Abstract
The ongoing pandemic of COVID-19 alongside the outbreaks of SARS in 2003 and MERS in 2012 underscore the significance to understand betacoronaviruses as a global health challenge. SARS-CoV-2, the etiological agent for COVID-19, has infected over 50 million individuals’ worldwide with more than ∼1 million fatalities. Autophagy modulators have emerged as potential therapeutic candidates against SARS-CoV-2 but recent clinical setbacks urge for better understanding of viral subversion of autophagy. Using MHV-A59 as a model betacoronavirus, time-course infections revealed significant loss in the protein level of ULK1, a canonical autophagy-regulating kinase, and the concomitant appearance of a possible cleavage fragment. To investigate whether virus-encoded proteases target ULK1, we conducted in-vitro and cellular cleavage assays and identified ULK1 as a novel bona fide substrate of SARS-CoV-2 papain-like protease (PLpro). Mutagenesis studies discovered that ULK1 is cleaved at a conserved PLpro recognition sequence (LGGG) after G499, separating its N-terminal kinase domain from a C-terminal substrate recognition region. Over-expression of SARS-CoV-2 PLpro is sufficient to impair starvation-induced autophagy and disrupt formation of ULK1-ATG13 complex. Finally, we demonstrated a dual role for ULK1 in MHV-A59 replication, serving a pro-viral functions during early replication that is inactivated at late stages of infection. In conclusion, our study identified a new mechanism by which PLpro of betacoronaviruses induces viral pathogenesis by targeting cellular autophagy.
Keywords: ULK1, Autophagy, Betacoronavirus, SARS-CoV-2, MHV, Papain-like protease
1. Introduction
The recent pandemic of coronavirus disease-2019 (COVID-19) highlights the health crisis worldwide. Although global efforts to restrict travelling and implement social distancing practices have mitigated the spread of Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2), the etiological agent of COVID-19, the virus remains a significant health threat with >50 million confirmed cases and 1.3 million fatalities.
Autophagy-modulating drugs such as chloroquine/hydroxychloroquine have emerged as anti-viral agents against SARS-CoV-2; however, recent placebo-controlled trials showing no clinical benefits and possible safety concerns are spearheading global efforts for better understanding [1]. Autophagy is an evolutionarily conserved process that acts to recycle cellular waste while also responding to invading pathogens. Cellular membranes termed phagophores first enwrap cargo inside double-membraned chambers, so called autophagosomes, after which cargo is degraded upon fusion of autophagosomes with digestive lysosomes. The process is tightly regulated and responsive to various stressors including nutrient, oxidative, and viral stress [2].
The serine/threonine unc-51-like kinase (ULK1) is an upstream regulator of autophagy. The role of ULK1 as nutrient-responsive orchestrator of autophagy has been well characterized [3]. ULK1 is recruited to sites of autophagosome biogenesis where it phosphorylates key autophagy regulatory proteins. Its central role in autophagy has implicated ULK1 in diverse human diseases from cancer and neurodegeneration to inflammatory disorders [[4], [5], [6]]. Structurally, ULK1 possesses an N-terminal kinase domain and a C-terminal early autophagy targeting (EAT) domain. The latter facilitates interaction of ULK1 with its various substrates. Autophagy-independent functions of ULK1 have also emerged including the regulation of ER-Golgi trafficking and innate immune signaling [[7], [8], [9], [10]]. For example, stimulator of interferon genes (STING), an adaptor of the DNA sensor cyclic GMP-AMP synthase (cGAS), was previously identified as a substrate of ULK1 [11]. Furthermore, the innate immune kinase TANK-binding kinase 1 (TBK1) is phosphorylated by ULK1 to participate in metabolic signaling [12].
Many positive-sense RNA viruses, including betacoronaviruses, have evolved strategies to co-opt autophagy by utilizing cellular double membrane vesicles as topological surfaces for viral RNA synthesis [[13], [14], [15], [16]]; however, the precise mechanisms of viral subversion of autophagy components are poorly defined. The current study uncovers the betacoronavirus-encoded papain-like protease (PLpro) as a pathogenic factor that disrupts autophagy in part by targeting the regulatory kinase ULK1.
2. Materials and methods
2.1. Cell culture, viral infection, and chemicals
Murine 17Cl1 fibroblast cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin (100 μg/mL). MHV-A59 and 17Cl1 cells were provided by Dr.Nerea Irigonen (Cambridge).
Cells were either sham-infected with DMEM or inoculated with MHV-A59 (MOI = 10). Starvation was performed for 2 h in HBSS medium. V-ATPase inhibitor, bafilomycin-A1 (BAF) and ULK1/2-kinase inhibitor, MRT68921-HCl, where used at 125 nM and 5 μM respectively.
2.2. Plasmids
3×Flag-ULK1 was cloned into CMV10 vector at EcorI/BamHI sites. Cleavage-resistant mutants, LGGG-499-QQQG and MRGG-531-MRDD were generated using gBLOCKS DNA synthesis (IDT) and cloned into CMV10 vector at BstEII/FseI and FseI/AflII sites respectively. EGFP-PLpro of SARS-COV-2 was cloned as outlined (Fig. 2A) using Wuhan Hu-1 Isolate [NC_045512.2; residues 713–1063]. Murine Ulk1 sgRNA(AATCTTGACTCGGATGTTGC) was cloned into pSpCas9-2A [addgene#48138]. Transfection with plasmid cDNA/sgRNA was performed using Lipofectamine 2000 following the manufacturer’s instructions.
