ABSTRACT
Up to 90% of the human genome is transcribed into Long-noncoding RNAs (lncRNAs) that longer than 200 nucleotides but do not code for proteins. LncRNAs play a vital role in a broad range of biological process, it’s dysregulations and mutations are linked to the development and progression of various complex human diseases. Given the dramatic changes and growing scientific outputs in lncRNAs field, using a quantitative measurement to analyze and characterize the existing studies has become imperative.Bibliometric analysis is a widely used tool to assess the academic influence of a publication or a country in a specific field. However, a bibliometric analysis of the top 100 most-cited papers in lncRNAs area has not been conducted. Thus, we executed a bibliometric study to identify the authors, journals, countries and institutions that contributed most to the top 100 lncRNAs list, characterize the key words and focus of top 100 most-cited papers, and detect the factors related to their successful citation. This study provides a comprehensive list of the most influential papers on lncRNAs research and demonstrates the important advances in this field, which might be benefit to researchers in their paper publication and scientific cooperation.
KEYWORDS: Long noncoding RNAs, lncRNAs, citation analysis, bibliometric study, top-cited
1. Introduction
Long-noncoding RNAs (lncRNAs) had been defined as non-protein-coding transcribed RNA molecules with more than 200 nucleotides in length1,2 and account for 90% of the RNAs transcribed by the human genome.3 In 1989, the first mammalian lncRNA H19 was discovered.4 Subsequently, the essential role of the lncRNA Xist in X chromosome inactivation was identified.5,6 Over the last decade, lncRNAs have aroused great attention from investigators worldwide due to their regulatory latent capacity in a diversity of physiological and pathological procedures.7 Advances in deep sequencing technology and computational prediction have enabled the extensive identification of a large number of lncRNAs. A plenty of evidence indicated that lncRNAs play a key role in a cluster of biological process,8 including chromatin remodeling or modification,9,10 epigenetic regulation,11 dosage compensation,12 genomic imprinting,13 allosteric regulation of proteins,14 methylation,15 cell development, and differentiation,12 cell cycle control,16 organ or tissue development,17 and metabolic processes.18 However, the mechanisms of action of these molecules remain incompletely understood.19 Some lncRNAs recruit transcriptional factors to their DNA targets, some act as baits to separate RNA conjugated proteins or miRNAs, and some interact with DNAs or other RNAs directly.20,21
Dysregulations and mutations of lncRNAs are associated with the development and progression of diverse human diseases,22,23 such as autism spectrum disorder24 and abdominal aortic aneurysm.25 LncRNAs also act as drivers joining in the process of tumor suppressive and oncogenic functions,26,27 and their differential expression is a symbolic feature in numerous cancer types,28 such as lung cancer,29 breast cancer,30 liver cancer,31 prostate cancer,26 and colon cancer.32
Considering growing outputs and dramatic changes in lncRNAs research, using a quantitative method to assess and analyze the available studies has become imperative.33 Bibliometric analysis, which was first introduced by Paul Otlet in 1934,34 is a popular tool to evaluate the academic influence of a publication or a country in a specific field. Bibliometric studies have been widely utilized to explore developing trends in several medical research, such as microRNAs,33 DNA repair,35 radiation-responsive genes,36 double helices,37 epigenetics,38 and cancer.39 Bibliometrics studies related to lncRNAs have also been published. For example, Chen X focused on lncRNAs and chemotherapeutic resistance,40 Zhai X mapped the expanding trend of global lncRNAs research from 1975 to 2017,41 Miao Y performed bibliometric analysis of lncRNAs trends over a short period (2007–2016),42 and Xing YH concentrated on Chinese progress in lncRNAs field.7
Nevertheless, a bibliometric analysis of the 100 most-cited papers in lncRNAs research has not been conducted. Thus, we executed a bibliometric study to identify and characterize the top 100 most-cited papers on lncRNAs and detect the factors related to their successful citation, which might be benefit to researchers in their paper publication and scientific cooperation.
2. Methods
Ethical approval from the institutional review board was not required because our study was a bibliometric analysis that did not involve human subjects.
