Abstract
Inflammatory biomarkers may be associated with disease severity and increased mortality in COVID-19 patients but have not been studied in North American populations. We sought to determine whether a set of commonly ordered inflammatory biomarkers can predict 28-day mortality. We analyzed a multi-centered (four) COVID-19 registry cohort from March 4th to December 7th, 2020. This cohort included COVID-19-positive patients admitted to medical wards or intensive care units. Patients presenting to the emergency department for COVID-19 symptoms and then subsequently discharged were also included. We performed Cox-regression analysis to measure whether commonly used biomarkers were associated with an increased 28-day mortality. Of 336 COVID-19-positive patients, 267 required hospital admission, and 69 were seen in the emergency room and discharged. The median age was 63 years (IQR 80–50) and the female-to-male ratio was 49:51. Derivation of internally validated cut-offs suggested that C-reactive protein ≥ 78.4 mg/L, neutrophil-to-lymphocyte ratio ≥ 6.1, lymphocyte-to-white blood cell ratio < 0.127, and a modified Glasgow prognostic score equal to 2 vs. 1 or 0 were associated with the highest increased risk of 28-day mortality. We provide early estimates of cut-off values for inflammatory biomarkers and indices measured at the time of admission that may be useful to clinicians for predicting 28-day mortality in North American COVID-19 patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11739-021-02637-8.
Keywords: Biomarkers, COVID-19, NLR, CRP, mGPS
Introduction
COVID-19 is a respiratory disease caused by the SARS-COV-2 infection. COVID-19 pathogenesis is associated with upregulation of inflammatory pathways and associated inflammatory biomarkers, which underlie the biological plausibility for their use clinically. For example, severe cases of COVID-19 are characterized by excessive expression of pro-inflammatory mediators with a relative paucity of anti-inflammatory mediators [1]. Infection causes the respiratory epithelial cells to release interleukins (IL) such as IL-6 [2]. IL-6 upregulates hepatic C-reactive protein (CRP) production and other acute phase reactants such as ferritin. COVID-19-infected individuals may have higher serum IL-6 levels due to an abnormal population of CD14 + CD16 + inflammatory monocytes. In addition to these pro-inflammatory mediators, severe COVID-19 cases present with lymphopenia, particularly of CD4 + T-cells, which includes regulatory T-cells important in the modulation of pro-inflammatory immune responses [3]. Therefore, both CRP and lymphopenia have been consistently referenced biomarkers for disease severity in COVID-19 patients. Other inflammatory biomarkers share similar biological plausibility. Low serum albumin for example has been postulated to be associated with increased mortality risk in COVID-19 patients due to its antioxidant and anticoagulant properties [4]. For this reason, the modified Glasgow Prognostic score (mGPS) is an interesting choice for investigation, as it is a composite score of CRP and albumin. The hypothesis being that if each of these markers were associated with increased mortality in COVID-19, the composite correction for each may provide a better estimate of its association with COVID-19 infection. Furthermore, ratios and composite scores of the complete blood count (CBC) such as neutrophil-to-lymphocyte (NLR) count have been hypothesized and studied in COVID-19. There is significant potential for these ratios, including previously understudied ones such as lymphocyte-to-white blood cell ratio, platelet-to-white blood cell ratio, and platelet-to-lymphocyte ratio.
Inflammatory biomarkers are used for diagnosis, prognosis, and theragnosis, have been proposed as a useful metric for prognosticating the disease course of patients with COVID-19 [4, 5]. Studies have examined the prognostic use of inflammatory biomarkers in prognosticating disease severity, 28-day mortality, and progression to Acute Respiratory Distress Syndrome (ARDS) [6–8]. A recent meta-analysis of 16 studies conducted in China, found consistent evidence of a positive association between inflammatory biomarkers and COVID-19 severity, however, noted the need for further research to clarify whether these results would be consistent across different countries and populations [5]. In addition, rigorously validated biomarker cutoffs, that could potentially be used by clinicians to risk stratify patients with COVID-19, have not been established.
Our objective was to examine the association between commonly ordered inflammatory biomarkers on admission, and 28-day mortality in a North American sample of COVID-19 patients, and to determine the optimal cut-off values on the relevant biomarkers for predicting mortality. We hypothesized that commonly ordered biomarkers and indices such as CRP and NLR would be useful to clinicians for estimating the risk of 28-day mortality.
Methods
Study design and population
We conducted an analysis of a registry cohort study of patients in three community hospital and one academic centre from the McMaster Multi-Regional Hospital Coronavirus Registry (COREG) between March 4th and December 7th 1st 2020. COREG is a multicentered data registry collecting information on positive SARS-CoV-2 PCR cases in Ontario, Canada. The region encompasses a catchment of approximately 1,000,000 persons, 3 community and 1 academic hospital. Our patient cohort includes emergency department (ED) visits and admissions into hospital including the intensive care units (ICU). Our study received ethics approval from the Hamilton Integrated Research Ethics Board (11,169-C) and Tri-Hospital Research Ethics Board (GRH RC 2020-166).
Study population
We selected all patients with a positive PCR for COVID-19 at three community centres and one academic centre, with records between March 4th, 2020 and December 7th, 2020, inclusive of all known hospitalized cases up to the time of analysis (n = 350). We excluded those with no known outcome at time of analysis (n = 4) and those without any of the target blood work (n = 10), for a final sample size of 336 (Fig. 1).
