Abstract
Comorbidities in COVID-19 patients often lead to more severe outcomes. The disease-specific molecular events, which may induce susceptibility to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection, are being investigated. To assess this, we retrieved array-based gene expression datasets from patients of 30 frequently occurring acute, chronic, or infectious diseases. Comparative analyses of the datasets were performed after quantile normalization and log2 transformation. Among the 78 host genes prominently implicated in COVID-19 infection, ACE2 (receptor for SARS-CoV-2) was positively regulated in several cases, namely, leukemia, psoriasis, lung cancer, non-alcoholic fatty liver disease (NAFLD), breast cancer, and pulmonary arterial hypertension (PAH). FURIN was positively regulated in some cases, such as leukemia, psoriasis, NAFLD, lung cancer, and type II diabetes (T2D), while TMPRSS2 was positively regulated in only 3 cases, namely, leukemia, lung cancer, and T2D. Genes encoding various interferons, cytokines, chemokines, and mediators of JAK-STAT pathway were positively regulated in leukemia, NAFLD, and T2D cases. Among the 161 genes that are positively regulated in the lungs of COVID-19 patients, 99–111 genes in leukemia (including various studied subtypes), 77 genes in NAFLD, and 48 genes in psoriasis were also positively regulated. Because of the high similarity in gene expression patterns, the patients of leukemia, NAFLD, T2D, psoriasis, and PAH may need additional preventive care against acquiring SARS-CoV-2 infections. Further, two genes CARBONIC ANHYDRASE 11 (CA11) and CLUSTERIN (CLU) were positively regulated in the lungs of patients infected with either SARS-CoV-2, or SARS-CoV or Middle East Respiratory Syndrome Coronavirus (MERS-CoV).
Keywords: COVID-19, Comorbidity, SARS-CoV-2, Leukemia, NAFLD, Psoriasis, Cancer, Type II diabetes
1. Introduction
Coronavirus disease (COVID-19) caused by Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the most dreaded pandemic of recent times. As per the global data released on December 25, 2020, by COVID-19 dashboard of the World health organization, SARS-CoV-2 has infected 77,920,564 people of which 1,731,901 people have died. A significant proportion of the COVID-19 patients have been reported to suffer from other pathophysiological conditions as well. For instance, in a cohort of 1590 COVID-19 patients from China, Guan et al. (2020) reported that 399 patients (25.1%) were having at least one comorbidity, while 130 patients (8.2%) had two or more comorbidities [1]. They reported that hypertension, diabetes, cardiovascular diseases, and chronic kidney diseases were among the most frequent comorbidities, which occurred in 16.9%, 8.2%, 3.7%, and 1.3% of all COVID-19 patients, respectively. Also, COPD and malignancy were identified as critical risk factors associated with severe COVID-19 conditions. Another study by Chen et al. (2020) reported that in a cohort of 99 COVID-19 patients in China, 50 patients (51%) suffered from chronic medical illnesses [2]. The reported comorbid diseases were cardiovascular or cerebrovascular diseases (40.4%), diabetes (12%), digestive system disease (11%), and malignant tumor (0.01%) that were identified in 40, 12, 11, and 1 patient, respectively. Similarly, others have also reported cancer of lungs [3] and of blood [4], NAFLD [5], and HIV infections (Human Immunodeficiency Virus) [6], as frequently occurring comorbidities that often worsen the outcome and increase the risk of mortality in COVID-19 patients.
Similarly Dolan et al. (2020) have reported comorbidities in severe forms of COVID-19 [7]. These included cardiovascular diseases, diabetes, hepatitis, lung disease, and kidney disease in addition to the previously mentioned comorbidities. However, the molecular mechanisms underlying COVID-19 associated comorbidities are poorly understood and are still being investigated. The aim of this study was to decipher such diseases and their associated gene expression patterns that may induce susceptibility to SARS-CoV-2 infection. We performed a meta-analysis with gene expression datasets in 30 widely prevalent acute, chronic, and infectious diseases to identify the gene expression signatures that could promote the pathogenesis of SARS-CoV-2. We found that the gene expression pattern in leukemia patients was most similar to SARS-CoV-2 cases, followed by the patients of chronic diseases, namely, non-alcoholic fatty liver disease (NAFLD), psoriasis, type II diabetes (T2D), and pulmonary arterial hypertension (PAH). Our study could serve as a guide for understanding the gene expression signatures underlying COVID-19 associated comorbidities.
2. Dataset, methods, and techniques
2.1. Data retrieval
Publicly available datasets of human acute, chronic, infectious diseases, and various types of cancer were retrieved from NCBI's GEO database [8]. We favored the expression profile GSEids with highest number of samples while focussing on the tissue of origin, and devoid of any treatments or other afflictions. We retrieved datasets of patients and controls for the following conditions: Asthma (GSE64913), Chronic Obstructive Pulmonary Disorder (COPD, GSE112811), Cardiovascular diseases (GSE109048), Hypertension (GSE113439), NAFLD (GSE49541, GSE107037), Atherosclerosis (Athero, GSE28829), T2D (GSE15653, GSE25462, GSE38642, GSE27949), Polycystic Ovary Syndrome (PCOS, GSE124226), Multiple Sclerosis (MS, GSE21942), Psoriasis (GSE78097), Blood Cancer or Leukemia (GSE51082, GSE9476), Breast Cancer (GSE65194), Cervical Cancer (GSE63514), Multiple Myeloma (MM, GSE85837), Lung Cancer (GSE136043), Lung adenocarcinoma or Non-small cell lung cancer (NSCLC, GSE118370), Liver Cancer (GSE88839), Pancreatic Ductal Adenocarcinoma (PDAC, GSE101448), AIDS (GSE73968), Tuberculosis (TB, GSE139825), Malaria (GSE119150), Acute Kidney Injury (AKI, GSE30718), and COVID-19 (GSE150316). In order to identify the common differentially expressed genes in different viral infections, we analyzed the datasets from the patients infected with either SARS-CoV (GSE1739), SARS-CoV-2 (GSE150316), MERS-CoV (GSE100496), H1N1 (GSE21802), or other influenza viruses (GSE22319; H7N1, H5N1, H3N2, H5N2). Furthermore, we studied six subtypes of leukemia that were retrieved from the GSE51082 and GSE9476 datasets, namely Acute myeloid leukemia (AML), B-cell chronic lymphocytic leukemia (BCLL), Chronic myelogenous leukemia (CML), Myelodysplastic syndrome (MDS), B-acute lymphoblastic leukemia (BALL), and T-cell acute lymphoblastic leukemia (TALL). The dataset on cardiovascular diseases included an equal number of patients suffering from coronary artery disease (CAD) and acute myocardial infarction (AMI), and the dataset of breast cancer included both breast cancer and triple-negative breast cancer tissues (TNBC) samples. Of the 29 samples in the GSE28829 dataset of atherosclerosis (Athero), 16 samples were from advanced atherosclerotic plaque (ATHERO-Adv), and 13 samples were from early atherosclerotic plaque (ATHERO-Early) regions. In the cases of leukemia and NAFLD, we obtained expression profiles of diseased and control samples from separate GSEids. Therefore, we used a data-integration strategy as previously explained by Hamid et al. (2009) for data analysis in leukemia and NAFLD cases [9,10]. The platform of microarray experiment, type of tissue sample, sample size, experimental design, and data processing strategy, are summarized in Table 1 .
Table 1.
Details of Expression Datasets taken from GEO for the study.
