Abstract
An acute respiratory syndrome (COVID-19), caused by a novel coronavirus (SARS-CoV-2) with a high rate of morbidity and elevate mortality, has emerged as one of the most important threats to humankind in the last centuries. Rigorous determination of SARS-CoV-2 infectivity is very difficult owing to the continuous evolution of the virus, with its single nucleotide polymorphism (SNP) variants and many lineages. However, it is urgently necessary to study the virus in depth, to understand the mechanism of its pathogenicity and virulence, and to develop effective therapeutic strategies. The present contribution summarizes in a succinct way the current knowledge on the evolutionary and structural features of the virus, with the aim of clarifying its mutational pattern and its possible role in the ongoing pandemic.
Keywords: SARS-CoV-2, Pandemic, Mutation, Evolution pattern
1. Introduction
The respiratory syndrome responsible for the current pandemic initially detected in Wuhan in late December 2019 (Na Z et al., 2020) is an infectious disease caused by a novel coronavirus (SARS-CoV-2). Coronaviruses (CoVs) are a group of enveloped viruses, with a positive single-stranded RNA genome of approximately 30,000 bases with 5′-cap structure and 3′-poly-A tail, belonging to the Coronaviridae family of the order Nidovirales [1]. They cause mainly respiratory and gastrointestinal tract infections and are genetically classified into four major genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. The Alpha and Beta-CoVs can infect humans, while the Gama and Delta-CoVs infect predominantly birds [2].
The SARS-CoV-2, which is responsible for the current Coronavirus disease (COVID-19), also belongs to the genus Beta-CoV and it is considered the third major coronavirus outbreak in the last 20 years, after Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) [2]. On March 11th, 2020, the World Health Organization (WHO) having established the spread (and severity) of the SARS-CoV-2 infection, declared that the COVID-19 outbreak recorded in the preceding months was a pandemic [3]. It has currently affected >200 countries. As on October 2020, about 35.6 million people have been infected, with more than 1.04 million deaths [4]. More than 24.8 million people have recovered completely, but a large number of the infected people end up in critical condition that require respiratory assistance. The Countries that have been affected most severely are the USA, Brazil, India, Russia, Mexico, South American Countries, most European Countries [4].
Adaptive mutations in the SARS-CoV-2 genome could alter its pathogenic potential, and at the same time would increase the difficulty of drug and vaccine development. This contribution will not deal in detail with the mass of molecular information now available for SARS-CoV-2. It will rapidly summarize the information on its evolutionary and structural features that could be useful for the development of vaccines.
1. SARS-CoV-2 genome composition, proximal origin and evolution patterns
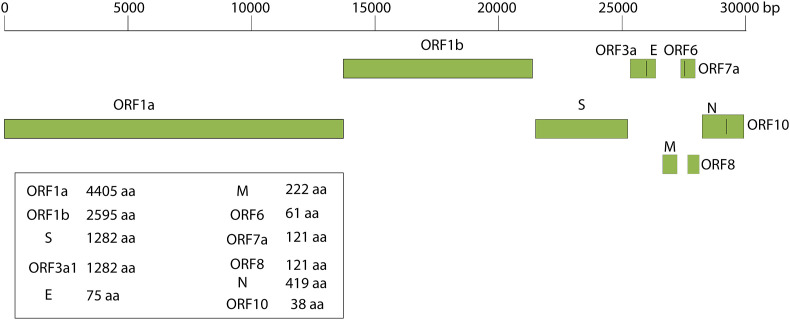
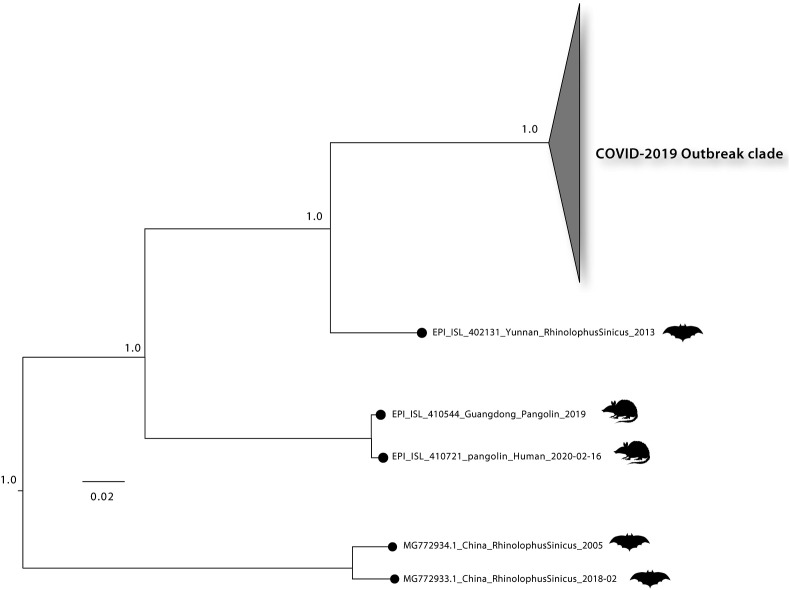
Genomes of Coronaviruses include a variable number of open reading frames (ORF). The first 5′ ORF (ORF1a/b) corresponds to about 2/3 of genome, and it is translated in the rough endoplasmic reticulum of the host cell into pp1a and pp1ab protein that are cleaved by proteases it yields 16 non-structural proteins (nsp1-16) [5]. The 3′ ORF, which corresponds to the remaining third of the genome consists of genes encoding accessory and structural proteins [5]. The four major structural proteins are: the surface Spike (S) protein, which recognizes the receptor of the host cell (the angiotensin converting enzyme 2 (ACE2)), binds to it, and mediates the penetration of the virus into the host cell the envelope E protein, the matrix protein (M), and the nucleocapsid (N) protein that binds the RNA and is fundamental for virion assembly [1,5] (Fig. 1 ). Additionally, the SARS-CoV-2 contains 6 accessory proteins, encoded by the ORF3a, ORF6, ORF7a, and ORF8, ORF10 genes: their functions are still largely unexplored. Thus, most of the proteins encoded by ORF1a and ORF1ab are essential for virus replication and at least for the adaption of the virus to a new host. In addition, a 5′ untranslated region (UTR) and a 3′-UTR have also been identified in the SARS-CoV-2 genome [1,5]. Some of the nsp form the replicase/transcriptase complex: nsp 12 is the RNA-dependent RNA polymerase that replicates the RNA of the virus, however, to function perfectly it also needs nsp 7 and nsp 8 and, possibly, of other non-structural proteins. The genomic organization of the SARS-CoV-2 shares about 89% sequence identity with that of other CoVs. Comparative sequence analysis of the SARS-CoV-2 genome indicates striking similarities to that of the Bat-CoV, suggesting a possible bat origin for that in the affected humans in Wuhan [6]. The possibility of other animal intermediate hosts from bats before the introduction of the virus into humans has been discussed. Initially, the Pangolin origin of COVID-19 in humans had received significant favour, particularly based on the finding that the virus of pangolins could use ACE2 as the receptor in the host cells. However, more recent results of various experimental approaches, e.g., on the very poor affinity of the pangolin virus for the human ACE2 receptor have apparently exempted pangolins from a possible role as intermediate host. The question of the intermediate host in the transmission of SARS-CoV-2 from bats to human is thus not settled, and it appears possible that the transmission occurs directly [7] (Fig. 2 ).
Fig. 1.
Genome architecture of SARS-CoV-2.
Fig. 2.
Maximum likelihood phylogeny was estimated with n = 143 complete genomes sequences from the current (2019–2020) SARS-CoV-2 epidemic plus n = 3 closely related bat strains plus n = 2 pangolin strains retrieved from GISAID. Number along branch represent bootstrap score. Scale bar represents expected substitutions per nucleotide site.
Generally, the rates of nucleotide substitution of RNA viruses are fast, and this rapid evolution is mainly shaped by natural selection. This high error rate and the consequent rapidly evolving virus populations [8], which could lead to the accumulation of amino acid mutations, might affect the transmissibility of the virus, its cell tropism and pathogenicity. It would unfortunately also present daunting challenges for the design of effective vaccines and diagnostic means. Fortunately, however, until now the observed diversity among SARS-CoV-2 sequences has been low. There has been an exception that has had dire consequences, i.e., the replacement of aspartic acid 614 of the Spike protein with a glycine, which has greatly increased the infectivity of the virus, but, in principle, the possibility of positive natural selection mutations exists. Considering its high transmissibility and the absence of pre-existing immunity in the general population its natural disappearance appears to be unlikely. Furthermore, it is not known whether SARS-CoV-2 is already fully adapted for efficient growth in human cells after its host-jump from bats or from a putative intermediate host [9]. Genomic epidemiology has revealed that the spillover from bats to humans has most likely occurred in late November or early December 2019. and that from that moment the spread of the virus occurred mainly by a human-to-human transmission [6].