Fig. 1.

Flow diagram for inclusion and exclusion process
Biomarkers
Our study examined several commonly ordered admission biomarkers: white blood cells (WBC), platelets, neutrophils, and lymphocytes, and ratios of these counts, and indices including NLR, platelet-to-lymphocyte ratio (PLR), lymphocyte–WBC ratio, and platelet–WBC ratio. These ratios were chosen to mirror previous research, as well as add to the potential list of inflammatory indices available for clinicians. We also included albumin, C-reactive protein (CRP), lactate dehydrogenase (LDH), ferritin, D-dimer, and creatine kinase (CK). We also calculated the modified Glasgow prognostic score, which is an ordinal measure combining albumin and CRP that has been validated in prognosticating various cancers [9]. It is calculated by attributing one point for each of CRP ≥ 10 mg/L and Albumin < 35 g/L. The range of the scale for mGPS is from 0 to 2, with 2 portending the worst prognosis.
Statistical methods
Means (standard deviation), median (interquartile range) and frequency (percentages) statistics were used to describe the study population characteristics, and biomarkers stratified by visit type. We classified patients based on their visit type: patients admitted to the emergency department with COVID-19 symptoms, but were later stable enough for discharge, admission to a medicine unit, and admission to an intensive care unit (ICU). A patient who was admitted to an ICU at any time during their hospital stay was classified in the ICU cohort. As many of the biomarkers have skewed distributions, we performed a logarithm transformation of all biomarkers except for the mGPS. We also standardized each biomarker (except for mGPS) to have a mean of zero and standard deviation of one to allow for comparison of hazard ratios across biomarkers. The primary outcome of this study was time to 28-day mortality.
We examined the association between each biomarker with mortality using Cox regression. Each biomarker was examined as a continuous measure both univariately and adjusted for age and sex. In addition to HRs and 95% confidence intervals, we reported the c-index for the Cox models as a measure of predictive discriminability [10]. We assessed the proportional hazards assumptions with a Schoenfeld residual test and by inspecting survival curves. A sensitivity analysis on the associations between biomarkers and hospital and ICU admission (disease severity) was conducted with binary logistic regression and can be found in supplementary Table 1.
Table 1.
Stratifying characteristics by visit types
| Characteristic | N | Overall, N = 336a | ED only, N = 69a | ICU, N = 63a | Ward, N = 204a | p valueb |
|---|---|---|---|---|---|---|
| Sex at birth | 0 | 0.074 | ||||
| Female | 166 (49%) | 37 (54%) | 23 (37%) | 106 (52%) | ||
| Male | 170 (51%) | 32 (46%) | 40 (63%) | 98 (48%) | ||
| Age | 0 | 63 (50, 80) | 47 (33, 54) | 57 (51, 70) | 72 (58, 85) | < 0.001 |
| Vitals and symptoms | ||||||
| Temperature | 21 | 37.10 (36.70, 38.10) | 37.10 (36.88, 37.50) | 37.75 (36.90, 38.90) | 37.10 (36.65, 38.10) | 0.030 |
| Oxygen saturation | 18 | 95.0 (93.0, 97.0) | 97.0 (96.0, 99.0) | 93.0 (90.0, 95.0) | 95.0 (93.0, 97.0) | < 0.001 |
| Fever | 0 | 199 (59%) | 44 (64%) | 49 (78%) | 106 (52%) | < 0.001 |
| Shortness of breath | 0 | 191 (57%) | 34 (49%) | 57 (90%) | 100 (49%) | < 0.001 |
| Cough | 0 | 214 (64%) | 48 (70%) | 49 (78%) | 117 (57%) | 0.007 |
| Fatigue/Malaise | 0 | 143 (43%) | 30 (43%) | 30 (48%) | 83 (41%) | 0.6 |
| Wheezing | 0 | 14 (4.2%) | 2 (2.9%) | 4 (6.3%) | 8 (3.9%) | 0.6 |
| Muscle aches (Myalgia) | 0 | 57 (17%) | 15 (22%) | 19 (30%) | 23 (11%) | 0.001 |
| Diarrhea | 0 | 55 (16%) | 6 (8.7%) | 13 (21%) | 36 (18%) | 0.13 |
| Comorbidities | ||||||
| Hypertension | 0 | 159 (47%) | 12 (17%) | 31 (49%) | 116 (57%) | < 0.001 |
| Coronary artery disease | 0 | 45 (13%) | 5 (7.2%) | 7 (11%) | 33 (16%) | 0.14 |
| Dementia | 0 | 48 (14%) | 1 (1.4%) | 0 (0%) | 47 (23%) | < 0.001 |
| COPD | 0 | 66 (20%) | 9 (13%) | 11 (17%) | 46 (23%) | 0.2 |
| Chronic kidney disease | 0 | 35 (10%) | 0 (0%) | 6 (9.5%) | 29 (14%) | 0.004 |
| Diabetes mellitus | 0 | 90 (27%) | 7 (10%) | 17 (27%) | 66 (32%) | 0.002 |
| Biomarkers | ||||||
| d-dimer | 214 | 0.8 (0.3, 1.6) | 0.2 (0.2, 0.2) | 1.0 (0.7, 3.0) | 0.8 (0.4, 1.6) | < 0.001 |
