Abstract
The recent detection of SARS-CoV-2 RNA in feces has led to speculation that it can be transmitted via the fecal-oral/ocular route. This review aims to critically evaluate the incidence of gastrointestinal (GI) symptoms, the quantity and infectivity of SARS-CoV-2 in feces and urine, and whether these pose an infection risk in sanitary settings, sewage networks, wastewater treatment plants, and the wider environment (e.g. rivers, lakes and marine waters). A review of 48 independent studies revealed that severe GI dysfunction is only evident in a small number of COVID-19 cases, with 11 ± 2% exhibiting diarrhea and 12 ± 3% exhibiting vomiting and nausea. In addition to these cases, SARS-CoV-2 RNA can be detected in feces from some asymptomatic, mildly- and pre-symptomatic individuals. Fecal shedding of the virus peaks in the symptomatic period and can persist for several weeks, but with declining abundances in the post-symptomatic phase. SARS-CoV-2 RNA is occasionally detected in urine, but reports in fecal samples are more frequent. The abundance of the virus genetic material in both urine (ca. 102–105 gc/ml) and feces (ca. 102–107 gc/ml) is much lower than in nasopharyngeal fluids (ca. 105–1011 gc/ml). There is strong evidence of multiplication of SARS-CoV-2 in the gut and infectious virus has occasionally been recovered from both urine and stool samples. The level and infectious capability of SARS-CoV-2 in vomit remain unknown. In comparison to enteric viruses transmitted via the fecal-oral route (e.g. norovirus, adenovirus), the likelihood of SARS-CoV-2 being transmitted via feces or urine appears much lower due to the lower relative amounts of virus present in feces/urine. The biggest risk of transmission will occur in clinical and care home settings where secondary handling of people and urine/fecal matter occurs. In addition, while SARS-CoV-2 RNA genetic material can be detected by in wastewater, this signal is greatly reduced by conventional treatment. Our analysis also suggests the likelihood of infection due to contact with sewage-contaminated water (e.g. swimming, surfing, angling) or food (e.g. salads, shellfish) is extremely low or negligible based on very low predicted abundances and limited environmental survival of SARS-CoV-2. These conclusions are corroborated by the fact that tens of million cases of COVID-19 have occurred globally, but exposure to feces or wastewater has never been implicated as a transmission vector.
Keywords: Bathing waters, Fecal-oral route, Infection risk, Sanitation, Waterborne illness
Graphical abstract
1. Introduction
In recent years, several viral epidemics have impacted human populations, resulting in substantial morbidity, mortality and a negative impact on the global economy [e.g. Zika virus (ZIKV), Ebola virus (EBOV), severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV)] (Peckham, 2013; Watkins, 2018). Of these, respiratory viruses such as coronaviruses (CoV) have proven particularly problematic to control due to their ease of human-to-human transmission and wide range of primary and secondary animal reservoirs (Assiri et al., 2013; Damas et al., 2020). They were also recently highlighted by the World Health Organization in 2018 as priority areas for research given their potential to cause a public health emergency and the absence of efficacious drugs and/or vaccines (WHO, 2018). To date, seven human coronaviruses (HCoV) have been identified that can induce a range of respiratory symptoms with variable case fatality rates. These include the circulating seasonal HCoVs (i.e. higher winter prevalence) that are generally considered to cause mild respiratory symptoms (αCoVs; HCoV-229E and HCoV-NL63, β-CoVs; HCoV-HKU1 and HCoV-OC43), through to novel CoVs that lead to severe and potentially fatal respiratory tract infections (β-CoVs; SARS-CoV-1, MERS-CoV and SARS-CoV-2) (Gaunt et al., 2010; Guo et al., 2020a; Pfefferle et al., 2011). The novel Coronavirus Disease 2019 (COVID-19), caused by SARS-CoV-2, presents with a range of respiratory symptoms which, in an estimated 14–17% of cases, leads to severe or critical disease such as severe pneumonia or acute respiratory distress syndrome (ARDS) (Petrosillo et al., 2020; Wu and McGoogan, 2020; Docherty et al., 2020). Although SARS-CoV-2 belongs to the same β-CoV genus as the CoVs responsible for the severe acute respiratory syndrome (SARS; caused by SARS-CoV) and Middle East respiratory syndrome (MERS; caused by MERS-CoV), this newly emerged virus tends to be associated with milder infections. For example, depending on the country, case fatality rates from COVID-19 have been estimated to be ca. 1–5%, significantly lower than the death rates for SARS (9.5%) and MERS (35%) (Wu and McGoogan, 2020; De Wit et al., 2016; Rajgor et al., 2020; CDC, 2020). In addition, SARS and MERS are predominantly associated with nosocomial spread, whereas SARS-CoV-2 is much more widely transmitted in the community, particularly in places with high population densities such as overcrowded housing, industrial processing plants, elderly care homes, and prisons (Abrams et al., 2020; Cloud et al., 2020; Franco-Paredes et al., 2020; Petrosillo et al., 2020; Razum et al., 2020; Waltenburg et al., 2020).
Coronaviruses are enveloped, positively charged (at neutral pH), single-stranded viruses that possess the largest genomes of all known RNA viruses (26.4 to 31.7 kb), giving them considerable plasticity to accommodate, acquire and modify genes, enabling jumps between animal hosts (Woo et al., 2010; Perlman and Netland, 2009). This is mainly evidenced by the observed spillover of SARS, MERS and now SARS-CoV-2, the emergence of new variants of SARS-CoV-2 and thus the possibility for antigenic drift (Koyama et al., 2020). The genome size of SARS-CoV-2 lies at the upper end of the coronavirus range (29.9 kB), encoding a total of 11 genes with 11 open reading frames (Yoshimoto, 2020). The direct ancestor of SARS-CoV-2 appears to have been circulating unnoticed for decades in bats and subsequently transmitted to pangolins and then humans (Boni et al., 2020). SARS-CoV-2 is 96.2% identical to the bat CoV RaTG13, and is far more distantly related to both SARS-CoV-1 (79.5% identity) and MERS-CoV (50% identity) (Guo et al., 2020a; Paraskevis et al., 2020; Rabaan et al., 2020; Andersen et al., 2020). The genetic differences between SARS-CoV-1 and SARS-CoV-2 (380 amino acid substitutions) are largely clustered in non-structural protein genes; however, 27 mutations also are present in genes encoding the viral spike protein S responsible for receptor binding and cell entry. These differences have resulted in contrasting patterns of human infection (e.g. antigen detection) and replication compared with both SARS-CoV-1 and MERS-CoV. Consequently, the use of past studies on SARS-CoV-1 and MERS-CoV to explain the transmission and environmental behavior of SARS-CoV-2 should be done with caution. Although SARS-CoV-2 is thought to be largely spread by the inhalation of contaminated respiratory droplets or via contact with fomites, the fecal-oral route also has been suggested in its spread due to the fact that infected persons can shed SARS-CoV-2 RNA in bodily fluids (e.g. feces and to a lesser extent urine; Peng et al., 2020). However, considerable debate exists about the relative importance of this pathway, partially because a comprehensive review does not yet exist.
Here we critically assess current and previous available evidence on (i) gastrointestinal (GI) symptoms associated with COVID-19, (ii) the behavior of SARS-CoV-2 in the GI tract, (iii) the abundance of SARS-CoV-2 in feces and urine, (iv) the evidence that SARS-CoV-2 remains infectious after release from the body, and (v) whether feces and urine in sanitary environments, sewage systems and wastewater consequently pose a risk to human health.
2. Proportion of COVID-19 cases showing gastrointestinal symptoms
Patients infected with SARS-CoV-2 typically exhibit a wide range of symptoms including fever, coughing, dyspnea, sore throat and headaches. In addition, GI symptoms including nausea, vomiting, loss of appetite, diarrhea, and abdominal pain have been reported (Lo et al., 2020; Adhikari et al., 2020). GI problems are also observed in other acute respiratory infections (e.g. influenza viruses, human rhinoviruses) and have been reported as a very common symptom of severe influenza in children (Poole et al., 2020). In some cases, this is due to co-infections with other organisms, but is frequently due to simultaneous viral replication in multiple organs, including the GI tract (Minodier et al., 2017; Rovida et al., 2013).
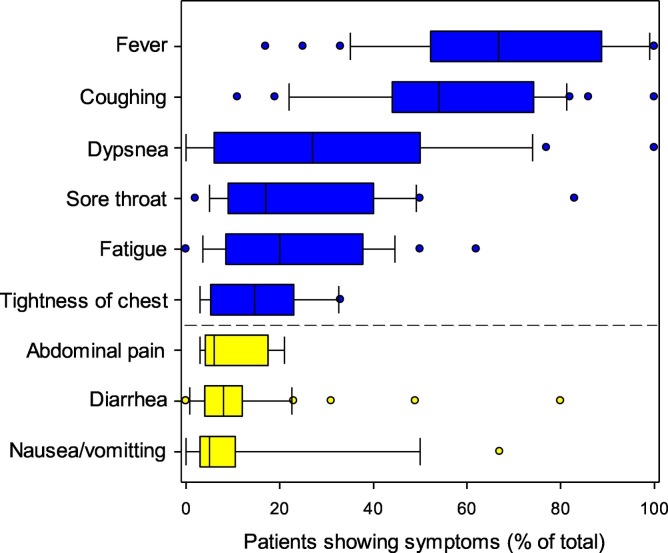
Most reports on the symptoms of COVID-19 are derived from clinical cases. From these, however, the number, range and severity of symptoms associated with COVID-19 can vary largely from person to person. Overall, our analysis of the symptoms from 48 independently published studies involving thousands of individuals has shown that a small, but significant number of patients experience gastrointestinal problems. Incidence of GI complaints, vomiting and diarrhea is similar to SARS-CoV-1 and MERS-CoV (Rabaan et al., 2020; Kanwar et al., 2017). Current evidence also suggests that rates of GI symptoms from SARS-CoV-2 are comparable in both children and adults in symptomatic cases. However, it should be noted that there is a greater proportion of asymptomatic carriage and mild infections in children in comparison to adults (Dong et al., 2020; Wang et al., 2020a). Further, other studies suggest the incidence of diarrhea is greatest in severely ill patients, while abdominal pain and vomiting are not (Yang et al., 2020; Tian et al., 2020). Our analysis suggests that, on average, the number of hospitalized cases experiencing diarrhea is 11% ± 2% while those exhibiting vomiting and nausea is 12% ± 3% (mean ± SEM, n = 48 independent studies). It is unknown from the reported data to what extent these symptoms co-occur. In a rare number of cases, diarrhea has been shown to be the only COVID-19 symptom, making these cases very difficult to diagnose (Li et al., 2020a; Taxonera et al., 2020). Although there are reports of renal organ failure from SARS-CoV-2 in severe infections (Martinez-Rojas et al., 2020), there are fewer reports of urinary dysfunction as a result of infection (Prabhu et al., 2020). It should be noted that injury to the renal system is common in COVID-19 cases, but that in most individuals these effects are subclinical (Martinez-Rojas et al., 2020). Further, the data presented in Fig. 1 does not account for SARS-CoV-2 infections that are either asymptomatic or very mild, and do not require hospitalization. Asymptomatic cases may account for ca. 40–45% of SARS-CoV-2 infections, with the potential to transmit the virus for extended periods, possibly longer than 14 days (Oran and Topol, 2020). It is therefore likely the incidence of these symptoms is greater than shown in Fig. 1. This underreporting is common for gastrointestinal infections (Fletcher et al., 2013; Gleizes et al., 2006). The variability in the data may also be associated with different reporting criteria for each condition used in the different studies (Kwan et al., 2005). Further, data may also be slightly confounded due to the administration of anti-viral drugs, antibiotics and traditional and alternative medicines to patients that also induce diarrhea and vomiting (Tian et al., 2020). Consequently, diarrhea in COVID-19 patients is not always associated with SARS-CoV-2 and may explain why GI symptoms do not correlate well with the severity of diseases or worse outcomes (Aguila et al., 2020; Cao et al., 2020). While self-reporting of SARS-CoV-2 infection and symptoms has been used in some countries to capture mild cases of COVID-19, these data have large uncertainties due to ‘hypochondriacal suspicion’ and the inclusion of symptoms from other diseases also circulating in the population (Gong et al., 2020). For this reason, this type of data was considered unreliable.
Fig. 1.
Summary of symptoms experienced in clinically reported SARS-CoV-2 infections. The data is the summary of 48 independent reports involving a total of 3706 patients. The yellow bars are those associated with gastrointestinal problems. In the box plots, the boundary of the box closest to zero indicates the 25th percentile, a black line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers above and below the box indicate the 10th and 90th percentiles. Points above and below the whiskers indicate outliers outside the 10th and 90th percentiles. The average size of the cohort studies was 79 ± 21 (n = 48). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As evidenced from Fig. 1, abdominal pain is a common symptom of COVID-19. The extent to which this is directly due to viral infection of the GI tract or from general anxiety, however, remains unknown. A range of studies have shown that the threat of contracting COVID-19 can induce a range of somatic symptoms (e.g. sleep dysfunction, GI pain, headaches; Liu et al., 2020a; Yuan et al., 2020; Shevlin et al., 2020). Somatic symptoms of nausea, vomiting, abdominal pain and diarrhea are also known to be common in society. In some cases, the levels of these GI-related symptoms in society are consistent with reports for symptom frequency in COVID-19 cases (Haug et al., 2002a, Haug et al., 2002b).
We conclude from our analysis that SARS-CoV-2 clearly causes gastrointestinal dysfunction in a small, but substantial proportion of COVID-19 cases (ca. 5–20%). However, the likelihood of prevalence could be much greater due to underreporting of mild infections. In addition, due to the prevalence of somatic symptoms, these symptoms should not be used as direct evidence for actual GI infection.
3. Fecal shedding patterns of SARS-CoV-2
Consistent with the symptoms presented in Fig. 1, SARS-CoV-2 RNA has been routinely detected in upper and lower respiratory tract fluids, sputum, saliva, stool, blood, and urine of infected persons (Yan et al., 2020; Lu et al., 2020). The presence of the virus in feces appears to be similar in patients both with and without GI symptoms (Lin et al., 2020). Overall, however, SARS-CoV-2 is mostly detected in respiratory tract samples (typical range 70–100%), to a lesser extent in stool (typical range 30–60%), and rarely in urine (<5%) (Lo et al., 2020; Huang et al., 2020a; Kashi et al., 2020). In a few cases, even though it cannot be detected in the upper respiratory tract, the virus can be found in stools (Zhang et al., 2020b; Ling et al., 2020). However, in these cases the potential for false-negatives cannot be discounted (Piras et al., 2020). This range of symptoms has led to speculation that there are two different subtypes of COVID-19 manifestations referred to as “gut-tropism” and “lung-tropism”, depending on where the virus enters the body (i.e. inhaled or ingested) and becomes established, and thus where symptoms develop (Lo et al., 2020). There is no evidence, however, to support this or that some strains of SARS-CoV-2 preferentially target the GI tract in comparison to the respiratory tract (Iwasaki and Grubaugh, 2020).
Shedding of the virus in feces and in respiratory droplets may occur ca. 3–5 days before other classic symptoms, such as fever or diarrhea manifest (i.e. pre-symptomatic; Buscarini et al., 2020; Wang et al., 2020b; He et al., 2020a). Current evidence suggests that despite showing no symptoms, asymptomatic, pre-symptomatic or post-symptomatic people may still be shedding the virus at appreciable levels, although asymptomatic individuals may not shed it for as long or in as high amounts as in severely infected individuals that require hospitalization (Lu et al., 2020; Su et al., 2020a; Shen et al., 2020; Chau et al., 2020; Byrne et al., 2020). Critically, however, it is not well established whether viral loads are similar between asymptomatic, and mild, moderate, or severe symptomatic cases, with conflicting reports present in the literature (Wang et al., 2020a; Lu et al., 2020; He et al., 2020a; Liu et al., 2020; Li et al., 2010; Schwierzeck et al., 2020; Zou et al., 2020). However, we note that if the viral loads are similar, the lack of coughing and diarrhea in asymptomatic cases should lower the risk of disease transmission.
