Abstract
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is rapidly spreading all over the world. A new quantifying reagent for detecting SARS-CoV-2 antigen was developed for early and accurate detection. We evaluated the novel quantitative reagent for detecting SARS-CoV-2 antigen using an automated laboratory device.
Methods
One-hundred nasopharyngeal samples were collected from 47 SARS-CoV-2-infected patients, and 200 samples were collected from healthy donners. We measured the SARS-CoV-2 antigen and nucleic acid using Lumipulse Presto SARS-CoV-2 Ag and the 2019 Novel Coronavirus Detection Kit, respectively.
Results
The sensitivity and specificity of the SARS-CoV-2 antigen test were 75.7% (56/74) and 96.0% (192/200), respectively. The concordance rate in the positive group between the antigen and nucleic acid tests was 66% (66/100). In addition, the correlation coefficient between the concentration of SARS-CoV-2 antigen and the level of SARS-CoV-2 RNA was 0.74. There were 19 discrepant samples in which SARS-CoV-2 RNA was detected without SARS-CoV-2 antigen. There was significant difference between the discrepant and matched samples in terms of the time since symptom onset: the 19 discrepant samples were collected a median of 33 days after onset, while the 55 matched samples were collected a median of 19 days after onset. In addition, the 19 discrepant samples were collected from patients who were immune against SARS-CoV-2.
Conclusions
This novel SARS-CoV-2 antigen detection assay is highly sensitive, rapid, accurate, easily diagnostic. It may be useful in both clinical diagnosis and in screening because it does not require special methods such as PCR.
Keywords: SARS-CoV-2, Antigen, Detection, Lumipulse
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes the atypical pneumonia known as coronavirus disease 2019 (COVID-19), emerged in late 2019 in Wuhan, China [1]. SARS-CoV-2 has lower pathogenicity than SARS-CoV, but higher transmissibility from human to human [2]. The World Health Organization has declared COVID-19 a public health emergency of international concern and given the disease a “very high” risk assessment at the global level [3,4].
Viral culture and nucleic acid tests are the gold standards to diagnose SARS-CoV-2 infection. Several methods have been developed to specifically detect viral nucleic acids, such as quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR). However, this technique takes hours to detect the nucleic acid, and it takes days to isolate the virus. Moreover, specialized instruments and expertise are required to carry out these tests [[5], [6], [7]]. Recently, new approaches have been developed to diagnose SARS-CoV-2 infection by targeting anti-SARS-CoV-2 antibodies or SARS-CoV-2 antigen. The advantage of antibody testing is that the sample species for testing is serum, which cannot be influenced by the difference of the amount of measurement target every sampling and carries a low risk of infection for healthcare workers [8]. However, the utility of antibody testing remains unclear. A positive antibody test may indicate past infection, rather than active infection, as it only shows whether the person has mounted an immune response against SARS-CoV-2. Moreover, the test has low sensitivity [[8], [9], [10]]. Conversely, antigen detection requires no special skills, although one study reported that antigen-based assays are 100,000-fold less sensitive than nucleic acid tests in patients suspected of SARS-CoV-2 infection, so they may produce more false negative results in clinical practice [11]. Recently, a new quantifying reagent for detecting SARS-CoV-2 antigen was developed to overcome these problems. In the present study, we evaluated this reagent in a clinical laboratory setting.
2. Materials and methods
2.1. Sample collection
We collected 100 positive nasopharyngeal specimens from patients diagnosed with COVID-19 and 200 negative nasopharyngeal specimens from healthy donors. Informed consent was obtained in the form of opt-out on the website. The details of this research were published on the website to provide an opportunity for patients to refuse. Those who rejected were excluded. The nasopharyngeal swabs from patients with COVID-19 were collected multiple times during hospitalization and treatment. All 300 samples were collected using a kit containing a nylon-flocked nasopharyngeal swab and a tube containing universal transport medium (UTM; Copan Diagnostics, Murrieta, CA, United States). All samples were tested using PCR at the time of collection to confirm whether they were positive or negative. All 200 healthy donors, defined as the negative group, had undergone SARS-CoV-2 nucleic acid tests and were negative for anti-SARS-CoV-2 antibodies. COVID-19-positive nasopharyngeal six swabs were purchased from Precision for Medicine (Bethesda, MD, United States). Six samples were diluted 4-fold in steps to prepare 17 samples All samples were preserved at −80 °C to await testing.
2.2. Test for SARS-CoV-2 antigen
We measured the SARS-CoV-2 antigen using Lumipulse Presto SARS-CoV-2 Ag and Espline SARS-CoV-2 (Fujirebio Inc., Tokyo, Japan). The Lumipulse Presto SARS-CoV-2 Ag was analyzed using the fully automated Lumipulse L2400 (Fujirebio Inc., Tokyo, Japan), while the Espline SARS-CoV-2 was analyzed by hand. All assays were performed according to the manufacturer's protocol.