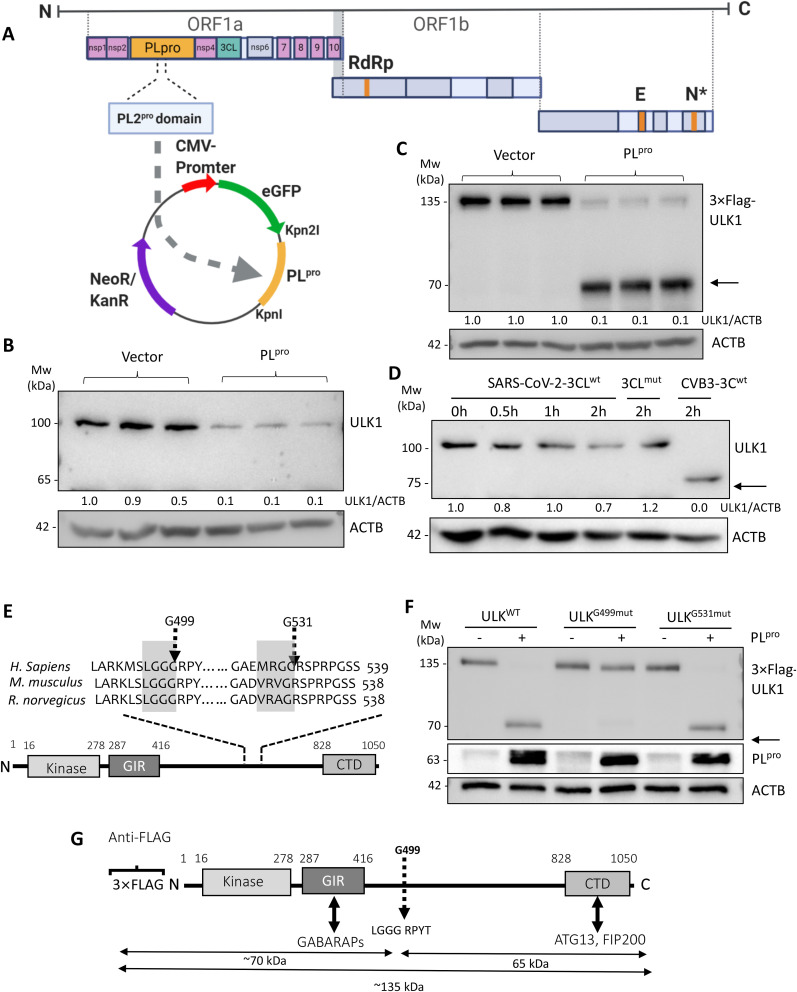
Fig. 2.
PLpro of SARS-CoV-2 cleaves ULK1 after G499.
(A) Sequence alignment of the PL protease domain of SARS-CoV-2 and PL protease domain 2 of MHV-A59 is shown. The SARS-CoV2-PLpro was cloned into EGFP-C1 vector. (B) HEK293T cells were transfected with either control or PLpro-expressing plasmid for 24 h. Lysates were probed by western blotting for endogenous ULK1 using the monoclonal anti-ULK1 antibody and normalized to ACTB as in Fig. 1A. (C) HEK293T cells were co-transfected with 3×Flag-ULK1 and either control vector or PLpro-expressing plasmid for 24 h. Lysates were probed with anti-Flag antibody to detect exogenous ULK1. Arrow denotes cleavage fragment observed at ∼70 kDa. Densitometry is provided as in Fig. 1A. (D) Purified SARS-CoV-2 3CLwt (4 μg) or catalytically-inactive C145A 3CLmut (4 μg) of SARS-CoV-2 and CVB3 3Cwt (0.1 μg) alongside HeLa lysates (30 μg) were used to perform in-vitro cleavage assay. Western blotting was performed to probe for endogenous ULK1 and normalized to ACTB as in Fig. 1A. Arrows denote cleavage fragments. (E) PLpro consensus cleavage motif in ULK1 from H. sapiens, M. musculus, and R. norvegicus is presented. Highlighted gray boxes denote consensus sites. Dashed arrows denote sites of potential cleavages. (F) HEK293T cells were co-transfected with PLpro and either 3×Flag-ULK1WT, 3×Flag-ULK1-G499mut or 3×Flag-ULK1-G531mut. Western blotting was conducted with anti-Flag and anti-GFP antibody for detection of ULK1 and PLpro, respectively. (G) Schematic diagram of ULK1 protein sequence with corresponding functional domains, antibody recognition epitopes and identified PLpro cleavage site.
