Abstract
Background and aim
Coronavirus Disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has now become the world pandemic. There is a race to develop suitable drugs and vaccines for the disease. The anti-HIV protease drugs are currently repurposed for the potential treatment of COVID-19. The drugs were primarily screened against the SARS-CoV-2 main protease. With an urgent need for safe and effective drugs to treat the virus, we have explored natural products isolated from edible and medicinal mushrooms that have been reported to possess anti-HIV protease.
Experimental procedures
We have examined 36 compounds for their potential to be SARS-CoV-2 main protease inhibitors using molecular docking study. Moreover, drug-likeness properties including absorption, distribution, metabolism, excretion and toxicity were evaluated by in silico ADMET analysis.
Results
Our AutoDock study showed that 25 of 36 candidate compounds have the potential to inhibit the main viral protease based on their binding affinity against the enzyme’s active site when compared to the standard drugs. Interestingly, ADMET analysis and toxicity prediction revealed that 6 out of 25 compounds are the best drug-like property candidates, including colossolactone VIII, colossolactone E, colossolactone G, ergosterol, heliantriol F and velutin.
Conclusion
Our study highlights the potential of existing mushroom-derived natural compounds for further investigation and possibly can be used to fight against SARS-CoV-2 infection.
Taxonomy (classification by evise)
Disease, Infectious Disease, Respiratory System Disease, Covid-19, Traditional Medicine, Traditional Herbal Medicine, Phamaceutical Analysis.
Keywords: COVID-19, Anti-HIV protease, Anti-SARS-CoV-2 main protease, Mushrooms, Molecular docking, ADMET analysis
Graphical abstract
Highlights
-
•
25 compounds from mushroom exhibited good binding affinities against the SARS-CoV-2 main protease using molecular docking.
-
•
ADMET analysis revealed the drug likeliness, toxicity, carcinogenicity and mutagenicity of studied compounds.
-
•
Ergosterol, heliantriol F, velutin, colossolactone VIII, E and G are promising SAR-CoV-2 main protease inhibitors.
List of abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
- COVID-19
Coronavirus Disease 2019
- HIV
Human immunodeficiency virus
- ADME
Absorption Distribution Metabolism and Excretion
- PDB
Protein Data Bank
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), also known as 2019 novel coronavirus (2019-nCoV), has quickly spread worldwide since late 2019. Coronavirus Disease 2019 (COVID-19) has emerged as a severe global pandemic. As per the recent situational report released by the World Health Organization (WHO) on 27 December 2020, the disease has caused 1.7 million deaths and 78 million infections worldwide.1 The disease’s clinical characteristics can range from asymptomatic infections, mild respiratory disease, severe pneumonia with respiratory failure, and even fatal respiratory diseases such as acute respiratory distress syndrome.2,3
SARS-CoV-2 belongs to the family of the coronavirus. It is an enveloped virus with a positive-sense single-stranded RNA and a single linear RNA segment.4 SARS-CoV-2 has been linked to two other strains, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), in the zoonotic origins and the cause of fatal pneumonia.4,5 The main viral protease has been proposed as a key therapeutic target for drug development against coronavirus and COVID-19 treatment.6, 7, 8, 9, 10, 11 The enzyme plays an essential role in the viral life cycle, mostly involving maturation cleavage events within several precursor proteins.8,12 The active sites of this enzyme are highly conserved among all coronaviruses.13 Furthermore, the main viral protease is a non-human homologue, making it an ideal antiviral target.13
Unfortunately, there is no anti-SARS-CoV-2 drug or vaccine sufficiently available to date. The urgent need for drugs to treat COVID-19 has led scientists to focus on protease inhibitors as potential drugs to cure the disease. Repurposing drugs currently used to treat HIV, HCV and SARS, taking advantage of drug safety, is one of the current anti-SARS-CoV-2 drug searching approaches. Repurposing HIV-drugs have been tested against SARS-CoV-2 main protease and shown effectiveness against the viral enzyme.14,15 Moreover, the combination of anti-HIV protease drugs, lopinavir and ritonavir, was currently employed in a clinical trial against COVID-19 in patients with mild and moderate COVID-19. 16
Lopinavir and Ritonavir, the approved drugs for HIV protease inhibitor, have been reported to inhibit the main protease of both SARS-CoV and MERS-CoV.17, 18, 19, 20, 21 With the urgent need to reduce the impact of COVID-19, a combination of these drugs was currently investigated to treat SARS-CoV-2-infected patients.20,22, 23, 24, 25 However, these drugs’ curative effect remains minimal, with potentially toxic side effects that might be harmful to the patients.25 Identifying bioactive compounds from natural sources that can inhibit SARS-CoV-2 main protease is considered an alternative approach to combat COVID-19. 26, 27, 28, 29 In silico technique, a computational approach, is the promising preliminary evidence for drug discovery.30, 31, 32, 33, 34 Several molecular docking studies have identified bioactive compounds from natural products as the potential SARS-CoV-2 main protease inhibitors derived from natural sources.35, 36, 37, 38, 39, 40
Mushrooms are a rich source of bioactive compounds with antiviral activity.41,42 Some of the compounds showed anti-inflammatory activity.43, 44, 45 These dual activities would be an effective treatment of COVID-19. A number of bioactive compounds have been shown to inhibit HIV protease, suggesting their potential activity against the main proteases of coronaviruses.42 In this work, we carried out an in silico investigation of bioactive compounds found in edible and medicinal mushrooms. These compounds have been shown to display anti-HIV protease activity. We examined 36 compounds for their potential candidates as SARS-CoV-2 main protease inhibitors and investigated their drug-like properties using molecular docking study.
2. Materials and methods
2.1. Ligand preparation
A total of 36 mushroom-isolated compounds were selected from the literature, including those listed by Suwannarach et al.42 (Table 1). Compounds No. 1–32 were reported for their anti-HIV protease property, whereas compound No. 33–36 showed anti-HIV activity via HIV reverse transcriptase inhibition. All chemical structures were obtained from the PubChem database. Prior to docking studies, each compound was loaded into Discovery Studio (DS) Visualizer (BIOVIA, CA, USA) and optimized using "Clean Geometry" function in DS. All compounds’ files were converted to PDBQT format using AutoDockTools-1.5.6 software (The Scripps Research Institute, USA) for docking studies.
Table 1.
List of anti- HIV bioactive compounds derived from mushrooms.