| Disease Type | Disease/Condition | GSE ID | Platform | Tissue | Experimental Design | Data Processing |
|---|---|---|---|---|---|---|
| Chronic | Asthma | GSE64913 | GPL570 [HG-U133_Plus_2] Affymetrix | Epithelial brushings from central and peripheral airways | 42 healthy volunteer, 28 asthmatic patients | Preprocessed: Normalization and log2 transformation by GCRMA method |
| Chronic Obstructive Pulmonary Disorder | GSE112811 | GPL570 [HG-U133_Plus_2] Affymetrix | Blood | 20 COPD patients, 22 healthy volunteers before administration of LPS or saline | Preprocessed: Normalization and log2 transformation by RMA | |
| Cardiovascular | GSE109048 | GPL17586 [HTA-2_0] Affymetrix | Blood platelets | 19 Healthy donors, 19 CAD patients, 19 AMI patients | Preprocessed: SST-RMA normalization and log2 transformation | |
| Hypertension | GSE113439 | GPL6244 [HuGene-1_0-st] Affymetrix | Lung | 15 patients with Pulmonary Arterial Hypertension and 11 normal controls | Preprocessed: Normalization and log2 transformation by RMA | |
| Non-Alcoholic Fatty Liver Disease | GSE49541 | GPL570 [HG-U133_Plus_2] Affymetrix | Liver | 72 patients with NAFLD | Preprocessed: Normalization and log2 transformation by GCRMA method | |
| GSE107037 | GPL570 [HG-U133_Plus_2] Affymetrix | Liver | 33 healthy liver donors | Preprocessed: Normalization and log2 transformation by RMA | ||
| Atherosclerosis | GSE28829 | GPL570 [HG-U133_Plus_2] Affymetrix | Carotid artery | Samples from atherosclerotic carotid artery segments of 29 patients | Preprocessed: Normalization and log2 transformation by RMA | |
| Type 2 Diabetes | GSE15653 | GPL96 [HG-U133A] Affymetrix | Liver | 4 type 2 diabetes and 5 control subjects | Preprocessed: MAS5.0 signal intensity. | |
| GSE25462 | GPL570 [HG-U133_Plus_2] Affymetrix | Muscle | 10 subjects with type 2 diabetes and 15 healthy subjects | Preprocessed: MAS5.0 signal intensity. | ||
| GSE38642 | GPL6244 [HuGene-1_0 -st] Affymetrix | Pancreas | 54 non-diabetic and 9 diabetic cadavers | Preprocessed: Normalization and log2 transformation by RMA | ||
| GSE27949 | GPL570 [HG-U133_Plus_2] Affymetrix | Adipose | 12 Normal and 11 T2D subjects | Preprocessed: Normalization and log2 transformation by RMA | ||
| Polycystic Ovary Syndrome | GSE124226 | GPL570 [HG-U133_Plus_2] Affymetrix | Adipose | 4 PCOS women and 4 control subjects | Preprocessed: Normalization and log2 transformation by RMA | |
| Multiple Sclerosis | GSE21942 | GPL570 [HG-U133_Plus_2] Affymetrix | PBMCs | 12 MS patients and 15 controls | Preprocessed: Normalization GCRMA method | |
| Psoriasis | GSE78097 | GPL570 [HG-U133_Plus_2] Affymetrix | Skin | 6 normal skin tissues and 27 psoriatic skin lesions | Preprocessed: Normalization GCRMA method | |
| Cancer | Blood Cancer (Leukemia) | GSE51082 | GPL96 [HG-U133A] Affymetrix | Bone Marrow | 37 AML, 41, BCLL1, 22 CML, 10 MDS, 17 B-ALL, 12 T-ALL | Preprocessed: Normalization and log2 transformation by RMA |
| GSE9476 | GPL96 [HG-U133A] Affymetrix | Bone Marrow | 38 healthy donors | Preprocessed: Normalization and log2 transformation by RMA | ||
| Breast Cancer | GSE65194 | GPL570 [HG-U133_Plus_2] Affymetrix | Breast sample | 11 control breast sample, 98 breast cancer samples, 55 TNBC samples | Preprocessed: Normalization and log2 transformation by GCRMA method | |
| Cervical Cancer | GSE63514 | GPL570 [HG-U133_Plus_2] Affymetrix | Cervix | 24 normal and 28 cancer specimens | Preprocessed: Normalization and log2 transformation by GCRMA method | |
| Multiple Myeloma | GSE85837 | GPL10558 Illumina HumanHT-12 V4.0 | Bone Marrow | 9 control and 9 multiple myeloma patients with bone lesion | Preprocessed: Robust spline normalization and log2 transformation by lumi R package | |
| Lung Cancer | GSE136043 | GPL13497 Agilent-026652 | Lung | 5 lung cancer tissue and 5 lung non-tumor tissues | Preprocessed: Normalization by Agilent Feature Extraction Software | |
| Lung adenocarcinoma (Non-small cell lung cancer) | GSE118370 | GPL570 [HG-U133_Plus_2] Affymetrix | Lung | 6 invasive lung adenocarcinoma tissues and 6 normal lung tissues | Preprocessed: Normalization and log2 transformation by MAS5.0 algorithm | |
| Liver Cancer | GSE88839 | GPL570 [HG-U133_Plus_2] Affymetrix | Liver | 35 HCA liver tumours and 3 normal liver samples | Preprocessed: Normalization by RMA | |
| Pancreatic Ductal Adenocarcinoma | GSE101448 | GPL10558 Illumina HumanHT-12 V4.0 | Pancreas | 18 with pancreatic tumor and 13 non-tumor pancreatic tissue samples | Preprocessed: Normalization and log2 transformation by Illumina's BeadStudio Data Analysis Software | |
| Infectious | AIDS | GSE73968 | GPL6244 [HuGene-1_0-st] Affymetrix | T Cells | 9 healthy control and 6 HIV positive patients | Preprocessed: Normalization and log2 transformation by RMA |
| Tuberculosis | GSE139825 | GPL10558 Illumina HumanHT-12 V4.0 | Alveolar Macrophages | Alveolar Macrophages from 5 TB patients and 5 control subjects | Preprocessed: Normalization and log2 transformation by lumi R package | |
| Malaria | GSE119150 | GPL15207 [Prime View] Affymetrix | Blood | 6 falciparum malaria and 6 normal subjects | Preprocessed: Normalization and log2 transformation by RMA | |
| Acute | Acute Kidney Injury | GSE30718 | GPL570 [HG-U133_Plus_2] Affymetrix | Kidney | 28 transplants with AKI to 11 pristine protocol biopsies of stable transplants | Preprocessed: Normalization and log2 transformation by RMA |
| COVID-19 | GSE150316 | GPL18573 | Lung | 16 lung samples with COV2 positive and 5 control lung samples | Preprocessed: DEseq2 normalized |
2.2. Data normalization
The datasets used in this study were all quantile normalized, and log2 transformed in R [11]. Briefly, raw expression values were quantile normalized using the normalize Quantiles function of LIMMA package in R, irrespective of their normalization status to maintain uniformity [12]. Subsequently, the average expression value of all probes for each gene in all the disease and control samples was obtained using collapse Rows function of WGCNA R package [13]. The normalized values were subsequently log2-transformed, provided the dataset was not already log2-transformed.
2.3. Data analysis
Principal Component Analysis (PCA) is a dimensionality reduction technique that identifies patterns in data, and highlights their similarities and differences. Elucidation of the principal components is based on identifying the variables most strongly correlated with each component. We used ‘prcomp’ function in R base package for PCA to analyze the segregation of datasets based on linear correlation and variance in gene expression values of 10,296 genes for all subjects in each disease [14]. The ‘ggbiplot’ function of ggplot2 package in R was used for graphical representation of PCA results [15].
Literature mining was carried out to identify 78 genes, which includes those encoding receptors, proteases, and others that are implicated in the replication and pathogenesis of one or other human infecting coronaviruses including SARS-CoV-2. The fold change values in the expression of these genes were computed and used to generate a clustered heatmap using pheatmap R package [16]. In addition, another heatmap was prepared using the gene expression values of 182 differentially expressed genes (at fold change > 2 or < 0.5 and p < 0.05) in COVID-19 patients compared to healthy controls. We used student's t-test (p < 0.05 as the level of significance) to analyze the gene expression data from SARS-CoV-2 infected human lung tissue. Additionally, the p-adjusted values (FDR<0.05) were used for identifying the highly significant dysregulated pathways in different disease cases. The data has been plotted as mean ± standard error from mean, and each dot represents individual reading. The graphical representation of gene expression values was obtained using GraphPad Prism (version 8.0.0) software. The overall methodology of this study is shown in Fig. 1 (Created with BioRender.com).
Fig. 1.
Flow diagram of the study.
2.4. Co-expression analysis
Co-expression analysis describes the correlation pattern in gene expression across different samples and it is frequently used for identifying the clusters (or modules) of highly correlated genes. We have used the weighted gene correlation network analysis (WGCNA) technique using WGCNA R package for identifying the highly correlated modules [13]. For co-expression analyses of individual disease cases, we have used the quantile normalized log2 values of gene expression in disease samples. For co-expression analysis of multiple diseases, we have used log2(FC) values. To ensure a scale-free topology of the network, soft-threshold power (ranged between 6 and 10) was chosen as per the Power Estimate value provided by pickSoft Threshold function in WGCNA R package. The pathway analysis for the genes in the identified modules were performed using DAVID [17]. The networks were drawn using Cytoscape 3.8 [18].
STAR METHODS:
LEAD CONTACT: The relevant information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Srinivasan Ramachandran (ramu@igib.res.in).
MATERIALS AVAILABILITY: This study did not generate any new material.
DATA AND CODE AVAILABILITY: This study did not generate any unique dataset or code.
