Abstract
The dysregulation of long non-coding RNAs (lncRNAs) serves a pivotal role in the pathogenesis and development of multiple types of human cancer, including gastric cancer (GC). MCM3AP-antisense 1 (MCM3AP-AS1) has been reported to function as a tumor promoter in various types of cancer. However, the biological function of MCM3AP-AS1 in the resistance of GC cells to cisplatin (CDDP) remains to be elucidated. The present study aimed to elucidate the mechanisms of MCM3AP-AS1 in the resistance of GC cells to CDDP. The expression levels of MCM3AP-AS1, miR-138 and FOXC1 were measured via reverse transcription-quantitative PCR. In addition, cell viability, migration and invasion were assessed via the Cell Counting Kit-8, wound healing and transwell assays, respectively. The interaction between genes was confirmed via the dual-luciferase reporter and pull-down assays. Western blot analysis was performed to detect FOXC1 protein expression. In the present study, it was demonstrated that MCM3AP-AS1 expression was upregulated in CDDP-resistant GC cells and that MCM3AP-AS1-knockdown suppressed CDDP resistance in GC cells. Moreover, the examination of the molecular mechanism indicated that MCM3AP-AS1 upregulated FOXC1 expression by sponging microRNA (miR)-138. Additionally, it was identified that the overexpression of FOXC1 abolished MCM3AP-AS1-knockdown- or miR-138 mimic-mediated inhibitory effects on CDDP resistance in GC cells. In conclusion, the present findings suggested that MCM3AP-AS1 enhanced CDDP resistance by sponging miR-138 to upregulate FOXC1 expression, indicating that MCM3AP-AS1 may be a novel promising biomarker for the diagnosis and treatment of patients with GC.
Keywords: MCM3AP-antisense 1, gastric cancer, cisplatin resistance, microRNA-138, FOXC1
Introduction
Gastric cancer (GC) is the third leading cause of cancer-associated mortality and the fifth most frequent type of cancer worldwide. According to the latest statistics, there were more than 1,000,000 new cases of GC and an estimated 783,000 deaths from GC worldwide in 2018 (1–3). Risk factors, including smoking and atrophic gastritis, have been identified to contribute to the incidence of GC (4). Despite improvements in the diagnosis and treatment of GC, the 5-year survival rate for patients with GC remains unsatisfactory (5). Chemotherapeutic cisplatin (CDDP) is one of the main options for the treatment of patients with various types of cancer (6). However, CDDP treatment often results in the development of chemoresistance, leading to therapeutic failure (7). Therefore, it is important to understand the molecular regulatory mechanism underlying chemoresistance and to identify effective tumor biomarkers to improve the prognosis of patients with GC.
Long non-coding RNAs (lncRNAs) are a type of transcript, with a length of >200 nucleotides, that lack protein-coding ability (8,9). Previous studies have reported that lncRNAs are involved in the regulation of human cancer by acting as oncogenes or tumor suppressor genes. For example, Wu et al (10) revealed that lncRNA MEG3 inhibited the progression of prostate cancer via binding to microRNA (miRNA/miR)-9-5p and regulating quaking I-5 protein expression. Moreover, Gao et al (11) indicated that lncRNA NORAD facilitated the progression of non-small cell lung cancer by enhancing cell proliferation and glycolysis. Additionally, Xu et al (12) demonstrated that lncRNA ROR1-antisense RNA 1 (AS1) sponged miR-375 to aggravate lung adenocarcinoma metastasis and to enhance epithelial-mesenchymal transition, while Zhang et al (13) reported that the lncRNA FOXD2-AS1/miR-25-3p/Sema4c axis increased colorectal cancer cell migration and invasion.
lncRNAs are reported to be involved in the regulation of CDDP resistance in GC. For instance, lncRNA MALAT1 enhances autophagy-associated CDDP resistance in GC by sponging miR-20b and by upregulating autophagy protein 5 (14). Furthermore, prostate cancer-associated ncRNA transcripts 1 promotes CDDP resistance in GC by regulating the miR-128/zinc-finger E-box binding homeobox 1 axis (15), while OIP5-AS1 suppression increases CDDP sensitivity by targeting the miR-377-3p/fos-like antigen 2 axis in osteosarcoma (16). MCM3AP-AS1 has been reported to promote tumorigenesis and progression in various types of cancer, such as pancreatic cancer, hepatocellular carcinoma and breast cancer (17–19). However, the regulatory effects of lncRNA MCM3AP-AS1 on CDDP resistance in GC remain to be determined.
The present study aimed to investigate the role of MCM3AP-AS1 and the regulatory mechanisms associated with this lncRNA in CDDP-resistant GC cells, which may provide a novel biomarker for the diagnosis and treatment of GC.
Materials and methods
Cell lines and cell culture
GC cell lines (AGS, MKN45, NCI-N87 and SNU638), the normal gastric mucosa epithelial GES-1 cell line and 293T cells were obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were incubated in RPMI-1640 medium supplemented with 10% FBS (both purchased from Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Thermo Fisher Scientific, Inc.). CDDP-resistant GC cell lines (NCI-N87/CDDP and AGS/CDDP) were induced in RPMI-1640 medium with increasing concentrations (0–10 µg/ml) of CDDP at 37°C for more than 6 months. Cell incubation conditions included a humidified environment with 5% CO2 at 37°C.