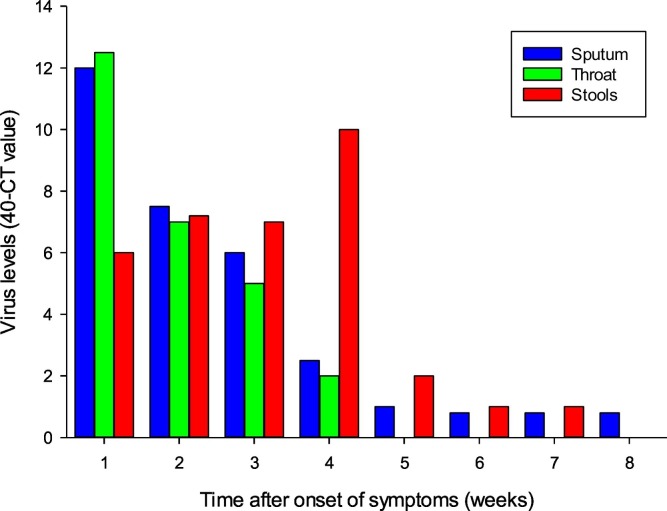
The information available so far from COVID-19 cases suggests the temporal dynamics of viral shedding in feces follows a classic infection cycle pattern (i.e. rapid build-up phase followed by a slow decline) (Sethuraman et al., 2020; Fig. 2 ). This is somewhat similar to that seen for SARS-CoV-1 where the rate of viral shedding in feces is low in the first five days of illness, but rises gradually to peak at days 9–14 with very high titers, often exceeding those of nasopharyngeal aspirates (Cheng et al., 2004). However, unlike SARS-CoV-1, it is known that shedding and transmission occurs with SARS-CoV-2 prior to symptom onset (Wei et al., 2020). In the case of SARS-CoV-2, initial reports provide good evidence of the rapid accumulation of viral loads in feces (Zhang et al., 2020b) and that it can be detected in stools of fecal-positive patients for at least two weeks after the decline of symptoms (Pan et al., 2020b). Since these early reports, the amount of fecal-positive cases in cohort-studies has been shown to be up to 75% of the total (Yan et al., 2020). Critically, however, it suggests that not all COVID-19 infections result in pronounced fecal shedding, consistent with the incidence of symptoms presented in Fig. 1. In addition, diarrhea is not always associated with viral shedding (Young et al., 2020). Taking all the available evidence on the temporal dynamics of viral shedding in feces suggests that shedding may occur for ca. five days prior to symptoms developing, ca. one week prior to hospitalization, and then for two weeks after symptoms have subsided (Lo et al., 2020; Byrne et al., 2020; Hosoda et al., 2020). Another diagnostic feature of COVID-19 cases is that SARS-CoV-2 can often be found in stool samples even after throat swabs appear negative in the post-symptomatic phase (Du et al., 2020a; Zhang et al., 2020a; Gupta et al., 2020; Xu et al., 2020; Jiang et al., 2020). For example, the median (IQR) time of detectable viral RNA was 18.5 (13−22) days for throat swabs, 22 (18–27) days for sputum, and 17 (11−32) days for stools (Fig. 2). In addition, viral loads in sputum and stool appear to decline slower than in throat swabs, with the longest shedding period recorded at 59 days (Huang et al., 2020b; Xiao et al., 2020b; Xu et al., 2020; Wu et al., 2020a). This has led to the suggestion that detection of SARS-CoV-2 in stool samples should be used alongside testing of viral presence in sputum and saliva samples (Ahamed Mim et al., 2020; Liu et al., 2020c; Ma et al., 2020; Mesoraca et al., 2020). However, in the late stages of infection it is possible that SARS-CoV-2 in feces may not be infectious and that RNA-based testing may result in unnecessary hospital bed-occupancy. Critically, this is supported by Hua et al. (2020) who found no transmission from fecal-positive children to family members.
Fig. 2.
Temporal dynamics of SARS-CoV-2 in the sputum, throat and stools. Data are from a cohort (n = 32) of COVID-19 patients in China. Adapted from Huang et al. (2020a) and Huang et al. (2020b).
4. Multiplication of SARS-CoV-2 in the gut
The evidence presented above has also led to the supposition that the fecal-oral route may be an opportunity for transmission of SARS-CoV-2 (Xu et al., 2020), as suggested previously also for SARS-CoV-1 and MERS-CoV (Yan et al., 2020). It is well established that stool samples contain an abundance of viruses in the human body and are an integral part of the transmission pathway for many pathogenic viruses (e.g. bocavirus, norovirus, rotavirus, astrovirus, sapovirus, adenovirus; Rovida et al., 2013; Drosten et al., 2013). Of the estimated 1.4 billion cases of diarrhea worldwide each year, viruses make up a considerable portion (Xie et al., 2013; Kotloff et al., 2019). Although seasonal HCoVs only make up a small proportion of these cases in comparison to viruses such as norovirus (NoV), rotavirus (RoV), rhinovirus (RhV) and adenovirus (AdV), it does imply that SARS-CoV-2 is not unusual in inducing GI problems and this symptom may represent a part of its infection cycle (Fig. 3 ) (Rovida et al., 2013; Drosten et al., 2013; Kheyami et al., 2010; Esper et al., 2010; Risku et al., 2010).
Fig. 3.
Prevalence of human pathogenic viruses in nasopharyngeal and stool samples from individuals (n = 331). The points represent individual viruses including Human Coronavirus (HCoV), Influenza A, Influenza B, Human Rhinovirus (HRV), Respiratory syncytial virus (RSV), Human Adenovirus (HAdV), Human Bocavirus (HBoV) and Human Parainfluenzavirus (HPIV). Data calculated from Minodier et al. (2017).
If sputum is swallowed, viral particles enveloped in mucus may pass down the GI tract in a semi-protected state and avoid degradation by gastric acid and bile/pancreatic juices (Hirose et al., 2017). This is likely to provide a primary route for infection of the GI tract, post-establishment of the virus in the upper respiratory tract. In addition, SARS-CoV-2 contained in sputum and saliva may also ultimately contribute to the viral load in feces, especially given the high viral load in these fluids and the large amounts (ca. 1.0–1.5 l person−1) produced and swallowed per day (Rudney et al., 1995; Iorgulescu, 2009). Although SARS-CoV-2 has been detected in blood, the prevalence rates are extremely low (ca. 1% of infections exhibit viremia; Lam et al., 2020), suggesting that this is not a primary route of infection of GI tract tissues and is a secondary manifestation of COVID-19. It is also possible that SARS-CoV-2 may reach the GI tract via contaminated food, however, there are no documented cases of food-borne transmission of SARS-CoV-2 (Li et al., 2021). A rare exception to this would be the handling and consumption of products from animals which have contracted the virus. The widespread risk of this, however, is likely to be extremely low based on evidence from previous SARS-CoV-1 and MERS-CoV outbreaks (Wang et al., 2005a; Todd, 2017; Rahman and Sarkar, 2019).
There is reasonable evidence to suggest that SARS-CoV-2 can replicate in the GI tract. Firstly, the GI tract contains an abundance of the metallopeptidase, angiotensin-converting enzyme 2 (ACE-2) which is the cell surface functional receptor (attachment site) for SARS-CoV-2 (Bertram et al., 2012; Lee et al., 2020; Li et al., 2020c). Secondly, it has been shown in vitro that HCoVs and SARS-CoV-2 can infect cells from the respiratory, gastrointestinal, hepatic and central nervous systems. Studies have indicated that SARS-CoV-2 has a 10–20 times greater affinity to ACE-2 receptors compared to SARS-CoV-1, with a potentially lower infectious dose (Galbadage et al., 2020; He et al., 2020b). It has been shown that the ACE-2 receptor protein is highly expressed not only in lung cells but also in esophageal epithelial cells and absorptive enterocytes (epithelial cells) present in the stomach, duodenum, ileum, colon and rectum (Xiao et al., 2020b; Lee et al., 2020; Li et al., 2020c; Zhang et al., 2020a, Zhang et al., 2020b, Zhang et al., 2020c, Zhang et al., 2020d, Zhang et al., 2020e; Guo et al., 2020b; Zang et al., 2020). Further, ACE-2 mRNA transcripts have been reported to be more abundant in intestinal cells than in lung tissues (Du et al., 2020b). Its expression in the small intestine has also been found to increase with age suggesting that it may help explain the increased severity of symptoms in eldery patients (Vuille-Dit-Bille et al., 2020). The ACE-2 receptor is also present in the kidney and bladder, suggesting the potential for viral replication in the urinary system (Du et al., 2020b; Li et al., 2020c; Martinez-Rojas et al., 2020) and potentially explaining the subsequent recovery of SARS-CoV-2 in urine (Ling et al., 2020). This is supported by autopsies of SARS-CoV-1 confirmed patients where presence of the virus has been demonstrated in tubular epithelial cells (Diao et al., 2020; Su et al., 2020). Gastrointestinal tissue samples obtained from esophageal, esophageal non-lesion, gastric, duodenum and rectum mucosa have also tested positive for the presence of SARS-CoV-2 in clinical cases (Xiao et al., 2020b).
Once in the GI tract, the spike (S) protein, which is abundant in the viral lipid membrane, induces binding of the virus to the ACE-2 receptor on the host cell surface, the main point of cell entry (Tian et al., 2020). The S glycoprotein has two key functional domains, S1 and S2. S1 contains the receptor-binding domain, which directly binds to the peptidase domain of ACE-2, whereas S2 is responsible for binding to the cell membrane (Mönkemüller et al., 2020). These two domains need to become physically separated to induce cell binding (i.e. activated). This process is initially mediated by the host cell protein convertase, furin, which acts on the S1/S2 site to break open the S protein structure to allow simultaneous binding to the ACE-2 receptor (via S1) and cell membrane (via S2; Bestle et al., 2020). This activation process is further facilitated by the host's type II transmembrane mucosa-specific serine protease (TMPRSS2) which acts on the S2ˊ domain to release the fusion peptide. TMPRSS2 is highly expressed in the lung, kidney, bladder, small intestine and colon relative to other tissues (Paniri et al., 2020). Fusion and subsequent entry of the viral genetic material into the host cell then occurs (Fig. 4 ; Hoffmann et al., 2020; Mönkemüller et al., 2020). Estimates suggest that this process takes from 10 to 15 min to complete (Ng et al., 2003). In addition to TMPRSS2, another mucosa-specific serine protease, TMPRSS4, also appears to enhance fusogenic activity and viral entry into the host cell (Zang et al., 2020). Once inside the cell, the uncoated viral RNA with 5′ cap structure and 3′ poly (A) tail, acts like mRNA, facilitating rapid translation of the replicase polyproteins (Pal et al., 2020). Once complete, viral replication proceeds, followed by RNA packing and envelope packaging as described in detail elsewhere (Guo et al., 2020a; Boopathi et al., 2020). The replicated virions are then released from the cell via exocytosis (i.e. continual budding rather than cell bursting) back into the GI tract to infect other cells (da Costa et al., 2020). This eclipse period (i.e. time taken from adsorption into the cell to the subsequent release of infectious progeny) is estimated to be 7–8 h (Harcourt et al., 2020; Schneider et al., 2012). Although not known for SARS-CoV-2, based on other viruses, each cell may produce up to 102–103 virions (Hirano et al., 1976). Given the number of epithelial cells with ACE-2 receptors in the GI tract, even a mild infection may therefore lead to a rapid multiplication of SARS-CoV-2, with the potential to produce a high abundance of viral RNA in fecal matter. Once released, however, the survival of these virions may be extremely low. For example, it has been shown that vesicular stomatitis virus chimeras expressing SARS-CoV-2 spike protein are rapidly inactivated by human colonic fluids with viral titers decreasing 100-fold in 1 h (Zang et al., 2020), however experiments using wild type SARS-CoV-2 are required to validate this finding. That said, this may help to explain why the capacity to recover infectious virus from stool specimens of COVID-19 patients is highly variable. It is also possible that transit time through the GI tract (i.e. greater in diarrhea cases; Roy et al., 1991) and pre-existing GI conditions (e.g. Crohn's disease, ulcerative colitis; An et al., 2020) may influence viral recovery in feces. This potentially poor survival contrasts with other human enteric viruses that primarily spread via the fecal-oral route (e.g. norovirus, rotavirus) and which are capable of withstanding the harsh environment in the GI tract, including the low pH of gastric fluids, bile and digestive enzymes in the small intestine and exposure to multiple bacterial by-products (Zang et al., 2020; Tung-Thompson et al., 2014) (Table 1 ).
Fig. 4.
Main routes by which SARS-CoV-2 leaves the body (left), and a summary of the mechanism of viral replication (right).
Table 1.
Comparison of the properties of SARS-CoV-2 with Norovirus, a virus with known fecal-oral transmission.
| SARS-CoV-2c | Norovirusc | |
|---|---|---|
| Family | Coronaviridae | Caliciviridae |
| Type | +ssRNA | +ssRNA |
| Shape | Spherical | Icosahedral |
| Genome size (kbp) | 29.9 | 7.5 |
| Size (nm) | 50–200 | 23–40 |
| Coating | Enveloped | Non-enveloped |
| Human infections per year | >7 million (Nov. 2019-Jun. 2020) | 685 million |
| Primary symptoms | Respiratory problems, fever, GI pain | Diarrhea, GI pain, vomiting |
| Prevalence of diarrhea (% of total cases) | 11 | 88 |
| Incubation period | 5–7 d | 1–3 d |
| Symptom duration | 7–14 d | 2–5 d |
| Death rate (% of total infections)a | 1.40 | 0.003 |
| Shedding rate in feces (gc/ml) | 102–107 | 108–1010 |
| Shedding duration after symptoms have subsided (d) | 14–28 | 14 |
| Infectious dose (PFU)b | Unknown (estimate 102–103) | 101–102 |
| Vaccine available | No | No |
| Cases directly linked to fecal-oral transmission | None | Frequent |
| Links to consuming contaminated water | None | Infrequent |
| Links to consuming contaminated food | None | Frequent |
| Individuals most at risk of complications | Elderly | Elderly |
| Environmental durability | Low | High |
| Sensitivity to low pH | High | Low |
| Sensitivity to alcohol | High | Low |
| Sensitivity to chlorine | High | Medium-high |
Deaths after accounting for both confirmed cases and estimates of asymptomatic carriage.
Infection mediated via the gastrointestinal tract. Only an estimate is available for SARS-CoV-2.
Values from the main text and from published values (Li et al., 2021; Robilotti et al., 2015; Hall et al., 2013; Pfeiffer, 2010; Kampf et al., 2020; Siddharta et al., 2017).
In mild COVID-19 infections, no significant damage to the mucous epithelium of esophagus, stomach, duodenum and rectum cells has been reported (Xiao et al., 2020b). However, it is also clear that severe infection can result in prolonged diarrhea and inflammation of the GI tract in a significant proportion of clinical cases (Fig. 1). Although tissue and organ damage may be precipitated by the body's immune response to SARS-CoV-2 infection (leading to the ‘cytokine storm’, viral sepsis and organ failure) (di Mauro et al., 2020; Li et al., 2020d), it might also be caused by direct viral attack of absorptive enterocytes which can induce diarrhea by destroying the cells resulting in malabsorption, unbalanced intestinal secretion and activated enteric nervous system (Tian et al., 2020; Zhang et al., 2020c). This is similar to that seen in porcine epidemic diarrhea (corona)virus (PEDV) infections where widespread histopathological damage to enterocytes occurs (Jung et al., 2014).
