Abstract
The surveillance and forecast of newly confirmed cases are important to mobilize medical resources and facilitate policymaking during a public health emergency. Digital surveillance using data available online has increasingly become a trend with the advancement of the Internet. In this study, we assessed the predictive value of multiple online medical behavioral data, including online medical consultation (OMC), online medical appointment (OMA), and online medical search (OMS) for the regional outbreak of coronavirus disease 2019 in Shenzhen, China during January 1, 2020 to March 5, 2020. Multivariate vector autoregression models were used for the prediction. The results identified a novel predictor, OMC, which can forecast the disease trend up to 2 days ahead of the official reports of confirmed cases from the local health department. OMS data had relatively weaker predictive power than OMC in our model, and OMA data failed to predict the confirmed cases. This study highlights the importance of OMC data and has implication in providing evidence-based guidelines for local authorities to evaluate risks and allocate resources during the pandemic.
Keywords: COVID-19, pandemic, online medical consultation, eHealth, telehealth
1. Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic has reemphasized the importance of the application of telemedicine worldwide because of the social distancing policies and shortage of medical resources. Hospital infections have been a wide concern for patients during the pandemic, as an investigation of 138 early confirmed cases found that nearly 41% of these cases might be related to hospital visits (Wang, Hu, Hu et al., 2020). Online healthcare services have become alternative choices to offline medical visits because of safety concerns during the pandemic and the perceived convenience of their usage. Online medical services allow people to access healthcare services remotely using digital information and telecommunication technologies (e.g., computers, mobile devices, interactive television) (Koch, 2006). The usage of online medical services during the pandemic has boosted because people are seeking for health information, syndrome diagnosis, and medical advices (Gong, Xu, Cai et al., 2020)
In addition to the benefits of serving patients with online medical services, the wide availability of online medical services has provided potentials for the digital monitoring of the epidemic. Public health professionals and epidemiologists have made great efforts to predict the trend of an epidemic using traditional laboratory, clinical, and epidemiological data. Nowadays, the popularity of using online medical services also enables scholars to use online medical behavioral data for epidemic counseling and surveillance (Kellermann, Isakov, Parker et al., 2010; Kjelsø, Galle, Bang et al., 2016). Online medical behavioral data refers to data recorded from multiple users’ behaviors with related to medical services in the online environment, such as medical information seeking (web query data), medical consultation and medical appointment. Massive amount of online medical behavioral data has been generated on a daily basis and exhibited great potential for epidemic surveillance (Chan, Sahai, Conrad et al., 2011; Althouse, Scarpino, Meyers et al., 2015). Comparatively, novel digital surveillance methods tend to be more cost-effective, energy saving, and efficient. However, the accuracy and predictive power of online medical service data are not yet clear. This study aimed to examine a variety of online medical behaviors and explore how medical service data predict the epidemic's trend. This study has implications for the surveillance of emerging diseases and public health policymaking.
2. Literature review
2.1. Digital surveillance
Digital surveillance or the so-called “infodemiology” and “infoveillance” are increasingly employed in detecting and identifying new and re-emerging infectious disease in public health practices (Eysenbach, 2006; 2011). Traditionally, national or local health authorities in the field of public health had relied on established systems to monitor known infectious diseases based on routine sentinel reports from doctors and laboratories (Velasco et al., 2014). In China, the China Information System for Disease Control and Prevention, the largest network-based infectious disease surveillance system reinforced after the severe acute respiratory syndrome (SARS) outbreak in 2003, helps detect and prevent communicable disease outbreak (Wang, Wang, Yang et al., 2013). However, the capacity of conventional forms (e.g., indicator-based reporting by medical institutions) to detect potential health threats is limited. In particular, delays in reporting and the lack of devices hinder the detection of unexpected disease occurrences (Velasco, Agheneza, Denecke et al., 2014). This case was true for the “pneumonia of unknown etiology” in the situation of COVID-19, which was not quickly detected in the early stage and thus took time to develop a tailored surveillance protocol to identify potential cases (Li, Guan, Wu et al., 2020).
During the COVID-19 pandemic, using digital methods for surveillance and guidance has become a trend. For epidemic management, many governments obtain and mine contact information, facial recognition, and location data to monitor and track people with COVID-19 and decide whether someone is allowed to access a public place or not (Calvo, Deterding, Ryan, 2020). In addition, for surveillance, digital data are important sources for early detection, continuously monitor disease levels, and assess disease-related sentiments relevant to disease control (Salathé, Freifeld, Mekaru et al., 2013). In coordination with the advanced computational approach, including machine learning method, digital data have shown great potentials to predict the spread of infectious diseases (Lenca & Effy, 2020). Studies have found that social media posts with related to COVID-19 and search queries can be used to predict the prevalence of disease (Li, Xu, Cuomo et al., 2020; Li, Chen, Chen et al., 2020). Specifically, the sick posts (contents related to users’ own or other people's symptoms and diagnoses) on social media can be a stronger factor for prediction (Shen, Chen, Luo et al., 2020).
Such trend of digital surveillance is prevalent, but it is not without pitfalls. Exploring Internet-based data and health information is becoming an important complement to traditional disease surveillance system as it can provide good opportunities to facilitate the nowcast and forecast of the trends of emerging infectious diseases (EIDs) (Aiello et al., 2020). The history of mining the Internet data for digital surveillance can be traced back to 1990s, when some health agencies employed some software to retrieve relevant information from news and chatroom discussions to monitor infectious diseases (Brownstein, Freifeld, & Madoff, 2009). The usage of web queries has ever since become a new direction for digital surveillance. One noted example of digital surveillance using search engine is Google Flu Trends, which uses historical influenza-like illness search terms to detect the flu epidemic to provide faster and lower-cost approaches than traditional official reports (Ginsberg, Mohebbi, Patel et al., 2009). On the basis of trends of Google queries, the Internet search-engine query data has been proven to be effective for the surveillance of other infectious diseases in other countries (Pelat, Turbelin, Bar-Hen et al., 2009; Verma, Kishore, Kumar et al., 2018). In China, the Baidu search index developed on the largest Chinese search engine similar to Google was used to predict the epidemics of dengue fever (Li, Liu, Zhu et al.,2017; Li, Liu, Feng et al., 2019) and seasonal influenza (Yuan, Nsoesie, Lv et al., 2013). Despite of their predicative power, applying these search engine data sources for digital surveillance also face multiple challenges. For example, the constantly changing feature of data (Salganik, 2019), the black boxes of algorithm (Lazer et al. 2014), and people's behavioral changes due to safety concerns during the pandemic (Cook, Conrad, Fowlkes et al., 2011) can affect the prediction accuracy of Google Flu Trends over time.
In addition to search engine data, health-relevant information shared on social media, such as Facebook posts and tweets, are used to monitor disease trends. For instance, some studies tried to anticipate the human trajectory and sustained spread of EIDs using geolocated Twitter activity data (Rocklöv, Tozan, Ramadona et al., 2019; Ramadona, Tozan, Lazuardi et al., 2019). Using other digital tools such as online educational tools, which allows parents to monitor the children's fever and influenza-like illness, can also forecast the outbreaks of influenza earlier than the traditional surveillance system (Hswen, Brownstein, Liu et al., 2017). In addition, employing aggregated activity patterns rather than focusing on a single predictor is promoted to further increase the accuracy of disease prediction (Broniatowski, Paul, Dredze et al., 2013). Researchers have built more accurate and robust models for flu predictions by combining multiple data sources, such as Google searches, Twitter posts, hospital visit records, and traditional participatory surveillance data (Santillana, Nguyen, Dredze et al., 2015; McGough, Brownstein, Hawkins et al., 2017).
Some early digital disease surveillance studies have used online medical search (OMS) data (Li, Chen, Chen et al., 2020; Qin, Sun, Wang et al., 2020) and social media posts (Li, Xu, Cuomo et al., 2020; Shen, Chen, Luo et al., 2020) to monitor and predict the outbreak of COVID-19 in China. These studies provide evidence-based practice by applying quantitative analysis to examine the correlation between real-time and lagged time series of web-based data with official reported COVID-19 case count. However, the association between diagnosed cases and search query or social media data is apparently indirect, and the mechanism remains unclear. Comparatively, the rapidly growing online medical services, such as online medical consultation (OMC) and appointments (OMA), tend to provide more directly correlated data for pandemic prediction. Moreover, existing studies have often relied on a single source of data for prediction (Husnayain, Fuad, & Su, 2020; Huang, Luo, Duan et al., 2020) and few studies have compared the predicative power of different sources (Santillana, Nguyen, Dredze et al., 2015) for disease surveillance to identify the most effective and efficient approach for disease prediction. This study thus aims to focusing on investigating the online medical behavioral data for prediction and comparing the predicative power of varied online medical behaviors for pandemic surveillance.
2.2. Online medical behaviors
The digital transformation of healthcare services has allowed citizens to access various online medical services, such as OMS, OMA, and OMC (Agarwal, Gao, DesRoches et al., 2010). These online medical services have become increasingly popular partly because of their ease of access, convenience, immediacy, and interactivity and partly because of the high medical burden and patient demands (Wang, Shi, & Kong, 2020). Such online medical services satisfy public needs from different perspectives.
OMS is a behavior driven by and responds to patients’ information need. Patients increasingly turn to the Internet for medical information to self-check symptoms, get prepared for an offline medical encounter, or make decisions for medical treatment (Hu, Bell, Kravitz et al., 2012; Wong & Cheung, 2019). The proportion of users who have used the Internet as health information source accounts for 87.44% in Hong Kong (Wong & Cheung, 2019), 71% in Germany (Wang et al., 2020), 62.9% in Kuwait, and 35.7% in Ghana (Nangsangna & Da-Costa Vroom, 2019). The usage of OMS has remarkably increased during the outbreak of EIDs. Studies found that searches for “coronavirus” increased by about 36% (95% confidence interval [CI]: 27%–44%) after the first case announcement in a state in the US (Bento et al., 2020).