| White blood cells | 26 | 6.6 (5.1, 9.5) | 6.1 (4.8, 7.5) | 7.7 (5.8, 11.3) | 6.6 (5.1, 9.6) | 0.003 |
| Neutrophils | 26 | 4.8 (3.5, 7.2) | 3.9 (3.0, 5.2) | 6.1 (4.3, 9.2) | 4.8 (3.4, 7.6) | < 0.001 |
| CRP | 129 | 66 (16, 137) | 3 (2, 16) | 126 (53, 195) | 66 (18, 124) | < 0.001 |
| Platelets | 28 | 212 (167, 279) | 198 (164, 252) | 191 (158, 252) | 228 (174, 287) | 0.047 |
| Ferritin | 219 | 456 (150, 978) | 95 (39, 546) | 808 (405, 1,355) | 333 (90, 605) | < 0.001 |
| Creatinine kinase | 171 | 90 (46, 220) | 52 (43, 71) | 132 (59, 351) | 94 (43, 209) | 0.002 |
| Lymphocyte | 26 | 1.00 (0.70, 1.50) | 1.30 (1.05, 1.70) | 0.80 (0.60, 1.20) | 1.00 (0.60, 1.50) | < 0.001 |
| LDH | 155 | 346 (214, 535) | 370 (186, 490) | 373 (281, 661) | 308 (206, 518) | 0.032 |
| NLR | 26 | 5.0 (2.8, 8.7) | 2.6 (1.9, 4.2) | 7.2 (5.0, 11.2) | 5.0 (3.0, 8.8) | < 0.001 |
| PLR | 28 | 219 (147, 310) | 151 (118, 210) | 228 (175, 301) | 231 (159, 334) | < 0.001 |
| P/WR | 28 | 31 (23, 42) | 33 (27, 44) | 27 (20, 34) | 33 (24, 46) | 0.002 |
| L/WR | 26 | 0.15 (0.09, 0.23) | 0.25 (0.18, 0.31) | 0.12 (0.08, 0.15) | 0.15 (0.09, 0.22) | < 0.001 |
| Albumin | 157 | 35.0 (32.0, 39.0) | 40.0 (37.5, 40.0) | 33.0 (30.0, 37.0) | 35.0 (32.0, 39.0) | < 0.001 |
| mGPS | 194 | < 0.001 | ||||
| 0 | 16 (11%) | 9 (64%) | 3 (6.2%) | 4 (5.0%) | ||
| 1 | 60 (42%) | 5 (36%) | 17 (35%) | 38 (48%) | ||
| 2 | 66 (46%) | 0 (0%) | 28 (58%) | 38 (48%) | ||
| Time since symptom onset | 10 | 5 (2, 10) | 6 (3, 11) | 7 (3, 10) | 5 (1, 9) | 0.062 |
| Died | 0 | 51 (15%) | 0 (0%) | 12 (19%) | 39 (19%) | < 0.001 |
aStatistics presented: n (%); Median (IQR)
bStatistical tests performed: Chi-square test of independence; Kruskal–Wallis test; Fisher's exact test
Biomarkers that were significantly associated with morality in the adjusted analysis were further examined using cutoffs on the original scale of the biomarker. A priori cutoffs were selected based on previous literature [8, 11–19], primarily COVID-19 biomarker studies. For biomarkers with no previous COVID-19 studies, cutoffs were used from other areas of research, such as cancer and heart disease [11, 12]. We also derived a novel cutoff for each biomarker, which aimed to maximize the predictive discriminability of the biomarker (c-index) for mortality. This cutoff was selected by splitting each biomarker into deciles and using tenfold cross-validation to select the decile cutoff with the highest c-index. We generated Kaplan–Meier curves on the original data for each of the derived cutoffs.
Results
Patient characteristics
Of the 336 patients included in the study, 69 (20.5%) visited the emergency department and were subsequently discharged, 150 (60.7%) were admitted to the medicine wards, and 63 (18.7%) were admitted to ICU (Table 1). The median age was 61 years old (IQR 50–80) and the female (166, 49%) to male (170, 51%) ratio was balanced. However, there were disproportionately more males admitted to the ICU than females [40 (63%) vs. 23 (37%)]. Symptoms of fever (n = 199, 59%) and cough (n = 214, 64%) were the most common presenting complaint for all admission types. Most patients (n = 235, 70.16%) had at least one comorbidity, and hypertension was the most prevalent comorbidity (n = 159, 47%). The median time to presentation from onset of symptoms was approximately 5 days (IQR = 10–2).
Biomarkers
There were significant differences between visit types in the median levels for all the biomarkers examined (Table 1). For example, the median admission NLR for patients discharged from the emergency department was 2.6 (IQR 1.9–4.2) compared to ICU 7.2 (IQR 5.0–11.2). Similarly, median CRP was 4.0 mg/L (IQR 2.0–16.0) versus 126.0 mg/L (IQR 53.0–195.0), respectively.
Missing biomarker data were handled with multivariate imputation by chained equations using 20 imputations [20] Our analysis of the missing data found that missingness was most strongly related to disease severity, which can be largely accounted for by analyzing the outcomes and other biomarker data collected (e.g., patients admitted to ICU were more likely to have the target biomarkers ordered than those admitted to a medicine unit or emergency department only visits). We performed an independent audit of the COREG study database to ensure no missing data were due to collection error. All analyses were done using R 4.0 [21].
