Abstract
Objectives
This study intended to investigate the dynamics of anti-spike (S) IgG and IgM antibodies in COVID-19 patients.
Methods
Anti-S IgG/IgM was determined by a semi-quantitative fluorescence immunoassay in the plasma of COVID-19 patients at the manifestation and rehabilitation stages. The immunoreactivity to full-length S proteins, C-terminal domain (CTD), and N-terminal domain (NTD) of S1 fragments were determined by an ELISA assay. Clinical properties at admission and discharge were collected simultaneously.
Results
The positive rates of anti-S IgG/IgM in COVID-19 patients were elevated after rehabilitation compared to the in-patients. Anti-S IgG and IgM were not apparent until day 14 and day ten, respectively, according to Simple Moving Average analysis with five days’ slide window deduction. More than 90% of the rehabilitation patients exhibited IgG and IgM responses targeting CTD-S1 fragments. Decreased total peripheral lymphocytes, CD4+ and CD8+ T cell counts were seen in COVID-19 patients at admission and recovered after the rehabilitation.
Conclusions
Anti-S IgG and IgM do not appear at the onset with the decrease in T cells, making early serological screening less significant. However, the presence of high IgG and IgM to S1-CTD in the recovered patients highlights humoral responses after SARS-CoV-2 infection, which might be associated with efficient immune protection in COVID-19 patients.
Keywords: COVID-19, SARS-CoV-2, Spike glycoprotein, IgG and IgM, Dynamics
Background
The outbreak of COVID-19 first occurred in December 2019 in Wuhan, China, and is now prevalent in 110 countries (Korber et al., 2020). It is caused by a newly identified human coronavirus SARS-CoV-2 transmitting mainly through an airborne pathway (Huang et al., 2020, Zhu et al., 2020). Clinical manifestations include fever, tiredness, dry cough, and sometimes diarrhea. One out of six patients becomes a severe case with shortness of breathing, rapid progression to acute respiratory distress syndrome (ARDS), septic shock, and difficult-to-tackle metabolic acidosis, which become the principal causes of death (Lan et al., 2020). A rapid increase in the number of patients caused the World Health Organization (WHO) to declare COVID-19 as a pandemic after the outbreak. Disease control thus became a challenge.
Virological investigations reveal that SARS-CoV-2 belongs to a positive-strand RNA virus with 29.8 kilo-bases of the whole genome, consisting of six major open-reading frames (ORFs) encoding the replicase (ORF1ab), the spike (S) glycoprotein, the envelope (E) protein, the membrane (M) protein and the nucleocapsid (N) protein (Wu et al., 2020). It shares nearly 80% identity with SARS-CoV and 96% with BatCoV RaTG13 at the whole-genome level (Zhou et al., 2020, Zhu et al., 2020). Similar to SARS-CoV (Li et al., 2003), the S protein on the surface of SARS-CoV-2 virion mediates receptor recognition and membrane fusion with human angiotensin-converting enzyme 2 (ACE2) molecules (Walls et al., 2020, Wan et al., 2020, Yan et al., 2020). It contains three fragments, including an ectodomain, a transmembrane domain, and a short intracellular segment. The ectodomain consists of a receptor-binding subunit (S1) and a membrane-fusion subunit (S2). During viral infection, the C-terminal domain of the S1 fragment (S1-CTD) binds to the extracellular peptidase domain (PD) of ACE2 to facilitate viral attachment to the surface of target cells. The N-terminal domain of the S1 fragment (S1-NTD) is reported to bind to the glycans. The attachment triggers the cleavage of S protein between S1 and S2 fragments by cellular proteases, which in turn initiates the fusion of viral and cellular membranes by the S2 unit (Li, 2016). In humans, ACE2 is mainly expressed on type II pneumocytes, epithelial cells of colon and kidney, etc. (Hamming et al., 2004). Although the life cycle of SARS-CoV-2 in the host cells is directly from RNA-to-RNA replication, molecular epidemiological investigations demonstrate that the mutation rate of SARS-CoV-2 among different geographic locations or individuals is low (Ahmed et al., 2020). D164G is the main reported mutation site in S protein (Korber et al., 2020), which is favorable for vaccine development as well as the dissection of immunological processes.
Antigen-specific antibodies targeting viral proteins are responsible for the clearance of the virus in the late stage, either exerting the neutralizing activity to eliminate the virus directly or killing the infected host cells to avoid long-term infection (De Silva and Klein, 2015). Although most of the viral proteins are able to induce specific antibodies after SARS-CoV-2 infection, antibodies targeting viral S protein are more noteworthy because they can block the entry of SARS-CoV-2 into the host cells. The generation of anti-S antibodies is also an important indicator of infection, the amelioration, and the rehabilitation of the disease. Serological characteristics of anti-S IgG in the serum in COVID-19 patients have been reported (Li et al., 2020). A previous study using seven confirmed cases reported an increase of viral antibodies from day five after illness onset (Zhang et al., 2020). Several reports described the properties of anti-S antibodies and their interaction with ACE2, based on crystal structures (Cao et al., 2020b, Lan et al., 2020, Wang et al., 2020b), which supported the concept that antibodies targeting S protein in the rehabilitation individuals are functional enough for plasma transfusion-based therapy (Shen et al., 2020a).