OMA is an instrumental service for patients to make an appointment with physicians via Internet-based technology. Chinese patients with high medical burden often suffer from difficulties to find appropriate physicians and schedule an appointment. OMA was developed based on the health information system and can ensure the efficiency of medical care delivery. The usage of OMA means that patients have relatively specific and severe symptoms and have decided to pay a visit to offline doctors. OMA data have been found to be an important factor for disease surveillance; for example, the number of patients who request appointment with influenza surges indicates a possibility of flu outbreak (Santillana et al., 2015).
OMC often refers to the dyadic dialogue between patient and physician. OMC allows patients to ask for medical advices from physicians through text, image, voice calls, or video calls. OMC moves the offline doctor visit process to the online scenario and thus remarkably reduces the patients’ time and effort in commuting and waiting to be treated. The number of online medical application users has exceeded 190 million in China as of 2019. More than 310 million consultations and inquiries have been conducted online (Kantar Consulting, 2019). OMC entails users to transcend physical barriers in time and space in seeking medical help (Lee & Zuercher, 2017). OMC also helps document the consultation process, and patients can receive a clear explanation of the symptoms and instructions from physicians (Lee & Zuercher, 2017). OMC is friendly to patients with stigmatized diseases, such as HIV and mental disorders (Dunn, 2012). The quick response received from OMC can comfort patients from being overly worried, especially in a resource-constrained environment (Wagg, Callanan, & Hassett, 2018).
These online medical services indicate the active role of patients in managing health issues and are especially preferred during the COVID-19 pandemic. The increasing usage of online medical services acts as one of the non-pharmaceutical interventions of COVID-19, which is conducive to the early identification and isolation of cases. Meanwhile, the digital footprints of online medical behavioral data generated by individuals raise the possibility of monitoring and tracing EIDs in the future. However, seldom research has examined the relationship between OMC and the transmission of EIDs largely because of the relatively low penetration rate of OMC and the lack of relevant and accessible data. The social distancing policies during the pandemic has made OMC more widely and actively adopted by users. During the epidemic, people who had used the OMC services of hospitals under the supervision of the National Health Commission increased 17 times compared with the same period last year. The volume of OMC on some third-party Internet service platforms has increased by more than 20 times during the same period (Wang, 2020). Thus, this study aimed to explore the possibility of applying the data of OMS, OMA, and OMC for the surveillance and monitoring of EIDs using vector autoregression (VAR) analysis method to model the identification of the newly confirmed cases of COVID-19.
OMS, OMA, and OMC may predict the spread of EIDs differently, as they are consumed differently by users depending on the perceived severity of illness and the personalization of medical service. The association between OMC and the spread of EIDs is likely to be stronger compared with OMS and OMA during the COVID-19 pandemic. First, OMS is a general behavior for information need. Seeking behaviors may be motivated by curiosity or fear and anxiety. The results and health information found through OMS are not personalized to solve patient-specific symptoms. Therefore, its association with pandemic prevalence can be largely contaminated by the “noise” of searching behaviors. Second, OMA is a pre-process for offline medical visit. However, OMA is restricted by the limited provision of medical services and the patients’ worries of being cross-infected through offline medical visits during the pandemic. Third, OMC is a more personalized and relevant behavior for medical advice, because people with symptoms are likely turn to OMC to search for help and receive personalized medical advices. Thus, our study focused on the predicting capability of these online medical behavior signals in the COVID-19 outbreak. In particular, we raised some research questions as follows:
Do online medical behaviors, including OMC, OMA and OMS, forecast the number of the newly confirmed COVID-19 cases? If yes, how does their predictive power differ from each other?
3. Methods
3.1. Research context
COVID-19 is an EID caused by the most recently discovered coronavirus, SARS-CoV-2. This disease has spread rapidly within China and all over the world since its outbreak in Wuhan, China in December 2019. A total of 823,626 confirmed cases and 40,598 deaths worldwide and 82,631 infected cases in China have been reported as of April 1, 2020 (WHO, 2020).
This study attempted to locate the context at a city level to control the data volume and ensure the data accuracy, because medical services are usually localized, and the data is aggregated at the city level. OMS has been a widely adopted behavior among Chinese, and digital tools such as Baidu index can provide the number of city-level queries. However, the adoption and data availability of OMA and OMC behaviors may vary from city to city. This study chose Shenzhen City as the research background. This city is a metropolis located in south southeastern China (near Hong Kong), with a population of more than 20 million. Shenzhen is a young Chinese city, known for reform practices in many domains. Shenzhen government is considered one of the best local governments in disclosing information on COVID-19 (Eichberger, 2020), as the city Health Committee released the newly confirmed cases of COVID-19 as a daily routine in the official websites and provided the detailed itineraries of the confirmed cases to keep the citizens informed.
Shenzhen is also one of the leading cities that actively adopt advanced technologies to promote medical services (Cheng, & Lu, 2018). Its wide adoption of online medical behaviors is one of the reasons we selected Shenzhen for this research. As of 2019, Shenzhen city has 114,882 health professionals from 4,513 medical institutions, including public hospitals, private clinics and community healthcare service (Shenzhen Municipal Health Commission, 2020). With the trend of digitalization of medical services, governmental departments, hospitals, and third parties in Shenzhen are all motivated to develop Internet hospitals to extend medical services. Under the supervision of provincial government, and many hospitals have been making trials to practice Internet-based medical services in Shenzhen.
Third-party companies have also entered the market to connect health professionals and patients. In particular, Health 160 (https://www.91160.com/), a pioneering third-party company as online medical service provider, has comprehensively provided OMA and OMC services for patients. More than 500,000 physicians from more than 7,000 hospitals in China can make online medical appointments through this platform (Health 160, 2020). In addition, although its services scope covers the whole country, Health 160 was established and based in Shenzhen, providing better localized and whole-process service for the city. In China, the era of traditional queuing methods for medical appointments has passed. Now most hospitals in Shenzhen requires their patients to make medical appointments through the online medical registration system.
In partnership with the municipal government, the Health 160 website and App serve as citizens’ "entry platform", which can provide OMA to 36,971 medical professionals from more than 1,120 Shenzhen medical institutions (Data provided by Health 160 Data Statistics Center). Invited by the Health 160 platform, many health professionals have also joined the platform to provide patients with OMC through online text exchange, telephone or video calls. During the pandemic, OMC cases in Shenzhen exceeded 2,300 people on a typical day (Data provided by Health 160 Data Statistics Center). These practices provide data accessibility and usability for current analysis, making Shenzhen an ideal place for in-depth mining of online medical behaviors to predict the number of confirmed COVID-19 cases.
3.2. Data collection
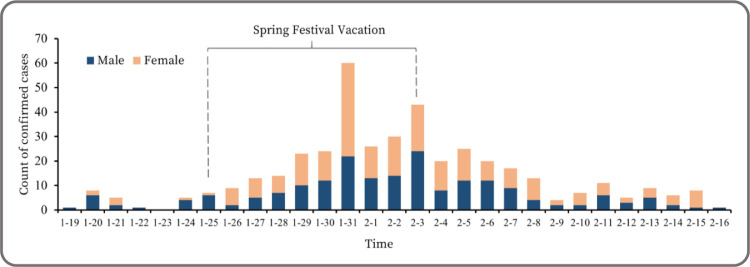
Data on the COVID-19 confirmed cases used in this study were solicited from the Shenzhen Municipal Government Data Opening Platform (https://opendata.sz.gov.cn), which has collated confirmed COVID-19 cases from January 19, 2020 when the first case appeared in Shenzhen. The number of confirmed cases peaked at the end of January and then gradually declined and reached zero for the first time in mid-February (Fig. 1 ). According briefings reported by Shenzhen Municipal Health Commission (http://wjw.sz.gov.cn/), the earliest onset time of the clinical case could date back to January 1, 2020, which was used as the starting point for prediction. The final daily new confirmed case (NCC) data was 416 within a 65-day time window from January 1, 2020 to March 5, 2020 (Fig. 3). An imported case from overseas was excluded during this study period.
Fig. 1.
Daily counts of new confirmed COVID-19 cases in Shenzhen, China (2020.1.19–2020.2.16).
Fig. 3.
Time plots of NCC, OMC, OMA, and OMS (2020.1.1–2020.3.5)
We obtained the OMC and OMA data from the abovementioned Health 160 platform. This study was approved by the researchers’ university ethical review committee (No: 2020028). Researchers’ institution has also signed data confidentiality agreement with the Health 160 company, and users on the Health 160 company have signed the informed consent about using data for scientific research. Statisticians only accessed the data in an aggregated format. No personal information was identifiable and visible during the data analysis.
