Abstract
Human coronaviruses (HCoV) are no stranger to the global environment. The etiology of previous outbreaks with reported symptoms of respiratory tract infections was attributed to different coronavirus strains, with the latest global pandemic in 2019 also belonging to the coronavirus family. Timely detection, effective therapeutics and future prevention are stake key holders in the management of coronavirus-induced infections. Apart from the gold standard clinical diagnostics, electrochemical techniques have also demonstrated their great potentials in the detection of different viruses and their correlated antibodies and antigens, showing high sensitivities and selectivities, and faster times for the analysis. This article aims to critically review the multifaceted electrochemical approaches, not only in the development of point-of-care portable devices but also as alternative detection strategies that can be coupled with traditional methods for the detection of various strains of coronaviruses.
Keywords: Coronavirus, Infections, SARS-CoV-2, COVID-19, Electrochemistry, Electrochemical methods, Biosensing, Biosensors, Diagnostic, Detection
Highlights
-
•
Electrochemical biosensing detection methods for coronavirus-induced infections.
-
•
Potential rapid and cost-effective point-of-care diagnostics for mass testing.
-
•
Electrochemistry helps to overcome limitations faced by gold standard diagnostics.
-
•
Experimental schemes, strengths, and drawbacks of various electrochemical strategies.
1. Introduction
1.1. Epidemiology and zoonotic origins of coronaviruses
The novel coronavirus outbreak in 2019, which associated disease has been named COVID-19, has once again placed coronavirus family under the spotlight. Comprising of four genera, Alphacoronavirus, Betacoronavirus, Deltacoronavirus and Gammacoronavirus [1,2], lineages of Alphacoronavirus and Betacoronavirus are commonly known to have originated from the viral gene reservoir of bats. They are responsible for the emergence of seven HCoV associated with mild to severe respiratory tract infections. Four of these identified strains were endemic in nature (HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1) [2], which accounts for 15–30% of common cold infections in humans [1]. On the other hand, the remaining three strains possess higher pathogenicity and virulence that have resulted in the development of past epidemics [2] like SARS-CoV in 2003 and Middle East respiratory syndrome (MERS-CoV) in 2012, and even including the recent historic pandemic (SARS-CoV-2/COVID-19) in 2019 that the world is currently fighting. The stronger pathogenicity and virulence of these strains are attributed to the zoonotic transmissions that humans lack immunity towards to. With bats acting as the primary host, SARS-CoV and MERS-CoV were transmitted to humans via intermediate species such as palm civets and dromedary camels respectively [3]. Similarly, COVID-19 was also speculated to be involved in the zoonotic transmission from bats to an intermediate host, however the intermediate's identity has not been validated yet [3]. Therefore, there always exists a continuous risk and future possibility of emerging coronaviruses that can originate from animal viral gene reservoirs to infect humans, which will create an unprecedented need for rapid diagnostic platforms for the early detection of these infectious diseases. Clinically similar but pathogenically different [4], the genomic sequence of SARS-CoV-2 shared around 96% similarity to a horseshoe bat (Rhinolophus affinis) coronavirus RaTG13 [5], approximately 80% similarity with SARS-CoV [3,5], and 50% similarity with MERS-CoV [3]. Since SARS-CoV, MERS-CoV and SARS-CoV-2 not only belong to the same genus but also share similarities in their genomic sequences and zoonotic transmission nature, our review article on the overview of electrochemistry detection methods on closely-related coronaviruses aim to shed light on ongoing efforts in the development of electrochemical devices for effective SARS-CoV-2 diagnostic assays.
SARS-CoV had affected 26 countries with more than 8000 cases reported in 2003 [2], while MERS-CoV had impacted 27 countries with 2494 laboratory-confirmed cases and 858 deaths recorded since September 2012 [2]. As of 17th August 2020, COVID-19 has spread to 216 countries, areas or territories with an alarming number of 21,549,706 confirmed cases and 767,158 reported deaths [6]. As a result of this exceptionally pernicious SARS-CoV-2 virus, global healthcare systems were maximized with social and economic activities being heavily disrupted as well. Furthermore, the data on COVID-19 infections and death toll still continue to evolve daily, where second waves of cluster infections were seen resurging already in countries like China, Iran, Japan and South Korea after easing of lockdown measures and reopening of economy [7]. Hence, there remains a dire need for accurate and scaled-up analytical testing to quickly isolate the infected individuals and ultimately stop the virus spread. However, in contrast to the past epidemics SARS-CoV and MERS-CoV, the wiliest characteristic of SARS-CoV-2 is the silent spread from asymptomatic infected individuals who, even without clinical symptoms, are capable of possibly shedding and transmitting the virus beyond the incubation period of 14 days, accounting for a significant percentage of 40–45% of the total infected cases [8]. Consequently, this high incidence rate of asymptomatic COVID-19 infections highlights the increasing demand and the challenge for highly sensitive diagnostics to detect these ‘silent’ carriers that might be tested negative due to the variability in viral loads. Besides, it was discovered that the reproduction number of SARS-CoV-2 (R0) was around 2.2 [9] which translates to one unknowingly infected individual can transmit the virus to 2.2 people, with the spreading effect increasing exponentially. With a commercial vaccine for COVID-19 still in the trial phase, there is an urgent need to develop rapid, cost-effective, sensitive and selective diagnostics for the early detection and fast isolation of infected people, in order to be able to flatten the virus propagation curve.
1.2. Challenges faced by current diagnostics in coronavirus detection
Comprising of a single-stranded positive-sense RNA genome enveloped inside, coronaviruses encode a RNA-dependent RNA polymerase gene (RdRP) and four structural proteins: spike surface glycoprotein (S), envelope protein (E), matrix protein (M) and nucleocapsid protein (N) [2]. These coronaviruses' structural features serve as the target sites in the development of detection strategies for coronavirus-induced infections.
Significant research efforts have been dedicated to the development of molecular (nucleic acids-based) and serological assays (antibodies-based) in the management of coronavirus detection [2,10,11]. Molecular assays help to directly identify infected individuals at the acute phase of the infection, while serological assays assist in the detection of antibodies produced by the individuals following after the viral exposure [11]. Real-time polymerase chain reaction (RT-PCR) has been the diagnostic workhorse in the detection of coronavirus-induced infections, being able to accurately identify and target the pathogens specifically based on their genomic sequences. Unfortunately, the time-consuming analysis [12], instrumentation costs [12], the need for trained personnel and sophisticated laboratory infrastructure [12], as well as the logistics involved for the reagents and extraction kits may impede the universal use of RT-PCR, especially in public health emergencies like COVID-19 infection with testing demand always increasing. In addition, the accuracy of RT-PCR could be compromised with improper sample collection and handling [13,14], degradation of viral RNA [13,15], presence of PCR inhibitors [13] and variability in viral load due to changes in viral shedding [13,16]. As a result of these factors that lowers analytical sensitivity, RT-PCR can be susceptible to false negatives, which will pose a risk of community transmission when infected individuals return to the community without isolation and treatment.
Reliable serological tests are mostly conducted by enzyme-linked immunosorbent assay (ELISA), which monitor seroconversion to determine past infections and possible viral immunity to aid in therapeutics development. However, a duration of 1–5 h is expected to obtain the results from ELISA, where skilled personnel and designated laboratories are also required [12,17]. All these factors add on to turnaround time and costs which decrease the effectiveness on the mass production of these diagnostic assays to cater to the detection demand. Also, the need for laboratories and trained labor for the operational aspects increases the complexity of the assay, where portable on-site detection and adoption by end users are considerably limited.
Given the challenges faced by these existing methods in the detection of coronavirus-induced infections, the opportunity may now lie in electrochemistry diagnostics to overcome these drawbacks with their benefits such as ease-of-use, on-the-spot detection, cost-effectiveness and fast turnaround time from sample to results.
