Abstract
An immunoinformatics-based approach was used to identify potential multivalent subunit CTL vaccine candidates for SARS-CoV-2. Criteria for computational screening included antigen processing, antigenicity, allergenicity, and toxicity. A total of 2604 epitopes were found to be strong binders to MHC class I molecules when analyzed using IEDB tools. Further testing for antigen processing yielded 826 peptides of which 451 were 9-mers that were analyzed for potential antigenicity. Antigenic properties were predicted for 102 of the 451 peptides. Further assessment for potential allergenicity and toxicity narrowed the number of candidate CTL epitopes to 50 peptide sequences, 45 of which were present in all strains of SARS-CoV-2 that were tested. The predicted CTL epitopes were then tested to eliminate those with MHC class II binding potential, a property that could induce hyperinflammatory responses mediated by TH2 cells in immunized hosts. Eighteen of the 50 epitopes did not show class II binding potential. To our knowledge this is the first comprehensive analysis on the proteome of SARS-CoV-2 for prediction of CTL epitopes lacking binding properties that could stimulate unwanted TH2 responses. Future studies will be needed to assess these epitopes as multivalent subunit vaccine candidates which stimulate protective CTL responses against SARS-COV-2.
Keywords: SARS-CoV-2, CTL epitopes, Epitope mapping
1. Introduction
Coronaviruses (CoVs), first reported in the 1930s, are a group of pathogenic viruses belonging to the family Coronoviridae and order Nidovirales. They are known to cause respiratory or intestinal infections in humans and in various animals (Cheng et al., 2007). Studies have suggested that up to 30% of all common colds could be attributed to coronaviruses (El-Sahly et al., 2000).
CoVs are enveloped and have a non-segmented, positive-sense, single strand RNA genome with size ranging from 26,000 to 37,000 bases (Young et al., 2020). This is the largest known genome among RNA viruses (Weiss and Navas-Martin, 2005). In 2003, a coronavirus named Severe Acute Respiratory Syndrome-associated Coronavirus (SARS-CoV) caused a global outbreak affecting more than 8000 people with a mortality rate of approximately 10% (Cheng et al., 2007; Parry, 2003). SARS-CoV was reported to cause more severe disease than the other previously known human coronaviruses including high fever, shortness of breath, and atypical pneumonia (Ksiazek et al., 2003).
The recent outbreak in China and global spread of a novel coronavirus similar to SARS-CoV named SARS Coronavirus 2 (SARS CoV-2) has caused a major pandemic. Initial comparisons revealed that SARS-CoV-2 was approximately 79% similar to SARS-CoV at the nucleotide level. Of course, patterns of similarity vary greatly between genes, and SARS-CoV and SARS-CoV-2 exhibit only ~72% nucleotide sequence similarity in the spike (S) protein, the key surface glycoprotein that interacts with host cell receptors (Zhang and Holmes, 2020).
Although the genome of SARS-CoV-2 exhibits a higher sequence homology to Bat-CoV-RaTG13 isolated from Rhinolophus affinis the origin of the virus is still controversial. While the intermediate host of SARS-CoV-2 has not been confirmed, reports suggest that pangolins are the likely host due to 99% sequence homology between the consensus sequence derived from metagenomic samples from the pangolin species and SARS-CoV-2 (Zhang et al., 2020). The SARS-CoV-2 genome contains 14 ORFs that encode 27 proteins. The orf1ab and orf1a genes are located at the 5′-terminus of the genome and encode 15 non-structural proteins (NSPs) from NSP1 to NSP10, and from NSP12 to NSP16. The 3′-terminus of the genome contains 4 structural proteins (S, E, M and N) and 8 accessory proteins (3a, 3b, p6, 7a, 7b, 8b, 9b and orf14). Though the two viruses, SARS-CoV and SARSCoV-2, are quite similar with respect to amino acid sequences among the proteins they share, there are some notable differences. The 8a protein is present in SARS-CoV and absent in SARS-CoV-2; the 3b protein is 154 amino acids in SARS-CoV but shorter in SARS-CoV-2 with only 22 amino acids. The S protein is responsible for target cell receptor-binding (angiotensin converting enzyme-2 [ACE2]) and subsequent viral entry into host cells. It comprises an N-terminal S1 subunit responsible for virus–receptor binding and a C-terminal S2 subunit responsible for virus–cell membrane fusion (Du et al., 2009, Du et al., 2017). The M and E proteins play important roles in viral assembly, and the N protein is necessary for RNA synthesis (Tian et al., 2020).
Neutralizing antibodies (nAb) specific for viral pathogens induced by vaccines or viral infections play crucial roles in preventing future disease. Currently developed SARS-CoV- and MERS-CoV-specific nAbs include monoclonal antibodies, their functional antigen-binding fragment, the single-chain variable region fragment, or single-domain antibodies (Jiang et al., 2020). They target receptor-binding domains (RBD) for these viruses and therefore interfere with S2-mediated membrane fusion or entry into the host cell, thus inhibiting viral infections. No SARS-CoV-2-specific nAbs have been confirmed although there are emerging reports that suggest SARS-CoV- and MERS-CoV-specific nAbs have potential cross-neutralizing activity against SARS-CoV-2 infection (Jiang et al., 2020).
In recent years, epitope prediction algorithms have been developed, validated and proven to be accurate tools for predicting the binding affinities of peptides to MHC molecules, with wide allelic coverage (Lundegaard et al., 2010; Nielsen and Andreatta, 2016). The discovery of T-cell epitopes derived from Vaccinia virus validated the power and value of prediction of computational in silico analysis for identifying CD8 + T-cell epitopes on a large scale (Moutaftsi et al., 2006). In that pioneering work, 49 CTL epitopes were found, and these were shown to account for 95% of the total Vaccinia virus CD8 response. Since then, enumerable studies have used in silico epitope discovery approaches to identify putative epitopes for inclusion in rationally designed multi-valent vaccines aimed at preventing infections caused by an array of viral and bacterial pathogens. For example, studies have shown that epitope-based vaccines can effectively elicit protective immune responses against various pathogens, such as HIV and influenza viruses (Oyarzún and Kobe, 2016; Ben-Yedidia and Arnon, 2007). Delivery of epitopes as DNA vaccines (Wilson et al., 2003) or DNA-launched nanoparticle vaccines (Xu et al., 2020) are known to induce strong CTL responses.
