Graphical abstract
Abstract
The spike protein is a focused target of COVID-19, a pandemic caused by SARS-CoV-2. A 12-nt insertion at S1/S2 in the spike coding sequence yields a furin cleavage site, which raised controversy views on origin of the virus. Here we analyzed the phylogenetic relationships of coronavirus spike proteins and mapped furin recognition motif on the tree. Furin cleavage sites occurred independently for multiple times in the evolution of the coronavirus family, supporting the natural occurring hypothesis of SARS-CoV-2.
1. Introduction
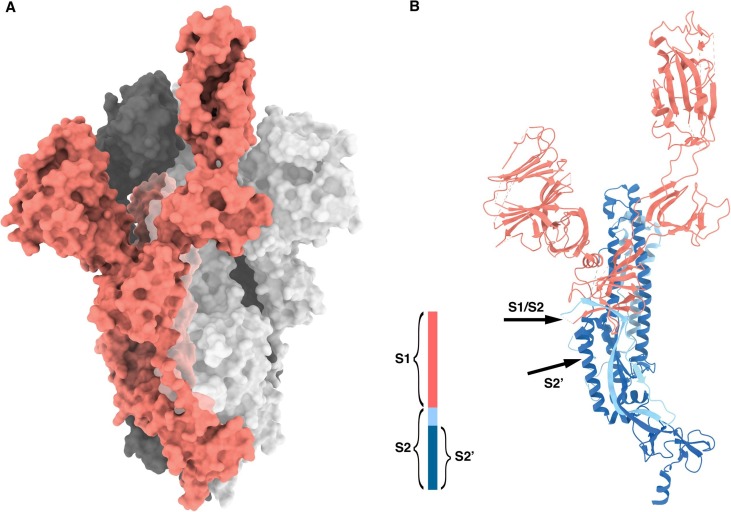
The coronavirus disease 2019 (COVID-19) pandemic, caused by coronavirus SARS-CoV-2, is still rapidly spreading. Scientists all around the world are studying the infection process of the virus and looking for therapeutic solutions to prevent and cure the disease. Spike protein is one of most important targets in COVID-19 research, not only in mechanism study but also in vaccine development and therapeutic antibody design (Premkumar et al., 2020, Abraham, 2020). The spike protein forms a homotrimer (Fig. 1 A) and protrudes outside the membrane of the virion (Li, 2016, Wrapp et al., 2020, Walls et al., 2020). It binds to angiotensin-converting enzyme 2 (ACE2) on human cell surface, undergoes conformational changes, and mediates the fusion of the virion to human cells (Li et al., 2003, Yan et al., 2020, Lan et al., 2020, Wang et al., 2020, Benton et al., 2020).
Fig. 1.
Structure of SARS-CoV-2 spike protein (PDB ID: 6VYB (Walls et al., 2020). A) Spike homotrimer in open conformation. B) Cleavage sites on spike protein (marked by arrows). Red cartoon, S1; light/dark blue cartoon, S2; dark blue cartoon, S2′). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The spike protein consists of two subunits: the N-terminal S1 subunit, containing the receptor binding domain that interacts with ACE2 and induces conformational change; while the C-terminal S2 subunit, containing the fusion peptide that is responsible for fusing the membranes of virion and human cells (Li, 2016). To function properly, the spike protein needs to be cleaved by host proteases to separate the two subunits (Simmons et al., 2013). Study of SARS-CoV (caused the severe acute respiratory syndrome in 2002–2003 and belonged to the same species as SARS-CoV-2) showed that cleavage at S1/S2 enhances fusogenicity of spike protein (Bosch et al., 2003). Another cleavage site producing S2′ (fusion peptide and the C-terminal region of S2) was also identified (Fig. 1B) (Belouzard et al., 2009). The S2′ site is not far from the S1/S2 site, and cleavage either or both them can yield the separation of two subunits of spike. For coronaviruses, multiple proteases were reported responsible for the cleavage, including TMPRSS2 (Hoffmann et al., 2020, Glowacka et al., 2011); cathepsin CTSL (Bosch et al., 2008, Ou et al., 2020), and trypsin (Belouzard et al., 2009, Bertram et al., 2011).
The spike protein of the Middle East respiratory syndrome coronavirus (MERS-CoV) was discovered to be more effectively cleaved by another protease, furin (Millet and Whittaker, 2014). In MERS-CoV, both S1/S2 and S2′ sites have sequence RXXR, the furin recognition motif. In human body, furin is ubiquitously expressed (Braun and Sauter, 2019). This may explain the highly lethal nature and high rate of multiple system failure caused by MERS (Millet and Whittaker, 2014).