In this study, with the advantages of serial collection of patient blood samples during treatment and in the rehabilitation period, we investigated the dynamics of IgM and IgG generation specifically targeting SARS-CoV-2 S protein and defined the property of the antibodies in the rehabilitation subjects.
Methods
Subjects
A total of 160 participants were enrolled in this study, including confirmed COVID-19 patients (N = 70) at the manifestation stage, convalescent COVID-19 patients (N = 33), non-COVID-19 pneumonia patients (N = 24), and healthy controls (HCs) (N = 33). All COVID-19 patients were in-hospital patients from Shanghai Public Health Clinical Center (Shanghai, China), and confirmed by at least one positivity of nucleotide acid detection using oral or nasal swabs recommended by the Chinese Center for Disease Control and Prevention (CCDC). The convalescent COVID-19 patients had at least two successive negativities of viral RNA in oral swabs with at least a 24 hours’ interval. They were followed-up for 1-2 weeks after the discharge. The non-COVID-19 pneumonia patients were recruited from Shanghai Ninth People’s Hospital. They displayed symptoms similar to COVID-19, including fever, dry cough, lowered lymphocyte count, and pulmonary CT image, whereas the negativity for virological detection in oral or nasal swabs occurred at least twice. HCs were healthy blood donors undergoing annual physical examinations in Shanghai Ninth People’s Hospital. They had no medical history or disease symptoms.
Blood sample collection and lymphocyte analysis
Whole blood was collected in Ethylene Diamine Tetraacetic Acid (EDTA) anticoagulation tubes (BD Biosciences, CA, USA). Plasma was collected after centrifugation at 2000 rpm for seven minutes, aliquoted, and stored at -80 °C. A blood Analyser (XE-5000, SYSMEX, Shanghai, China) was used to count the total peripheral lymphocytes and flow cytometry (BD FACSCanto II, BD, NJ, USA) for CD4+, CD8+, and CD3+ T cells.
Fluorescence Immunoassay (FIA) for the detection of anti-S IgG and Ig
An FIA assay was performed using the detection cards coated with fluorescence-labeled S protein (Sino Biological, Beijing, China) for IgG and IgM detection according to the manufacturer’s instructions (Dialab ZJG Biotech Co, Suzhou, China). Briefly, 10 μL plasma was mixed in 990 μL dilution buffers. 80 μL diluted solution was added to the sampling well of the detection cards. The fluorescence signal was captured by DL300 Quantitative Immunofluorescent Analyzer within 15 min. The cutoff value for IgG positivity was 15, while 3.4 was the cutoff value for IgM positivity. Anti-S IgG and IgM levels were represented by the values of the fluorescence signal.
ELISA assay
An enzyme-linked immunosorbent assay (ELISA) that was developed by Wuxi Diagnostics (Shanghai, China) and was carried out according to the manufacturer’s instructions. Briefly, 5 μL plasma and 95 μL Sample Dilutent was added to 96-well polystyrene plates (Corning, NY, USA), coated with full-length S proteins (both from Sino Biological Inc., Beijing, China), S1-CTD and S1-NTD fragments (both from Shanghai Tolo Biotech, Shanghai, China), and incubated at 37 °C for 30 min. After washing three times with Wash Buffer (1×), the wells were incubated with Enzyme Conjugate for 30 min at 37 °C. After washing three times with Wash Buffer (1×), 50 μL of Chromogenic Reagent A and B were added respectively to each well and incubated at 37 °C for ten min. 50 μL of Stop Solution was added to each well, and the absorbance at 450 nm was detected within five min by using PowerWaveXS2 microplate spectrophotometer (BioTek Instruments, Inc., VT, USA).
Statistical analysis
The descriptive data were represented by mean ± S.E.M. or median (range). All statistical analyses were performed using SPSS 20.0 statistics software (IBM Corp., NY, USA) or Graphpad Prism 5.0 (Graphpad Software Inc., CA, USA). Statistical significance was calculated using paired or unpaired t-tests for the comparisons between two groups, while the One-way ANOVA test was used for at least three groups. The Chi-square test was performed to analyze the significance of clinicopathological characteristics and the comparisons of positivity in ELISA data. The correlation between the days of hospital stays and anti-S IgG levels was analyzed by the Pearson test. Results were considered statistically significant when the two-tailed P value was < 0.05.
A Simple Moving Average (SMA) with a five days’ slide window was calculated, based on anti-S IgG and IgM's fluorescence signal values to reduce the variations of individual fluorescence values. The SMA values were plotted with the sampling days (the day between the disease onset and sample collection) using Excel (Microsoft, USA).