To date, few studies have analyzed or tested the possibility of using CoV proteins as vaccine targets for cytotoxic T lymphocyte (CTL)-based cellular immunity which is essential for combating any virus infection such as the one caused by SARS-CoV-2 (Baruah and Bose, 2020; Bhattacharya et al., 2020; Grifoni et al., 2020a). These studies only analyzed potential epitopes present in the surface glycoprotein (spike). In this study, we undertook a comprehensive evaluation of the SARS-CoV-2 proteome. Such an approach for screening genome-wide SARS-CoV-2 epitopes could lead to the design and development of an effective CTL-based multivalent subunit vaccine (Rappuoli et al., 2016; Bruno et al., 2015). Identification of candidate CTL epitopes was carried out using immune epitope database (IEDB) tools for the major population distribution of class I and class II HLA loci (Fleri et al., 2017; Yao et al., 2013; Mohan et al., 2018). The results of the present study identify 18 potential CTL epitopes that lack predicted ability to bind MHC class II molecules that could promote harmful hyperinflammatory respiratory pathogenesis (cytokine storms) seen in some COVID-19 patients. Results support the idea that immunoinformatic-driven immunogen screening is a promising strategy to accelerate vaccine development for emerging highly pathogenic pathogens.
2. Materials and methods
2.1. Collection of proteome data
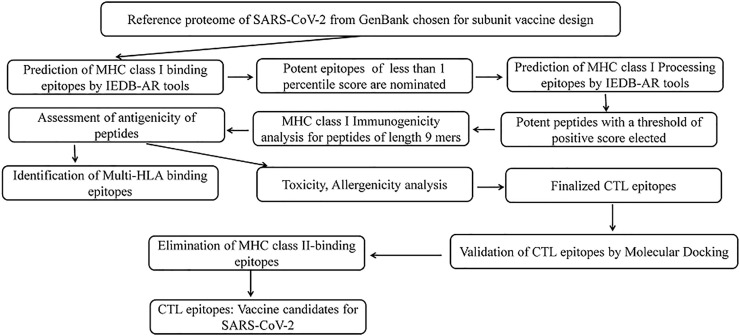
The complete proteome of SARS-CoV-2 was retrieved from NCBI-GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). The accession number of reference proteome is NC_045512. All the proteins of the proteome were individually subjected to the reverse vaccinology workflow for prediction of CTL epitopes as shown in Fig. 1 .
Fig. 1.
Illustration of systematic reverse vaccinology work flow for prediction of SARS-CoV-2 CTL epitopes.
2.2. CTL epitope prediction
MHC class I-binding SARS-CoV-2 epitopes of 8, 9, 10 and 11 amino acids in length were predicted using the IEDB-AR resource (http://tools.iedb.org/mhci/) (Fleri et al., 2017). The HLA-A alleles selected for the study included A*0101, A*2402, A*2601, A*3101, A*3303, A*0201, A*0206, A*0301, A*1101. These were chosen because they are known to be widely distributed in the population worldwide (Yao et al., 2013; Mohan et al., 2018). CTL epitopes were predicted using various methods such as Artificial Neural Network (ANN), Stabilized Matrix Method (SMM, Scoring Matrices derived from Combinatorial Peptide Libraries; Comblib_Sidney 2008), Consensus, NetMHCpan (Fleri et al., 2017). The epitopes that scored a percentile rank of 1% or less were considered as strong binders of MHC class I and were selected for further studies (Mohan et al., 2018; Solanki and Tiwari, 2018).
2.3. MHC class I epitope processing analysis
Antigens to be presented by MHC class I molecules undergo a proteolytic cascade known as antigen processing within antigen-presenting cells (APCs). Proteosomal cleavage is followed by peptide transport using transporter associated with antigen presentation (TAP) and ultimately, MHC class I binding. The NetMHCpan method of IEDB-AR database processing predictor tool (http://tools.iedb.org/processing/) was used to measure the overall score for each of the SARS-CoV-2 CTL epitopes identified (Peters et al., 2003; Tenzer et al., 2005). Peptides with positive total score were chosen for subsequent studies (Mohan et al., 2018; Solanki and Tiwari, 2018).
2.4. MHC class I immunogenicity prediction
Activation of CTLs expressing T cell receptors for peptides depends upon how strongly the processed peptides (epitopes) bind to class I MHC molecules. Once activated, a cascade of activation events that elicit an immune response that results in killing of target cells expressing the antigenic epitope. Immunogenicity of peptides is also influenced by properties including the size of associated side chains and position of each amino acid in the peptide. The pMHC immunogenicity prediction tool of IEDB server (http://tools.iedb.org/immunogenicity/) was employed for immunogenic analysis of nine-mer selected epitopes (Calis et al., 2013). Epitopes that yielded negative scores were excluded and those with positive scores were only selected for further analysis. Only nine-mer peptides can be validated by pMHC tools and hence only 9-mer peptides were analyzed using this tool and were used for further analyses. The higher the score, the greater the probability that a given peptide is capable of eliciting an immune response (Mohan et al., 2018; Solanki and Tiwari, 2018).
2.5. Assessment of antigenic epitopes
Selected potent epitopes predicted for immunogenicity were evaluated for antigenicity using VaxiJen server with a threshold of 0.4. Probable protective antigenic epitopes are selected for subsequent study (Doytchinova and Flower, 2007).
2.6. Identification of multiple allele binders
All the promiscuous peptides identified as antigenic for each MHC class I allele were manually compared with one another for the prediction of multiple allele binders.
2.7. Toxicity and allergenicity analysis
Toxicity levels of predicted potent epitopes was investigated using the online server, ToxinPred (https://webs.iiitd.edu.in/raghava/toxinpred/multi_submit.php) with default parameters (Gupta et al., 2013). In addition, allergenicity of potent CTL epitopes was also analyzed using AllerTop (https://www.ddg-pharmfac.net/AllerTOP/) to identify epitopes that might induce allergic reactions (Dimitrov et al., 2013). Screening of non-toxic and non-allergen epitopes that can elicit strong immune response are suitable as they will not harm the host. The peptides which are found to be toxic or allergenic were excluded from the study and the remaining peptides were used for molecular docking studies.
2.8. Molecular docking
Validation of CTL epitopes was done using an open-source program named AutoDock Vina (Trott and Olson, 2010). Molecular docking was performed with the known crystal structures of HLA alleles (A*0101, A*0201, A*0301, A*1101, A*2402), PDB IDs of which are, 4NQV,3UTQ, 3RL1, 2HN7, 2BCK respectively. The top binding epitopes were identified for each allele. Epitopes which bind to more than one allele were also analyzed. The insights of interaction of epitopes with the amino acids of HLA molecules were also examined.