Surprisingly, SARS-CoV-2 has the furin recognition motif at S1/S2, causing by a 12-nucleotide insertion not presented even in its closest relatives (Walls et al., 2020, Coutard et al., 2020). This stimulates a conspiracy that this furin site can only be manual work, thus SARS-CoV-2 must be created in a laboratory. Here, we analyzed the sequences of coronaviruses and found furin sites occurred independently for multiple times during evolution. This exhibits natural occurrence of furin cleavage site in SARS-CoV-2 spike protein is highly possible. Thus, the insertion of furin cleavage site into SARS-CoV-2 spike protein is not necessarily a result of manual work.
2. Results
2.1. Phylogenetic tree of spike protein sequences reveals major groups of coronaviruses
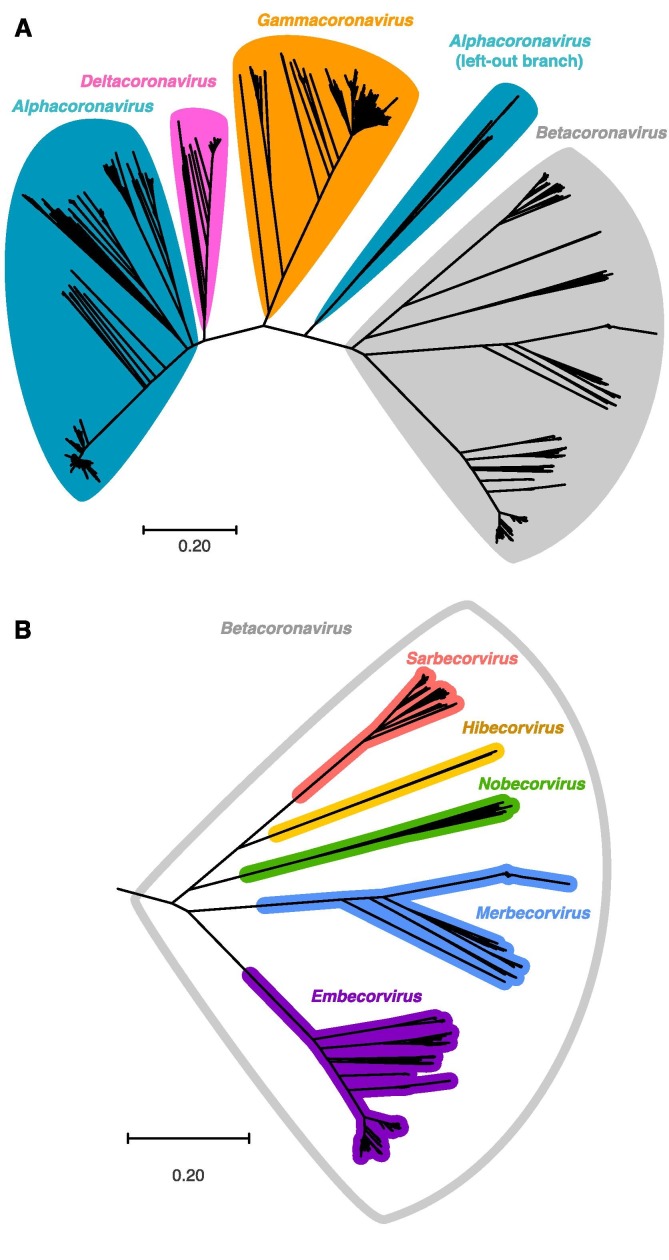
Coronaviruses are members of the subfamily Orthocoronavinae (also named Coronavinae) in the family Coronavidae. There are four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. The first two genera only infect mammals, while the last two primarily infect birds but also mammals (Cui et al., 2019). We collected sequences of coronavirus spike proteins from the InterPro database (Mitchell et al., 2019) and performed phylogenetic analysis. The phylogenetic tree (Fig. 2 A) generally matches reported relationship of coronaviruses (Cui et al., 2019): three genera (Betacoronavirus; Gammacoronavirus, and Deltacoronavirus) each forms a single clade, while only Alphacoronavirus has a small group left outside (see Fig. 5 for representative sequences). The left-out Alphacoronavirus group contains Rhinolophus bat coronavirus HKU2 that was reported to be closely related to two typical Alphacoronavirus, human coronavirus NL63 and human coronavirus 229E, in a phylogenetic analysis based on the RNA-dependent RNA polymerase coding region (Cui et al., 2019). We aligned the whole genomes of the three viruses and found Rhinolophus bat coronavirus HKU2 is quite unique in its spike protein-coding region (Fig. S1). Such result explains why this clade was separated from other Alphacoronavirus in our phylogenetic analysis.