1.3. Trends in recent literature diagnostics for coronavirus-induced infections
To circumvent the bottleneck and improve on the analytical figures of merit (sensitivity, selectivity and reproducibility) of gold standard diagnostics, in response to the recent COVID-19 pandemic, a new frontier of diagnostics has been fueled with emergence of novel viable alternatives. Molecular assays recently published were CRISPR-based method DETECTR Cas12 that recognizes SARS-CoV-2 RNA [18], and single-step nested quantitative RT-PCR (qRT-PCR) in one closed tube that targets the ORF1ab and N genes of SARS-CoV-2 [19]. In the DETECTR assay, the extracted RNA undergoes reverse transcription and isothermal amplification via loop-mediated amplification (RT-LAMP) [18]. The Cas12 guide RNAs (gRNA) specifically target SARS-CoV-2 sequences, in this case here is the N and E genes [18]. Upon recognition of the target probe, the Cas12-gRNA cleaves the FAM-biotinylated reporter molecule attached, where the generated signal can be detected by a fluorescent reader or a lateral flow strip readout [18]. Reporter molecules that are not cleaved will be captured at the control line (first detection line) while Cas12 cleavage activity will produce a signal at the test line (second detection line) [18]. For the single-step nested quantitative qRT-PCR assay, the working principle is still primary based on qRT-PCR followed by the detection of fluorescence signals [19]. The difference from the conventional qRT-PCR is that the two PCR steps at the optimum annealing temperature are distinct from each other in a closed single tube [19]. Also, five oligonucleotides are now employed for each gene region [19]. Since the single-step nested quantitative qRT-PCR assay targets two gene regions, ORF1ab and N genes, the total number of primer sets for the two regions can help to improve the analytical sensitivity over the conventional qRT-PCR [19].
Some of the novel optical assays include a selective colorimetric assay with thiol-modified antisense oligonucleotides (ASOs) capped gold nanoparticles (AuNPs) that is specific to the N protein of SARS-CoV-2 [20], and a biosensor with an integration of plasmonic photothermal effect (PPT) and localized surface plasmon resonance (LSPR) in the discrimination of SARS-CoV-2 viral sequences [21]. Without the use of any sophisticated instruments, the colorimetric assay with ASO-capped AuNPs could enable a selective visual detection of SARS-CoV-2 with the naked-eye [20]. In the presence of the target RNA extracted from SARS-CoV-2, the previously well-dispersed ASO-capped AuNPs agglomerate and result in a color change from purple to blue and surface plasmon resonance (SPR) [20]. Upon addition of RNase H, precipitation of AuNPs occurs as RNase H specifically recognizes and breaks the phosphodiester bonds of the SARS-CoV-2 N gene, thus amplifying the SPR signal [20]. The dual-functional biosensor (PPT and LSPR) is made up of complementary DNA sequences (receptors) of SARS-CoV-2 immobilized onto the surface of two-dimensional gold nanoislands (AuNIs) [21]. As the target analyte binds to the receptors functionalized on the AuNIs, the change in refractive index is proportional to the analyte concentration in the sample [21]. Owing to the enhanced in-situ photothermal effect of the AuNIs, the hybridization kinetics between the target analyte and the receptor is enhanced and spurious binding of non-target sequences can be minimized [21].
1.4. Potential of electrochemistry to overcome the limitations faced by current diagnostic strategies
Besides fluorescence, chemiluminescence and colorimetric based detection, electrochemical methods have also emerged as valid alternatives to bridge the clinical gap for widespread accessibility, especially in low-income and resource-limited settings. Electrochemistry including potentiometry, voltammetry, coulometry, and electrochemical impedance spectroscopy, is a branch of physical chemistry in which the electrical parameters are correlated to the chemical changes of the system under study [22]. The past few decades have witnessed an upsurge of electroanalytical methods for interdisciplinary applications, together with the exceptional advancement in the design and performance of electrochemical devices due to the rise of nanotechnology. With the electrode material acting as the transducer, an electrochemical sensor is able to measure the signal changes in current, potential, conductivity and impedance due to the recognition process happening on the sensing surface [23]. When compared to traditional standard methods based on optical detection, electrochemical sensors and biosensors can show an improved selectivity and sensitivity, a lower amount of sample needed (μL to nL), faster response times (several seconds to minutes), operational simplicity, cost-effectiveness, capability for multiplexing, and possibility for miniaturization [23,24]. These desirable attributes duly fulfill most of the ASSURED criteria (Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free and Deliverable to end-users) set by the World Health Organization (WHO) to qualify as efficient diagnostic testing [25]. On that account, electrochemistry can offer attractive strategies in the fight against the COVID-19 pandemic and possible emergence of other coronaviruses surfacing in the future, and contribute to overcoming the limitations of present clinical diagnostics employed for COVID-19 detection (see Table 1, Table 2 ).
Table 1.
Summary table on the various literature electrochemical methods based on their applications, LOD and analysis time.
| Detection Approach | Electrochemical Platforms | Target Analyte Detected | LOD | Analysis Time |
References |
|---|---|---|---|---|---|
| Detection of coronavirus-related proteins | Metal-Oxide-Semiconductor FET (MOSFET) | SARS-CoV N protein |
Sub-nanomolar concentrations | 10 min | [30] |
| Carbon nanotube FET (CNTFET) | SARS-CoV N protein |
5 nM | 10 min | [33] | |
| High-Electron-Mobility Transistor FET (HEMT) | SARS-CoV N protein |
0.003 nM (0.4 pg) | – | [34] | |
| Graphene-based FET (GFET) | SARS-CoV-2 S protein |
2.42 × 102 copies/ml (clinical samples) | – | [36] | |
| Array of AuNPs-modified carbon electrodes | MERS-CoV, OC43 |
0.4 pg/mL (OC43); 1.0 pg/mL (MERS-CoV) |
– | [37] | |
| Eight-channel AuSPEs | SARS-CoV-2 S protein |
1 fg/mL | 3 min | [35] | |
| Detection of viral DNA | Gold films | SARS-CoV 30-mer sequence |
6 pmol/L | – | [42] |
| Polylysine | SARS-CoV | 5 pmol/L | – | [44] | |
| modified SPCEs | 30-mer sequence | 8 pmol/L | – | [45] | |
| Au nanostructured SPCEs | SARS-CoV 30-mer sequence |
2.5 pmol/L | – | [43] | |
| Au/Au/Au electrode system | SARS-CoV | up to 400,000 copies | – | [46] | |
| Glassy carbon | SARS-CoV 30-mer sequence |
5.0 pg/μL | – | [41] | |
| AuSPEs modified with CNTs | SARS-CoV 15-/30-mer sequence |
800 nM | – | [47] | |
| Gold screen-printed electrodes (AuSPEs) | Coronavirus Aviair (174 bp) |
5 ng/mL | 30 min | [49] | |
| AuNPs electrodeposited on Ti surface | SARS-CoV-2 | – | – | [50] | |
| Detection of PCR Products | NanoChip 400 system Gold |
OC43, 229E and HKU1 | 760 ng/μL (OC43); 128 ng/μL (229E) |
– | [54] |
| SARS-CoV-2 | 1 × 105 copies/mL | Under 2 h | [51] | ||
| Detection of ROS | Steel tips modified with MWCNTs | SARS-CoV-2 ROS level |
– | 30 s | [59] |
Table 2.