The role of the gut microbiome in the potential colonization of the GI tract by SARS-CoV-2 remains unknown. Evidence from the upper respiratory tract, however, suggests that some commensal bacteria in the mucosal biofilm (e.g. Proteobacteria) express proteins which can bind to the viral S-protein. This may prevent viral interactions with cell surface ACE-2 receptors and which may help prevent severe infections from developing (i.e. bacterial decoys; Honarmand Ebrahimi, 2020). Whether this occurs in the GI tract remains unknown, however, it should be noted that the overabundance of Proteobacteria in the GI tract is normally associated with dysbiosis (Shin et al., 2015; Rizzatti et al., 2017). It should also be noted that microbial diversity in the GI tract decreases with age, leading to suggestions that this may play a role in increased disease severity in elderly and type-II diabetic patients (Dhar and Mohanty, 2020; Gurung et al., 2020). It does suggest that further investigations of the gut microbiome are needed to establish its role is viral infection and the development of symptoms. Ultimately, this may also lead to the development of therapies to reduce the severity of COVID-19 (Kalantar-Zadeh et al., 2020). For example, fecal transplants have been considered as a treatment for critically ill COVID-19 patients, however, the success of such interventions remains unknown (Thalanayar Muthukrishnan and Faillace, 2020). Alternatively, changes in diet and the use of probiotics/prebiotics have also been proposed as a strategy to build immunity and reduce disease severity in the GI tract (Dhar and Mohanty, 2020; Ejtahed et al., 2020).
5. Levels of SARS-CoV-2 in urine and feces
A range of PCR-based technologies (e.g. RT-qPCR, digital PCR) are available to quantify the amount of SARS-CoV-2 RNA present in tissue, fluid and stool samples with very high sensitivity (≤10 gc in a sample). These assays typically target genes encoding the S, E and N structural proteins, the RdRp gene which encodes the RNA-dependent RNA polymerase or the replicase protein ORF1ab gene (van Kasteren et al., 2020). These quantitative assays, however, also have limitations that must be considered. For example, differences in sensitivity can occur depending on the PCR primer and probe sets used (Jung et al., 2020; Pillonel et al., 2020). Poor sensitivity and PCR inhibitors in fecal material (e.g. bile salts, lipids) may also lead to underestimation of viral abundance, or the reporting of false negatives (Schrader et al., 2012). Loss of viral signal during sample pre-treatment (e.g. heat inactivation) may also occur (Pan et al., 2020a). Further, the lack of extraction controls (i.e. surrogate CoVs to look at viral recovery from the sample) may lead to underestimates of viral abundance. The lack of standards has meant that only semi-quantitative results (i.e. Ct values) have been reported in most early studies, especially those examining the temporal dynamics of viral shedding. Lastly, these Ct values vary between platform, gene target assay, and template concentrations used, which causes issues of comparability between studies (Seong et al., 2016; Rahman et al., 2020). These issues seem to be most apparent in samples with high Ct values (Ct ≥ 34; Lowe et al., 2020). It is also important to state that quantification of viral RNA by RT-qPCR or digital PCR does not necessarily equate to infectious viral particles (Atkinson and Petersen, 2020), as it is likely that a large proportion of viral particles are damaged during passage through the GI tract and are thus non-infectious (Pfeiffer, 2010; Zhou et al., 2017; Zang et al., 2020). Despite these limitations, there is strong evidence to suggest that feces contain high viral RNA loads. For example, one study has shown that levels of SARS-CoV-2 RNA in stools can range from 5.5 × 102 to 1.2 × 105 copies/ml (Pan et al., 2020b), while another has reported levels of 6 × 105 to 7 × 106 gc/ml in three patients (Zang et al., 2020) and two studies reporting fecal shedding of up maximum of 1.0 × 107 gc/ml (Han et al., 2020; Wölfel et al., 2020). This wide variation in fecal viral RNA load (102–107 gc/ml) reflects differences in the severity of disease between patients and also the temporal dynamics of the disease (To et al., 2020b). It should be noted, however, that the abundance of SARS-CoV-2 RNA in feces are much lower than for other non-enveloped enteric viruses, such as norovirus (ssRNA virus; 108 to 1010/g; Lai et al., 2013; Lee et al., 2007), rotavirus (dsRNA virus; up to 109/g; Bennett et al., 2019) and adenovirus (dsDNA virus; 106 to 1011/g; Srinivasan et al., 2015).
In comparison with feces, at the peak of infection, levels of SARS-CoV-2 in saliva have been shown to typically range from 103 to 108 gc/ml with averages of 3.3 × 106 gc/ml (To et al., 2020a), 5.7 × 105 gc/ml (To et al., 2020b), 8.4 × 106 gc/ml (Yoon et al., 2020) and 5.0 × 105 gc/ml (Han et al., 2020). Analysis of nasopharyngeal fluid has reported values ranging from 6.4 × 102 gc/ml to 1.3 × 1011 gc/ml (median of 8.0 × 104 in throat samples and 7.5 × 105 in sputum samples) (Han et al., 2020; Pan et al., 2020b; Yoon et al., 2020), while others have reported viral loads ranging from 106 to 108 gc/ml in pharyngeal mucosa and endotracheal aspirate (To et al., 2020b; Fitzek et al., 2020). This implies that swallowing of sputum, saliva and nasopharyngeal fluids may contribute to the fecal SARS-CoV-2 RNA signal in some individuals. However, the fact that SARS-CoV-2 RNA cannot be found in feces from all infections (i.e. nasopharyngeal positive, fecal negative) suggests that its contribution might be small.
There are few reports of SARS-CoV-2 RNA in urine as this is not a common manifestation of COVID-19, even in severe infections (Lo et al., 2020; Wang et al., 2020c; Wölfel et al., 2020); however, one study has reported levels of 3.2 × 102 gc/ml (Peng et al., 2020) and in another a very short-lived peak of 6.1 × 105 gc/ml (Yoon et al., 2020). It should be noted that most of the reports of viral loads are for hospitalized patients with mild to severe COVID-19 symptoms and that this may not accurately reflect viral abundance in asymptomatic, pre-symptomatic or very mild cases where levels in feces are likely to be much lower. It is also expected that renal infections will not occur in these mild or asymptomatic cases, suggesting that urine is not a vehicle for disease transmission outside of clinical settings, or at all.
The between-person variability in viral load, even within severe cases, appears to be very large (To et al., 2020b). This likely reflects the wide variation in symptoms experienced by individuals and organs targeted by the virus (Fig. 1). Overall, evidence suggests that high levels of SARS-CoV-2 RNA in feces are consistent with a GI tract infection in some individuals. However, the possibility that GI tract symptoms in COVID-19 cases are caused by other organisms cannot be discounted. For example, antibiotics are often prescribed during treatment of severely ill patients, creating a niche for opportunistic GI bacterial pathogens, and have been directly linked to the incidence of diarrhea in some COVID-19 studies (Lin et al., 2020). Accumulating evidence also indicates that microbial co-infection may increase the risk of disease severity in humans by suppression of the immune system or by overcoming antibiotics used in disease therapies (Zhu et al., 2020). The evidence on co-infections on the outcome of COVID-19 patients appears contradictory (Pinky and Dobrovolny, 2020). What is clear, however, is that co-infections are commonplace. For example, an analysis of nasopharyngeal swabs showed that 20% of the individuals (n = 116), who tested positive for SARS-CoV-2 also tested positive for other respiratory pathogens (Kim et al., 2020). The most common co-infections being rhinovirus/enterovirus (6.9%), respiratory syncytial virus (5.2%), and non-SARS-CoV-2 coronaviridae (4.3%). Similarly, Zhu et al. (2020) found that 32% had viral co-infection, 92% had bacterial co-infection, and 23% had fungal co-infections with respiratory pathogens. This level of co-infection is similar to other HCoV strains (Gaunt et al., 2010). A similar study reported co-infection of the respiratory tract by SARS-CoV-2 and influenza A and B (Ding et al., 2020a). Similar work is therefore required to determine the level of co-infections in the GI tract, especially as this might impact on the severity of infection by SARS-CoV-2. The quantities of SARS-CoV-2 RNA in feces are also within the range reported for other respiratory viruses such as influenza H1N1 (swine flu) which has been detected in respiratory, stool, and urine samples at levels of 2.7 × 109, 7.2 × 106, and 7.24 × 104 copies/ml, respectively (To et al., 2010), and in the case of MERS-CoV where levels in urine ranged from 102 to 103 gc/ml, feces from 103 to 104 gc/ml and those in the respiratory tract from 106 to 107 gc/ml (Corman et al., 2015; Drosten et al., 2013) (Fig. 3). In contrast, the levels of SARS-CoV-1 in feces, however, has been reported to be much higher than for SARS-CoV-2, ranging from 103 to 109 gc/ml (Cheng et al., 2004; Hung et al., 2009). This latter result suggests that conclusions on fecal-oral transmission risk from SARS-CoV-1 should be extrapolated to SARS-CoV-2 with extreme caution.
6. Is SARS-CoV-2 in stool and urine infectious?
Of critical concern in evaluating the risk of a fecal/urine-oral or fecal/urine-ocular transmission pathway for SARS-CoV-2 is the degree of infectivity of fecal- and urine-derived virus particles. These studies require tissue culture with human (or other) cell lines where addition of SARS-CoV-2 leads to an increase in viral titer from 102 particles/ml in the culture medium to 106 particles/ml within 12 h (Lamers et al., 2020; Matsuyama et al., 2020; Ogando et al., 2020). One of the first infectivity studies was undertaken from stool samples taken from a laboratory-confirmed COVID-19 severe pneumonia case, 15 days after the onset of symptoms. After viral isolation, VERO cell cultures were inoculated and virus multiplication was subsequently detected, suggesting that feces have the potential to transmit the disease (Zhang et al., 2020d). In a subsequent, more comprehensive study of COVID-19 cases, it was found that of the 153 stool specimens analyzed, 29% tested positive for SARS-CoV-2, from which infectious virus was recovered from 2 samples (Wang et al., 2020c). Similar studies have also confirmed the recovery of infectious virus from stools, VERO cells and human in intestinal organoid cultures (Lamers et al., 2020; Xiao et al., 2020a; Zhou et al., 2020).
Other comprehensive studies have suggested that no infectious viral particles can be recovered from feces at the peak of infection, despite infectious virus being recovered from respiratory specimens (Wölfel et al., 2020). Similar findings have been reported in animal models (Richard et al., 2020; Sia et al., 2020). The recent isolation of infectious virus from urine raises the possibility for urine-based transmission (Sun et al., 2020a), although given the low prevalence of this phenomenon, its significance outside of clinical settings is probably extremely low. Although these studies confirm that feces and urine may contain infectious viral particles, they also have various drawbacks. Firstly, it is evident that while viral recovery is possible from some samples, interestingly it is not from others, despite all the feces testing RT-qPCR or digital PCR positive for SARS-CoV-2 RNA. Similar observations have also been made for nasopharyngeal swabs from patients with lower viral load, suggesting viral nucleic acids might be detected for longer periods than infectious virus in different sample types (NCIC-AMS, 2020). In addition, studies have only focused on feces with high viral loads (based on Ct values) and these may not be reflective of pre- or asymptomatic cases. The levels of SARS-CoV-2 RNA in the samples used in these infectivity assays are also not reported, preventing realistic quantitative risk assessments to be made for fecal/urine-oral transmission (and to account for the levels added in the source material itself). The lack of inclusion of positive controls is also problematic where no infectious virus is recovered from any samples; i.e. problems with local culturing protocols cannot be eliminated (Wang et al., 2005b). Further, in plaque-based assays, co-contaminating (non-CoV) viruses may also lead to false-positive results, although metagenomics could be used to identify this. In such cases, it is essential that a quantitative increase in SARS-CoV-2 beyond the inoculum dose is confirmed by qPCR. It would also be advantageous to undertake dose response curves (i.e. serial dilution of fecal extracts) to allow determination of comparative levels of infectivity between samples with known viral titers (Matsuyama et al., 2020). Further, the virus is known to propagate poorly in some cell lines currently being used to assay the infectivity of SARS-CoV-2 (Harcourt et al., 2020; Matsuyama et al., 2020; Ogando et al., 2020). Therefore, it is unclear whether negative infectivity results indicate a lack of infectious particles or just a poor choice of screening assay. Based on this we conclude that further work is needed to better evaluate the temporal dynamics of viral shedding and its infectious nature in feces and urine.
For disease transmission in the community it is important to know whether feces and urine contain infectious virus in the pre- and post-symptomatic phase. This is particularly pertinent given that clinical cases may still be shedding the virus after the relieving of symptoms and their discharge back into the community. However, current evidence suggests that the infectious viral count will decline rapidly within a week of symptoms starting. Drawing on evidence from nasopharyngeal samples, which has shown a close correlation between viral abundance and infectivity, it is likely that viral shedding in feces in the post-symptomatic phase poses a much lower transmission risk (La Scola et al., 2020; Wölfel et al., 2020). In addition, even if infectious virus is detected in cell culture, it doesn't necessarily imply that it will cause infection in the upper respiratory tract of humans at the same dose, as physicochemical barriers (e.g. mucus, low pH) can further limit virus infectivity (NIS-PHE, 2020).
Overall, we conclude that while virus particles contained in respiratory droplets are known to be highly infectious, evidence suggests that feces and urine probably contain low levels to no infectious particles. In comparison to respiratory particles, they are also less likely to be spread during daily life, being confined largely to toilets and other enclosed environments. This may subsequently lead to contamination of hands, surfaces, food and water; however, in most cases the levels of contamination are likely to be low where good hygiene and sanitation is practiced. Despite this, the possibility of infection by contamination of the oral cavity, respiratory mucosa and eyes cannot be entirely discounted. This risk of infection spread is most likely associated with those experiencing co-infections or frequent watery diarrhea (Peiris et al., 2003; Tsang et al., 2003). As shedding rates appear to be correlated with symptom severity and the peak of the infection cycle, this risk would be greatest firstly in intensive care units (i.e. nosocomial spread), followed by care facilities (e.g. elderly care homes) where residents with diarrhea need secondary assistance, and heavily used and poorly maintained public toilets. The potential for the virus to spread from domestic toilets is likely to be very low as these have restricted use, probably involve individuals with mild infections and those with the capacity to practice good personal hygiene unassisted. Subsequently, in developing regions, where access to safe and hygienic sanitation is limited, the risks associated with fecal transmission routes may be higher (Anser et al., 2020; Patel, 2020). For example, an estimated 9% (673 million) of the global population defecate in the open and another 8% (627 million) use a facility shared with at least one other household as their primary sanitation location (Caruso and Freeman, 2020). This risk is perceived to be highest in urban sub-Saharan Africa where an estimated 32% of sanitation is shared (UNICEF-WHO, 2019). Further, women might be at increased risk due to more frequent use, both for meeting their own needs, including menstruation, and assisting dependent family members (Caruso et al., 2017). Another exemplar is India, where ca. 15% of households lack access to improved sanitation. The availability of soap for effective handwashing and elimination of SARS-CoV-2 from the face and hands is also problematic in many countries (Patel, 2020; Coetzee and Kagee, 2020). Sanitary workers in less economically developed countries may also be at higher risk of contracting COVID-19, due to underlying respiratory problems associated with exposure to various hazardous materials and lack of personal protection equipment (Salve and Jungari, 2020).