Fig. 2.
Phylogenetic tree of coronavirus spike protein sequences. A) Noting genera of coronavirus. B) Subtree of Betacoronavirus, noting subgenera.
Fig. 5.
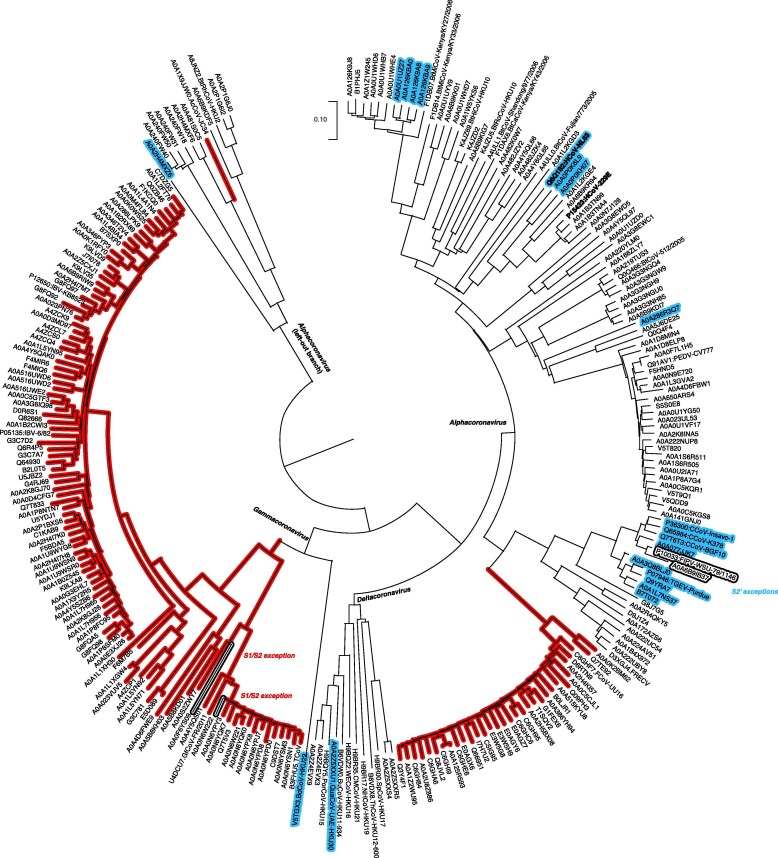
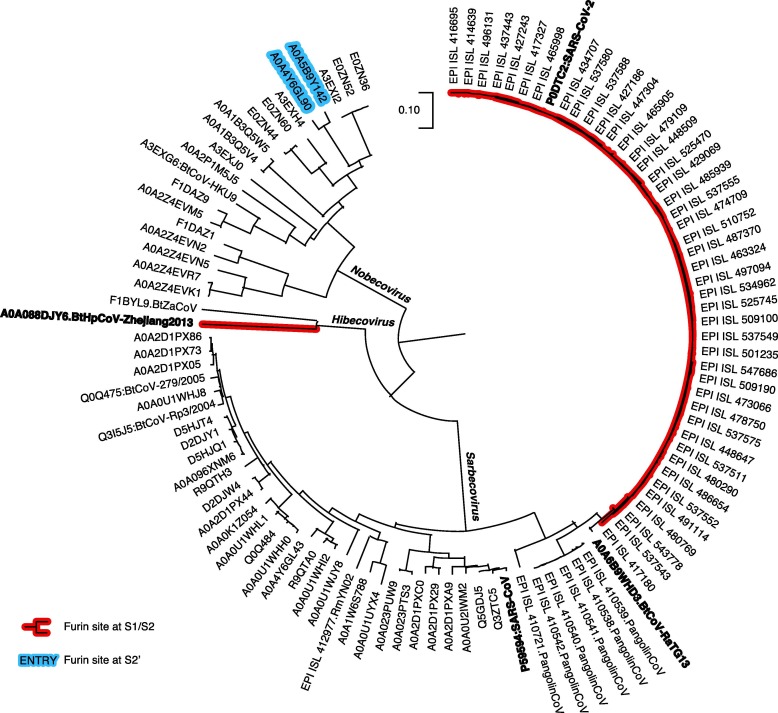
Mapping of furin recognition motif on phylogenetic tree of spike protein sequences, the Alphacoronavirus + Gammacoronavirus + Deltacoronavirus clade. Sequences clustered with 95% identity threshold.