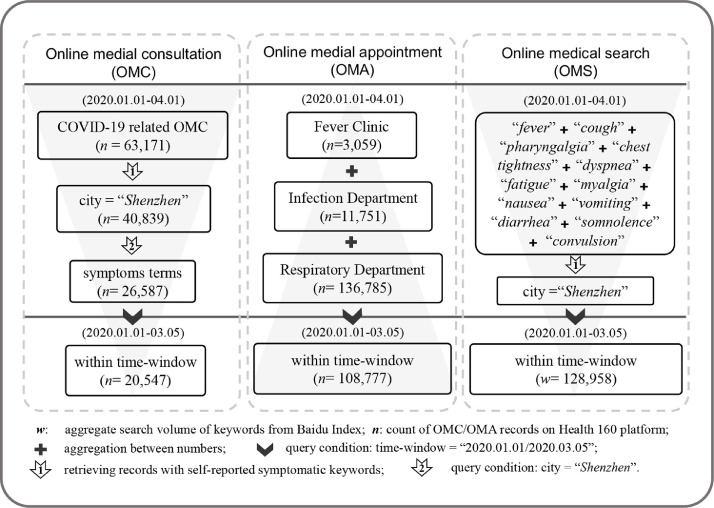
Relevant patient consulting records were retrieved from the Health 160’s database on May 31, 2020 using a number of keywords related to COVID-19 to solicit the OMC data. These keywords included common terms, such as diseases (e.g., “pneumonia,” “coronavirus”), symptoms (e.g., “fever,” “cough”), exposure history (e.g., “Wuhan,” “droplets”), and diagnosis measures (e.g., “CT,” “NAT”). The search returned over 63,171 records from January 1, 2020 to April 1, 2020 (Fig. 2).
Fig. 2.
Data collection process for the three types of online medical behaviors
The COVID-19 related symptoms keywords set derived from early retrospective studies (e.g.,Wang et al., 2020) and Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) released by National Health Commission and State Administration of Traditional Chinese Medicine (http://www.gov.cn/zhengce/zhengceku/2020-03/04/content_5486705.htm). Twelve main manifestations terms and their synonyms (26 keywords in total, see Table A in Appendix), such as “发热” (“fever” in Chinese) and “发烧”, “肌痛” (i.e., “myalgia”) and “肌肉酸痛”, were selected and used to retrieve from those consulting records above. Only 20,547 records, which contain the self-reported symptoms terms of patients whose location were Shenzhen, remained after excluding the doctors’ replies and other cities’ records (Fig. 2).
Patients’ OMA to the fever clinic (3,059, 2.02%), infection department (11,751, 7.75%), and respiratory department (136,785, 90.23%) of local hospitals in Shenzhen were also collected through the same platform to obtain the OMA data from January 1, 2020 to April 1, 2020. A total of 108,777 appointment records within the 65-day observation period (2020.01.01-03.05) were included for analysis. Finally, we computed the average of daily counts of all three kinds of appointments closely related to COVID-19 from different hospitals in the city. The processes of data collection and filtering are shown in Fig. 2 .
In addition, we collected OMS data from a Chinese counterpart of Google Trends, Baidu Index (http://index.baidu.com), which is a keyword research tool based on the Baidu search engine at www.baidu.com. Baidu Index would return search volume data of a certain keyword or phrase at different time frames and regions at the city level. In the present study, the original time window was also specified from January 1, 2020 to March 5, 2020, and the region was constrained to Shenzhen. Compared with OMC data, 12 core keywords for each symptom were submitted to Baidu Index and retrieved their search trends (see Table A in Appendix). As Baidu Index provided absolute search volume data instead of relative volume data reported by Google Trends (Vaughan & Chen, 2015), all 12 search results were straightforwardly aggregated to the daily level by taking the arithmetic mean, which was used to representt the index of OMS.
3.3. VAR model
In present study, we applied the VAR model for multivariate time series analysis to capture their dynamic interdependence by taking each variable as the linear function of past lags of itself and the past lags of the other explanatory variables (Hamilton, 1994; Lütkepohl, 2006). Recent studies used VAR models to predict the trend of infection diseases such as sexually transmitted diseases (Huang, Luo, Duan et al., 2020), Dengue (Ramadona, Lazuardi, Hii et al., 2016) and the ongoing COVID-19 (Khan, Saeed, Ali et al., 2020; Khan, Saeed, Ali et al., 2020; Fantazzini, 2020). A basic VAR model contains a set of n endogenous variables yt = (y 1 t, y 2 t, …, y n t). The p-lag vector autoregressive process (i.e., VAR(p)) is denoted as:
where Ai are (n × n) coefficient matrices for i = 1, …, p, and εt is an (n × 1) unobservable white noise vector process with E(εt) = 0 and time invariant covariance matrix. This model also allows us to estimate coefficient of exogenous variables.
Time plot and unit root test were applied before modeling to illustrate the characteristics of the time series and test whether they were stationary for model construction. Fig. 3 shows the time plots of NCC, OMC, OMA, and OMS. In the case of OMC, COVID-19 symptom-related consultations increased from about 100 to over 500 per day between late January and mid-February, when the majority of confirmed cases were diagnosed. OMS followed a similar trend with most Baidu Index values peaked on January 25, although its peak arrived 4 days earlier than that of OMC on January 29. OMA remained at a lower level and presented an opposite trend compared with OMC, and its daily count reached the minimum on January 24. Notably, a dummy variable was included in the VAR model as an exogenous variable, in which 1 represented vacation and 0 represented workday to reduce the confounding influence of the Spring Festival vacation (SFV) when human activities are different from their routine in China (Figs. 1 and 3).
Augmented Dickey Fuller (ADF) tests should be applied to each series to avoid the problem of spurious regressions, and the null hypothesis is that a unit root is present in the time series (Dickey & Fuller, 1981). In our study, the new COVID-19 cases and three types of behavioral data were included simultaneously in the VAR model after calculating their difference scores, that is, yt = (∆NCCt, ∆OMCt, ∆OMAt, ∆OMSt). The basic statistics and stationarity test results for the multiple time series of NCC, OMC, OMA, OMS and their differences are presented in Table 1 . The results show that all variables reject the null hypothesis of non-stationary in their first difference at 1% significance level and they are all suitable to perform the VAR model.
Table 1.
Basic statistics of variables and unit root test results
| total | ADF | ADF 1st difference | Critical values |
||||
|---|---|---|---|---|---|---|---|
| 1% | 5% | 10% | |||||
| NCC | 416 | 6.42±11.41 | -1.67 | -8.32 | -4.04 | -3.45 | -3.15 |
| OMC | 20547 | 316.11±170.98 | -1.08 | -7.74 | -4.04 | -3.45 | -3.15 |
| OMA | 108777 | 557.83 ± 372.93 | -0.90 | -5.87 | -4.04 | -3.45 | -3.15 |
| OMS | 128958 | 165.33 ± 23.19 | -2.09 | -5.66 | -4.04 | -3.45 | -3.15 |
The VAR approach also provides a set of methods for evaluating whether the relationship being tested actually exists. Granger causality test was used to determine the direction of causality between two series. The tools used to interpret the VAR model included impulse response functions (IRFs), which described the impact of shocks on the model's variable, and variance decompositions, which assessed the contribution of various shocks to the variance of prediction error. We used the package “vars” of R to conduct these analyses, as well as the model's lag selection, estimation, and diagnostic testing.
4. Results
4.1. Model identification and validation
Determining the model order is necessary for model identification, since the lag order of a VAR(p) process is usually unknown. Applying information criteria is useful for selecting the adequate lag order p and ensuring high forecast precision. The Akaike information criterion (AIC), Hannan–Quinn criterion (HQ), Schwarz criterion (SC), and final prediction error criterion (FPE) were used for model order selection. Table 2 indicates the estimation of the four criteria for the largest length of order from 1 to 6. AIC, HQ, SC, and PFE indicated that the optimal lag length appropriate for modeling was p = 1 and p = 2.
Table 2.
Selection of optimal lag order of VAR model
| Lag | AIC | HQ | SC | FPE |
|---|---|---|---|---|
| lag-1 | 25.90 | 26.23 | 26.75* | 1.77E+11 |
| lag-2 | 25.49* | 26.05* | 26.91 | 1.19E+11* |
| lag-3 | 25.61 | 26.38 | 27.60 | 1.38E+11 |
| lag-4 | 25.55 | 26.55 | 28.11 | 1.36E+11 |
| lag-5 | 25.57 | 26.79 | 28.70 | 1.50E+11 |
| lag-6 | 25.78 | 27.22 | 29.48 | 2.07E+11 |
Optimal lag order. +P<0.10, * P<0.05, ** P<0.01, *** P<0.001.
We exploratively chose the lag lengths p = 1 and p = 2 to capture the longer-term relationship between variables and avoid unnecessary reduction in the degrees of freedom. Ordinary least squares regression were used to estimate the parameters of the VAR(1) and VAR(2) models. We focused on the impact of lagged online medical behavior variables on ∆NCCt (see Table B in Appendix) because we were interested in predicting the daily changes in confirmed COVID-19 cases. The results indicate a good reliability of forecast and goodness of fit for VAR(1) (R 2 = 0.325, adjusted R 2 = 0.265) and VAR(2) models (R 2 = 0.658, adjusted R 2 = 0.559).
We performed some diagnostic tests to consider the robustness of the models’ results. First, multivariate portmanteau tests were implemented to test serial correlation. The residuals of the VAR(2) model were not distinguishable from a white noise series (χ 2 = 190.51, P = 0.050), whereas VAR(1) model's residuals have some remaining autocorrelation (χ 2 = 232.06, P = 0.002) at the significance level of 0.01. In addition, we verified that the two models satisfied the stationary condition, that is, all the moduli of AR(p) characteristic polynomial should be less than 1 (Lütkepohl, 2006). In sum, the better goodness of fit and the absence of autocorrelation makes the VAR(2) model more suitable than the VAR(1) model.