2.3. Test for SARS-CoV-2 nucleic acid
The SARS-CoV-2 nucleic acid test was performed on a LightCycler480 System (Roche, Basel, Switzerland) using the 2019 Novel Coronavirus Detection Kit (Shimadzu Corporation, Kyoto, Japan). All assays were performed according to the manufacturer's protocol, and the samples were judged as positive or negative based on the threshold cycle (Ct) value—when the measured Ct value of the sample was 40 or less, it was judged as positive.
2.4. Comparing detection levels between the SARS-CoV-2 antigen and nucleic acid tests
Detection levels were compared using COVID-19-positive nasopharyngeal swabs prepared by repeated 4-fold dilution.
2.5. Test for anti-SARS-CoV-2 antibody
We measured anti-SARS-CoV-2 antibodies using Elecsys anti-SARS-CoV-2 (Roche Diagnostics GmbH, Mannheim, Germany) and ARCHITECT SARS-CoV-2 IgG (Abbott, Illinois, United States). These two reagents were analyzed using Cobas e801(Roche Diagnostics GmbH, Mannheim, Germany) and ARCHITECT i2000SR (Abbott, Illinois, United States), respectively. All assays were performed according to the manufacturer's protocol.
2.6. Statistical analysis
Statistical analyses such as receiver operating characteristic (ROC) curve analysis and p-value calculation were performed using SAS Platform JMP Pro version 15.1.0. (SAS Institute Inc., Cary, NC, USA).
2.7. Ethical statement
This study was approved by the ethics committee of Sapporo Medical University Hospital (reference number 2-1-48) and the ethics committee of Sapporo Medical University. https://web.sapmed.ac.jp/byoin/chiken/index.html (reference number 322-144, 322-146).
3. Results
The 100 samples in the positive sample group were sourced from 47 patients with COVID-19 who had a median age of 61 years (range: 22–90 years); 61.7% of them were male. According to the patients’ electronic medical records, the samples were collected a median of 17 days (range: 1–88 days) after symptom onset in cases where such information was available.