The survival of SARS-CoV-2 in feces after release from the body is poorly understood. However, this information is important to evaluate the potential for environmental transmission. The fecal-oral route has also been implicated in disease transmission during sexual contact, however, this risk is believed to be very low in comparison to disease transmission via respiratory droplets and the oral-oral route (Pan et al., 2020c; Cui et al., 2020; Li et al., 2020e). From the available evidence on SARS-CoV-1 it has been shown that the virus can survive for 3 h to 5 days depending on the watery nature of the diarrhea (positively related to water content), but numbers fall exponentially with time and survival rate is less than in nasopharyngeal or tracheal aspirate (Chan et al., 2004; Lai et al., 2005). More work is needed to understand the factors that influence the survival of fecal-derived SARS-CoV-2 on different matrices after release (e.g. bed sheets, towels, clothes, toilets).
7. Persistence of SARS-CoV-2 in sanitation facilities
One of the most likely points of disease transmission from feces and urine is via shared toilets (e.g. hospitals, workplaces). Based on the use of surrogate viruses and 106 viral particles per fecal event, work has shown that is unlikely that SARS-CoV-2 would reach high levels on contact surfaces via the aerosol route after flushing (e.g. <103 particles on either the seat, handle, floor, walls) (Sassi et al., 2018). In contrast, repeated use by people infected by SARS-CoV-2 might lead to a progressive accumulation of virus to higher levels, assuming infrequent cleaning. This is supported by studies in a dedicated SARS-CoV-2 outbreak center in Singapore where SARS-CoV-2 RNA was recovered from the toilet bowl, sink and door handle (Ong et al., 2020). Another study also found elevated levels of the virus in a patient-dedicated mobile toilet in China (Liu et al., 2020d), while others have detected contamination of toilet seats, exhaust grilles and taps in a COVID-19 dedicated hospital (Ding et al., 2020b; Chia et al., 2020; Lei et al., 2020) and in households (Döhla et al., 2020). The source of contamination could have been from urine and feces in the toilet, particularly in facilities used by patients with diarrhea (Chia et al., 2020). This spread is likely to be highly dependent on the operational design of the toilet (Li et al., 2020f). It is also likely that contamination on touch surfaces and walls was caused via respiratory droplets during coughing, or from transfer to surfaces from hands contaminated with nasopharyngeal fluids. Although each episode of diarrhea or vomit may spread low levels of virus, patients with GI symptoms often have several/frequent episodes of these symptoms, potentially increasing the virus load on those surfaces.
Vomiting also has the potential to spread the virus more widely than either defecation or urination events (i.e. vomiting onto floors, toilets and sinks) due to the greater potential for droplet formation and aerosolization (Kirby et al., 2016; Makison Booth and Frost, 2019). For example, projectile vomit can contaminate an area of up to 8 m2 (Makison Booth, 2014). Unfortunately, the levels of infectious SARS-CoV-2 in vomit remain unknown, but are likely to be low based on the low pH of vomit (mean pH of 3.8, range 2.5–5.0) and studies in other CoVs (Kirby et al., 2016; Willumsen et al., 2004; Cowen and Hitchner, 1975; Panon et al., 1988). Vomit is also likely to contain SARS-CoV-2 from nasopharyngeal fluids as well as from the GI tract. The potential for vomit-, fecal- and urine-derived SARS-CoV-2 to remain infectious on sanitation surfaces for long periods of time remains unclear and is probably highly dependent on the receiving surface (toilet bowl, walls, floor etc.), prevailing climatic conditions (e.g. temperature, humidity, UV exposure; Ren et al., 2020), and cleaning regime (Kampf et al., 2020). Studies on other matrices, however, have shown that viable SARS-CoV-2 might persist for at least 3 h in aerosols after their formation (Smither et al., 2020), and for up to 2 or 4 days on plastic and stainless steel surfaces (van Doremalen et al., 2020; Chin et al., 2020). This has led to guidance suggesting that toilets in communal areas should be disinfected with sodium hypochlorite or other virucidal disinfectants at least daily (ECDC, 2020). In conclusion, there is evidence to suggest that viral contamination of toilet environments may occur, although levels of contamination are expected to be very low in most settings based on infectious viral loads in feces and urine. An exception to this could be very high occupancy toilets where a progressive accumulation of the virus may occur, no sanitary cleaning is undertaken, and personal hygiene practices are poor. Although we cannot discount the potential for fecal-mucosal transmission when individuals touch their mouth, nose or eyes with contaminated hands, this would be largely preventable through handwashing and regular disinfection of sanitation facilities.
The discussion above mainly relates to countries with good levels of domestic sanitation; however, over 2.5 billion people worldwide lack access to improved water and sanitation (e.g. urban slums, rural locations, refugee camps) (Sommer et al., 2015). In these settings, infection control may be more challenging due to the lack of handwashing facilities and cultural issues (e.g. gender violence; Poole et al., 2020; Sommer et al., 2015; Truelove et al., 2020). Additionally, existing toilet and sanitation facilities tend to be less private, which leads to greater personal congregation near central facilities. Similar is true for community potable water sources, which often are only in a handful of locations, such as community water taps, for whole neighborhoods. To date, very little is known about the persistence and infectivity of SARS-CoV-2 in these contexts and further work is clearly needed in this area.
8. Amount and persistence of SARS-CoV-2 in the sewer network
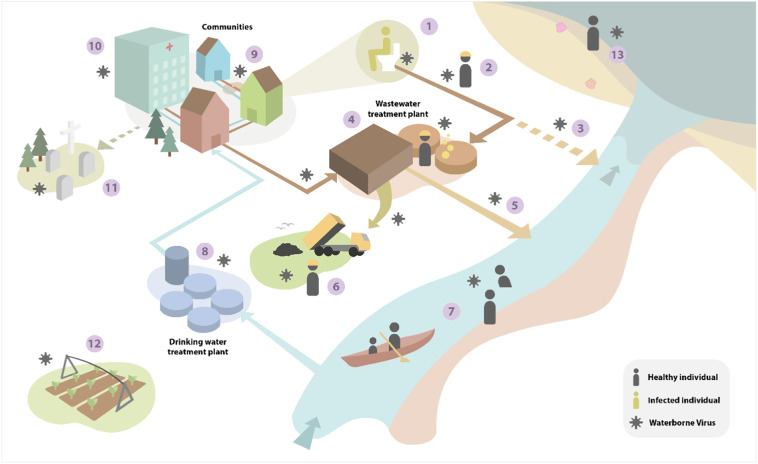
Once feces and urine enter the sewer network there are several points at which human exposure may occur (Fig. 5 ). However, significant dilution will occur in the drainage network due to inflow of water from other domestic and industrial sources. For example, at the peak of a severe infection, based on our analysis, an adult may be expected to lose ca. 1.0 l of fluid in diarrhea (during 3–6 events) and 0.8 l in urine per day (Aranda-Michel and Giannella, 1999; Pan et al., 2020d). Assuming a SARS-CoV-2 load of 8 × 106 gc/ml in feces and 3.2 × 102 gc/ml in urine and a flushing volume of 6.8 l per defecation/urination event (6 per d), this equates to a viral concentration in water leaving the toilet of 1.9 × 108 gc/l. In a single occupancy household setting, and assuming a total water use of 135 l/person/d, this will be further diluted, giving a maximum final effluent concentration of 5.9 × 107 gc/l and total viral excretion load of 8.0 × 109 gc/person/d. It is important to note that these calculations are based on genome copy numbers, which are significantly higher than infectious virus particle numbers, due to the production of defective viral genomes during RNA virus replication (Vignuzzi and López, 2019). Studies of wastewater have yet to recover infectious virus, despite its genetic material being readily detected by PCR (Döhla et al., 2020).
Fig. 5.
Summary of the main infection pathways by which SARS-CoV-2 can theoretically contaminate the environment and cause secondary infections. The numbers denote the major pathogen transport routes and exposure points: (1) contamination of toilets by infected individuals, aerosolization of feces/urine, faulty plumbing in buildings (2) pathogen transfer in the sewer network and potential exposure to sanitation workers in the sewer network, (3) discharge of untreated contaminated wastewater to rivers (sewer overflows), (4) release in bioaerosols from wastewater treatment plants and exposure of workers to potentially contaminated wastewater, (5) release of treated wastewater to rivers, (6) disposal of wastewater-derived biosolids to land, (7) transport in freshwater and exposure of individuals during recreational activities, (8) abstraction of river water for human consumption, (9) breaks in sewage pipes leading to groundwater contamination (10) hospital/medical centre release of wastewater, (11) contamination of groundwater from burial of infected bodies, (12) irrigation of crops with potentially contaminated water abstracted from rivers, (13) contamination of marine waters, dispersal in the coastal zone and potential contamination of fish/shellfish and people engaging in recreational activities.
The human minimal infectious dose of SARS-CoV-2 is not currently known. Estimates for SARS-CoV-1 range from 16 to 280 plaque forming units (PFU) (Watanabe et al., 2010). Unfortunately, the relationship between genome copies and PFU is also unknown for SARS-CoV-2, however, it is interesting to note that viable SARS-CoV-2 could not be isolated from clinical respiratory tract samples containing fewer than 106 gc/ml (Wölfel et al., 2020). For influenza virus, the ratio between TCID50 (TCID50 = PFU/0.7) and particle count is 1:100 to 1:1000 (Yezli and Otter, 2011), whilst work with clinical influenza samples has demonstrated a 100–10,000 fold difference between TCID50 and genome copy number (van Wesenbeeck et al., 2015). On this basis, it is likely that the human minimal infectious dose of aerosolized SARS-CoV-2 is in the order of 103–104 gc. The route of infection is also critical when considering the infectious dose. In influenza, the infectious dose of aerosolized virus appears to be several orders of magnitude lower than for virus that is deposited in droplets on the upper respiratory tract (Yezli and Otter, 2011). The infectious dose of SARS-CoV-2 if transmitted via the feco-oral route is therefore likely significantly higher than 103–104 gc. On this basis, exposure to raw sewerage from an infected household, elderly care home, or medical center could theoretically pose a small infection risk to sanitation workers, assuming the virus is still infectious. Parallels from SARS-CoV-1 investigations can also be drawn here. In the classic Amoy Gardens case study, raw sewage from one household entered vertically connected neighboring households, resulting in a localized infection hotspot (McKinney et al., 2006; Yu et al., 2014; Stein, 2011). It should be noted, however, that this sanitation network was poorly maintained and would not represent those in most municipal buildings and should not be used to infer the risk of fecal-oral transmission of SARS-CoV-2. Furthermore, transmission in the Amoy Gardens case study was believed to be via the aerosolization and inhalation of infectious fecal matter, rather than via the feco-oral route.
Beyond the immediate point of entry into the sewer system point, the wastewater will be further diluted in the drainage network by the addition of sewerage from non-infected households. At the peak of infection in the UK in April 2020, it was estimated that 0.25% of the population was infected (ONS, 2020). This would equate to an average community sewerage load of 1.75 × 105 gc/l reaching a centralized wastewater treatment plant. This is consistent with typical concentrations being reported in wastewater in many regions of the world ranging from 102 to 106 gc/l (Ahmed et al., 2020; Foladori et al., 2020; Randazzo et al., 2020; Wu et al., 2020b; Wurtzer et al., 2020a, Wurtzer et al., 2020b). At present, there are many uncertainties in the survival of SARS-CoV-2 during its passage through the sewer pipe network. CoVs are not thought to survive well in aqueous environments, especially in comparison with other viruses which can persist for months (e.g. poliovirus, norovirus; Seitz et al., 2011). This is supported by studies in which SARS-CoV-2 RNA can be readily detected by qPCR in wastewater leaving hospitals, but which has yet to be found to contain infectious virus (Wang et al., 2005b; Zhang et al., 2020e; Wang et al., 2020d). In fact, a recent study suggests that levels of infectious virus were not significant in wastewater and receiving rivers, indicating the effectiveness of wastewater treatment, combined with the natural loss of viral integrity (Rimoldi et al., 2020). Additionally, viral particles are likely to become bound to biofilms in the pipes, degraded by other microorganisms and inactivated by xenobiotics (e.g. surfactants, disinfectants), all of which will lead to a progressive loss of qPCR RNA signal and degrade infectious virus (if any is present at all) (Cheng et al., 2004; Wigginton et al., 2015). However, when SARS-CoV-1 was inoculated into sewage at high titers (105–106 gc/l) it was found to still contain infectious material after 14 days at 4 °C and 2 days at 20 °C (Wang et al., 2005c). These conflicting laboratory and field-based studies may reflect the different nature of the starting inoculum and failure of the lab conditions to reflect those in the field. This, however, may suggest that, if any live virus is present in the wastewater, some could survive during passage through the sewage network, based on typical transit times from households to the wastewater treatment plant (1 to 24 h). Current evidence suggests that the levels of SARS-CoV-2 are greatly lowered during wastewater treatment, suggesting that the virus is either degraded or becomes associated with the solids fraction during flocculation (Wang et al., 2020d). This is consistent with studies showing a 2 to 3 log10 removal efficiency in viral RNA abundance when comparing viral levels in influent and effluent (Wurtzer et al., 2020a, Wurtzer et al., 2020b) and the accumulation of SARS-CoV-2 in the sludge fraction (Peccia et al., 2020; Alpaslan Kocamemi et al., 2020). If the sludge (biosolids) fraction is treated (e.g. pasteurized, heat-dried, alkali-lime treated), as per the legislative requirement in many countries, this should pose no further risk to human health. One potential area where a heightened risk of exposure may occur is during the release of bioaerosols from wastewater aeration ranks. However, based on current estimates of the infectious dose of SARS-CoV-2, the likelihood that this poses a risk to workers is extremely low based on the amount of sewage that would need to be inhaled by this route to cause infection (assuming appropriate use of personal protection equipment). In addition, there is no evidence to suggest that wastewater plant operatives are at any greater risk to SARS-CoV-2 exposure via this route than that of the general population, particularly when standard issue personal protective equipment is worn (WHO, 2020). In theory, it is possible that local residents can be exposed to bioaerosols emitted from wastewater plants (Brisebois et al., 2018; Yang et al., 2019), however, there are few documented examples where direct viral transmission has been linked back to a wastewater treatment facility. In the case of SARS-CoV-2, parallels should not be drawn with other viruses (e.g. norovirus, rotavirus) whose concentrations in wastewater are typically much higher (Pasalari et al., 2019).
9. Amount and persistence of SARS-CoV-2 in the wider environment
Given the reduced evidence on infectious virus in sewers at present and the possible degradation and treatment processes explained above, detection in the wider environment most likely reflects viral RNA, not infectious virus. Based on the available evidence and our own measurements, the quantity of SARS-CoV-2 RNA in the effluent from wastewater treatment plants at the peak of a community infection (< 0.5% of the total population) is unlikely to exceed 104 gc/l (Wurtzer et al., 2020a, Wurtzer et al., 2020b). Assuming that levels of viral infection decline in the community due to the implementation of successful control measures (e.g. ‘lock down’ and social distancing) then levels in wastewater are expected to fall below <102 gc/l. Based on the large dilutions of treated wastewater after discharge into adjacent freshwaters (ca. 5–100 fold dilution under low river flow conditions when the risk is greatest) or the coastal zone (ca. 105 fold dilution), it is highly likely that SARS-CoV-2 will pose very little threat to human health (e.g. during watersports, bathing, angling, consumption of shellfish etc.; Keller et al., 2014). This is supported by measurements of typical levels of water ingestion during recreational activities of 3–30 ml/person in rivers and lakes (Dorevitch et al., 2011), 34 ml/person during surfing (Stone et al., 2008), and 10–50 ml/person during swimming and bathing (Dufour et al., 2017; Schets et al., 2011). Assuming a worst case human feco-oral infectious dose of 103 gc/person, this would necessitate that levels of infectious SARS-CoV-2 greater than 3.3 × 104 gc/l would be needed to cause concern. It should also be noted that while the eyes are often in contact with water during recreational activities, this route of SARS-CoV-2 entry into the body is thought to be minimal, particularly in comparison to ingestion of water and oral/nasopharynx mucosal exposure (Sun et al., 2020b; Deng et al., 2020). This analysis for SARS-CoV-2 contrasts with other viruses transmitted by the fecal-oral route (e.g. norovirus) where the infectious dose is very low (ca. 10 viral particles), levels in wastewater are higher and water-borne outbreaks have been reported (Parkkali et al., 2017; Russo et al., 2020).