2.3. Purification of SARS-CoV-2 3CLpro
pET-41c encoding wild-type or mutant(C145A) SARS-CoV-2 3CLpro were transformed into C41(DE3) E. coli. Starter-culture from a single colony was grown overnight and diluted 100-fold in Terrific-Broth. Expression was induced with 1 mM IPTG after cultures reached an OD600 of 0.6–0.8 and proceeded at 25 °C for 5 h. Protein was purified using Ni-NTA according to the manufacturer’s instructions.
2.4. In-vitro cleavage assay
HeLa lysates (20 μg) were incubated with WT or inactive (C145A) SARS-CoV-2 3CLpro (4 μg) in cleavage assay buffer (20 mM HEPES pH 7.4, 150 mM KOAc, 1 mM DTT) for indicated times at 37 °C. Reactions were terminated with 6×sample buffer and subjected to western analysis.
2.5. Western blot analysis
Cells were lysed in MOSLB buffer and subjected to western analysis using the following primary antibodies: LC3(NB100-2220), ACTB(A5316), monoclonal-ULK1 (sc-390904), polyclonal-ULK1 (ab167139), NSP9-(SAB3701435), FLAG-(F1804), and GFP-(A-6455).
2.6. Immunoprecipitation
Immunoprecipitation of Flag-ULK1 was performed using ANTI-FLAG®M2 Affinity Gel (F2426) according to manufacturer’s instructions.
2.7. Viral titer measurement
Samples were serially diluted and overlaid on Terasaki plates pre-seeded with 17Cl1 cells. 48 h post-incubation, TCID50 was calculated by the statistical method of Reed/Muench [17]. Titers were expressed as plaque forming unit (PFU)/mL as previously described [18].
2.8. Real-time quantitative RT-PCR
Gene expression was quantified as previously described [19], using primers for Ulk1 (forward:GCAGCAAAGACTCCTGTGACAC; reverse:CCACTACACAGCAGGCTATCAG), and N-gene (forward:CAAAGAAAAGGGCGTAGACAGG; reverse:CGCCATCATCAAGGATCTGAG) and normalized to Actb.
2.9. Statistical analysis
Statistical analysis was performed with unpaired Student’s t-test or analysis of variance (ANOVA). P-value <0.05 was considered statistically significant. Results are presented as mean ± SD and representative of 3 independent experiments.
3. Results
Protein expression of ULK1 is decreased while RNA level is upregulated following mouse coronavirus (M-CoV) infection.
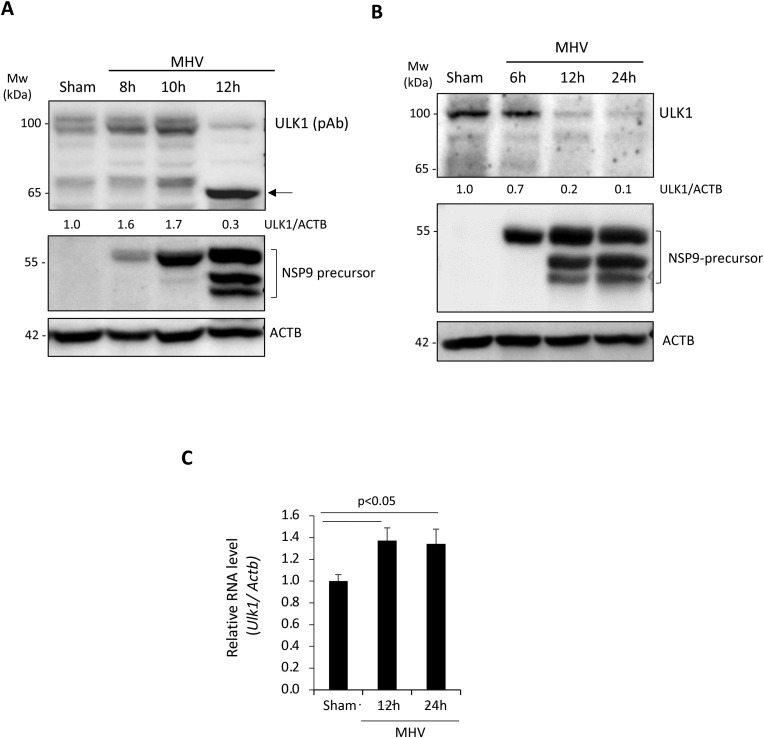
To understand the molecular underpinnings of betacoronavirus subversion of cellular autophagy, we utilized mouse hepatitis virus-A59 (MHV-A59) as a model and examined its effects on critical components of the canonical autophagy pathway. The serine-threonine kinase ULK1 is an important regulator of cellular autophagy and has recently emerged as an innate immune signaling factor [3,[7], [8], [9]]. The significance of autophagy and innate immune signaling as critical facets of host anti-viral defense, prompted us to investigate the regulation and function of ULK1 during betacoronavirus infection. A time-course infection was conducted with MHV-A59 at a multiplicity of infection (MOI) of 10 in the murine fibroblast cell line 17Cl1. Viral replication was verified by immunoprobing for viral non-structural protein 9 (NSP9) precursor. Coincident with active viral polyprotein processing of NSP9 precursor, we observed a significant loss of ULK1 protein starting at 12 h post-infection with both anti-ULK1 antibodies tested in this study (Fig. 1A & B). Interestingly, using the polyclonal anti-ULK1 antibody that recognizes amino acids 351–400, we observed an additional band at ∼65 kDa (Fig. 1A). However, this potential cleavage fragment was undetected with monoclonal anti-ULK1 antibody that recognizes amino acids 511–750 (Fig. 1B), indicating a cleavage may occur within this region. To investigate whether virus-mediated loss of ULK1 is through transcriptional regulation, we performed RT-qPCR. Fig. 1C showed that mRNA levels of Ulk1 were elevated following 12 h and 24 h infection, suggesting that reduced protein expression of ULK1 is not a result of decreased mRNA level. In contrast, elevated gene expression may indicate a compensatory mechanism following reduced protein expression.