28-day mortality
In this cohort, there were 51 (15.0%) deaths, 39 (19%) of which were patients on the wards, 12 (19%) in ICU and no deaths were recorded in patients who were discharged from the emergency department. The relative high mortality rate in our ward patients is due to the high numbers of long-term care patients and elderly patients with whom ICU care was not within their goals of care. Seven of the 14 biomarkers examined demonstrated a significant association with higher risk of 28-day mortality adjusted for age and sex. The largest effect size for a one standard deviation increase was observed for log-CRP with an adjusted HR of 2.43 (95% CI 1.44, 4.07) (Table 2). Other significant but smaller effect sizes were found for log-WBC aHR 1.77 (95% CI 1.32, 2.37); log-NLR 1.76 (95% CI 1.37, 2.26); log-lymphocyte count 0.77 (95% CI 0.59, 0.99); log-platelet–WBC ratio 0.56 (95% CI 0.45, 0.69); and log-lymphocyte–WBC ratio 0.60 (95% CI 0.48, 0.76). A one-unit increase in mGPS was also significantly associated with 28 day with an aHR of 2.94 (95% CI 1.41, 6.13).
Table 2.
Cox regression analysis for both unadjusted and adjusted 28-day mortality by biomarker
| Biomarker | HRa (95% CI) | p value | c-index | aHRa (95% CI) | p value | c-index |
|---|---|---|---|---|---|---|
| CBC | ||||||
| WBC | 1.79 (1.37, 2.35) | < 0.001 | 0.66 | 1.77 (1.32, 2.37) | < 0.001 | 0.82 |
| Platelets | 0.90 (0.68, 1.20) | 0.480 | 0.51 | 0.83 (0.63, 1.11) | 0.204 | 0.80 |
| Lymphocytes | 0.68 (0.52, 0.90) | 0.007 | 0.61 | 0.77 (0.59, 0.99) | 0.044 | 0.81 |
| CBC ratios | ||||||
| NLR | 2.07 (1.58, 2.71) | < 0.001 | 0.71 | 1.76 (1.37, 2.26) | < 0.001 | 0.85 |
| PLR | 1.34 (1.03, 1.76) | 0.032 | 0.59 | 1.16 (0.90, 1.50) | 0.255 | 0.80 |
| P/WBC | 0.57 (0.46, 0.71) | < 0.001 | 0.66 | 0.56 (0.45, 0.69) | < 0.001 | 0.84 |
| L/WBC | 0.51 (0.40, 0.66) | < 0.001 | 0.72 | 0.60 (0.48, 0.76) | < 0.001 | 0.85 |
| Acute phase reactants | ||||||
| CRP | 2.42 (1.46, 4.01) | 0.001 | 0.68 | 2.43 (1.44, 4.07) | 0.001 | 0.84 |
| CK | 1.54 (1.01, 2.35) | 0.045 | 0.62 | 1.37 (0.90, 2.10) | 0.134 | 0.81 |
| Ferritin | 1.41 (0.82, 2.40) | 0.192 | 0.59 | 1.48 (0.92, 2.37) | 0.103 | 0.81 |
| d-dimer | 1.42 (1.02, 1.96) | 0.040 | 0.63 | 1.20 (0.82, 1.76) | 0.336 | 0.80 |
| LDH | 1.06 (0.70, 1.59) | 0.787 | 0.51 | 1.16 (0.78, 1.73) | 0.453 | 0.80 |
| Albumin | 0.49 (0.37, 0.66) | < 0.001 | 0.71 | 0.63 (0.46, 0.84) | 0.003 | 0.82 |
| Composite scores | ||||||
| mGPSb | 4.02 (1.98, 8.16) | < 0.001 | 0.70 | 2.94 (1.41, 6.13) | 0.005 | 0.83 |
aFor each increase in one standard deviation of the natural logarithm of the biomarker
bmGPS is an ordinal measure and was, therefore, not transformed. Hazard ratios represent change in risk for each increase of one unit of the modified Glasgow Prognostic Score
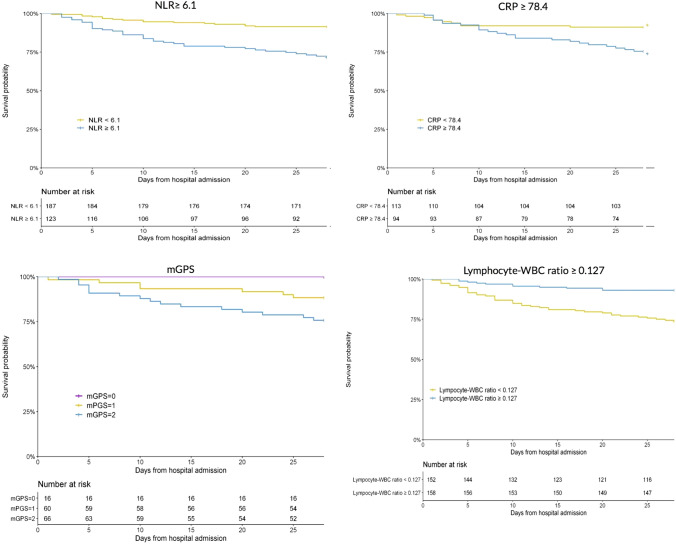
The Schoenfeld-residual test indicated a potential violation of the proportional hazard’s assumption for CRP. Inspection of survival curves showed overlapping survival curves for the first 5 days, which we did not consider a significant enough violation to warrant changing our analytic approach (Fig. 2).