In comparison to wastewater entering waterbodies, a greater source of potential risk to infection could be the presence of an infected individual within the water itself. It is likely that during swimming, a person may release ca. 30–60 ml of saliva into the water (Bretz and Carrilho, 2013). Contamination may also occur from ocular fluids (Güemes-Villahoz et al., 2020). Given the highest recorded levels of virus in saliva (108 gc/ml), a swimming volume of 375,000 l (25 × 10 × 1.5 m), then the levels of SARS-CoV-2 in the water would be 1.2 × 104 gc/l. Assuming the inadvertent ingestion of 20 ml by an individual during swimming, this would result in a SARS-CoV-2 exposure dose of 2.4 × 102 gc/person. This risk would be most relevant in non-chlorinated waters as standard disinfection procedures (e.g. chlorination and UV treatment in swimming pools) should rapidly reduce levels of infectious virus in the water (WHO, 2020). It should be noted that natural UV irradiation is also likely to eliminate the virus in water (Lytle and Sagripanti, 2005), however, the effect of this on SARS-CoV-2 in aqueous media remains unknown. Work on aerosolized SARS-CoV-2 has shown that it is inactivated relatively quickly (within hours) by solar UV irradiation (Sagripanti and Lytle, 2020). Further work is required to model the dispersal of SARS-CoV-2 in a range of aqueous environments (e.g. lidos, swimming pools, rivers, estuaries, coastal waters). Fundamental to this is a better knowledge of (i) the persistence and infectivity of SARS-CoV-2 in these environments, (ii) the potential for zoonotic infection (secondary hosts for SARS-CoV-2), and (iii) establishing the infectious dose of the virus. Using these data, and currently known information on SARS-CoV-2, quantitative microbial risk assessments could be undertaken to inform on human health risks in different environmental exposure scenarios based on dose-response models (Beaudequin et al., 2015).
Unlike other viruses (e.g. norovirus), there is no evidence to suggest that SARS-CoV-2 can accumulate in marine and freshwater organisms destined for human consumption (e.g. fish, oysters, mussels). The low likelihood of SARS-CoV-2 accumulation in fish is supported by the low levels of ACE-2 receptors in these organisms (Damas et al., 2020). In the case of shellfish, it is known that norovirus readily accumulates in shellfish as it binds to a human-like intestinal type A histo-blood group antigen in the shellfish tissue (Tian et al., 2007). Evidence also suggest that oysters possess an ACE-2-like receptor (CgACE) suggesting that bioaccumulation may be possible, however, whether SARS-CoV-2 can bind to CgACE, and whether the receptor is present in sufficient amounts to induce bioaccumulation remains unknown (Riviere et al., 2011).
10. Conclusions and implications for public health
Our critical analysis of the available evidence and potential transmission routes suggests that the possibility of fecal/urine-oral/ocular transmission of SARS-CoV-2 is extremely low to negligible except where direct person-to-person contact occurs. This is consistent with the many millions of documented cases of COVID-19 worldwide, and the fact that none of these have implicated feces or fecal contaminated material as part of the infection pathway. Feces have been implicated in contamination of the healthcare environment/surfaces, however, the role of those in infection remains unclear. It should be noted that our conclusions are based on western-style sanitation networks and wastewater treatment. The risks may be higher in less economically developed countries and areas with poor sanitation; however, there is insufficient evidence to enable this to be critically evaluated. This is clearly an area that warrants further research. Assuming levels of SARS-CoV-2 remain relatively low in the population (<1%), our analysis also suggests that the risk of contracting COVID-19 from water supplies, wastewater, food, bathing/recreational waters, and the coastal zone remains extremely low. This is particularly the case if personal hygiene measures are maintained (e.g. handwashing) and communal sanitary facilities are regularly cleaned and disinfected (Lotfinejad et al., 2020; Brauer et al., 2020). Following a precautionary principle, we would also recommend that households with an on-going infection, and particularly those exhibiting diarrhea, add sodium-hypochlorite or similar disinfectant prior to flushing to reduce further downstream risk of infection.
CRediT authorship contribution statement
David L. Jones: Conceptualization, Writing - original draft. Marcos Quintela Baluja: Writing - original draft. David W. Graham: Writing - original draft. Alexander Corbishley: Writing - original draft. James E. McDonald: Writing - original draft. Shelagh K. Malham: Writing - original draft. Luke S. Hillary: Writing - original draft. Thomas R. Connor: Writing - original draft. William H. Gaze: Writing - original draft. Ines B. Moura: Writing - original draft. Mark H. Wilcox: Writing - original draft. Kata Farkas: Writing - original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank Neil Dickson, Emily Cooledge (Bangor University) and Andrew Singer (UK Centre for Ecology & Hydrology) for comments on the manuscript and Eadington Graham and Ooid Scientific for the illustrations. This work was funded by the UK Research and Innovation (UKRI) project NE/M009106/1, NE/V004883/1 and NE/V010441/1 under the NERC-COVID-19 programme and the Centre for Environmental Biotechnology Project funded though the European Regional Development Fund (ERDF) by Welsh Government. LSH was supported by a Soils Training and Research Studentship (STARS) grant from the Biotechnology and Biological Sciences Research Council (BBSRC) and Natural Environment Research Council (NE/M009106/1). AC was supported by funding from the Centre of Expertise for Waters (CREW).
Author contributions
DLJ conceived the project and led the writing. All other authors contributed to drafts of the article.
Editor: Dr. Damia Barcelo
References
- Abrams H.R., Loomer L., Gandhi A., Grabowski D.C. Characteristics of U.S. nursing homes with COVID-19 cases. J. Am. Geriatr. Soci. 2020 doi: 10.1111/jgs.16661. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari S.P., Meng S., Wu Y., Mao Y., Ye R., Wang Q., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9:29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguila E., Cua I., Dumagpi J. When do you say it's SARS-CoV-2-associated diarrhea? J. Gastroenterol. Hepatol. 2020 doi: 10.1111/jgh.15141. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahamed Mim M., Naznin Rakhi N., Saha O., Rahaman M.M. Recommendation of fecal specimen for routine molecular detection of SARS-CoV-2 and for COVID-19 discharge criteria. Pathog. Glob. Health. 2020;00:1–2. doi: 10.1080/20477724.2020.1765651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpaslan Kocamemi B., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. 2020. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. medRxiv. [DOI] [Google Scholar]
- An P., Ji M., Ren H., Su J., Ding N.S., Kang J., Yin A., Zhou Q., Linyong Shen, Zhao L., Jiang X., Xiao Y., Tan W., Lv X., Li J., Liu S., Zhou J., Chen H., Xu Y., Liu J., Chen M., Cao J., Zhou Z., Lei Shen, Tan S., Yu H., Dong W., Ding Y. Prevention of COVID-19 in patients with inflammatory bowel disease in Wuhan, China. Lancet Gastroenterol. Hepatol. 2020;5:525–527. doi: 10.1016/S2468-1253(20)30121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anser M.K., Yousaf Z., Khan M.A., Nassani A.A., Alotaibi S.M., Abro M.M.Q., Vo X.V., Zaman K. Does communicable diseases (including COVID-19) may increase global poverty risk? A cloud on the horizon. Environ. Res. 2020;187 doi: 10.1016/j.envres.2020.109668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Michel J., Giannella R.A. Acute diarrhea: a practical review. Am. J. Med. 1999;106:670–676. doi: 10.1016/S0002-9343(99)00128-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A.T., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A. Hospital outbreak of middle east respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson B., Petersen E. SARS-CoV-2 shedding and infectivity. Lancet. 2020;395:1339–1340. doi: 10.1016/S0140-6736(20)30868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudequin D., Harden F., Roiko A., Stratton H., Lemckert C., Mengersen K. Beyond QMRA: modelling microbial health risk as a complex system using Bayesian networks. Environ. Int. 2015;80:8–18. doi: 10.1016/j.envint.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Bennett A., Pollock L., Jere K.C., Pitzer V.E., Lopman B., Bar-Zeev N., Iturriza-Gomara M., Cunliffe N.A. Duration and density of fecal rotavirus shedding in vaccinated Malawian children with rotavirus gastroenteritis. J. Infect. Dis. 2019;1–6 doi: 10.1093/infdis/jiz612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., Lucas J.M., Nelson P.S., Pöhlmann S., Soilleux E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7:1–8. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., van T.V.L., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Becker S., Klenk H.-D., Garten W., Steinmetzer T., Böttcher-Friebertshäuser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B., Castoe T., Rambaut A., Robertson D.L. 2020. Evolutionary Origins of the SARS-CoV-2 Sarbecovirus Lineage Responsible for the COVID-19 Pandemic. bioRxiv 2020.03.30.015008. [DOI] [PubMed] [Google Scholar]
- Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2020;0:1–14. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M., Zhao J.T., Bennitt F.B., Stanaway J.D. Global access to handwashing: implications for COVID-19 control in low-income countries. Environ. Health Perspect. 2020;128 doi: 10.1289/ehp7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretz W.A., Carrilho M.R. Salivary parameters of competitive swimmers at gas-chlorinated swimming-pools. J. Sport. Sci. Med. 2013;12:207–208. [PMC free article] [PubMed] [Google Scholar]
- Brisebois E., Veillette M., Dion-Dupont V., Lavoie J., Corbeil J., Culley A., Duchaine C. Human viral pathogens are pervasive in wastewater treatment center aerosols. J. Environ. Sci. 2018;67:45–53. doi: 10.1016/j.jes.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarini E., Manfredi G., Brambilla G., Menozzi F., Londoni C., Alicante S., Iiritano E., Romeo S., Pedaci M., Benelli G., Canetta C., La Piana G., Merli G., Scartabellati A., Viganò G., Sfogliarini R., Melilli G., Assandri R., Cazzato D., Rossi D.S., Usai S., Tramacere I., Pellegata G., Lauria G. GI symptoms as early signs of COVID-19 in hospitalised Italian patients. Gut. 2020;69:1547–1548. doi: 10.1136/gutjnl-2020-321434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A.W., McEvoy D., Collins A., Hunt K., Casey M., Barber A., Butler F., Griffin J., Lane E., McAloon C., O’Brien K., Wall P., Walsh K., More S. 2020. Inferred Duration of Infectious Period of SARS-CoV-2: Rapid Scoping Review and Analysis of Available Evidence for Asymptomatic and Symptomatic COVID-19 Cases, medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Chen M., He L., Xie J., Chen X. Clinical features and outcomes of COVID-19 patients with gastrointestinal symptoms. Crit. Care. 2020;24:340. doi: 10.1186/s13054-020-03034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso B.A., Freeman M.C. Shared sanitation and the spread of COVID-19: risks and next steps. The Lancet. Planetary Health. 2020;4:e173. doi: 10.1016/S2542-5196(20)30086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso B.A., Clasen T.F., Hadley C. Understanding and defining sanitation insecurity: women's gendered experiences of urination, defecation and menstruation in rural Odisha, India. BMJ Glob. Health. 2017;2 doi: 10.1136/bmjgh-2017-000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Centers for Disease Control and Prevention; Atlanta, GA, USA: 2020. COVID-19 Pandemic Planning Scenarios. [Google Scholar]
- Chan P.K.S., To W.K., Ng K.C., Lam R.K.Y., Ng T.K., Chan R.C.W., Wu A., Yu W.C., Lee N., Hui D.S.C., Lai S.T., Hon E.K.L., Li C.K., Sung J.J.Y., Tam J.S. Laboratory diagnosis of SARS. Emerg. Infect. Dis. 2004;10:825–831. doi: 10.3201/eid1005.030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau N.V.V., Lam V.T., Dung Nguyen Thanh, Yen L.M., Minh N.N.Q., Hung L.M., Ngoc N.M., Dung Nguyen Tri, Man D.N.H., Nguyet L.A., Nhat L.T.H., Nhu L.N.T., Ny N.T.H., Hong N.T.T., Kestelyn E., Dung N.T.P., Phong N.T., Xuan T.C., Hien T.T., Tu T.N.H., Geskus R.B., Thanh T.T., Truong N.T., Binh N.T., Thuong T.C., Thwaites G., Tan L. Van. The natural history and transmission potential of severe acute respiratory syndrome coronavirus 2 infection. Clin. Infect. Dis. 2020;ciaa711 doi: 10.1093/cid/ciaa711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.K.C., Wong D.A., Tong L.K.L., Ip S.M., Lo A.C.T., Lau C.S., Yeung E.Y.H., Lim W.W.L. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K., Lim X.F., Lim A.S., Sutjipto S., Lee P.H., Son T.T., Young B.E., Milton D.K., Gray G.C., Schuster S., Barkham T., De P.P., Vasoo S., Chan M., Ang B.S.P., Tan B.H., Leo Y.-S., Ng O.-T., Wong M.S.Y., Marimuthu K. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020;11:2800. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020;1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud D.H., Ahalt C., Augustine D., Sears D., Williams B. Medical isolation and solitary confinement: balancing health and humanity in US jails and prisons during COVID-19. J. Gen. Intern. Med. 2020 doi: 10.1007/s11606-020-05968-y. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee B.J., Kagee A. Structural barriers to adhering to health behaviours in the context of the COVID-19 crisis: considerations for low- and middle-income countries. Glob. Public Health. 2020 doi: 10.1080/17441692.2020.1779331. in press. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M., Muth D., Sieberg A., Meyer B., Assiri A.M., Binger T., Steinhagen K., Lattwein E., Al-Tawfiq J., Müller M.A., Drosten C., Memish Z.A. Viral shedding and antibody response in 37 patients with Middle East Respiratory Syndrome coronavirus infection. Clin. Infect. Dis. 2015;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa V.G., Moreli M.L., Saivish M.V. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020 doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen B.S., Hitchner S.B. pH stability studies with avian infectious bronchitis virus (coronavirus) strains. J. Virol. 1975;15:430–432. doi: 10.1128/jvi.15.2.430-432.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P., Chen Z., Wang T., Dai J., Zhang J., Ding T., Jiang J., Liu J., Zhang C., Shan W., Wang Sheng, Rong Y., Chang J., Miao X., Ma X., Wang Shixuan. 2020. Clinical Features and Sexual Transmission Potential of SARS-CoV-2 Infected Female Patients: A Descriptive Study in Wuhan, China. medRxiv 2020.02.26.20028225. [DOI] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.-P., Pfenning A.R., Zhao H., Genereux D.P., Swofford R., Pollard K.S., Ryder O.A., Nweeia M.T., Lindblad-Toh K., Teeling E.C., Karlsson E.K., Lewin H.A. 2020. Broad Host Range of SARS-CoV-2 Predicted by Comparative and Structural Analysis of ACE2 in Vertebrates. bioRxiv 2020.04.16.045302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Yang Y., Chen H., Chen W., Chen Z., Ma K., Wang J. Low risk of SARS-CoV-2 transmission through the ocular surface. Acta Ophthalmol. 