Results
Demographic characteristics and the treatments of COVID-19 patients
A total of 160 subjects were enrolled in this study. All 103 COVID-19 patients were confirmed in the Shanghai Public Health Clinical Center and received further treatments (Ling et al., 2020). Twenty-four non-COVID-19 pneumonia patients recruited for this study were excluded from the COVID-19 group after two nucleic acid tests yielded negative results. Thirty-three healthy donors were enrolled as the controls. The median ages were comparable between the confirmed COVID-19 patients (44y, 21y–83y) and non-COVID-19 patients (36y, 21y–81y), whereas the healthy controls were younger (26y, 20y–56y). All the COVID-19 patients received supportive treatments included oxygen therapy and subcutaneous injection of thymosin twice a week. Antibiotics and glucocorticoids were strictly adapted during the treatment.
Antibody profiles against S protein in COVID-19 patients
A fluorescence immunoassay (FIA) was performed to determine the positive rates of S protein-specific IgG and IgM in 186 samples, including 129 confirmed COVID-19 samples (33 convalescent samples and 96 manifestation samples), 24 non-COVID-19 samples, and 33 HC samples. The assay we utilized exhibited high specificity with no positivity in non-COVID-19 pneumonia patients and HCs (Table 1 ). Of 33 convalescent samples, 96.97% (32/33) of the samples were positive for anti-S IgG and 87.88% (29/33) for IgM. Among 96 manifestation samples from 70 individual patients (median sampling day: 14 (2–37 days)), 66.7% (64/96) were IgG positive, and 57.3% (55/96) were IgM positive, which were both significantly lower than those in the convalescent group (P = 0.0003 and 0.0013, respectively). Additionally, there was a large difference in IgG/IgM double-negative samples between two groups (manifestation: 30.2% (29/96) vs convalescent: 0%; P < 0.0001) (Table 2 ). Thus, our findings indicated that the positive rates of both anti-S IgG and IgM were significantly higher at the convalescent stage than the manifestation stage.
Table 1.
Positive rates of anti-spike IgG and IgM in the enrolled subjects.
| COVID-19 | Non-COVID-19 | Healthy controls | |
|---|---|---|---|
| Total | 129 | 24 | 33 |
| IgG+ | 74.41% (96/129) | 0 | 0 |
| IgM+ | 65.12% (84/129) | 0 | 0 |
| IgG-IgM- | 22.48%(29/129) | 100% (24/24) | 100% (33/33) |
Table 2.
Comparison of anti-S IgG and IgM positivity in COVID-19 patients between manifestation and convalescent periods.
| COVID-19 Manifestation | COVID-19 Convalescent |
P valueb | |
|---|---|---|---|
| Total | 96 | 33 | |
| Sampling daya (Median: range) | 14 (2–37) | 27 (18–43) | |
| IgG+ | 66.7% (64/96) | 96.97% (32/33) | 0.0003 |
| IgM+ | 57.3% (55/96) | 87.88% (29/33) | 0.0013 |
| IgG- IgM- | 30.2% (29/96) | 0% (0/33) | <0.0001 |
The day between the disease onset and sample collection.
By Chi-square test.
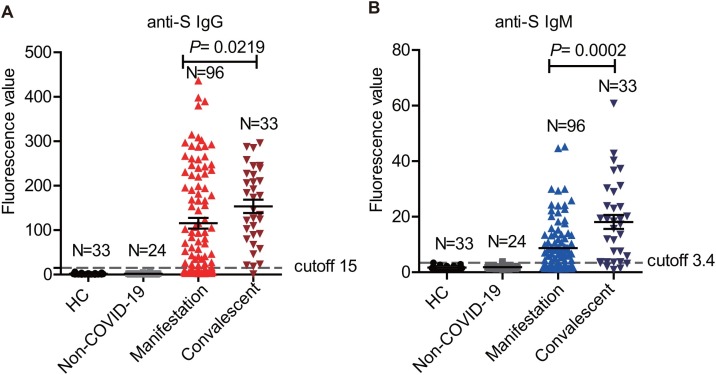
We directly compared the anti-S lgG and IgM levels among the four groups based on semi-quantitative fluorescence signal values. It was evident that IgG and IgM’s average signals in non-COVID-19 pneumonia patients and HCs were lower than cutoff values of 15 and 3.4, respectively. Consistent with the positive rates, IgG and IgM’s average signals in the convalescent group were 153.50 ± 15.03 and 18.13 ± 2.53, respectively. Both were higher than those in the manifestation group (115.46 ± 12.08 for anti-S IgG, P = 0.0219; 8.69 ± 0.95 for anti-S IgM, P = 0.0002), in which the difference in IgM was more prominent (Figure 1 A and B).
Figure 1.
The positive rates of anti-spike protein IgG and IgM in COVID-19 confirmed samples during the manifestation and the convalescent periods.
(A) Comparison of the anti-spike protein IgG levels in the convalescent samples (n = 33), the manifestation samples (n = 96), the non-COVID-19 samples (n = 24) and healthy controls (n = 33) (P = 0.0219, Unpaired t-test).
(B) Comparison of the anti-spike protein IgM levels in the convalescent samples (n = 33), the manifestation samples (n = 96), the non-COVID-19 samples (n = 24) and healthy controls (n = 33) (P = 0.0002, Unpaired t-test).