Fig. 2.
Kaplan–Meier curves for NLR, CRP, mGPS and L/WBC ratio with non-imputed data
Inflammatory biomarker cutoffs for 28-day mortality
The newly derived cutoffs which maximized the predictive discriminability of each biomarker for in hospital mortality were as follows: WBC ≥ 7.50 × 109/L, NLR ≥ 6.1, CRP ≥ 78.4 mg/L, Lymphocytes ≤ 0.8 × 109/L, P–WBC ratio ≥ 31.0, L–WBC ratio ≤ 0.127. All a priori and derived cutoffs were significantly associated with increased 28-day mortality. In the unadjusted analysis, the cutoff with the largest effect size was mGPS ≥ 2 (HR, 4.88 (95% CI 2.17, 10.99) followed by L–WBC ratio ≥ 0.228 (0.25 (95% CI 0.13, 0.5) (Table 3). In adjusted analysis, the cutoff with the largest effect size was L–WBC ratio ≥ 0.228 (aHR 0.12 (0.03, 0.53), followed by NLR ratio ≥ 3 (aHR, 5.69 (1.7, 19.03). Kaplan–Meier curves were generated for the derived cutoffs and for the mGPS (Fig. 2).
Table 3.
Unadjusted and adjusted hazard ratios for literature driven and ROC cross-validated cutoffs for significant biomarkers for 28-day mortalityd
| Biomarker | HR (95% CI) | p value | c-index | aHR (95% CI) | p value | c-index |
|---|---|---|---|---|---|---|
| WBC | ||||||
| ≥ 11.1 × 10^9/L | 3.14 (1.71, 5.75) | < 0.001 | 0.60 | 2.54 (1.39, 4.65) | 0.003 | 0.82 |
| ≥ 7.5 × 10^9/La | 3.39 (1.86, 6.18) | < 0.001 | 0.65 | 2.90 (1.59, 5.29) | 0.008 | 0.83 |
| NLR | ||||||
| ≥ 3 | 5.65 (1.71, 18.73) | 0.005 | 0.60 | 5.69 (1.7, 19.03) | 0.006 | 0.82 |
| ≥ 5 | 3.25 (1.65, 6.37) | < 0.001 | 0.63 | 3.52 (1.8, 6.87) | < 0.001 | 0.84 |
| ≥ 6.1a | 3.66 (1.97, 6.78) | < 0.001 | 0.66 | 3.15 (1.71, 5.82) | < 0.001 | 0.83 |
| CRP | ||||||
| ≥ 41.8 mg/L | 3.16 (1.37, 7.29) | 0.009 | 0.62 | 3.15 (1.36, 7.32) | 0.009 | 0.82 |
| ≥ 78.4 mg/La | 3.08 (1.47, 6.45) | 0.004 | 0.63 | 2.75 (1.34, 5.66) | 0.008 | 0.82 |
| Lymphocytes | ||||||
| ≥ 0.8 × 10^9/L | 0.43 (0.24, 0.77) | 0.005 | 0.60 | 0.43 (0.24, 0.78) | 0.006 | 0.82 |
| ≥ 1.0 × 10^9/L | 0.46 (0.25, 0.83) | 0.011 | 0.59 | 0.45 (0.24, 0.82) | 0.011 | 0.82 |
| ≥ 0.8 × 10^9/Lb | 0.43 (0.24, 0.77) | 0.005 | 0.60 | 0.43 (0.24, 0.78) | 0.006 | 0.82 |
| mGPS | ||||||
| ≥ 2 | 4.88 (2.17, 10.99) | < 0.001 | 0.69 | 3.33 (1.45, 7.66) | 0.006 | 0.83 |
| P/WBC | ||||||
| ≥ 20 | 0.31 (0.17, 0.57) | < 0.001 | 0.60 | 0.29 (0.16, 0.53) | < 0.001 | 0.82 |
| ≥ 30 | 0.47 (0.26, 0.85) | 0.014 | 0.59 | 0.40 (0.22, 0.72) | 0.003 | 0.82 |
| ≥ 31c | 0.32 (0.17, 0.62) | 0.001 | 0.63 | 0.30 (0.16, 0.58) | < 0.001 | 0.83 |
| L/WBC | ||||||
| ≥ 0.228 | 0.12 (0.03, 0.5) | 0.005 | 0.61 | 0.12 (0.03, 0.53) | 0.006 | 0.83 |
| ≥ 0.127c | 0.25 (0.13, 0.5) | < 0.001 | 0.65 | 0.26 (0.13, 0.51) | < 0.001 | 0.84 |
aIndicates cut off of 60th percentile
bIndicates cut off of 40th percentile
cIndicates cut off of 50th percentile
dROC cross-validated cut offs are designated by percentile and all else are literature-driven cut offs
Sensitivity analysis
Analysis of the outcomes of hospital and ICU admission produced generally similar results to the main analysis (supplemental table). A notable difference was that CK and d-dimer (positively) and albumin (negatively) were associated with ICU admission but were not associated with 28-day mortality.