3. Results
3.1. Human genes implicated in the pathogenesis of COVID-19 are upregulated in leukemia, psoriasis, NAFLD, and type II diabetes cases
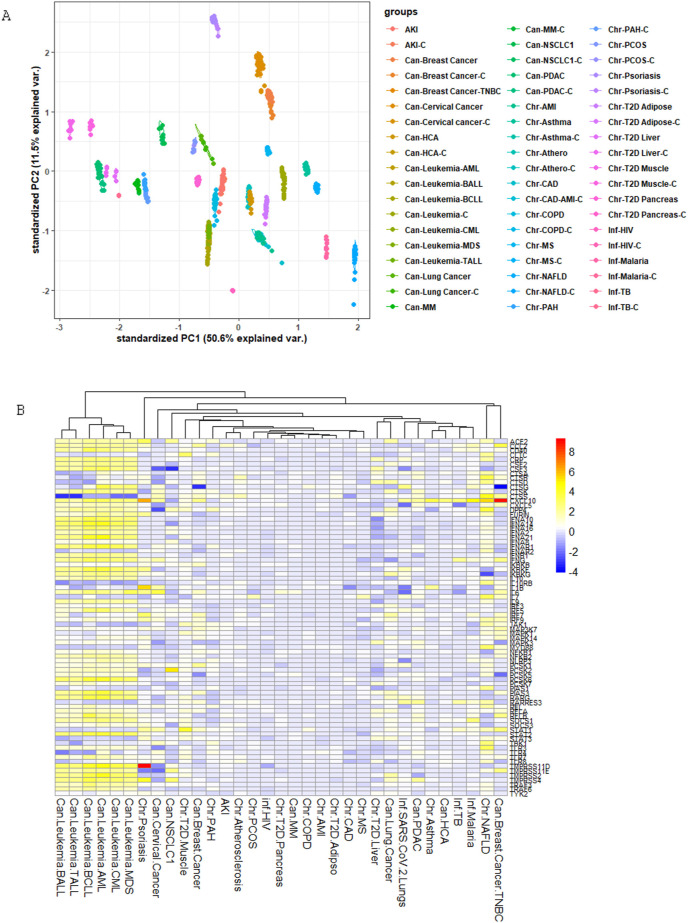
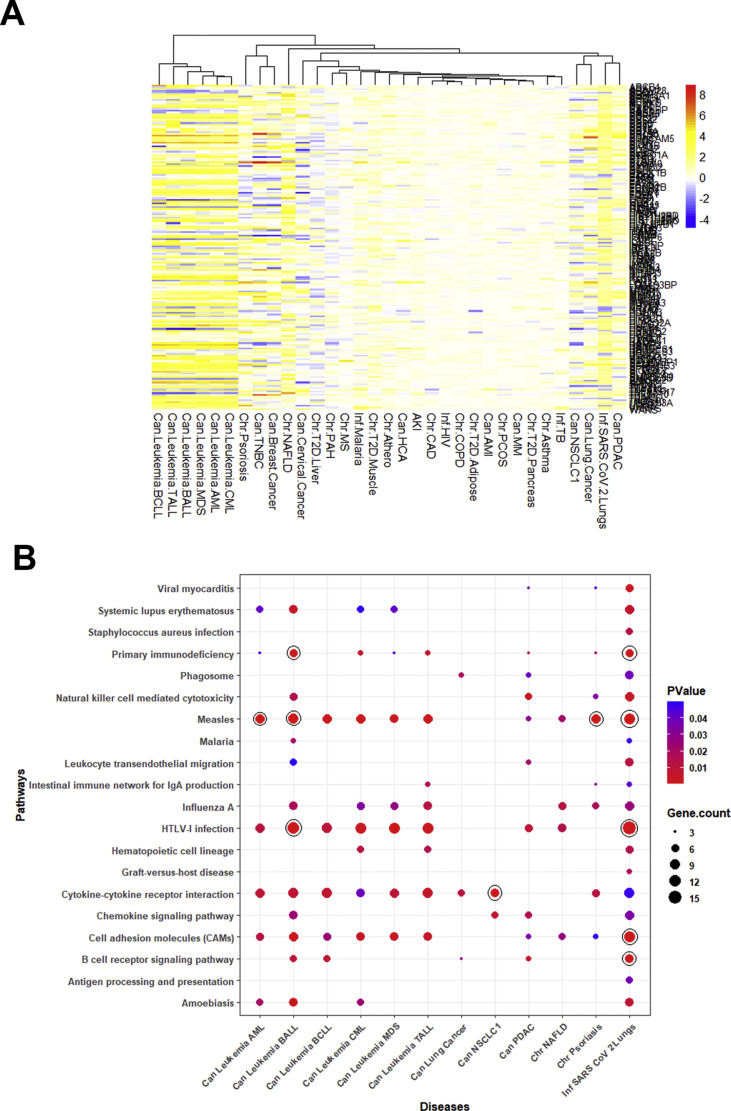
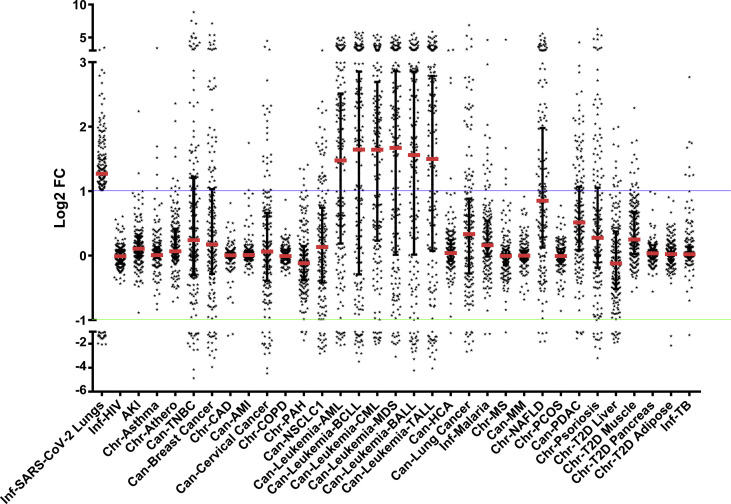
In leukemia and NAFLD condition, there were more than one dataset, namely one GSEid for samples and another GSEid for controls. In addition, in leukemia and breast cancer, the sample size of individual datasets was large. Therefore, we performed a principal component analysis (PCA) on the expression values of 10296 genes to explore the variability between datasets. We observed that the gene expression profile of datasets in a given disease correlated with each other, and each disease produced isolated galaxies of points closely spaced to each other (Fig. 2 A). We then investigated the differential gene expression in a set of chronic, acute, and infectious disease conditions to identify the gene expression patterns that may induce susceptibility to SARS-CoV-2 infection. The molecular details of SARS-CoV-2 infection and spread are still under active research, and some steps in the pathogenesis of SARS-CoV-2 have been reported as either identical or similar to that of other pathogenic human coronaviruses (HCoVs), namely, SARS-CoV and MERS-CoV. Since gene expression pattern is characteristically correlated with the pathogenesis of diseases [19,20], we examined the expression patterns of human genes, which are implicated in the replication and pathogenesis of SARS-CoV-2 or other HCoVs, in different disease conditions. To this end, we performed a literature mining exercise and identified 78 genes that were reported to have important implications in the entry and pathogenesis of HCoVs. These genes are enlisted in Supplementary Table 1. Some of these genes have been identified with key roles in promoting the pathogenesis of SARS-CoV-2, namely ACE2, FURIN, and TMPRSS2. The heatmap of log2(FC) fold changes in expression values of these 78 genes, in all 30 disease cases including COVID-19, is shown in Fig. 2B. It is evident that several of these genes are upregulated in patients with SARS-CoV-2 infection and patients of all the studied subtypes of leukemia (hereafter, collectively referred to as leukemia; 45–50 genes), NAFLD (32 genes), psoriasis (22 genes), breast cancer (17 genes), cervical cancer (12 genes), NSCLC1 (7 genes), and type II diabetes liver (7 genes). It is noteworthy that the differential gene expression pattern was particularly pronounced in leukemia (log2(FC) in the range 2–6) and NAFLD (log2(FC) in the range 2–5).
Fig. 2.
(A) Principal Component Analysis of 10,296 gene expression values in the datasets used in this study for 30 disease conditions and their respective controls. Individual datasets are represented by separate points. To categorize and differentially color different disease types, we have inserted the prefixes “Can,” “Chr,” and “Inf” to identify the clusters of various types of cancer, chronic diseases, and infectious diseases, respectively. (B) Clustered heatmap depicting fold change (log2(FC)) in the expression of 78 host genes in COVID-19 patients and in other 30 studied disease patients. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
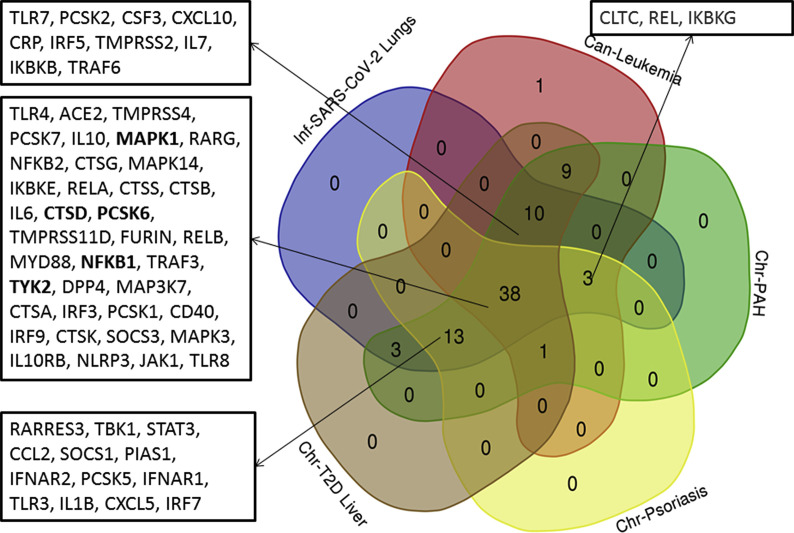