Summary table on the present diagnostics used for SARS-CoV-2 on their limits of detection (LOD) and analysis time.
| Detection Approach | Diagnostic Platforms | Target Analyte Detected | LOD | Analysis Time | References |
|---|---|---|---|---|---|
| Nucleic acid amplification | qRT-PCR | SARS-CoV-2 E and RdRp genes | 3.9 copies/reaction (E gene); 3.6 copies/reaction (RdRp gene) |
2–4 h | [61] |
| RT-LAMP with colorimetric detection | SARS-CoV-2 | 100 copies/reaction | 30–60 min | [62] | |
| Detection of viral antibodies and antigens | Capture ELISA immunoassay | anti-SARS-CoV-2 IgG and IgM | 2.384a (IgG); 2.187a (IgM) |
2.5 h | [63] |
| ELISA immunoassay | SARS-CoV-2 antigens (N and S proteins) | 100 pg/mL | 3–5 h | [64] |
Fold change over average reading of negative controls.
Besides electrochemical sensors and biosensors for point-of-care diagnosis, electrochemistry was also recently used as an alternative to conventional optical PCR, whereby electroactive indicators or detection labels were employed to monitor the progress of the PCR reaction, with the measured oxidation or reduction signal being correlated to the amount of the PCR-amplified product [26]. Without the need for fluorescent labels and optical detection, electrochemical PCR can represent the cost-effective and portable option sought for COVID-19 intensive testing.
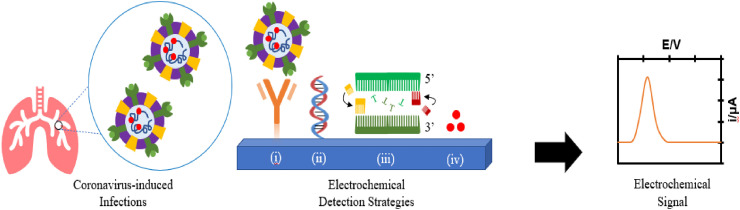
To shed light on ongoing efforts in the development of electrochemical tools for effective coronaviruses diagnostic assays, this review article aims at providing an overview of the state-of-the-art electrochemical detection strategies on closely related coronaviruses. The recognition of viral proteins, DNA, PCR products and reactive oxygen species (ROS) (see Fig. 1 ) correlated to coronavirus induced diseases will be reviewed. Peer-reviewed publications on electrochemical methodologies developed in both the academic and commercial research laboratories will be covered (see Table 1), as well as s summary table on present the present diagnostics used for SARS-CoV-2 will be provided (see Table 2).
Fig. 1.
Overview of the electrochemical approaches on the detection of coronavirus-induced infections: (i) Detection of proteins, (ii) Detection of viral DNA, (iii) Detection of PCR products, (iv) Detection of ROS.
The experimental schemes, advantages and limitations will be presented in this critical review with future directions proposed on the potential of electrochemical techniques for point-of-care diagnosis in the medical field. We believe that this review could help to facilitate the scientific community in the innovation and implementation of the electrochemical diagnostics for the detection of coronavirus-induced infections.
2. Electrochemistry for the detection of coronavirus-induced infections
2.1. Electrochemical detection of coronavirus-related proteins
Serological assays that are based on the detection of recombinant proteins as antigens are extensively used in clinical diagnostics [27]. Protein-based biomarkers show a higher availability and can be more easily isolated as compared to both nucleic acid and cell-based biomarkers [28]. It has been proven that CoV genome encodes four structural proteins namely spike (S), nucleocapsid (N), envelope (E), and matrix (M), with the N and S proteins being the major immunogenic ones in SARS-CoV [29].
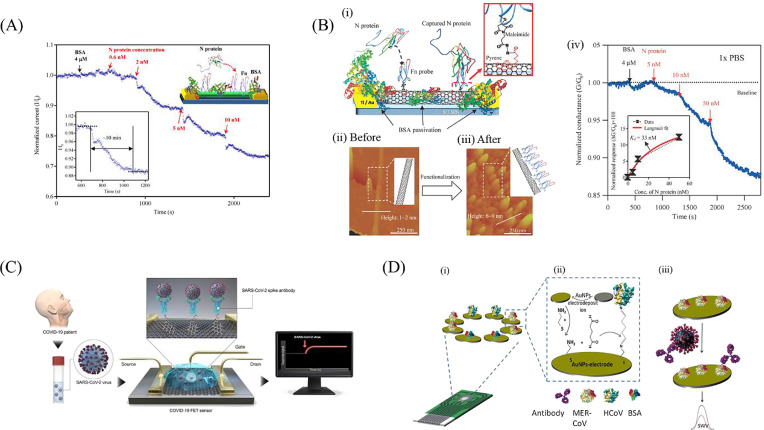
Zhou's group successfully fabricated an In2O3 nanowire field effect transistor (FET) modified with the antibody mimic protein fibronectin (Fn) for the selective detection of the nucleocapsid (N) protein of SARS-CoV [30]. FET-based electrochemical sensors consist of a field effect transistor as the transducer component and a sensing surface [31], which consits of a dielectric layer functionalized with specific receptors that have selective affinity for the target analyte [31,32]. The current flow between the source and the drain is electrostatically regulated by the gate through an applied voltage [32]. As the target analytes are being captured on the sensing surface, a gating (electrostatic) effect is then converted into a measurable electronic signal due to the change in FET electrical properties in the form of a drain-to-source current or channel conductance [32]. Given the nature of the signal output of field effect transistors, detection with FETs has been included in the electrochemical detection of coronavirus-related proteins. In Zhou's work, the Si/SiO2 substrate was first functionalized with In2O3 nanowires via laser ablation chemical vapor deposition (CVD). Subsequently, the carboxyl groups on the surface of In2O3 nanowires were activated for further crosslinking with the thiol group on Fn capture probe (see Fig. 2 A). The electronic properties of the nanowire are affected upon binding of N protein to the Fn biorecognition element, with the normalized current response being proportional to the analyte concentration. With this strategy, the detection of N protein was achieved at sub-nanomolar concentration, with a sensitivity comparable to other immunological assays. In an attempt to enhance the detection performance for N protein of SARS-CoV, the same group later designed a low density carbon nanotube FET (CNTFET) [33]. In contrast to the previous sensing approach of In2O3 nanowires [30], the density of CNTFET had stronger evident effects on the voltage shift in the presence of the target analyte, which contributed to the improved sensitivity that is vital for reliable coronavirus diagnostics [33]. The amplified voltage shift observed was attributed to its small to negligible off-current, lower capacitance and strong gate dependence [33]. Owing to these features of low density CNTFET, the assay's sensitivity and selectivity were enhanced, with a reported detection limit of 5 nM for SARS-CoV N protein (see Fig. 2B).
Fig. 2.
Experimental setups for electrochemical detection of coronavirus related proteins: (A) In2O3 nanowire FET modified with fibronectin for the detection of N protein. The current response is measured after the addition of increasing concentrations of N protein [30]. Reproduced with permission from Ref. [30]; (B) (i) low density CNTFET, (ii, iii) atomic force microscopy images of CNT before and after immobilization of Fn probe (scan area of 1 μm × 1 μm), (iv) plot of Ids versus time due to addition of increasing N protein concentrations of 5, 10 and 50 nM, data points (in black), Langmuir isotherm model fitting (in red) [33]. Reproduced with permission from Ref. [33]; (C) GFET in the sensing of SARS-CoV-2 S protein [36]. Reproduced with permission from Ref. [36]; (D) (i) electrochemical immunosensor for MERS-CoV with an assembled array of gold nanoparticle-modified carbon electrodes, (ii) immunosensor fabrication, (iii) change in peak current due to addition of MERS-CoV antigen observed via square-wave voltammetry [37]. Reproduced with permission from Ref. [37].