4.2. Granger causality test
We used Granger causality test (Granger, 1969) to determine whether the past value of a variable was able to forecast the change of others in the VAR model. Table 3 shows the estimated results of the Granger causality test for bivariate and multivariate models. The null hypothesis that ∆OMC does not Granger cause ∆NCC, ∆OMA and ∆OMS must be rejected (F = 5.48, P < 0. 001) when SFV was used as exogenous variable in the VAR model. And the hypothesis that ∆OMS does not Granger cause ∆NCC, ∆OMC and ∆OMA must also be rejected (F = 2.55, P < 0.05). More specifically, ∆OMC (F = 23.12, P < 0.001) and ∆OMS (F = 3.14, P< 0.05) are the Granger causality of ∆NCC when ∆NCC was used as dependent variable in the bivariate VAR Granger causality test. However, no Granger causality ran from ∆OMA to any other variables.
Table 3.
Results of Granger causality test for the VAR(2) model
| Granger | Bivariate test |
Multivariate test | |||
|---|---|---|---|---|---|
| Cause | ∆NCC | ∆OMC | ∆OMA | ∆OMS | |
| ∆NCC | - | 1.50(0.230) | 0.34(0.713) | 0.49(0.613) | 0.68(0.664) |
| ∆OMC | 23.12(0.000) | - | 0.28(0.755) | 3.03(0.055)+ | 5.48(0.000) |
| ∆OMA | 0.07 (0.928) | 1.02(0.366) | - | 0.92(0.404) | 0.43(0.862) |
| ∆OMS | 3.41(0.040) | 0.28(0.758) | 5.75(0.005) | - | 2.55(0.021) |
+P<0.10, * P<0.05, ** P<0.01, *** P<0.001.
In turn, ∆NCC did not Granger cause any indexes of online medical behaviors. Interestingly, between the three types of online medical behaviors have Granger causations, in which ∆OMC Granger causes ∆OMS (F = 3.03, P = 0.055) at 10% significance level and ∆OMS Granger causes ∆OMA (F = 5.75, P = 0.005) at 1% significance level.
4.3. Response of NCC
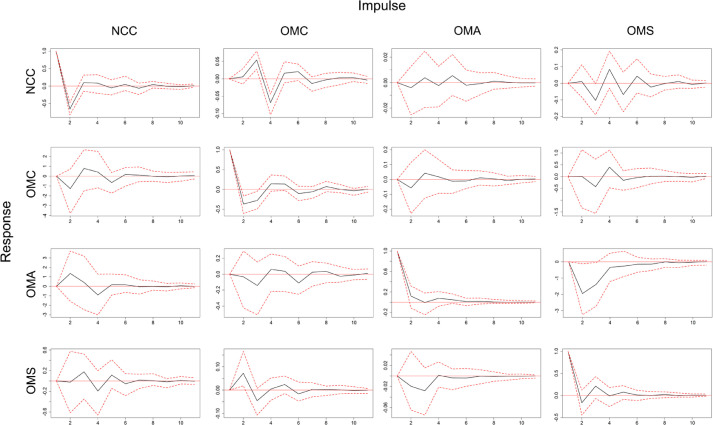
Individual estimated values (see in Table B in Appendix) provide limited information about the variable system because all variables in the VAR model are interdependent. IRF analysis was used to obtain a better picture of the dynamic reaction of every single variable on an exogenous shock to the model. For the VAR process, an impulse response is the reaction of a certain variable in response to the external shock of other variables (see Fig. A in Appendix). We aimed to investigate the responses of the NCC variable to the shocks of online medical behaviors (Fig. 4 ).
Fig. A.
Impulse response function for the four variables of the VAR(2) model (10 days).
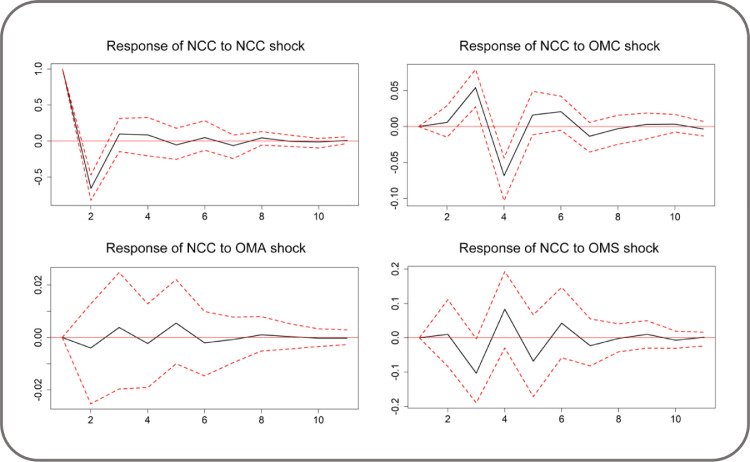
Fig. 4.
IRF for the response of NCC to other variables (95% CI).
Considering the use of first-difference variables, a one-time impulse on a variable has a permanent impact on its first difference level. The response of NCC to a one standard deviation shock from itself was substantially positive on the 1st day and negative on the 2nd day; then, the shock weakened and reached zero within a period of 10 days. Starting with zero level of response on the 1st day, NCC responded to OMC positively on the 3rd day, negatively on the 4th day with 95% Confidence Interval (CI); and then the shock stabilized toward zero over time. Thus, combining with the optimal order of VAR mode, we can conclude that to same extent the spreading trend of COVID-19 could be captured by the change in OMC before two days.
NCC responded faintly and negatively to OMA for most of the time; however, zero was included in a 95% CI throughout the whole process and implies that the impact from OMA to NCC is not that obvious and statistically substantial. NCC responded positively to OMS on the 2nd, 4th, 6th, and 9th days. This trend went negatively on the other days with the largest and significant coefficient on the 3th day during a 10-day forecasting period.
4.4. Variance decomposition
Forecast error variance decomposition (FEVD) was used to quantify the amount of information each variable contributes to the other variables in the VAR model. Based on orthogonal impulse response, FEVD computes innovation (i.e., the part that cannot be predicted in the series) by decomposing the forecast error variance. As a complement to the Granger causality test and IRF analysis, FEVD allowed us to evaluate the relative importance of predictor variables with percentage terms in explaining the innovation to the dependent variables over time. The shock of a variable contributed the most fraction of the overall forecast on itself (see Table C in Appendix).
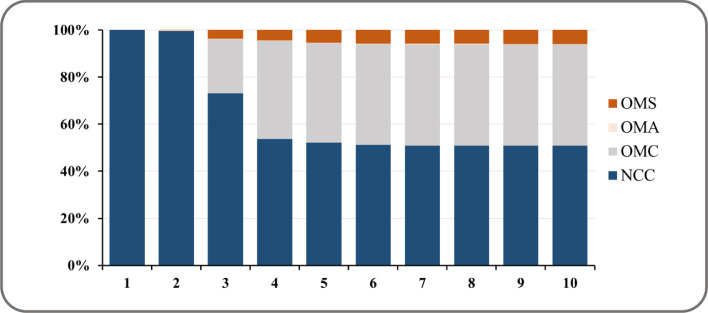
Fig. 5 presents the result of variance decomposition with respect to NCC. Initially, almost all variations (100% and 99.55%) for the first two days in NCC were explained by its own shock and followed by OMC with 22.99% on the third day, along with other two variables’ shocks were negligible. OMC explained 43.11% of the variation in NCC and became the most important variable in the system by the 10th day, if NCC own innovation is excluded (50.73%). By the end of 10 days, only 5.95% of the variations in NCC were explained by OMS. This finding further confirmed that OMS has less important shock on NCC. Again, OMA was proven to have the weakest effect, as it explained no more than 1% of the variation in NCC by the 10th day.
Fig. 5.
Variance decomposition in NCC by other variables in the VAR(2) model.
4.5. Robustness analyses
Several alternative models with subset or additional data were used to establish other VAR studies to check the robustness of the results. In particular, we investigated four alternative models as follows. First, to examine the problem of “false positive” which due to the OMC data being used contain the COVID-19 pandemic, we run studies with extra OMC data that were irrelevant to pandemic to see if it falsely predict the trend of NCC. Second, in terms of time-windows, we looked into labeled “time effect” with a longer or shorter period of observation than a 65-day timeframe based on the benchmark model. Furthermore, compared with aggregate data, OMA data retrieved from respiratory department, fever clinic and infection department were used to be the variable of OMA respectively. Finally, in the aspect of OMS, the Baidu Index data based on multiple keywords were recalculated at daily level with their quartile 1(Q1), median (Q2), quartile 3 (Q3) and maximum values.
As shown in Table 4 , additionally, OMC data were retrieved from the same platform within the corresponding period in year of 2018 and 2019, as well as the same length (i.e., 65 days) of period before pandemic at the end of 2019. For different lengths of lag from lag-1 to lag-6, the NCC did not Granger caused by those extra OMC datasets (i.e., OMC_18, OMC_19a and OMC_19b in Table 4), which did not contain the COVID-19 for Shenzhen city. By contrast with OMC_20b in benchmark model, OMC_20a data were retrieved excluding symptom keywords we used indicated that it did not Granger cause NCC for lag 1 to 6. Thus, the result demonstrated that there was no a spurious relationship between NCC and other OMC data that not related to COVID-19 or not based on specific keywords.
Table 4.