2020:1–2. doi: 10.1111/aos.14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D., Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Feng Z., Wang C., Wang H., Liu L., Wang C., Wang R., Liu Y., Liu Y., Wang G., Yuan Z., Wu Y., Chen Y. 2020. Human Kidney Is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. medRxiv: 2020.03.04.20031120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Lu P., Fan Y., Xia Y., Liu M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol. 2020:1–7. doi: 10.1002/jmv.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Qian H., Xu B., Huang Y., Miao T., Yen H.-L., Xiao S., Cui L., Wu X., Shao W., Song Y., Sha L., Zhou L., Xu Y., Zhu B., Li Y. 2020. Toilets Dominate Environmental Detection of SARS-CoV-2 Virus in a Hospital. medRxiv 2020.04.03.20052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L., Holden K.A., Read J.M., Dondelinger F., Carson G., Merson L., Lee J., Plotkin D., Sigfrid L., Halpin S., Jackson C., Gamble C., Horby P.W., Nguyen-Van-Tam J.S., Ho A., Russell C.D., Dunning J., Openshaw P.J., Baillie J.K., Semple M.G. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ. 2020;369:m1985. doi: 10.1136/bmj. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M., Wilbring G., Schulte B., Kümmerer B.M., Diegmann C., Sib E., Richter E., Haag A., Engelhart S., Eis-Hübinger A.M., Exner M., Streeck H., Schmithausen R.M. vol. 49. 2020. SARS-CoV-2 in Environmental Samples of Quarantined Households. medRxiv; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z., Tong S. Epidemiological characteristics of 2143 pediatric patients with 2019 coronavirus disease in China. J. Emerg. Med. 2020;58:712–713. doi: 10.1016/j.jemermed.2020.04.006. [DOI] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorevitch S., Panthi S., Huang Y., Li H., Michalek A.M., Pratap P., Wroblewski M., Liu L., Scheff P.A., Li A. Water ingestion during water recreation. Water Res. 2011;45:2020–2028. doi: 10.1016/j.watres.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S., Guggemos W., Kallies R., Muth D., Junglen S., Müller M.A., Haas W., Guberina H., Röhnisch T., Schmid-Wendtner M., Aldabbagh S., Dittmer U., Gold H., Graf P., Bonin F., Rambaut A., Wendtner C.M. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., Cai G., Chen F., Christiani D.C., Zhang Z., Wang M. Multiomics evaluation of gastrointestinal and other clinical characteristics of severe acute respiratory syndrome coronavirus 2 and coronavirus disease 2019. Gastroenterology. 2020;158:2298–2301. doi: 10.1053/j.gastro.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Yu J., Liu X., Chen H., Lin L., Li Q. Persistence of SARS-CoV-2 virus RNA in feces: a case series of children. J. Infect. Public Health. 2020;13:926–931. doi: 10.1016/j.jiph.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A.P., Behymer T.D., Cantú R., Magnuson M., Wymer L.J. Ingestion of swimming pool water by recreational swimmers. J. Water Health. 2017;15:429–437. doi: 10.2166/wh.2017.255. [DOI] [PubMed] [Google Scholar]
- ECDC . European Centre for Disease Prevention and Control; Solna Sweden: 2020. Disinfection of Environments in Healthcare and Nonhealthcare Settings Potentially Contaminated with SARS-CoV-2. [Google Scholar]
- Ejtahed H.S., Hasani-Ranjbar S., Siadat S.D., Larijani B. The most important challenges ahead of microbiome pattern in the post era of the COVID-19 pandemic. J. Diabetes Metab. Disord. 2020 doi: 10.1007/s40200-020-00579-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F., Ou Z., Huang Y.T. Human coronaviruses are uncommon in patients with gastrointestinal illness. J. Clin. Virol. 2010;48:131–133. doi: 10.1016/j.jcv.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzek A., Sperhake J., Edler C., Schröder A.S., Heinemann A., Heinrich F., Ron A., Mushumba H., Lütgehetmann M., Püschel K. Evidence for systematic autopsies in COVID-19 positive deceased: case report of the first German investigated COVID-19 death. Rechtsmedizin. 2020;30:184–189. doi: 10.1007/s00194-020-00401-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S.M., Lewis-Fuller E., Williams H., Miller Z., Scarlett H.P., Cooper C., Gordon-Johnson K.A., Vickers I., Shaw K., Wellington I., Thame J., Pérez E., Indar L. Magnitude, distribution, and estimated level of underreporting of acute gastroenteritis in Jamaica. J. Health Popul. Nutr. 2013;31:69–80. doi: 10.3329/jhpn.v31i4.2310. [DOI] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Paredes C., Jankousky K., Schultz J., Bernfeld J., Cullen K., Quan N.G., Kon S., Hotez P., Henao-Martínez A.F., Krsak M. COVID-19 in jails and prisons: a neglected infection in a marginalized population. PLoS Negl. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbadage T., Peterson B.M., Gunasekera R.S. Does COVID-19 spread through droplets alone? Front. Public Heal. 2020;8:1–4. doi: 10.3389/fpubh.2020.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes O., Desselberger U., Tatochenko V., Rodrigo C., Salman N., Mezner Z., Giaquinto C., Grimprel E. Nosocomial rotavirus infection in European countries: a review of the epidemiology, severity and economic burden of hospital-acquired rotavirus disease. Pediatr. Infect. Dis. J. 2006;25:12–21. doi: 10.1097/01.inf.0000197563.03895.91. [DOI] [PubMed] [Google Scholar]
- Gong K., Xu Z., Cai Z., Chen Y., Wang Z. Internet hospitals help prevent and control the epidemic of COVID-19 in China: multicenter user profiling study. J. Med. Internet Res. 2020;22 doi: 10.2196/18908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güemes-Villahoz N., Burgos-Blasco B., Arribi-Vilela A., Arriola-Villalobos P., Rico-Luna C.M., Cuiña-Sardiña R., Delgado-Iribarren A., García-Feijoó J. Detecting SARS-CoV-2 RNA in conjunctival secretions: is it a valuable diagnostic method of COVID-19? J. Med. Virol. In press. 2020 doi: 10.1002/jmv.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A.X., Cui J.J., OuYang Q.Y., He L., Guo C.X., Yin J.Y. 2020. The Clinical Characteristics and Mortal Causes Analysis of COVID-19 Death Patients. medRxiv 2020.04.12.20062380. [DOI] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K. Sen, Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak: an update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces - a rapid review. Color. Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M., Li Z.P., You H., Rodrigues R., Jump D.B., Morgun A., Shulzhenko N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:UNSP102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A.J., Lopman B.A., Payne D.C., Patel M.M., Gastañaduy P.A., Vinjé J., Parashar U.D. Norovirus disease in the United States. Emerg. Infect. Dis. 2013;19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Seong M.W., Heo E.Y., Park J.H., Kim N., Shin S., Cho S.I., Park S.S., Choi E.H. Sequential analysis of viral load in a neonate and her mother infected with SARS-CoV-2. Clin. Infect. Dis. 2020:1–4. doi: 10.1093/cid/ciaa447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K.G., Scharton D., Plante J.A., Mirchandani D., Widen S.G., Narayanan K., Makino S., Ksiazek T.G., Plante K.S., Weaver S.C., Lindstrom S., Tong S., Menachery V.D., Thornburg N.J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug T.T., Mykletun A., Dahl A.A. The prevalence of nausea in the community: psychological, social and somatic factors. Gen. Hosp. Psychiatry. 2002;24:81–86. doi: 10.1016/S0163-8343(01)00184-0. [DOI] [PubMed] [Google Scholar]
- Haug T.Tangen., Mykletun A., Dahl A.A. Are anxiety and depression related to gastrointestinal symptoms in the general population? Scand. J. Gastroenterol. 2002;37:294–298. doi: 10.1080/003655202317284192. [DOI] [PubMed] [Google Scholar]
- He J., Tao H., Yan Y., Huang S.Y., Xiao Y. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Matumoto M. Mouse hepatitis virus (MHV-2). Plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 1976;20:219–225. [PubMed] [Google Scholar]
- Hirose R., Nakaya T., Naito Y., Daidoji T., Watanabe Y., Yasuda H., Konishi H., Itoh Y. Mechanism of human influenza virus RNA persistence and virion survival in feces: mucus protects virions from acid and digestive juices. J. Infect. Dis. 2017;216:105–109. doi: 10.1093/infdis/jix224. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honarmand Ebrahimi K. SARS-CoV-2 spike glycoprotein-binding proteins expressed by upper respiratory tract bacteria may prevent severe viral infection. FEBS Lett. 2020;594:1651–1660. doi: 10.1002/1873-3468.13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda T., Sakamoto M., Shimizu H., Okabe N. SARS-CoV-2 enterocolitis with persisting to excrete the virus for about two weeks after recovering from diarrhea: a case report. Infect. Control Hosp. Epidemiol. 2020:753–754. doi: 10.1017/ice.2020.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua C.Z., Miao Z.P., Zheng J.S., Huang Q., Sun Q.F., Lu H.P., Su F.F., Wang W.H., Huang L.P., Chen D.Q., Xu Z.W., Ji L.D., Zhang H.P., Yang X.W., Li M.H., Mao Y.Y., Ying M.Z., Ye S., Shu Q., Chen E.F., Fu J.F. Epidemiological features and viral shedding in children with SARS-CoV-2 infection. J. Med. Virol. 2020 doi: 10.1002/jmv.26180. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Mao T., Li S., Wu L., Xu X., Li H., Xu C., Su F., Dai J., Shi J., Cai J., Huang C., Lin X., Chen D., Lin X., Sun B., Tang S. 2020. Long Period Dynamics of Viral Load and Antibodies for SARS-CoV-2 Infection: An Observational Cohort Study. medRxiv 2020.04.22.20071258. [DOI] [Google Scholar]
- Huang Y., Chen S., Yang Z., Guan W., Liu D., Lin Z., Zhang Y., Xu Z., Liu X., Li Y. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am. J. Respir. Crit. Care Med. 2020;201:1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.N., Lau S.K.P., Woo P.C.Y., Yuen K.Y. Viral loads in clinical specimens and SARS manifestations. Hong Kong Med. J. 2009;15:20–22. doi: 10.3201/eid1009.040058. [DOI] [PubMed] [Google Scholar]
- Iorgulescu G. Saliva between normal and pathological. Important factors in determining systemic and oral health. J. Med. Life. 2009;2:303–307. [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Grubaugh N.D. Why does Japan have so few cases of COVID-19? EMBO Mol. Med. 2020;12:10–12. doi: 10.15252/emmm.202012481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Luo M., Zou Z., Wang X., Chen C., Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020 doi: 10.1002/jmv.25941. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim Seil, Park E.C., Park D., Lee J.-H., Byeon C.W., Lee J.J., Maeng J., Kim S.J., Kim Seung Il, Kim B.-T., Lee M.J., Kim H.G. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. BioRxiv. 2020;2020(02):25.964775. doi: 10.1101/2020.02.25.964775. [DOI] [Google Scholar]
- Kalantar-Zadeh Kourosh, Ward S.A., Kalantar-Zadeh Kamyar, El-Omar E.M. Considering the effects of microbiome and diet on SARS-CoV-2 infection: nanotechnology roles. ACS Nano. 2020;14:5179–5182. doi: 10.1021/acsnano.0c03402. [DOI] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar A., Selvaraju S., Esper F. Human coronavirus-HKU1 infection among adults in Cleveland, Ohio. Open Forum Infect. Dis. 2017;4:1–6. doi: 10.1093/ofid/ofx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashi A.H., Fallah-karkan M., Amini E., Vaezjalali M. 2020. The Presence of COVID-19 in Urine: A Systematic Review and Meta-analysis of the Literature. medRxiv 2020.05.15.20094920. [DOI] [Google Scholar]
- van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller V.D.J., Williams R.J., Lofthouse C., Johnson A.C. Worldwide estimation of river concentrations of any chemical originating from sewage-treatment plants using dilution factors. Environ. Toxicol. Chem. 2014;33:447–452. doi: 10.1002/etc.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheyami A.M., Nakagomi T., Nakagomi O., Getty B., Hart C.A., Cunliffe N.A. Detection of coronaviruses in children with acute gastroenteritis in Maddina, Saudi Arabia. Ann. Trop. Paediatr. 2010;30:45–50. doi: 10.1179/146532810X12637745451997. [DOI] [PubMed] [Google Scholar]
- Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. J. Am. Med. Assoc. 2020;323:2085–2086. doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A.E., Streby A., Moe C.L. Vomiting as a symptom and transmission risk in norovirus illness: evidence from human challenge studies. PLoS One. 2016;11:1–10. doi: 10.1371/journal.pone.0143759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K.L., Nasrin D., Blackwelder W.C., Wu Y., Farag T., Panchalingham S., Sow S.O., Sur D., Zaidi A.K.M., Faruque A.S.G., Saha D., Alonso P.L., Tamboura B., Sanogo D., Onwuchekwa U., Manna B., Ramamurthy T., Kanungo S., Ahmed S., Qureshi S., Quadri F., Hossain A., Das S.K., Antonio M., Hossain M.J., Mandomando I., Acácio S., Biswas K., Tennant S.M., Verweij J.J., Sommerfelt H., Nataro J.P., Robins-Browne R.M., Levine M.M. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS) Lancet Glob. Heal. 2019;7:e568–e584. doi: 10.1016/S2214-109X(19)30076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Weeraratne D., Snowdon J.L., Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020;9:324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan A.C., Chau T., Tong W., Tsang O.T., Tso E.Y., Chiu M., Yu W., Lai T.S. Severe acute respiratory syndrome-related diarrhea. J. Gastroenterol. Hepatol. 2005;20:606–610. doi: 10.1111/j.1400-1746.2005.03775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Wang Y.H., Wu C.Y., Hung C.H., Jiang D.D.S., Wu F.T. A norovirus outbreak in a nursing home: norovirus shedding time associated with age. J. Clin. Virol. 2013;56:96–101. doi: 10.1016/j.jcv.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Lai M.Y.Y., Cheng P.K.C., Lim W.W.L. Survival of severe acute respiratory syndrome coronavirus. Clin. Infect. Dis. 2005;41:e67–e71. doi: 10.1086/433186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J.C.M., Moshi G.B., Ang S.H., Chew H.M., Ng Q.H., Madjukie A., Logeswary M. Management of COVID-19 related paediatric blood samples in a clinical haematology laboratory. Br. J. Haematol. 2020;189:848–851. doi: 10.1111/bjh.16721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Chan M.C.W., Wong B., Choi K.W., Sin W., Lui G., Chan P.K.S., Lai R.W.M., Cockram C.S., Sung J.J.Y., Leung W.K. Fecal viral concentration and diarrhea in norovirus gastroenteritis. Emerg. Infect. Dis. 2007;13:1399–1401. doi: 10.3201/eid1309.061535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.C., Wang L., Eng H.L., You H.L., Chang L.S., Tang K.S., Lin Y.J., Kuo H.C., Lee I.K., Liu J.W., Huang E.Y., Yang K.D. Correlation of pandemic (H1N1) 2009 viral load with disease severity and prolonged viral shedding in children. Emerg. Infect. Dis. 2010;16:1265–1272. doi: 10.3201/eid1608.091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Jin M., Bao P., Zhao W., Zhang S. Clinical characteristics and results of semen tests among men with coronavirus disease 2019. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhao M.Y., Tan T.H.M. What makes a foodborne virus: comparing coronaviruses with human noroviruses. Curr. Opin. Food Sci. 2021;42:1–7. doi: 10.1016/j.cofs.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., Su X., Cao B. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.Y., Li L., Zhang Y., Wang X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9:1–7. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.J., Kopetz S., Vilar E., Shen J.P., Chen K., Maitra A. Relative abundance of SARS-CoV-2 entry genes in the enterocytes of the lower gastrointestinal tract. Genes. 2020;11:645. doi: 10.3390/genes11060645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Ye F., Liu X., Huang Z., Ling S., Jiang Z., Cheng J., Huang X., Wu Q., Wu S., Xie Y., Xiao C., Ye D., Yang Z., Li Y., Leung N.H.L., Cowling B.J., He J., Wong S.S., Zanin M. SARS-CoV-2 environmental contamination associated with persistently infected COVID-19 patients. Influenza Other Respi. Viruses. 2020 doi: 10.1111/irv.12783. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.-L., Chu S.-G., Luo Y., Huang Z.-H., Hao Y., Fan C.-H. Atypical presentation of SARS-CoV-2 infection: a case report. World J. Clin. Cases. 2020;8:1265–1270. doi: 10.12998/wjcc.v8.i7.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Y., Wang J.X., Chen X. Can a toilet promote virus transmission? From a fluid dynamics perspective. Phys. Fluids. 2020;32:065107. doi: 10.1063/5.0013318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Zhenling, Huang S., Zhang Zhenyi, Fang Z., Gu Z., Gao L., Shi H., Mai L., Liu Y., Lin X., Lai R., Yan Z., Li X., Shan H. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., Wu F., Song Z.G., Huang W., Chen J., Hu B.J., Wang S., Mao E.Q., Zhu L., Zhang W.H., Lu H.Z. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133:1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Xiao Y., Shen Y., Shi C., Chen Y., Shi P., Gao Y., Wang Y., Lu B. Detection of SARS-CoV-2 by RT-PCR in anal from patients who have recovered from coronavirus disease 2019. J. Med. Virol. 2020:2019–2021. doi: 10.1002/jmv.25875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu Y., Liu Y. Somatic symptoms and concern regarding COVID-19 among Chinese college and primary school students: a cross-sectional survey. Psychiatry Res. 2020;289:113070. doi: 10.1016/j.psychres.2020.113070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.Y., Ning Z., Chen Y., Guo M., Liu Y.Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Ho K., Kan H., Fu Q., Lan K. 2020. Aerodynamic Characteristics and RNA Concentration of SARS-CoV-2 Aerosol in Wuhan Hospitals during COVID-19 Outbreak. bioRxiv 86, 2020.03.08.982637. [DOI] [Google Scholar]
- Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X., Tian Y., Sin N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfinejad N., Peters A., Pittet D. Hand hygiene and the novel coronavirus pandemic: the role of healthcare workers. J. Hosp. Infect. 2020 doi: 10.1016/j.jhin.2020.03.017. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C.F., Matic N., Ritchie G., Lawson T., Stefanovic A., Champagne S., Leung V., Romney M.G. Detection of low levels of SARS-CoV-2 RNA from nasopharyngeal swabs using three commercial molecular assays. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., Sun J., François S., Kraemer M.U.G., Faria N.R., McCrone J.T., Peng J., Xiong Q., Yuan R., Zeng L., Zhou P., Liang C., Yi L., Liu J., Xiao J., Hu J., Liu T., Ma W., Li W., Su J., Zheng H., Peng B., Fang S., Su W., Li K., Sun R., Bai R., Tang X., Liang M., Quick J., Song T., Rambaut A., Loman N., Raghwani J., Pybus O.G., Ke C. Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020;181:997–1003. doi: 10.1016/j.cell.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle C.D., Sagripanti J.L. Predicted inactivation of viruses of relevance to biodefense by solar radiation. J. Virol. 2005;79:14244–14252. doi: 10.1128/JVI.79.22.14244-14252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Su L., Zhang Y., Zhang X., Gai Z., Zhang Z. Do children need a longer time to shed SARS-CoV-2 in stool than adults? J. Microbiol. Immunol. Infect. 2020;53:373–376. doi: 10.1016/j.jmii.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makison Booth C. Vomiting Larry: a simulated vomiting system for assessing environmental contamination from projectile vomiting related to norovirus infection. J. Infect. Prev. 2014;15:176–180. doi: 10.1177/1757177414545390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makison Booth C., Frost G. Potential distribution of viable norovirus after simulated vomiting. J. Hosp. Infect. 2019;102:304–310. doi: 10.1016/j.jhin.2019.02.010. [DOI] [PubMed] [Google Scholar]
- Martinez-Rojas M.A., Vega-Vega O., Bobadilla N.A. Is the kidney a target of SARS-CoV-2? Am. J. Physiol. Renal Physiol. 2020;318:F1454–F1462. doi: 10.1152/ajprenal.00160.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2- expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Mauro Gabriella, Cristina S., Concetta R., Francesco R., Annalisa C. SARS-Cov-2 infection: response of human immune system and possible implications for the rapid test and treatment. Int. Immunopharmacol. 2020;84 doi: 10.1016/j.intimp.2020.106519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K.R., Gong Y.Y., Lewis T.G. Environmental transmission of SARS at Amoy Gardens. J. Environ. Health. 2006;68:26–30. [PubMed] [Google Scholar]
- Mesoraca A., Margiotti K., Viola A., Cima A., Sparacino D., Giorlandino C. Evaluation of SARS-CoV-2 viral RNA in fecal samples. Virol. J. 2020;17:86. doi: 10.1186/s12985-020-01359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minodier L., Masse S., Capai L., Blanchon T., Ceccaldi P.E., van der Werf S., Hanslik T., Charrel R., Falchi A. Clinical and virological factors associated with gastrointestinal symptoms in patients with acute respiratory infection: a two-year prospective study in general practice medicine. BMC Infect. Dis. 2017;17:1–11. doi: 10.1186/s12879-017-2823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönkemüller K., Fry L., Rickes S. Covid-19, coronavirus, SARS-CoV-2 and the small bowel. Rev. Esp. Enferm. Dig. 2020;112:383–388. doi: 10.17235/reed.2020.7137/2020. [DOI] [PubMed] [Google Scholar]
- NCIC-AMS . National Centre for Infectious Diseases and the Chapter of Infectious Disease Physicians, Academy of Medicine; Singapore: 2020. Period of Infectivity to Inform Strategies for De-isolation for COVID-19 Patients: Position Statement. [Google Scholar]
- Ng M.L., Tan S.H., See E.E., Ooi E.E., Ling A.E. Early events of SARS coronavirus infection in vero cells. J. Med. Virol. 2003;71:323–331. doi: 10.1002/jmv.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIS-PHE . Virology Cell for the Scientific Advisory Group for Emergencies (SAGE), UK Government; London, UK: 2020. Review of Data on Persistence of SARS-CoV-2 in the Environment and Potential Infection Risk. [Google Scholar]
- Ogando N., Dalebout T., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J., Kikkert M., Barcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020 doi: 10.1099/jgv.0.001453. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y., Marimuthu K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. J. Am. Med. Assoc. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONS . Office for National Statistics; London, UK: 2020. Coronavirus (COVID-19) Infection Survey Pilot: England, 24th July 2020. [Google Scholar]
- Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 2020;M20-3012 doi: 10.7326/M20-3012. [DOI] [PubMed] [Google Scholar]
- Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12:e7423. doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Xiao X., Guo J., Song Y., Li H., Patel D.P., Spivak A.M., Alukal J.P., Zhang X., Xiong C., Li P.S., Hotaling J.M. No evidence of SARS-CoV-2 in semen of males recovering from COVID-19. Fertil. Steril. 2020;113:1135–1139. doi: 10.1016/j.fertnstert.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C., Jin Y., Niu X., Ping R., Du Y., Li T., Xu G., Hu Q., Tu L. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Long L., Zhang D., Yuan T., Cui S., Yang P., Wang Q., Ren S. Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clin. Chem. 2020;66:794–801. doi: 10.1093/clinchem/hvaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniri A., Hosseini M.M., Akhavan-Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1767690. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panon G., Tache S., Labie C. Respective stability of rotavirus and coronavirus in bovine milk. Lait. 1988;68:49–64. doi: 10.1051/lait:198814. [DOI] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkkali S., Joosten R., Fanoy E., Pijnacker R., VAN Beek J., Brandwagt D., van Pelt W. Outbreak of diarrhoea among participants of a triathlon and a duathlon on 12 July 2015 in Utrecht, the Netherlands. Epidemiol. Infect. 2017;145:2176–2184. doi: 10.1017/S0950268817001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasalari H., Ataei-Pirkooh A., Aminikhah M., Jafari A.J., Farzadkia M. Assessment of airborne enteric viruses emitted from wastewater treatment plant: atmospheric dispersion model, quantitative microbial risk assessment, disease burden. Environ. Poll. 2019;253:464–473. doi: 10.1016/j.envpol.2019.07.010. [DOI] [PubMed] [Google Scholar]
- Patel A. Preventing COVID-19 amid public health and urban planning failures in slums of Indian cities. World Medical & Health Policy. 2020 doi: 10.1002/wmh3.351. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Weinberger D., Omer S. 2020. SARS-CoV-2 RNA Concentrations in Primary Municipal Sewage Sludge as a Leading Indicator of COVID-19 Outbreak Dynamics. medRxiv. [DOI] [Google Scholar]
- Peckham R. Economies of contagion: financial crisis and pandemic. Econ. Soc. 2013;42:226–248. doi: 10.1080/03085147.2012.718626. [DOI] [Google Scholar]
- Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M., Law K.I., Tang B.S.F., Hon T.Y.W., Chan C.S., Chan K.H., Ng J.S.C., Zheng B.J., Ng W.L., Lai R.W.M., Guan Y., Yuen K.Y. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Liu J., Xu W., Luo Q., Chen D., Lei Z., Huang Z., Li X., Deng K., Lin B., Gao Z. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020 doi: 10.1002/jmv.25936. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;20:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Schöpf J., Kögl M., Friedel C.C., Müller M.A., Carbajo-Lozoya J., Stellberger T., von Dall’Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Züst R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Weßels C., Pöhlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer J.K. Innate host barriers to viral trafficking and population diversity: lessons learned from poliovirus. Adv. Virus Res. 2010;77:85–118. doi: 10.1016/B978-0-12-385034-8.00004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillonel T., Scherz V., Jaton K., Greub G., Bertelli C. Letter to the editor: SARS-CoV-2 detection by real-time RT-PCR. Euro Surveill. 2020;25:1–2. doi: 10.2807/1560-7917.ES.2020.25.21.2000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinky L., Dobrovolny H.M. SARS-CoV-2 coinfections: could influenza and the common cold be beneficial? J. Med. Virol. 2020 doi: 10.1002/jmv.26098. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras A., Rizzo D., Uzzau S., De Riu G., Rubino S., Bussu F. Inappropriate nasopharyngeal sampling for SARS-CoV-2 detection is a relevant cause of false-negative reports. Otolaryngol. Head. Neck Surg. 2020;194599820931793 doi: 10.1177/0194599820931793. [DOI] [PubMed] [Google Scholar]
- Poole D.N., Escudero D.J., Gostin L.O., Leblang D., Talbot E.A. Responding to the COVID-19 pandemic in complex humanitarian crises. Int. J. Equity Health. 2020;19:1–2. doi: 10.1186/s12939-020-01162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu V., Hsu E., Lestin S., Soltanianzadeh Y., Hadi S. Bradycardia, renal failure, atrioventricular nodal blockade, shock, and hyperkalemia (BRASH) syndrome as a presentation of coronavirus disease 2019. Cureus. 2020;12:e7816. doi: 10.7759/cureus.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez. Med. 2020;28:174–184. [PubMed] [Google Scholar]
- Rahman A., Sarkar A. Risk factors for fatal Middle East respiratory syndrome coronavirus infections in Saudi Arabia: analysis of the WHO line list, 2013–2018. Am. J. Public Health. 2019;109:1288–1293. doi: 10.2105/AJPH.2019.305186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman H., Carter I., Basile K., Donovan L., Kumar S., Tran T., Ko D., Alderson S., Sivaruban T., Eden J.S., Rockett R., O’Sullivan M.V., Sintchenko V., Chen S.C., Maddocks S., Dwyer D.E., Kok J. Interpret with caution: An evaluation of the commercial AusDiagnostics versus in-house developed assays for the detection of SARS-CoV-2 virus. J. Clin. Vir. 2020;127 doi: 10.1016/j.jcv.2020.104374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D.D., Lee M.H., Archuleta S., Bagdasarian N., Quek S.C. The many estimates of the COVID-19 case fatality rate. Lancet Infect. Dis. 2020;3099:30244. doi: 10.1016/S1473-3099(20)30244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razum O., Penning V., Mohsenpour A., Bozorgmehr K. Covid-19 in refugee shelters: the German public health service needs strengthening now. Gesundheitswesen. 2020;82:392–396. doi: 10.1055/a-1154-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S.Y., Wang W.B., Hao Y.G., Zhang H.R., Wang Z.C., Chen Y.L., Gao R.D. Stability and infectivity of coronaviruses in inanimate environments. World J. Clin. Cases. 2020;8:1391–1399. doi: 10.12998/wjcc.v8.i8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Kok A., de Meulder D., Bestebroer T.M., Lamers M.M., Okba N.M.A., van Vlissingen M.F., Rockx B., Haagmans B.L., Koopmans M.P.G., Fouchier R.A.M., Herfst S. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020;11:3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Moja L., Gismondo M.R., Salerno F. 2020. Presence and Vitality of SARS-CoV-2 Virus in Wastewaters and Rivers. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risku M., Lappalainen S., Räsänen S., Vesikari T. Detection of human coronaviruses in children with acute gastroenteritis. J. Clin. Virol. 2010;48:27–30. doi: 10.1016/j.jcv.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere G., Fellous A., Franco A., Bernay B., Favrel P. A crucial role in fertility for the oyster angiotensin-converting enzyme orthologue CgACE. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzatti G., Lopetuso L.R., Gibiino G., Binda C., Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed. Res. Int. 2017;2017:9351507. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robilotti E., Deresinski S., Pinsky B.A. Norovirus. Clin. Microbiol. Rev. 2015;28:134–164. doi: 10.1128/CMR.00075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida F., Campanini G., Piralla A., Adzasehoun K.M.G., Sarasini A., Baldanti F. Molecular detection of gastrointestinal viral infections in hospitalized patients. Diagn. Microbiol. Infect. Dis. 2013;77:231–235. doi: 10.1016/j.diagmicrobio.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.K., Akramuzzaman S.M., Akbar M.S. Persistent diarrhea: total gut transit time and its relationship with nutrient absorption and clinical response. J. Pediatr. Gastroenterol. Nutr. 1991;13:409–414. [PubMed] [Google Scholar]
- Rudney J.D., Ji Z., Larson C.J. The prediction of saliva swallowing frequency in humans from estimates of salivary flow-rate and the volume of saliva swallowed. Arch. Oral Biol. 1995;40:507–512. doi: 10.1016/0003-9969(95)00004-9. [DOI] [PubMed] [Google Scholar]
- Russo G.S., Eftim S.E., Goldstone A.E., Dufour A.P., Nappier S.P., Wade T.J. Evaluating health risks associated with exposure to ambient surface waters during recreational activities: a systematic review and meta-analysis. Water Res. 2020 doi: 10.1016/j.watres.2020.115729. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagripanti J., Lytle C.D. Estimated inactivation of coronaviruses by solar radiation with special reference to COVID-19. Photochem. Photobiol. 2020 doi: 10.1111/php.13293. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salve P.S., Jungari S. Sanitation workers at the frontline: work and vulnerability in response to COVID-19. Local Environ. 2020 doi: 10.1080/13549839.2020.1792430. In press. [DOI] [Google Scholar]