A different platform was employed by Hsu's group who investigated on the applicability of AlGaN/GaN HEMT-based sensor to be used as a cost-effective diagnostic strategy for coronavirus detection [34]. Similarly, this work also targeted the N protein of SARS-CoV. The AlGaN/GaN layers were formed through metal-organic CVD, before immobilizing a double stranded DNA sequence from the SARS-CoV genome as the biorecognition element onto the gate regions. Upon binding of the N protein (SARS-CoV CTD), a real-time current response was generated, which is proportional to the analyte concentration. The developed AlGaN/GaN HEMT-based sensor was able to detect a minimum of 0.003 nM (0.4 pg) of the SARS-CoV N protein [34].
It has been reported that the S protein is responsible for inducing the infections as it helps to bind the virus to the cellular receptor in humans [35]. For this reason, the detection via S protein would be more specific in differentiating between the various types of coronavirus-induced infections.
In the latest research work by Park's and Kim's group [4], a graphene-based FET (GFET) biosensor was successfully developed to detect SARS-CoV-2 in clinical samples. The spike (S) protein was targeted as the antigen due to its high immunogenicity [36]. Using coupling agents like 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBASE), the specific SARS-CoV-2 spike antibody was conjugated to the graphene sheets (see Fig. 2C). The S protein (antigen) will specifically bind to the spike antibody functionalized GFET, resulting in a change in the channel surface potential, which in turn produces a real-time electrical response that is dose-dependent. The performance of the fabricated functionalized GFET was evaluated with the patients' nasopharyngeal swab samples, antigen protein, and cultured virus. The recently developed spike antibody functionalized GFET was able to detect SARS-CoV-2 S protein effectively for concentrations as low as 1 fg/mL in phosphate-buffered saline, 100 fg/mL in universal transport medium (UTM), 1.6 × 101 pfu/mL in culture medium and 2.42 × 102 copies/mL in patients' samples. In addition, the high sensitivities accomplished in this GFET removed the need for sample pre-treatment and processing, thus reducing the amount of analysis time, which is crucial for fast results from diagnostic assays. Besides, this functionalized GFET device harnessed the ability to discriminate the S protein of MERS-CoV from SARS-CoV-2 due to the absence of cross-reactivity observed, thus enhancing its selectivity and specificity as a COVID-19 diagnostic approach.
In the work from Eissa et al. [37], a competitive immunosensor was developed based on the indirect competition between the unbound MERS-CoV virus found in the sample and the immobilized MERS-CoV recombinant spike protein, S1 at a specific antibody concentration (see Fig. 2D). The immunosensor was fabricated by electrodepositing gold nanoparticles (AuNPs) on an array of carbon disposable electrodes. As varying concentrations of antigen against the MERS-CoV S1 protein were added, the reduction peak currents of the ferro/ferricyanide redox couple were measured via square-wave voltammetry (SWV). Due to the binding of the antibody to the immobilized MERS-CoV S1 protein, the ferro/ferricyanide electron transfer efficiency was reduced, thus resulting in a lower reduction peak observed. Due to the increased electrode surface area and electron transfer efficiency for the presence of AuNPs, the developed immunosensor was able to achieve a detection limit as low as 0.4 pg/mL and 1.0 pg/mL for HCoV and MERS-CoV respectively.
Very recently, Kintzios et al. demonstrated a portable label-free cell-based bioassay to detect the spike protein antigen S1 of SARS-CoV-2 virus [35]. As a result of the electro-insertion of SARS-CoV-2 spike S1 antibodies into the membrane-engineered mammalian kidney cells, the membrane potential was altered, thus generating a potentiometric signal on the eight-channel gold screen-printed electrode that is proportional to the concentration of the added S1 antibodies. The developed cell-based bioassay was able to attain a detection limit of 1 fg/mL [35] for S1 protein detection, with no cross-reactivity reported against the viral N protein [35]. Kintzios's group also proposed on the possibility of interfacing the developed device with smartphone/tablet-based read-out systems [35], which can help to extend its application to a portable detection tool with smart healthcare surveillance.
It is noteworthy that besides the detection of coronavirus structural proteins, immunoassays are also being developed for the identification of the production of immunoglobulins from the human body after the onset of symptoms. Immunoglobulin M (IgM) are usually first identified in serum after 4–5 days before seroconversion to immunoglobulin G (IgG) (10–14 days later or longer) [12,14,15,38]. As such, the antibody titers at the acute phase of infection might be insufficient to be detectable, and be mistakenly diagnosed as seronegative case, where in fact the individual could already be infected with the virus. In this context, electrochemical detection should be used as a tool to improve the sensitivity of the assay, for the development of an early-detection portable device for the cost-effective and rapid identification of COVID-19 past and current infections.
2.2. Electrochemical detection of viral DNA
Electrochemical detection of specific DNA sequences is mostly carried out using hybridization sensors, also known as DNA sensors or genosensors. A DNA sensor is constructed via the immobilization of a short single-stranded DNA (ssDNA), also called DNA probe, as the biorecognition element on the surface of a transducer [39]. In the presence of the target DNA (a complementary DNA strand), hybridization occurs to form a more stable double-stranded DNA (dsDNA) hybrid, with the biorecognition event being converted by the transducer into a measurable electrochemical signal [40]. Sequence-specific hybridization events can be detected either by indirect measurement with the help of electroactive labels, or directly based on the inherent electroactivity of the DNA nucleobases [40,41].
2.2.1. Label-based assays
The label-based detection of coronavirus specific sequences has been mostly carried out by using non-electroactive markers (such as enzymatic labels) or employing electroactive markers (namely methylene blue and metal complexes) [41].
DNA signaling probes modified with enzymatic labels like alkaline phosphatase (AP) and horseradish peroxidase (HRP) are able to catalyze the hydrolysis of the enzyme substrate into a soluble electroactive product, which redox signal can be measured and correlated to the presence of the target DNA. Due to the inherent amplification efficiency of the enzymatic reaction [42], PCR amplification process of the specific sequence could be avoided, while not compromising the sensitivity of the assay.
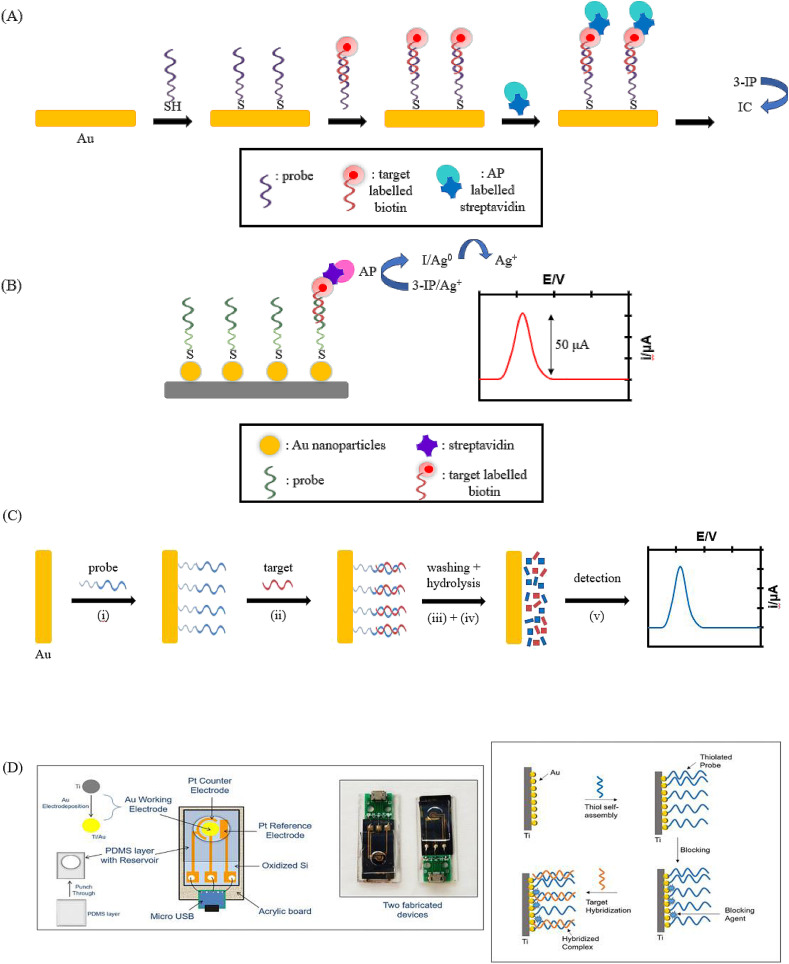
In the detection of SARS-CoV genomic DNA, Costa-García's group made use of the strong thiol-gold interaction to immobilize SH-DNA probes on a 100 nm sputtered gold film (see Fig. 3 A) [[42], [43], [44]]. Upon hybridization with the biotinylated 30-mer SARS-CoV sequence, conjugation with AP-labelled streptavidin occurred, thus promoting the conversion of the enzymatic substrate 3-Indoxy phosphate (3-IP) into the electroactive indigo carmine (IC). The voltammetric signal of the latter was then measured and correlated to the presence and amount of the target analyte. The selectivity of the developed biosensor was assessed by using a 3-base mismatch DNA target as the negative control. Due to the nature of the SARS-CoV virus to undergo potential mutation in order to adapt to the environment, the same group showed that the developed biosensor was also able to distinguish among the complementary target and sequences carrying one and two base mismatches respectively [44]. Later on, the same approach was also employed for the immobilization of the biotinylated probe via electrostatic interactions on screen-printed carbon electrodes (SPCEs) modified with positively charged polylysine [45]. The merits of disposable, single-use and cost-effective make SPCEs an attractive electrochemical detection tool especially for infectious disease to meet the demand of extensive testing, avoid contamination and protect the healthcare workers during sample collection.