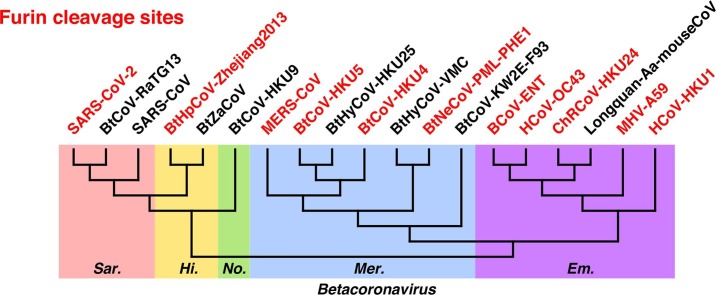
Furthermore, the five subgenera of Betacoronavirus were nicely displayed as monophyletic (Fig. 2B): Sarbecovirus (e.g. SARS-CoV-2 and SARS-CoV), Merbecovirus (e.g. MERS-CoV), Embecovirus (e.g. human coronavirus OC43 and human coronavirus HKU1, both causing common cold), and two small subgenera Hibecovirus and Nobecovirus. Hibecovirus and Nobecovirus are only discovered in bats so far, and our phylogenetic analysis shows they are related to Sarbecovirus.
2.2. Furin cleavage site at spike S1/S2 also occurs in a virus close to Sarbecovirus
We mapped the furin recognition motif RXXR at both S1/S2 and S2′ positions in phylogenetic trees of spike protein sequences (Fig. 3, Fig. 4, Fig. 5 and S2, S3). In the Sarbecovirus + Hibecovirus + Nobecovirus clade (Fig. 3), furin cleavage sites at either position occur only in limited ranges. Strains of SARS-CoV-2 (we also added sequences from the GISAID database) have furin cleavage sites at spike S1/S2. Moreover, SARS-CoV-2 is the only virus in subgenus Sarbecovirus having this feature, while even its closest relatives, bat coronavirus RaTG13 (sequence identity 97.7%) and pangolin coronaviruses (92.9%–90.7%), do not have furin site. However, in Hibecovirus, the sister clade of Sarbecovirus, a Hipposideros bat coronavirus collected in 2013 at Zhejiang Province in China has furin site at S1/S2. Interestingly, the other member in Hibecovirus lacks such site, similar to the situation of SARS-CoV-2 and its close relatives. The SARS-CoV-2 strains and Hipposideros bat coronavirus (Zhejiang 2013) are not sister groups, in agreement with the distinct sequence patterns of their furin cleavage sites at spike S1/S2 (Fig. 6 A). Besides, two strains of bat coronavirus HKU9, belonging to Nobecovirus, are the only members in the Sarbecovirus + Hibecovirus + Nobecovirus clade having furin cleavage site at spike S2′.