Assessment of the occurrence duration of anti-S IgG and IgM upon SARS-CoV-2 infection
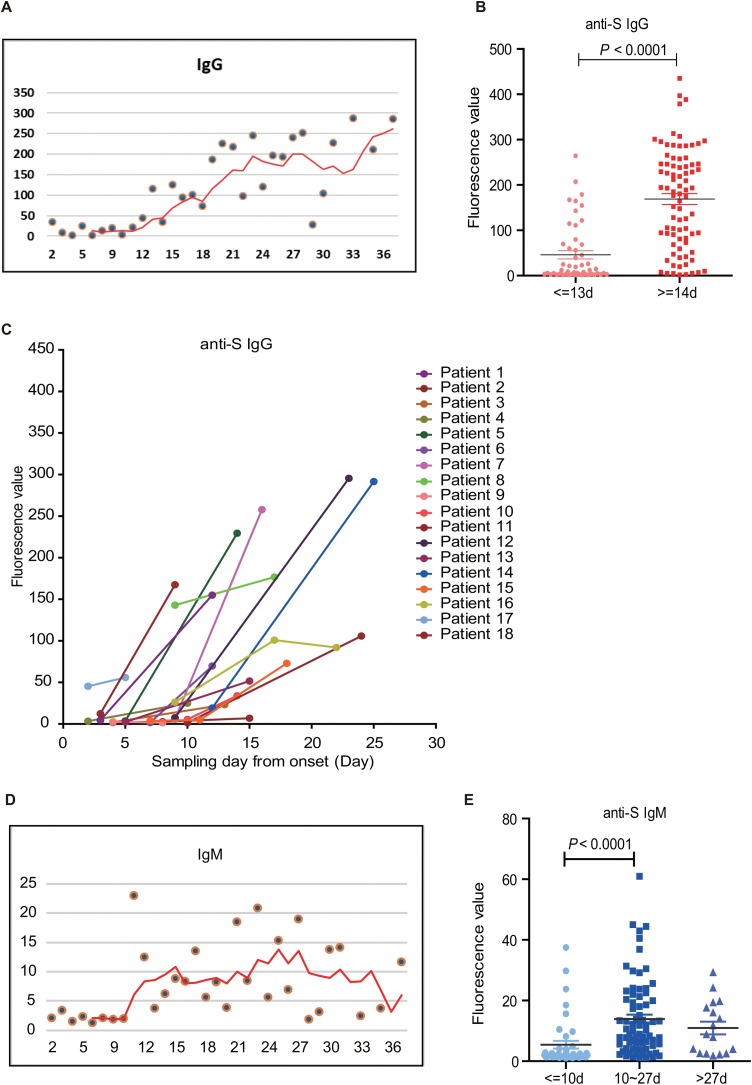
Since SARS-CoV-2 is a newly emerging virus, determination of the occurrence periods of anti-S IgG and IgM is very informative both for early diagnosis and vaccine evaluation. Owing to the corresponding anti-S IgG and IgM signal values with the time points of sample collection, we intended to deduce their antibody production window periods. Using the 5-day Simple Moving Average (SMA) trajectory of anti-S IgG median values, it was found that anti-S IgG remained at a very low level from the onset of the disease until day 12, where there exhibited an apparent increase in fluorescence signals. The signal values increased until day 22 and remained at that plateau later on (Figure 2 A). When we sub-grouped the samples by day 14, a typical time point for antibody generation after a viral infection, it showed that the average value of anti-S IgG since day 14 was dramatically higher than that before day 14 (P < 0.0001) (Figure 2B). When testing blood samples at two or three time points of individual COVID-19 patients, it was apparent that anti-S IgG in most patients also exhibited a dramatic increase after 14 days (Figure 2C).
Figure 2.
Assessment of the window periods for the occurrence of anti-spike protein IgG and IgM upon SARS-CoV-2 infection.
(A) The Simple Moving Average (SMA) of anti-spike protein IgG in COVID-19 patients with five days’ slide window. Red line: the regression curve of anti-spike IgG with the time moving in the duration from two days to 37 days.
(B) Comparison of anti-spike protein IgG levels in COVID-19 patients between the duration of 0–13 days and more than 14 days (P < 0.0001, Unpaired t-test).
(C) Anti-S IgG in 18 COVID-19 patients with two or three samples at different time points.
(D) The Simple Moving Average (SMA) of anti-spike protein IgM in COVID-19 patients with five days’ slide window. Red line: the regression curve of anti-spike IgM with the time moving in the duration from two days to 37 days.
(E) Comparison of anti-S IgM levels in COVID-19 patients at different sampling days: 0–10 days, 10–27 days, and more than 27 days (P < 0.0001, Unpaired t-test).
Similarly, anti-S IgM also exhibited very low fluorescence values at the onset until day ten with a rapid increase. The plateau peaked on day 15 and maintained until day 27, then slowly descended afterward (Figure 2D). When we compared the fluorescent values based on the time points deduced from the SMA analysis, it was evident that the average fluorescence value before day ten was the lowest and that between day ten and day 27 was the highest. After day 27, the average level of anti-S IgM started to descend (Figure 2E).