Interpretation
We found that commonly available clinical inflammatory biomarkers were significantly associated with 28-day mortality. Specifically, C-reactive protein, neutrophil-to-lymphocyte ratio, and lymphocyte-to-white blood count ratio showed the most consistent and strong associations with mortality in our study. The modified Glasgow prognostic score also demonstrated a consistently strong association with elevated risk for mortality. Our findings align with previous published biomarker studies, albeit in populations outside of North America. Our work further extends this prior work by providing newly derived optimal cutoff values for predicting mortality for the associated biomarkers. Furthermore, we examined new biomarkers (L/WBC ratio and mGPS) which had not been previously studied in COVID-19 and found they were significantly associated with mortality.
NLR, CRP, and L/W ratio as predictors of 28-day mortality
To our knowledge, there have been no prior studies examining the association between blood and clinical biomarkers with clinical outcomes in COVID-19 in populations outside of China. A recent meta-analysis which included 16 Chinese studies found an association between several important blood biomarkers and their association with disease severity in COVID-19 hospitalized patients. [5] These included NLR, which was associated with increased odds of developing COVID-19 pneumonitis and worse disease severity [14, 15, 22, 23]. Similarly, CRP above values ≥ 41.8 mg/L, were shown to significantly correlate with worse disease severity, particularly on radiological CT chest findings. [8, 9] There is also an overall paucity of studies examining the predictors of in hospital mortality. Only CRP has been investigated for its association with COVID-19-related mortality and so far, the results have been mixed and inconclusive [18, 19]. To date, L/WBC ratio has not been examined in COVID-19, even though it has predictive implications in distinguishing between bacterial tonsillitis and glandular fever. [24] The biologic plausibility for CRP has been previously discussed. NLR may be associated with fundamental pathogenesis as well. Elevated neutrophils are associated with elevated secretion of IL-6 and TNF-alpha, causing a general inflammatory responses syndrome that may lead to underlying acute respiratory distress syndrome when present systematically. The ratio of NLR encompasses both this general inflammatory state and the underlying disease severity represented by lymphopenia. Similarly, L/WBC may represent the inverse of NLR, whereas the lymphopenia represents underlying disease severity from depleted lymphocytes to the general WBC count, which is mostly made up of neutrophils.
Largely, our results are consistent with biomarker studies in COVID-19 from China, but notably our data are from a North American cohort and focuses on 28-day mortality as the primary outcome. Our findings indicate that NLR is a useful biomarker for predicting elevated risk of mortality among patients with COVID-19 within the ward and ICU settings. In particular, at a cut off of greater than or equal to 6.1 and, there was an absolute increase in 28 day in hospital mortality of 19.9% (28.5% vs. 8.6%). This is an easily accessible biomarker available as a part of routine laboratory investigations ordered by clinicians, which can be used to risk stratify COVID-19 patients admitted to the ward or ICU. We also found CRP was a strong predictor, and admission to hospital and ICU. At our derived cutoff of 78.4 mg/L, CRP was associated with an absolute increase in mortality of 15.6% (24.5% vs 8.9%). However, unlike NLR, CRP may not be routinely performed in all admitting patients and, therefore, is likely not as easily accessible. Lastly, we are the first to investigate the role of L/WBC ratio as a biomarker in COVID-19 patients. L/WBC ratio had the largest effect with an absolute decrease in mortality of 17.2.4% (7.0% vs. 26.3%) with a cut off at 0.127. Like NLR, L/WBC is readily available as a part of the complete blood count. Of note, our reported optimal cut-off values are consistent with studies in other disease populations, increasing the external validity of our results.
Modified Glasgow prognostic score
The mGPS is a composite of two common inflammatory biomarkers: CRP and albumin, both of which have individually been investigated in this analysis. The mGPS is validated for prognostication of relapse and increased mortality in different types of cancer [25–27], and can also predict clinical outcomes in patients with heart failure with preserved ejection fraction [28] While no studies to date have investigated its usefulness in COVID-19, we have demonstrated that mGPS is a potential tool for predicting risk of 28-day mortality. For a score of 2 vs. 1 or 0, we found an absolute increase of 26.3% (26.3% vs. 0.0%) mortality risk. This was the largest increase amongst our biomarkers but needs to be interpreted more cautiously given the high proportion of missing mGPS data. Also, while an mGPS score of 2 versus 1 or 0 conferred the greatest relative risk of death compared to cutoffs in other biomarkers in the unadjusted analysis, this did not hold in the adjusted analysis. The relationship between mGPS and COVID-19 needs to be studied further to validate its role in prognostication.
Other inflammatory biomarkers
The remainder of the analysis showed mixed results for many other biomarkers that are mostly in keeping with the available literature on the topic, particularly for LDH, CK and Albumin. [8, 12, 18, 19, 29–31]. Notably, our analysis did not support D-dimer’s use for prognosticating mortality risk, however, it must note that we are limited in our recommendation due to the high degree of missing d-dimer data. However, our analysis did find that it was significant in predicting admission to hospital. Previous studies have been inconsistent in reporting the role of d-dimer, varying from reports to support its role in predicting disease severity, to its association with modestly increased mortality risk and even extreme high risk of mortality [8, 11, 17–19]. Other biomarkers studied include platelets and lymphocytes. These biomarkers have had very little literature written regarding associations with disease severity in COVID-19. However, our findings are mostly in keeping with these earlier findings. [8, 32].