Fig. 3.
Mapping of furin recognition motif on phylogenetic tree of spike protein sequences, the Sarbecovirus + Hibecovirus + Nobecovirus clade. Sequences in the SARS-CoV-2 clade were clustered with 95% identity threshold; in the SARS-CoV-2 clade were clustered with 99% identity threshold.
Fig. 4.
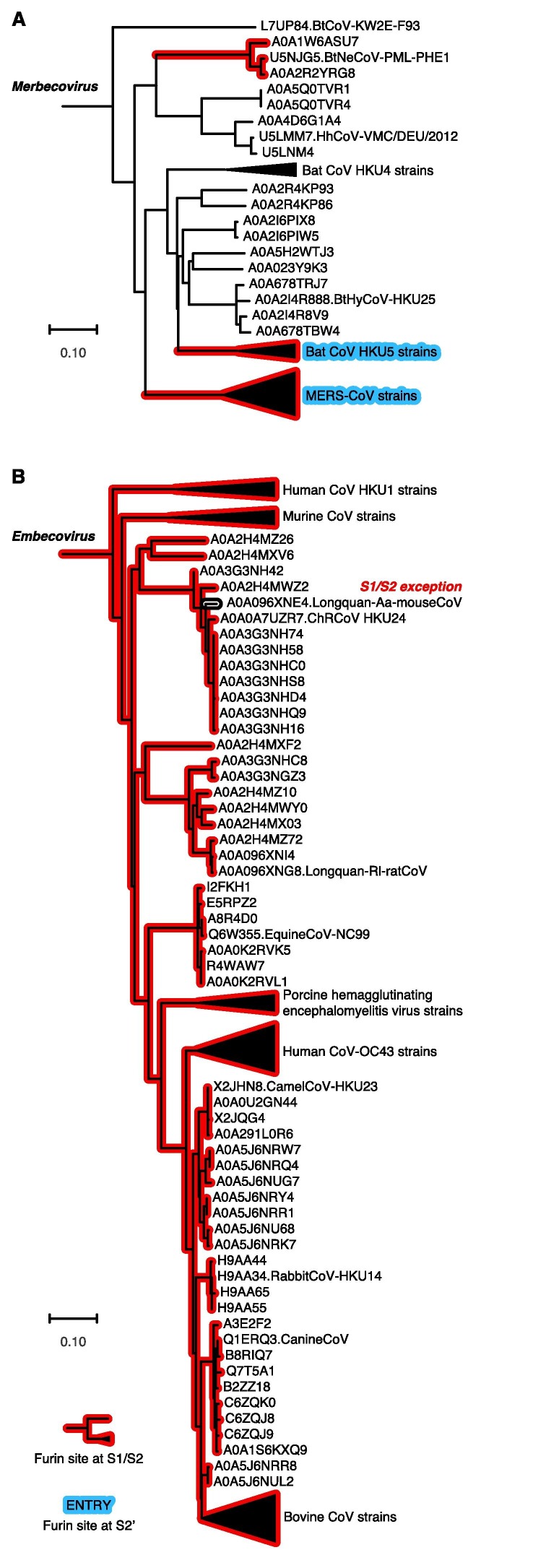
Mapping of furin recognition motif on phylogenetic tree of spike protein sequences, A) Merbecovirus (for uncompressed tree see Fig. S2) and B) Embecovirus (for uncompressed tree see Fig. S3).
Fig. 6.
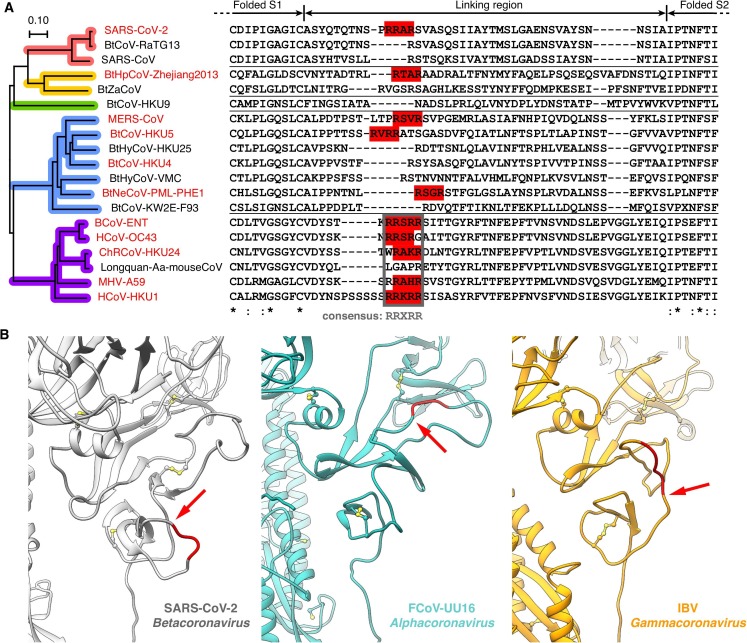
Positions of furin cleavage sites at the linking region of S1 and S2. A) Multiple sequence alignment of representative Betacoronavirus spike protein S1/S2 region, with furin recognition motifs highlighted (red colorboxes in sequence alignment). Phylogenetic tree of spike protein sequences is colored to indicate subgenera (coloring scheme the same as in Fig. 2B). B) Positions of furin cleavage sites in different coronavirus genera (red cartoon, furin recognition motif; red arrow, cleavage site); structures: SARS-CoV-2, PDB ID 6VYB (Walls et al., 2020), with missing loop added; feline coronavirus (FCoV) UU16, homology model based on PDB ID 5SZS (Walls et al., 2016); infectious bronchitis coronavirus (IBV), PDB ID 6CV0 (Shang et al., 2018). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Furin cleavage sites are common in Betacoronavirus
Our mapping results showed that the furin recognition motif is more common in Merbecovirus and Embecovirus (Figs. 4 and S2, S3). In Merbecovirus, furin sites at spike S1/S2 occur in three clades: MERS-CoV strains, the bat coronavirus HKU5 strains, and coronavirus Neoromicia/PML-PHE1/RSA/2011 with its relatives (Figs. 4A and S2). Besides, MERS-CoV and bat coronavirus HKU5 are the only clades in Merbecovirus having furin cleavage site at S2′.