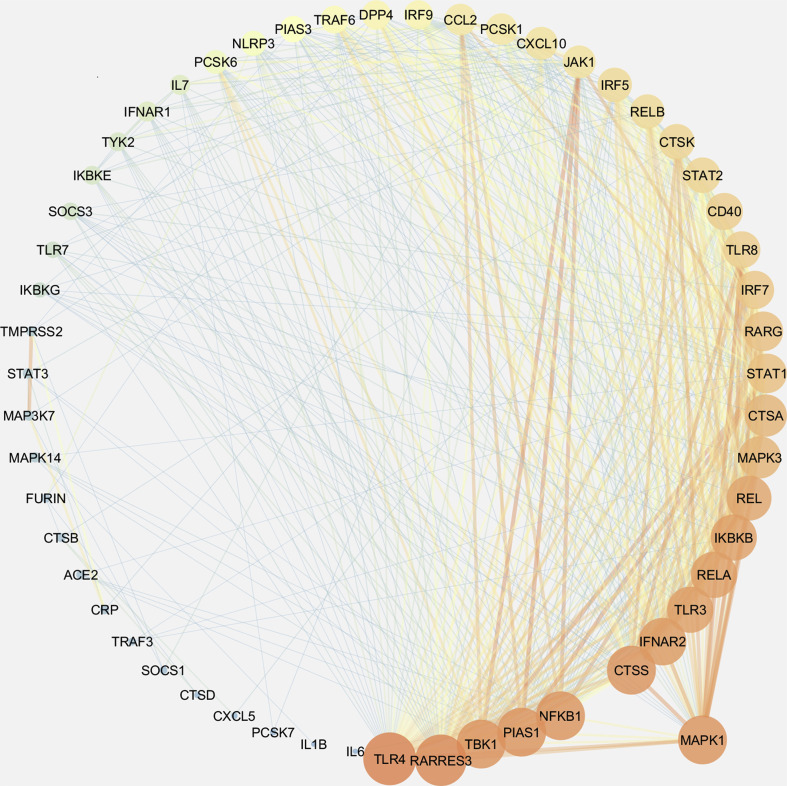
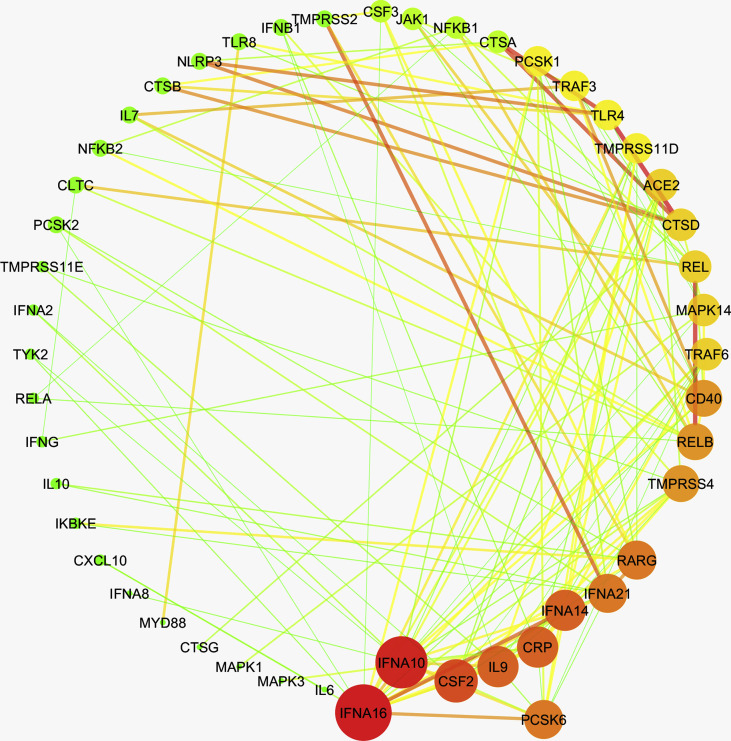
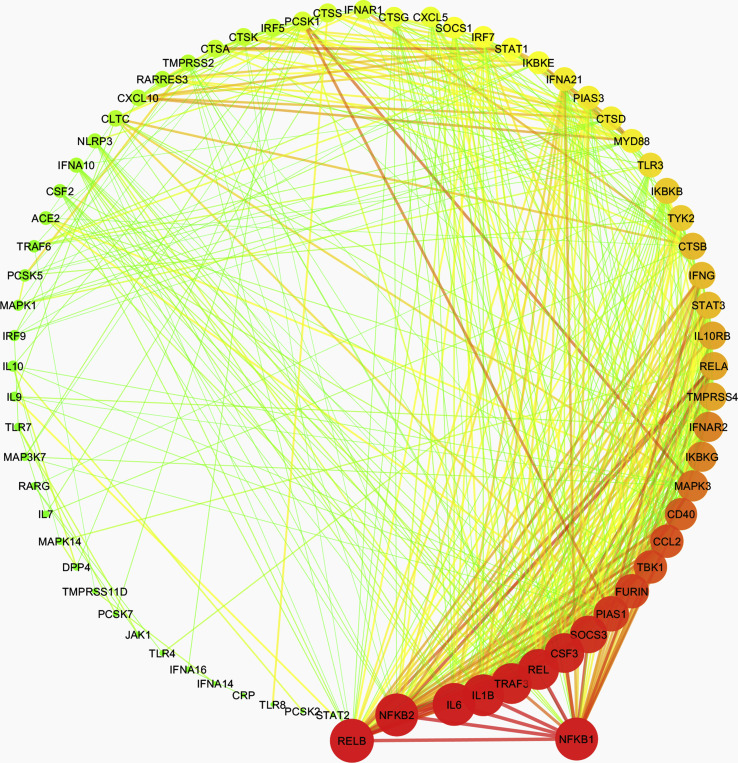
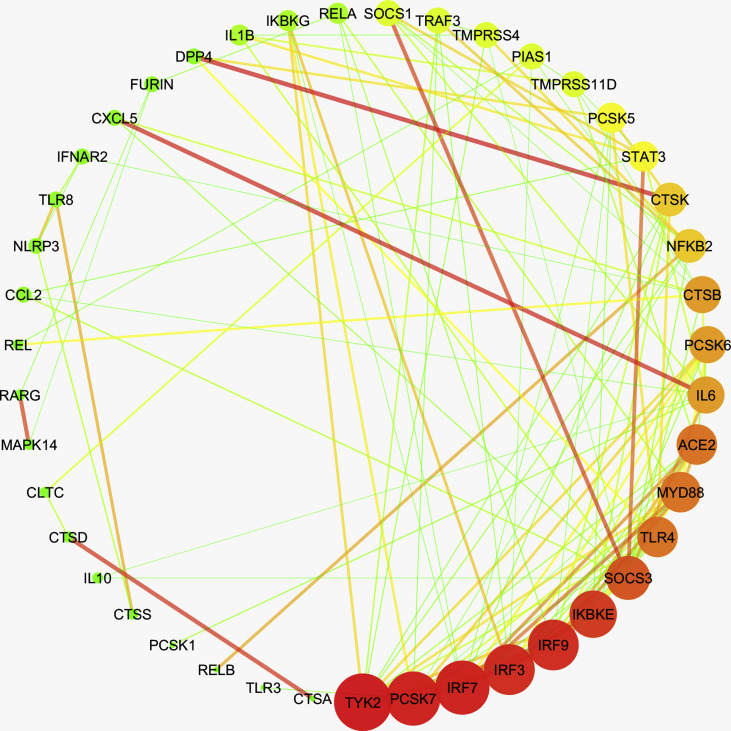
Further, to investigate any prospective covet genetic feature, we performed the co-expression analysis of 78 genes, which are implicated in the pathogenesis of coronaviruses, in these diseases. First, we examined the disease samples separately for each disease and identified the co-expressed genes in each case. Herein, we obtained a single module (turquoise) each for SARS-CoV-2, leukemia, psoriasis, PAH, and T2D Liver (Supplementary Figs. 1–5). 38 genes were observed to co-express in each of these disease types (Supplementary Fig. 6). We obtained different hub genes in each of these diseases, namely, MAPK1 in SARS-CoV-2, PCSK6 in leukemia, NFΚB1 in PAH, TYK2 in psoriasis, and CTSD in T2D liver (Supplementary Figs. 7–11). However, in the case of NAFLD, we did not obtain any module of co-expressed genes.
Thereafter, we performed another co-expression analysis using integrated log2(FC) values for SARS-CoV-2, leukemia (6 subtypes were considered separately), NAFLD, psoriasis, PAH, and T2D Liver (Supplementary Fig. 12). We obtained two modules with 62 and 16 genes, however, only one module was significant wherein PCSK6 was identified as the hub gene (Supplementary Fig. 13). Pathway analysis with these genes showed the prominent enrichment of Toll-like receptor signaling, JAK/STAT signaling, TNF signaling and NF-κB signaling pathways (Supplementary Table 2).
3.2. The expression of ACE2, FURIN, and TMPRSS2 is increased in leukemia, NAFLD, and psoriasis patients
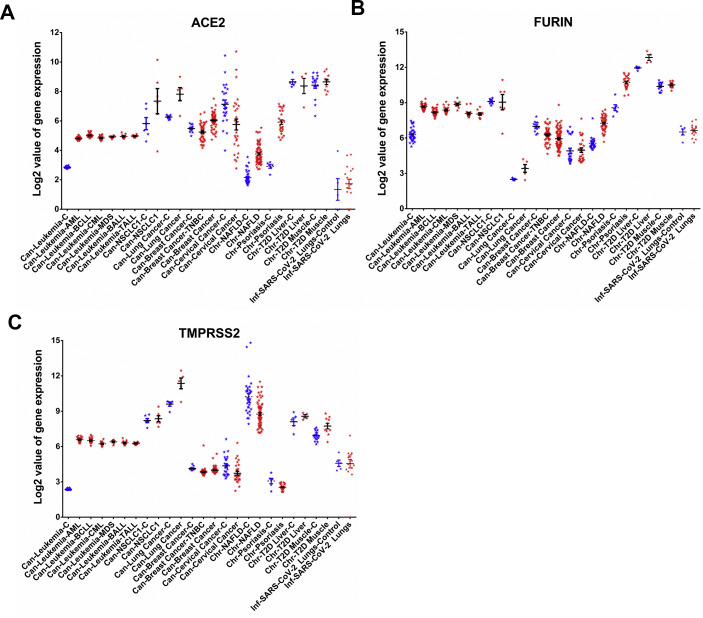
The earliest steps in establishing COVID-19 include cellular entry of SARS-CoV-2, which is critically dependent on the host's ACE2 receptor and serine proteases FURIN and TMPRSS2. ACE2 functions as the receptor for the entry of SARS-CoV-2 by binding to the viral spike protein, whereas, the FURIN and TMPRSS2 proteases are essential for processing the spike protein that facilitates viral entry into the cells [21,22]. Therefore, we investigated the expression patterns of ACE2, FURIN, and TMPRSS2 in the distinct disease cases (as identified in Fig. 2B), namely breast cancer, cervical cancer, leukemia, NAFLD, NSCLC1, psoriasis, and T2D. The expression of ACE2 was upregulated in leukemia, psoriasis, NAFLD, lung cancer, breast cancer, and cervical cancer patients (Fig. 3 A). The expression of FURIN was upregulated in leukemia, psoriasis, NAFLD, lung cancer, and in T2D liver whereas it was downregulated in breast cancer (Fig. 3B). We observed that TMPRSS2 was upregulated in leukemia, lung cancer, and T2D, but it was downregulated in psoriasis, NAFLD, lung cancer, breast cancer, and cervical cancer (Fig. 3C). It is worthwhile to mention that after the interaction of the viral spike protein with ACE2 receptor, the host's FURIN protease cleaves the spike protein at the interface of two subunits of the trimeric spike. Thus, the protease activity of FURIN is critical in promoting spike mediated entry of SARS-CoV-2 and it is also known to be crucial for protein processing in other infectious diseases and in cancer [23]. Similar to FURIN, the proteolytic cleavage of spike protein by TMPRSS2 is critical for its fusogenic activity.
Fig. 3.
