Statistical comparison between two datasets was conducted using the two-tailed, unpaired Student’s t test (Fig. 1 B; Fig. 2 E; Fig. 4, F and G; Fig. S1 H; Fig. S2 C; Fig. S3, B and D; and Fig. S4 I). Multiple comparison was conducted using Tukey’s multiple comparisons test (Fig. 4 D; Fig. 8, D and E; Fig. S4, E and F; and Fig. 6 D) or Dunnett’s multiple comparisons test (Fig. 1, D, E, F, and H; and Fig. 7 B). The P values are shown in the figures as ***, P < 0.001; **, P < 0.01; *, P < 0.05; or NS, P > 0.05. Means and errors (SD) were calculated using Excel software. Student’s t tests were performed using Excel software. Kolmogorov-Smirnov tests, Dunnett’s multiple comparisons tests, and Tukey’s multiple comparisons tests were performed using R.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Online supplemental material
Fig. S1 shows the confirmation of the phenotype in Cep57/Cep57L1-depleted cells. Fig. S2 shows the phenotypic analyses of the cell cycle in Cep57/Cep57L1-depleted cells. Fig. S3 shows the localization analyses of Cep57 and Cep57L1. Fig. S4 shows the molecular properties of Cep57 and Cep57L1. Fig. S5 shows the phenotypic analyses of the control and Cep57-, Cep57L1-, and Cep57/Cep57L1-depleted cells in mitosis. Video 1 shows the precocious centriole disengagement in Cep57/Cep57L1-depleted cells. Video 2 shows the movement of the disengaged centrioles in Cep57/Cep57L1-depleted cells. Video 3 shows the increase in the centriole number after precocious centriole disengagement in Cep57/Cep57L1-depleted cells. Video 4, Video 5, Video 6, and Video 7 show the chromosome segregation errors in Cep57/Cep57L1-depleted cells with (Video 6 and Video 7) and without (Video 4 and Video 5) amplified centrioles.
Supplementary Material
Acknowledgments
We gratefully acknowledge A. Sekigawa for technical support in structured illumination microscopy and the members of the Kitagawa laboratory for technical support and discussion.
This work was supported by a grant-in-aid for scientific research (S, 19H05651) and research activity start-up from the Ministry of Education, Culture, Sports, Science and Technology; by the Takeda Science Foundation; by the Japan Science Society (Sasakawa Scientific Research Grant); and by the Daiichi Sankyo Foundation of Life Science.
The authors declare no competing financial interests.
Author contributions: K.K. Ito, K. Watanabe, and D. Kitagawa designed the study; K.K. Ito, K. Watanabe, K. Matsuhashi, T. Chinen, S. Hata, and D. Kitagawa designed experiments; K.K. Ito, K. Watanabe, and H. Ishida performed experiments; K.K. Ito, K. Watanabe, and H. Ishida analyzed data; K.K. Ito, K. Watanabe, and D. Kitagawa wrote the manuscript, which was commented on by all authors.
References
- Azimzadeh, J., Hergert P., Delouvée A., Euteneuer U., Formstecher E., Khodjakov A., and Bornens M.. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185:101–114. 10.1083/jcb.200808082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, K., Sieben C.J., Jeganathan K.B., Hamada M., Davies B.A., Velasco R.O.F., Rahman N., Katzmann D.J., and van Deursen J.M.. 2018. Mosaic-variegated aneuploidy syndrome mutation or haploinsufficiency in Cep57 impairs tumor suppression. J. Clin. Invest. 128:3517–3534. 10.1172/JCI120316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., Hildebrandt F., Pellman D., Woods G., and Godinho S.A.. 2011. Centrosomes and cilia in human disease. Trends Genet. 27:307–315. 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N.J., Marjanović M., Lüders J., Stracker T.H., and Costanzo V.. 2013. Cep63 and Cep152 cooperate to ensure centriole duplication. PLoS One. 8:e69986. 10.1371/journal.pone.0069986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.-W., Hsu W.-B., Tsai J.-J., Tang C.-J.C., and Tang T.K.. 2016. CEP295 interacts with microtubules and is required for centriole elongation. J. Cell Sci. 129:2501–2513. 10.1242/jcs.186338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Wainman A., and Raff J.W.. 2015. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16:611–624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- Douthwright, S., and Sluder G.. 2014. Link between DNA damage and centriole disengagement/reduplication in untransformed human cells. J. Cell. Physiol. 229:1427–1436. 10.1002/jcp.24579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., Lipinszki Z., Rangone H., Min M., Mykura C., Chao-Chu J., Schneider S., Dzhindzhev N.S., Gottardo M., Riparbelli M.G., et al. 2016. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 18:87–99. 10.1038/ncb3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy, P., and Hatzopoulos G.N.. 2019. Centriole assembly at a glance. J. Cell Sci. 132:jcs228833. 10.1242/jcs.228833 [DOI] [PubMed] [Google Scholar]
- Hatch, E.M., Kulukian A., Holland A.J., Cleveland D.W., and Stearns T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, A.L., Wise S.S., Pelsue S.C., Aboueissa A.-M., Lingle W., Salisbury J., Gallagher J., and Wise J.P. Sr. 2010. Chronic exposure to zinc chromate induces centrosome amplification and spindle assembly checkpoint bypass in human lung fibroblasts. Chem. Res. Toxicol. 23:386–395. 10.1021/tx900360w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanç, B., Dodson H., and Morrison C.G.. 2010. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol. Biol. Cell. 21:3866–3877. 10.1091/mbc.e10-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and Rieder C.L.. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. 10.1083/jcb.146.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Lee K., and Rhee K.. 2015. PLK1 regulation of PCNT cleavage ensures fidelity of centriole separation during mitotic exit. Nat. Commun. 