| No. | Compound | Structure | Source | Reference |
|---|---|---|---|---|
| 1 | 20(21)-dehydrolucidenic acid N | 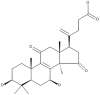 |
Ganoderma sinense | 46 |
| 2 | 20-hydroxylucidenic acid N | 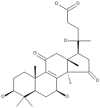 |
Ganoderma sinense | 46 |
| 3 | Agrocybin | 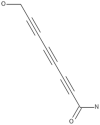 |
Agrocybe cylindracea | 47 |
| 4 | Betulinic acid | 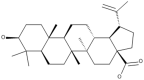 |
Trametes versicolor | 48,49 |
| 5 | Colossolactone A |  |
Ganoderma colosum | 50 |
| 6 | Colossolactone E |  |
Ganoderma colosum | 50 |
| 7 | Colossolactone G |  |
Ganoderma colosum | 50 |
| 8 | Colossolactone V |  |
Ganoderma colosum | 50 |
| 9 | Colossolactone VII | 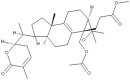 |
Ganoderma colosum | 50 |
| 10 | Colossolactone VIII | 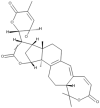 |
Ganoderma colosum | 50 |
| 11 | Ellagic acid | 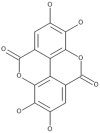 |
Flammulina velutipes, Phellinus linteus, Pleurotus eryngii |
51, 52, 53, 54 |
| 12 | Ergosterol | 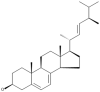 |
Auricularia polytricha, Flammulina velutipes, Lentinula edodes |
55, 56, 57 |
| 13 | Gallic acid | 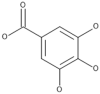 |
Agaricus bisporus, Flammulina velutipes, Ganoderma lucidum, Laetiporus sulphureus, Lentinus lepideus, Leucoagaricus leucothites, Macrocybe gigantea, Pleurotus ostreatus |
51,58, 59, 60, 61, 62 |
| 14 | Ganoderic acid alpha | 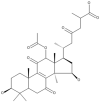 |
Ganoderma lucidum | 63 |
| 15 | Ganoderic acid B | 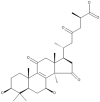 |
Ganoderma lucidum | 63,64 |
| 16 | Ganoderic acid beta | 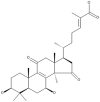 |
Ganoderma lucidum | 65 |
| 17 | Ganoderic acid C1 | 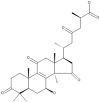 |
Ganoderma lucidum | 63 |
| 18 | Ganoderic acid GS-2 | 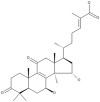 |
Ganoderma sinense | 46 |
| 19 | Ganoderic acid H | 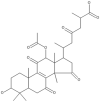 |
Ganoderma lucidum | 63 |
| 20 | Ganoderiol A | 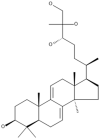 |
Ganoderma lucidum | 63 |
| 21 | Ganoderiol B |  |
Ganoderma lucidum | 63 |
| 22 | Ganoderiol F |  |
Ganoderma lucidum, Ganoderma sinense | 46,63 |
| 23 | Ganodermanondiol |  |
Ganoderma lucidum | 65 |
| 24 | Ganodermanontriol |  |
Ganoderma lucidum | 63,65 |
| 25 | Ganolucidic acid A |  |
Ganoderma lucidum | 65 |
| 26 | Ganomycin B |  |
Ganoderma colosum | 50,66 |
| 27 | Ganomycin I |  |
Ganoderma colosum | 50,66 |
| 28 | Heliantriol F |  |
Lignosus rhinocerus | 67 |
| 29 | Linoleic acid |  |
Auricularia polytricha, Lentinula edodes |
55,57 |
| 30 | Lucidumol B |  |
Ganoderma lucidum | 65 |
| 31 | Methyl gallate |  |
Pholiota adiposa | 68 |
| 32 | Oleanolic acid |  |
Hericium erinaceus, Lactarius deliciosus, Lactarius sanguifluus, Lactarius semisanguifluus Russula delica, Suillus bellinii |
69, 70, 71 |
| 33 | Semicochliodinol A | 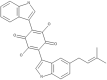 |
Chrysosporium merdarium | 72 |
| 34 | Semicochliodinol B | 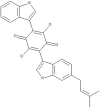 |
Chrysosporium merdarium | 72 |
| 35 | Ursolic acid | 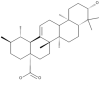 |
Cynomorium songaricum, Hericium erinaceus, Russula delica, Suillus bellinii |
70,71,73 |
| 36 | Velutin | 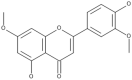 |
Flammulina velutipes | 74 |
2.2. Protein preparation
The crystal structure of COVID-19 main protease in complex with an inhibitor N3 (PDB ID: 6LU7)6 was obtained from RCSB Protein Data Bank. Before the docking studies, the protein structure was first prepared using the Prepare protein set up in AutoDock. The protein preparation is an optimization step that corrects structure, atomic and bond length and charges anomalies. Water molecules and the original inhibitor were removed from the protein structure. Then any missing hydrogen atoms were added. The optimization step was then employed to provide the stable conformation before converting to PDBQT format for docking analysis.
2.3. Molecular docking
Molecular docking studies were performed using the default protocol in Autodock tools 1.5.6 (AutoDock 4.2 software, The Scripps Research Institute, USA). The active site of SARS-CoV-2 main protease 6LU7 was set as a grid box. The grid box was set to 50 x 50 x 50 points in xyz-dimension that equated to a grid box spacing of 0.375 Å3, and the coordinate of the x, y and z centers of the box are at −9.1, 11.0 and 68.0, respectively. The docking simulations were performed using the Lamarckian Genetic Algorithm with default parameters, including 10 Genetic Algorithm runs. The docking results of 10 runs were ranked by energy. The compound with the best energy ranking was further closely analyzed for its protein-ligand interactions using the DS Visualizer (BIOVIA, San Diego, CA, USA).
2.4. Drug-likeness prediction
The structures of compounds presented in mushrooms (Table 1) were obtained from PubChem database in SMILE files. The compounds were then evaluated for drug-likeness using the Lipinski’s rule of five to predict their pharmacokinetic properties such as the Absorption, Distribution, Metabolism and Excretion (ADME) of the compounds using SwissADME (http://www.swissadme.ch/).75
2.5. Toxicity, carcinogenicity and mutagenicity prediction
The canonical SMILES of the list of compounds in Table 1 was further submitted to the pkCSM database and Toxtree software to predict their toxicity and carcinogenicity, respectively. The toxicity was predicted via toxicity mode in pkCSM database,76 while carcinogenicity and mutagenicity prediction was analyzed using Toxtree 3.1.0 software based on the Benigni-Bossa rule.77,78
3. Results and discussion
3.1. Investigation of mushroom-derived bioactive compounds against SARS-CoV-2 main protease by molecular docking
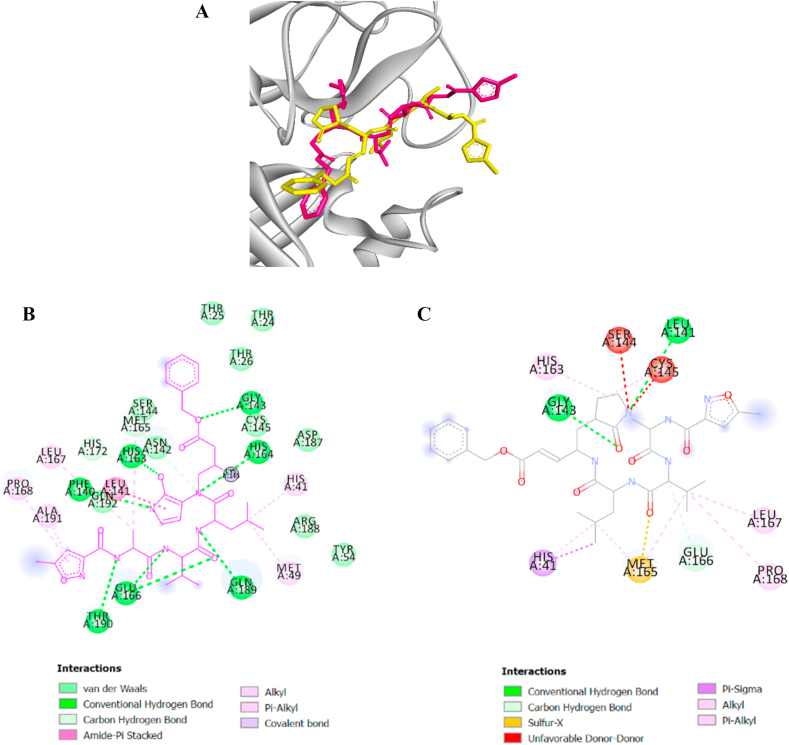
The crystal structure of COVID-19 main protease in complex with the inhibitor N3 (PDB ID: 6LU7 obtained on 21 June 2020) 6 was obtained from RCSB Protein Data Bank. The protein structure was prepared for the docking studies by removing water molecules and the inhibitor before optimization step to provide the stable conformation. The docking protocol was validated using the standard set up in AutoDock. The minimized structure of the inhibitor N3 was re-docked into the original binding site of the protein. The results of 10 Genetic Algorithm runs of the re-docking were ranked and shown in Table 2. The best docking score of N3 was re-docked structure 1 which showed binding energy at −7.97 kcal/mol. The conformation, orientation, position and interactions of the redocked N3 at the binding site were analyzed and compared to that found in the original X-ray structure (PDB ID: 6LU7). Superimposing of the redocked protein-N3 over the original X-ray structure indicated that N3 was able to seek out the location of the receptor site. It is noteworthy that the re-docked N3 has adopted a slightly different conformation, specifically at the 5-methylisoxazole-3-carboxamide part of the molecule compared to that found in the X-ray structure, as shown in Fig. 1B. The variation in conformation, perhaps due to the stable conformation of N3 adopted in silico.The best re-docked N3 was found to interact with only two active site amino acid residues (GLY143 and GLU166) out of nine different residues observed in the X-ray structures. The common key interactions are shown in Table 2. This indicates that N3 was able to recognize its original receptor site in the X-ray structure of 6LU7, using this docking protocol. The results above validated the docking protocol as a suitable method for this study.