In Embecovirus, furin recognition motif at spike S1/S2 is universal: All strains but a few exceptions have furin cleavage sites at spike S1/S2 (Figs. 4B and S3). Interestingly, the Longquan Aa mouse coronavirus (Wang et al., 2015) loses this furin site, while its close relatives (e.g. China Rattus coronavirus HKU24, sequence identity 96.0%) maintains the furin cleavage site. This provides an example of naturally occurred sequence variation at spike S1/S2 among closely related coronaviruses. Besides, for spike S2′, only several single strains have furin recognition motif (Fig. S3).
2.4. Furin cleavage sites also occur in other genera of coronavirus
Our mapping results showed furin cleavage sites are widely present in the whole coronavirus family (Fig. 5). For spike S1/S2, furin recognition motif is universal in Gammacoronavirus, and also occurs in two clades of Alphacorovanvirus: feline coronavirus and relatives, and Chevrier's field mouse coronavirus. For spike S2′, furin recognition motif occurs in several independent clades, covering all the three genera. Notably, in the two human coronaviruses in Alphacoronavirus causing common cold, HCoV NL63 has furin cleavage site at spike S2′, while the HCoV 229E (protein sequence identity 63.8%) lacks such feature.
2.5. Furin cleavages sites occurred independently for six times in Betacoronavirus
The alignment of linking regions of spike S1 and S2 domains in representative Betacoronavirus (Fig. 6A) shows this region is less conserved than the neighboring folded S1 and S2 segments. Within a subgenus the sequences are well aligned, but among subgenera the similarity is low. The furin cleavage site of SARS-CoV-2 spike S1/S2 is formed by a insertion of PRRA in comparison to other Sarbecovirus including close relative RaTG13, showing it occurred very recently and independently. Similarly, Hipposideros bat coronavirus (Zhejiang 2013) in Hibecovirus has furin site of independent origin, though the occurring time is hard to decide for in this subgenera only two sequences were published.
Merbecovirus and Embecovirus both have multiple coronavirus species with furin cleavage sites at spike S1/S2, but their situations are different: In Merbecovirus, furin cleavage sites prevail in three non-sister clades (Figs. 4A and 6A). Moreover, the positions of furin recognition motifs in the linking regions are unique to each clade, as exhibited in alignments of both protein sequences (Fig. 6A) and nucleotide sequences (Fig. S4A). These indicated for of the three clades in Merbecovirus, furin cleavage sites have an independent origin. In Embecovirus, to the contrast, all the furin cleavage sites are variations based on a 5-residue region with consensus sequence RRXRR. The region is well aligned in both protein and nucleotide sequences (Figs. 6A and S4B). This suggested the furin cleavage sites of Embecovirus share a common ancestor.
In addition, in Alphacoronavirus and Gammacoronavirus, S1/S2 cleavage sites reside at a different loop comparing to the site in Betacoronavirus (Fig. 6B), therefore furin cleavage sites at spike S1/S2 in these two genera occurred independently from those in Betacoronavirus in evolution.
3. Discussion
Furin cleavage is critical to many viral diseases, including HIV, Ebola, and influenza H5 and H7 (Becker et al., 2012). Furin is a ubiquitously expressed protease. In human body, it has a wider distribution range than the major protease responsible for cleaving spike, TMPRSS2 (Fig. S5). Therefore, coronaviruses with spike containing furin cleavage site may have advantage in spreading. Deletions of furin cleavage site in SARS-CoV-2 attenuates replication on respiratory cells (Johnson et al., 2020) and pathogenesis in hamster (Johnson et al., 2020, Lau et al., 2020). Furin inhibitors suppress virus production and cytopathic effects in kidney cells (Cheng et al., 2020). Natural polymorphisms losing furin recognition motif in SARS-CoV-2 spike S1/S2 are observed, but very rare (Xing et al., 2020). Variations in this region are more common in viruses cultured in vitro than viruses isolated from clinical samples, suggesting this cleavage site is under selection pressure in human body (Lau et al., 2020, Liu et al., 2020).
Our analysis exhibits furin cleavage sites at spike S1/S2 occurred independently for several times in coronavirus. Consequently, natural occurring of the site in SARS-CoV-2 is highly possible. This is further supported by other observed natural variations at the linking region of S1 and S2: A natural insertion in SARS-CoV spike though not related to furin recognition motif was reported (Zhou et al., 2020). In Embecovirus; Longquan Aa mouse coronavirus (Wang et al., 2015) has a frameshift mutation led to the loss of furin recognition motif (Fig. S4B); Some strains of murine coronavirus lose furin recognition motif through substitution mutations (Fig. S3), e.g. in MHV-2 (Yamada et al., 1997). Further study of losing the furin cleavage site in Embecovirus would help to interpret the S1/S2 cleavage of Betacoronaviruses. Besides, independent occurrences of furin cleavage sites in surface glycoproteins are not unique to coronavirus: for the hemagglutinin of influenza, only H5 and H7 have furin cleavage sites (Bottcher-Friebertshauser et al., 2013); and these subtypes are distant in phylogenetic tree (Fig. S6).