Fig. 1.
Protein level of ULK1 is decreased following MHV-A59 infection.
(A–B) Murine 17Cl1 fibroblasts were infected with MHV-A59 (MOI = 10) for various times as indicated or sham-infected with DMEM. MHV replication was assessed via western blot analysis of NSP9 precursor processing. Cell lysates were harvested and probed for ULK1 using polyclonal anti-ULK1 (A, which recognizes amino acids 351–400) or monoclonal anti-ULK1 (B, which was raised against amino acids 511–750), NSP9, and ACTB. Densitometric analysis of ULK1 protein level was performed with NIH ImageJ, normalized to ACTB and presented underneath as fold changes where the first lane was arbitrarily set to 1. Arrow denotes the cleavage fragment detected at ∼65 kDa. (C) Murine 17Cl1 cells were sham-infected or infected with MHV-A59 (MOI = 10) for 12 h or 24 h. Gene level of Ulk1 was measured by real-time quantitative RT-PCR and normalized to Actb (mean ± SD, n = 3).
3.1. PLpro cleaves ULK1 after G499
The discovery of a ULK1 cleavage product in MHV-infected lysates prompted us to investigate whether betacoronaviral proteases are responsible for ULK1 cleavage. Betacoronaviruses encode two proteases, a papain-like cysteine protease (PLpro) and a 3-chymotrypsin-like cysteine protease (3CLpro, also known as Main protease, Mpro) that process viral polyproteins into individual functional proteins [20]. We generated a construct expressing PLpro of SARS-CoV-2, which shares 63% similarity with PLpro domain 2 of MHV-A59. To determine whether PLpro targets ULK1, HEK293T cells were transfected with either a control vector or a plasmid expressing PLpro for 24 h. Expression of PLpro alone was sufficient to recapitulate the reduction in full-length ULK1 observed following MHV-A59 infection using the monoclonal anti-ULK1 antibody (Fig. 2 B). PLpro-mediated cleavage of ULK1 was verified using N-terminal 3×Flag-ULK1 construct (Fig. 2C). HEK293T cells were co-transfected with PLpro together with 3×Flag-ULK1. Lysates probed with anti-Flag antibody revealed significant reduction of ULK1 with the concomitant detection of a lower-molecular-weight fragment of ∼70 kDa. To exclude the involvement of 3CLpro, we performed in-vitro cleavage assay. Time-course treatment revealed that ULK1 was not targeted by SARS-CoV-2 3CLpro, whereas coxsackievirus 3Cpro, the positive control [21], demonstrated efficient cleavage of ULK1 (Fig. 2D). Together, our data suggest that PLpro, but not 3CLpro, targets ULK1 for cleavage.
To identify the precise location within ULK1 that is targeted by PLpro, we closely investigated the protein sequence of ULK1 and identified two potential cleavage sites in the central region of ULK1 (Fig. 2E) [22]. Using Flag-tagged ULK1 construct, we mutated the sites and co-transfected wild-type (WT) or mutant ULK1 with either control or PLpro construct into HEK293T cells. Expression of PLpro resulted in the loss of the 3×Flag-ULK1 and the appearance of a faster migrating band at ∼70 kDa in cells expressing WT-ULK1 and G531 mutant ULK1, but not in cells expressing G499 mutant (Fig. 2F), indicating that PLpro cleaves ULK1 after G499. The observed N-terminal fragment matches the molecular weight of the predicted cleavage fragment. The resulting cleavage of ULK1 by PLpro separates its N-terminal kinase domain from a C-terminal substrate binding domain (Fig. 2G).