Cell transfection
Specific short hairpin RNAs (shRNAs) against MCM3AP-AS1 (shMCM3AP-AS1#1; 5′-GGGAGUAAGUGAAAGUAAU-3′ and shMCM3AP-AS1#2; 5′-GCUUCGAUGUGUUACUUAA-3′) were used to knock down MCM3AP-AS1 expression. A sh-negative control (shNC; 10 nM; 5′-UUCUCCGAACGUGUCACGU-3′) was used as the NC. shRNAs were used for stable transfection. The pcDNA3.1 plasmids targeting FOXC1 or MCM3AP-AS1 were used to overexpress FOXC1 or MCM3AP-AS1, respectively, with empty pcDNA3.1 as the NC. miR-138 mimics (5′-AGCUGGUGUUGUGAAUCAGGCCG-3′) and miR-138 inhibitor (5′-CGGCCUGAUUCACAACACCAGCU-3′) were utilized to overexpress and knock down miR-138 expression, respectively. NC mimics (NC miR-mimics; 10 nM; 5′-ACUCUAUCUGCACGCUGACUU-3′) and NC inhibitor (NC miR-inhibitor; 10 nM; 5′-CAGUACUUUUGUGUAGUACAA-3′) served as scrambled NCs. The aforementioned vectors were provided by Shanghai GenePharma Co., Ltd.. Transfection was conducted using Lipofectamine® 2000 for 48 h at 37°C (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were subjected to subsequent experimentation 48 h following transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from NCI-N87 and AGS cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A PrimeScript RT reagent kit (cat. no. RR047A; Takara Biotechnology Co., Ltd.) or a TaqMan™ Advanced miRNA cDNA Synthesis kit (cat. no. 4366596; Thermo Fisher Scientific, Inc.) were used to reverse transcribe RNAs into cDNA according to the manufacturer's protocol. RT-qPCR was performed on the ABI 7500 real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using a SYBR-Green PCR Master Mix kit (Takara Biotechnology Co., Ltd.). The following thermocycling conditions were used: Pre-denaturation at 95°C for 15 sec, denaturation at 94°C for 30 sec, annealing at 60°C for 20 sec, and extension at 72°C for 40 sec for 40 cycles. The expression levels of genes were calculated utilizing the 2−ΔΔCq method (20), with GAPDH or U6 as the internal controls The primer (Shanghai GeneChem) sequences used in the present study were as follows: MCM3AP-AS1 forward, 5′-CCTATCCCTTTCTCTAAGA-3′, reverse, 5′-ACTTCTGCAAAAACGTGCTG-3′; miR-138 forward, 5′-GTTAGGGCAGGTGGGATG-3′, reverse, 5′-TGTATGCGGCTGGTAAGTAG-3′; FOXC1 forward, 5′-AACATCGCCTGCGTTATCCTC-3′, reverse, 5′-ACGTCCCGGATGATCCCAA-3′; U6 forward, 5′-TTATGGGTCCTAGCCTGAC-3′, reverse, 5′-CACTATTGCGGGTCTGC-3′; and GAPDH forward, 5′-CCACATCGCTCAGACACCAT-3′ and reverse, 5′-ACCAGGCGCCCAATACG-3′.
Cell viability and drug-sensitivity assay
The viability of NCI-N87/CDDP and AGS/CDDP cells was assessed using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay. Cells (1×104 cells/well) were seeded into 96-well plates. Following incubation at 37°C for 0, 24, 48 and 72 h, each well was supplemented with 10 µl CCK-8 reagent and the cells were incubated for 4 h at room temperature to examine cell viability. Finally, cell viability was represented by absorbance values at 450 nm, which were measured using the MRX II microplate reader (Dynex Technologies).
To determine the IC50, NCI-N87/CDDP and AGS/CDDP cells were treated with varying concentrations (0, 1, 2, 4, 8, 16 and 32 µg/ml) of CDDP for 48 h.
RNA pull-down
An RNA pull-down assay was conducted using the Pierce Magnetic RNA-Protein Pull-Down kit (cat. no. 20164; Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. GC cells were transfected with 50 µM biotinylated-MCM3AP-AS1 (Biotin-MCM3AP-AS1) and biotinylated NC. Cells were collected 48 h after transfection and subsequently lysed in RNase-free cell lysis solution (1 mM HEPES, 200 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 200 U/ml RNase Inhibitor) at 4°C. Cell lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen; Thermo Fisher Scientific, Inc.) overnight at 4°C, according to the manufacturer's protocol. Next, the beads were washed with high salt buffer (1% Triton X-100; 0.1% SDS; 20 mM Tris-HCl, pH 8.0; 2 mM EDTA; 500 mM NaCl). After washing and centrifugation (1,500 × g; 10 min, 4°C), the pellet was lysed with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), the binding of miR-138 to MCM3AP-AS1 was assessed via RT-qPCR, as aforementioned.
Wound healing assay
NCI-N87 and AGS cells (5×104 cells) were plated in 6-well plates and incubated with RPMI-1640 medium without serum for 24 h at 37°C. When cells reached 100% confluence, they were used forthe wound healing assay. A 200-µl pipette tip was used to generate scratches. The wound width at 0 and 24 h was observed under an light microscope (magnification, ×200; Olympus Corporation) and measured using ImageJ software version 1.8 (National Institutes of Health).
Transwell assay
NCI-N87 and AGS cells (1×104 cells) suspended in serum-free medium were added into the upper chambers, which were precoated with Matrigel® at 37°C for 1 h. The lower chamber was filled with RPMI-1640 medium (600 µl) containing 10% FBS. After 48 h of incubation at 37°C, invasive cells were fixed with methanol for 20 min at room temperature and stained with 0.1% crystal violet for 20 min at room temperature. The number of invasive cells was counted under a light microscope (magnification, ×200; Olympus Corporation).
Luciferase reporter assay
The starBase 2.0 database (http://starbase.sysu.edu.cn/) was used to predict the potential target genes of MCM3AP-AS1. The pmirGLO vectors (Shanghai GenePharma Co., Ltd.) of MCM3AP-AS1-wild-type (Wt) or MCM3AP-AS1-mutant (Mut) reporters were co-transfected with NC mimics, miR-138 mimics, NC inhibitor or miR-138 inhibitor into 293T cells using Lipofectamine 2000. In addition, pmirGLO-FOXC1-Wt or pmirGLO-FOXC1-Mut reporters were co-transfected with NC mimics, miR-138 mimics, NC inhibitor or miR-138 inhibitor into 293T cells using Lipofectamine 2000. After 48 h, the relative luciferase activity was measured using a Dual Luciferase Reporter assay system (Promega Corporation) and normalized to Renilla luciferase activity.