4. Conclusion
Furin cleavage sites in spike proteins naturally occurred independently for multiple times in coronaviruses. Such feature of SARS-CoV-2 spike protein is not necessarily a product of manual intervention, though our observation does not rule out the lab-engineered scenario.
5. Methods
5.1. Sequences of spike proteins in coronaviruses
Most sequences were obtained from the InterPro database (Mitchell et al., 2019). All the sequences sharing the same domain annotation SAR2-CoV-2 or SAR2-CoV (entries: IPR002552, IPR018548, IPR036326, and IPR042578) were selected and then filtered in three steps: 1) Sequences shorter than 1000 amino acids or not starting with methionine were dropped; 2) The rest sequences were put into multiple sequence alignment and phylogenetic analysis, and a few sequences were found to be out of the coronavirus family thus were dropped; 3) Duplicate sequences were removed using the software CD-HIT version 4.8.1 (Fu et al., 2012, Li and Godzik, 2006). Finally spike protein sequences from pangolin coronaviruses, closely related to SARS-CoV-2 but not included in InterPro database, were added in to the sequence set. Genomes of bat coronavirus RmYN02 and pangolin covonaviruses were obtained from GISAID database (https://www.gisaid.org/), and spike protein sequences were generated using NCBI ORFfinder server (https://www.ncbi.nlm.nih.gov/orffinder/). Additional sequences of SARS-CoV-2 spike proteins were also obtained from GISAID database (at Sep. 29, 2020); Sequences with 25 or more continuous unreadable amino acids were dropped, and 93,420 spike protein sequences were used for analysis. Nucleotide sequences of representative spike sequences were obtained from NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/).
5.2. Sequence clustering
Sequence set were clustered using the software CD-HIT version 4.8.1 (Fu et al., 2012, Li and Godzik, 2006).
5.3. Multiple sequence alignment
Multiple sequence alignment jobs were performed using the MAFFT software version 7.299b (Katoh and Standley, 2013). The strategy FFT-NS-2 was selected. For other parameters, the default values were used. The alignment figures in Fig. S4 were generated using the ESPript 3.0 website (http://espript.ibcp.fr/ESPript/ESPript/) (Robert and Gouet, 2014).
5.4. Phylogenetic analysis
The phylogenetic trees were generated from multiple sequence alignment results of spike proteins using the MegaX software version 10.1.7 (Kumar et al., 2018). The algorithm Neighbor Joining was selected, and for all parameters the default values were used.
5.5. Molecular modeling
Homology model of feline coronavirus UU16 was build based on electron-microscopy structure of human coronavirus NL63 spike protein (PDB ID: 5SZS (Walls et al., 2016) using the Prime program version 4.2 (Schrödinger, 2015) in Schrodinger software platform. Missing loops of SARS-CoV-2 spike protein was modeled also using the Prime program based on structure with PDB ID: 6VYB (Walls et al., 2020).
CRediT authorship contribution statement
Yiran Wu: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing - original draft, Visualization. Suwen Zhao: Conceptualization, Resources, Writing - review & editing, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank ShanghaiTech University for supporting this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2020.102115.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Slide-window Simplot graph of Rhinolophus bat coronavirus HKU2, human coronavirus NL63, and human coronavirus 229E.
Mapping of furin recognition motif on uncompressed phylogenetic tree of spike protein sequences, Merbecovirus.
Mapping of furin recognition motif on uncompressed phylogenetic tree of spike protein sequences, Embecovirus.
Multiple sequence alignments of nucleotide sequences for linking regions of S1 and S2 in representative Merbecovirus (A) and Embecovirus (B). Furin recognition motif are highlighted with red colorboxes.
Tissue distribution of representative proteases responsible for cleavaging of coronavirus spike proteins (from The Human Protein Atlas website, https://www.proteinatlas.org, IDs: ENSG00000140564, ENSG00000184012). A) Furin; B) TMPRSS2.
Mapping of furin recognition motif on phylogenetic tree of influenza A/B spike proteins. Tree topology taken from reference (Jang and Seong, 2014).