Table 2.
Hydrogen bond interaction between N3 and 6LU7.
| 6LU7-N3 interaction | Binding energy (kcal/mol) | Hydrogen bonding |
3D diagrams | |
|---|---|---|---|---|
| Number | Amino acid interaction | |||
| X-ray structure | – | 10 | GLY143, THR190, GLN189, PHE140, HIS163, HIS164, MET165, HIS172, GLU166 (2) | |
| Re-docked structure 1 | −7.97 | 3 | GLY143, LEU141, GLU166 | 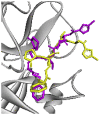 |
| Re-docked structure 2 | −6.28 | 2 | GLN189, THR190 | 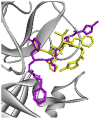 |
| Re-docked structure 3 | −5.89 | 5 | ASN142, HIS163, GLU166, GLY143, CYS145 | 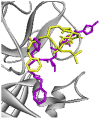 |
| Re-docked structure 4 | −5.86 | 2 | CYS145, THR190 | 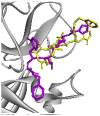 |
| Re-docked structure 5 | −5.85 | 2 | GLU166, GLN189 | 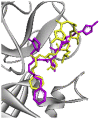 |
| Re-docked structure 6 | −5.65 | 5 | HIS41, CYS145, GLU166, GLN189, MET165 |
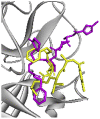 |
| Re-docked structure 7 | −5.03 | 3 | GLN189, PRO168, GLY143 | 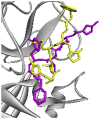 |
| Re-docked structure 8 | −4.36 | 6 | ASN142, CYS145, HIS163, SER46, MET165, HIS172 | 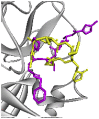 |
| Re-docked structure 9 | −4.30 | 4 | GLU166, PRO168, GLU166(2) |  |
| Re-docked structure 10 | −3.55 | 3 | ASN142, GLU166(2) |  |
The 10 Genetic Algorithm runs on the re-docking of 6LU7-N3 using the validated method were shown. The results were ranked by binding energy. The lowest binding energy showed the best overlapping between N3 ligand from crystal (violet) and re-docking via AutoDock (yellow).
Fig. 1.
The results from validation method for molecular docking study of SARS-CoV-2 main protease (6LU7). (A) The 3D diagrams of interaction between N3 and 6LU7 at the active site demonstrated overlapping between N3 ligand from crystal (magenta) and re-docking via AutoDock (yellow). (B) and (C) displayed the 2D ligands-receptor interactions of N3 from crystal and re-docking ligand via AutoDock structure with 6LU7, respectively.
The validated docking protocol was subsequently used for in silico screening of potential compounds as inhibitors against SARS-CoV-2 main protease. The in silico inhibition of SARS-CoV-2 main protease is gauged on the potential binding affinity of the compounds. The binding energy expressed the binding affinity as Gibbs free energy by which the compounds displayed the higher negative binding energy were considered betters.79 The in silico binding affinities of Lopinavir and Ritonavir to 6LU7 were determined. Since Lopinavir and Ritonavir are anti-HIV protease drugs that have been currently used in hospitals for COVID-19 treatment.22, 23, 24, 25 Therefore, it is appropriate to use them as standard drugs in addition to N3 for comparative study. Lopinavir and Ritonavir showed binding energy at −7.41 and −6.19 kcal/mol against 6LU7, respectively. Andrographolide and kaempferol have been previously demonstrated the potential inhibition against 6LU7 via docking studies.36,80 They were also docked in this study to confirm our validated method. Our docking results agreed with recent reports that both andrographolide and kaempferol had lower binding energy (−7.93 and −7.99 kcal/mol, respectively) than that of anti-HIV agents, indicating their potency to inhibit SARS-CoV-2 main protease.
The docking scores of all compounds are presented in Table 3. Out of 36 compounds, seventeen compounds (No.1–17) showed lower binding energy (<−7.97 kcal/mol) binding to 6LU7 than that of original inhibitor (N3), Lopinavir and Ritonavir. Notably, 25 out of 36 mushroom-derived compounds (No.1–25) showed lower binding energy against 6LU7 than that of lopinavir, while 30 out of 36 compounds (No.1–30) showed lower binding energy than that of Ritonavir. From these results, colossolactone VIII exhibited the lowest binding energy against SARS-CoV-2 protease (−10.55 kcal/mol) followed by colossolactone E, colossolactone G, ergosterol, semicochliodinol A and heliantriol F (−10.10, −9.86, −9.49, −8.91 and −8.91 kcal/mol, respectively). Consistent with previous reports,81, 82, 83 our results from molecular docking found that ursolic acid, oleanolic acid and betulinic acid also exhibited the potential to inhibit SARS-CoV-2 main protease.
Table 3.
Docking results of all mushroom-derived compounds with the SARS-CoV-2 main protease.
| No | Compound | Binding energy (kcal/mol) | Ligand Efficiency |
Inhibition constant (Kpi) |
|---|---|---|---|---|
| N3 (original inhibitor) | −7.97 | −0.16 | 1.44 μM | |
| Lopinavir | −7.41 | −0.16 | 3.32 μM | |
| Ritonavir | −6.19 | −0.12 | 28.94 μM | |
| Andrographolide | −7.93 | −0.32 | 1.55 μM | |
| Kaempferol | −7.99 | −0.38 | 1.38 μM | |
| 1 | Colossolactone VIII | −10.55 | −0.27 | 18.58 nM |
| 2 | Colossolactone E | −10.10 | −0.27 | 39.58 nM |
| 3 | Colossolactone G | −9.86 | −0.25 | 58.86 nM |
| 4 | Ergosterol | −9.49 | −0.33 | 111.07 nM |
| 5 | Semicochliodinol A | −8.91 | −0.27 | 294.8 nM |
| 6 | Heliantriol F | −8.91 | −0.27 | 292.54 nM |
| 7 | Semicochliodinol B | −8.71 | −0.26 | 412.95 nM |
| 8 | Ganoderic acid GS-2 | −8.68 | −0.24 | 433.02 nM |
| 9 | Lucidumol B | −8.64 | −0.26 | 464.56 nM |
| 10 | Ganodermanondiol | −8.60 | −0.26 | 500.55 nM |
| 11 | Ursolic acid | −8.50 | −0.26 | 598.68 nM |
| 12 | Colossolactone A | −8.49 | −0.23 | 594.72 nM |
| 13 | Oleanolic acid | −8.35 | −0.25 | 756.26 nM |
| 14 | Ganodermanontriol | −8.30 | −0.24 | 818.74 nM |
| 15 | Ganolucidic acid A | −8.27 | −0.23 | 865.36 nM |
| 16 | Ganoderic acid beta | −8.17 | −0.23 | 1.02 μM |
| 17 | Ganoderic acid B | −7.99 | −0.22 | 1.39 μM |
| 18 | Velutin | −7.97 | −0.35 | 1.45 μM |
| 19 | Ganoderiol F | −7.95 | −0.24 | 1.48 μM |
| 20 | 20(21)-dehydrolucidenic acid N | −7.88 | −0.24 | 1.67 μM |
| 21 | Ganoderic acid C1 | −7.88 | −0.21 | 1.66 μM |
| 22 | Ganoderiol A | −7.87 | −0.23 | 1.72 μM |
| 23 | Ganoderiol B | −7.68 | −0.23 | 2.34 μM |
| 24 | Ganoderic acid H | −7.65 | −0.19 | 2.48 μM |
| 25 | Betulinic acid | −7.62 | −0.23 | 2.61 μM |
| 26 | Ganomycin I | −7.37 | −0.29 | 3.96 μM |
| 27 | Colossolactone VII | −7.33 | −0.18 | 4.24 μM |
| 28 | 20-hydroxylucidenic acid N | −6.98 | −0.21 | 7.71 μM |
| 29 | Ganoderic acid alpha | −6.79 | −0.17 | 10.55 μM |
| 30 | Ellagic acid | −6.76 | −0.31 | 11.04 μM |
| 31 | Ganomycin B | −6.02 | −0.24 | 38.57 μM |
| 32 | Linoleic acid | −5.37 | −0.27 | 116.76 μM |
| 33 | Colossolactone V | −4.86 | −0.11 | 274.96 μM |
| 34 | Agrocybin | −4.81 | −0.44 | 300.41 μM |
| 35 | Methyl gallate | −4.79 | −0.37 | 308.49 μM |
| 36 | Gallic acid | −4.26 | −0.36 | 757.15 μM |
3.2. ADME analysis
The ADME (Adsorption, Distribution, Metabolism and Excretion) analysis was performed to evaluate the drug-likeness of 36 compounds found in mushrooms. The prediction was performed using SwissADME database. Drug-likeness was indicated by Lipinski’s rule of five. With respect to the criteria, molecular weight (MW) ≤ 500, the number of hydrogen bond acceptor ≤ 10, the number of hydrogen bond donors ≤ 5 and the Log Po/w ≤ 5 84, no more than 1 violation is allowed. We found compounds listed 1–25 with lower binding energy than those of both drug standards as ranked in Table 4. The drug-like property’s prediction was then evaluated.