Dominant domain of S protein recognized by specific antibodies in rehabilitation patients
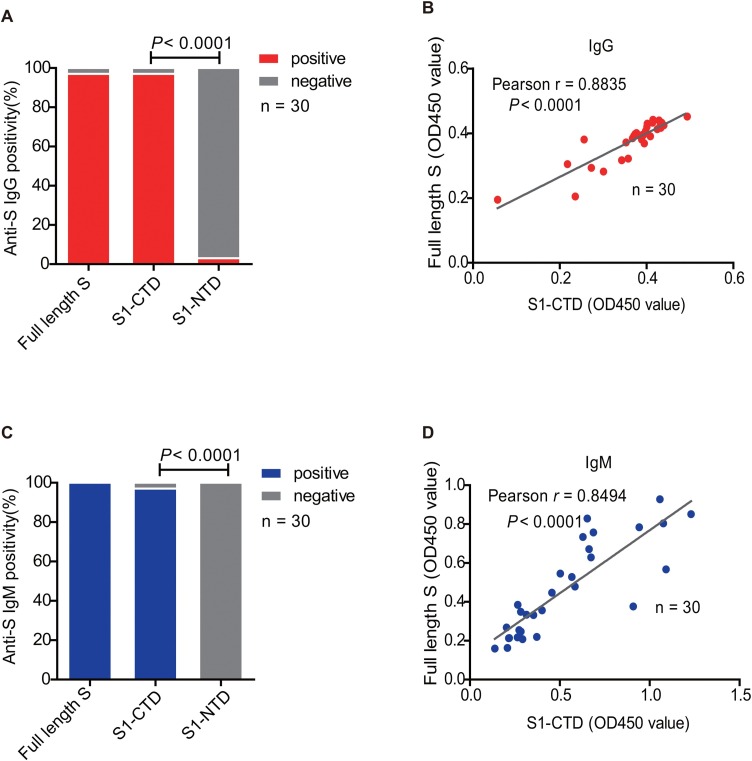
SARS-CoV-2 S protein binds to human ACE2 molecule in a very similar pattern to SARS-CoV according to the crystal structures (Gui et al., 2017, Song et al., 2018, Yan et al., 2020). To investigate the targeting fragments of anti-S protein IgG and IgM, full-length S protein, S1-NTD, and S1-CTD fragments were prepared and subjected to ELISA assays. Our results revealed that 96.67% (29/30) of the convalescent COVID-19 patients exhibited IgG positivity to full-length S protein (Figure 3 A), which were all immunoreactive to S1-CTD fragment (96.67%, 29/30) (Pearson r = 0.8835, P < 0.0001) (Figure 3B). Only 3.33% (1/30) of the convalescent patients displayed IgG immunoreactivity to S1-NTD in our assay (Figure 3A). Similar IgM immunoreactivity patterns to S protein fragments were observed (Figure 3C and D). Therefore, both anti-S IgG and IgM in the plasma of the convalescent COVID-19 patients mainly target the S1-CTD fragment rather than S1-NTD.
Figure 3.
Determination of the Immunoreactivity to full-length spike protein and two S1 fragments in the plasma of the convalescent COVID-19 patients.
An ELISA assay was performed using full-length, S1-NTD, and S1-CTD as coating antigens to determine the samples' immunoreactivity from convalescent COVID-19 patients.
(A) Positive rates of IgG to full-length spike protein and two S1 fragments in the plasma of the convalescent COVID-19 patients (P < 0.0001, Chi-square).
(B) Correlation between IgG reactivity to full-length spike protein and S1-CTD fragment (P < 0.0001, Pearson r = 0.8835).
(C) Positive rates of IgM to full-length spike protein and two S1 fragments in the plasma of the convalescent COVID-19 patients (P < 0.0001, Chi-square).
(D) Correlations between IgM reactivity to full-length spike protein and S1-CTD fragment (P < 0.0001, Pearson r = 0.8494).
S: spike protein; S1-CTD: C-terminal domain of S1 fragment; S1-NTD: N-terminal domain of S1 fragment.