Results of Granger causality test between OMC and NCC
| Lag | ∆OMC does not Granger cause ∆NCC |
||||
|---|---|---|---|---|---|
| ∆OMC_18 | ∆OMC_19a | ∆OMC_19b | ∆OMC_20a | ∆OMC_20b | |
| lag-1 | 0.43(0.514) | 1.02(0.318) | 0.11(0.734) | 0.53(0.470) | 0.52(0.470) |
| lag-2 | 0.37(0.694) | 1.00(0.374) | 0.09(0.913) | 1.87(0.162) | 23.12(0.000)⁎⁎⁎ |
| lag-3 | 0.55(0.647) | 0.66(0.578) | 0.73(0.539) | 1.23(0.308) | 19.56(0.000)⁎⁎⁎ |
| lag-4 | 0.39(0.808) | 0.48(0.753) | 0.92(0.455) | 1.00(0.416) | 14.34(0.000)⁎⁎⁎ |
| lag-5 | 0.23(0.948) | 0.49(0.785) | 0.82(0.535) | 0.98(0.436) | 12.97(0.000)⁎⁎⁎ |
| lag-6 | 0.17(0.981) | 0.45(0.843) | 0.69(0.655) | 1.50(0.201) | 9.91(0.000)⁎⁎⁎ |
OMC_18: the daily amount of OMC containing symptom keywords within 65 days (2018.1.1-2018.3.6);
OMC_19a: the daily amount of OMC containing symptom keywords within 65 days (2019.1.1-2019.3.6);
OMC_19b: the daily amount of OMC containing symptom keywords within 65 days (2019.10.28-2019.12.31);
OMC_20a: the daily amount of OMC excluding symptom keywords within 65 days (2020.1.1-2020.3.5);
OMC_20b: the daily amount of OMC containing symptom keywords within 65 days (2020.1.1-2020.3.5).
⁎⁎⁎P<0.001, ⁎⁎P<0.01, * P<0.05, +P<0.10.
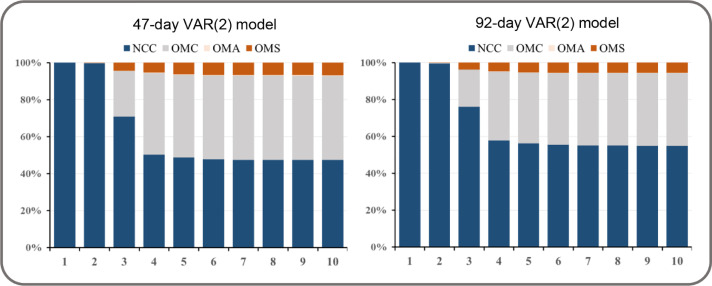
We carried out alternative model analysis exercise based on the different lengths of time window selection as a next step in checking the robustness of the VAR model in our study. We sliced a 47-day segment from January 10, 2020 to February 25, 2020 during the peak time (Fig. 1) of COVID-19 breakout in Shenzhen as a shorter-term series of variables. The longer-term time window of over 92 days was selected from January 1, 2020 to April 1, 2020. Fig. 6 reports the results of the FEVD for the 47-VAR(2) model and the 92-VAR(2) model whose stationary conditions were satisfied. The results both indicated that except “own shock,” OMC had the most predictive power by explaining about 40% to 50% of the variation in NCC, and OMS explained around 5% of the innovation of NCC for 10 days. The shock of OMA on NCC was relatively weak. We thus concluded that the findings are quite robust compared with the results of variance decomposition.
Fig. 6.
Variance decomposition plots for alternative VAR models in different timeframes.
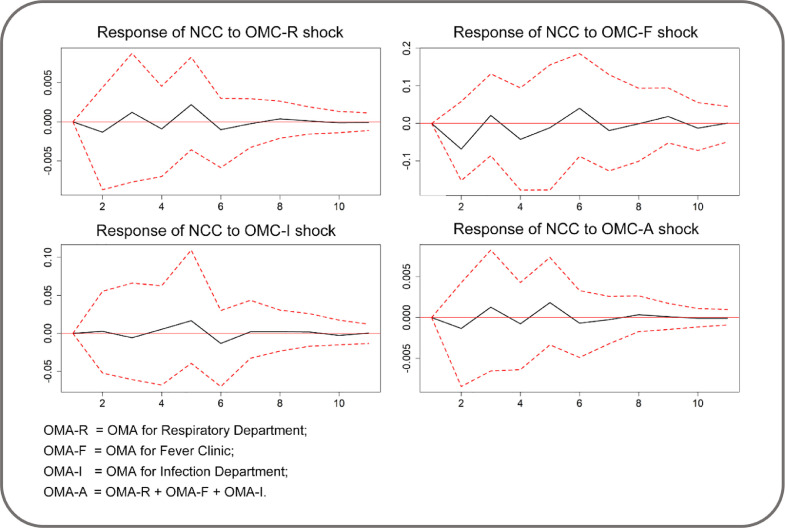
We also assessed whether changes in OMA variable with respiratory department, fever clinic and infection department data provide different results. Fig. 7 indicates that the IRF results obtained using individual level datasets (i.e., OMD-R, OMD-F and OMA-I) and their aggregate (i.e., OMA-All) to measure OMA were consistent with the result obtained from their arithmetic mean. In any case, the estimated response of NCC to the variant of OMA shock did not present any substantial change, because zero was always included in the 95% CI during the 10-day period.
Fig. 7.
IRF for the response of NCC to OMA in alternative models (95% CI).
For the last robustness check, we studied the effects of using different statistics for Baidu Index search volume on predicting NCC. As Table 4 shows, the result of Granger causality test using aggregate data for OMS variable is equivalent to arithmetic mean used in the benchmark model, that is, OMS dose Granger cause the change in NCC for lags 1 to 4. However for alternative statistics (i.e., quantiles and maximum) of OMS variable, it cannot rejected the hypothesis that no Granger causality between OMS and ONC throughout all order levels from lag-1 to lag-4. This means that in the case of OMS, computing variable by using the aggregate or average of Baidu Index at daily level has a relatively better approximation, which was also a general approach for Google Trend research (Qin & Peng, 2016).
5. Discussion
The proactive identification and prediction of the spread of novel infectious diseases are an urgent need for public health, especially when unexpected EIDs are not recorded by traditional surveillance systems. People are increasingly seeking medical help through online medical platforms, which afford the opportunity of digital surveillance as a supplement approach to traditional surveillance. We believe that this study is the first research to apply OMC data for disease surveillances. In this study, a new approach based on VAR model was proposed to investigate the dynamic relationship between confirmed COVID-19 cases and three online medical behaviors. This study specifically highlights the potential of OMC in predicting the trend of EIDS and has implications to facilitate public health policymaking.
This study proposed and verified the potentials of using online medical behaviors to predict the trends of infection pandemic. In line with the digital surveillance studies using other data sources, this study used the digital footprints of online medical behaviors to forecast the newly confirmed infectious cases. Different from other studies mainly using web queries and social media contents (Li, Xu, Cuomo et al., 2020; Li, Chen, Chen et al., 2020; Shen, Chen, Luo et al., 2020), this research emphasizes the usage of online users’ medical behaviors. As other studies addressed, the dynamics of “information spread” are inherently different from the dynamics of disease spread (Salathé, Freifeld, Mekaru et al., 2013). Both web queries and user-generated social media contents are representations of information spread, but the online medical behaviors data indicates the behavioral trend, which can be closely connected with the spread of infectious diseases. With the rapid development of digital medicalization, people may turn to the Internet for help first before paying an offline visit to doctors, when the diseases start transmitting. Therefore, it is highly likely that using online medical data can capture the trend of pandemics earlier than other surveillance system.
This study found that OMC and OMS were the Granger causes of confirmed cases during the outbreak of COVID-19, whereas OMA was not. The predictive power of OMS was not surprising, as it is in line with the Google Flu studies (Dugas, Jalalpour, Gel et al., 2013; Martin, Lee, Yasui, 2016) and other Baidu search index studies (He, Chen, Chen et al., 2018; Huang et al., 2020). However, the predictive value of OMS in this study was lower than that of OMC – an underexplored online medical behavior. OMC is a rapidly expanding service that bridges the communication between patients and physicians during the social distancing period. OMC displays more accurate information of health situations among citizens compared with OMS, as OMC users often have specific medical appeals or syndromes. OMA failed to predict the trends of COVID-19 in this study. Apparently, one reason is that people are worried of being cross-infected and reluctant to visit hospitals in-person during the pandemic. Another reason for the decrease in the number of hospital visits is the SFV and travel restrictions. In addition, a non-Granger causality exists between OMC and OMA. This result indicates that the trends of online consultation and in-person doctor visits were exclusive in the short term and people who chose to consult online tend not to go to the hospital at least without using online appointment, vice versa. But, presumably, the shrink of OMA might due to patients sought medical care directly at hospitals rather than making online appointments at first. For example, as specialist clinics that affiliated with the emergency departments, online appointment to fever clinics may not be easily and immediately accessed to from a commercial medical platform, and this is evident from the amount (only 3059) of OMA at fever clinic. The results also showed that the future trend of OMA to the hospital was captured by OMS instead of OMC. This finding implies that perhaps those who sought medical help via search engine also turned to offline medical consultation.
The present study showed the dynamic relationship among online medical behaviors and the prediction of NCC along with time variation. We investigated the dynamic consequences of shocks on the future behavior of NCC. The impulse responses to the “own shock” of NCC appeared instantaneous on the 1st and 2nd days. The relationship between NCC and OMC and between NCC and OMS was statistically substantial in the subsequent periods. And they both provided a short-term prediction about two days in advance. However, their impacts tended to disappear progressively. The size of the relationship between explanatory variables and independent variable was also studied. FEVD analysis revealed that OMC behaviors played the most important role in predicting the future trends of pandemic by explaining nearly half of the variation in NCC, except NCC itself, followed by OMS. The effect of OMA was minimal and explained less than 1% of the innovation in NCC. The conclusions drawn from the Granger causality test and IRF analysis were further verified.