3.2. PLpro-mediated cleavage of ULK1 disrupts autophagy
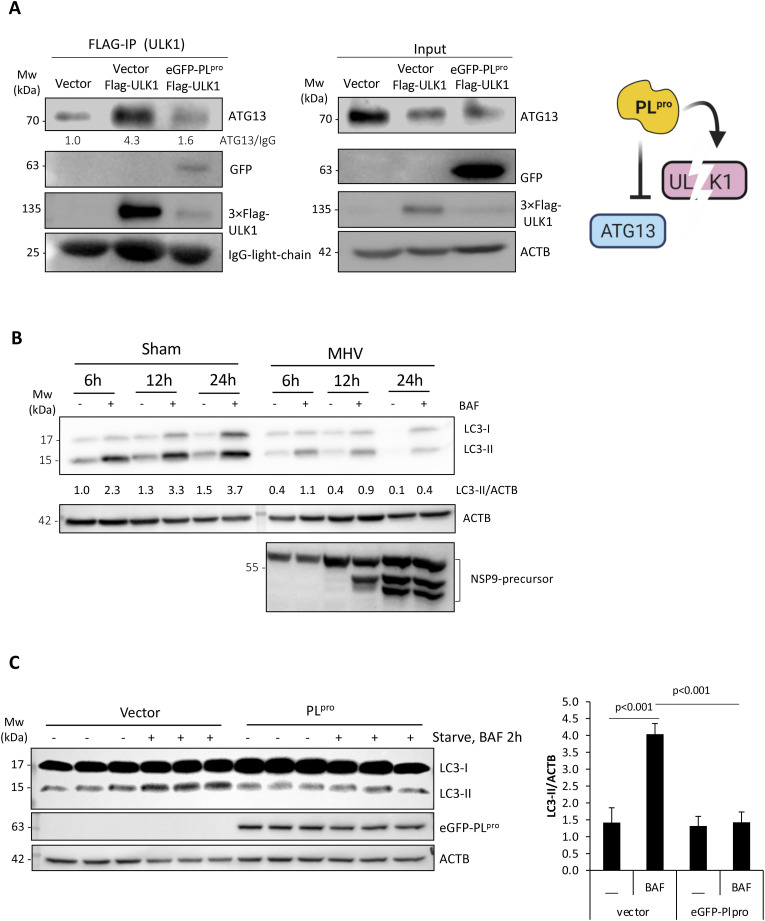
To understand the functional consequence of PLpro-mediated cleavage of ULK1, we first examined whether the ability of ULK1 to interact with binding partners is disrupted. ATG13 is a bridging molecule that directly interacts with ULK1 and the scaffold protein FIP200 in the autophagy-initiating ULK1 complex [3]. Expression of PLpro resulted in reduced co-immunoprecipitation of ULK1 with its binding partner ATG13 (Fig. 3 A). Given that ULK1 complex is required for starvation-induced autophagy, we inquired the functional significance of its disruption. The rate of autophagic degradation, designated as ‘autophagy-flux’, can be measured by comparing levels of LC3-II, a substrate of autophagy, in the presence or absence of lysosomal fusion inhibitors [23]. 17Cl1 cells were sham- or MHV-A59-infected for 6 h, 12 h, and 24 h either in the presence of DMSO or lysosomal fusion inhibitor, bafilomycin-A1 (BAF). Consistent with impaired autophagy, MHV-infected cells had reduced accumulation of LC3-II in the presence of BAF (Fig. 3B). We next tested whether PLpro expression can recapitulate the impaired autophagy observed following MHV-A59 infection. HEK293T cells were transfected with either a control vector or plasmid expressing PLpro for 24 h and then subjected to starvation in HBSS medium for 2 h in the presence or absence of BAF. Control cells responded efficiently to starvation by increasing autophagy-flux; however, cells expressing PLpro did not initiate autophagy (Fig. 3C), suggesting that PLpro expression disrupts cellular autophagy.
Fig. 3.
PLpro-mediated cleavage of ULK1 disrupts autophagy.
(A) HEK293T cells were co-transfected with 3×Flag-ULK1 together with control vector or PLpro for 24 h. Immunoprecipitation of exogenous 3×Flag-ULK1 was performed using anti-Flag M2 agarose beads. IgG light chain and ACTB served as loading controls for immunoprecipitation and inputs, respectively. Schematic depiction of disrupted formation of ULK1-ATG13 complex (right). (B) Disruption of autophagy-flux in MHV-A59-infected cells. Autophagy-flux was assayed in the presence or absence of BAF (125 nM) following sham or viral infection. Cell lysates were analyzed by western blotting for LC3 and normalized to ACTB. NSP9 was used as marker of active viral replication. (C) HEK293T cells expressing either empty vector or PLpro construct were starved for 2 h in the presence or absence of lysosomal inhibitor BAF (125 nM) for 2 h. Lysates were probed for LC3 and normalized to ACTB. Quantification of LC3-II was conducted as in Fig. 1A and presented as (mean ± SD, n = 3) in the right panel.