Lymphocyte features in COVID-19 patients
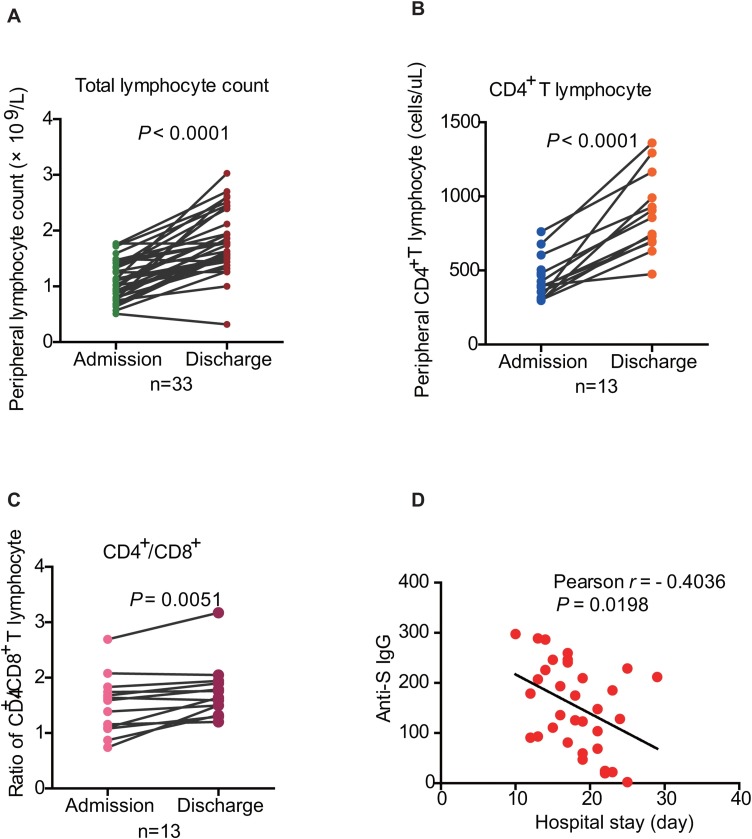
Clinical reports indicate that lymphopenia is one of the common symptoms at the early stage of COVID-19 infection (Wang et al., 2020a). The decrease in peripheral total lymphocyte counts has been considered an important indicator for the disease and its severity in COVID-19 patients (Ling et al., 2020). We also tracked the peripheral total lymphocyte dynamics at admission and discharge in 33 convalescent patients (Table 3 ). The average days of hospital stay were 18.15 ± 4.47 days. At admission, the average total lymphocyte counts were (1.11 ± 0.06) × 109/L. 51.52% of the onset patients (17/33) had lymphocyte counts below the lower limit (1.10–3.20 × 109/L). A significant increase in lymphocyte counts was observed up to the average (1.81 ± 0.10) × 109/L at discharge when compared to the admission point (P < 0.0001) (Table 3 and Figure 4 A). Among 17 patients with lowered lymphocyte counts at admission, 15 (15/17, 88.90%) were recovered to the normal levels, whereas two patients remained below the cutoff value (1.00 × 109/L and 0.32 × 109/L, respectively) (Figure 4A). There exhibited dramatic decreases in the average of CD3+ T cell counts (721.88 ± 45.12 cells/μL; cutoff value: 1141 cells/μL), CD4+ T cell count (395.09 ± 29.06 cells/μL; cutoff value: 478 cells/μL) as well as CD8+ T cell counts (291.91 ± 20.47 cells/μL; cutoff value: 393 cells/μL). The CD4/CD8 ratio was 1.48 ± 0.11, which was at the normal levels (normal ratio: 0.87–2.29) (Table 3). CD3+, CD4+, and CD8+ T lymphocyte counts were compared in 13 convalescent patients between admission and discharge time points. It was shown that the counts of CD4+ T lymphocyte at discharge increased dramatically in 13 patients (average: 882.31 ± 73.10 cells/μL) when compared to that at admission (average: 445.40 ± 36.51 cells/μL) (P < 0.0001) except for one patient (from 398 to 475 cells/μL) (Figure 4B). While total CD3+ (P < 0.0001) and CD8+ T cell counts (P = 0.0003) of 13 patients also increased at discharge (Figure S1A and S1B), the increase in CD4/CD8 ratio (P = 0.0051) (Figure 4C) was apparent, indicating that the restoration of the CD4+ T cell pool was more significant. Interestingly, there was a negative correlation between the days of hospital stays and the patients’ anti-S IgG titer (Pearson r = −0.4036, P = 0.0198) (Figure 4D). These data revealed that although COVID-19 patients exhibited a decrease in lymphocyte pools at the onset of the disease, most of them could restore T cell pools after recovery. The moderate increase in CD4/CD8 ratio implies the expansion of CD4+ T cells to a greater extent.
Table 3.
Lymphocyte features of COVID-19 patients at admission and at discharge.
| Admission | Discharge | P-valueb | Cutoff value | |
|---|---|---|---|---|
| Hospital stay (day): | 18.15 ± 4.47 | |||
| WBC(×109/L) | 5.36 ± 0.60 | 5.80 ± 0.30 | 0.3568 | 3.95–9.5 |
| N(×109/L) | 3.75 ± 0.57 | 3.31 ± 0.24 | 0.3358 | 1.8–6.3 |
| L(×109/L) | 1.11 ± 0.06 | 1.81 ± 0.10 | < 0.0001 | 1.1–3.2 |
| CD4 (cell/μL) | 395.09 ± 29.06 | 882.31 ± 73.10a | < 0.0001 | 478–1440 |
| CD8 (cell/μL) | 291.91 ± 20.47 | 525.92 ± 42.93a | 0.0003 | 393–1250 |
| CD3 (cell/μL) | 721.88 ± 45.12 | 1488.46 ± 116.28a | < 0.0001 | 1141–1920 |
| CD4/CD8 | 1.48 ± 0.11 | 1.74 ± 0.14a | 0.0051 | 0.87–2.29 |
n = 13.
By paired t-test.
Figure 4.
Lymphocyte features in the COVID-19 patients.
(A) The comparative analysis of total lymphocyte counts at admission and discharge of 33 patients (P < 0.0001, paired t-test).