2.1. Search strategy
The papers were searched from the Science Citation Index Expanded (SCI-Expanded) of Web of Science (WOS), which contains over 5,700 major journals across 164 scientific disciplines.43 We performed our literature search on August 15, 2019 to avoid changes in the online activity of papers. The search keywords were referred to some academic papers41,42 and MESH terms from PubMed: (TI = (“lncRNA*” OR “lnc RNA*” OR “long ncRNA*” OR “long noncoding RNA*” OR “long non coding RNA*” OR “long non translated RNA*” OR “long non protein coding RNA*” OR “linc RNA* ”OR “lincRNA*”)) and Language = English, and publishing year was set from 1998 to 2018.
2.2. Inclusion criteria
The primary search results were sorted based on the citation counts in descending order. Then, the papers cited no less than 200 times were downloaded for further analysis; Figure 1 shows the selection process. First, in terms of document types, only peer-reviewed articles, and reviews were included; conference abstracts, conference presentations, and book chapters were excluded. Second, we read titles and abstracts to remove studies that were unrelated to lncRNAs research. Finally, the 100 most-cited papers were exported to Microsoft Excel 2016 to create tables and figures. To enhance our search sensitivity, data extraction was conducted by two independent reviewers (Mengsi Peng and Juan Wang evaluated), and a third researcher (XueQiang Wang) was consulted to deal with discrepancies.
Figure 1.
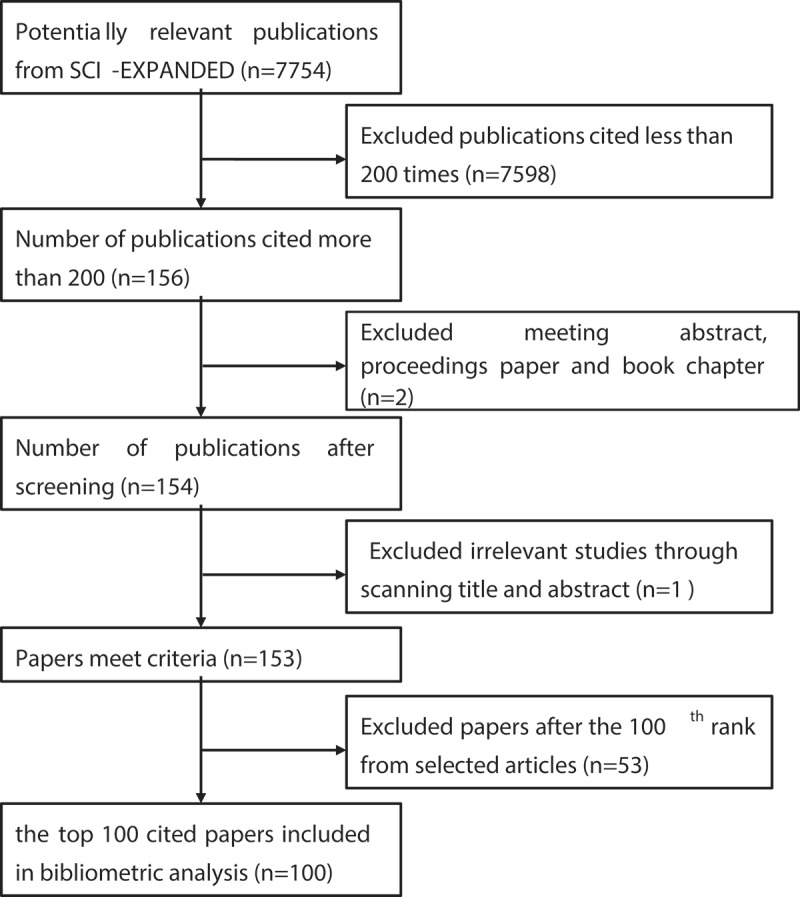
Data extraction process
2.3. Data selection
The following information was extracted from the top-cited papers: publication year, citation count, citation per year (total citations/the number of years since publication), author, journal, country or region (based on the correspondent author’s address), institution, documental type, research field, and key words. The journal impact factor 2018 (IF 2018) and five-year IF were obtained from the Journal Citation Report (2018 edition). Moreover, the latest statistics on gross domestic product (GDP) was obtained from the Word Bank.41
An inherent limitation of this study is that recent important papers may not have been captured because of insufficient time to accumulate citations. To address this issue, we performed the same search within a narrower time range (2017–2018) to select the top 20 most-cited papers.