3.3. ULK1 plays a dual role in MCoV replication
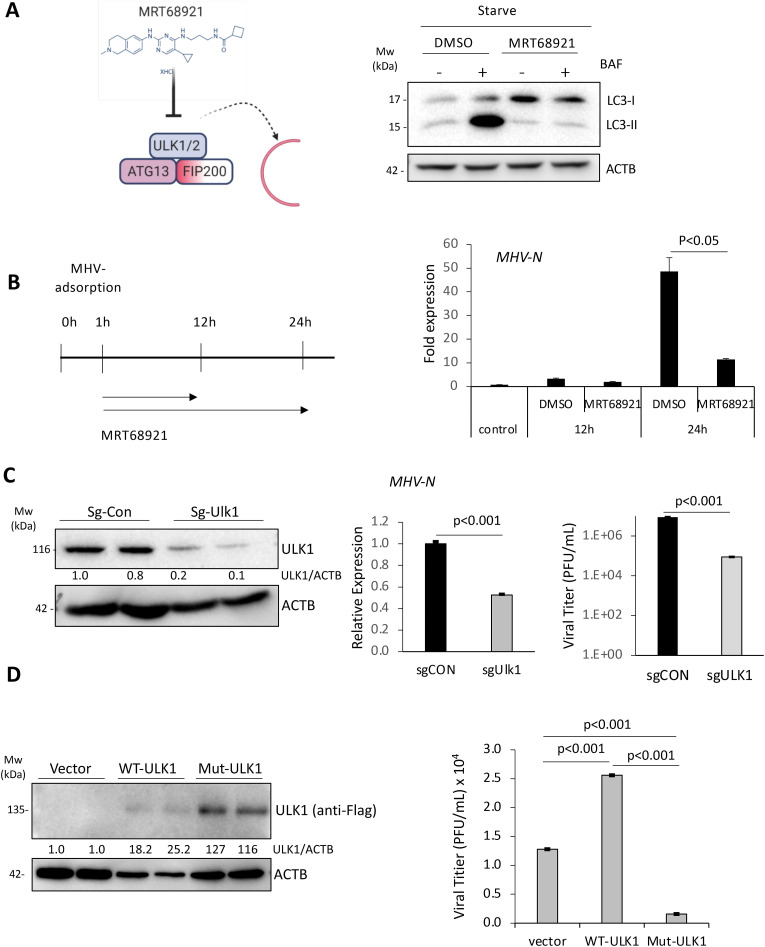
The role of ULK1 in betacoronavirus replication is currently unclear. To clarify this, we utilized chemical and genetic approaches to target ULK1 prior to infection with MHV-A59. The chemical inhibitor, MRT68921, potently targets ULK1/2 kinase activity [24]. Functional validation of the inhibitor showed the complete blockage of starvation-induced autophagy (ie. reduced LC3-II accumulation in starvation-induced, BAF-treated cells (Fig. 4 A). To investigate the consequence of ULK1 inhibition, 17Cl1 cells were inoculated with MHV-A59 for 1 h, followed by replenishment with medium containing either DMSO or MRT68921 (5 μM). After 12 or 24 h, the N-gene levels of MHV-A59 were measured by RT-qPCR. Compared to control treatment, MRT68921 significantly attenuated viral RNA replication at 24 h post-infection (Fig. 4B). Consistent with a pro-viral role for ULK1, RNA levels and viral titers of MHV-A59 were significantly reduced following gene-silencing of Ulk1 (Fig. 4C). We also assessed the effects of expression of WT- or non-cleavable mutant-ULK1 on viral replication. We showed that cells expressing WT-ULK1 displayed significantly enhanced viral titers (Fig. 4D), supporting a pro-viral function for ULK1. However, a non-cleavable ULK1 mutant demonstrated diminished viral titers (Fig. 4D). Collectively, these data support a requirement of ULK1 for replication prior to its cleavage.
Fig. 4.
ULK1 plays a dual role in MHV-A59 replication.
(A) Schematic diagram of the chemical structure and mechanism of action of MRT68921 (left). HEK293T cells were starved for 2 h in the presence or absence of MRT68921 (5 μM) and BAF (125 nM) as indicated. Western blotting was carried out for detection of LC3 and ACTB. (B) Schematic illustration of MRT68921 treatment of 17Cl1 cells following MHV-A59 infection (left). 17Cl1 cells were infected with MHV-A59 (MOI = 10) for indicated time-points in the presence of DMSO (vehicle control) or MRT68921 (5 μM). Relative MHV-N RNA level was determined by RT-qPCR and normalized to Actb (mean ± SD, n = 3).(C) Murine 17Cl1 cells were transfected Cas9 and sg-CON or sg-ULK1 for 48 h. Cells were then infected with MHV-A59 (MOI = 10) for an additional 24 h. ULK1 knockout efficiency (left), viral RNA levels in the cells (middle) and viral titers in the supernatant (right) were measured by western blotting, RT-qPCR, and TCID50 assay, respectively. (D) 17Cl1 cells were transfected with vector, 3× Flag-ULK1WT, or 3× Flag-ULK1MUT for 24 h. Cells were then infected with MHV-A59 (MOI = 10) for 24 h. Cell lysates were analyzed by western analysis for exogenous ULK1 with anti-Flag antibody. Viral titers in culture medium were determined by TCID50 assay (mean ± SD, n = 3).
4. Discussions
Diverse viruses have evolved strategies to circumvent the inherently anti-viral defense capacity of autophagy [25]. In particular, RNA viruses that replicate in the cytoplasm can utilize autophagic membranes as topological surfaces for viral replication [15,26,27]. However, viruses must evade selective targeting and degradation via autophagy, termed virophagy, a process mediated by autophagy receptors that recognize and sequester viral components inside autophagosomes [28]. The autophagy receptor SQSMT1 was previously shown to mediate virophagy of the positive-strand RNA virus Sindbus [29]. To counteract this, enterovirus such as coxsackievirus B3 and enterovirus-D68 have evolved strategies to subvert the host virophagy efforts by engaging virus-encoded proteases to cleave SQSTM1 [19,30]. Similarly, enteroviral proteases impede the degradation capacity of autophagy by disrupting autophagosome-lysosome fusion through the selective cleavage of autophagic fusion SNARES [23,31]. Although recent studies are beginning to unravel the subversion strategies utilized by other RNA viruses, including betacoronaviruses, it remains unclear whether betacoronavirus-encoded proteases target host substrates.