Quantitative representation of gene expression values of (A) ACE2, (B) FURIN, and (C) TMPRSS2 in patients and in their respective controls from 14 selected diseases wherein these genes were differentially regulated. Each point represents the gene expression values of controls (Blue) and of patients (Red) in individual disease cases. Bars depict standard error of mean. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Disease-associated dysregulation of innate and adaptive immune response in patients with other diseases
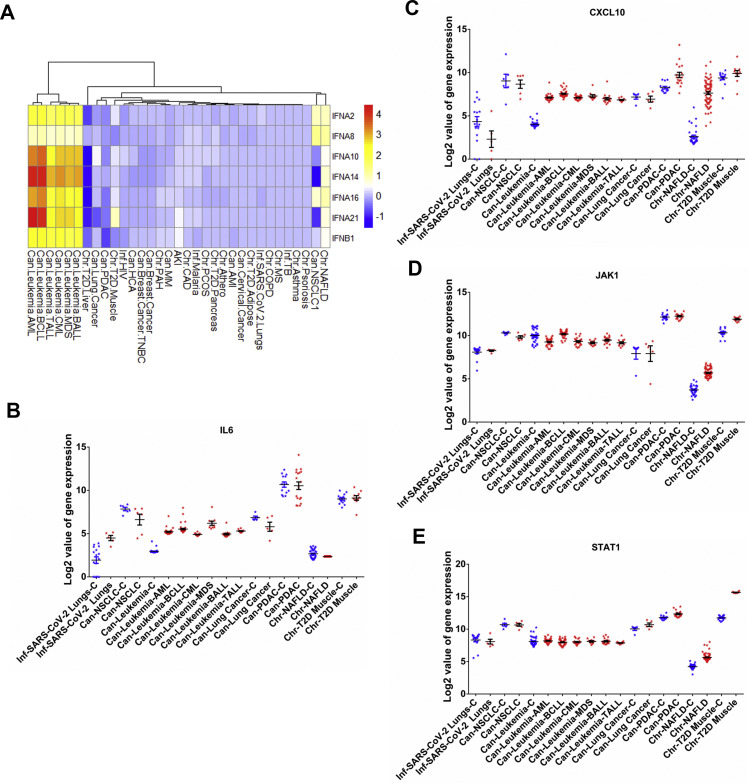
Following entry, the presence of viral RNA in cellular milieu evokes an immune response in the host. Apart from the receptors and proteases, the heatmap in Fig. 2B also shows the differential expression of genes, which are involved in the innate and acquired immune responses to SARS-CoV-2 invasion. The genes Interferon-alpha and Interferon-beta (IFNA2, IFNA8, IFNA10, IFNA14, IFNA16, IFNA21, and IFNB1) are the initial response elements of the innate immune signaling pathway. These responses activate several interferon-stimulated genes (ISGs) via JAK1/STAT1 pathway, which leads to early clearance of the viral load [24]. We prepared a heatmap of the log2(FC) of the expression of the interferons that were differentially expressed in any of the 30 diseases (p < 0.05, Fig. 4 A). We also examined the expression of genes encoding cytokines that underlie the anti-viral immune responses, namely IL6, CXCL10, JAK1, and STAT1 (Fig. 4B–E). The genes IFNA2, IFNA8, IFNA10, IFNA14, IFNA16, IFNA21, and IFNB1 were upregulated in leukemia (log2(FC) ranged from 1.003 to 4.63) and in NAFLD (log2(FC) ranged from 1.009 to 1.93), whereas they were downregulated in T2D liver (log2(FC) ranged from −1.09 to −1.61). The expression of JAK1 was slightly decreased (log2(FC) ranged from −0.63 to −0.94) in leukemia except in BCLL cases, whereas STAT1 was slightly decreased in TALL (log2(FC) −0.41) and was unchanged in other types of leukemia. Both JAK1 and STAT1 were increased in NAFLD (log2(FC) ranged from 1.52 to 1.96) and in T2D muscles (log2(FC) ranged from 1.52 to 3.88). The initial interferon-mediated response is followed by a specific cell-mediated adaptive immune response to clear viral invasion. To this end, the cytokines IL6 and CXCL10 are produced by helper T cells and macrophages that promote the migration of immune cells to the site of infection. They are also associated with the cytokine storm observed in COVID-19 associated mortalities [25]. We observed that the expression of IL6 and CXCL10 was upregulated in leukemia (log2(FC) ranged from 1.92 to 3.55). But the expression of IL6 was slightly decreased in NAFLD (log2(FC) −0.4). The expression of CXCL10 was increased in NAFLD (log2(FC) 5.23) and PDAC (log2(FC) 2.095).
Fig. 4.
(A) Clustered heatmap of log2(FC) fold change values in the expression of genes encoding interferons that are differentially expressed in COVID-19 and 30 other studied disease patients. Quantification of (B) IL6, (C) CXCL10, (D) JAK1, and (E) STAT1 expression in patients and in their respective controls from 12 selected diseases including COVID-19 wherein these genes were found to be differentially regulated. Each point represents fold changes from individual patient or control. Bars depict standard error of mean.
3.4. The patterns of differential gene expression are similar in SARS-CoV-2, leukemia, and NAFLD
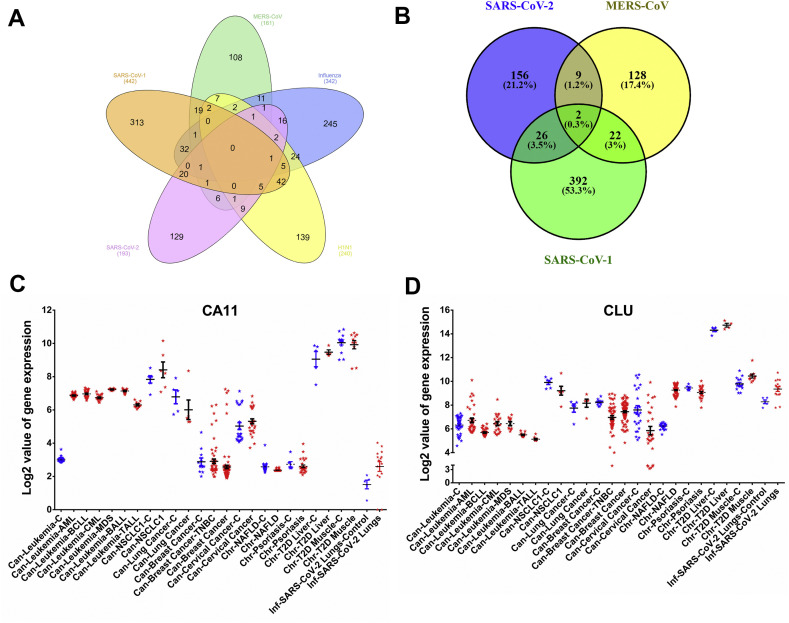
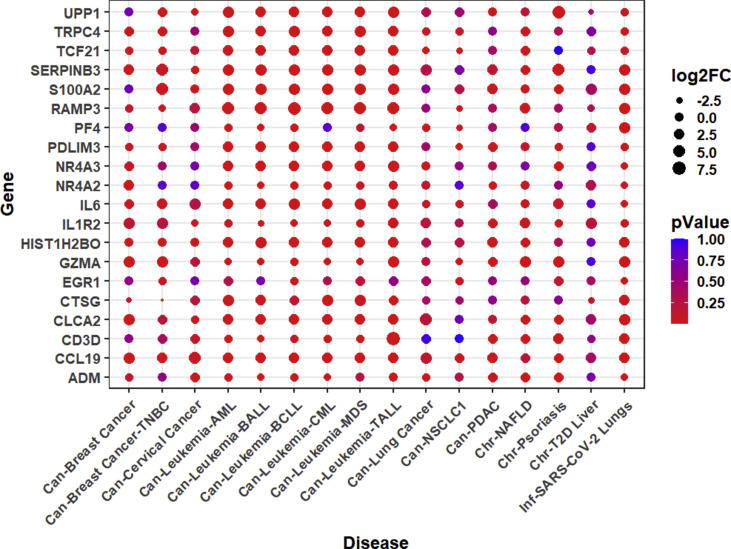
We analyzed the expression pattern of 193 differentially expressed genes from 16 SARS-CoV-2 infected patients. Out of these 193 differentially expressed genes, we found that the expression values for only 182 genes were available in the datasets of all disease types included in our study. Therefore, we generated a clustered heatmap (Fig. 5 A) and a scatter plot (Supplementary Fig. 14) depicting the expression of these 182 differentially expressed genes in 30 diseases and COVID-19 conditions. It is evident from the heatmap that the pattern of gene expression is similar in COVID-19 and PDAC, which are segregated together in the heatmap. Similarly, lung cancer and NSCLC also clustered together. Among the 161 genes upregulated in the lungs of COVID-19 patients (log2(FC) ranged from 1 to 3.45), 99–111 genes in leukemia (log2(FC) ranged from 1 to 5.86), 77 genes in NAFLD (log2(FC) ranged from 1 to 5.58), and 48 genes in psoriasis (log2(FC) ranged from 1 to 6.27) were upregulated. Pathways enrichment analysis of these 182 genes using DAVID [26,27] showed significant enrichment of the host's immune response to viral infection or infection-related immune pathways (Fig. 5B). Furthermore, we inferred the expression of 10 topmost significantly altered up- and down-regulated genes in COVID-19 patients and compared them to the disease cases showing similar pattern of gene expression, namely, T2D liver, NAFLD, psoriasis, leukemia, PDAC, lung cancer, NSCLC, TNBC, breast cancer, and cervical cancer (Supplementary Fig. 15). We observed that three genes, namely RAMP3 (Receptor Activity Modifying Protein 3), S100A2 (S100 Calcium Binding Protein A2), and CLCA2 (Chloride Channel Accessory 2) with a functional role in calcium signaling, were prominently upregulated in at least 7 studied disease types (including all subtypes of leukemia).
Fig. 5.
(A) Clustered heat map of log2(FC) fold change in the expression of 182 genes significantly altered in SARS-CoV-2 infected patients and in the 30 studied disease patients. (B) Bubble plot depicting the Pathways using 182 genes altered in COVID-19, leukemia, lung cancer, psoriasis, NAFLD, NSCLC, PDAC, and in T2D liver disease patients. Highly significant pathways in these diseases are highlighted with encircled bubbles according to p-adjusted values (FDR < 0.05).