2.9. Identification of multi-strain consensus epitopes
Epitopes found in more than one isolate of SARS-CoV-2 were also identified. For this the proteome of ten different isolates of SARS-CoV-2 were obtained from NCBI GenBank database, the accession numbers of which are: MT126808.1, LC528233.1, MT123291.1, MT066176.1, MT152824.1, MT019533.1, MT007544.1, MN985325.1, MT039873.1 and MN997409.1. Epitopes present in more than one of these isolates were designated as consensus epitopes and consensus vaccine candidates.
2.10. Prediction of MHC class II-binding peptides
The predicted SARS-CoV-2 CTL epitopes were analyzed for MHC class II-binding. Peptides of 15 amino acids in length were predicted using a novel artificial neural network-based method, NN-align (http://tools.iedb.org/mhcii/) that allows for simultaneous identification of the MHC class II binding core and binding affinity. MHC class II HLA alleles selected for analysis included 27 alleles: DRB1*01:01, DRB1*03:01, DRB1*04:01, DRB1*04:05, DRB1*07:01, DRB1*08:02, DRB1*09:01, DRB1*11:01, DRB1*12:01, DRB1*13:02, DRB1*15:01, DRB3*01:01, DRB3*02:02, DRB4*01:01, DRB5*01:01, DQA1*05:01/DQB1*02:01, DQA1*05:01/DQB1*03:01, DQA1*03:01/DQB1*03:02, DQA1*04:01/DQB1*04:02, DQA1*01:01/DQB1*05:01, HLA-DQA1*01:02/DQB1*06:02, DPA1*02:01/DPB1*01:01, DPA1*01:03/DPB1*02:01, DPA1*01:03/DPB1*04:01, DPA1*03:01/DPB1*04:02, DPA1*02:01/DPB1*05:01, and, DPA1*02:01/DPB1*14:01 (Greenbaum et al., 2011). IEDB recommended 2.20 tool was used for prediction. The non-binding epitopes were selected for further analysis.
3. Results
3.1. MHC class I binding CTL epitopes prediction
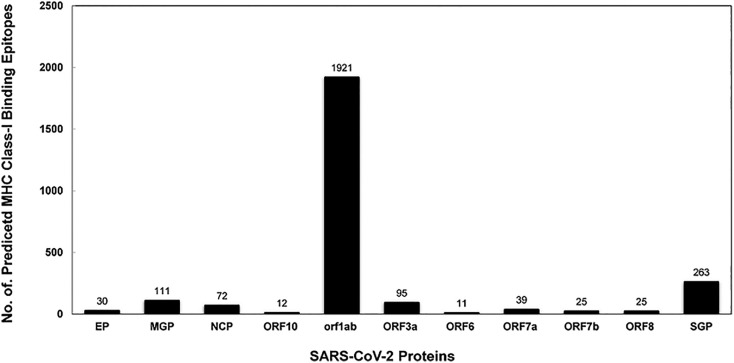
The complete SARS-CoV-2 proteome was analyzed for the presence of possible T-cell epitopes using a variety of IEDB-AR tools. Epitopes with lower percentile rankings (1.0) were selected and designated as best binders. A total of 2604 epitopes which are 8, 9, 10 and 11 mers in length, were found to be strong binders to MHC class I molecules. The highest number of epitopes were predicted for HLA A*3303 (515) and lowest peptides for HLA A*3101 (191) (Table 1 ). The number of mapped CTL epitopes in proteins: Envelope Protein, Membrane Glycoprotein, Nucleocapsid Phosphoprotein, ORF10 protein, ORF1ab polyprotein, ORF3a protein, ORF6 protein, ORF7a protein, ORF7b protein, ORF8 protein, Surface Glycoprotein were 30(1.15%), 111(4.26%), 72(2.76%), 12(0.46%), 1921(73.77%), 95(3.65%), 11(0.42%), 39(1.50%), 25(0.96%), 25(0.96%), 263(10.10%) respectively (Fig. 2 ). Epitopes showing a percentile rank of 1% and less were considered as best binding epitopes. A total of 2604 predicted CTL epitopes were then subjected to further analysis (Supplementary Table 1).
Table 1.
Prediction of potential CTL epitopes for nine HLA alleles.
| HLA Alleles |
Length of peptides |
Number of peptides | |||
|---|---|---|---|---|---|
| 8 | 9 | 10 | 11 | ||
| HLA A*0101 | 2 | 103 | 112 | 34 | 251 |
| HLA A*0201 | 3 | 124 | 104 | 14 | 245 |
| HLA A*0206 | 0 | 145 | 102 | 3 | 250 |
| HLA A*0301 | 1 | 126 | 127 | 17 | 271 |
| HLA A*1101 | 5 | 164 | 133 | 55 | 357 |
| HLA A*2402 | 1 | 136 | 122 | 21 | 280 |
| HLA A*2601 | 1 | 146 | 96 | 1 | 244 |
| HLA A*3101 | 21 | 94 | 73 | 3 | 191 |
| HLA A*3303 | 42 | 288 | 156 | 29 | 515 |
| Total number of peptides | 76 | 1326 | 1025 | 177 | 2604 |
Fig. 2.
MHC binding analysis of peptides of SARS-CoV-2.
3.2. Assessment of epitope processing
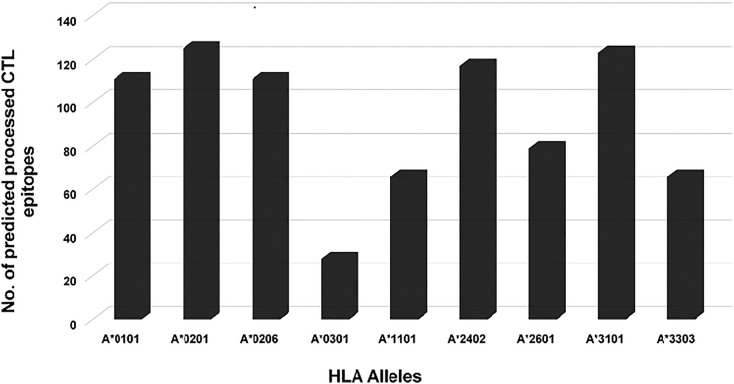
Each of the 2604 CTL epitopes was analyzed for their ability to undergo antigen processing using the processing tool available on the IEDB server. Only 826 (31.72%) of the candidate CTL epitopes were found to possess high processing potential (Fig. 3 ). Peptides predicted as binders to HLA A*0201 (125), HLA A*3101 (123) and HLA-A*0206 (111) had the strongest predicted ability to undergo processing, while minimum processing potential was observed for HLA A*0301 (Nielsen and Andreatta, 2016), HLA A*1101 (66) and HLA A*3303 (66) alleles. In the case of HLA A*3303 and HLA A*0301, approximately 90% of the peptides were eliminated.