References
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C.E., Weimer E., Scherer E.M., Rouphael N., Edupuganti S., Weiskopf D., Tse L.V., Hou Y.J., Margolis D., Sette A., Collins M.H., Schmitz J., Baric R.S., de Silva A.M. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham J. Passive antibody therapy in COVID-19. Nat. Rev. Immunol. 2020;20(7):401–403. doi: 10.1038/s41577-020-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904 e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020 doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L. functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82(17):8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pohlmann S. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85(24):13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019;8(8) doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.Y., El-Gebali S., Fraser M.I., Gough J., Haft D.R., Huang H., Letunic I., Lopez R., Luciani A., Madeira F., Marchler-Bauer A., Mi H., Natale D.A., Necci M., Nuka G., Orengo C., Pandurangan A.P., Paysan-Lafosse T., Pesseat S., Potter S.C., Qureshi M.A., Rawlings N.D., Redaschi N., Richardson L.J., Rivoire C., Salazar G.A., Sangrador-Vegas A., Sigrist C.J.A., Sillitoe I., Sutton G.G., Thanki N., Thomas P.D., Tosatto S.C.E., Yong S.Y., Finn R.D. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47(D1):D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.D., Guo W.P., Zhou R.H., Wang M.R., Wang C.Q., Ge S., Mei S.H., Li M.H., Shi M., Holmes E.C., Zhang Y.Z. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker G.L., Lu Y., Hardes K., Strehlow B., Levesque C., Lindberg I., Sandvig K., Bakowsky U., Day R., Garten W., Steinmetzer T. Highly potent inhibitors of proprotein convertase furin as potential drugs for treatment of infectious diseases. J. Biol. Chem. 2012;287(26):21992–22003. doi: 10.1074/jbc.M111.332643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Xie X., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., Zhang L., Bopp N., Schindewolf C., Vu M., Vanderheiden A., Swetnam D., Plante J.A., Aguilar P., Plante K.S., Lee B., Weaver S.C., Suthar M.S., Routh A.L., Ren P., Ku Z., An Z., Debbink K., Shi P.Y., Freiberg A.N., Menachery V.D. Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv. 2020 doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.Y., Wang P., Mok B.W., Zhang A.J., Chu H., Lee A.C., Deng S., Chen P., Chan K.H., Song W., Chen Z., To K.K., Chan J.F., Yuen K.Y., Chen H. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020;9(1):837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.W., Chao T.L., Li C.L., Chiu M.F., Kao H.C., Wang S.H., Pang Y.H., Lin C.H., Tsai Y.M., Lee W.H., Tao M.H., Ho T.C., Wu P.Y., Jang L.T., Chen P.J., Chang S.Y., Yeh S.H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2) doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Li X., Gao X., Dong Q. Natural polymorphisms are present in the furin cleavage site of the SARS-CoV-2 spike glycoprotein. Front. Genet. 2020;11:783. doi: 10.3389/fgene.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J., Xiong Q., Sun J., Li B., Wu J., Yi L., Peng X., Zhang H., Zhang W., Hulswit R.J.G., Loman N., Rambaut A., Ke C., Bowden T.A., Pybus O.G., Lu J. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94(17) doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., Hughes A.C., Bi Y., Shi W. A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the Spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020 [Google Scholar]
- Yamada Y.K., Takimoto K., Yabe M., Taguchi F. Virology. 1997 Jan 6;227(1):215–219. doi: 10.1006/viro.1996.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher-Friebertshauser E., Klenk H.D., Garten W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 2013;69(2):87–100. doi: 10.1111/2049-632X.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23(10):899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Prime version 4.2, Schrödinger, LLC, New York, NY, 2015.
- Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Luo C., Zhang W., Li F. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog. 2018;14(4) doi: 10.1371/journal.ppat.1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y.H., Seong B.L. Options and obstacles for designing a universal influenza vaccine. Viruses. 2014;6(8):3159–3180. doi: 10.3390/v6083159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Slide-window Simplot graph of Rhinolophus bat coronavirus HKU2, human coronavirus NL63, and human coronavirus 229E.
Mapping of furin recognition motif on uncompressed phylogenetic tree of spike protein sequences, Merbecovirus.
Mapping of furin recognition motif on uncompressed phylogenetic tree of spike protein sequences, Embecovirus.
Multiple sequence alignments of nucleotide sequences for linking regions of S1 and S2 in representative Merbecovirus (A) and Embecovirus (B). Furin recognition motif are highlighted with red colorboxes.
Tissue distribution of representative proteases responsible for cleavaging of coronavirus spike proteins (from The Human Protein Atlas website, https://www.proteinatlas.org, IDs: ENSG00000140564, ENSG00000184012). A) Furin; B) TMPRSS2.
Mapping of furin recognition motif on phylogenetic tree of influenza A/B spike proteins. Tree topology taken from reference (Jang and Seong, 2014).