Firstly, we compared SARS-CoV-2 antigen detection ability between the Lumipulse Presto SARS-CoV-2 Ag and the Espline SARS-CoV-2. The concordance rate between the Lumipulse Presto SARS-CoV-2 Ag and Espline SARS-CoV-2 was 52% (52/100; Table 1 ). The Lumipulse Presto SARS-CoV-2 Ag judged 72 of the 100 patient samples as positive. The 2019 Novel Coronavirus Detection Kit judged 74 of the 100 samples as positive at the same time as antigen measurement. Therefore, the sensitivity of the Lumipulse Presto SARS-CoV-2 Ag was 75.7% (56/74; Table 2 a). On the other hand, the specificity when testing the 200 negative samples was 96% (192/200; Table 2b).
Table 1.
Concordance between Espline SARS-CoV-2 and Lumipulse Presto SARS-CoV-2 Ag in the positive group.
| Lumipulse Presto SARS-CoV-2 Ag |
Total | |||
|---|---|---|---|---|
| (+) | (−) | |||
| Espline SARS-CoV-2 | (+) | 24 | 0 | 24 |
| (−) | 48 | 28 | 76 | |
| Total | 72 | 28 | 100 | |
Table 2.
Concordance between the SARS-CoV-2 antigen and nucleic acid tests.
|
a | ||||
| 2019 Novel Coronavirus Detection Kit |
Total |
|||
| (+) |
(-) |
|||
| Lumipulse Presto SARS-CoV-2 Ag ((+)≥1.00 pg/mL) |
(+) |
56 |
16 |
72 |
| (-) |
18 |
10 |
28 |
|
| Total | 74 | 26 | 100 | |
| Sensitivity: 75.7% (95% confidence interval: 65.0%–86.5%) Concordance rate: 66.0% | ||||
|
b | ||||
| 2019 Novel Coronavirus Detection Kit |
Total |
|||
| (+) |
(-) |
|||
| Lumipulse Presto SARS-CoV-2 Ag ((+)≥1.00 pg/mL) |
(+) |
0 |
8 |
8 |
| (-) |
0 |
192 |
192 |
|
| Total | 0 | 200 | 200 | |
| Specificity: 96.0% (95% confidence interval: 93.0%–99.0%) Concordance rate: 96.0% Concordance in the positive group (n = 100) (a), and in the negative group (n=200) (b) | ||||
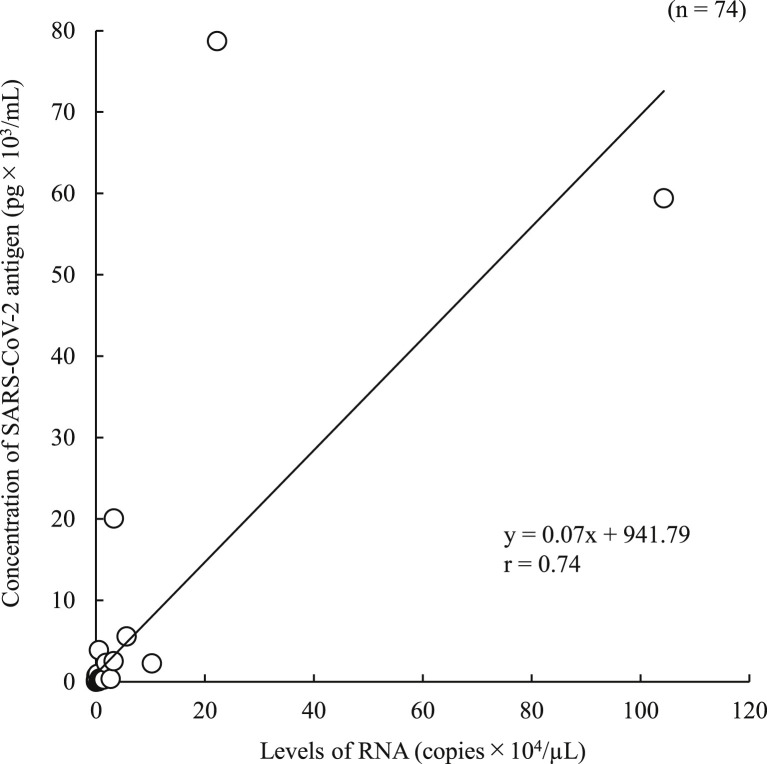
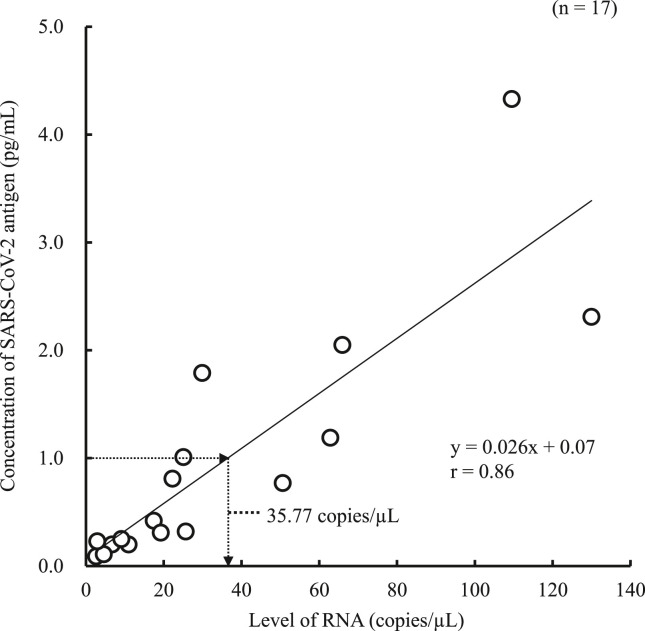