The murine betacoronavirus MHV-A59 was reported to induce double membrane vesicles reminiscent of autophagosomes in the absence of intact autophagy [32], while infection of porcine betacoronavirus, PHEV, demonstrated significant reduction of ULK1 [33]. Findings from the current study suggest that betacoronaviruses may actively target canonical autophagy factors. We provide evidence that SARS-CoV-2-endoced PLpro can cleave ULK1 to disrupt formation of the autophagy-initiating ULK1-complex and functionally impair starvation-induced cellular autophagy/degradative capacity. SARS-CoV-1 PLpro was previously reported to have de-ubiquitinase activity by recognizing the consensus LXGG motif similar to other host deubiquitinating enzymes [34]. Of note, we identified the SARS-CoV-2 PLpro cleavage site of ULK1 after glycine(G) 499, follows precisely the consensus sequence recognized by the de-ubiquitinase. The interferon regulatory factor 3 (IRF3) was also recently reported as a substrate of PLpro following an LGGG consensus motif similar to ULK1 [20]. The targeting of ULK1 and possibly other host proteins harboring an LXGG motif therefore underscores a novel mechanism through which betacoronaviruses subvert cellular autophagy. In addition to disrupting ULK1-mediated autophagy, the deubiquitinase activity of PLpro may favor viral pathogenesis by disrupting selective autophagy, a process that relies on autophagy receptors recognizing ubiquitin-modified pathogens or cellular cargo [35].
The role of ULK1 in betacoronaviral replication is poorly defined. We found ULK1 protein levels are relatively intact during the initial 6 h of MHV infection. Furthermore, loss-of-function studies prior to infection either through chemical inhibition of ULK1/2 kinase activity or genetic silencing of ULK1 revealed a significant reduction in MHV viral replication. In contrast, ULK1 protein levels are significantly decreased during late infection, coinciding with the emergence of lower-molecular-weight cleavage fragment. Additionally, the expression of WT-ULK1 that can undergo proteolytic processing by PLpro enhances viral titers unlike the expression of a non-cleavable ULK1. Collectively, these data suggest that ULK1 may serve a hitherto uncharacterized pro-viral role during early replication. The cleavage of ULK1 during late infection likely suggests that (1) ULK1 function may be dispensable at late stages; (2) ULK1 may serve late-stage anti-viral function that needs to be inactivated; and/or (3) ULK1 cleavage fragments may serve novel pro-viral function. The attenuation of viral titers in cells expressing non-cleavable ULK1 suggests that persistent ULK1 activity throughout the course of infection may be undesirable for virus. Indeed, ULK1 has previously been reported to regulate anti-viral innate immune signaling [11,12].
In summary, we uncover a novel function for the PLpro of SARS-CoV-2 in cleaving the autophagy-regulating kinase ULK1 to clarify betacoronaviral pathogenesis and subversion of cellular autophagy.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by NSERC (RGPIN-2016-03811), CIHR (PJT-159546 and PJT-173318), and Heart & Stroke Foundation (G-18-0022051) to HL. YM is the recipient of ALS Canada-Brain Canada Doctoral-Fellowship. YM, YCX, and AB are recipients of a four-year PhD Fellowship from UBC. YCX is recipient of CIHR Doctoral-Fellowship. CSN and HTL are supported by MITACS Accelerate.