2.4. SPSS
Statistical analysis was performed using SPSS 22.0. P value<.05 was the criterion for statistically significant. Descriptive statistics were quantified as average or counts(percentages) of parameters. The Pearson product moment correlation coefficient was employed to test the correlations between IF(2018)and paper counts, IF(2018)and citation counts, GDP and paper counts, GDP and citation counts, as well as correlations between annual citations and total citations. The Mann–Whitney test was used to exam whether there was any significant difference in citation counts between articles and reviews. The differences in some quantitative indicators (the total citations, citations per year, citations 2018, citations per article, citations per review and IF 2018) before and after 2011 were also analyzed by Mann–Whitney test. One-way analysis of variance (ANOVA) was performed to test qualitative indicators, including the distribution differences in paper count among country, type of paper, open access, and highly cited before and after 2011.
3. Result
3.1. Citation
Table 1 lists the top 100 most-cited papers in lncRNAs research. The median number of citations was 414.5 with a range of 249–2,828 (mean: 597.33 ± 489.37). For annual citations, the median number was 57.47 (mean: 76.41 ± 51.28) with a range of 27.3–282.8. When comparing the ranking of annual citations to that of total citations, the change in position ranged from −30 to +42 with a mean absolute rank change of 10.08 ± 9. Papers with higher annual citations tended to have more total citations, and the correlation between these factors was significantly strong (r = 0.927, P = .000).
Table 1.
The 100 most-cited papers in long non-coding RNAs field
| Rank | First author | Paper | Citations WOS | Citations per year | Citations in 2018 |
|---|---|---|---|---|---|
| 1 | Gupta, RA | Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6. | 2828 | 282.8 | 440 |
| 2 | Mercer, TR | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10(3):155–9. | 2636 | 239.64 | 472 |
| 3 | Ponting, CP | Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41. | 2301 | 209.18 | 441 |
| 4 | Derrien, T | Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22(9):1775–89. | 2075 | 259.38 | 381 |
| 5 | Tsai, MC | Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–93. | 1752 | 175.2 | 233 |
| 6 | Wang, KC | Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell. 2011;43(6):904–14. | 1676 | 186.22 | 329 |
| 7 | Cesana, M | Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69. | 1277 | 141.89 | 221 |
| 8 | Wilusz, JE | Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes & development. 2009;23(13):1494–504. | 1183 | 107.55 | 194 |
| 9 | Guttman, M | Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. | 1156 | 128.44 | 146 |
| 10 | Fatica, A | Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature reviews Genetics. 2014;15(1):7–21. | 1119 | 186.5 | 284 |
| 11 | Orom, UA | Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. | 1095 | 109.5 | 137 |
| 12 | Wapinski, O | Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–61. | 1061 | 117.89 | 202 |
| 13 | Wang, KC | Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–4. | 1030 | 114.44 | 144 |
| 14 | Batista, PJ | Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–307. | 1012 | 144.57 | 258 |
| 15 | Ulitsky, I | Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. | 989 | 141.29 | 212 |
| 16 | Gibb, EA | Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. | 902 | 100.22 | 178 |
| 17 | Prensner, JR | Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1(5):391–407. | 894 | 99.33 | 180 |
| 18 | Gutschner, T | Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA biology. 2012;9(6):703–19. | 883 | 110.38 | 144 |
| 19 | Iyer, MK | Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47(3):199–208. | 819 | 163.8 | 248 |
| 20 | Kogo, R | Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6. | 791 | 87.89 | 133 |
| 21 | Guttman, M | Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature biotechnology. 2010;28(5):503–10. | 771 | 77.1 | 78 |
| 22 | Yuan, JH | Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25(5):666–81. | 731 | 121.83 | 197 |
| 23 | Kung, JT | Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–69. | 684 | 97.71 | 175 |
| 24 | Mercer, TR | Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(2):716–21. | 679 | 56.58 | 66 |
| 25 | Mercer, TR | Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20(3):300–7. | 668 | 95.43 | 126 |
| 26 | Lee, JT | Lee JT. Epigenetic Regulation by Long Noncoding RNAs. Science. 2012;338(6113):1435–9. | 656 | 82 | 115 |
| 27 | Quinn, JJ | Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature reviews Genetics. 2016;17(1):47–62. | 654 | 163.5 | 248 |
| 28 | Schmitt, AM | Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29(4):452–63. | 652 | 163 | 249 |
| 29 | Ulitsky, I | Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–50. | 592 | 65.78 | 97 |
| 30 | Geisler, S | Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nature reviews Molecular cell biology. 2013;14(11):699–712. | 585 | 83.57 | 151 |