Fig. 3.
MHC allele specific distribution of SARS-CoV-2 predicted peptides.
3.3. Analysis of epitope immunogenicity and antigenicity
Among the 826 peptides found to have processing potential, 451 were found to be 9-mers. For each SARS-CoV-2 protein studied, the percentage of CTL epitopes which are nine-mers in length were found to be in the range of 40 to 81.82%. These were analyzed for immunogenicity using the IEDB pMHC immunogenicity predictor tool which identified 260 with immunogenic properties. It is noteworthy that none of the peptides contained in ORF10 were found to have immunogenic potential (Table 2 ). Among the 260 positively-scored epitopes, potential antigenicity was assessed using the VaxiJen server which identified 102 fulfilling the criteria used to define potential antigenicity.
Table 2.
Immunogenicity and antigenicity prediction of putative CTL epitopes of SARS-CoV-2.
| Protein Name | Length (mer) | Potent Processed epitopes | % of 9 mer epitopes | Potent immunogenic epitopes | Antigenicity (Threshold ≥ 4) |
|---|---|---|---|---|---|
| Envelope protein | 8 | 0 | |||
| 9 | 9 | 81.82 | 7 | 3 | |
| 10 | 2 | ||||
| 11 | 0 | ||||
| Membrane glycoprotein | 8 | 2 | |||
| 9 | 27 | 58.70 | 23 | 7 | |
| 10 | 16 | ||||
| 11 | 1 | ||||
| Nucleocapsid phosphoprotein | 8 | 0 | |||
| 9 | 9 | 52.94 | 6 | 2 | |
| 10 | 7 | ||||
| 11 | 1 | ||||
| ORF10 protein | 8 | 0 | |||
| 9 | 2 | 40 | 0 | 0 | |
| 10 | 3 | ||||
| 11 | 0 | ||||
| ORF1ab polyprotein | 8 | 5 | |||
| 9 | 328 | 55.03 | 182 | 72 | |
| 10 | 248 | ||||
| 11 | 15 | ||||
| ORF3a protein | 8 | 0 | |||
| 9 | 16 | 50.00 | 10 | 3 | |
| 10 | 14 | ||||
| 11 | 2 | ||||
| ORF6 protein | 8 | 0 | |||
| 9 | 1 | 50.00 | 1 | 1 | |
| 10 | 1 | ||||
| 11 | 0 | ||||
| ORF7a protein | 8 | 0 | |||
| 9 | 7 | 70.00 | 2 | 1 | |
| 10 | 3 | ||||
| 11 | 0 | ||||
| ORF7b | 8 | 0 | |||
| 9 | 3 | 42.86 | 3 | 2 | |
| 10 | 4 | ||||
| 11 | 0 | ||||
| ORF8 protein | 8 | 0 | |||
| 9 | 7 | 63.63 | 3 | 2 | |
| 10 | 4 | ||||
| 11 | 0 | ||||
| Surface glycoprotein | 8 | 0 | |||
| 9 | 42 | 47.19 | 23 | 9 | |
| 10 | 41 | ||||
| 11 | 6 |
3.4. Identification of multiple allele binders
A total of 25 out of 102 selected peptides were found to have the potential to bind more than one HLA allele and, thus, were defined as consensus epitopes. Among these consensus epitopes, CSLSHRFYR would bind with more than 2 HLA alleles (HLA-A*11:01, HLA-A*31:01 and HLA-A*33:03).
3.5. Prediction of toxic and allergenic epitopes
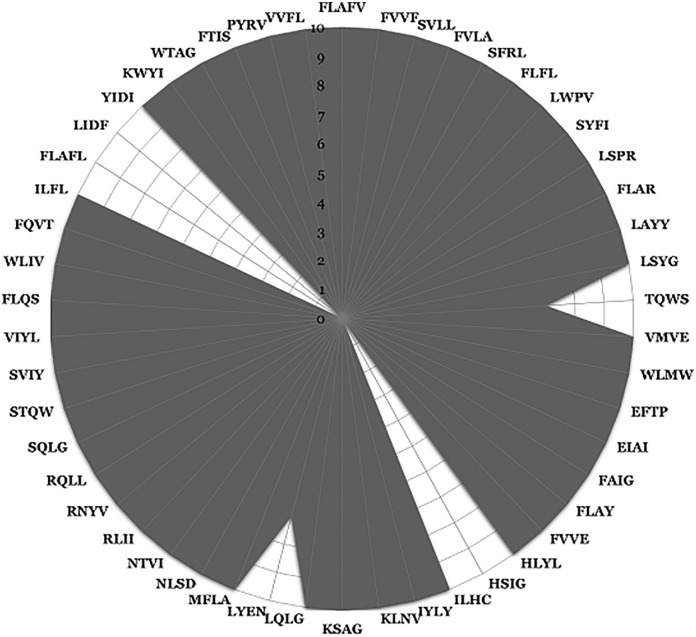
Epitopes with predicted toxicity and allergenicity have the potential to trigger serious adverse reactions if used in multivariant subunit vaccines (Chung, 2014). In the evaluation of 102 epitopes identified as SARS-CoV-2 candidate CTL epitopes, one peptide (TYKPNTWCI) was predicted to be both toxic and allergenic. In addition, 51 other peptides were found to have potential allergenicity. Excluding these for further analysis yielded a total of 50 epitopes defined as promiscuous CTL epitopes with a potential to serve as CTL vaccine candidates for SARS-CoV-2. Most of these peptides are present in each of the 10 selected SARS-CoV-2 isolates analyzed whereas LYENAFLPF and TQWSLFFFL were observed in 7 out of 10 isolates. Three predicted epitopes, FLAFLLFLV, LIDFYLCFL and HSIGFDYVY, were not conserved in any of the selected proteomes (Fig. 4 ; Supplementary Table 2).
Fig. 4.