Western blot analysis
The proteins from transfected AGS/CDDP cells were isolated using RIPA lysis buffer (Beyotime Institute of Biotechnology) and then quantified using a BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). After separating via 10% SDS-PAGE (Bio-Rad Laboratories, Inc.), a total of 30 µg proteins were transferred onto PVDF membranes (Invitrogen; Thermo Fisher Scientific, Inc.). After blocking with 5% skimmed milk for 2 h at room temperature, membranes were incubated with primary antibodies against FOXC1 (1:1,000; cat. no. ab227977; Abcam) and GAPDH (1:1,000; cat. no. ab9485; Abcam) overnight at 4°C. Subsequently, HRP-conjugated goat anti-rabbit IgG secondary antibodies (1:1,000; cat. no. ab205718; Abcam) were incubated with the membranes for 2 h at room temperature. The blots were visualized using an ECL detection system (Thermo Fisher Scientific, Inc.).
Nuclear/cytoplasmic RNA fractionation
Nuclear and cytoplasmic fractions were acquired from NCI-N87/CDDP and AGS/CDDP cells using nucleoplasmic fractionation buffer (140 mmol/l NaCl, 1.5 mmol/l MgCl2, 10 mmol/l Tris-HCl pH 8.5, 0.5% NP-40). First, cell pellet was resuspended through nucleoplasmic fractionation buffer and incubated for 5 min on ice. After centrifugation, the supernatant and pellet were collected as the cytoplasmic and nuclear fractions, respectively. RNA was isolated from nuclear/cytoplasmic fractions, and RT-qPCR was then used to assess the levels of MCM3AP-AS1, GAPDH and U6. GAPDH served as the cytoplasmic endogenous control, and U6 served as the nuclear endogenous control.
Statistical analysis
Data are presented as the mean ± SD, and were analyzed using SPSS 20.0 Software (IBM Corp.). Each experiment was repeated ≥3 times. A one-way ANOVA followed by Tukey's post hoc test and unpaired Student's t-test were used to compare the differences among >2 and 2 groups, respectively. P<0.05 was considered to indicate a statistically significant difference.
Results
MCM3AP-AS1-knockdown weakens CDDP resistance in GC cells
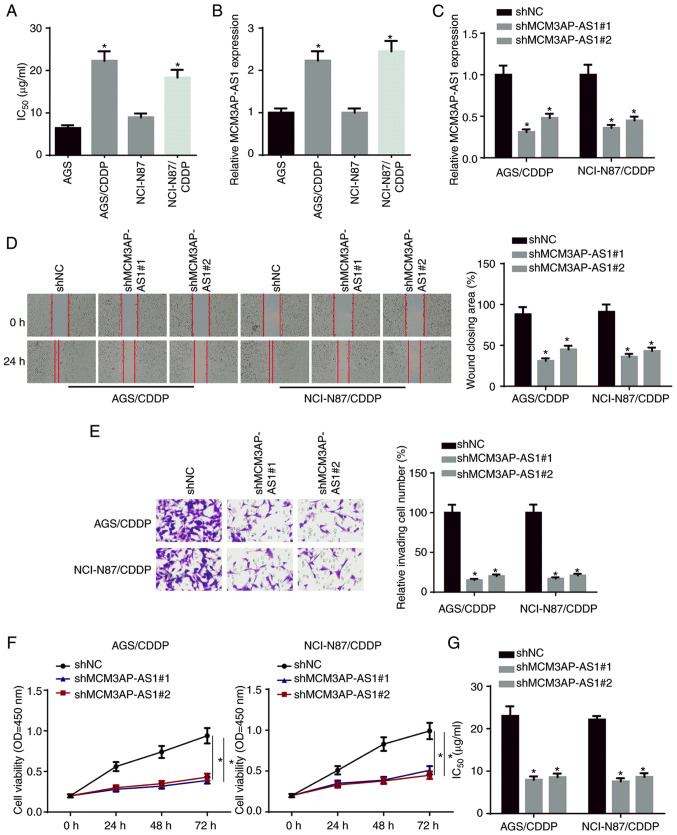
To examine the biological role of MCM3AP-AS1 in the CDDP resistance of GC cells, CDDP-resistant NCI-N87 cells (NCI-N87/CDDP) and AGS cells (AGS/CDDP) were established. As presented in Fig. 1A, the IC50 of CDDP was significantly increased in NCI-N87/CDDP and AGS/CDDP cells compared with in their parental cell lines. Moreover, MCM3AP-AS1 expression was upregulated in CDDP-resistant GC cells compared with in their parental cell lines (Fig. 1B). Subsequently, NCI-N87/CDDP and AGS/CDDP cells were transfected with shMCM3AP-AS1#1/2. The transfection efficiency was confirmed via RT-qPCR (Fig. 1C). Wound healing, Transwell and CCK-8 assay results indicated that the knockdown of MCM3AP-AS1 significantly inhibited the migration, invasion and proliferation of NCI-N87/CDDP and AGS/CDDP cells (Fig. 1D-F). In addition, it was found that the IC50 in NCI-N87/CDDP and AGS/CDDP cells was significantly decreased following MCM3AP-AS1-knockdown (Fig. 1G). These findings indicated that MCM3AP-AS1-knockdown potentiated the CDDP sensitivity of GC cells.
Figure 1.