2. Recent genomic changes in SARS-CoV-2
Coronaviruses such as SARS-CoV-2 are relatively stable thanks to a proofreading mechanism that operates during replication. Many genomic studies have nevertheless revealed changes in their genomes, including mutations and deletions. The D614G point-mutation in the Spike protein of SARS-CoV-2, which rapidly became the most widespread variant of SARS-CoV-2, has just been mentioned: we were among the first to observe it [[10], [11], [12]], but we had also observed that this mutation clustered with a series of other point mutations, including one in the polymerase gene [10]. A series of other mutations were then identified, allowing the classification of several SARS-CoV-2 lineages [13]. At the same time, additional profound changes in the genome, i.e. deletions, started to be reported. In particular, an extensive deletion in the ORF7a gene [14] and a deletion in the nsp2 gene [15]. More recently, analysing a more comprehensive dataset of more than 17.000 sequences obtained from GISAID we identified the emergence of a strain with a deletion of 9 nucleotides in the nsp1 gene (nucleotides 686–694 corresponding to amino acids 241–243) in patients infected with COVID-19 from different areas of the world. The overall frequency of the genome deletion was 0.44%, but was not distributed homogeneously: for instance, we did not find it in Italy, Germany, and Austria, whereas it was more frequent in Sweden, Israel, and the USA. Structural analysis suggest that this deletion might affect the C-terminal region of the protein that appear to be important for the regulation of viral replication and appear to have negative effect on host’s gene expression [16]. Those results were also confirmed by other groups that highlighted that SARS-CoV-2 appear to be undergoing profound genomic changes [17,18].
While the D614G mutation confers a selective advantage for SARS-CoV-2 fitness [11,12], the exact biological relevance of the other mutations is still unknown. However, nsp 1, which is also known as the leader protein, is central in the inhibition of the anti-viral innate immune response, in particular the expression of interferon-alpha [19] and is possibly the most important determinant of the viral pathogenicity. We feel it is appropriate to mention that a viral isolate from an asymptomatic SARS-CoV-2-positive subject had an unprecedented replication ability in VeroE6 cells in the absence of any clear cytopathic effect [9]. Even though it was a single observation, and even if the precise molecular mechanism that explains the absence of cytopathic effect has not yet been identified, we believe that the observation is significant. Such results could indicate the evolution of a possible new viral quasi-specie, but further data will be necessary in order to confirm this hypothesis. On this respect we believe that priority in this moment will be the monitoring over time of asymptomatic and pauci-symptomatic subjects to confirm the spreading of this particular viral strain with a possible decreased viral pathogenicity.
3. Parameters on viral mutations and spreading and their effect
Environmental factors, e.g., temperature, population density and air pollution, seem to affect viral spreading and mortality rate [[20], [21], [22], [23], [24]]. In addition, interventions aimed at limiting people movement and interactions have been implemented in several areas of the globe to curb the pandemic. The overall effect of these measures has been a reduction of the number of infections and the decrease of death rates. However, while it is intuitively clear that, for instance, full lockdowns by themselves are very effective, they are economically so burdensome to become impractical and unsustainable in the long run. Consequently, it becomes important to implement a series of concomitant and complementary measures to limit viral spread. One important point to be considered concerns the virus itself: at the moment there are no reasons to believe that changes of the virus have occurred in the direction of decreased pathogenicity. The decrease in the seriousness of the infection which is now generally observed is prima facie related to the containment measures: they certainly influence one parameter that is important, i.e. de magnitude of the viral load which is transmitted, thus in a sense they alter, quantitatively if not qualitatively the virus. Whether the measures above also have an effect on the viability of the viral particles has been suggested as a possibility [25,26]. But, as we have discussed above, the chances that the virus itself may change (i.e., mutate) in the direction of decreased pathogenicity could still be considered.