Table 4.
The physiochemical properties of the top 25 potential compounds derived from mushrooms as SARS-CoV-2 main protease inhibitors based on Lipinski’s rule of five.
| No | Compound | MW (g/mol) (≤ 500 g/mol) |
Num. H-bond acceptors (≤ 10) | Num. H-bond donors (≤ 5) | Log Po/w (≤ 5) | Violation (≤ 1) |
|---|---|---|---|---|---|---|
| 1 | Colossolactone VIIIa | 558.75 | 7 | 1 | 5.49 | 2 |
| 2 | Colossolactone Ea | 522.67 | 4 | 0 | 5.27 | 2 |
| 3 | Colossolactone G | 538.67 | 7 | 1 | 4.57 | 1 |
| 4 | Ergosterol | 396.65 | 1 | 1 | 6.49 | 1 |
| 5 | Heliantriol F | 458.72 | 3 | 3 | 5.28 | 1 |
| 6 | Semicochliodinol A | 438.47 | 4 | 4 | 4.15 | 0 |
| 7 | Semicochliodinol B | 438.47 | 4 | 4 | 4.13 | 0 |
| 8 | Ganoderic acid GS-2 | 500.67 | 6 | 3 | 4.07 | 1 |
| 9 | Lucidumol B | 458.72 | 3 | 3 | 5.63 | 1 |
| 10 | Ganodermanondiol | 456.70 | 5 | 2 | 5.71 | 1 |
| 11 | Ursolic acid | 456.70 | 3 | 2 | 5.94 | 1 |
| 12 | Colossolactone Aa | 516.75 | 5 | 3 | 5.46 | 2 |
| 13 | Oleanolic acid | 456.70 | 3 | 2 | 6.06 | 1 |
| 14 | Ganodermanontriol | 472.70 | 4 | 3 | 4.90 | 0 |
| 15 | Ganolucidic acid A | 500.67 | 6 | 2 | 4.10 | 1 |
| 16 | Ganoderic acid beta | 500.67 | 6 | 3 | 4.02 | 1 |
| 17 | Ganoderic acid B | 516.67 | 7 | 3 | 3.34 | 1 |
| 18 | Velutin | 314.29 | 6 | 2 | 2.60 | 0 |
| 19 | Ganoderiol F | 454.68 | 3 | 2 | 5.63 | 1 |
| 20 | Ganoderic acid C1 | 514.65 | 7 | 2 | 3.27 | 1 |
| 21 | 20(21)-dehydrolucidenic acid N | 476.60 | 7 | 4 | 2.41 | 0 |
| 22 | Ganoderiol A | 474.72 | 4 | 4 | 4.89 | 0 |
| 23 | Ganoderiol B | 470.68 | 4 | 3 | 4.90 | 0 |
| 24 | Ganoderic acid H | 572.69 | 9 | 2 | 3.20 | 1 |
| 25 | Betulinic acid | 456.70 | 3 | 2 | 6.11 | 1 |
Note.
Non-drug-likeness compounds based on Lipinski’s rule of five.
ADME is an essential tool for analyzing the proposed molecule’s oral bioavailability as possible drugs. As shown in Table 4, the prediction results showed that compounds list 1–22 passed the Lipinski’s rule of five. The best five compounds based on docking score and Lipinski’s rule of five for SARS-CoV-2 main protease inhibition are colossolactone G, ergosterol, heliantriol F, semicochliodinol A and semicochliodinol B. Although three compounds, i.e. colossolactone VIII, colossolactone E and colossolactone A, violated more than one rule, there is an exception to Lipinski’s rule for natural products. Therefore, these three are acceptable as druglikness compounds.
3.3. Toxicity, carcinogenicity and mutagenicity prediction
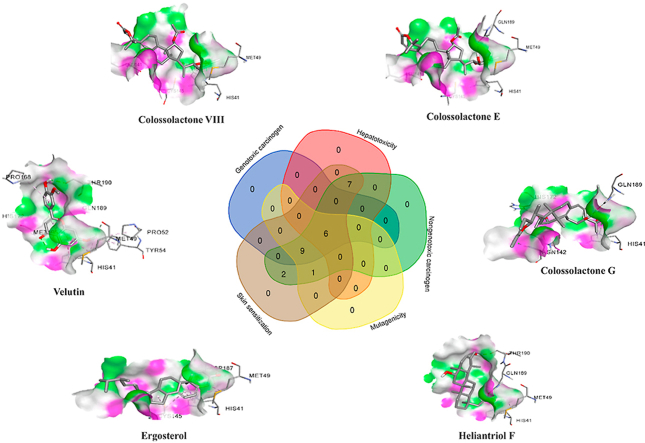
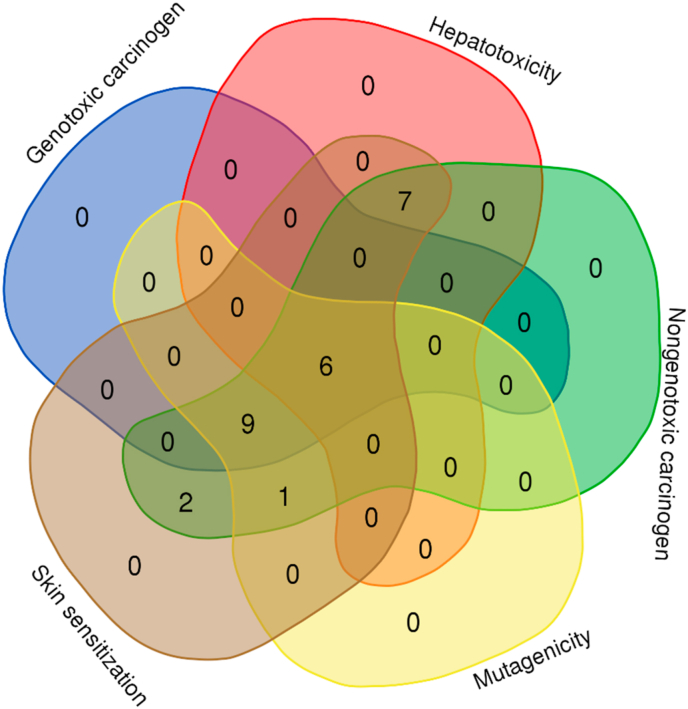
The toxicity of the potential compounds was predicted via toxicity mode in pkCSM database.76 The toxicity prediction was described in Supplementary data 1. The top 25 compounds’ carcinogenicity and mutagenicity were determined using Toxtree 3.1.0 software based on the Benigni-Bossa rule. The rule determines the compound’s carcinogenic and mutagenic potency based on the compound’s functional groups. The functional groups confer as either carcinogenic or mutagenic are acyl halide, haloalkene, epoxide, aliphatic halogen, alkyl nitrate, aldehyde, hydrazine, isocyanate, polyaromatic hydrocarbon, azide, alkyl/aromatic nitro, coumarin, diazo aromatic, benzyl sulfinyl ether, alkyl halide and thiocarbonyl.77,78,85 Predicted genotoxic carcinogenicity, nongenotoxic carcinogenicity and mutagenic potential of the compounds were shown in Supplementary data 2. Overall, six compounds are shown in Fig. 2, including colossolactone VIII, colossolactone E, colossolactone G, ergosterol, heliantriol F and velutin were predicted as non-toxic, non-carcinogenic and non-mutagenic compounds derived from mushrooms.