6:10076. 10.1038/ncomms10076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.-S., Zhang L., Il Ahn J., Meng L., Chen Y., Lee E., Bang J.K., Lim J.M., Ghirlando R., Fan L., et al. 2019. Molecular architecture of a cylindrical self-assembly at human centrosomes. Nat. Commun. 10:1151. 10.1038/s41467-019-08838-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo, S., Hasegan M., Gupta G.D., and Pelletier L.. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158. 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Lee, K., and Rhee K.. 2012. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle. 11:2476–2485. 10.4161/cc.20878 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Li Y., March M.E., Nguyen K., Xu K., Wang F., Guo Y., Keating B., Glessner J., Li J., et al. 2015. Copy number variation in CEP57L1 predisposes to congenital absence of bilateral ACL and PCL ligaments. Hum. Genomics. 9:31. 10.1186/s40246-015-0053-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lončarek, J., Hergert P., and Khodjakov A.. 2010. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 20:1277–1282. 10.1016/j.cub.2010.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius, G., Lavogina D., Orpinell M., Umezawa K., Reymond L., Garin N., Gönczy P., and Johnsson K.. 2013. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr. Biol. 23:265–270. 10.1016/j.cub.2012.12.030 [DOI] [PubMed] [Google Scholar]
- Martino, J., Holmes A.L., Xie H., Wise S.S., and Wise J.P. Sr. 2015. Chronic exposure to particulate chromate induces premature centrosome separation and centriole disengagement in human lung cells. Toxicol. Sci. 147:490–499. 10.1093/toxsci/kfv146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, K., Ohsumi K., Iwabuchi M., Kawamata T., Ono Y., and Takahashi M.. 2012. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr. Biol. 22:915–921. 10.1016/j.cub.2012.03.048 [DOI] [PubMed] [Google Scholar]
- Mennella, V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168. 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momotani, K., Khromov A.S., Miyake T., Stukenberg P.T., and Somlyo A.V.. 2008. Cep57, a multidomain protein with unique microtubule and centrosomal localization domains. Biochem. J. 412:265–273. 10.1042/BJ20071501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A., and Holland A.J.. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19:297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry, S. 2016. Mechanisms of mitotic spindle assembly. Annu. Rev. Biochem. 85:659–683. 10.1146/annurev-biochem-060815-014528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser, S.L., Samant M.D., Baxter J.E., Morrison C.G., and Fry A.M.. 2012. Oscillation of APC/C activity during cell cycle arrest promotes centrosome amplification. J. Cell Sci. 125:5353–5368. 10.1242/jcs.106096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker, S., and Sumara I.. 2014. Molecular dynamics of PLK1 during mitosis. Mol. Cell. Oncol. 1:e954507. 10.1080/23723548.2014.954507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, M.Y., Jang W., and Rhee K.. 2015. Integrity of the pericentriolar material is essential for maintaining centriole association during m phase. PLoS One. 10:e0138905. 10.1371/journal.pone.0138905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, A., Kong D., Sharma M., Magidson V., and Loncarek J.. 2015. Plk1 relieves centriole block to reduplication by promoting daughter centriole maturation. Nat. Commun. 6:8077. 10.1038/ncomms9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen, K.F., Gabryjonczyk A.-M., Anselm E., Stierhof Y.-D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Takahashi, M., Yamagiwa A., Nishimura T., Mukai H., and Ono Y.. 2002. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring γ-tubulin ring complex. Mol. Biol. Cell. 13:3235–3245. 10.1091/mbc.e02-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou, M.-F.B., Wang W.-J., George K.A., Uryu K., Stearns T., and Jallepalli P.V.. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 17:344–354. 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, Y., Yoshiba S., Gupta A., Watanabe K., and Kitagawa D.. 2016. Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat. Commun. 7:12567. 10.1038/ncomms12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.-J., Soni R.K., Uryu K., and Tsou M.F.. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193:727–739. 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., Takao D., Ito K.K., Takahashi M., and Kitagawa D.. 2019. The Cep57-pericentrin module organizes PCM expansion and centriole engagement. Nat. Commun. 10:931. 10.1038/s41467-019-08862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z., Kim T.-S., Ahn J.I., Meng L., Chen Y., Ryu E.K., Ku B., Zhou M., Kim S.J., Bang J.K., et al. 2020. Requirement of the Cep57-Cep63 interaction for proper Cep152 recruitment and centriole duplication. Mol. Cell. Biol. 40:e00535-19. 10.1128/MCB.00535-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q., He R., Zhou H., Yu A.C.H., Zhang B., Teng J., and Chen J.. 2012. Cep57, a NEDD1-binding pericentriolar material component, is essential for spindle pole integrity. Cell Res. 22:1390–1401. 10.1038/cr.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Yang S., Chen Q., Duan X., Li G., Huang Q., Zhu X., and Yan X.. 2020. Cep57 and Cep57l1 function redundantly to recruit the Cep63-Cep152 complex for centriole biogenesis. J. Cell Sci. 133:jcs241836. 10.1242/jcs.241836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.