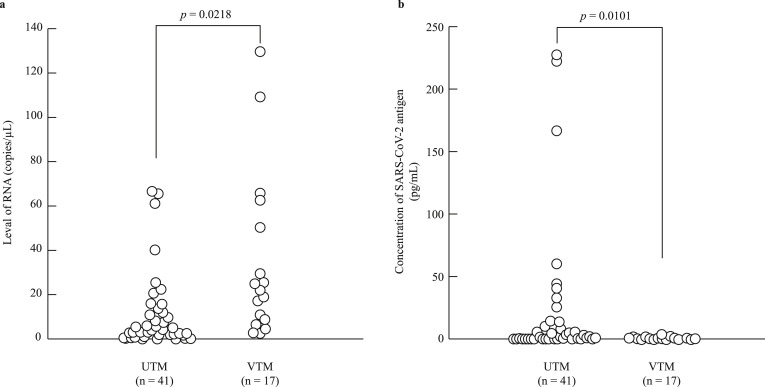
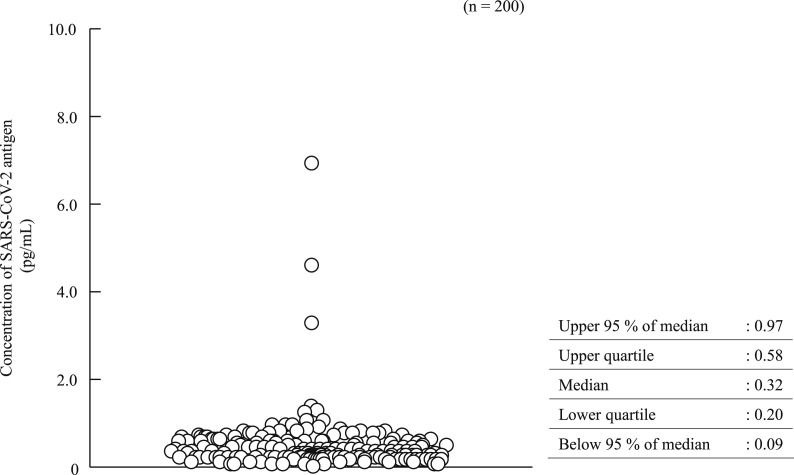
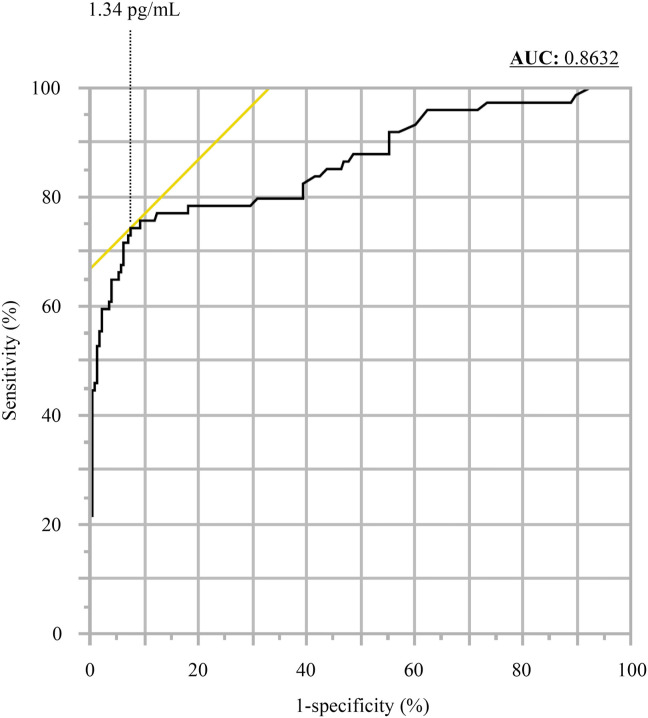
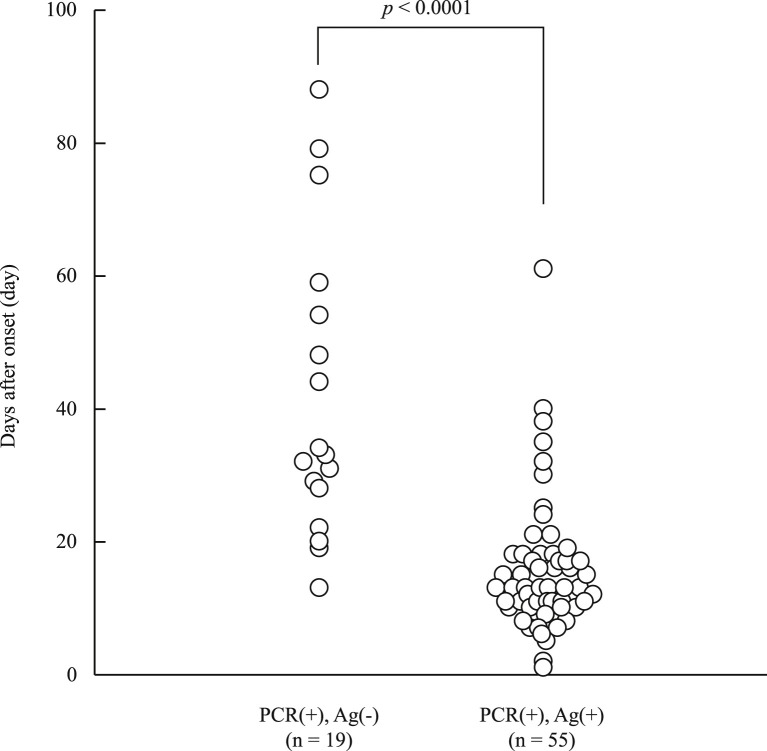
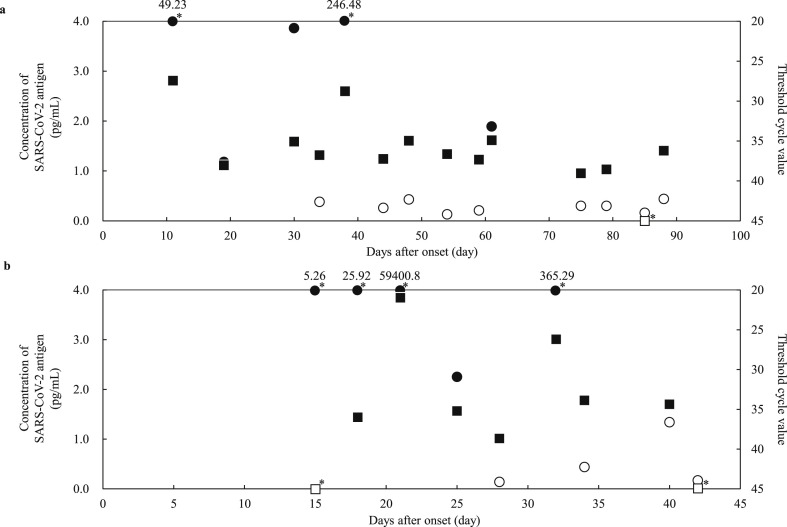
In the 100 positive samples, the correlation coefficient between the concentration of SARS-CoV-2 antigen and the level of SARS-CoV-2 RNA was 0.74 (Fig. 1 ). Seventeen samples from six commercial swabs were prepared by dilution, which were then measured using both the Lumipulse Presto SARS-CoV-2 Ag and 2019 Novel Coronavirus Detection Kit to compare the detection capacity. The level of RNA corresponding to an antigen concentration of 1.0 pg/mL was 35.77 copies/μL, as determined from the regression equation (Fig. 2 ). On the other hand, 74 of the 100 positive samples had <35.77 copies/μL, and 37 of these were judged as positive by the Lumipulse Presto SARS-CoV-2 Ag. However, the median levels of RNA in the 74 samples preserved in UTM were lower than those of the purchased samples preserved in viral transport medium (VTM) (p = 0.02), while the median concentration of SARS-CoV-2 antigen in the 74 samples was higher than that of the purchased samples (p = 0.01) (Fig. 3 a and b). The antigen concentration in the samples from the 200 healthy donors was lower than 1.0 pg/mL in almost all cases, although it showed wide variation. A histogram analysis of the 200 negative samples indicated that the most appropriate cut-off value was 0.97 pg/mL (Fig. 4 ), while ROC curve analysis of the 74 positive and 226 negative samples indicated that the most effective cut-off value was 1.34 pg/mL, based on the sensitivity and specificity (Fig. 5 ). We investigated the clinical performance of the two calculated cut-off values in all 300 samples (Table 3 a–d). There were 34 discrepant samples between the results of the antigen and nucleic acid tests in the positive group. Nineteen of these