4. Conclusions
The COVID-19 pandemic has stressed our health care systems in an unprecedented way and underlined once more the important role of the molecular evolution succinctly described in this review. Within a few days from the first reported cases of anomalous pneumonia, significant progress was made in the fight against it: the virus was isolated, sequenced, identified and genetically characterized. It was named SARS-CoV-2 because of its phylogenetic relationship with SARS-CoV and bat SARS-like coronaviruses. Based on its genetic features, molecular and serological assays were developed and have been introduced in routine diagnostics. Pharmacological means have been gradually discovered and introduced, and general vaccine strategies have been developed or are in development. Trials are ongoing or are about to start to determine their effectiveness. This contribution has pictured the current research on the molecular evolution of the SARS-CoV-2 after its epidemic outbreak. Phylogenetic analysis and homology modeling have added knowledge to the fine details of the virus, and so have done the studies exploring the genome of the virus and the structure of its proteins. The search for viral variants with decreased or no pathogenic potential would be a significant step.
Funding
MG is supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;10(10):25–38. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;28(2):21–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Organization WHO . 2020. Coronavirus disease (COVID-19) pandemic. Accessed 2 Jul, 2020. [Google Scholar]
- 4.Organization WHO . 2020. Coronavirus Disease (COVID-19) Report. [Google Scholar]
- 5.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS- CoV-2. Gene Reports. 2020;16(19):100–682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benvenuto D., Giovanetti M., Ciccozzi M., Spoto S., Angeletti S., Ciccozzi M. The 2019- new coronavirus epidemic: evidence for virus evolution. J. Med. Virol. 2020;9(4):455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siew Woh C. Are pangolins scapegoats of the COVID-19 outbreak-CoV transmission and pathology evidence? J of the Society for conservation Biology. 2020;(8):9–19. 02. [Google Scholar]
- 8.Moya A., Elena S.F., Bracho A., Miralles R., Barrio E. The evolution of RNA viruses: a population genetics view. Proc. Natl. Acad. Sci. U.S.A. 2000;97(2):6967–6973. doi: 10.1073/pnas.97.13.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caccuri F. A persistently replicating SARS-CoV-2 variant derived from an asymptomatic individual. J. Transl. Med. 2020;2(9):13–98. doi: 10.1186/s12967-020-02535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pachetti M M. Emerging SARS-CoV-2 mutation hot spots include a novel RNA- dependent-RNA polymerase variant. J. Transl. Med. 2020;2(18):1–79. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korber B. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 04.29.069054. [Google Scholar]
- 12.Korber B. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(7):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(7):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland L.A. An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (january to March 2020) J. Virol. 2020;20(9):84–94. doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bal A. Molecular characterization of SARS-CoV-2 in the first COVID-19 cluster in France reveals an amino acid deletion in nsp2 (Asp268del) Clin. Microbiol. Infect. 2020;26(1):960–962. doi: 10.1016/j.cmi.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benedetti F., Snyder G.A., Giovanetti M., Angeletti S., Gallo R.C., Ciccozzi M., Zella D. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 2020;18(11):3–29. doi: 10.1186/s12967-020-02507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81(11):104–260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam M.R. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10(12):14–40. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jauregui A.R., Savalia D., Lowry V.K., Farrell C.M., Wathelet M.G. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PloS One. 2013;29(4):62–416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sajadi M. Temperature and latitude analysis to predict potential spread and seasonality for COVID-19. SSRN Electronic Journal. 2020;2(33):10–20. [Google Scholar]
- 21.Carleton T., Meng K.C. Causal empirical estimates suggest COVID-19 transmission rates are highly seasonal. medRxiv. 2020 03.26.20044420. [Google Scholar]
- 22.Ficetola G.F., Rubolini D. Climate affects global patterns of COVID-19 early outbreak dynamics. MedRxiv. 2020 03.23.20040501. [Google Scholar]
- 23.Wu X. Exposure to air pollution and COVID-19 mortality in the United States: a nationwide cross-sectional study. medRxiv. 2020 04.05.20054502. [Google Scholar]
- 24.Benedetti F., Pachetti M., Marini B., Ippodrino R., Gallo R.C., Ciccozzi M., Zella D. Inverse correlation between average monthly high temperatures and COVID-19-related death rates in different geographical areas. J. Transl. Med. 2020;18(22):22–51. doi: 10.1186/s12967-020-02418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandhi M., Rutherford G.W. Facial masking for covid-19 — potential for “variolation” as we await a vaccine. N. Engl. J. Med. 2020;9(1):8–10. doi: 10.1056/NEJMp2026913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abba Y. Antiviral activity of resveratrol against human and animal viruses. Adv Virol. 2005;18(12) doi: 10.1155/2015/184241. 4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]