Radar graph indicating the presence of predicted epitopes in 10 selected strains of SARS-CoV-2. The graph illustrates the ten SARS-CoV-2 strains as concentric rings in the following order from the center to outer circles: MT123291.1, MT066176.1, MT152824.1, MT019533.1, MT007544.1, MN985325.1, MT039873.1, MN997409.1, MT126808.1, LC528233.1 respectively.” First four-letter amino acid sequences are shown for each of the 9-mer peptides listed in Supplementary Table 2.
3.6. Molecular docking
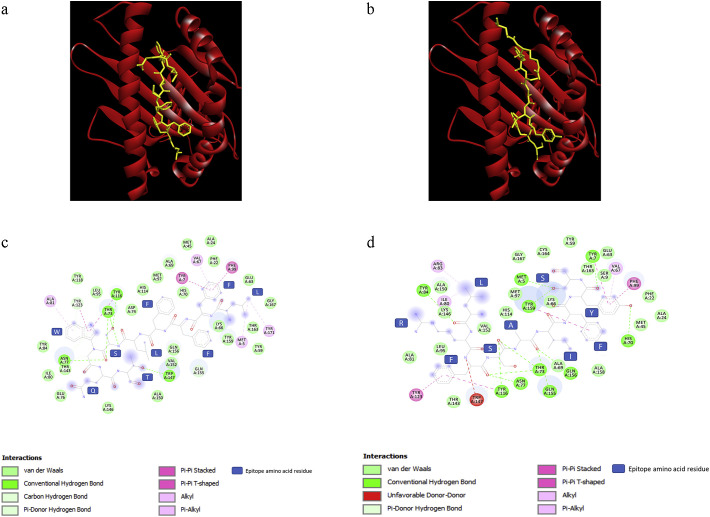
We modeled 3D structures of selected promiscuous CTL epitopes using ChemSketch and Discovery Studio Visualizer software. These epitopes were docked into binding cavities of 5 HLA A-alleles (A*0101, A*0201, A*0301, A*1101, A*2402) using AutoDock Vina software. The top binding peptides for each HLA molecule were investigated. Epitopes HSIGFDYVY and STQWSLFFF were found to have the lowest binding energy (−9.2 kcal/mol) with HLA A*0101. The top binder for HLA A*0201 was TQWSLFFFL, with a binding energy of −8.8 kcal/mol. The epitopes EFTPFDVVR, FAIGLALYY and TQWSLFFFL were found to be the top binders for HLA molecules HLA A*0301, HLA A*1101 and HLA A*2402 with the binding energy values of −8.5, −8.9 and − 10 kcal/mol respectively (Table 3 ). The molecular interactions of epitopes TQW and STQ are presented in Fig. 5a and c. The 2D ligand interaction maps for epitopes TQW and STQ are Fig. 5b, Fig. 5d respectively.
Table 3.
Validation of predicted epitopes using molecular docking analysis.
| S. No. | Predicted Epitopes |
Binding Energy (kcal/mol) against |
||||
|---|---|---|---|---|---|---|
| HLA-A*0101 | HLA-A*0201 | HLA-A*0301 | HLA-A*1101 | HLA-A*2402 | ||
| 1 | FLAFVVFLL | −7.5 | −7.1 | −7.9 | −7.2 | −9.3 |
| 2 | FVVFLLVTL | −8 | −7.1 | −7.5 | −8.8 | −9.4 |
| 3 | SVLLFLAFV | −8.1 | −7.9 | −7.4 | −7.8 | −8.2 |
| 4 | FVLAAVYRI | −7.7 | −7.7 | −6.5 | −6.6 | −8.2 |
| 5 | SFRLFARTR | −7.4 | −7 | −7.5 | −7 | −8 |
| 6 | FLFLTWICL | −8 | −7.4 | −8 | NB | −9.4 |
| 7 | LWPVTLACF | −7.9 | −7.1 | −7.5 | −8 | −8.6 |
| 8 | SYFIASFRL | −7.6 | −7.5 | −8.2 | −7.3 | −9.9 |
| 9 | LSPRWYFYY | −7.4 | −7.2 | −7.1 | −8.4 | −9.8 |
| 10 | FLARGIVFM | −5.5 | −6.6 | −6.9 | −6.2 | −7.1 |
| 11 | LAYYFMRFR | −6.7 | −6.7 | −7.3 | −7.9 | −8.8 |
| 12 | LSYGIATVR | −6.4 | −6.7 | −6.3 | −6.9 | −7.9 |
| 13 | TQWSLFFFL | −8.2 | −8.8 | −8.2 | −8.4 | −10 |
| 14 | VMVELVAEL | −6 | −7.4 | −6.4 | −6.9 | −6.9 |
| 15 | WLMWLIINL | −7.2 | −5.6 | −7.2 | −6.9 | −8.7 |
| 16 | EFTPFDVVR | −8.1 | −6.6 | −8.5 | −8.8 | −7.9 |
| 17 | EIAIILASF | −7.3 | −5.8 | −7 | −7.5 | −8.7 |
| 18 | FAIGLALYY | −6 | −7.8 | −8 | −8.9 | −9.8 |
| 19 | FLAYILFTR | −7.5 | −6.8 | −7 | −8.5 | −8.4 |
| 20 | FVVEVVDKY | −7 | −7.3 | −7.4 | −7.3 | −8.7 |
| 21 | HLYLQYIRK | −7.7 | −7.5 | −7.6 | −7.8 | −9.1 |
| 22 | HSIGFDYVY | −9.2 | −7 | −8.3 | −7.7 | −9.4 |
| 23 | ILHCANFNV | −7.2 | −7.6 | −7.4 | −7.4 | −8.8 |
| 24 | IYLYLTFYL | −7.8 | −7.1 | −7.9 | −7.4 | −9.3 |
| 25 | KLNVGDYFV | NB | NB | NB | NB | NB |
| 26 | KSAGFPFNK | −8.1 | −7.3 | −8.2 | −7.7 | −8.6 |
| 27 | LQLGFSTGV | −7 | −6 | −6.9 | −7 | −8.4 |
| 28 | LYENAFLPF | −7.4 | −8.1 | −7.1 | −8.6 | −8.4 |
| 29 | MFLARGIVF | −6.2 | −7.3 | −7.3 | −6.8 | −9.2 |
| 30 | NLSDRVVFV | −7.2 | −6.6 | −7.7 | −6.9 | −8.5 |
| 31 | NTVIWDYKR | −6.7 | −6.5 | −8 | −7.8 | −7.7 |
| 32 | RLIIRENNR | NB | NB | NB | NB | NB |
| 33 | RNYVFTGYR | −8 | −7 | −7.8 | −8.1 | −8.3 |
| 34 | RQLLFVVEV | −7.4 | −7.3 | −7.1 | −8 | −7.5 |
| 35 | SQLGGLHLL | −7 | −6.7 | −6.6 | −6.7 | −7.7 |
| 36 | STQWSLFFF | −9.2 | −7.1 | −7.6 | −6.6 | −9.6 |
| 37 | SVIYLYLTF | −8 | −6.9 | −6.8 | −7.6 | −8.3 |
| 38 | VIYLYLTFY | −7.4 | −7.4 | −8 | −8 | −9.9 |
| 39 | FLQSINFVR | −7.5 | −6.3 | −7.5 | −7.6 | −9.5 |
| 40 | WLIVGVALL | −7.1 | −7.3 | −7.1 | −6.7 | −8.5 |
| 41 | FQVTIAEIL | −6.8 | −6.9 | −7.3 | −7.9 | −8 |
| 42 | ILFLALITL | −6.9 | −6.7 | −7.2 | −8 | −8 |
| 43 | FLAFLLFLV | −7.1 | −6.4 | −7.3 | −6.8 | −8.7 |
| 44 | LIDFYLCFL | −8.1 | −6.8 | −8.3 | −7.7 | −7.8 |
| 45 | YIDIGNYTV | −7.2 | −7.2 | −7.2 | −7.9 | −7.8 |
| 46 | KWYIRVGAR | −7 | −7.3 | −7.1 | −8 | −7.9 |
| 47 | WTAGAAAYY | −6.1 | −7.3 | −7.3 | −6.9 | −6.6 |
| 48 | FTISVTTEI | −6.9 | −6.8 | −7.3 | −7.5 | −8.7 |
| 49 | PYRVVVLSF | NB | NB | NB | NB | NB |
| 50 | VVFLHVTYV | −6.9 | −7.2 | −7.3 | −8.1 | −8.1 |
NB - Ligand not binding to the target MHC allele at the peptide binding groove.