This study applied solid and multiple statistical analyses to verify the results to increase the creditability and robustness of our findings. We made cautious decisions for false positive, data collection, processing, and measurement. For example, applying extra and irrelevant data to verify the predictive power of OMA on NCC instead of spurious correlation; and using comparable methods to examine the sampling and measurement effects for variables on prediction can entail robust and validated results.
Although the medical systems of various countries/regions are different and the data availability of online medical behaviors may vary greatly, this study shows the potentials of using aggregated online medical consultation behaviors at a city level for digital surveillance. Such surveillance is more directly related to the pandemic outbreak and thus can more precisely predict the trend (Shen, Chen, Luo et al., 2020). This study can call for the integration of disruptive online medical consultation data into the surveillance system. Different countries may consider the possibility and feasibility of such integration depending on their existing medical systems. Nevertheless, online medical behavioral data is also closely related to patient data privacy, and it is never too much to be cautious when using these data, even if it is used for public health surveillance and social good (Bari & O'Neill, 2019). The present study accessed the aggregated and de-identified data to predict the NCC of COVID-19. More discussions are needed to examine how to strike a balance between protecting the patients’ privacy and using online medical behavioral records for public good.
5.1. Contribution and implication
The main contributions of our study are as follows. First, we found a novel indicator, OMC, as a remarkable predictor for the outbreak of EIDs. Compared with the best-known approaches based on search engines, OMC is more directly related to disease management but has been neglected before. This study hopes to bring scholarly and health professional attention to such data and its potential of predicting EIDs. Online consultation data should be considered an important digital surveillance channel supplementary to the traditional surveillance system.
Second, this study provides a solid methodology to examine the predictive power of online medical behavioral data. Many studies were concerned on digital surveillance based on single data type instead of validating two or more indicators against each other, and neglected the importance of integration of disparate sources of data (Bansal et al., 2016). This study proposes the necessity to investigate the interdependent relationship between competitive variables in the dynamic system. A VAR model for multivariate analysis was performed in this study to explore the dynamic relationship between the different variables of interest and test the hypothesis of (non-)Granger causality, impulse response processes, and their FEVD. In addition, developing a flexible VAR model focusing on a certain target variable at different lag lengths is suitable for a data-driven exploratory research for epidemiology information in the absence of sufficient theoretical models. Taking the “counterfactual” data, time-windows and statistical methods into account, we still observed the similar predictive power of OMC on COVID-19 trend; hence, our findings were robust against alternative models.
Lastly, the results obtained from our model were verified in specific for the city-level data of Shenzhen, China. Different from data used in web-based and social media surveillance studies, OMC data recodes patients’ digital footprints more granularly to allow the use of more specific and accurate location-based information for regional epidemic surveillance in real time.
This study has broad public health implications. Paying attention to OMC data and deep mining data are important to foresee the trend of infectious diseases. Given that the data of OMC might be fragmented in the hands of different stakeholders and OMC providers, local and national governments should make efforts to collate the data and integrate them into the surveillance system. This effort can help prevent the outbreak of EIDs in the future and also respond to the needs of chronic diseases and other known diseases.
6. Limitations
This study has some limitations. First, similar to other existing studies that relied on web-based data and query data based on keywords, choosing the appropriate combination of keywords is challenging. This study adopted the terms of symptoms of COVID-19, such as “fever,” “cough” and “fatigue” instead of direct “novel coronavirus” or “novel coronavirus pneumonia” , to avoid the errors of overfitting caused by self-correction in keyword selection (Cook et al., 2011; Lazer et al., 2014), because the strong link between search queries and NCC might result from the fear and panic during pandemic. However, keywords selection and filtering methods based on different inclusion criteria and correlation analysis (Yuan et al., 2013; Santillana, Zhang, Althouse, et al., 2014) can be considered in the future.
Second, although COVID-19 cases were categorized as confirmed, suspected, clinically diagnosed, and asymptomatic in China, NCC only included confirmed cases. Symptoms and exposures are related to suspected cases and clinically diagnosed patients, whereas asymptomatic cases are those whose viral nucleic acid test results turned positive but without any symptoms (NCPERE Team, 2020). We may omit asymptomatic cases in the prediction by using symptoms terms to retrieve data. Thus, the bias in the predictor model might be caused by the self-reported symptoms of suspected and clinically diagnosed cases (Shen et al., 2020; Qin et al., 2020) and the absence of a small number of asymptomatic cases. However, the estimated parameters of our model reflected the trends and changes in confirmed cases rather than the actual numbers; thus, OMC might be a good predictor of a potential outbreak of COVID-19 but not a definitive factor for case counts.
Finally, our data collection was limited to one local online medical platform. Using local medical data can be more relevant and increase the accuracy and efficacy of prediction because of the diverse landscapes of medical services. However, our work may not be generalizable to all OMC and OMA behaviors in different online medical platforms because of regional variation in COVID-19 outbreak over periods of time. In addition, collecting adequate data on nationwide platforms is challenging, as few platforms provide OMC and OMA services at the same time. Meanwhile, other local platforms lack sufficient conditions, such as infrastructures of medical networks and broad usage of mobile devices, to take full advantage of online medical platforms. Future studies may duplicate this study to validate the predictive power of each service with the advancement of online medical services.
Table 5.
Results of Granger causality test from OMS to NCC by keywords.
| A Granger causes B |
F (p-value) |
|||
|---|---|---|---|---|
| lag-1 | lag-2 | lag-3 | lag-4 | |
| ∆OMS-S→∆NCC | 3.40(0.070)+ | 3.41(0.040)* | 2.76(0.051)+ | 2.32(0.069)+ |
| ∆OMS-Q1→∆NCC | 0.47(0.498) | 0.48(0.624) | 0.98(0.408) | 1.06(0.386) |
| ∆OMS-Q2→∆NCC | 1.80(0.185) | 2.79(0.070)+ | 1.94(0.134) | 1.46(0.223) |
| ∆OMS-Q3→∆NCC | 0.60(0.440) | 0.36(0.700) | 0.58(0.628) | 0.50(0.735) |
| ∆OMS-M→∆NCC | 2.12(0.151) | 2.89(0.063)+ | 1.91(0.139) | 1.37(0.257) |
+P<0.10, * P<0.05, ** P<0.01, *** P<0.001; -S= sum, -Q1=quantile 1, - Q3 =quantile 2, - Q3= quantile 3, -M= maximum.
7. Conclusion
Echoing the trend of digital disease surveillance in the era of the Internet, this study used three different online medical behavioral data to predict the confirmed cases of COVID-19 in China. The findings highlight the strong predictive value of OMC in forecasting the trend of the pandemic and also confirmed OMS as a predictive factor. Online medical service is rapidly expanding as people are used to turn to the Internet before paying a visit to a doctor in-person nowadays. Integrating online medical behavioral data, particularly OMC, into existing disease surveillance systems is highly needed to prevent future EIDs and respond to other known diseases.
Declaration of Competing Interest
We declare no competing interest.
Acknowledgements
This study was funded by the National Social Science Fund of China [grant numbers: 19ZDA324 & 18CXW017] and Planning Project of Guangdong Philosophy and Social Science Fund [grant numbers: GD19YXW03].
Appendix
Table A.
Keywords list used to collect data for OMC and OMS
| Chinese | English | |
|---|---|---|
| 1 | 发热*/发烧/高烧/低热/低烧 | Fever |
| 2 | 咳嗽*/干咳 | Cough |
| 3 | 咽痛*/喉咙痛 | Pharyngalgia |
| 4 | 胸闷*/胸痛 | Chest tightness |
| 5 | 呼吸困难*/呼吸急促/气短/气促 | Dyspnea |
| 6 | 乏力*/四肢无力 | Fatigue |
| 7 | 肌肉酸痛*/肌痛/四肢酸痛 | Myalgia |
| 8 | 恶心* | Nausea |
| 9 | 呕吐* | Vomiting |
| 10 | 腹泻*/拉肚子 | Diarrhea |
| 11 | 嗜睡* | Somnolence |
| 12 | 惊厥* | Convulsion |
Core keyword was used to query with Baidu Index.
Table B.
Estimation results for the equation of ∆NCCt in VAR(1) and VAR(2).
| ∆NCCt for VAR(1) |
∆NCCt for VAR(2) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Estimate | s.e. | t | Pr (>|t|) | Estimate | s.e. | t | Pr (>|t|) | |
| ∆NCCt−1 | −0.484⁎⁎⁎ | 0.111 | −4.372 | 0.000 | −0.658⁎⁎⁎ | 0.104 | −6.313 | 0.000 |
| ∆OMCt−1 | 0.006 | 0.014 | 0.426 | 0.672 | 0.006 | 0.012 | 0.497 | 0.621 |
| ∆OMAt−1 | −0.002 | 0.011 | −0.213 | 0.832 | −0.004 | 0.010 | −0.418 | 0.678 |
| ∆OMSt−1 | 0.125* | 0.061 | 2.050 | 0.045 | 0.010 | 0.051 | 0.197 | 0.845 |
| ∆NCCt−2 | −0.324⁎⁎ | 0.111 | −2.919 | 0.005 | ||||
| ∆OMCt−2 | 0.059⁎⁎⁎ | 0.012 | 5.073 | 0.000 | ||||
| ∆OMAt−2 | 0.002 | 0.009 | 0.244 | 0.809 | ||||
| ∆OMSt−2 | −0.103* | 0.051 | −2.025 | 0.048 | ||||
| SPVt | 5.049* | 2.424 | 2.083 | 0.042 | 5.291* | 2.326 | 2.274 | 0.027 |
| Intercept | −0.918 | 0.913 | −1.006 | 0.319 | −1.001 | 0.709 | −1.411 | 0.164 |
| R2 | 0.325 | 0.658 | ||||||
| Adjusted R2 | 0.265 | 0.599 | ||||||
+P<0.10, * P<0.05, ** P<0.01, *** P<0.001.