were judged as negative by the antigen test and positive by the nucleic acid test. These 19 samples were collected a median of 33 days (range: 13–88 days) after symptom onset, while the 55 samples for which the antigen and nucleic acid tests matched were collected a median of 13 days (range: 1–60 days) after symptom onset (Fig. 6 ). Additionally, the antibody against SARS-CoV-2 was detected in the serum of the 19 patients who showed a discrepancy, which was collected at a similar time (Table 4 ). Eleven of these 19 samples were derived from two patients. Therefore, the antigen load seemed to be lower when more time had passed since symptom onset, while RNA remained in the nasopharynx (Fig. 7 a and b). However, the antigen and nucleic acid loads showed corresponding trends of increasing and decreasing concentration.
Fig. 1.
Correlation between SARS-CoV-2 antigen concentration and level of RNA.
Fig. 2.
Predicting the level of RNA in the cut-off range of the antigen test. The level of RNA was estimated based on the intersection of the horizontal arrow at an antigen concentration of 1.0 pg/mL and the regression equation, as well as on the intersection of the down arrow and the X-axis.
Fig. 3.
Comparing universal transport medium (UTM) with viral transport medium (VTM) in terms of the SARS-CoV-2 antigen and RNA levels. The number of samples in UTM and VTM were 41 and 17, respectively. The median levels of RNA in UTM and VTM were 4.21 copies/μL and 22.25 copies/μL, respectively (a). The median SARS-CoV-2 concentrations in UTM and VTM were 1.9 pg/mL and 0.42 pg/mL, respectively (b). All p-values were calculated using the Wilcoxon rank-sum test.
Fig. 4.
Measurement distribution in the negative group.
Fig. 5.
Receiver operating characteristic curve analysis.
Table 3.
Concordance between the results of SARS-CoV-2 antigen and nucleic acid tests by each cut-off value.