MCM3AP-AS1-knockdown weakens CDDP resistance in GC cells. (A) The IC50 was tested in NCI-N87, NCI-N87/CDDP, AGS and AGS/CDDP. (B) MCM3AP-AS1 expression in GC cells and CDDP-resistant GC cells was measured by RT-qPCR. (C) The knockdown efficiency of MCM3AP-AS1 in NCI-N87/CDDP and AGS/CDDP cells was assessed by RT-qPCR. (D) Migratory (magnification, ×200) and (E) invasive abilities (magnification, ×200) of CDDP-resistant GC cells were detected after repressing MCM3AP-AS1 by wound healing and Transwell assays, respectively. (F) Cell viability of NCI-N87/CDDP and AGS/CDDP cells was measured after repressing MCM3AP-AS1 by Cell Counting Kit-8 assay. (G) The IC50 of NCI-N87/CDDP and AGS/CDDP was tested after repressing MCM3AP-AS1 by drug-sensitivity assay. *P<0.05 vs. AGS, NCI-N87 or shNC. MCM3AP-AS1, MCM3AP antisense RNA 1; CDDP, cisplatin; GC, gastric cancer; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin RNA; NC, negative control; OD, optical density.
MCM3AP-AS1 overexpression enhances CDDP resistance in GC cells
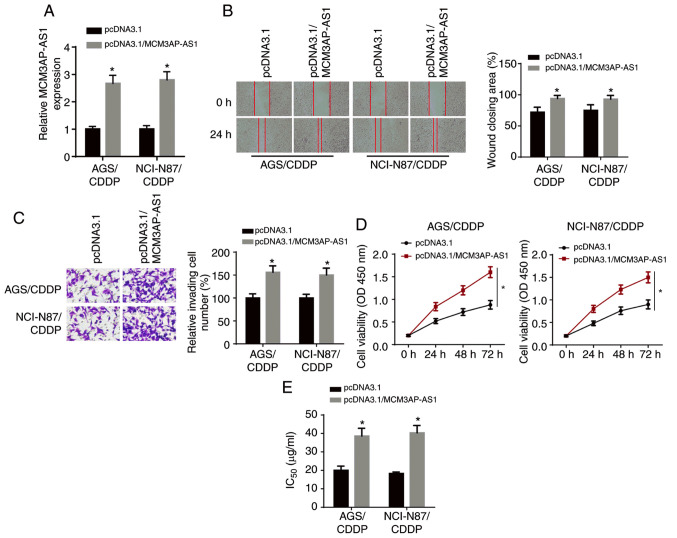
The overexpression efficiency of MCM3AP-AS1 was verified via RT-qPCR analysis (Fig. 2A). Wound healing and Transwell assay results demonstrated that the overexpression of MCM3AP-AS1 significantly enhanced the migratory and invasive abilities of NCI-N87/CDDP and AGS/CDDP cells (Fig. 2B and C). Moreover, CCK-8 assay results identified that the proliferative ability of NCI-N87/CDDP and AGS/CDDP cells was increased after MCM3AP-AS1 overexpression (Fig. 2D). It was found that the IC50 was significantly enhanced in NCI-N87/CDDP and AGS/CDDP cells after MCM3AP-AS1 overexpression (Fig. 2E). Thus, MCM3AP-AS1 overexpression decreased the CDDP sensitivity of GC cells.
Figure 2.
MCM3AP-AS1 overexpression enhances CDDP resistance in GC cells. (A) The overexpression efficiency of MCM3AP-AS1 in NCI-N87/CDDP and AGS/CDDP cells was assessed by reverse transcription-quantitative PCR. (B) Migratory (magnification, ×200) and (C) invasive abilities (magnification, ×200) of CDDP-resistant GC cells was detected after overexpressing MCM3AP-AS1 by wound healing and Transwell assays, respectively. (D) Cell viability of NCI-N87/CDDP and AGS/CDDP cells was measured after overexpressing MCM3AP-AS1 by Cell Counting Kit-8 assay. (E) The IC50 of NCI-N87/CDDP and AGS/CDDP cells was tested after overexpressing MCM3AP-AS1 by drug-sensitivity assay. *P<0.05 vs. pcDNA3.1. MCM3AP-AS1, MCM3AP antisense RNA 1; CDDP, cisplatin; GC, gastric cancer; OD, optical density.