Fig. 2.
Venn diagram describes the number of compounds predicted no toxicity, no carcinogenicity and no mutagenicity. There are six compounds predicted non-toxicity to skin and liver as well as non-mutagenicity and non-carcinogenicity.
3.4. Hydrogen bond analysis
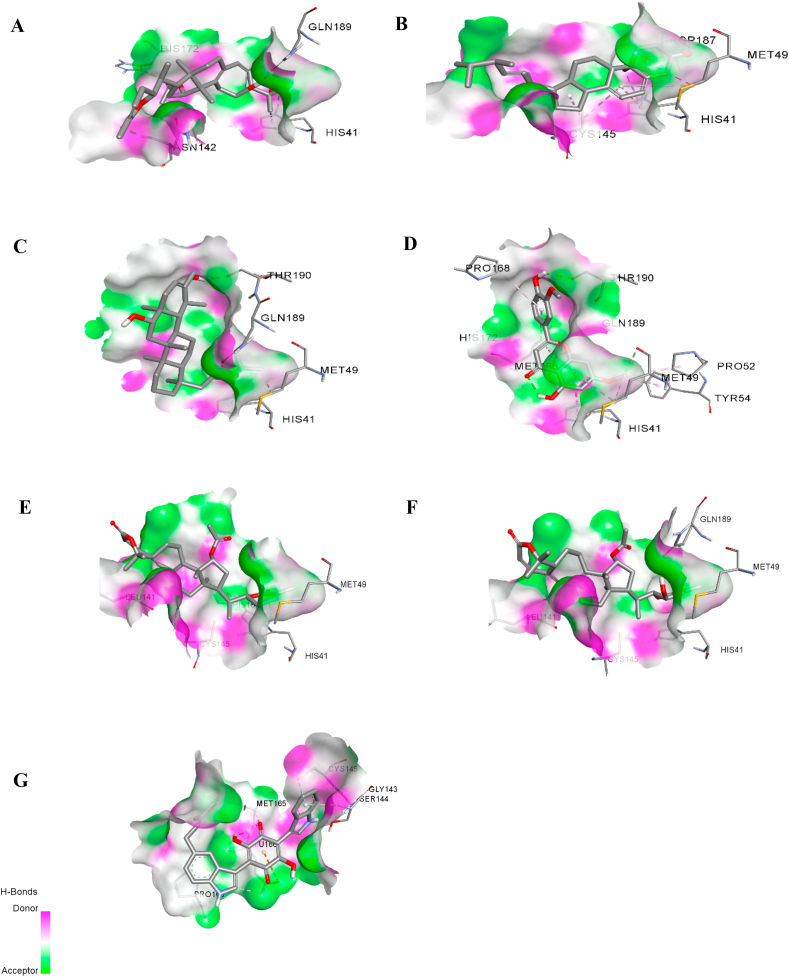
The complex interactions between the SARS-CoV-2 protease and compounds listed 1–25 were visualized, and hydrogen bond analysis was conducted using Discovery Studio Visualizer. The number of hydrogen bonds and the residues involved in hydrogen bond interactions are summarized in Table 6. Colossolactone G, ergosterol, heliantriol F and velutin are the potential candidate compounds for SARS-CoV-2 inhibitors with drug-likeness property, no toxicity, no carcinogenicity and no mutagenicity. Colossolactone G has four hydrogen bonds interacting with the active site of SARS-CoV-2 main protease at the residue HIS41, ASN142, GLN189 and HIS172 (Fig. 3A). Ergosterol shows a hydrogen bond binding to viral protease at the residue ASP187 (Fig. 3B). Hydrogen bond interaction of heliantriol F against the main protease are two interactions at the residue THR190 and GLN189, while velutin exhibited two interactions at the residue THR190 and MET49 (Fig. 3D-E). It is noteworthy that colossolactone G, heliantriol F and velutin exhibited the common interactions when compared to X-ray structure of 6LU7-N3. The common interaction of colossolactone G and velutin is GLN189 residue, while heliantriol F is THR190 residue. Even though colossolactone VIII and colossolactone E were predicted as non-druglikness compounds; it was acceptable to include them as the potential anti-SARS-CoV-2 main protease candidates as we previously mentioned. Colossolactone VIII has two hydrogen bonds interacting with the viral protease’s active site at the residue HIS41 and GLN189, while colossolactone E has a hydrogen bond interaction at the residue HIS41 (Fig. 3E-F). Semicochliodinol A was predicted as a hepatotoxicity agent; it has shown the significant six interactions at the receptor site. Those involved residues are SER144, MET165, GLY143, CYS145, GLU166 and GLU166 (Fig. 3G), in which MET165, GLY143 and GLU166 are the common interactions when compared to X-ray structure of 6LU7-N3. Compared to the original inhibitor, the studied compounds’ common interactions could demonstrate mushroom-derived compounds as the potential inhibitors against SARS-CoV-2 main protease.
Table 6.
H-bond interactions of top 25 potential compounds derived from mushrooms against SARS-CoV-2 main protease inhibitor.