Our analysis found varying associations between biomarkers and disease severity (admission to ICU or hospital) and 28-day mortality. The reason for why specific biomarkers may predict disease severity (admission to hospital or ICU) but not overall mortality or vice-versa may be explained by correlations between these biomarkers and clinician decision making.
Strengths and limitations
Our study has many strengths, including the relatively large size of our patient cohort and the fact that this cohort is specifically North American. Furthermore, we tested previously established COVID-19 biomarkers, as well as novel inflammatory indices including L/WBC ratio and mGPS. Our analysis of the previously studied biomarkers bolsters evidence for generalizability in North American populations and provides specific cut-off values for clinicians to utilize for predicting mortality. We believe that these preliminary North American studies provide clinicians an important piece of information that helps determine objective risk stratification. Interestingly, there is often a discrepancy of underlying pathophysiology that portends a worst outcome and the initial presentation of COVID-19 patients. Inflammatory biomarkers and useful cut offs can be used to help make objective risk stratification of patients. This can be useful in patients who come in initially well but have elevated inflammatory markers, which may prompt for more initial aggressive monitoring and treatment. Furthermore, this study is the first North American study that may serve as the basis for integration clinical prediction models validated for specific questions, including treatment decisions.
Missing data are a notable limitation. The missingness of our data varied by biomarker and was associated with disease severity, namely biomarkers such as CRP or Ferritin were more likely to be ordered in patients in the ICU. Although we used multiple imputation to account for the missing data, there are known limitations to the method [33]. Furthermore, it is difficult to deal with potential superimposed infection that may confound whether the biomarkers truly represent COVID-19 infection or the unknown superimposed infection. We think that using admission blood at least reduces the risk of bias but avoids potential infection that often occurs in COVID-19 patients. However, it would be difficult to rule out this confounder if potential infections occurred prior to admission. Lastly, given that all the biomarkers tested exhibited significant correlation, multivariate models were not produced, as the models would experience high degrees of multicollinearity.
Conclusion and relevance
We have demonstrated that several commonly used inflammatory biomarkers can be used to predict 28-day mortality, including C-reactive protein, neutrophil-to-lymphocyte ratio, and lymphocyte-to-white blood count ratio. We also demonstrated the predictive value of less commonly used composite measures such as the modified Glasgow prognostic score. The validation of these specific biomarkers may be useful in monitoring response to therapies, especially as more and more drug therapies are developed and tested. We derived novel cutoff values for these biomarkers which can be used at the time of admission to help predict mortality and inform clinicians’ decision-making in North American populations. Because components of the complete blood count are readily available, we would recommend the routine use of these biomarkers in prognosticating disease courses. Further study needs to be done to determine whether clinicians can use these biomarkers to guide therapies in patients with COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study leveraged the McMaster Coronavirus (COVID-19) Registry (COREG) led by Drs. Andrew Costa (NPI), Marla Beauchamp (co-PI), MyLinh Duong (co-PI), Rebecca Kruisselbrink (co-PI), and Terence Ho (co-PI) and including the following institutions: Grand River Hospital, St. Mary’s General Hospital, Hamilton Health Sciences, St. Joseph’s Healthcare Hamilton, and the Niagara Health System. COREG is supported by a grant from the Canadian Institutes of Health Research (CIHR) (172754) and from the Hamilton Academic Health Sciences Organization (HAHSO) (HAH-21-04). The COREG acknowledges the following individuals for their contributions: Darly Dash, Laura Dawson, Michael Dennis, Megan Donaghy-Hughes, Hannah Farnworth, Edward Feng, Stefan Jevtic, Candice Luo, Michael Mallender, Rina Patel, Vivek Patel, Nivedh Patro, Noam Raiter, Pranali Raval, Brittany Salter, Laura Spatafora, Xinxin Tang, Cooper Webb, Kristin Wright, and Zaka Ahmad Zia. The members of the MPH on behalf of the COREG Investigators are, William Ciccotelli, Sophie Corriveau, George Farjou, Stephen Giilck, Carla Girolametto, Lauren Griffith, Brent Guy, Shariq Haider, Rajendar Hanmiah, Paul Hosek, Mats Lyndon Junek, Jessica Kapralik, Cindy Cin Yee Law, Theresa T. Liu, Maura Marcucci, Leslie Martin, John Neary, Ameen Patel, Natya Raghavan, Parminder Raina, Samir Raza, Connie Schumacher, Catherine Tong, Jennifer Tsnlrang, and Joshua Wald.
Funding
Canadian Institutes of Health Research (CIHR) (172,754) and from the Hamilton Academic Health Sciences Organization (HAHSO) (HAH-21-04).
Data availability
Request can be made to the corresponding author.
Code availability
Request can be made to the corresponding author.
Compliance with ethical standards
Conflict of interest
The authors declared that they have no conflict of interest.
Statement of human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
For this type of study, formal consent is not required.