MCM3AP-AS1 facilitates CDDP resistance in GC cells via targeting miR-138
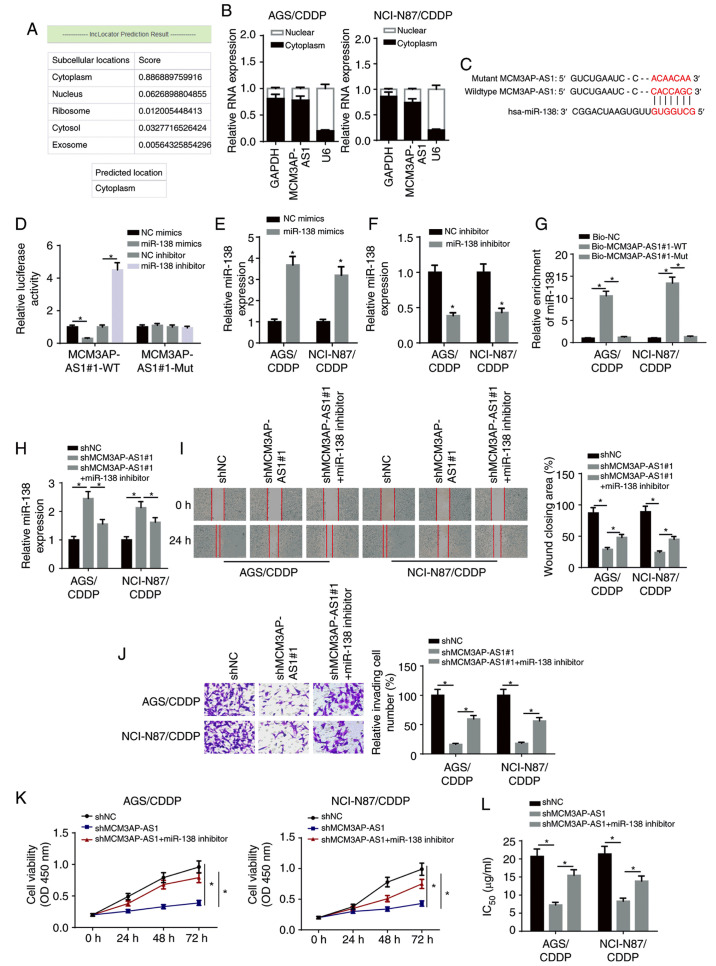
Accumulating evidence has revealed that lncRNAs function as competitive endogenous (ce)RNAs in the regulation of multiple types of human cancer by sponging miRNAs (21,22). Therefore, it was hypothesized that MCM3AP-AS1 may be involved in the regulation of CDDP resistance in GC via the ceRNA network. First, the LncLocator program (http://www.csbio.sjtu.edu.cn/bioinf/LncLocator/) predicted that MCM3AP-AS1 was mainly expressed in the cytoplasm (Fig. 3A). Subsequently, nuclear-cytoplasmic fractionation assay results suggested that the majority of MCM3AP-AS1 was distributed in the cytoplasm of GC cells (Fig. 3B). Using the starBase v2.0 database, the potential binding sites between MCM3AP-AS1 and miR-138 were predicted (Fig. 3C). A luciferase reporter assay demonstrated that the luciferase activity of pmirGLO-MCM3AP-AS1-Wt reporters was significantly decreased in the miR-138 mimics group, but was enhanced in the miR-138 inhibitor group; moreover, no significant differences were identified in pmirGLO-MCM3AP-AS1-Mut reporters (Fig. 3D).
Figure 3.
MCM3AP-AS1 facilitates CDDP resistance of GC cells by targeting miR-138. (A) LncLocator program predicted the subcellular localization of MCM3AP-AS1. (B) MCM3AP-AS1 expression in the cytoplasmic and nuclear fractions was measured in NCI-N87/CDDP and AGS/CDDP cells by subcellular fraction assay. (C) The binding site between MCM3AP-AS1 and miR-138 was predicted using the starBase website. (D) The binding ability of MCM3AP-AS1 for miR-138 was detected in 293T cells by luciferase reporter assay. miR-138 expression in NCI-N87/CDDP and AGS/CDDP cells transfected with (E) NC mimics and miR-138 mimics, and (F) NC inhibitor and miR-138 inhibitor. (G) The binding capacity between MCM3AP-AS1 and miR-138 was verified by RNA pull-down assay. (H) miR-138 expression was assessed in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by reverse transcription-quantitative PCR. (I) Migratory (magnification, ×200) and (J) invasive abilities (magnification, ×200) of NCI-N87/CDDP and AGS/CDDP cells were detected in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by wound healing and Transwell assay, respectively. (K) Cell viability of NCI-N87/CDDP and AGS/CDDP cells was measured in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by Cell Counting Kit-8 assay. (L) The IC50 values of CDDP-resistant GC cells was tested in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by drug-sensitivity assay. *P<0.05. miR, microRNA; MCM3AP-AS1, MCM3AP antisense RNA 1; CDDP, cisplatin; GC, gastric cancer; sh, short hairpin RNA; NC, negative control; OD, optical density; WT, wild-type; Mut, mutant; Bio, biotinylated.
RT-qPCR revealed that miR-138 expression was upregulated in NCI-N87/CDDP and AGS/CDDP cells transfected with miR-138 mimics, and downregulated in NCI-N87/CDDP and AGS/CDDP cells transfected with miR-138 inhibitor (Fig. 3E and F). RNA-pull down assay results indicated that biotinylated-MCM3AP-AS1 (Biotin-MCM3AP-AS1-WT) was able to directly precipitate miR-138, while biotinylated-MCM3AP-AS1 with predicted mutant binding sites (Biotin-MCM3AP-AS1-Mut) could not precipitate miR-138 (Fig. 3G). In addition, miR-138 expression was significantly increased by knocking down MCM3AP-AS1, but this effect was abrogated by the miR-138 inhibitor (Fig. 3H). Additionally, MCM3AP-AS1-knockdown inhibited the migration, invasion and proliferation of CDDP-resistant GC cells, which was then reversed by miR-138 silencing (Fig. 3I-K). Additionally, the miR-138 inhibitor partially abolished the inhibitory effect of MCM3AP-AS1-knockdown on the IC50 in NCI-N87/CDDP and AGS/CDDP cells (Fig. 3L). Therefore, it was suggested that MCM3AP-AS1 increased the CDDP resistance of CDDP-resistant GC cells via sponging miR-138.