3.5. Pathogenic HCoVs differentially regulate the expression of CARBONIC ANHYDRASE 11 and CLUSTERIN gene
We extended our investigation to identify the genes, whose expression may be commonly altered by the dreaded viruses of recent times. We analyzed the differential gene expression in patients with different viral infections, namely, SARS-CoV, SARS-CoV-2, MERS-CoV, H1N1, and other influenza viruses (H7N1, H5N1, H3N2, and H5N2). We observed that no gene was commonly altered in these viral infections (at a fold change > ±2, p < 0.05; Fig. 6 A). However, two genes, namely CARBONIC ANHYDRASE 11 (CA11) and CLUSTERIN (CLU) were commonly altered in the patients infected by pathogenic HCoVs, namely SARS-CoV, SARS-CoV-2, and MERS-CoV (at a fold change > ±2, p < 0.05; Fig. 6B). Based on the similarities in the patterns of differential gene expression in COVID-19 and other 30 diseases, we examined the expression of CA11 and CLU in breast cancer, cervical cancer, leukemia, NAFLD, NSCLC1, psoriasis, and T2D patients. The expression of CA11 was significantly upregulated in COVID-19 and leukemia (Fig. 6C), whereas CLU was upregulated in COVID-19 only (Fig. 6D).
Fig. 6.
Venn diagram depicting the number of differentially regulated genes in common to various viral infections, namely SARS-CoV, SARS-CoV-2, MERS-CoV, H1N1, and other influenza viruses (H7N1, H5N1, H3N2, and H5N2). (A) and to post-infection by pathogenic human infecting coronaviruses, namely SARS-CoV, SARS-CoV-2, and MERS-CoV (B). Quantification of expression of Carbonic anhydrase 11 (CA11) (C) and Clusterin (Clu) (D) in patients and their respective controls from 14 selected diseases including COVID-19 wherein these genes were found to be differentially regulated. Each point represents fold changes from individual patients or controls. Bars depict standard error of mean.
4. Discussion
COVID-19 associated comorbidities have been reported with several acute and chronic diseases, which lead to poor outcomes. For instance, diabetes, cardiovascular disease, renal and pulmonary diseases, are frequently observed comorbidities that increase the case fatality rate in acute respiratory diseases caused by SARS-CoV [28], MERS-CoV [29,30], and SARS-CoV-2. Our study revealed that the characteristic gene expression patterns in the disease cases, namely, leukemia, NAFLD, psoriasis, T2D, and PAH are highly similar to that of COVID-19. It is likely that the similarities in gene expression pattern offer a favorable environment to SARS-CoV-2 infection.
ACE2 is the functional receptor of three human infecting coronaviruses, namely NL-63, SARS-CoV, and SARS-CoV-2 [21,31]. Previous reports suggest that the expression of ACE2 is moderate in lung alveolar epithelium cells, high in enterocytes of the small intestine, and to a lesser extent in the vascular endothelial and smooth muscle in several organs including kidney, liver, bone marrow, skin, and brain [32]. Through ACE2, these organs may provide an easy port of entry for SARS-CoV-2, and the gene expression data suggests that more severe symptoms could develop upon SARS-CoV-2 infection particularly in the respiratory tract and the gut. The degree of ACE2 expression, in association with a 10–20 fold higher binding affinity of SARS-CoV-2 spike compared to SARS-CoV spike, could underlie the efficient cellular entry and higher infectivity of SARS-CoV-2 compared to SARS-CoV [33,34]. We observed that the basal expression of ACE2 was significantly high in many pathological conditions, namely leukemia, cancer of lungs, breasts, and cervix, NAFLD, psoriasis, and PAH. Hence, the increased number of available cellular receptors that facilitate viral entry can account for the increased susceptibility of these disease cases to SARS-CoV-2 infection.
Following the initial interaction of viral spike with ACE2 receptors, the pre-activation of viral spike by cleavage at polybasic S1/S2 site in the spike is mediated by proprotein convertase FURIN that enables a second cleavage by the cellular serine protease TMPRSS2. Both these proteolytic cleavages are important in facilitating viral entry. Inactivation of either FURIN [22] or TMPRSS2 [21] has been reported to inhibit cell-cell fusion and entry of SARS-CoV-2 in lung cells. Other cellular proteases such as TMPRSS4 [35] Cathepsin B and L [34] may have a cumulative effect on the FURIN-mediated promotion of SARS-CoV-2 entry into enterocytes or liver or lungs cells. Except in CLL, BALL, and TALL, wherein CATHEPSIN A, B, and D were downregulated, we observed that majority of the host proteases were highly upregulated in other subtypes of leukemia. Although cathepsins have an additive effect, they may not be indispensable for viral entry. Yet their increased expression along with that of FURIN and TMPRSS2 can promote the processing of viral spike and enhance cellular entry of SARS-CoV-2 [21]. Taken together these data suggest that the patients of leukemia are highly prone to SARS-CoV-2 infection. Similarly, the expression of FURIN was observed to be higher in NAFLD and psoriasis patients. Recently the abundance of either FURIN or TMPRSS2 was shown to be sufficient in promoting the cellular entry of SARS-CoV-2 [21]. Although the relevant information on psoriasis is missing in the literature, Dong et al. (2020) have recently observed NAFLD comorbidity in COVID-19 patients [5]. Therefore, we propose that the increased expression of ACE2 and FURIN could result in COVID-19 associated comorbidities. However, studies aimed at testing the redundancy of proteases are required to arrive at a definite conclusion.
Following the proteolytic cleavage of viral spike, the viral envelope fuses with host membrane and subsequently evokes the primary defense response of the host. This response is composed of the interferon-mediated innate immune response [36]. The production and binding of type I and type III interferons to their respective cellular receptors culminate into activating JAK1/STAT1 mediated transcription of several anti-viral interferon-stimulated genes [37]. It has been reported that the JAK1 deficient mice exhibit poor lymphoid development, and defective response to cytokines and interferons and die perinatally [38]. Similarly, mice with disrupted expression of STAT1 have compromised innate immunity [39] and are prone to viral infections [40]. Somatic mutations and dysregulation of JAK1 mediated signaling have been frequently observed in acute lymphoblastic leukemia [41,42]. Also, the deficiency of TYK2 (tyrosine kinase 2) in humans, which constitutes a key component of type I and type III interferon response, was shown to induce cytokine signaling defects and susceptibility to infection [43]. We observed that the expression of IFNA2, IFNA8, IFNA10, IFNA14, IFNA16, IFNA21 and IFNB1 were increased, whereas that of JAK1, STAT1, and TYK2 did not change significantly in leukemia patients. Thus, the increased interferon response in leukemia patients may involve components other than JAK1, STAT1, and TYK2. In contrast, in NAFLD patients the increased expression of JAK1 and STAT1 corresponds well with the increased expression of the interferons encoding genes, namely, IFNA2, IFNA8, IFNA10, IFNA14, IFNA16, IFNA21 and IFNB1. On the other hand, the expression of JAK1 and STAT1 decreased in T2D muscle and the expression of interferons decreased in T2D liver. However, p-STAT1 levels must be quantified to conclusively reveal the correlation between STAT1 expression in COVID-19 associated comorbidities. The higher levels of interferons may be one of the reasons for the previously observed reports describing relatively milder symptomatic COVID-19 in CLL patients [44]. However, SARS-CoV-2 may use several escape or immune-suppression strategies including the formation of a replication organelle and 2′-O-methylated capping of viral RNA to proliferate despite of increased basal interferon levels in leukemia and NAFLD patients [45]. Thereafter, the specific adaptive immune response comes into effect for curbing the viral invasion. An optimal secretion of cytokines and chemokines (such as IL6 and CXCL10) from immune cells is essential to adjust the host's immune response against foreign invaders. However, the excess release of cytokines, also known as cytokine storm, is associated with an increased severity of disease and poorer outcomes in SARS-CoV [46,47] and SARS-CoV-2 [48] infected patients. Earlier, the inhibition of NF-κB mediated production of IL6 was found to increase the survival in SARS-CoV infected mice [49] and IL6 blockade has been thought of as a mechanism to manage cytokine storm and save COVID-19 patients [50]. We observed a higher basal expression of CXCL10 in leukemia, NAFLD, and PDAC patients that may subsequently lead to cytokine storm upon SARS-CoV-2 infection. Recently Malard et al. (2020) have also reported that patients with hematologic malignancies are at higher risk of developing a severe form of COVID-19 [51]. Thus, we propose that the inhibitors of IL6 and CXCL10 could be examined for clinical interventions in leukemia and NAFLD patients who have been tested positive for SARS-CoV-2.
Furthermore, calcium signaling was found to be perturbed in COVID-19 and at least 6 other studied disease types including leukemia, NAFLD, and psoriasis that manifested in the form of altered expression of RAMP3, S100A2, and CLCA2 genes. RAMP3 is a co-activator that targets the calcium-sensing receptor to cell surface [52,53]. CLCA2 regulates the calcium-activated chloride channel currents and enhances the store-operated cellular entry of calcium [54]. S100A2 encodes a cytoplasmic calcium-binding protein and is known to be dysregulated in human cancers [55]. Together, these three genes modulate the cellular calcium levels in response to various stimuli and were distinctly upregulated in leukemia, NAFLD, and psoriasis (Supplementary Fig. 1). Recently, Sun et al. (2020) showed that calcium channel blockers inhibit the replication of SARS-CoV-2 in the cellular milieu and reduce the COVID-19 associated case-fatality rate [56]. Thus, cellular calcium levels may play a significant role in inducing susceptibility to SARS-CoV-2 infection. Furthermore, we found that the expression of CA11 and CLU genes were commonly altered in SARS-CoV, SARS-CoV-2, and MERS-CoV infected cases. These data indicate the uniqueness of the host gene expression patterns, thereby supporting the distinctive nature of these infections. Although CA11 was upregulated in leukemia, no trend was observed in the expression of CLU in the studied disease types. As of now, no direct correlation has been identified between the expression of either CA11 or CLU with the pathogenesis of SARS-CoV-2. However, several authors have recently identified that COVID-19 may lead to ketosis, ketoacidosis [57], and altered glucose metabolism [58]. Because CA11 plays an important role in hepatic gluconeogenesis [59], it could be interesting to investigate the potential relationship between SARS-CoV-2 infection, differential CA11 expression, and the onset of diabetes in COVID-19 patients.