Fig. 3.
Experimental principles of label-based and label-free assays in the detection of viral DNA: (A) Schematic diagram of DNA hybridization sensor on gold films with enzymatic electrochemical detection of SARS-CoV gene sequence [42]. Adapted with permission from Ref. [42]; (B) Schematic diagram of DNA hybridization sensor using gold nanostructured screen-printed carbon electrodes for SARS-CoV virus detection [43]. Adapted with permission from Ref. [43]; (C) Schematic diagram on a screen-printed electrochemical biosensor for the detection of released purine bases of coronavirus aviair (i) immobilization of ssDNA probe onto the gold electrode, (ii) hybridization with the complementary target sequence, (iii) washing of the electrode surface, (iv) hydrolysis of dsDNA, (v) electrochemical signal detection [49]. Adapted with permission from Ref. [49]; (D) Schematic diagram on a label-free electrochemical genosensor for COVID-19 diagnosis [50]. Reproduced with permission from Ref. [50].
Higher assay sensitivity is paramount in developing diagnostic tools for coronavirus-induced infections to increase the accuracy of the analysis and reduce the rate of false-negatives during testing. To improve the sensitivity of the label-based assays for the detection of SARS-CoV, Costa-García's group then took on a different approach whereby the product of AP enzymatic reaction was able to reduce silver ions (Ag+) to metallic silver (Ag) [43]. The latter was then detected via anodic stripping voltammetry (see Fig. 3B) for a quantitative correlation to the target analyte. Since the sensitivity of anodic stripping voltammetry of Ag+ was increased by 14-fold, the detection limit of this assay could reach as low as 2.5 pmol/L [43].
In addition to employing enzymatic labels like alkaline phosphatase, horseradish peroxidase (HRP) was used by Gau's group for the development of a single-use electrochemical platform with multi-assay sensor chips in the detection of SARS-CoV [46]. The self-assembled monolayer was constructed with a spacing stem and a recognition stem. The recognition stem consists of the target/probe complex labelled with the HRP immobilized onto the sensor surface (Au/Au/Au electrode system). On the other hand, the spacing stem serves to block the conductive samples like blood and urine from interfering with the electrochemical signal recorded in the presence of the target analyte [46].
Besides enzymatic labels, a label-based genosensor can also make use of electroactive markers such as metal complexes and heterocyclic dyes for signal generation. This allows the use of simplified protocols with a reduced number of steps, together with a shorter time for the analysis at a much-reduced costs [41,45].
In 2006, Costa-García's group reported on a DNA hybridization sensor based on sodium aurothiomalate as the electroactive label [41]. After the physical adsorption of the DNA probes, hybridization was carried out with a 30-mer SARS-CoV sequence modified with ionic gold (Au+). After which, gold electrodeposition was carried out by the application of anodic potentials. The further silver catalytic electrodeposition by anodic stripping occurring on the gold surface was correlated to the amount of the gold-labelled SARS-CoV sequence that has bound onto the complementary probe. The limit of detection obtained for this work was 5.0 pg/μL [41].
The same group also employed methylene blue (MB) as the electroactive label for the detection of a SARS-CoV genome sequence [47], due to MB's high binding affinity to ssDNA and dsDNA and its reversible electrochemistry [48]. A limit of detection of 800 nM was achieved on a gold platform modified with carbon nanotubes (CNTs-AuSPE) as the transducer [47].
2.2.2. Label-free assays
By making use of the intrinsic electroactive properties of nucleobases, a more simplified experimental procedure, together with reduced assay time can be achieved. Being rapid diagnostic essentials, especially in widespread testing to quickly isolate the affected patients from the community and to control the virus propagation, label-free protocols are one of the more promising solutions for the detection of coronavirus induced infections.
Amine's group reported on the label-free electrochemical detection of a double stranded coronavirus aviair short sequences (174 bp) [49]. Upon hybridization of the target analyte, the voltammetric current generated from the oxidation of purines (guanine and adenine bases) released after acidic hydrolysis is correlated to the presence and amount of the coronavirus aviair short sequences (Fig. 3C). A detection limit of 5 ng/mL was achieved with this approach [49].
More recently, Singh's group speculated on the integration of a miniaturized electrochemical system in a device that can be interfaced with smartphone-based read-out systems, for COVID-19 diagnosis in a smart healthcare fashion [50]. In order to achieve that, a label-free electrochemical detection strategy should be adopted, whereby the electrochemical signal (i.e. voltammetric or impedimetric) can be generated and correlated to the concentration of the target DNA (Fig. 3D) [50].