MCM3AP-AS1 serves as a ceRNA for miR-138 to upregulate FOXC1 expression
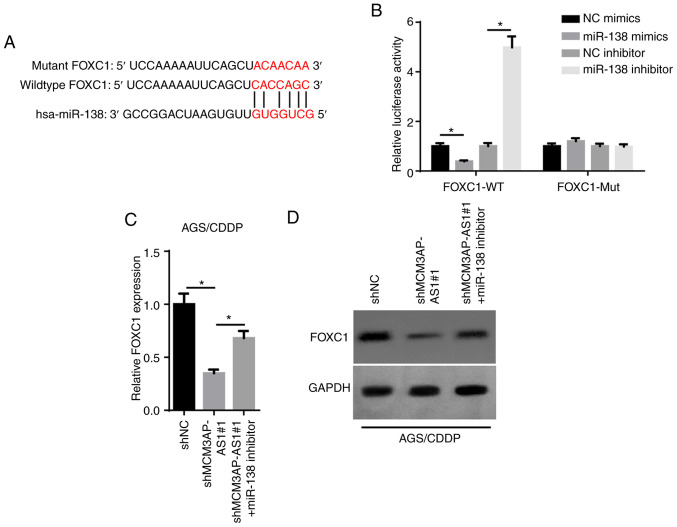
Upregulated FOXC1 has been reported to be associated with a poor prognosis in patients with GC (23). The starBase database was used to predict the binding sites between miR-138 and FOXC1 (Fig. 4A). As presented in Fig. 4B, the luciferase activity of the FOXC1-WT reporters was significantly decreased by overexpressing miR-138, while it was enhanced by inhibiting miR-138. Additionally, no significant change was observed in the FOXC1-Mut group (Fig. 4B). Furthermore, FOXC1 mRNA and protein expression was decreased by MCM3AP-AS1-knockdown, but this effect was counteracted by the miR-138 inhibitor in AGS/CDDP cells (Fig. 4C and D). Overall, these findings demonstrated that MCM3AP-AS1 positively regulated FOXC1 expression by directly sponging miR-138.
Figure 4.
MCM3AP-AS1 serves as a competitive endogenous RNA for miR-138 to upregulate FOXC1 expression. (A) The binding site between FOXC1 and miR-138 was obtained from the starBase website. (B) The binding ability of MCM3AP-AS1 for miR-138 was detected by luciferase reporter assay in 293T cells. FOXC1 expression was measured in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by (C) reverse transcription-quantitative PCR and (D) western blot assays. *P<0.05. miR, microRNA; MCM3AP-AS1, MCM3AP antisense RNA 1; CDDP, cisplatin; sh, short hairpin RNA; NC, negative control; WT, wild-type; Mut, mutant.
MCM3AP-AS1/miR-138 axis increases the resistance of GC cells to CDDP by regulating FOXC1
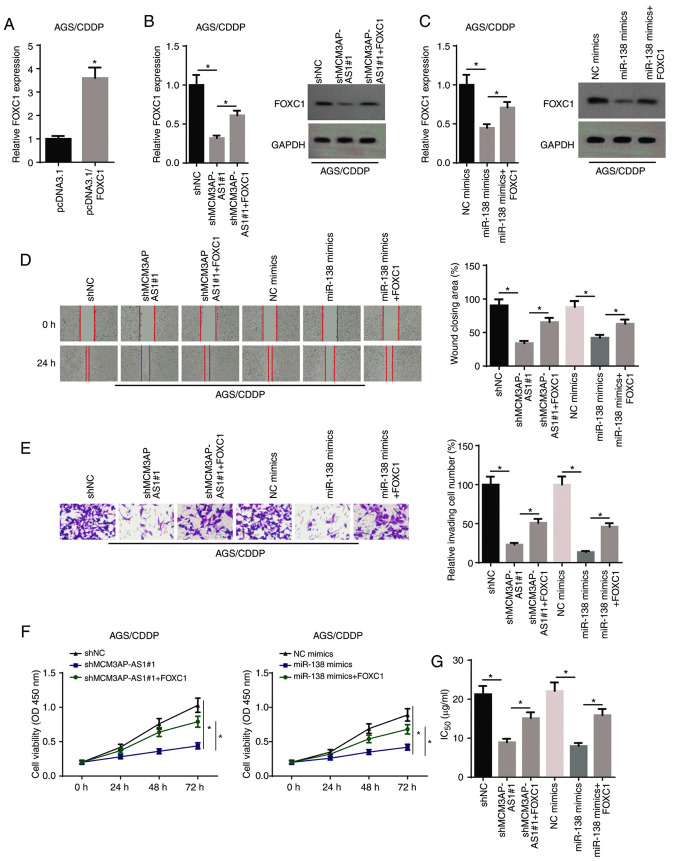
RT-qPCR revealed that transfection of the FOXC1 overexpression plasmid significantly increased FOXC1 expression in AGS/CDDP cells (Fig. 5A). As illustrated in Fig. 5B and C, FOXC1 expression was decreased in AGS/CDDP cells transfected with shMCM3AP-AS1#1 or miR-138 mimics, but this effect was counteracted by overexpressing FOXC1. Rescue assays were performed to examine whether the MCM3AP-AS1/miR-138/FOXC1 axis affected the resistance of GC cells to CDDP. Subsequently, wound healing, Transwell and CCK-8 assay results identified that the overexpression of FOXC1 could reverse the decrease in cellular migration, invasion and proliferation caused by MCM3AP-AS1-knockdown or miR-138 mimics in AGS/CDDP cells (Fig. 5D-F). In addition, FOXC1 overexpression partially abrogated the inhibitory effect of MCM3AP-AS1-knockdown or miR-138 overexpression on the IC50 in AGS/CDDP cells (Fig. 5G). Therefore, it was indicated that the MCM3AP-AS1/miR-138 axis regulated CDDP resistance of GC cells via FOXC1.
Figure 5.
MCM3AP-AS1/miR-138 increases gastric cancer cell resistance to CDDP via regulating FOXC1. (A) FOXC1 expression in AGS/CDDP cells transfected with pcDNA3.1 and pcDNA3.1-FOXC1. (B) FOXC1 expression was assessed in AGS/CDDP cells transfected with shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+FOXC1 groups by RT-qPCR and western blot assays. (C) FOXC1 expression was assessed in AGS/CDDP cells transfected with NC mimics, miR-138 mimics and miR-138 mimics+FOXC1 groups by RT-qPCR and western blot assays. (D) Migratory (magnification, ×200) and (E) invasive abilities (magnification, ×200) of AGS/CDDP cells were detected in different groups by wound healing and Transwell assays, respectively. (F) Cell viability of AGS/CDDP cells was measured in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+FOXC1 groups, as well as NC, mimics, miR-138 mimics and miR-138 mimics+FOXC1 groups by Cell Counting Kit-8 assay. (G) The IC50 of AGS/CDDP cells was tested in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+FOXC1 groups, as well as NC, mimics, miR-138 mimics and miR-138 mimics+FOXC1 groups by drug-sensitivity assay. *P<0.05. miR, microRNA; MCM3AP-AS1, MCM3AP antisense RNA 1; CDDP, cisplatin; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin RNA; NC, negative control; OD, optical density.