Table C.
Results of FEVD for the four variables of the VAR(2) model (10 steps).
| Period | NCC | OMC | ONA | OMS | |
|---|---|---|---|---|---|
| NCC | 1 | 100.00% | 0.00% | 0.00% | 0.00% |
| 2 | 99.55% | 0.21% | 0.19% | 0.05% | |
| 3 | 73.08% | 22.99% | 0.14% | 3.78% | |
| 4 | 53.73% | 41.63% | 0.10% | 4.54% | |
| 5 | 52.22% | 42.06% | 0.18% | 5.54% | |
| 6 | 51.16% | 42.80% | 0.18% | 5.86% | |
| 7 | 50.78% | 43.10% | 0.20% | 5.93% | |
| 8 | 50.79% | 43.08% | 0.20% | 5.93% | |
| 9 | 50.76% | 43.09% | 0.21% | 5.95% | |
| 10 | 50.73% | 43.11% | 0.21% | 5.95% | |
| OMC | 1 | 0.08% | 99.92% | 0.00% | 0.00% |
| 2 | 0.94% | 98.64% | 0.41% | 0.00% | |
| 3 | 1.28% | 97.45% | 0.49% | 0.78% | |
| 4 | 1.36% | 96.63% | 0.58% | 1.43% | |
| 5 | 1.64% | 96.26% | 0.60% | 1.50% | |
| 6 | 1.65% | 96.24% | 0.61% | 1.49% | |
| 7 | 1.66% | 96.23% | 0.62% | 1.49% | |
| 8 | 1.65% | 96.24% | 0.62% | 1.49% | |
| 9 | 1.65% | 96.23% | 0.63% | 1.49% | |
| 10 | 1.65% | 96.23% | 0.63% | 1.49% | |
| OMA | 1 | 0.01% | 1.47% | 98.52% | 0.00% |
| 2 | 0.48% | 1.44% | 86.54% | 11.54% | |
| 3 | 0.47% | 1.84% | 81.43% | 16.26% | |
| 4 | 0.85% | 2.19% | 80.60% | 16.35% | |
| 5 | 0.86% | 2.33% | 80.35% | 16.47% | |
| 6 | 0.86% | 2.88% | 79.83% | 16.42% | |
| 7 | 0.86% | 2.94% | 79.74% | 16.46% | |
| 8 | 0.86% | 3.00% | 79.68% | 16.45% | |
| 9 | 0.86% | 3.03% | 79.65% | 16.45% | |
| 10 | 0.87% | 3.03% | 79.65% | 16.45% | |
| OMS | 1 | 0.45% | 2.21% | 2.43% | 94.91% |
| 2 | 0.47% | 10.82% | 3.53% | 85.18% | |
| 3 | 0.98% | 14.38% | 4.20% | 80.44% | |
| 4 | 1.40% | 14.35% | 4.18% | 80.07% | |
| 5 | 1.53% | 14.80% | 4.14% | 79.54% | |
| 6 | 1.55% | 15.13% | 4.14% | 79.18% | |
| 7 | 1.55% | 15.14% | 4.14% | 79.16% | |
| 8 | 1.55% | 15.14% | 4.14% | 79.17% | |
| 9 | 1.56% | 15.14% | 4.14% | 79.17% | |
| 10 | 1.56% | 15.14% | 4.14% | 79.16% |
References
- Agarwal R., Gao G., DesRoches C., Jha A.K. The digital transformation of healthcare: Current status and the road ahead. Information Systems Research. 2010;21(4):796–809. [Google Scholar]
- Aiello A.E., Renson A., Zivich P.N. Social Media–and Internet-Based Disease Surveillance for Public Health. Annual Review of Public Health. 2020;41:101–118. doi: 10.1146/annurev-publhealth-040119-094402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althouse B.M., Scarpino S.V., Meyers L.A., et al. Enhancing disease surveillance with novel data streams: challenges and opportunities. EPJ Data Sci. 2015;4:17. doi: 10.1140/epjds/s13688-015-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S., Chowell G., Simonsen L., Vespignani A., Viboud C. Big data for infectious disease surveillance and modeling. The Journal of infectious diseases. 2016;214(suppl_4):S375–S379. doi: 10.1093/infdis/jiw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari L., O'Neill D.P. Health Affairs; 2019. Rethinking Patient Data Privacy In The Era Of Digital Health.https://www.healthaffairs.org/do/10.1377/hblog20191210.216658/full/ accessed from. [Google Scholar]
- Bento A.I., Nguyen T., Wing C., Lozano-Rojas F., Ahn Y.Y., Simon K. Evidence from internet search data shows information-seeking responses to news of local COVID-19 cases. Proceedings of the National Academy of Sciences. 2020;117(21):11220–11222. doi: 10.1073/pnas.2005335117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broniatowski D.A., Paul M.J, Dredze M. National and Local Influenza Surveillance through Twitter: An Analysis of the 2012-2013 Influenza Epidemic. PLoS ONE. 2013;8(12):e83672. doi: 10.1371/journal.pone.0083672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein J.S., Freifeld C.C., Madoff L.C. Digital disease detection–harnessing the Web for public health surveillance. The New England journal of medicine. 2009;360(21):2153–2157. doi: 10.1056/NEJMp0900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo Rafael A, Deterding Sebastian, Ryan Richard M. Health surveillance during covid-19 pandemic. BMJ. 2020;369:m1373. doi: 10.1136/bmj.m1373. [DOI] [PubMed] [Google Scholar]
- Chan E.H., Sahai V., Conrad C, Brownstein JS. Using Web Search Query Data to Monitor Dengue Epidemics: A New Model for Neglected Tropical Disease Surveillance. PLoS Negl Trop Dis. 2011;5(5):e1206. doi: 10.1371/journal.pntd.0001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Lu H. China Daily; 2018. Shenzhen aims to be global technology innovation hub.https://www.chinadaily.com.cn May 21. Retrieved from. [Google Scholar]
- Cook S., Conrad C., Fowlkes A.L., Mohebbi M.H. Assessing Google flu trends performance in the United States during the 2009 influenza virus A (H1N1) pandemic. PloS one. 2011;6(8):e23610. doi: 10.1371/journal.pone.0023610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey D.A., Fuller W.A. Likelihood ratio statistics for autoregressive time series with a unit root. Econometrica: journal of the Econometric Society. 1981:1057–1072. [Google Scholar]
- Dugas A.F., Jalalpour M., Gel Y., Levin S., Torcaso F., Igusa T., Rothman R.E. Influenza forecasting with Google flu trends. PloS one. 2013;8(2):e56176. doi: 10.1371/journal.pone.0056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K. A qualitative investigation into the online counselling relationship: To meet or not to meet, that is the question. Counselling and Psychotherapy Research. 2012;12(4):316–326. [Google Scholar]
- Eichberger J. Johns Hopkins University website; 2020. Study of data from Shenzhen, china, provides key covid-19 insights.https://hub.jhu.edu/ Retrieved April 28, 2010, from. [Google Scholar]
- Eysenbach G. AMIA ... Annual Symposium proceedings. AMIA Symposium, 2006. 2006. Infodemiology: tracking flu-related searches on the web for syndromic surveillance; pp. 244–248. [PMC free article] [PubMed] [Google Scholar]
- Eysenbach G. Infodemiology and infoveillance: tracking online health information and cyberbehavior for public health. American journal of preventive medicine. 2011;40(5):S154–S158. doi: 10.1016/j.amepre.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Fantazzini D. Short-term forecasting of the COVID-19 pandemic using Google Trends data: Evidence from 158 countries. Applied Econometrics. 2020;59:33–54. [Google Scholar]
- Ginsberg J., Mohebbi M.H., Patel R.S., Brammer L., Smolinski M.S., Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457(7232):1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- Gong K., Xu Z., Cai Z., Chen Y., Wang Z. Internet hospitals help prevent and control the epidemic of COVID-19 in China: Multicenter user profiling study. Journal of medical Internet research. 2020;22(4):e18908. doi: 10.2196/18908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger C.W.J. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:424–438. [Google Scholar]
- Hamilton J.D. Princeton University Press; Princeton: 1994. Time Series Analysis. [Google Scholar]
- He G., Chen Y., Chen B., Wang H., Shen L., Liu L., Du S. Using the Baidu search index to predict the incidence of HIV/AIDS in China. Scientific reports. 2018;8(1):1–10. doi: 10.1038/s41598-018-27413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health 160 (2020). The Introduction to Health 160. Retreived from https://news.91160.com/about/index.htmlonDec152020.