2.3. Electrochemistry for the detection of PCR products
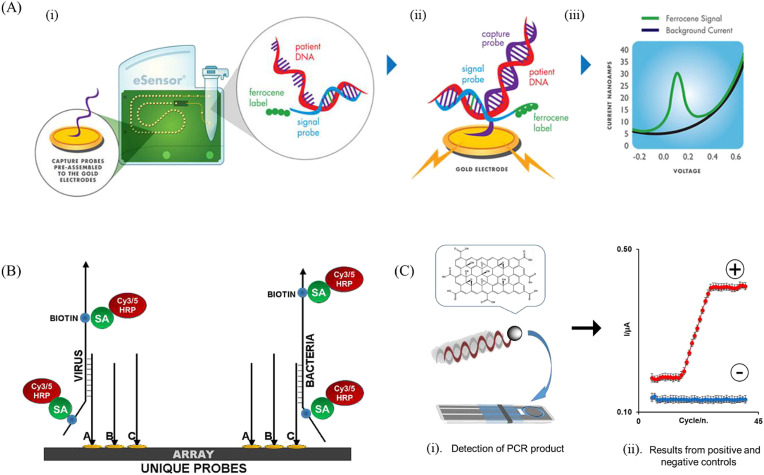
It has been recently shown that electrochemical biosensors can be employed in combination with RT-PCR assays to detect PCR amplified products. In 2020 GenMark Diagnostics Inc. has developed a commercial electrochemical ePlex® SARS-CoV-2 test approved by the US Food and Drug Administration (FDA) for emergency use authorization (EUA) for the qualitative detection of SARS-CoV-2 in nasopharyngeal swab specimens [51]. The single-use ePlex® SARS-CoV-2 cartridge incorporates both GenMark's eSensor® technology and surface electrowetting technique. Firstly, viral RNA is extracted from the swab samples and reverse-transcribed to complementary DNA (cDNA) before PCR amplification. Next, a complementary ferrocene-labelled signaling probe is combined with the amplified target DNA to form a target DNA/signaling probe complex (Fig. 4 A). Upon hybridization with the specific capture probes immobilized onto the surfaces of the gold electrode microarray, the electrochemical signal from the ferrocene label can be correlated to the presence of the target DNA (Fig. 4A). Trial studies showed the clinical performance of the ePlex® SARS-CoV-2 test to be comparable to the approved RT-PCR for EUA [52], with less susceptibility to contamination and reduction in sample preparation time required [53]. The sensitivity of the commercial device was 1 × 105 copies/mL for the target DNA of SARS-CoV-2 [52]. Cross-reactivity analyses were also carried out on other coronavirus-induced infections like SARS-CoV, MERS-CoV and human coronavirus strains 229E, OC43, HKU1 and NL63. At high titers, the only cross-reactivity observed was for SARS-CoV [52].
Fig. 4.
Electrochemical detection of PCR amplification products: (A) Experimental principle of Genmark's eSensor® technology (i) hybridization of signal probe to the target DNA, (ii) reaction of target DNA/signal probe with the captured probe in the cartridge's microfluidic chamber, (iii) collection of the voltammetric response from the target DNA [53]. Adapted with permission from Ref. [53]; (B) Schematic diagram on the microarray for detection of target viral and bacterial analytes. Single-stranded biotinylated target sequence is first hybridized to the immobilized complementary probes (A, B and C) before labelling with streptavidin (SA)-horseradish peroxidase (HRP) to enable electrochemical detection [54]. Adapted with permission from Ref. [54]; (C) Electrochemical PCR scheme using graphene oxide nanoparticles as labels for DNA primers: (i) Electrochemical detection on miniaturized electrode; (ii) nanographene oxide reduction current vs PCR cycle number for positive (red) and negative control (blue) [26]. Reproduced with permission from Ref. [26].
Previous to that, Michael J. Lodes's group developed an oligonucleotide microarray for multi-analyte detection of nine viral pathogens including coronavirus OC43, 229E and HKU1 [54]. Multiple probes per pathogens were used to cover for the potential genomic variability of the different viruses. RT-PCR was designed such that biotinylated single stranded DNA fragments were obtained after the amplification step. Upon hybridization with the biotinylated target sequence specific to coronavirus OC43, 229E, HKU1 and the further conjugation with the streptavidin modified HRP, the current generated by the electrochemical detection of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate confirmed the presence of the target analyte (Fig. 4B). The assay sensitivities were 760 ng/μL and 128 ng/μL for coronavirus OC43 and 229E respectively.
Another approach to introduce electrochemical detection into PCR amplification would be to incorporate electrochemical species of different nature into the PCR mixture, and which yield an electrochemically detectable PCR product [[55], [56], [57]]. This approach, also known as electrochemical PCR, benefits from a substantial cost reduction as compared to optical PCR, together with the possibility of miniaturization which is typical of electrochemical systems.
Our group has recently made use of electrochemical PCR to successfully detect genetically modified organisms by amplifying the Cauliflower Mosaic Virus 35S promoter sequence with the help of electroactive nanographene oxide nanoparticles as labels for the DNA primers used in PCR amplification step [26]. Electrochemical PCR has been already employed for the detection of Influenza A (AH1pdm) virus [58] using methylene blue as electroactive intercalator, which electrochemical signal is inversely correlated to the amount of PCR product [58]. The same approach could be easily extended to the detection of coronavirus-induced infections for a more rapid and widespread testing.
2.4. Electrochemical detection of reactive oxygen species (ROS)
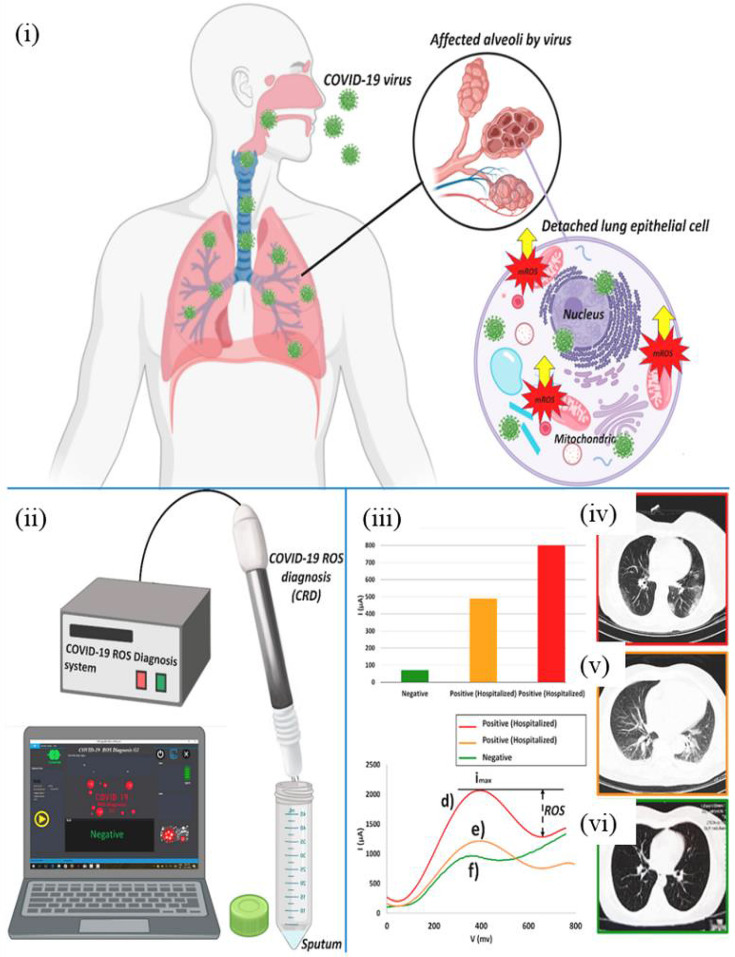
Adopting a novel approach other than the detection of structural proteins and genomic sequences of SARS-CoV-2, Abdolahad's group devised a non-invasive real-time electrochemical ROS/H2O2 system to detect mitochondrial ROS level in patients' sputum samples [59]. It was discovered that, upon infection with SARS-CoV-2, ROS were released from the damaged mitochondria which in turn activate the SARS-CoV 3a NLRP3 (Nod-like receptor family, pyrin domain-containing 3) inflammasome in order to induce virus replication [59]. Correspondingly, there was a rise in ROS level in SARS-CoV3L pro-expressing cells [60]. The electrochemical sensor was created by growing multi-walled carbon nanotubes (MWCNTs) on the needles' tip of a steel electrode [59]. The electrochemical signal detected by cyclic voltammetry was then correlated to the amount of ROS released from the viral-infected lung epithelial host cells (Fig. 5 (ii)-(iii)). Oxidation peak currents that were less than 190 μA were determined to be negative results of SARS-CoV-2, while peak currents that were more than 230 μA were diagnosed as positive cases. The accuracy and sensitivity of the sensor is 94% and 92% respectively when compared with the confirmatory diagnostic method used for clinically suspicious cases, Computed Tomography (CT). Additionally, the MWCNTs sensor can be combined to a computerized platform to process electrochemical readouts automatically. The non-invasive, disposable, and portable system offers fast response time of 30 s to receive conclusive diagnosis results, showing potential to be implemented as a promising point-of-care diagnostic tool for the detection of coronavirus induced infections.