Fig. 5.
Molecular and 2D ligand interaction maps of predicted epitopes TQWS (a, c) and SYFI (b, d) with HLA allele A*2402.
3.7. Elimination of MHC class II-binding epitopes
In light of the need to identify SARS-CoV-2 T cell epitopes that will optimally serve as vaccine candidates while having little or no potential to promote pulmonary immunopathology caused by TH2 responses, we examined each of the candidate CTL epitopes' ability to bind MHC class II molecules across 27 alleles. The cytokine release syndrome (CRS) induced by excessive TH2 responses has been shown to accentuate the pathogenesis of SARS corona viruses (Nicholls et al., 2003; Zheng et al., 2020). Late T cell responses may instead amplify pathogenic inflammatory outcomes in the presence of sustained high viral loads in the lungs, by multiple hypothetical possible mechanisms (Guo et al., 2020; Li et al., 2008; Liu et al., 2019). Severe and fatal COVID-19 (and SARS) outcomes are associated with elevated levels of inflammatory cytokines and chemokines, including IL-6 (Giamarellos-Bourboulis et al., 2020; Wong et al., 2004; Zhou et al., 2020).
MHC class II molecules bind peptides larger than those that bind to class I molecules due to their open-ended binding grove. We therefore analyzed the ability of 15-mer SARS-CoV-2 peptide sequences that contained the predicted CTL epitopes for binding to class II molecules. Among the 50 CTL epitopes identified, 18 were found to be MHC class II non-binders (Table 4 ). These epitopes would be expected to stimulate CTL responses without promoting TH2 responses capable of promoting CRS.
Table 4.
Predicted non-binders to MHC Class II.
| S. No | Peptide | Protein | Binding Allele(s) |
|---|---|---|---|
| 1 | LSPRWYFYY | Nucleocapsid phosphoprotein | HLA-A*01:01 |
| 2 | FLARGIVFM | nsp6 | HLA-A*02:01; HLA-A*02:06 |
| 3 | MFLARGIVF | nsp6 | HLA-A*24:02 |
| 4 | EFTPFDVVR | 3C-like proteinase | HLA-A*33:03 |
| 5 | FAIGLALYY | nsp13 | HLA-A*26:01 |
| 6 | RNYVFTGYR | nsp13 | HLA-A*31:01 |
| 7 | FVVEVVDKY | nsp12 | HLA-A*26:01 |
| 8 | ILHCANFNV | nsp12 | HLA-A*02:01 |
| 9 | KSAGFPFNK | nsp12 | HLA-A*11:01 |
| 10 | RQLLFVVEV | nsp12 | HLA-A*02:06 |
| 11 | HSIGFDYVY | nsp14 | HLA-A*26:01 |
| 12 | LQLGFSTGV | nsp14 | HLA-A*02:06 |
| 13 | NLSDRVVFV | nsp14 | HLA-A*02:01 |
| 14 | NTVIWDYKR | nsp15 | HLA-A*33:03 |
| 15 | SQLGGLHLL | nsp15 | HLA-A*02:06 |
| 16 | WLIVGVALL | ORF3a protein | HLA-A*02:01 |
| 17 | YIDIGNYTV | ORF8 protein | HLA-A*02:01; HLA-A*02:06 |
| 18 | VVFLHVTYV | Surface glycoprotein | HLA-A*02:06 |
4. Discussion
At the end of 2019, infections caused by a novel coronavirus named SARS-CoV-2 led to the COVID-19 pandemic affecting millions of people world-wide. At the time of the writing of this article, the global health challenge accounts for ~9.65 million confirmed cases and ~ 491,128 fatalities (WHO, 2020). No effective therapies to treat patients suffering from COVID-19 have been identified. Moreover, despite the initiation of several vaccine strategies to prevent SARS-CoV-2 infections, none have been authorized for use at this time. Most of these investigations are focused on the generation of SARS-CoV-2-specific nAb responses in subjects participating in vaccine clinical trials.
Computational genomic approaches to vaccine development allow prediction of immunogenic epitopes without the need to grow the pathogen in vitro, a process known as reverse vaccinology. It employs a wide range of bioinformatics tools to predict immunogenic epitopes that have the potential to stimulate protective B- and T-cell responses mediated by nAbs and CTLs, respectively. When applied to the identification of conventional vaccine candidates that exploit B cell responses, reverse vaccinology relies on the principle that the target antigen of a protective antibody will elicit the same antibody response when used as an immunogen. In contrast, in silico analysis of pathogen genomes to identify T cell epitopes must consider epitope processing and presentation associated with activation of CD4+ TH cells and CD8+ CTLs. In the present study, we first identified potential SARS-CoV-2 CTL epitopes and then computationally analyzed the ability of peptide sequences containing those epitopes to bind MHC class II alleles. Our goal was to determine whether any of the former (CTL epitopes) also had MHC class II binding potential. The latter property is one that may be considered harmful in light of the need to minimize SARS-CoV-2-induced cytokine release syndrome (CRS) seen in some patients.