- Sassi H.P., Reynolds K.A., Pepper I.L., Gerba C.P. Evaluation of hospital-grade disinfectants on viral deposition on surfaces after toilet flushing. Am. J. Infect. Control. 2018;46:507–511. doi: 10.1016/j.ajic.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Schets F.M., Schijven J.F., de Roda Husman A.M. Exposure assessment for swimmers in bathing waters and swimming pools. Water Res. 2011;45:2392–2400. doi: 10.1016/j.watres.2011.01.025. [DOI] [PubMed] [Google Scholar]
- Schneider M., Ackermann K., Stuart M., Wex C., Protzer U., Schätzl H.M., Gilch S. Severe acute respiratory syndrome coronavirus replication is severely impaired by MG132 due to proteasome-independent inhibition of M-calpain. J. Virol. 2012;86:10112–10122. doi: 10.1128/jvi.01001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- Schwierzeck V., König J.C., Kühn J., Mellmann A., Correa-Martínez C.L., Omran H., Konrad M., Kaiser T., Kampmeier S. First reported nosocomial outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a pediatric dialysis unit. Clin. Infect. Dis. 2020;2:1–21. doi: 10.1093/cid/ciaa491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz S.R., Leon J.S., Schwab K.J., Lyon G.M., Dowd M., McDaniels M., Abdulhafid G., Fernandez M.L., Lindesmith L.C., Baric R.S., Moe C.L. Norovirus infectivity in humans and persistence in water. Appl. Environ. Microbiol. 2011;77:6884–6888. doi: 10.1128/AEM.05806-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong M.-W., Lee S.J., Cho S.I., Ko K., Kim M.-N., Sung H., Kim J.-S., Ahn J.S., Yu B.S., Kim T.S., Kim E.C., Park S.S. External quality assessment of MERS-CoV molecular diagnostics during the 2015 Korean outbreak. Ann. Lab. Med. 2016;36:230. doi: 10.3343/alm.2016.36.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. J. Am. Med. Assoc. 2020;2019:2019–2021. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Shen Q., Guo W., Guo T., Li J., He W., Ni S., Ouyang X., Liu J., Xie Y., Tan X., Zhou Z., Peng H. Novel coronavirus infection in children outside of Wuhan, China. Pediatr. Pulmonol. 2020:1–6. doi: 10.1002/ppul.24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevlin M., Nolan E., Owczarek M., McBride O., Murphy J., Gibson Miller J., Hartman T.K., Levita L., Mason L., Martinez A.P., McKay R., Stocks T.V.A., Bennett K.M., Hyland P., Bentall R.P. COVID-19-related anxiety predicts somatic symptoms in the UK population. Br. J. Health Psychol. 2020 doi: 10.1111/bjhp.12430. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddharta A., Pfaender S., Vielle N.J., Dijkman R., Friesland M., Becker B., Yang J., Engelmann M., Todt D., Windisch M.P., Brill F.H., Steinmann Joerg, Steinmann Jochen, Becker S., Alves M.P., Pietschmann T., Eickmann M., Thiel V., Steinmann E. Virucidal activity of world health organization-recommended formulations against enveloped viruses, including zika, ebola, and emerging coronaviruses. J. Infect. Dis. 2017;215:902–906. doi: 10.1093/infdis/jix046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smither S.J., Eastaugh L.S., Findlay J.S., Lever M.S. Experimental aerosol survival of SARS-CoV-2 in artificial saliva and tissue culture media at medium and high humidity. Emerg. Microbes Infect. 2020;9:1415–1417. doi: 10.1080/22221751.2020.1777906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M., Ferron S., Cavill S., House S. Violence, gender and WASH: spurring action on a complex, under-documented and sensitive topic. Environ. Urban. 2015;27:105–116. doi: 10.1177/0956247814564528. [DOI] [Google Scholar]
- Srinivasan A., Klepper C., Sunkara A., Kang G., Carr J., Gu Z., Leung W., Hayden R.T. Impact of adenoviral stool load on adenoviremia in pediatric hematopoietic stem cell transplant recipients. Pediatr. Infect. Dis. J. 2015;34:562–565. doi: 10.1097/INF.0000000000000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R.A. Super-spreaders in infectious diseases. Int. J. Infect. Dis. 2011;15:e510–e513. doi: 10.1016/j.ijid.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D.L., Harding A.K., Hope B.K., Slaughter-Mason S. Exposure assessment and risk of gastrointestinal illness among surfers. J. Toxicol. Environ. Heal. A. 2008;71:1603–1615. doi: 10.1080/15287390802414406. [DOI] [PubMed] [Google Scholar]
- Su H., Yang M., Wan C., Yi L.X., Tang F., Zhu H.Y., Yi F., Yang H.C., Fogo A.B., Nie X., Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Ma X., Yu H., Zhang Zhaohua, Bian P., Han Y., Sun J., Liu Y., Yang C., Geng J., Zhang Zhongfa, Gai Z. The different clinical characteristics of corona virus disease cases between children and their families in China–the character of children with COVID-19. Emerg. Microbes Infect. 2020;9:707–713. doi: 10.1080/22221751.2020.1744483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.B., Wang Y.Y., Liu G.H., Liu Z. Role of the eye in transmitting human coronavirus: what we know and what we do not know. Front. Public Heal. 2020;8:1–7. doi: 10.3389/fpubh.2020.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., Shi Y., Zhang Z., Chen S.B., Liu X., Dai J., Li X., Huang S., Huang X., Luo L., Wen L., Zhuo J., Li Y., Wang Y., Zhang L., Zhang Y., Li F., Feng L., Chen X., Zhong N., Yang Z., Huang J., Zhao J., Li Y.M. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020;1751:1–8. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxonera C., Sagastagoitia I., Alba C., Mañas N., Olivares D., Rey E. 2019 novel coronavirus disease (COVID-19) in patients with inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2020;52:276–283. doi: 10.1111/apt.15804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalanayar Muthukrishnan P., Faillace R. Compassionate use of others’ immunity - understanding gut microbiome in Covid-19. Crit. Care. 2020;24:358. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P., Engelbrektson A.L., Jiang X., Zhong W., Mandrell R.E. Norovirus recognizes histo-blood group antigens on gastrointestinal cells of clams, mussels, and oysters: a possible mechanism of bioaccumulation. J. Food Prot. 2007;70:2140–2147. doi: 10.4315/0362-028X-70.9.2140. [DOI] [PubMed] [Google Scholar]
- Tian Y., Rong L., Nian W., He Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment. Pharmacol. Ther. 2020;51:843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Chan K.-H., Li I.W.S., Tsang T.-Y., Tse H., Chan J.F.W., Hung I.F.N., Lai S.-T., Leung C.-W., Kwan Y.-W., Lau Y.-L., Ng T.-K., Cheng V.C.C., Peiris J.S.M., Yuen K.-Y. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J. Med. Virol. 2010;82:1–7. doi: 10.1002/jmv.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Tsang O.T.Y., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C., Leung W.S., Chik T.S.H., Choi C.Y.C., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W.S., Fung A.Y.F., Hung I.F.N., Cheng V.C.C., Chan J.F.W., Yuen K.Y. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020:ciaa149. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C.Y., Cai J.P., Chan J.M.C., Chik T.S.H., Lau D.P.L., Choi C.Y.C., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C.K., Poon R.W.S., Luo C.T., Cheng V.C.C., Chan J.F.W., Hung I.F.N., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd E.C.D. Foodborne disease and food control in the Gulf States. Food Control. 2017;73:341–366. doi: 10.1016/j.foodcont.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truelove S., Abrahim O., Altare C., Lauer S.A., Grantz K.H., Azman A.S., Spiegel P. The potential impact of COVID-19 in refugee camps in Bangladesh and beyond: a modeling study. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., Wong P.C., Lam B., Ip M.S., Chan J., Yuen K.Y., Lai K.N. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- Tung-Thompson G., Gentry-Shields J., Fraser A., Jaykus L.A. Persistence of human Norovirus RT-qPCR signals in simulated gastric fluid. Food Environ. Virol. 2014;7:32–40. doi: 10.1007/s12560-014-9170-4. [DOI] [PubMed] [Google Scholar]
- UNICEF-WHO . UN Children’s Fund, World Health Organization; New York, USA: 2019. Progress on Household Drinking Water, Sanitation and Hygiene 2000–2017: Special Focus on Inequalities. [Google Scholar]
- Vignuzzi M., López C.B. Defective viral genomes are key drivers of the virus–host interaction. Nat. Microbiol. 2019;4:1075–1087. doi: 10.1038/s41564-019-0465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuille-Dit-Bille R.N., Liechty K.W., Verrey F., Guglielmetti L.C. SARS-CoV-2 receptor ACE2 gene expression in small intestine correlates with age. Amino Acids. 2020 doi: 10.1007/s00726-020-02870-z. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltenburg M.A., Victoroff T., Rose C.E. Update: COVID-19 Among Workers in Meat and Poultry Processing Facilities ― United States, April–May 2020. MMWR Morb Mortal Wkly Rep. 2020;69:887–892. doi: 10.15585/mmwr.mm6927e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Yan M., Xu H., Liang W., Kan B., Zheng B., Chen H., Zheng H., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Liu Y.F., Guo R.T., Liu X.N., Zhan L.H., Zhou D.H., Zhao A., Hai R., Yu D., Guan Y., Xu J. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Guo T.K., Zhen B., Kong Q.X., Yi B., Li Z., Song N., Jin M., Wu X.M., Xiao W.J., Zhu X.M., Gu C.Q., Yin J., Wei W., Yao W., Liu C., Li J.F., Ou G.R., Wang M.N., Fang T.Y., Wang G.J., Qiu Y.H., Wu H.H., Chao F.H., Li J.W. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J. Gastroenterol. 2005;11:4390–4395. doi: 10.3748/wjg.v11.i28.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J., Guo T., Zhen B., Kong Q., Yi B., Li Z., Song N., Jin M., Xiao W., Zhu X., Gu C., Yin J., Wei W., Yao W., Liu C., Li J., Ou G., Wang M., Fang T., Wang G., Qiu Y., Wu H., Chao F., Li J. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Sci. Technol. 2005;52:213–221. doi: 10.2166/wst.2005.0266. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kang H., Liu X., Tong Z. Asymptomatic cases with SARS-CoV-2 infection. J. Med. Virol. 2020 doi: 10.1002/jmv.25990. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Bartrand T.A., Weir M.H., Omura T., Haas C.N. Development of a dose-response model for SARS coronavirus. Risk Anal. 2010;30:1129–1138. doi: 10.1111/j.1539-6924.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K. Emerging infectious diseases: a review. Curr. Emerg. Hosp. Med. Rep. 2018;6:86–93. doi: 10.1007/s40138-018-0162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W.E., Li Z., Chiew C.J., Yong S.E., Toh M.P., Lee V.J. Presymptomatic transmission of SARS-CoV-2-Singapore. Morb. Mortal. Wkly Rep. 2020;69:411–415. doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wesenbeeck L., D’Haese D., Tolboom J., Meeuws H., Dwyer D.E., Holmes M., Ison M.G., Katz K., McGeer A., Sadoff J., Weverling G.J., Stuyver L. A downward trend of the ratio of Influenza RNA copy number to infectious viral titer in hospitalized influenza A-infected patients. Open forum Infect. Dis. 2015;2:ofv166. doi: 10.1093/ofid/ofv166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organisation; Geneva, Switzerland: 2018. 2018 Review of Diseases Prioritized under the Research and Development Blueprint. [Google Scholar]
- WHO . Who Global, World Health Organisation; Geneva, Switzerland: 2020. Water, Sanitation, Hygiene and Waste Management for the COVID-19 Virus: Interim Guidance. [DOI] [Google Scholar]
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1:735–746. doi: 10.1039/c5ew00125k. [DOI] [Google Scholar]
- Willumsen T., Øgaard B., Hansen B.F., Rølla G. Effects from pretreatment of stannous fluoride versus sodium fluoride on enamel exposed to 0.1 M or 0.01 M hydrochloric acid. Acta Odontol. Scand. 2004;62:278–281. doi: 10.1080/00016350410000174. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1805–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. lancet. Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China. J. Am. Med. Assoc. 2020;323:1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics through Viral Genome Quantification in Paris Wastewaters. medRxiv 2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Moulin L. 2020. Time Course Quantitative Detection of SARS-CoV-2 in Parisian Wastewaters Correlates with COVID-19 Confirmed Cases; pp. 10–13. medRxiv. [DOI] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao Jingxian, Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G.C., Yu J.M., Duan Z.J. New strategy for virus discovery: viruses identified in human feces in the last decade. Sci. China Life Sci. 2013;56:688–696. doi: 10.1007/s11427-013-4516-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30:1–14. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Shi S., Zhu J., Shi J., Dai K., Chen X. Clinical characteristics and outcomes of cancer patients with COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.25972. [DOI] [PubMed] [Google Scholar]
- Yang K.X., Li L., Wang Y.J., Xue S., Han Y.P., Liu J.X. Airborne bacteria in a wastewater treatment plant: emission characterization, source analysis and health risk assessment. Water Res. 2019;149:596–606. doi: 10.1016/j.watres.2018.11.027. [DOI] [PubMed] [Google Scholar]
- Yezli S., Otter J.A. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ. Virol. 2011;3:1–30. doi: 10.1007/s12560-011-9056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.G., Yoon J., Song J.Y., Yoon S.Y., Lim C.S., Seong H., Noh J.Y., Cheong H.J., Kim W.J. Clinical significance of a high SARS-CoV-2 viral load in the saliva. J. Korean Med. Sci. 2020;35:e195. doi: 10.3346/jkms.2020.35.e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto F.K. The proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I.C., Chan M., Vasoo S., Wang L.F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.S., Lye D.C. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. J. Am. Med. Assoc. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.T.S., Qiu H., Tse L.A., Wong T.W. Severe acute respiratory syndrome beyond amoy gardens: completing the incomplete legacy. Clin. Infect. Dis. 2014;58:683–686. doi: 10.1093/cid/cit797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Liao Z., Huang H., Jiang B., Zhang X., Wang Y., Zhao M. Comparison of the indicators of psychological stress in the population of Hubei Province and non-endemic provinces in China during two weeks during the Coronavirus Disease 2019 (COVID-19) outbreak in February 2020. Med. Sci. Monit. 2020;26:e923767. doi: 10.12659/MSM.923767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Cui X., Xiao J., Meng T., Zhou W., Liu J., Xu H. Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69:1010–1018. doi: 10.1136/gutjnl-2020-320953. [DOI] [Google Scholar]
- Zhang T., Cui X., Zhao X., Wang J., Zheng J., Zheng G., Guo W., Cai C., He S., Xu Y. Detectable SARS-CoV-2 viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J. Med. Virol. 2020 doi: 10.1002/jmv.25795. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X. Lou, Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the Coronavirus Disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. doi: 10.46234/ccdcw2020.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Zhao G., Chu H., Wang D., Yan H.H.N., Poon V.K.M., Wen L., Wong B.H.Y., Zhao X., Chiu M.C., Yang D., Wang Y., Au-Yeung R.K.H., Chan I.H.Y., Sun S., Chan J.F.W., To K.K.W., Memish Z.A., Corman V.M., Drosten C., Hung I.F.N., Zhou Y., Leung S.Y., Yuen K.Y. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3:eaao4966. doi: 10.1126/sciadv.aao49660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H., Cai J.P., Yip C.C.Y., Chan I.H.Y., Wong K.K.Y., Tsang O.T.Y., Chan K.H., Chan J.F.W., To K.K.W., Chen H., Yuen K.Y. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Zhu X., Ge Y., Wu T., Zhao K., Chen Y., Wu B., Zhu F., Zhu B., Cui L. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]