| No | Compound | Hydrogen bonding |
|
|---|---|---|---|
| Number | Amino acid interaction: bond length | ||
| X-ray structure (N3) | 10 | GLY143: 2.86529, THR190:2.84976 , GLN189: 2.88951, PHE140: 3.18679, HIS163: 2.36734, HIS164: 2.80304, MET165: 3.69210, HIS172: 3.72440, GLU166: 2.97927, GLU166: 2.83433 | |
| 1 | Colossolactone VIII | 2 | HIS41: 2.72763, GLN189: 1.96169 |
| 2 | Colossolactone E | 1 | HIS41: 3.02314 |
| 3 | Colossolactone G | 4 | HIS41: 2.58995, ASN142: 1.88729, GLN189: 1.65515, HIS172: 2.84417 |
| 4 | Ergosterol | 1 | ASP187: 2.18813 |
| 5 | Heliantriol F | 2 | THR190: 1.85702, GLN189: 2.18737 |
| 6 | Semicochliodinol A | 6 | SER144: 1.82069, MET165: 3.03681, GLY143: 2.7737, CYS145: 3.69761, GLU166: 2.01863, GLU166: 3.36082 |
| 7 | Semicochliodinol B | 3 | ARG188: 1.86701, CYS145: 3.18962, CYS145: 3.89699 |
| 8 | Ganoderic acid GS-2 | 4 | HIS41: 2.83185, GLU166: 2.24025, GLN189: 2.41463, ASN142: 2.11084 |
| 9 | Lucidumol B | 3 | GLN189: 1.96259, GLN189: 2.14332, GLU166: 2.83998 |
| 10 | Ganodermanondiol | 8 | GLY143: 2.49923, CYS145: 2.76771, HIS163: 2.08786, SER144: 1.90278, LEU141: 1.64009, SER144: 2.43463, LEU167: 3.51115, PRO168: 2.91688 |
| 11 | Ursolic acid | 3 | SER144: 2.87305, CYS145: 2.02309, CYS145: 2.97091 |
| 12 | Colossolactone A | 3 | HIS163: 1.91024, CYS145: 2.52781, THR190: 1.85473 |
| 13 | Oleanolic acid | 3 | SER144: 2.97806, CYS145: 1.97439, CYS145: 2.65435 |
| 14 | Ganodermanontriol | 8 | GLY143: 2.6619, HIS163: 2.25078, LEU141: 2.11468, SER144: 2.86257, LEU141: 1.79129, ASN142: 2.96571, LEU167: 3.47167, PRO168: 2.72296 |
| 15 | Ganolucidic acid A | 3 | GLU166: 2.24559, GLN189: 2.44479, ASN142: 1.94471 |
| 16 | Ganoderic acid beta | 4 | TYR54: 2.24534, GLU166: 2.24309, MET49: 2.05161, ARG188: 2.99000 |
| 17 | Ganoderic acid B | 6 | HIS41: 2.52071, GLY143: 2.65843, CYS145: 3.63501, GLU166: 2.42919, MET49: 2.0714, ASN142: 1.77428 |
| 18 | Velutin | 2 | THR190: 2.15553, MET49: 3.56078 |
| 19 | Ganoderiol F | 1 | ASP187: 1.81627 |
| 20 | Ganoderic acid C1 | 4 | TYR54: 2.29094, GLN189: 2.96608, ASN142: 2.04706, ARG188: 3.33477 |
| 21 | 20(21)-dehydrolucidenic acid N | 5 | GLY143: 2.52618, GLU166: 2.2808, CYS145: 2.70312, MET49: 2.20979, ASN142: 1.81534 |
| 22 | Ganoderiol A | 4 | MET49: 2.2339, GLU166: 1.94189, GLU166: 1.89131, GLU166: 3.10886 |
| 23 | Ganoderiol B | 6 | CYS145: 2.73888, CYS145: 3.60588, MET165: 2.75963, LEU141: 2.23062, LEU141: 2.48645, SER144: 2.06885 |
| 24 | Ganoderic acid H | 5 | ASN142: 1.83347, ASN142: 2.34095, GLU166: 2.10428, GLU166: 2.68895, THR190: 1.78362 |
| 25 | Betulinic acid | 2 | PRO168: 2.29376, GLN189: 1.67449 |
Fig. 3.
Hydrogen bond interactions between 6LU7 and (A) colossolactone G, (B) ergosterol, (C) heliantriol F, (D) velutin, (E) colossolactone VIII, (F) colossolactone E and (G) semicochliodinol A.
Our project plan’s next step is to confirm the anti- SARS-CoV-2 of the identified compounds using SARS-CoV-2 assay kit (BPS Bioscience, CA, USA). The compounds that show inhibitory activity will be screened for their toxicity against normal human cell models such as human peripheral blood mononuclear cells (PBMCs).
4. Conclusions
The current therapeutic drugs against COVID-19 is a group of anti-HIV protease drugs. However, the compound with more efficiency and less toxicity is needed. Moreover, the most severe cases of COVID-19 are caused by infection-induced hyper-inflammation of the lungs and followed by acute respiratory distress, resulting in death. Therefore, effective treatment of COVID-19 would likely be a combination of therapies, using both antiviral and anti-inflammatory drugs. Several bioactive compounds derived from mushrooms have been promisingly demonstrated to exhibit anti-HIV protease and anti-inflammatory activities. Those compounds may have the potential to inhibit SARS-CoV-2 main protease and become useful for the treatment of COVID-19 patients. In this study, mushroom-derived compounds were docked against the main viral protease. In addition, the in silico ADME analysis and toxicity of these compounds were conducted to predict their drug-like properties, toxicity, carcinogenicity and mutagenicity. Our study revealed six compounds found in mushrooms, namely colossolactone VIII, colossolactone E colossolactone G, ergosterol, heliantriol F and velutin the best potential candidates for anti-SAR-CoV-2 agents. Those potential compounds could be used and developed as an alternative or complementary medicine for COVID-19 treatment. Moreover, ergosterol has been reported as an anti-inflammatory agent.86,87 These could support the idea of ergosterol being a useful compound for COVID-19 treatment due to its dual activities. Strategies are in place to confirm theirs in vitro inhibitory activity against SARS-CoV-2 protease and toxicity in healthy mammalian cells such as human PBMCs. The discovery of such inhibitors with low or no toxicity would provide us with further opportunity to develop them into anti-COVID-19 in monotherapy or combination drugs.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research project is supported by the Second Century Fund (C2F), Chulalongkorn University, Bangkok, Thailand for Panthakarn Rangsinth’s postdoctoral fellowship.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtcme.2020.12.002.
Contributor Information
Alison T. Ung, Email: Alison.Ung@uts.edu.au.
Siriporn Chuchawankul, Email: Siriporn.Ch@chula.ac.th.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Health Organization . World Health Organization; 27 December 2020. Coronavirus disease (COVID-19) Situation Report. [Google Scholar]
- 2.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Z., Du X., Xu Y. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 7.Jin Z., Zhao Y., Sun Y. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 8.Dai W., Zhang B., Su H. 6497th. Vol. 368. Science; New York, NY: 2020. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease; pp. 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y. Jung S-HJJomc. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors. Peptidomimetics Small Mol. Chemother. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar V., Shin J.S., Shie J.J. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CL(Pro) inhibitors. Antivir Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar V., Tan K.P., Wang Y.M., Lin S.W., Liang P.H. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg Med Chem. 2016;24(13):3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 13.Yang H., Xie W., Xue X. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10) doi: 10.1371/journal.pbio.0030324. e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuji M. Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS open bio. 2020;10(6):995–1004. doi: 10.1002/2211-5463.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serafin M.B., Bottega A., Foletto V.S., da Rosa T.F., Hörner A., Hörner R. Drug repositioning is an alternative for the treatment of coronavirus COVID-19. Int J Antimicrob Agents. 2020:105969. doi: 10.1016/j.ijantimicag.2020.105969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu Y.F., Chien C.S., Yarmishyn A.A. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci. 2020;21(7) doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu C.M., Cheng V.C., Hung I.F. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arabi Y.M., Alothman A., Balkhy H.H. Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-β1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19(1):81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nutho B., Mahalapbutr P., Hengphasatporn K. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry. 2020;59(18):1769–1779. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- 21.Sheahan T.P., Sims A.C., Leist S.R. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J.Y., Choe P.G., Oh Y. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: implication for infection prevention and control measures. J Kor Med Sci. 2020;35(5) doi: 10.3346/jkms.2020.35.e61. e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. The Lancet Infectious disea. 2020;20(6):689–696. doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye X.T., Luo Y.L., Xia S.C. Clinical efficacy of lopinavir/ritonavir in the treatment of Coronavirus disease 2019. Eur Rev Med Pharmacol Sci. 2020;24(6):3390–3396. doi: 10.26355/eurrev_202003_20706. [DOI] [PubMed] [Google Scholar]