Fig. 5.
Electrochemical ROS/H2O2 system for the detection of SARS-CoV-2 virus: (i) production of mitochondrial ROS in human lung epithelial cells due to SARS-CoV-2 infection; (ii) fabrication of electrochemical system with three needle electrodes functionalized with MWCNTs; (iii) reduction peaks obtained due to selective responses of the released ROS on MWCNTs obtained from two hospitalized patients with COVID-19 infection (d and e in the voltammogram) versus a confirmed negative case (f in the voltammogram) where the intensities of the reduction peak currents correspond to the amount of mitochondrial ROS released as a result of the viral infection; (iv, v) CT scans of COVID-19 patients showing infected lungs; (vi) CT scan of a confirmed negative patient showing no signs of COVID-19 effect on lungs [59]. Reproduced with permission from Ref. [59].
3. Summary and future perspectives
Electrochemical methods have shown their great potential in the detection of infections induced by coronaviruses such as SARS-CoV, MERS-CoV, SARS-CoV-2 and the lesser known OC43, 229E, HKU1. In the current review, we have first provided an introduction on the epidemiology and zoonotic origins of coronaviruses and the gold standard methods employed for their detection, including their current limitations. After which, we have reported the multifaceted electrochemical approaches for the identification of coronavirus structural proteins, as well as the detection of antibodies induced by COVID-19 infection. Fields effect transistors were mostly employed for N and S protein assays, due to their higher sensitivity for label-free detection of charged biomolecules. In addition, an overview on electrochemical DNA sensors for the detection of coronavirus related genome has been provided, with a differentiation between label-free and label-based approaches. While label-free detection was performed by exploiting the intrinsic electroactivity of the DNA nucleobases, label-based assays contributed to an enhanced sensitivity with the use of enzymes with their electroactive substrates, or metal complexes and heterocyclic dyes as the electroactive labels.
The use of electrochemical PCR as an alternative to optical PCR was also reviewed, showing how electroactive indicators or detection labels can be added to the PCR mixture to obtain an electroactive PCR product. Lastly, besides detecting structural proteins, antibodies and nucleic acids that are specific to coronaviruses, a real-time electrochemical ROS/H2O2 sensor functionalized with MWCNTs was shown, which was able to successfully detect mitochondrial ROS level in COVID-19 patients' sputum samples, achieving a sensitivity and an accuracy that were comparable to those obtained with gold standard diagnostics.
Overall, the use of electrochemical methods combined with different nanostructured platforms for the detection of coronavirus-induced infections have demonstrated that electrochemistry could be the analytical solution for the development of simple, rapid, miniaturized, portable and cost-effective diagnostic tools for the decentralized and early-detection of infected patients. This could help in the current emergency for COVID-19 intensive testing, and in other emerging coronavirus-induced infections in the future.
The ultimate challenge would be to integrate electrochemical detection into a commercial point-of-care (POC) device for clinical use. The ideal envision for the practical application would be to develop miniaturized testing with a multi-integrated system for patient sampling, together with microfluidics chambers for the processing and the detection of the sought analyte. To meet the increase in demand for testing, samples from different individuals can be pooled and multiplexed through electrochemical platforms to minimize the number of individual tests collected, making coronaviruses detection more efficient and effective in identifying the infected individuals based on the unique barcode assigned. In this way, electrochemical devices as POC can provide rapid, highly sensitive and accessibility to places outside of laboratory settings, such as to self-isolated people at home or drive-through stations for ramp-up testing.
A decentralized healthcare would also be beneficial in combating coronavirus-induced infections through the connection of these electrochemical sensing devices to smartphones and phone applications. On-site detection and data acquisition can occur simultaneously so that healthcare personnel can monitor the patient's health status from a single database established. In addition, if multi-analyte detection can be effectively performed on electrochemical platforms, the applicability of the electrochemical sensors can be further extended to cover past, present and possibly other emerging coronavirus-induced infections as well as to differentiate them from other non-coronaviruses that might present similar clinical symptoms in humans. Also, on top of the spike and nucleocapsid proteins that are commonly used as target analytes for detection, the sensitivity of electrochemical sensors should be improved in case of potential mutation in the viral gene regions by modifying the sensing surface to target other structural proteins, including envelope and membrane proteins. In this way, electrochemical sensing platforms will be a more commercially attractive and viable technology for the detection of coronavirus-induced infections.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
The authors gratefully acknowledge Singapore Ministry of Education (MOE), AcRF Tier 1 grant (Reference No: RG18/17) for the funding of this research.
References
- 1.Liu D.X., Liang J.Q., Fung T.S. Elsevier; 2020. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. [Google Scholar]
- 2.Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg. Microb. Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., Schipper D., van Run P., Leijten L., Sikkema R., Verschoor E., Verstrepen B., Bogers W., Langermans J., Drosten C., Fentener van Vlissingen M., Fouchier R., de Swart R., Koopmans M., Haagmans B.L. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronavirus Disease (COVID-19) Pandemic. WHO; 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- 7.Yong J.Y. 2020. Cautionary Tales: 4 Countries Hit by a Resurgence in Covid-19 Cases after Reopening Economies.https://www.todayonline.com/singapore/cautionary-tales-4-countries-hit-resurgence-covid-19-cases-after-reopening-economies TODAYOnline. [Google Scholar]
- 8.Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann. Intern. Med. 2020 doi: 10.7326/M20-3012. M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I., Gutierrez R.A., Gwee S.X.W., Chua P.E.Y., Yang Q., Ng X.Y., Yap R.K., Tan H.Y., Teo Y.Y., Tan C.C., Cook A.R., Yap J.C., Hsu L.Y. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J. Clin. Med. 2020;9:623. doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 12.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng W., Newbigging A., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.B., Cui M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 14.Morales-Narvaez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19, Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Gharizadeh B., Lu D., Yue J., Yu M., Liu Y., Zhou M. Navigating the pandemic response life cycle: molecular diagnostics and immunoassays in the context of COVID-19 management. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2991444. PMID: 32356761. [DOI] [PubMed] [Google Scholar]
- 16.Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: a narrative review. Ann. Intern. Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., Andre E., Van Ranst M., Lagrou K., Vermeersch P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020:1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Cai K., Zhang R., He X., Shen X., Liu J., Xu J., Qiu F., Lei W., Wang J., Li X., Gao Y., Jiang Y., Xu W., Ma X. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal. Chem. 2020;92:9399–9404. doi: 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- 20.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 22.Li G. Elsevier; 2019. Nano-Inspired Biosensors for Protein Assay with Clinical Applications. [Google Scholar]