(B) The comparative analysis of CD4+ T lymphocytes at admission and discharge of 13 patients (P < 0.0001, paired t-test).
(C) The comparative analysis of the ratio of CD4+/CD8+ T lymphocytes at admission and discharge of 13 patients (P = 0.0051, paired t-test).
(D) The correlation between hospital stays and anti-S IgG titers in COVID-19 patients (P = 0.0198, Pearson r = −0.4036).
Discussion
Unlike low transmission and high mortality of SARS-CoV and MERS-CoV infections that occurred some years ago (WHO), SARS-CoV-2 spreads more prevalently, leading to substantial more infected cases in China as well as worldwide. The virological property of SARS-CoV-2 facilitates the development of serological assays upon SARS-CoV-2 infection. In the present study, we have explicitly analyzed anti-S IgG and IgM generation dynamics in COVID-19 patients. At the early stage of the illness, both IgG and IgM levels were very low and reached a high level after recovery. When we analyzed the clinical information collected in parallel (Supplementary Tables S1–S4), it was found that impaired cell counts of peripheral lymphocytes including CD3+, CD8+, and CD4+ T cells were the notable features of SARS-CoV-2 patients, which is similar to HIV infection (Gottlieb et al., 1981). Together with the restoration of periphery lymphocyte pools, the anti-S IgG and IgM increased and reached a certain level after treatments. Therefore, the recovery of lymphocyte numbers might be a significant event associated with the induction of viral-specific anti-IgG and IgM antibodies. This is entirely rational. It is well known that CD4+ T cells play an essential role in mediating adaptive humoral immunity. Upon viral infection, host CD4+ T cells are activated and differentiated into functional subsets, including type 2 help T cells (Th2) to assist B cell activation and trigger humoral immunity. With a deficiency of CD4+ T cells, humoral immunity is impaired (Xin et al., 2018). In fact, our previous study reported that CD4+ T lymphocyte count at admission was inclined to predict the duration of SARS-CoV-2 viral RNA detection (Ling et al., 2020). In this study, total lymphocyte and CD4+ T cell counts and the CD4/CD8 ratio increased in almost all patients, with the clearance of SARS-CoV-2 at discharge. Interestingly, we also observed that the hospital stay was negatively correlated with the anti-S IgG titer, suggesting that anti-S IgG might be protective in COVID-19 patients. Therefore, the recovery of T cell homeostasis with antibody induction might be indicative of the clearance of the virus in the rehabilitation of COVID-19 patients.
Being a newly emerging virus, the generation kinetics of SARS-CoV-2-specific IgM and IgG is very important for diagnosis and treatment. According to the WHO's recommendation, the estimated latent period for COVID-19 infection is suggested to be around five days ranging from 1–14 days (WHO). We have observed that the durations to induce high levels of both anti-S IgG and IgM upon SARS-CoV-2 infection were 14 days and ten days, respectively. Another interesting issue is whether viral-specific IgA levels are generated earlier due to the airborne infection, which can be investigated (He et al., 2015). How long antiviral antibodies will exist after the recovery is more important. A cohort study of 56 convalescent patients with SARS-CoV has shown that virus antigen-specific IgG antibodies peaked at month four after the onset of disease but decreased after that (Liu et al., 2006). For SARS-CoV-2-specific antibodies, a recent investigation reveals that a similar duration of antiviral responses exists after the recovery (Gudbjartsson et al., 2020). Because of the occurrence of re-infection cases in Europe and the USA (Hoang et al., 2020, Yuan et al., 2020), whether the existing antiviral antibodies exert protection against re-infection needs to be urgently investigated in the future through a long-term follow-up.
At present, no antiviral drug is demonstrated to be completely effective in treating COVID-19 (Cao et al., 2020a). Antibody-based treatment is one of the promising ways with more specificity and success in several studies, such as Zika virus (Bailey et al., 2018, Heffron et al., 2018), Hepatitis Virus C (Brown et al., 2007), SARS-CoV as well as SARS-CoV-2 infection (Butler, 2015, Hui et al., 2018, Mo and Fisher, 2016, Shang et al., 2007). Since patient plasma contains polyclonal antibodies targeting multiple epitopes of different viral proteins, convalescent plasma therapy's success mostly relies on the broad spectrum in different individuals. Fortunately, the SARS-CoV-2 virus remains genome-stable among infected populations (Shen et al., 2020b). When we investigated the possible targets of convalescent plasma, especially in the S protein, it was found that the fragment targeted by anti-S IgG and IgM was mainly S1-CTD, whereas very low immunoreactivity was observed to S1-NTD. S1-CTD is mainly responsible for binding to ACE2, while S1-NTD is responsible for binding sugar (Wrapp et al., 2020). The possible reasons for the dramatic difference in immune reactivity to two S1 fragments may lie in the fact that the S1-NTD fragment displays a ceiling-like structure that would protect their sugar-binding site from host immune surveillance (Li, 2016). However, the exact mechanisms of the difference in the diverse immunoreactivity to S1-CTD and S1-NTD needs to be further investigated. Nevertheless, the high positivity of anti-S IgG and IgM to the S1-CTD fragment in convalescent patients provides strong support for the plasma transfusion therapy.