A total of 2604 best binding epitopes were predicted for the SARS-CoV-2 proteome. Peptides with low percentile ranking scores (0.1) were defined as best (strong) class I binding molecules and the percentile ranking criterion of <1.00 was used as the basis for identifying epitopes for nine alleles of HLA-A loci. Percentile rank is generated by comparing the peptide's IC50 against those of a set of random peptides from SWISSPROT database. To our knowledge, this is the first study reporting a comprehensive analysis of T cell epitope mapping for SARS-CoV-2. Recent studies reported the prediction of T cell epitopes in the SARS-CoV-2 spike glycoprotein (Baruah and Bose, 2020; Bhattacharya et al., 2020). Another used the B cell epitopes available in the IEDB server for the prediction of both CTL and T-helper epitopes (Grifoni et al., 2020a).
The predicted best binders observed in this study were derived from an analytical workflow that used highly stringent selection criteria including analysis of processing, allergenicity, antigenicity, toxicity and physiochemical properties (Mohan et al., 2018). Application of these selection parameters is important because, ultimately, they help to ensure that future use of the predicted immunogenic peptides identified will stimulate CTL immunity while minimizing or eliminating adverse reactions to the host. The results indicated that 826 (31.72%) of the predicted epitopes will effectively undergo antigen processing. These peptides were further evaluated for their immunogenicity using the pMHC immunogenicity predictor tool. This tool analyzes the interactions of the peptide-MHC complex present on the antigen presenting cells with the T cell receptor (TCR). This bioinformatics screening step yielded a total of 260 (31.48%) predicted immunogenic 9-mer length epitopes. The selected peptides were further evaluated for their antigenicity using an algorithm available on the VaxiJen server which predicts the physical interaction of immunogenic peptides within TCR synapses as well as their predicted ability to trigger intracellular signals leading to elicitation of T-cell activation (Doytchinova and Flower, 2007). This screening step resulted in the elimination of 130 peptide sequences, leaving 130 nine-mers which scored positive for antigenicity. After the exclusion of toxic and allergenic epitopes, 50 out of 2604 best binding peptides were found to be potential CTL epitopes which could be used as possible vaccine candidates for SARS-CoV-2.
Molecular modeling simulation software (AutoDock) was then used to predict non-covalent binding affinities of each of the identified peptides to MHC class I loci. The following HLA molecules that we tested were predicted to form a stable complex with the predicted binders: A*0101, A*0201, A*0301, A*1101, A*2402. All the peptides exhibited a stronger binding affinity towards HLA A*2402 with low binding energies. The epitopes HSIGFDYVY and STQWSLFFF were found to be binding with a lower binding energy (−9.2 kcal/mol) with HLA A*0101. HLA A*0201 was found to have the epitope TQWSLFFFL as the top binder, with a binding energy of −8.8 kcal/mol. The epitopes EFTPFDVVR, FAIGLALYY and TQWSLFFFL were found to be the top binders of the HLA molecules HLA A*0301, HLA A*1101 and HLA A*2402 with the binding energy values of −8.5, −8.9 and − 10 kcal/mol respectively (Table 3; Fig. 5a,b).
Out of all the epitopes docked, TQWSLFFFL was showing a good binding energy (−10 kcal/mol) on binding towards HLA A*2402. Also, it is the top binder of HLA A*0201, making this epitope a potential candidate to be studied further. The eight epitopes, HSIGFDYVY, STQWSLFFF, TQWSLFFFL, SVLLFLAFV, EFTPFDVVR, FAIGLALYY, FVVFLLVTL, SYFIASFRL were found to be the top 5 binders for at least two HLA molecules indicating their potential as epitopes for the 5 HLA alleles that were tested here.
Of the 50 epitopes that were identified in this study only one of the peptides was reported earlier. Peptide, WTAGAAAYY, from surface glycoprotein that binds to HLA-A*0101 and HLA-A*2601 alleles was already reported (Baruah and Bose, 2020). The remaining 49 peptides are novel epitopes that are reported for the first time. Thirty-eight of these were found to possess binding affinities for single HLA alleles. The remaining candidate epitopes had binding affinity for more than one HLA class I allele. Overall, the array of predicted peptides showed the greatest predisposition for binding to HLA-A*0206 (16 out of 50, 32%),
It is widely believed that the SARS-CoV-2 surface glycoprotein (spike) is the major antigenic protein that can be exploited for vaccine strategies aimed at inducing nAb responses. While virus-stimulated natural immunity to SARS-CoV-2 as well as future vaccine-induced immunity will largely rely on humoral immune responses to prevent infection, vaccine efforts aimed at inducing SARS-CoV2-specific T cell immunity should also be considered as part of the overall public health arsenal to prevent COVID-19. A recent study using peripheral blood mononuclear cells from convalescent COVID-19 donors demonstrated circulating SARS-CoV-2 − specific CD8+ and CD4+ T cells in all subjects tested (Grifoni et al., 2020b). CD4+ T cell responses to spike protein were robust and correlated with the magnitude of the anti-SARS-CoV-2 IgG and IgA titers. The M, spike and N proteins each accounted for 11–27% of the total CD4+ response. For CD8+ T cells, spike and M were recognized. Knowledge of the T cell responses to COVID-19 can guide selection of appropriate immunological endpoints for COVID-19 candidate vaccine clinical trials.
Vaccine development against viral infections classically focuses on vaccine-elicited recapitulation of the type of protective immune response elicited by natural infection. Such foundational knowledge is currently missing for SARS-CoV-2. In particular, we lack knowledge concerning how the balance and the phenotypes of responding immune cells vary as a function of disease course and severity. Such knowledge can guide selection of vaccine strategies most likely to elicit protective immunity to prevent COVID-19 disease. Furthermore, knowledge of the T cell responses can guide selection of appropriate immunological endpoints for COVID-19 candidate vaccine clinical trials.