- 23.Fang W., Lillehoj P.B., Ho C.-M. DNA diagnostics: nanotechnology-enhanced electrochemical detection of nucleic acids. Pediatr. Res. 2010;67:458–468. doi: 10.1203/PDR.0b013e3181d361c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhalla N., Pan Y., Yang Z., Payam A.F. Opportunities and challenges for biosensors and nanoscale Analytical tools for pandemics: COVID-19. ACS Nano. 2020:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokhrel P., Hu C., Mao H. Detecting the coronavirus (COVID-19) ACS Sens. 2020;5(8):2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ang W.L., Seah X.Y., Koh P.C., Caroline C., Bonanni A. Electrochemical polymerase chain reaction using electroactive graphene oxide nanoparticles as detection labels. ACS Appl. Nano Mater. 2020;3:5489–5498. [Google Scholar]
- 27.Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur M., Tiwari S., Jain R. Protein based biomarkers for non-invasive Covid-19 detection. Sens. Biosensing Res. 2020;29:100362. doi: 10.1016/j.sbsr.2020.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B., Song Y., Li J., Yang L., Wang J., Wang H., Pang X., Zhai J., Du Z., Liu Y., Zhang Y., Li L., Wang J., Sun B., Yang R. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microb. Infect. 2005;7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishikawa F.N., Chang H.K., Curreli M., Liao H.I., Olson C.A., Chen P.C., Zhang R., Roberts R.W., Sun R., Cote R.J., Thompson M.E., Zhou C. Label-free, electrical detection of the SARS virus N-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano. 2009;3:1219–1224. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zafar S., Lu M., Jagtiani A. Comparison between field effect transistors and bipolar junction transistors as transducers in electrochemical sensors. Sci. Rep. 2017;7:41430. doi: 10.1038/srep41430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar D., Liu W., Xie X., Anselmo A.C., Mitragotri S., Banerjee K. MoS2 field-effect transistor for next-generation label-free biosensors. ACS Nano. 2014;8:3992–4003. doi: 10.1021/nn5009148. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa F.N., Curreli M., Olson C.A., Liao H.I., Sun R., Roberts R.W., Cote R.J., Thompson M.E., Zhou C. Importance of controlling nanotube density for highly sensitive and reliable biosensors functional in physiological conditions. ACS Nano. 2010;4:6914–6922. doi: 10.1021/nn101198u. [DOI] [PubMed] [Google Scholar]
- 34.Hsu Y.-R., Lee G.-Y., Chyi J.-I., Chang C.-k., Huang C.-C., Hsu C.-P., Huang T.-h., Ren F., Wang Y.-L. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein using AlGaN/GaN high electron mobility transistors. ECS Trans. 2013;50:239–243. doi: 10.1149/05006.0239ecst. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors (Basel) 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 37.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuccetelli M., Pieri M., Grelli S., Ciotti M., Miano R., Andreoni M., Bernardini S. SARS-CoV-2 infection serology: a useful tool to overcome lockdown? Cell Death Dis. 2020;6 doi: 10.1038/s41420-020-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bănică F.G. Wiley; 2012. Chemical Sensors and Biosensors: Fundamentals and Applications. [Google Scholar]
- 40.Teles F., Fonseca L. Trends in DNA biosensors. Talanta. 2008;77:606–623. [Google Scholar]
- 41.de la Escosura-Muniz A., Gonzalez-Garcia M.B., Costa-Garcia A. DNA hybridization sensor based on aurothiomalate electroactive label on glassy carbon electrodes. Biosens. Bioelectron. 2007;22:1048–1054. doi: 10.1016/j.bios.2006.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abad-Valle P., Fernandez-Abedul M.T., Costa-Garcia A. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Biosens. Bioelectron. 2005;20:2251–2260. doi: 10.1016/j.bios.2004.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martínez-Paredes G., González-García M.B., Costa-García A. Genosensor for SARS virus detection based on gold nanostructured screen-printed carbon electrodes. Electroanalysis. 2009;21:379–385. [Google Scholar]
- 44.Abad-Valle P., Fernandez-Abedul M.T., Costa-Garcia A. DNA single-base mismatch study with an electrochemical enzymatic genosensor. Biosens. Bioelectron. 2007;22:1642–1650. doi: 10.1016/j.bios.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz-Gonzalez M., de la Escosura-Muniz A., Gonzalez-Garcia M.B., Costa-Garcia A. DNA hybridization biosensors using polylysine modified SPCEs. Biosens. Bioelectron. 2008;23:1340–1346. doi: 10.1016/j.bios.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gau V., Ma S.C., Wang H., Tsukuda J., Kibler J., Haake D.A. Electrochemical molecular analysis without nucleic acid amplification. Methods. 2005;37:73–83. doi: 10.1016/j.ymeth.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Gonzalez R., Costa-Garcia A., Fernandez-Abedul M.T. Methylene blue covalently attached to single stranded DNA as electroactive label for potential bioassays. Sensor. Actuator. B Chem. 2014;191:784–790. doi: 10.1016/j.snb.2013.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rashid J.I.A., Yusof N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: a review. Sens. Biosensing Res. 2017;16:19–31. [Google Scholar]
- 49.Zari N., Mohammedi H., Amine A., Ennaji M.M. DNA hydrolysis and voltammetric determination of guanine and adenine using different electrodes. Anal. Lett. 2007;40:1698–1713. [Google Scholar]
- 50.Tripathy S., Singh S.G. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Trans. Indian Natl. Acad. Eng. 2020:1–5. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GenMark Receives FDA Emergency Use Authorization for its ePlex® SARS-CoV-2 Test. GenMark Diagnostics, Inc; 2020. http://ir.genmarkdx.com/news-releases/news-release-details/genmark-receives-fda-emergency-use-authorization-its-eplexr-sars [Google Scholar]
- 52.ePlex® SARS-CoV-2 Test Assay Manual. GenMark Diagnostics, Inc.; 2020. https://www.fda.gov/media/136282/download [Google Scholar]
- 53.eSensor Technology . 2020. GenMark Diagnostics, Inc.https://www.genmarkdx.com/solutions/technology/esensor/ [Google Scholar]
- 54.Lodes M.J., Suciu D., Wilmoth J.L., Ross M., Munro S., Dix K., Bernards K., Stover A.G., Quintana M., Iihoshi N., Lyon W.J., Danley D.L., McShea A. Identification of upper respiratory tract pathogens using electrochemical detection on an oligonucleotide microarray. PloS One. 2007;2:e924. doi: 10.1371/journal.pone.0000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deféver T., Druet M., Rochelet-Dequaire M., Joannes M., Grossiord C., Limoges B., Marchal D. Real-time electrochemical monitoring of the polymerase chain reaction by mediated redox catalysis. J. Am. Chem. Soc. 2009;131:11433–11441. doi: 10.1021/ja901368m. [DOI] [PubMed] [Google Scholar]
- 56.Defever T., Druet M., Evrard D., Marchal D., Limoges B. Real-time electrochemical PCR with a DNA intercalating redox probe. Anal. Chem. 2011;83:1815–1821. doi: 10.1021/ac1033374. [DOI] [PubMed] [Google Scholar]
- 57.de la Escosura-Muniz A., Baptista-Pires L., Serrano L., Altet L., Francino O., Sanchez A., Merkoci A. Magnetic bead/gold nanoparticle double-labeled primers for electrochemical detection of isothermal amplified leishmania DNA. Small. 2016;12:205–213. doi: 10.1002/smll.201502350. [DOI] [PubMed] [Google Scholar]
- 58.Yamanaka K., Saito M., Kondoh K., Hossain M.M., Koketsu R., Sasaki T., Nagatani N., Ikuta K., Tamiya E. Rapid detection for primary screening of influenza A virus: microfluidic RT-PCR chip and electrochemical DNA sensor. Analyst. 2011;136:2064–2068. doi: 10.1039/c1an15066a. [DOI] [PubMed] [Google Scholar]
- 59.Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin C.W., Lin K.H., Hsieh T.H., Shiu S.Y., Li J.Y. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol. Med. Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., Maeng J.S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chia W.N., Tan C.W., Foo R., Kang A.E.Z., Peng Y., Sivalingam V., Tiu C., Ong X.M., Zhu F., Young B.E., Chen M.I., Tan Y.J., Lye D.C., Anderson D.E., Wang L.F. Serological differentiation between COVID-19 and SARS infections, Emerg. Microb. Infect. 2020;9:1497–1505. doi: 10.1080/22221751.2020.1780951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.SARS-CoV-2 Serological Analysis Kit. Sino Biological Inc; 2020. https://www.sinobiological.com/research/virus/sars-cov-2-antigen-detection-assay [Google Scholar]