Our results collectively demonstrated that both anti-S IgG and IgM do not appear at the onset of the disease. However, the higher titers of anti-S IgG in the rehabilitation COVID-19 patients might be useful to determine the latently infected population by using serological screening after the pandemic. The dominant recognition of the convalescent plasma to S1-CTD fragment of the S protein provides supportive evidence of the plasma transfusion application for the treatment of severe COVID-19 patients.
Funding
This work was supported by the Chinese National Mega Science and Technology Program on Infectious Diseases [grant number 2018ZX10731301-001-004]; Shanghai Academic Research Leader Project [grant number 2018XD1403300]; and the Emergency Project of Shanghai Institute of Immunology. The Development of the Diagnostic Kit for 2019 Novel Coronavirus granted by Shanghai Ecnomic and Informatization Commission.
Conflicts of interest
The funders had no role in study design, data collection, analysis, decision to publish, or manuscript preparation. All the authors declare no conflict of interest.
Author contributions
Conceived the project: YW and JX. Designed the experiments: YW, JX, and GPZ. Performed the experiments: YJB, YYC, DT, YL, XHZ, and HH. Collected the samples and clinical data: YL, DT, and HZL. Analyzed the data: YJB, YYC, YW, JX, GPZ, and BS. Wrote the manuscript: YJB, YYC, JX, and YW. Obtained funding: YW. YW, HZL, and JX are senior authors.
Ethical approval
This study was approved by the Ethical Committee of Shanghai Ninth People's Hospital (SH9H-2020-TK9-1) and the Shanghai Ethics Committee for Clinical Research (SECCR/2020-04-01).
Acknowledgments
We appreciated Dr. Feng Wang, Dr. Lei Chen, and Dr. Fu-bing Li from the Shanghai Institute of Immunology for their informative discussion.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.12.014.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M.J., Duehr J., Dulin H., Broecker F., Brown J.A., Arumemi F.O., et al. Human antibodies targeting Zika virus NS1 provide protection against disease in a mouse model. Nat Commun. 2018;9(1):4560. doi: 10.1038/s41467-018-07008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.S., Ryder S.D., Irving W.L., Sim R.B., Hickling T.P. Mannan binding lectin and viral hepatitis. Immunol Lett. 2007;108(1):34–44. doi: 10.1016/j.imlet.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Butler D. Ebola raises profile of blood-based therapy. Nature. 2015;517(7532):9–10. doi: 10.1038/517009a. [DOI] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of Lopinavir-Ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva N.S., Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15(3):137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb M.S., Schroff R., Schanker H.M., Weisman J.D., Fan P.T., Wolf R.A., et al. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N Engl J Med. 1981;305(24):1425–1431. doi: 10.1056/NEJM198112103052401. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383(18):1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Mullarkey C.E., Duty J.A., Moran T.M., Palese P., Miller M.S. Broadly neutralizing anti-influenza virus antibodies: enhancement of neutralizing potency in polyclonal mixtures and IgA backbones. J Virol. 2015;89(7):3610–3618. doi: 10.1128/JVI.03099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron A.S., Mohr E.L., Baker D., Haj A.K., Buechler C.R., Bailey A., et al. Antibody responses to Zika virus proteins in pregnant and non-pregnant macaques. PLoS Negl Trop Dis. 2018;12(11) doi: 10.1371/journal.pntd.0006903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang V.T., Dao T.L., Gautret P. Recurrence of positive SARS-CoV-2 in patients recovered from COVID-19. J Med Virol. 2020;92(11):2366–2367. doi: 10.1002/jmv.26056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., Lee N., Chan P.K., Beigel J.H. The role of adjuvant immunomodulatory agents for treatment of severe influenza. Antiviral Res. 2018;150:202–216. doi: 10.1016/j.antiviral.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., et al. Development and clinical application of A rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Fontanet A., Zhang P.H., Zhan L., Xin Z.T., Baril L., et al. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis. 2006;193(6):792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y., Fisher D. A review of treatment modalities for Middle East Respiratory Syndrome. J Antimicrob Chemother. 2016;71(12):3340–3350. doi: 10.1093/jac/dkw338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G., Biggerstaff B.J., Yang B., Shao C., Farrugia A. Theoretically estimated risk of severe acute respiratory syndrome transmission through blood transfusion during an epidemic in Shenzhen, Guangdong, China in 2003. Transfus Apher Sci. 2007;37(3):233–240. doi: 10.1016/j.transci.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., et al. Genomic diversity of SARS-CoV-2 in Coronavirus Disease 2019 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin G., Zander R., Schauder D.M., Chen Y., Weinstein J.S., Drobyski W.R., et al. Single-cell RNA sequencing unveils an IL-10-producing helper subset that sustains humoral immunity during persistent infection. Nat Commun. 2018;9(1):5037. doi: 10.1038/s41467-018-07492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B., Liu H.Q., Yang Z.R., Chen Y.X., Liu Z.Y., Zhang K., et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci Rep. 2020;10(1):11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.