Our comprehensive evaluation of putative immunogenic CTL epitopes within the orf1ab polyprotein demonstrates the presence of many non-structural SARS-CoV-2 proteins that contain such candidate epitopes. Many epitopes with the predicted ability to stimulate protective cellular immunity were also observed in structural proteins of the virus including its envelope, membrane glycoprotein and surface glycoprotein. Future studies will need to be undertaken using in vitro and in vivo strategies to evaluate potential efficacy of the full array of predicted CTL epitopes that can be used in conjunction with those that induce nAb against SARS-CoV-2.
Author contribution
CFR and KS designed the study. CFR performed the experiments with the help of JCR. RC, KS and MM analyzed the results. JCR, RC and KS wrote the manuscript with input from all authors.
Credit author statement
Clayton Fernando Rencilin: Conceptualization, Methodology, Investigation. Joseph Christina Rosy: Methodology, Data curation, Writing-original draft. Manikandan Mohan: Data curation, Formal analysis. Richard Coico: Supervision, Writing Review & Editing. Krishnan Sundar: Conceptualization, Supervision, Writing Review & Editing, Funding acquisition.
Declaration of Competing Interest
All the authors declare that there is no conflict of interest.
Acknowledgements
KS would like to acknowledge the financial support by Science and Engineering Research Board of India (EMR/2016/003035). CFR acknowledges Kalasalingam Academy of Research and Education for financial support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104712.
Appendix A. Supplementary data
Prediction of MHC-I Binding Peptides using IEDB-AR.
Presence of promiscuous CTL epitopes of SARS-CoV-2 in other strains.
References
- Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yedidia T., Arnon R. Epitope-based vaccine against influenza. Expert Rev. Vaccines. 2007;6:939–948. doi: 10.1586/14760584.6.6.939. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno L., Cortese M., Rappuoli R., Merola M. Lessons from reverse vaccinology for viral vaccine design. Curr. Opin. Virol. 2015;11:89–97. doi: 10.1016/j.coviro.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Calis J.J.A., Maybeno M., Greenbaum J.A., Weiskopf D., De Silva A.D., Sette A., Keşmir C., Peters B. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C.C., Lau S.K.P., Woo P.C.Y., Kwok Y.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and re-emerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E.H. Vaccine allergies. Clin. Exp. Vaccine Res. 2014;3:50. doi: 10.7774/cevr.2014.3.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I., Flower D.R., Doytchinova I. AllerTOP - a server for in silico prediction of allergens. BMC Bioinformatics. 2013;14:S4. doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sahly H.M., Atmar R.L., Glezen W.P., Greenberg S.B. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin. Infect. Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleri W., Paul S., Dhanda S.K., Mahajan S., Xu X., Peters B., Sette A. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and Bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P.S. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M., Douek D.C., Mongkolsapaya J., Tran B.-H., Lin C.S., Screaton G.R., Hou J., McMichael A.J., Xu X.-N. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., Wu T., Cheung K.-W., Chan K.-H., Alvarez X., Qin C., Lackner A., Perlman S., Yuen K.-Y., Chen Z. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C., Lund O., Buus S., Nielsen M. Major histocompatibility complex class I binding predictions as a tool in epitope discovery. Immunology. 2010;130:309–318. doi: 10.1111/j.1365-2567.2010.03300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M., Haribalaganesh R., Coico R., Sundar K. HLA-directed bioinformatics approach for genome-wide mapping of dengue CTL epitopes. Futur. Virol. 2018;13:331–342. doi: 10.2217/fvl-2017-0157. [DOI] [Google Scholar]
- Moutaftsi M., Peters B., Pasquetto V., Tscharke D.C., Sidney J., Bui H.-H., Grey H., Sette A. A consensus epitope prediction approach identifies the breadth of murine TCD8+-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., Yan K.W., Chan K.H., Tsang N.C., Guan Y., Yuen K.Y., Malik Peiris J. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarzún P., Kobe B. Recombinant and epitope-based vaccines on the road to the market and implications for vaccine design and production. Hum. Vaccin. Immunother. 2016;12:763–767. doi: 10.1080/21645515.2015.1094595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry J. WHO issues guidelines to manage any future SARS outbreak. BMJ. 2003;327 doi: 10.1136/bmj.327.7412.411-a. 411-a-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B., Bulik S., Tampe R., van Endert P.M., Holzhütter H.-G. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J. Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- Rappuoli R., Bottomley M.J., D’Oro U., Finco O., De Gregorio E. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J. Exp. Med. 2016;213:469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018;8:9044. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzer S., Peters B., Bulik S., Schoor O., Lemmel C., Schatz M.M., Kloetzel P.-M., Rammensee H.-G., Schild H., Holzhütter H.-G. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage. TAP transport and MHC class I binding. C. Cell. Mol. Life Sci. 2005;62:1025–1037. doi: 10.1007/s00018-005-4528-2. [DOI] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO COVID-19 situation dash board at https://covid19.who.int/ accessed on June 27, 2020.
- Wilson C.C., McKinney D., Anders M., MaWhinney S., Forster J., Crimi C., Southwood S., Sette A., Chesnut R., Newman M.J., Livingston B.D. Development of a DNA vaccine designed to induce cytotoxic T lymphocyte responses to multiple conserved epitopes in HIV-1. J. Immunol. 2003;171:5611–5623. doi: 10.4049/jimmunol.171.10.5611. [DOI] [PubMed] [Google Scholar]
- Wong C.K., Lam C.W.K., Wu A.K.L., Ip W.K., Lee N.L.S., Chan I.H.S., Lit L.C.W., Hui D.S.C., Chang M.H.M., Chung S.S.C., Sung J.J.Y. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Chokkalingam N., Tello-Ruiz E., Wise M.C., Bah M.A., Walker S., Tursi N.J., Fisher P.D., Schultheis K., Broderick K.E., Humeau L., Kulp D.W., Weiner D.B. A DNA-launched nanoparticle vaccine elicits CD8+ T-cell immunity to Promote In Vivo Tumor Control. Cancer Immunol. Res. 2020;8:1354–1364. doi: 10.1158/2326-6066.CIR-20-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Huang W., Yang X., Sun W., Liu X., Cun W., Ma Y. HPV-16 E6 and E7 protein T cell epitopes prediction analysis based on distributions of HLA-A loci across populations: an in silico approach. Vaccine. 2013;31:2289–2294. doi: 10.1016/j.vaccine.2013.02.065. [DOI] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I.C., Chan M., Vasoo S., Wang L.F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.S., Lye D.C. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA - J. Am. Med. Assoc. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prediction of MHC-I Binding Peptides using IEDB-AR.
Presence of promiscuous CTL epitopes of SARS-CoV-2 in other strains.