|
a | ||||
| 2019 Novel Coronavirus Detection Kit |
Total |
|||
| (+) |
(-) |
|||
| Lumipulse Presto SARS-CoV-2 Ag ((+)≥1.34 pg/mL) |
(+) |
55 |
13 |
68 |
| (-) |
19 |
13 |
32 |
|
| Total | 74 | 26 | 100 | |
| Sensitivity: 74.3% (95% confidence interval: 64.0%–83.6%) Concordance rate: 68.0% | ||||
|
b | ||||
| 2019 Novel Coronavirus Detection Kit |
Total |
|||
| (+) |
(-) |
|||
| Lumipulse Presto SARS-CoV-2 Ag ((+)≥1.34 pg/mL) |
(+) |
0 |
4 |
4 |
| (-) |
0 |
196 |
196 |
|
| Total | 0 | 200 | 200 | |
| Specificity: 98.0% (95% confidence interval: 95.0%–100.0%) Concordance rate: 98.0% | ||||
|
c | ||||
| 2019 Novel Coronavirus Detection Kit |
Total |
|||
| (+) |
(-) |
|||
| Lumipulse Presto SARS-CoV-2 Ag ((+)≧0.97 pg/mL) |
(+) |
56 |
16 |
72 |
| (-) |
18 |
10 |
28 |
|
| Total | 74 | 26 | 100 | |
| Sensitivity: 75.7% (95% confidence interval: 65.5%–85.9%) Concordance rate: 66.0% | ||||
|
d | ||||
| 2019 Novel Coronavirus Detection Kit |
Total |
|||
| (+) |
(-) |
|||
| Lumipulse Presto SARS-CoV-2 Ag ((+)≧0.97 pg/mL) |
(+) |
0 |
10 |
10 |
| (-) |
0 |
190 |
190 |
|
| Total | 0 | 200 | 200 | |
| Specificity: 95.0% (95% confidence interval: 92.5%–97.5%) Concordance rate : 95.0% When 1.34 pg/mL was set as the cut-off value in the positive (3a) and negative groups (3b) When 0.97 pg/mL was set as the cut-off value in the positive (3c) and negative groups (3d) | ||||
Fig. 6.
Comparing days after onset at the time of sample collection between the positive matched and antigen-only positive groups. The median number of days after onset was 33 days in the 19 samples that only tested positive in the antigen test. The median number of days after onset was 19 days in the 55 samples that tested positive in both the antigen and nucleic acid tests. All p-values were calculated using the Wilcoxon rank-sum test.
Table 4.
Discrepant samples that only tested positive for SARS-CoV-2 antigen.
| Divergence Sample ID | Days after onset (day) | Concentration of SARS-CoV-2 antigen (pg/mL) | Ct value | ARCHITECT SARS CoV-2 IgG ((+) ≥1.40 pg/mL) | Elecsys Anti-SARS-CoV-2 ((+) ≥1.00 C.O.I.) | |||
|---|---|---|---|---|---|---|---|---|
| 1 | 13 | 0.78 | 38.15 | (+) | 7.90 | (+) | 12.00 | |
| 2 | 13 | 0.30 | 39.15 | (+) | 126.00 | (+) | 10.40 | |
| 3 | 19 | 1.18 | 38.05 | (+) | 31.80 | (+) | 16.50 | |
| 4 | 20 | 0.26 | 35.4 | (+) | 2.14 | (+) | 8.18 | |
| 5 | 22 | 0.58 | 39.82 | (+) | 2.68 | (+) | 8.30 | |
| 6 | 28 | 0.14 | 38.66 | (+) | 9.47 | (+) | 350.00 | |
| 7 | 29 | 0.95 | 35.39 | (+) | 86.00 | (+) | 134.80 | |
| 8 | 31 | 0.34 | 36.36 | (+) | 169.00 | (+) | 194.00 | |
| 9 | 32 | 0.41 | 36.32 | (+) | 50.00 | (+) | 62.80 | |
| 10 | 33 | 0.27 | 37.44 | (+) | 52.50 | (+) | 91.30 | |
| 11 | 34 | 0.38 | 36.1 | (+) | 15.40 | (+) | 24.15 | |
| 12 | 34 | 0.44 | 33.04 | (+) | 55.20 | (+) | 236.00 | |
| 13 | 44 | 0.26 | 37.25 | (+) | 3.34 | (+) | 18.85 | |
| 14 | 48 | 0.43 | 34.97 | (+) | 3.27 | (+) | 24.40 | |
| 15 | 54 | 0.13 | 36.65 | (+) | 2.54 | (+) | 16.60 | |
| 16 | 59 | 0.21 | 37.34 | (+) | 2.78 | (+) | 19.55 | |
| 17 | 75 | 0.30 | 38.51 | (+) | 1.45 | (+) | 9.76 | |
| 18 | 79 | 0.30 | 38.56 | (−) | 1.31 | (+) | 8.18 | |
| 19 | 88 | 0.44 | 36.22 | (−) | 0.79 | (+) | 3.82 | |
Fig. 7.
Changes over time in the antigen concentration and Ct value in two cases. Open circle (〇): SARS-CoV-2 antigen negative, closed circle (●): SARS-CoV-2 antigen positive, opened square (□): PCR negative, closed square (■): PCR positive, open square with asterisk (□∗): not detected, closed circle with asterisk (●∗): out of Y-axis range.