Footnotes
The COREG Investigators are listed in acknowledgements.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tyler Pitre, Email: tyler.pitre@medportal.ca.
on behalf of the COREG Investigators:
William Ciccotelli, Sophie Corriveau, George Farjou, Stephen Giilck, Carla Girolametto, Lauren Griffith, Brent Guy, Shariq Haider, Rajendar Hanmiah, Paul Hosek, Mats Lyndon Junek, Jessica Kapralik, Cindy Cin Yee Law, Theresa T. Liu, Maura Marcucci, Leslie Martin, John Neary, Ameen Patel, Natya Raghavan, Parminder Raina, Samir Raza, Connie Schumacher, Catherine Tong, Jennifer Tsnlrang, and Joshua Wald
References
- 1.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 2.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: A review. Clin Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Violi F, Cangemi R, Department of Translational and Precision Medicine, Romiti GF, Ceccarelli G, et al. (2020) Is Albumin Predictor of Mortality in COVID-19?. Mary Ann Liebert, Inc., publishers. 2020 [cited 2020Dec11]. Available from: https://www.liebertpub.com/doi/full/10.1089/ars.2020.8142
- 5.Zeng F, Huang Y, Guo Y, et al. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis. 2020;96:467–474. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. 2020;50(4):332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F, Li L, Xu M, et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proctor MJ, Morrison DS, Talwar D, et al. An inflammation-based prognostic score (mGPS) predicts cancer survival independent of tumour site: a Glasgow inflammation outcome study. Br J Cancer. 2011;104(4):726–734. doi: 10.1038/sj.bjc.6606087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Yan X, Fan Q, et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J Thromb Haemost. 2020;18(6):1324–1329. doi: 10.1111/jth.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Qin L, Li K, et al. A novel scoring system for prediction of disease severity in COVID-19. Front Cell Infect Microbiol. 2020 doi: 10.3389/fcimb.2020.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Zhou P, Liu Y, et al. Platelet-to-lymphocyte ratio in advanced cancer: review and meta-analysis. Clin Chim Acta. 2018;483:48–56. doi: 10.1016/j.cca.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Valdes M, Villeda J, Mithoowani H, Pitre T, Chasen M. Inflammatory markers as prognostic factors of recurrence in advanced stage squamous cell carcinoma of the head and neck. Curr Oncol. 2020;27(3):135–141. doi: 10.3747/co.27.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang A-P, Liu J-P, Tao W-Q, Li H-M. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol. 2020;84:106504. doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Ma J, Jiang Z, Ming L. Prognostic value of neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio in acute pulmonary embolism: a systematic review and meta-analysis. Int Angiol. 2018;37(1):4–11. doi: 10.23736/S0392-9590.17.03848-2. [DOI] [PubMed] [Google Scholar]
- 17.Tang N, Bai H, Chen X, Gong J, Li D, Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemost. 2020;18(5):1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan China. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Zheutlin AB, Kao Y-H, et al. Analysis of hospitalized COVID-19 patients in the Mount Sinai health system using electronic medical records (EMR) reveals important prognostic factors for improved clinical outcomes. medRxiv. 2020 doi: 10.1101/2020.04.28.20075788. [DOI] [Google Scholar]
- 20.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. J Stat Softw. 2011;45(1):1–67. doi: 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 21.R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/. Published in 2018. Accessed June 30, 2020
- 22.Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang W, Dong J, Ren Y, et al. The value of clinical parameters in predicting the severity of COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf DM, Friedrichs I, Toma AG. Lymphocyte-white blood cell count ratio: a quickly available screening tool to differentiate acute purulent tonsillitis from glandular fever. Arch Otolaryngol Head Neck Surg. 2007;133(1):61–64. doi: 10.1001/archotol.133.1.61. [DOI] [PubMed] [Google Scholar]
- 25.Nozoe T, Matono R, Ijichi H, Ohga T, Ezaki T. Glasgow prognostic score (GPS) can be a useful indicator to determine prognosis of patients with colorectal carcinoma. Int Surg. 2014;99(5):512–517. doi: 10.9738/INTSURG-D-13-00118.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jomrich G, Hollenstein M, John M, et al. The modified glasgow prognostic score is an independent prognostic indicator in neoadjuvantly treated adenocarcinoma of the esophagogastric junction. Oncotarget. 2018;9(6):6968–6976. doi: 10.18632/oncotarget.24087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanai N, Sawabe M, Kimura T, et al. The high-sensitivity modified Glasgow prognostic score is superior to the modified Glasgow prognostic score as a prognostic predictor for head and neck cancer. Oncotarget. 2018;9(97):37008–37016. doi: 10.18632/oncotarget.26438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolat I, Biteker M. Modified Glasgow Prognostic Score is a novel predictor of clinical outcome in heart failure with preserved ejection fraction. Scand Cardiovasc J. 2020;54(3):174–178. doi: 10.1080/14017431.2019.1709656. [DOI] [PubMed] [Google Scholar]
- 29.Giacomelli A, Ridolfo AL, Milazzo L, et al. 30-day mortality in patients hospitalized with COVID-19 during the first wave of the Italian epidemic: a prospective cohort study. Pharmacol Res. 2020;158:104931. doi: 10.1016/j.phrs.2020.104931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aziz M, Fatima R, Lee-Smith W, Assaly R. The association of low serum albumin level with severe COVID-19: a systematic review and meta-analysis. Crit Care. 2020 doi: 10.1186/s13054-020-02995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Li M, Zheng S, et al. Plasma albumin levels predict risk for nonsurvivors in critically ill patients with COVID-19. Biomark Med. 2020 doi: 10.2217/bmm-2020-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terpos E, Ntanasis-Stathopoulos I, Elalamy I, et al. Hematological findings and complications of COVID-19. Am J Hematol. 2020;95(7):834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sterne JAC, White IR, Carlin JB, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009 doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Request can be made to the corresponding author.
Request can be made to the corresponding author.