Our study has few associated caveats. At first, we have selected the gene expression datasets from 30 diseases with strict criteria, namely, human samples with disease-specific tissues. The datasets were generated through expression profiling by array and the cases were devoid of any other treatments or afflictions. Given the reasonable number of samples used in these studies, we believe that our observations could be generally applicable. However, the limited sample size could impose limitations on confirming these observations in other samples including patients from other populations. This study concludes that the patients of leukemia are relatively more susceptible to SARS-CoV-2 infection followed by NAFLD, psoriasis, T2D, and PAH. It has been reported that STAT1 signaling promotes the proliferation of leukemia [60] and non-alcoholic steatohepatitis [61], and the inhibition of JAK/STAT signaling has shown protective activity in leukemia [60] and type II diabetes [62]. Complementarily, recent reports have observed downregulation of STAT1 [63] and upregulation of CXCL10 [64] post SARS-CoV-2 infection. These reports suggest a potential target avenue [65]. Furthermore, it has been reported that the expression of IL-6 is upregulated in NAFLD [66], type II diabetes [67], and COVID-19 [68] patients. Previously, the IL-6 overexpression following SARS-CoV-2 infection was reported to occur via NF-κB [46] and inhibition of NF-κB signaling was reported to increase the survival in SARS-CoV infected mice [49]. Therefore, the strategy of inhibition of inflammatory cascade appears important for curbing SARS-CoV-2 infection with a concomitant increase in survival rates and for the added benefit of management of associated comorbidities. Therefore, our study indicates that disease-specific inhibition of IL6, CXCL10, JAK1, and STAT1 either alone or in various combinations could benefit in curbing COVID-19 associated comorbidities. Our report could support the healthcare systems across the globe in devising better management practices for preventing the complications of COVID-19 associated comorbidities.
Author contribution
MKS, AM, and AC: Conceptualization, Data curation, Formal analysis, Investigation, methodology, validation, visualization, writing – original draft, review & editing. SJ: Investigation, methodology, validation, visualization, writing – original draft, review & editing. SR: Data curation, Funding acquisition, Project administration, supervision, writing – review & editing.
Funding statement
No disclosures to make.
Declaration of competing interest
Authors declare that they have no conflict of interest.
Acknowledgment
MKS and AC acknowledge CSIR for a research fellowship, AM acknowledges ICMR for a research fellowship. SR acknowledges financial support from Department of Biotechnology, India [Grant No. BT/PR16472/BID/7/629/2016].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104219.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
The clustered dendrogram for SARS-CoV-2. The nodes with similar height depict co-expression.
Supplementary Fig. 2.
The clustered dendrogram and heatmap for Leukemia. The nodes with similar height depict co-expression.
Supplementary Fig. 3.
The clustered dendrogram and heatmap for Pulmonary arterial hypertension (PAH). The nodes with similar height depict co-expression.
Supplementary Fig. 4.
The clustered dendrogram and heatmap for Psoriasis. The nodes with similar height depict co-expression.
Supplementary Fig. 5.
The clustered dendrogram and heatmap for Type 2 diabetes liver. The nodes with similar height in the dendrogram depict co-expression.
Supplementary Fig. 6.
Venn diagram representing co-expressed genes in SARS-CoV-2, leukemia, psoriasis, pulmonary arterial hypertension, T2D liver disease patients. Genes that were common (38 genes) in all of these diseases and in four other diseases are depicted in the boxes. Hub genes are highlighted in bold.
Supplementary Fig. 7.
The co-expression network in SARS-CoV-2. The edge weight values > 0.05 for each node were filtered to obtain a meaningful interaction network and was drawn in Cytoscape. Dark color (Red - high, Orange - medium, Blue - low) represents the degree of connection for each node, while color and width of the edges represent the interaction confidence (red and thick edge - high confidence; blue and narrow edge - low weight). The hub gene MAPK1 is highlighted by dragging.
Supplementary Fig. 8.
The co-expression network in leukemia. The edge weight values > 0.05 for each node were filtered to obtain a meaningful interaction network and was drawn in Cytoscape. Dark color (Red - high, Orange - medium, Green - low) represents the degree of connection for each node, while color and width of the edges represent the interaction confidence (red and thick edge - high confidence; green and narrow edge - low weight). The hub gene PCSK6 is highlighted by dragging.
Supplementary Fig. 9.
The co-expression network in pulmonary atrial hypertension. The edge weight values > 0.05 for each node were filtered to obtain a meaningful interaction network and was drawn in Cytoscape. Dark color (Red - high, Orange - medium, Green - low) represents the degree of connection for each node, while color and width of the edges represent the interaction confidence (red and thick edge - high confidence; green and narrow edge - low confidence). The hub gene NFKB1 is highlighted by dragging.
Supplementary Fig. 10.
The co-expression network in psoriasis. The edge weight values > 0.05 for each node were filtered to obtain a meaningful interaction network and was drawn in Cytoscape. Dark color (Red - high, Orange - medium, Green - low) represents the degree of each node, while color and width of the edges represent the interaction confidence (red and thick edge - high confidence; green and narrow edge - low confidence). The hub gene TYK2 is highlighted.
Supplementary Fig. 11.
The co-expression network in T2D liver. The edge weight values > 0.05 for each node were filtered to obtain a meaningful interaction network and was drawn in Cytoscape. Dark color (Red - high, Orange - medium, Blue - low) represents the degree of connection for each node, while color and width of the edges represent the interaction confidence (red and thick edge - high confidence; blue and narrow edge - low weight). The hub gene CTSD is highlighted by dragging.
Supplementary Fig. 12.
The clustered dendrogram and heatmap for 6 selected diseases (SARS-CoV-2, 6 studied subtypes of leukemia considered individually, NAFLD, psoriasis, PAH, and T2D Liver). The nodes with similar height depict co-expression.
Supplementary Fig. 13.
The co-expression network in SARS-CoV-2, leukemia, psoriasis, pulmonary arterial hypertension, T2D Liver disease. The edge weight values > 0.05 for each node were filtered to obtain a meaningful interaction network and was drawn in Cytoscape. Dark color (Red - high, Orange - medium, Green - low) represents the degree of connection for each node, while color and width of the edges represent the interaction confidence (red and thick edge - high confidence; green and narrow edge - low confidence). The hub gene PCSK6 is highlighted by dragging.
Supplementary Fig. 14.
Scatter plot of log2(FC) fold change values in the expression of 182 genes significantly altered post SARS-CoV-2 infection and in the 30 studied disease conditions. Red bar depicts median.
Supplementary Fig. 15.
Bubble plot of log2(FC) fold change of 10 topmost significantly altered up- and down-regulated genes in COVID-19 patients. Also shown are the expression of these genes in 30 other studied disease cases. Dark red bubbles represent significant fold changes, light pink bubbles are statistically insignificant changes.
References
- 1.Guan W.J., Liang W.H., Zhao Y., Liang H.R., Chen Z.S., Li Y.M., Liu X.Q., Chen R.C., Tang C.L., Wang T., Ou C.Q., Li L., Chen P.Y., Sang L., Wang W., Li J.F., Li C.C., Ou L.M., Cheng B., Xiong S., Ni Z.Y., Xiang J., Hu Y., Liu L., Shan H., Lei C.L., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Cheng L.L., Ye F., Li S.Y., Zheng J.P., Zhang N.F., Zhong N.S., He J.X., China Medical Treatment C. Expert Group for, Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang F., Shi S., Zhu J., Shi J., Dai K., Chen X. Clinical characteristics and outcomes of cancer patients with COVID-19. J. Med. Virol. 2020;92(10):2067–2073. doi: 10.1002/jmv.25972. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Wang D., Guo J., Yuan G., Yang Z., Gale R.P., You Y., Chen Z., Chen S., Wan C., Zhu X., Chang W., Sheng L., Cheng H., Zhang Y., Li Q., Qin J., Hubei Anti-Cancer A., Meng L., Jiang Q. COVID-19 in persons with chronic myeloid leukaemia. Leukemia. 2020 doi: 10.1038/s41375-020-0853-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji D., Qin E., Xu J., Zhang D., Cheng G., Wang Y., Lau G. Non-alcoholic fatty liver diseases in patients with COVID-19: a retrospective study. J. Hepatol. 2020;73(2):451–453. doi: 10.1016/j.jhep.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco J.L., Ambrosioni J., Garcia F., Martinez E., Soriano A., Mallolas J., Miro J.M., Investigators C.-i.H. COVID-19 in patients with HIV: clinical case series. Lancet HIV. 2020;7:e314–e316. doi: 10.1016/S2352-3018(20)30111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolan M.E., Hill D.P., Mukherjee G., McAndrews M.S., Chesler E.J., Blake J.A. Investigation of COVID-19 comorbidities reveals genes and pathways coincident with the SARS-CoV-2 viral disease. Sci. Rep. 2020;10:20848. doi: 10.1038/s41598-020-77632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh C.J., Hu P., Batt J., Santos C.C. Microarray meta-analysis and cross-platform normalization: integrative genomics for robust biomarker discovery. Microarrays (Basel) 2015;4:389–406. doi: 10.3390/microarrays4030389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamid J.S., Hu P., Roslin N.M., Ling V., Greenwood C.M., Beyene J. Data integration in genetics and genomics: methods and challenges. Hum. Genom. Proteonomics. 2009:2009. doi: 10.4061/2009/869093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Team R.C. 2013. R: A Language and Environment for Statistical Computing. Vienna, Austria. [Google Scholar]