4. Discussion
In the present study, we evaluated the Lumipulse Presto SARS-CoV-2 Ag. The concordance rate between Lumipulse Presto SARS-CoV-2 Ag and the Espline SARS-CoV-2 was 52%. The 48 samples that showed a discrepancy between the Lumipulse Presto SARS-CoV-2 Ag and Espline SARS-CoV-2 were all judged as positive by the Lumipulse Presto SARS-CoV-2 Ag, and all concentrations were <96.08 pg/mL, as measured by the Lumipulse Presto SARS-CoV-2 Ag (data not shown). Previous studies have reported that antigen reagents have low detection capacity [11]. Such reagents, including the Espline SARS-CoV-2, employ the immunochromatography (IC) method. In particular, the Lumipulse Presto SARS-CoV-2 Ag used the chemiluminescence enzyme immunoassay (CLEIA), which has a reported detection capacity equivalent to that of the nucleic amplification test [12]. Therefore, because the sensitivity of antigen detection had improved, we believed that the reported problems with sensitivity had been overcome.
The sensitivity and specificity of the SARS-CoV-2 antigen test, using the nucleic acid test as the gold standard, were 75.7% and 96%, respectively, in the present study. In the positive group, the concordance rate between the antigen and nucleic acid tests was 66%. The concentration of SARS-CoV-2 antigen correlated well with the level of SARS-CoV-2 RNA (r = 0.74), as reported for other RNA viruses [13].
Next, we investigated the capability of antigen detection. The level of RNA corresponding to 1.0 pg/mL of the antigen concentration was 35.77 copies/μL, determined using the obtained regression equation. The detection performance of this reagent was sufficient compared to previous reports involving RT-qPCR [14]. However, in 57 of the positive samples with an antigen concentration <10 pg/mL, as measured using the Lumipulse Presto SARS-CoV-2 Ag, the level of RNA corresponding to 1.0 pg/mL of antigen concentration was 2.56 copies/μL, determined using the obtained regression equation (data not shown). We assumed that the difference was caused by different preservation solutions. The purchased samples were preserved in Centers for Disease Control and Prevention protocol VTM [15], while our samples were preserved in UTM. Analysis of preservation medium showed that the concentration of SARS-CoV-2 antigen in UTM was higher than that in VTM, perhaps because VTM contains fetal bovine serum, which affects antigen-antibody reactivity, but not nucleic acid test reactions. Therefore, VTM may not be suitable as a solution for sample collection when conducting antigen tests [16].
The negative group samples showed variation, although almost all were under 1.0 pg/mL, which was the cut-off value according to the manufacturer's protocol. Because enzyme immunoassays involve a non-specific reaction [17], we next validated the cut-off value. By ROC and histogram analysis, the effective cut-off values were 1.34 and 0.97 pg/mL, respectively. However, no significant advantage was identified using three cut-off values (0.97, 1.00, and 1.34 pg/mL) in the present study.
Among the 100 positive samples, 34 showed a discrepancy between the results of the antigen test and those of the nucleic acid analysis. Sixteen samples were judged as positive by the antigen test and negative by the nucleic acid test. Because SARS-CoV-2 nucleic acid was detected in all samples at the time of collection, the discrepancy was likely caused by RNA degradation due to freezing and thawing, or by variations in the results due to low levels of RNA. Conversely, we analyzed the discrepancies among samples that were judged as negative by the antigen test and positive by the nucleic acid test. There were 19 such samples, and we checked the clinical information and measured antibodies against SARS-CoV-2 in serum collected at a similar time. The time since onset in the discrepant samples was significantly longer than in the matched samples. The nineteen samples were from seven patients, all of whom had antibodies against SARS-CoV-2 in their serum. Next, we analyzed the details of the two patients whose clinical course could be investigated. Antigen levels seemed to reduce gradually after the date of onset as antibody was produced. However, RNA remained in the nasopharynx, indicating that submitting a nasopharyngeal swab for antigen testing will not produce useful results 13 days after onset. The antibody against SARS-CoV-2 is still not well understood. Researchers do not know whether it is a neutralization antibody. We concluded that antibody detection was not useful for detecting active SARS-CoV-2 infection because the antibody is not detectable in the serum until 3–5 days after onset [18]. As shown in our results, the sensitivity problem of the antigen tests has been resolved, and the sensitivity of the Lumipulse SARS-CoV-2 Ag was equivalent to that of the nucleic acid test. Therefore, this new reagent has sufficient capability to diagnose initial infection. Moreover, because the results of antigen detection almost behaved as the results of PCR in the investigation of two clinical courses, this highly sensitive antigen test can be used as a surrogate for nucleic acid testing. That said, antigens may not be detected long after onset, when antibodies have been acquired.
The nucleic acid test is the current gold standard for the diagnosis of COVID-19 globally. The antigen test, mainly qualitative test, is also widely used due to the rapidity, low cost, and simple technique; however, it has been less sensitive than the nucleic acid test [11,19]. Our study indicated that the quantitative antigen test, which has not been commonly available yet, would be highly sensitive. This novel reagent for the SARS-CoV-2 test has the potential to play a role in suppressing the spread of SARS-CoV-2 infection [20].