Discussion
Chemoresistance is considered a major obstacle for cancer therapy in the clinic (24). lncRNAs are reported to be closely associated with chemoresistance in human cancer. For example, lncRNA HOXD-AS1 contributes to CDDP resistance in GC by epigenetically suppressing programmed cell death 4 expression by recruiting enhancer of zeste homolog 2 (25). Moreover, lncRNA NEAT1 enhances paclitaxel resistance of ovarian cancer via the miR-194/ZEB1 axis (26). Although MCM3AP-AS1 has been reported to act as an oncogene in various types of cancer, the biological role of lncRNA MCM3AP-AS1 in the resistance of GC cells to CDDP remains unknown. The present study demonstrated that MCM3AP-AS1 expression was upregulated in CDDP-resistant GC cells. In addition, MCM3AP-AS1-knockdown inhibited the migration, invasion and proliferation of CDDP-resistant GC cells, suggesting that MCM3AP-AS1 enhanced CDDP resistance in GC cells.
miRNAs are a class of endogenous non-coding RNAs with a length of 22–25 nucleotides (27,28) and they serve vital roles in various types of cancer, including GC. For example, Hu et al (29) reported that miR-4317 inhibited the proliferation of GC cells by targeting zinc finger 322, while He and Zou (30) revealed that miR-96 promoted the proliferation of GC cells by inhibiting FOXO3 expression. miR-138 has been shown to function as a tumor suppressor in various types of cancer, such as breast, ovarian and non-small cell lung cancer (31–33). In addition, several studies have indicated that miR-138 is involved in the drug resistance of multiple types of human cancer. For instance, Tang et al (34) reported that miR-138 inhibited gefitinib resistance in non-small cell lung. Furthermore, Li et al (35) observed that LINC00174-knockdown decreased chemoresistance in glioma cells by downregulating miR-138 expression. The present study identified that miR-138 was a downstream target of MCM3AP-AS1, and that a miR-138 inhibitor could abolish the inhibitory effect of MCM3AP-AS1-knockdown on the CDDP resistance of GC cells.
Previous studies have reported that lncRNAs may act as ceRNAs by sponging miRNAs to release downstream mRNAs (36–38). FOXC1, a member of the FOX transcription factor family, serves a vital role in numerous types of cancer, such as colorectal, non-small cell lung and prostate cancer (39–41). With regards to GC, Xu et al (23) found that FOXC1 overexpression contributed to poor prognosis in patients with GC. The present results indicated that MCM3AP-AS1-knockdown inhibited FOXC1 expression by targeting miR-138 in GC. In addition, it was identified that the overexpression of FOXC1 abolished the inhibitory effects of MCM3AP-AS1-knockdown (or miR-138 mimics) on CDDP resistance in GC.
In conclusion, to the best of our knowledge, the present study was the first to investigate the role and potential mechanism of MCM3AP-AS1 in the resistance of GC cells to CDDP. The current findings demonstrated that MCM3AP-AS1 promoted CDDP resistance in GC by sponging miR-138 to upregulate FOXC1 expression, which may provide a novel promising therapeutic approach for GC treatment. However, in vivo studies and associated clinical trials are required to verify and elucidate this molecular mechanism.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
HS, PW and WC designed the present study. PW, BZ and XW performed all the experiments, analyzed the data and prepared the figures. HS and WC drafted the initial manuscript. WC and PW reviewed and revised the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Wadhwa R, Taketa T, Sudo K, Blum MA, Ajani JA. Modern oncological approaches to gastric adenocarcinoma. Gastroenterol Clin North Am. 2013;42:359–369. doi: 10.1016/j.gtc.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Ashraf N, Hoffe S, Kim R. Adjuvant treatment for gastric cancer: Chemotherapy versus radiation. Oncologist. 2013;18:1013–1021. doi: 10.1634/theoncologist.2012-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh S. Cisplatin: The first metal based anticancer drug. Bioorg Chem. 2019;88:102925. doi: 10.1016/j.bioorg.2019.102925. [DOI] [PubMed] [Google Scholar]
- 7.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 8.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu M, Huang Y, Chen T, Wang W, Yang S, Ye Z, Xi X. lncRNA MEG3 inhibits the progression of prostate cancer by modulating miR-9-5p/QKI-5 axis. J Cell Mol Med. 2019;23:29–38. doi: 10.1111/jcmm.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao W, Weng T, Wang L, Shi B, Meng W, Wang X, Wu Y, Jin L, Fei L. Long non-coding RNA NORAD promotes cell proliferation and glycolysis in non-small cell lung cancer by acting as a sponge for miR1365p. Mol Med Rep. 2019;19:5397–5405. doi: 10.3892/mmr.2019.10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu N, Qiao L, Yin L, Li H. Long noncoding RNA ROR1-AS1 enhances lung adenocarcinoma metastasis and induces epithelial-mesenchymal transition by sponging miR-375. J BUON. 2019;24:2273–2279. [PubMed] [Google Scholar]