- 12.Smyth G.K. Springer; 2005. Limma: Linear Models for Microarray Data, Bioinformatics and Computational Biology Solutions Using R and Bioconductor; pp. 397–420. [Google Scholar]
- 13.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ripley B., Venables B., Bates D.M., Hornik K., Gebhardt A., Firth D., Ripley M.B. Cran R; 2013. Package ‘mass’; p. 538. [Google Scholar]
- 15.Vu V.Q. ggbiplot: a ggplot2 based biplot. R package. 2011:342. [Google Scholar]
- 16.Kolde R., Kolde M.R. Package ‘pheatmap’. R Package. 2015;1:790. [Google Scholar]
- 17.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning W., Li C.J., Kaminski N., Feghali-Bostwick C.A., Alber S.M., Di Y.P., Otterbein S.L., Song R., Hayashi S., Zhou Z., Pinsky D.J., Watkins S.C., Pilewski J.M., Sciurba F.C., Peters D.G., Hogg J.C., Choi A.M. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14895–14900. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunology. 2019;8 doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Page C., Genin P., Baines M.G., Hiscott J. Interferon activation and innate immunity. Rev. Immunogenet. 2000;2:374–386. [PubMed] [Google Scholar]
- 25.Zhang N., Zhao Y.D., Wang X.M. CXCL10 an important chemokine associated with cytokine storm in COVID-19 infected patients. Eur. Rev. Med. Pharmacol. Sci. 2020;24:7497–7505. doi: 10.26355/eurrev_202007_21922. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau E.H., Hsiung C.A., Cowling B.J., Chen C.H., Ho L.M., Tsang T., Chang C.W., Donnelly C.A., Leung G.M. A comparative epidemiologic analysis of SARS in Hong Kong, Beijing and Taiwan. BMC Infect. Dis. 2010;10:50. doi: 10.1186/1471-2334-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulcsar K.A., Coleman C.M., Beck S.E., Frieman M.B. Comorbid diabetes results in immune dysregulation and enhanced disease severity following MERS-CoV infection. JCI Insight. 2019:4. doi: 10.1172/jci.insight.131774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y.M., Hsu C.Y., Lai C.C., Yen M.F., Wikramaratna P.S., Chen H.H., Wang T.H. Impact of comorbidity on fatality rate of patients with Middle East respiratory syndrome. Sci. Rep. 2017;7:11307. doi: 10.1038/s41598-017-10402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F., Eichler J., Drosten C., Pohlmann S. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M., Wang T., Zhou Y., Zhao Y., Zhang Y., Li J. Potential role of ACE2 in coronavirus disease 2019 (COVID-19) prevention and management. J Transl Int Med. 2020;8:9–19. doi: 10.2478/jtim-2020-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majoros A., Platanitis E., Kernbauer-Holzl E., Rosebrock F., Muller M., Decker T. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front. Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Igaz P., Toth S., Falus A. Biological and clinical significance of the JAK-STAT pathway; lessons from knockout mice. Inflamm. Res. 2001;50:435–441. doi: 10.1007/PL00000267. [DOI] [PubMed] [Google Scholar]
- 39.Durbin J.E., Hackenmiller R., Simon M.C., Levy D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 40.Jung S.R., Ashhurst T.M., West P.K., Viengkhou B., King N.J.C., Campbell I.L., Hofer M.J. Contribution of STAT1 to innate and adaptive immunity during type I interferon-mediated lethal virus infection. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flex E., Petrangeli V., Stella L., Chiaretti S., Hornakova T., Knoops L., Ariola C., Fodale V., Clappier E., Paoloni F., Martinelli S., Fragale A., Sanchez M., Tavolaro S., Messina M., Cazzaniga G., Camera A., Pizzolo G., Tornesello A., Vignetti M., Battistini A., Cave H., Gelb B.D., Renauld J.C., Biondi A., Constantinescu S.N., Foa R., Tartaglia M. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J. Exp. Med. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hornakova T., Staerk J., Royer Y., Flex E., Tartaglia M., Constantinescu S.N., Knoops L., Renauld J.C. Acute lymphoblastic leukemia-associated JAK1 mutants activate the Janus kinase/STAT pathway via interleukin-9 receptor alpha homodimers. J. Biol. Chem. 2009;284:6773–6781. doi: 10.1074/jbc.M807531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minegishi Y., Saito M., Morio T., Watanabe K., Agematsu K., Tsuchiya S., Takada H., Hara T., Kawamura N., Ariga T., Kaneko H., Kondo N., Tsuge I., Yachie A., Sakiyama Y., Iwata T., Bessho F., Ohishi T., Joh K., Imai K., Kogawa K., Shinohara M., Fujieda M., Wakiguchi H., Pasic S., Abinun M., Ochs H.D., Renner E.D., Jansson A., Belohradsky B.H., Metin A., Shimizu N., Mizutani S., Miyawaki T., Nonoyama S., Karasuyama H. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Baumann T., Delgado J., Montserrat E. 2020. CLL and COVID-19 at the Hospital Clinic of Barcelona: an Interim Report. Leukemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., Kong L., Fang X., Zheng H., Wu Z., She Y. Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res. 2007;128:1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castilletti C., Bordi L., Lalle E., Rozera G., Poccia F., Agrati C., Abbate I., Capobianchi M.R. Coordinate induction of IFN-alpha and -gamma by SARS-CoV also in the absence of virus replication. Virology. 2005;341:163–169. doi: 10.1016/j.virol.2005.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., Castano-Rodriguez C., Perlman S., Enjuanes L. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020;111:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malard F., Genthon A., Brissot E., van de Wyngaert Z., Marjanovic Z., Ikhlef S., Banet A., Lapusan S., Sestilli S., Corre E., Paviglianiti A., Adaeva R., Hammedi-Bouzina F.M., Labopin M., Legrand O., Dulery R., Mohty M. COVID-19 outcomes in patients with hematologic disease. Bone Marrow Transplant. 2020 doi: 10.1038/s41409-020-0931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouschet T., Martin S., Henley J.M. Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J. Cell Sci. 2005;118:4709–4720. doi: 10.1242/jcs.02598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D'Souza-Li L. The calcium-sensing receptor and related diseases. Arq. Bras. Endocrinol. Metabol. 2006;50:628–639. doi: 10.1590/s0004-27302006000400008. [DOI] [PubMed] [Google Scholar]
- 54.Sharma A., Ramena G., Yin Y., Premkumar L., Elble R.C. CLCA2 is a positive regulator of store-operated calcium entry and TMEM16A. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bresnick A.R., Weber D.J., Zimmer D.B. S100 proteins in cancer. Nat. Rev. Canc. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Sun Y., Zeng H.-L., Peng Y., Jiang X., Shang W.-J., Wu Y., Li S., Zhang Y.-L., Yang L. 2020. Calcium Channel Blocker Amlodipine Besylate Is Associated with Reduced Case Fatality Rate of COVID-19 Patients with Hypertension. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Wang X., Chen J., Zuo X., Zhang H., Deng A. COVID-19 infection may cause ketosis and ketoacidosis. Diabetes Obes Metab. 2020 doi: 10.1111/dom.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubino F., Amiel S.A., Zimmet P., Alberti G., Bornstein S., Eckel R.H., Mingrone G., Boehm B., Cooper M.E., Chai Z., Del Prato S., Ji L., Hopkins D., Herman W.H., Khunti K., Mbanya J.C., Renard E. New-onseT diabetes in Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMc2018688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ismail I.S. The role of carbonic anhydrase in hepatic glucose production. Curr. Diabetes Rev. 2018;14:108–112. doi: 10.2174/1573399812666161214122351. [DOI] [PubMed] [Google Scholar]
- 60.Habbel J., Arnold L., Chen Y., Mollmann M., Bruderek K., Brandau S., Duhrsen U., Hanoun M. Inflammation-driven activation of JAK/STAT signaling reversibly accelerates acute myeloid leukemia in vitro. Blood Adv. 2020;4:3000–3010. doi: 10.1182/bloodadvances.2019001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grohmann M., Wiede F., Dodd G.T., Gurzov E.N., Ooi G.J., Butt T., Rasmiena A.A., Kaur S., Gulati T., Goh P.K., Treloar A.E., Archer S., Brown W.A., Muller M., Watt M.J., Ohara O., McLean C.A., Tiganis T. Obesity drives STAT-1-dependent NASH and STAT-3-dependent HCC. Cell. 2018;175:1289–1306 e1220. doi: 10.1016/j.cell.2018.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Couto F.M., Minn A.H., Pise-Masison C.A., Radonovich M., Brady J.N., Hanson M., Fernandez L.A., Wang P., Kendziorski C., Shalev A. Exenatide blocks JAK1-STAT1 in pancreatic beta cells. Metabolism. 2007;56:915–918. doi: 10.1016/j.metabol.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K.M., Rathnasinghe R., Uccellini M., Gao S., Aydillo T., Mena I., Yin X., Martin-Sancho L., Krogan N.J., Chanda S.K., Schotsaert M., Wozniak R.W., Ren Y., Rosenberg B.R., Fontoura B.M.A., Garcia-Sastre A. SARS-CoV-2 Orf 6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blot M., Jacquier M., Aho Glele L.S., Beltramo G., Nguyen M., Bonniaud P., Prin S., Andreu P., Bouhemad B., Bour J.B., Binquet C., Piroth L., Pais de Barros J.P., Masson D., Quenot J.P., Charles P.E., Pneumochondrie study g. CXCL10 could drive longer duration of mechanical ventilation during COVID-19 ARDS. Crit. Care. 2020;24:632. doi: 10.1186/s13054-020-03328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang Y.S., Ko B.H., Ju J.C., Chang H.H., Huang S.H., Lin C.W. SARS unique domain (SUD) of severe acute respiratory syndrome coronavirus induces NLRP3 inflammasome-dependent CXCL10-mediated pulmonary inflammation. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21093179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das S.K., Balakrishnan V. Role of cytokines in the pathogenesis of non-alcoholic Fatty liver disease. Indian J. Clin. Biochem. 2011;26:202–209. doi: 10.1007/s12291-011-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kristiansen O.P., Mandrup-Poulsen T. Interleukin-6 and diabetes: the good, the bad, or the indifferent? Diabetes. 2005;54(Suppl 2):S114–S124. doi: 10.2337/diabetes.54.suppl_2.s114. [DOI] [PubMed] [Google Scholar]
- 68.Grifoni E., Valoriani A., Cei F., Lamanna R., Gelli A.M.G., Ciambotti B., Vannucchi V., Moroni F., Pelagatti L., Tarquini R., Landini G., Vanni S., Masotti L. Interleukin-6 as prognosticator in patients with COVID-19. J. Infect. 2020;81:452–482. doi: 10.1016/j.jinf.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.