The major limitation of the present study was that asymptomatic individuals were not included in the positive group. Additionally, further research is needed to determine whether patients with COVID-19 can infect others when no SARS-CoV-2 antigen is detected in the nasopharynx. Nonetheless, SARS-CoV-2 antigen detection is an excellent clinical test because it does not require any special methods or techniques, such as PCR, and it takes less time to report the results than PCR.
In conclusion, the Lumipulse Presto SARS-CoV-2 Ag may an important role in clinical laboratory testing and help us to prevent the spread of infection all over the world. We hope that new highly sensitive antigen tests such as Lumipulse Presto SARS-CoV-2 Ag will be developed and evaluated sufficiently.
Authorship statement
All authors meet the ICMJE authorship criteria. Contributors R.K., K.A., Y.F., and S.T. were responsible for the organization and coordination of the trial. S.T. was the chief investigator responsible for the data analysis. R.K. and R.M. developed the trial design and conducted an investigation. All authors contributed to the writing of the final manuscript.
Declaration of competing interest
Satoshi Takahashi received speaker honoraria from MSD K.K. and research grants from Shino-Test Corporation, Roche Diagnostics K.K., FUJIREBIO Inc. and Abbott Japan Co., Ltd.
Funding source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgments
None.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization Novel coronavirus. nCoV) situation report– 10. 2019. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200130-sitrep-10-ncov.pdf?sfvrsn=d0b2e480_2 Access date: 20 October, 2020.
- 4.Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 7.WHO Laboratory testing for coronavirus Disease 2019 (COVID-19) in suspected human cases. Interim guidance. 2020 https://apps.who.int/iris/handle/10665/331329 Available from: (2020) (Accessed on 15/11/2020. [Google Scholar]
- 8.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeks J.J., Dinnes J., Takwoingi Y., Davenport C., Spijker R., Taylor-Phillips S., et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;6:CD013652. doi: 10.1002/14651858.CD013652. https://doi:10.1002:CD013652 Pubmed: 32584464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv H., Wu N.C., Tsang O.T., Yuan M., Perera R.A.P.M., Leung W.S., et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107725. https://doi:10.1016/j.celrep.2020.107725 107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. https://doi:10.1016/j.jcv.2020.104500:104500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kesli R., Polat H., Terzi Y., Kurtoglu M.G., Uyar Y. Comparison of a newly developed automated and quantitative hepatitis C virus (HCV) core antigen test with the HCV RNA assay for clinical usefulness in confirming anti-HCV results. J Clin Microbiol. 2011;49:4089–4093. doi: 10.1128/JCM.05292-11. https://doi:10.1128/JCM.05292-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long L., Shen T., Gao J., Duan Z., Liang H., Lu F. Effectiveness of HCV core antigen and RNA quantification in HCV-infected and HCV/HIV-1- coinfected patients. BMC Infect Dis. 2014;14:577. doi: 10.1186/s12879-014-0577-1. https://doi:10.1186/s12879-014-0577-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B., Hu M., Ren Y., Xu X., Wang Z., Lyu X., et al. Evaluation of seven commercial SARS-CoV-2 RNA detection kits based on real-time polymerase chain reaction (PCR) in China. Clin Chem Lab Med. 2020;58:e149–e153. doi: 10.1515/cclm-2020-0271. https://doi:10.1515/cclm-2020-0271 [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. Standard operating procedure (SOP) for laboratories to create their own VTM in accordance with CDC's protocol, https://www.cdc.gov/coronavirus/2019-ncov/downloads/Viral-Transport-Medium.pdf, (Access date: 20 October, 2020).
- 16.Kim H.J., Park J.S., Kwon S.R. Development of a stringent ELISA protocol to evaluate anti-viral hemorrhagic septicemia virus-specific antibodies in olive flounder Paralichthys olivaceus with improved specificity. J Microbiol. 2015;53:481–485. doi: 10.1007/s12275-015-5101-9. https://doi:10.1007/s12275-015-5101-9 [DOI] [PubMed] [Google Scholar]
- 17.Watanabe T., Hashida S. The immune complex transfer enzyme immunoassay: mechanism of improved sensitivity compared with conventional sandwich enzyme immunoassay. J Immunol Methods. 2018;459:76–80. doi: 10.1016/j.jim.2018.05.010. https://doi:10.1016/j.jim.2018.05.010 [DOI] [PubMed] [Google Scholar]
- 18.Chinese Centre for Disease Control and Prevention Diagnosis and treatment COVID-19 prevention and Control. 2020. http://www.chinacdc.cn/en/COVID19/202011/P020201119530272386786.pdf Available from:
- 19.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Imöhl M., Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J Virol Methods. 2021;218:114024. doi: 10.1016/j.jviromet.2020.114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mina M.J., Parker R., Larremore D.B. Rethinking Covid-19 test sensitivity -A strategy for containment. N Engl J Med. 2020;383 doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]