- 25.Cao B., Wang Y., Wen D. A trial of lopinavir-ritonavirses in adults hospitalized with severe COVID-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuzimoto A.D., Isidoro C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds - additional weapons in the fight against the COVID-19 pandemic? J Tradit, Complementary Med. 2020;10(4):405–419. doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benarba B., Pandiella A. Vol. 11. Front Pharmacol.; 2020. Medicinal plants as sources of active molecules against COVID-19; pp. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhuiyan F.R., Howlader S., Raihan T., Hasan M. Vol. 7. Front Med (Lausanne); 2020. Plants metabolites: possibility of natural therapeutics against the COVID-19 pandemic; pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasansuklab A., Theerasri A., Rangsinth P., Sillapachaiyaporn C., Chuchawankul S., Tencomnao T. Anti-COVID-19 drug candidates: a review on potential biological activities of natural products in the management of new coronavirus infection. J Tradit Complement Med. 2020 doi: 10.1016/j.jtcme.2020.12.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R., Bhardwaj V., Das P., Purohit R. Natural analogues inhibiting selective cyclin-dependent kinase protein isoforms: a computational perspective. J Biomol Struct Dynam. 2020;38(17):5126–5135. doi: 10.1080/07391102.2019.1696709. [DOI] [PubMed] [Google Scholar]
- 31.Bhardwaj V., Singh R., Singh P., Purohit R., Kumar S. Elimination of bitter-off taste of stevioside through structure modification and computational interventions. J Theor Biol. 2020;486:110094. doi: 10.1016/j.jtbi.2019.110094. [DOI] [PubMed] [Google Scholar]
- 32.Bhardwaj V.K., Purohit R. Targeting the protein-protein interface pocket of Aurora-A-TPX2 complex: rational drug design and validation. J Biomol Struct Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1772109. [DOI] [PubMed] [Google Scholar]
- 33.Sharma J., Bhardwaj V.K., Das P., Purohit R. Identification of naturally originated molecules as γ-aminobutyric acid receptor antagonist. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1720818. [DOI] [PubMed] [Google Scholar]
- 34.Bhardwaj V.K., Singh R., Sharma J., Das P., Purohit R. Structural based study to identify new potential inhibitors for dual specificity tyrosine-phosphorylation- regulated kinase. Comput Methods Progr Biomed. 2020;194:105494. doi: 10.1016/j.cmpb.2020.105494. [DOI] [PubMed] [Google Scholar]
- 35.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dynam. 2020:1–7. doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar A., Choudhir G., Shukla S.K. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1772112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gogoi N., Chowdhury P., Goswami A.K., Das A., Chetia D., Gogoi B. Computational guided identification of a citrus flavonoid as potential inhibitor of SARS-CoV-2 main protease. Mol Divers. 2020:1–15. doi: 10.1007/s11030-020-10150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uniyal A., Mahapatra M.K., Tiwari V., Sandhir R., Kumar R. Targeting SARS-CoV-2 main protease: structure based virtual screening, in silico ADMET studies and molecular dynamics simulation for identification of potential inhibitors. J Biomol Struct Dynam. 2020:1–17. doi: 10.1080/07391102.2020.1848636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajagopal K., Varakumar P., Baliwada A., Byran G. Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach. Future J Pharmaceut Sci. 2020;6(1):104. doi: 10.1186/s43094-020-00126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linnakoski R., Reshamwala D., Veteli P., Cortina-Escribano M., Vanhanen H., Marjomäki V. Antiviral agents from fungi: diversity, mechanisms and potential applications. Front Microbiol. 2018;9:2325. doi: 10.3389/fmicb.2018.02325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suwannarach N., Kumla J., Sujarit K., Pattananandecha T., Saenjum C., Lumyong S. Natural bioactive compounds from fungi as potential candidates for protease inhibitors and immunomodulators to apply for coronaviruses. Molecules (Basel, Switzerland) 2020;25(8) doi: 10.3390/molecules25081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elsayed E.A., El Enshasy H., Wadaan M.A., Aziz R. Mushrooms: a potential natural source of anti-inflammatory compounds for medical applications. Mediat Inflamm. 2014;2014:805841. doi: 10.1155/2014/805841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muszyńska B., Grzywacz-Kisielewska A., Kała K., Gdula-Argasińska J. Anti-inflammatory properties of edible mushrooms: a review. Food Chem. 2018;243:373–381. doi: 10.1016/j.foodchem.2017.09.149. [DOI] [PubMed] [Google Scholar]
- 45.Nallathamby N., Phan C.W., Seow S.L. A status review of the bioactive activities of tiger milk mushroom Lignosus rhinocerotis (cooke) Ryvarden. Front Pharmacol. 2017;8:998. doi: 10.3389/fphar.2017.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato N., Zhang Q., Ma C.-M., Hattori M. Anti-human immunodeficiency virus-1 protease activity of new lanostane-type triterpenoids from Ganoderma sinense. Chem Pharm Bull. 2009;57(10):1076–1080. doi: 10.1248/cpb.57.1076. [DOI] [PubMed] [Google Scholar]
- 47.Ngai P.H., Zhao Z., Ng T.B. Agrocybin, an antifungal peptide from the edible mushroom Agrocybe cylindracea. Peptides. 2005;26(2):191–196. doi: 10.1016/j.peptides.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Kashiwada Y., Hashimoto F., Cosentino L.M., Chen C.H., Garrett P.E., Lee K.H. Betulinic acid and dihydrobetulinic acid derivatives as potent anti-HIV agents. J Med Chem. 1996;39(5):1016–1017. doi: 10.1021/jm950922q. [DOI] [PubMed] [Google Scholar]
- 49.Habibi E., Sadat-Ebrahimi S.E., Mousazadeh S.A., Amanzadeh Y. Mycochemical investigation of the Turkey tail medicinal mushroom Trametes versicolor (higher basidiomycetes): a potential application of the isolated compounds in documented pharmacological studies. Int J Med Mushrooms. 2015;17(3):255–265. doi: 10.1615/intjmedmushrooms.v17.i3.50. [DOI] [PubMed] [Google Scholar]
- 50.El Dine R.S., El Halawany A.M., Ma C.-M., Hattori M. Anti-HIV-1 protease activity of lanostane triterpenes from the Vietnamese mushroom Ganoderma colossum. J Nat Prod. 2008;71(6):1022–1026. doi: 10.1021/np8001139. [DOI] [PubMed] [Google Scholar]
- 51.Modi M., Goel T., Das T. Ellagic acid & gallic acid from Lagerstroemia speciosa L. inhibit HIV-1 infection through inhibition of HIV-1 protease & reverse transcriptase activity. Indian J Med Res. 2013;137(3):540–548. [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Q., Yuan B., Xiao H. Polyphenols-rich extract from Pleurotus eryngii with growth inhibitory of HCT116 colon cancer cells and anti-inflammatory function in RAW264.7 cells. Food & function. 2018;9(3):1601–1611. doi: 10.1039/c7fo01794d. [DOI] [PubMed] [Google Scholar]
- 53.Rahman M.A., Abdullah N., Aminudin N. Antioxidative effects and inhibition of human low density lipoprotein oxidation in vitro of polyphenolic compounds in Flammulina velutipes (golden needle mushroom) Oxidative Medicine and Cellular Longevity. 2015;2015:1–10. doi: 10.1155/2015/403023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee Y.S., Kang Y.H., Jung J.Y. Inhibitory constituents of aldose reductase in the fruiting body of Phellinus linteus. Biol Pharm Bull. 2008;31(4):765–768. doi: 10.1248/bpb.31.765. [DOI] [PubMed] [Google Scholar]