References
- 1.Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., Skipper C.P., Nascene A.A., Nicol M.R., Abassi M., Engen N.W., Cheng M.P., LaBar D., Lother S.A., MacKenzie L.J., Drobot G., Marten N., Zarychanski R., Kelly L.E., Schwartz I.S., McDonald E.G., Rajasingham R., Lee T.C., Hullsiek K.H. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N. Engl. J. Med. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.Y., Kim J., Kim H., Neufeld T.P., Dillin A., Guan K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egan D.F., Chun M.G., Vamos M., Zou H., Rong J., Miller C.J., Lou H.J., Raveendra-Panickar D., Yang C.C., Sheffler D.J., Teriete P., Asara J.M., Turk B.E., Cosford N.D., Shaw R.J. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henckaerts L., Cleynen I., Brinar M., John J.M., Van Steen K., Rutgeerts P., Vermeire S. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm. Bowel Dis. 2011;17:1392–1397. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 6.Yang M., Liang C., Swaminathan K., Herrlinger S., Lai F., Shiekhattar R., Chen J.F. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joo J.H., Wang B., Frankel E., Ge L., Xu L., Iyengar R., Li-Harms X., Wright C., Shaw T.I., Lindsten T., Green D.R., Peng J., Hendershot L.M., Kilic F., Sze J.Y., Audhya A., Kundu M. The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol. Cell. 2016;62:982. doi: 10.1016/j.molcel.2016.05.030. [DOI] [PubMed] [Google Scholar]
- 8.Saleiro D., Blyth G.T., Kosciuczuk E.M., Ozark P.A., Majchrzak-Kita B., Arslan A.D., Fischietti M., Reddy N.K., Horvath C.M., Davis R.J., Fish E.N., Platanias L.C. IFN-gamma-inducible antiviral responses require ULK1-mediated activation of MLK3 and ERK5. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aap9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleiro D., Mehrotra S., Kroczynska B., Beauchamp E.M., Lisowski P., Majchrzak-Kita B., Bhagat T.D., Stein B.L., McMahon B., Altman J.K., Kosciuczuk E.M., Baker D.P., Jie C., Jafari N., Thompson C.B., Levine R.L., Fish E.N., Verma A.K., Platanias L.C. Central role of ULK1 in type I interferon signaling. Cell Rep. 2015;11:605–617. doi: 10.1016/j.celrep.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saul V.V., Seibert M., Kruger M., Jeratsch S., Kracht M., Schmitz M.L. ULK1/2 restricts the formation of inducible SINT-speckles, membraneless organelles controlling the threshold of TBK1 activation. iScience. 2019;19:527–544. doi: 10.1016/j.isci.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konno H., Konno K., Barber G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao P., Wong K.I., Sun X., Reilly S.M., Uhm M., Liao Z., Skorobogatko Y., Saltiel A.R. TBK1 at the crossroads of inflammation and energy homeostasis in adipose tissue. Cell. 2018;172:731–743 e712. doi: 10.1016/j.cell.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkegaard K. Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 2009;335:323–333. doi: 10.1007/978-3-642-00302-8_16. [DOI] [PubMed] [Google Scholar]
- 16.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hygiene. 1938;27:493–497. [Google Scholar]
- 18.Davis B.D., Dulbecco R., Eisen H.N., Ginsberg H.S., Wood W.B. Microbiology. Harper and Row; New York: 1972. Nature of viruses. [Google Scholar]
- 19.Mohamud Y., Qu J., Xue Y.C., Liu H., Deng H., Luo H. CALCOCO2/NDP52 and SQSTM1/p62 differentially regulate coxsackievirus B3 propagation. Cell Death Differ. 2019;26:1062–1076. doi: 10.1038/s41418-018-0185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moustaqil M., Olivier E., Chiu H., Van Tol S., Rudolffi-Soto P., Stevens C., Bhumkar A., Hunter D.J.B., Freiberg A., Jacques D., Lee B., Sierecki E., Gambin Y. 2020. SARS-CoV-2 Proteases Cleave IRF3 and Critical Modulators of Inflammatory Pathways (NLRP12 and TAB1) : Implications for Disease Presentation across Species and the Search for Reservoir Hosts. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamud Y., Shi J., Tang H., Xiang P., Xue Y.C., Liu H., Ng C.S., Luo H. Coxsackievirus infection induces a non-canonical autophagy independent of the ULK and PI3K complexes. Sci. Rep. 2020;10:19068. doi: 10.1038/s41598-020-76227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohamud Y., Shi J., Qu J., Poon T., Xue Y.C., Deng H., Zhang J., Luo H. Enteroviral infection inhibits autophagic flux via disruption of the SNARE complex to enhance viral replication. Cell Rep. 2018;22:3292–3303. doi: 10.1016/j.celrep.2018.02.090. [DOI] [PubMed] [Google Scholar]
- 24.Petherick K.J., Conway O.J.L., Mpamhanga C., Osborne S.A., Kamal A., Saxty B., Ganley I.G. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy (vol 290, pg 11376, 2015) J. Biol. Chem. 2015;290 doi: 10.1074/jbc.C114.627778. 28726-28726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong X., Levine B. Autophagy and viruses: adversaries or allies? J. Innate Immun. 2013;5:480–493. doi: 10.1159/000346388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson W.T., Giddings T.H., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:861–871. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohamud Y., Luo H. The intertwined life cycles of enterovirus and autophagy. Virulence. 2019;10:470–480. doi: 10.1080/21505594.2018.1551010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudchodkar S.B., Levine B. Viruses and autophagy. Rev. Med. Virol. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orvedahl A., MacPherson S., Sumpter R., Jr., Talloczy Z., Zou Z., Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corona A.K., Saulsbery H.M., Corona Velazquez A.F., Jackson W.T. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. 2018;22:3304–3314. doi: 10.1016/j.celrep.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corona A.K., Saulsbery H.M., Velazquez A.F.C., Jackson W.T. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. 2018;22:3304–3314. doi: 10.1016/j.celrep.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maier H.J., Britton P. Involvement of autophagy in coronavirus replication. Viruses. 2012;4:3440–3451. doi: 10.3390/v4123440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Zhao K., Lv X., Lan Y., Hu S., Shi J., Guan J., Yang Y., Lu H., He H., Gao F., He W. Ulk1 governs nerve growth factor/TrkA signaling by mediating Rab5 GTPase activation in porcine hemagglutinating encephalomyelitis virus-induced neurodegenerative disorders. J. Virol. 2018;92 doi: 10.1128/JVI.00325-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]