- Hswen Y., Brownstein J.S., Liu J., Hawkins J.B. Use of a Digital Health Application for Influenza Surveillance in China. American journal of public health. 2017;107(7):1130–1136. doi: 10.2105/AJPH.2017.303767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Bell R.A., Kravitz R.L., Orrange S. The prepared patient: information seeking of online support group members before their medical appointments. Journal of health communication. 2012;17(8):960–978. doi: 10.1080/10810730.2011.650828. [DOI] [PubMed] [Google Scholar]
- Huang R., Luo G., Duan Q., Zhang L., Zhang Q., Tang W., Zou H. Using Baidu search index to monitor and predict newly diagnosed cases of HIV/AIDS, syphilis and gonorrhea in China: estimates from a vector autoregressive (VAR) model. BMJ open. 2020;10(3) doi: 10.1136/bmjopen-2019-036098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnayain A., Fuad A., Su E.C.Y. Applications of google search trends for risk communication in infectious disease management: A case study of COVID-19 outbreak in Taiwan. International Journal of Infectious Diseases. 2020 doi: 10.1016/j.ijid.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantar Consulting . 2019. Kantar Consulting: Internet + Medical Health White Papers.http://www.199it.com/archives/823842.html [Google Scholar]
- Kellermann A.L., Isakov A.P., Parker R., Handrigan M.T., Foldy S. Web-based self-triage of influenza-like illness during the 2009 H1N1 influenza pandemic. Annals of emergency medicine. 2010;56(3):288–294. doi: 10.1016/j.annemergmed.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F., Saeed A., Ali S. Modelling and forecasting of new cases, deaths and recover cases of COVID-19 by using Vector Autoregressive model in Pakistan. Chaos, Solitons & Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelsø C., Galle M., Bang H., Ethelberg S., Krause T.G. Influmeter–an online tool for self-reporting of influenza-like illness in Denmark. Infectious Diseases. 2016;48(4):322–327. doi: 10.3109/23744235.2015.1122224. [DOI] [PubMed] [Google Scholar]
- Koch S. Home telehealth—current state and future trends. International journal of medical informatics. 2006;75(8):565–576. doi: 10.1016/j.ijmedinf.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Lazer D., Kennedy R., King G., Vespignani A. The parable of Google Flu: traps in big data analysis. Science. 2014;343(6176):1203–1205. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- Lee S.A., Zuercher R.J. A current review of doctor–patient computer-mediated communication. Journal of Communication in Healthcare. 2017;10(1):22–30. [Google Scholar]
- Lenca Marcello, Vayena Effy. On the responsible use of digital data to tackle the COVID-19 pandemic. Nature medicine. 2020;26(4):463–464. doi: 10.1038/s41591-020-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Xu Q., Cuomo R, Purushothaman V, Mackey T. Data Mining and Content Analysis of the Chinese Social Media Platform Weibo During the Early COVID-19 Outbreak: Retrospective Observational Infoveillance Study. JMIR Public Health Surveill. 2020;6(2):e18700. doi: 10.2196/18700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Chen L.J., Chen X., Zhang M., Pang C.P., Chen H. Retrospective analysis of the possibility of predicting the COVID-19 outbreak from Internet searches and social media data, China, 2020. Eurosurveillance. 2020;25(10) doi: 10.2807/1560-7917.ES.2020.25.10.2000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Liu M., Feng Y., Ning C., Ou W., Sun J., Shao Y. Using Baidu search engine to monitor AIDS epidemics inform for targeted intervention of HIV/AIDS in China. Scientific reports. 2019;9(1):1–12. doi: 10.1038/s41598-018-35685-w. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Xing X. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2001316. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu T., Zhu G., Lin H., Zhang Y., He J., Xie R. Dengue Baidu Search Index data can improve the prediction of local dengue epidemic: A case study in Guangzhou, China. PLOS neglected tropical diseases. 2017;11(3) doi: 10.1371/journal.pntd.0005354. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütkepohl H. Springer; New York: 2006. New Introduction to Multiple Time Series Analysis. [Google Scholar]
- Martin L.J., Lee B.E., Yasui Y. Google Flu Trends in Canada: a comparison of digital disease surveillance data with physician consultations and respiratory virus surveillance data, 2010–2014. Epidemiology & Infection. 2016;144(2):325–332. doi: 10.1017/S0950268815001478. [DOI] [PubMed] [Google Scholar]
- McGough S.F, Brownstein JS, Hawkins JB, Santillana M. Forecasting Zika Incidence in the 2016 Latin America Outbreak Combining Traditional Disease Surveillance with Search, Social Media, and News Report Data. PLOS Neglected Tropical Diseases. 2017;11(1) doi: 10.1371/journal.pntd.0005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangsangna R.D., Vroom F.D.C. Factors influencing online health information seeking behaviour among patients in Kwahu West Municipal, Nkawkaw, Ghana. Online journal of public health informatics. 2019;11(2) doi: 10.5210/ojphi.v11i2.10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelat, C., Turbelin, C., Bar-Hen, A., Flahault, A., & Valleron, A. (2009). More diseases tracked by using Google Trends. Emerging infectious diseases, 15(8), 1327–1328. https://doi.org/10.3201/eid1508.090299. [DOI] [PMC free article] [PubMed]
- Qin J., Peng T.Q. Googling environmental issues: Web search queries as a measurement of public attention on environmental issues. Internet research. 2016;26(1):57–73. [Google Scholar]
- Qin L., Sun Q., Wang Y., Wu K.F., Chen M., Shia B.C., Wu S.Y. Prediction of number of cases of 2019 novel coronavirus (COVID-19) using social media search index. International journal of environmental research and public health. 2020;17(7):2365. doi: 10.3390/ijerph17072365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadona A.L, Tozan Y, Lazuardi L, Rocklöv J. A combination of incidence data and mobility proxies from social media predicts the intra-urban spread of dengue in Yogyakarta, Indonesia. PLOS Neglected Tropical Diseases. 2019;13(4) doi: 10.1371/journal.pntd.0007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadona A.L., Lazuardi L., Hii Y.L., Holmner Å., Kusnanto H., Rocklöv J. Prediction of dengue outbreaks based on disease surveillance and meteorological data. PloS one. 2016;11(3) doi: 10.1371/journal.pone.0152688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocklöv J., Tozan Y., Ramadona A., Sewe M.O., Sudre B., Garrido J., de Saint Lary C.B., Lohr W., Semenza J.C. Using Big Data to Monitor the Introduction and Spread of Chikungunya, Europe, 2017. Emerging infectious diseases. 2019;25(6):1041–1049. doi: 10.3201/eid2506.180138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathé M., Freifeld C.C., Mekaru S.R., Tomasulo A.F., Brownstein J.S. Influenza A (H7N9) and the importance of digital epidemiology. The New England journal of medicine. 2013;369(5):401–404. doi: 10.1056/NEJMp1307752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salganik M.J. Princeton University Press; 2019. Bit by bit: Social research in the digital age.https://www.bitbybitbook.com/en/1st-ed/observing-behavior/strategies/forecasting/ [Google Scholar]
- Santillana M., Nguyen AT, Dredze M, Paul MJ, Nsoesie EO, et al. Combining Search, Social Media, and Traditional Data Sources to Improve Influenza Surveillance. PLOS Computational Biology. 2015;11(10) doi: 10.1371/journal.pcbi.1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santillana M., Zhang D.W., Althouse B.M., Ayers J.W. What can digital disease detection learn from (an external revision to) Google Flu Trends? American journal of preventive medicine. 2014;47(3):341–347. doi: 10.1016/j.amepre.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Shen C., Chen A., Luo C., Zhang J., Feng B., Liao W. Using Reports of Symptoms and Diagnoses on Social Media to Predict COVID-19 Case Counts in Mainland China: Observational Infoveillance Study. Journal of Medical Internet Research. 2020;22(5):e19421. doi: 10.2196/19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenzhen Municipal Health Commission (2020). Summary of health statistics of Shenzhen city in 2019. http://wjw.sz.gov.cn/xxgk/tjsj/zxtjxx/content/post_7786068.html.
- The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)—China, 2020. China CDC Weekly, 2020;2(8):113–122. [PMC free article] [PubMed] [Google Scholar]
- Vaughan L., Chen Y. Data mining from web search queries: A comparison of google trends and baidu index. Journal of the Association for Information Science and Technology. 2015;66(1):13–22. [Google Scholar]
- Velasco E., Agheneza T., Denecke K., Kirchner G., Eckmanns T. Social media and internet-based data in global systems for public health surveillance: a systematic review. The Milbank quarterly. 2014;92(1):7–33. doi: 10.1111/1468-0009.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M., Kishore K., Kumar M., Sondh A.R., Aggarwal G., Kathirvel S. Google Search Trends Predicting Disease Outbreaks: An Analysis from India. Healthcare informatics research. 2018;24(4):300–308. doi: 10.4258/hir.2018.24.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagg A.J., Callanan M.M., Hassett A. The use of computer mediated communication in providing patient support: A review of the research literature. International journal of nursing studies. 2018;82:68–78. doi: 10.1016/j.ijnurstu.2018.03.010. [DOI] [PubMed] [Google Scholar]
- Wang H. 04. People’s Daily; 2020. p. 17.http://paper.people.com.cn/rmrb/html/2020-04/17/nw.D110000renmrb_20200417_1-13.htm (Internet diagnosis and treatment accelerates in this spring). [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Y., Yang G., Ma J., Wang L., Qi X. China information system for disease control and prevention (CISDCP) HIT briefing book. 2013:101–108. [Google Scholar]
- Wang X., Shi J., Kong H. Online Health Information Seeking: A Review and Meta-Analysis. Health Communication. 2020:1–13. doi: 10.1080/10410236.2020.1748829. [DOI] [PubMed] [Google Scholar]
- Wong D.K.K., Cheung M.K. Online health information seeking and ehealth literacy among patients attending a primary care clinic in Hong Kong: A cross-sectional survey. Journal of medical Internet research. 2019;21(3):e10831. doi: 10.2196/10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2020). Coronavirus disease 2019 (COVID-19): situation report, 72.
- Yuan Q., Nsoesie E.O., Lv B., Peng G., Chunara R., Brownstein J.S. Monitoring influenza epidemics in china with search query from baidu. PloS one. 2013;8(5):e64323. doi: 10.1371/journal.pone.0064323. [DOI] [PMC free article] [PubMed] [Google Scholar]