- 13.Zhang M, Jiang X, Jiang S, Guo Z, Zhou Q, He J. lncRNA FOXD2-AS1 regulates miR-25-3p/Sema4c axis to promote the invasion and migration of colorectal cancer cells. Cancer Manag Res. 2019;11:10633–10639. doi: 10.2147/CMAR.S228628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi Z, Si J, Nan J. lncRNA MALAT1 potentiates autophagy-associated cisplatin resistance by regulating the microRNA-30b/autophagy-related gene 5 axis in gastric cancer. Int J Oncol. 2019;54:239–248. doi: 10.3892/ijo.2018.4609. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Yue P, Wang Y, Chen G, Li Y. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother. 2019;118:109255. doi: 10.1016/j.biopha.2019.109255. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Wang S. Long non-coding RNA OIP5-AS1 knockdown enhances CDDP sensitivity in osteosarcoma via miR-377-3p/FOSL2 axis. Onco Targets Ther. 2020;13:3853–3866. doi: 10.2147/OTT.S232918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang M, Sun S, Guo Y, Qin J, Liu G. Long non-coding RNA MCM3AP-AS1 promotes growth and migration through modulating FOXK1 by sponging miR-138-5p in pancreatic cancer. Mol Med. 2019;25:55. doi: 10.1186/s10020-019-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Luo C, Zhang G. lncRNA MCM3AP-AS1 regulates epidermal growth factor receptor and autophagy to promote hepatocellular carcinoma metastasis by interacting with miR-455. DNA Cell Biol. 2019;38:857–864. doi: 10.1089/dna.2019.4770. [DOI] [PubMed] [Google Scholar]
- 19.Chen Q, Xu H, Zhu J, Feng K, Hu C. lncRNA MCM3AP-AS1 promotes breast cancer progression via modulating miR-28-5p/CENPF axis. Biomed Pharmacother. 2020;128:110289. doi: 10.1016/j.biopha.2020.110289. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, Liu Y, Zhang N, Yang Q. lncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. lncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129. doi: 10.1016/j.lfs.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Shao QS, Yao HB, Jin Y, Ma YY, Jia LH. Overexpression of FOXC1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2014;64:963–970. doi: 10.1111/his.12347. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Zhu QN, Deng JL, Li ZX, Wang G, Zhu YS. Emerging role of long non-coding RNAs in cisplatin resistance. Onco Targets Ther. 2018;11:3185–3194. doi: 10.2147/OTT.S158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, Yang S, Han Y, Sun J, Xv L, Wu L, Ming L. HOXD-AS1 confers cisplatin resistance in gastric cancer through epigenetically silencing PDCD4 via recruiting EZH2. Open Biol. 2019;9:190068. doi: 10.1098/rsob.190068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.An J, Lv W, Zhang Y. lncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther. 2017;10:5377–5390. doi: 10.2147/OTT.S147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendell JT. MicroRNAs: Critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 28.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 29.Hu X, Zhang M, Miao J, Wang X, Huang C. miRNA-4317 suppresses human gastric cancer cell proliferation by targeting ZNF322. Cell Biol Int. 2018;42:923–930. doi: 10.1002/cbin.10870. [DOI] [PubMed] [Google Scholar]
- 30.He X, Zou K. miRNA-96-5p contributed to the proliferation of gastric cancer cells by targeting FOXO3. J Biochem. 2020;167:101–108. doi: 10.1093/jb/mvz080. [DOI] [PubMed] [Google Scholar]
- 31.Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1α. Int J Cancer. 2013;133:867–878. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Liu D, Feng Z, Mao J, Zhang C, Lu Y, Li J, Zhang Q, Li Q, Li L. MicroRNA-138 modulates metastasis and EMT in breast cancer cells by targeting vimentin. Biomed Pharmacother. 2016;77:135–141. doi: 10.1016/j.biopha.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Zhu D, Gu L, Li Z, Jin W, Lu Q, Ren T. miR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol Res Pract. 2019;215:861–872. doi: 10.1016/j.prp.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Tang X, Jiang J, Zhu J, He N, Tan J. HOXA4-regulated miR-138 suppresses proliferation and gefitinib resistance in non-small cell lung cancer. Mol Genet Genomics. 2019;294:85–93. doi: 10.1007/s00438-018-1489-3. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Zhao H, Song J, Wang F, Chen M. LINC00174 down-regulation decreases chemoresistance to temozolomide in human glioma cells by regulating miR-138-5p/SOX9 axis. Hum Cell. 2020;33:159–174. doi: 10.1007/s13577-019-00281-1. [DOI] [PubMed] [Google Scholar]
- 36.Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M, Zhang R, Lu L, Du J, Chen C, Ding K, Wei X, Zhang G, Huang Y, Hou J. lncRNA PVT1 accelerates malignant phenotypes of bladder cancer cells by modulating miR-194-5p/BCLAF1 axis as a ceRNA. Aging (Albany NY) 2020;12:22291–22312. doi: 10.18632/aging.202203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Li W, Lin Z, Hu J, Wang J, Ren Y, Wei B, Fan Y, Yang Y. The long noncoding RNA Linc01833 enhances lung adenocarcinoma progression via miR-519e-3p/S100A4 axis. Cancer Manag Res. 2020;12:11157–11167. doi: 10.2147/CMAR.S279623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun SH, Han SH, Park JI. COUP-TFII knock-down promotes proliferation and invasion in colorectal cancer cells via activation of Akt pathway and up-regulation of FOXC1. Anticancer Res. 2020;40:177–190. doi: 10.21873/anticanres.13939. [DOI] [PubMed] [Google Scholar]
- 40.Gong R, Lin W, Gao A, Liu Y, Li J, Sun M, Chen X, Han S, Men C, Sun Y, Liu J. Forkhead box C1 promotes metastasis and invasion of non-small cell lung cancer by binding directly to the lysyl oxidase promoter. Cancer Sci. 2019;110:3663–3676. doi: 10.1111/cas.14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang H, Xiong Y, Wu Z, He Y, Gao X, Zhou Z, Wang T. MIR-138-5P inhibits the progression of prostate cancer by targeting FOXC1. Mol Genet Genomic Med. 2020;8:e1193. doi: 10.1002/mgg3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.