- 55.Sillapachaiyaporn C., Nilkhet S., Ung A.T., Chuchawankul S. Anti-HIV-1 protease activity of the crude extracts and isolated compounds from Auricularia polytricha. BMC Compl Alternative Med. 2019;19(1):351. doi: 10.1186/s12906-019-2766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong S., Zhong H., Yi C. Simultaneous HPLC determination of ergosterol and 22,23-dihydroergosterol in Flammulina velutipes sterol-loaded microemulsion. Biomed Chromatogr : BMC (Biomed Chromatogr) 2014;28(2):247–254. doi: 10.1002/bmc.3012. [DOI] [PubMed] [Google Scholar]
- 57.Rahman M.A., Abdullah N., Aminudin N. Lentinula edodes (shiitake mushroom): an assessment of in vitro anti-atherosclerotic bio-functionality. Saudi J Biol Sci. 2018;25(8):1515–1523. doi: 10.1016/j.sjbs.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sevindik M., Rasul A., Hussain G. Determination of anti-oxidative, anti-microbial activity and heavy metal contents of Leucoagaricus leucothites. Pak J Pharm Sci. 2018;31(5):2163–2168. [PubMed] [Google Scholar]
- 59.Gaur T., Rao P.B. Analysis of antibacterial activity and bioactive compounds of the giant mushroom, Macrocybe gigantea (agaricomycetes), from India. Int J Med Mushrooms. 2017;19(12):1083–1092. doi: 10.1615/IntJMedMushrooms.2017024559. [DOI] [PubMed] [Google Scholar]
- 60.Liu J., Jia L., Kan J., Jin C.H. In vitro and in vivo antioxidant activity of ethanolic extract of white button mushroom (Agaricus bisporus) Food Chem Toxicol : Int J Publ Br Ind Biol Res Assoc. 2013;51:310–316. doi: 10.1016/j.fct.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Yoon K.N., Alam N., Lee K.R. Antioxidant and antityrosinase activities of various extracts from the fruiting bodies of Lentinus lepideus. Molecules (Basel, Switzerland) 2011;16(3):2334–2347. doi: 10.3390/molecules16032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karaman M., Jovin E., Malbasa R., Matavuly M., Popović M. Medicinal and edible lignicolous fungi as natural sources of antioxidative and antibacterial agents. Phytother Res : PTR. 2010;24(10):1473–1481. doi: 10.1002/ptr.2969. [DOI] [PubMed] [Google Scholar]
- 63.el-Mekkawy S., Meselhy M.R., Nakamura N. Anti-HIV-1 and anti-HIV-1-protease substances from Ganoderma lucidum. Phytochemistry. 1998;49(6):1651–1657. doi: 10.1016/s0031-9422(98)00254-4. [DOI] [PubMed] [Google Scholar]
- 64.Akbar R., Yam W.K. Interaction of ganoderic acid on HIV related target: molecular docking studies. Bioinformation. 2011;7(8):413–417. doi: 10.6026/97320630007413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Min B.S., Nakamura N., Miyashiro H., Bae K.W., Hattori M. Triterpenes from the spores of Ganoderma lucidum and their inhibitory activity against HIV-1 protease. Chem Pharm Bull (Tokyo) 1998;46(10):1607–1612. doi: 10.1248/cpb.46.1607. [DOI] [PubMed] [Google Scholar]
- 66.El Dine R.S., El Halawany A.M., Ma C.-M., Hattori M. Inhibition of the dimerization and active site of HIV-1 protease by secondary metabolites from the Vietnamese mushroom Ganoderma colossum. J Nat Prod. 2009;72(11):2019–2023. doi: 10.1021/np900279u. [DOI] [PubMed] [Google Scholar]
- 67.Sillapachaiyaporn C., Chuchawankul S. HIV-1 protease and reverse transcriptase inhibition by tiger milk mushroom (Lignosus rhinocerus) sclerotium extracts: in vitro and in silico studies. J Tradit Complement Med. 2019;10(4):396–404. doi: 10.1016/j.jtcme.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C.R., Zhou R., Ng T.B., Wong J.H., Qiao W.T., Liu F. First report on isolation of methyl gallate with antioxidant, anti-HIV-1 and HIV-1 enzyme inhibitory activities from a mushroom (Pholiota adiposa) Environ Toxicol Pharmacol. 2014;37(2):626–637. doi: 10.1016/j.etap.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 69.Mengoni F., Lichtner M., Battinelli L. In vitro anti-HIV activity of oleanolic acid on infected human mononuclear cells. Planta Med. 2002;68(2):111–114. doi: 10.1055/s-2002-20256. [DOI] [PubMed] [Google Scholar]
- 70.Zhang C.C., Yin X., Cao C.Y., Wei J., Zhang Q., Gao J.M. Chemical constituents from Hericium erinaceus and their ability to stimulate NGF-mediated neurite outgrowth on PC12 cells. Bioorg Med Chem Lett. 2015;25(22):5078–5082. doi: 10.1016/j.bmcl.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 71.Kalogeropoulos N., Yanni A.E., Koutrotsios G., Aloupi M. Bioactive microconstituents and antioxidant properties of wild edible mushrooms from the island of Lesvos, Greece. Food Chem Toxicol : Int J Publ Br Ind Biol Res Assoc. 2013;55:378–385. doi: 10.1016/j.fct.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 72.Fredenhagen A., Petersen F., Tintelnot-Blomley M., Rösel J., Mett H., Hug P. Semicochliodinol A and B: inhibitors of HIV-1 protease and EGF-R protein tyrosine kinase related to asterriquinones produced by the fungus Chrysosporium merdarium. J Antibiot. 1997;50(5):395–401. doi: 10.7164/antibiotics.50.395. [DOI] [PubMed] [Google Scholar]
- 73.Ma C., Nakamura N., Miyashiro H., Hattori M., Shimotohno K. Inhibitory effects of constituents from Cynomorium songaricum and related triterpene derivatives on HIV-1 protease. Chem Pharm Bull (Tokyo) 1999;47(2):141–145. doi: 10.1248/cpb.47.141. [DOI] [PubMed] [Google Scholar]
- 74.Wang H., Ng T.B. Isolation and characterization of velutin, a novel low-molecular-weight ribosome-inactivating protein from winter mushroom (Flammulina velutipes) fruiting bodies. Life Sci. 2001;68(18):2151–2158. doi: 10.1016/s0024-3205(01)01023-2. [DOI] [PubMed] [Google Scholar]
- 75.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benigni R, Bossa C, Jeliazkova N, Netzeva T, Worth A. OPOCE; 2008. The Benigni/Bossa rulebase for mutagenicity and carcinogenicity – a module of Toxtree; pp. 1–78. [Google Scholar]
- 78.Benigni R., Bossa C., Netzeva T., Rodomonte A., Tsakovska I. Mechanistic QSAR of aromatic amines: new models for discriminating between homocyclic mutagens and nonmutagens, and validation of models for carcinogens. Environ Mol Mutagen. 2007;48(9):754–771. doi: 10.1002/em.20355. [DOI] [PubMed] [Google Scholar]
- 79.Gibbs J.W. Transactions of the Connecticut Academy of Arts and Sciences. Vol. 2. New Haven; 1873. A Method of Geometrical Representation of the Thermodynamic Properties by Means of Surfaces; pp. 382–404. [Google Scholar]
- 80.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., Soetjipto S. Vol. 1. Preprints; 2020. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study; pp. 1–14. [Google Scholar]
- 81.Islam R., Parves M.R., Paul A.S. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J Biomol Struct Dynam. 2020:1–12. doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CL(pro)): an in silico screening of alkaloids and terpenoids from African medicinal plants. J Biomol Struct Dynam. 2020:1–13. doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar A., Choudhir G., Shukla S.K. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J Biomol Struct Dynam. 2020:1–21. doi: 10.1080/07391102.2020.1772112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 85.Nasution M.A.F., Toepak E.P., Alkaff A.H., Tambunan U.S.F. Flexible docking-based molecular dynamics simulation of natural product compounds and Ebola virus Nucleocapsid (EBOV NP): a computational approach to discover new drug for combating Ebola. BMC Bioinf. 2018;19(Suppl 14):419. doi: 10.1186/s12859-018-2387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu C., Zhao S., Zhu C. Ergosterol ameliorates renal inflammatory responses in mice model of diabetic nephropathy. Biomedicine & pharmacotherapy. 2020;128:110252. doi: 10.1016/j.biopha.2020.110252. [DOI] [PubMed] [Google Scholar]
- 87.Sun X., Feng X., Zheng D. Ergosterol attenuates cigarette smoke extract-induced COPD by modulating inflammation, oxidative stress and apoptosis in vitro and in vivo. Clinical science (London, England: 1979) 2019;133(13):1523–1536. doi: 10.1042/CS20190331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.