| 31 | Prensner, JR | Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29(8):742–9. | 578 | 64.22 | 74 |
| 32 | Chu, C | Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–78. | 574 | 63.78 | 68 |
| 33 | Gong, C | Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3ʹ UTRs via Alu elements. Nature. 2011;470(7333):284–8. | 573 | 63.67 | 77 |
| 34 | Kotake, Y | Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–62. | 564 | 62.67 | 83 |
| 35 | Wang, J | Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–83. | 554 | 55.4 | 80 |
| 36 | Shi, X | Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–66. | 539 | 77 | 138 |
| 37 | Zhang, Y | Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. | 530 | 75.71 | 186 |
| 38 | Spizzo, R | Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–87. | 521 | 65.13 | 105 |
| 39 | Kallen, AN | Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–12. | 520 | 74.29 | 108 |
| 40 | Dinger, ME | Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome research. 2008;18(9):1433–45. | 498 | 41.5 | 44 |
| 41 | Nagano, T | Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–81. | 497 | 55.22 | 96 |
| 42 | Klattenhoff, CA | Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–83. | 493 | 70.43 | 91 |
| 43 | Yoon, JH | Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–55. | 460 | 57.5 | 76 |
| 44 | Yang, Z | Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Annals of surgical oncology. 2011;18(5):1243–50. | 459 | 51 | 72 |
| 45 | Liu, XH | Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. | 454 | 75.67 | 109 |
| 46 | Pauli, A | Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome research. 2012;22(3):577–91. | 437 | 54.63 | 80 |
| 47 | Cheetham, SW | Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. British journal of cancer. 2013;108(12):2419–25. | 432 | 61.71 | 97 |
| 48 | Wang, P | Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–3. | 431 | 71.83 | 104 |
| 49 | Kretz, M | Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–5. | 420 | 60 | 64 |
| 50 | Yang, F | Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology (Baltimore, Md). 2011;54(5):1679–89. | 420 | 46.67 | 65 |
| 51 | Necsulea, A | Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–40. | 409 | 68.17 | 72 |
| 52 | Carpenter, S | Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–92. | 406 | 58 | 84 |
| 53 | Chen, G | Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41(Database issue):D983-6. | 404 | 57.71 | 88 |
| 54 | Wang, Y | Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Developmental cell. 2013;25(1):69–80. | 402 | 57.43 | 80 |
| 55 | Engreitz, JM | Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. | 395 | 56.43 | 63 |
| 56 | Michalik, KM | Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circulation research. 2014;114(9):1389–97. | 389 | 64.83 | 100 |
| 57 | Keniry, A | Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature cell biology. 2012;14(7):659–65. | 389 | 48.63 | 66 |
| 58 | Braconi, C | Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–6. | 387 | 43 | 57 |
| 59 | Ponjavic, J | Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome research. 2007;17(5):556–65. | 385 | 29.62 | 30 |
| 60 | Tripathi, V | Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS genetics. 2013;9(3):e1003368. | 372 | 53.14 | 71 |
| 61 | Gutschner, T | Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. Journal of molecular medicine (Berlin, Germany). 2013;91(7):791–801. | 364 | 52 | 84 |
| 62 | Sauvageau, M | Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. | 355 | 50.71 | 59 |
| 63 | McHugh, CA | McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–6. | 353 | 70.6 | 78 |
| 64 | Lai, MC | Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Medical oncology (Northwood, London, England). 2012;29(3):1810–6. | 352 | 44 | 44 |
| 65 | Lee, JT | Lee JT, Bartolomei MS. X–inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–23. | 349 | 49.86 | 41 |
| 66 | Prensner, JR | Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature genetics. 2013;45(11):1392–8. | 341 | 48.71 | 89 |
| 67 | Anderson, DM | Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. | 336 | 67.2 | 100 |
| 68 | Luo, M | Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–21. | 324 | 46.29 | 62 |
| 69 | Yang, GD | Yang GD, Lu XZ, Yuan LJ. LncRNA: A link between RNA and cancer. Biochim Biophys Acta-Gene Regul Mech. 2014;1839(11):1097–109. | 320 | 53.33 | 95 |
| 70 | Yang, L | Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. | 320 | 45.71 | 44 |
| 71 | Amaral, PP | Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39(Database issue):D146-51. | 319 | 35.44 | 35 |
| 72 | Liu, B | Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27(3):370–81. | 314 | 62.8 | 114 |
| 73 | Moran, VA | Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–400. | 314 | 39.25 | 43 |
| 74 | Yang, F | Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49(6):1083–96. | 309 | 44.14 | 38 |
| 75 | Liao, Q | Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39(9):3864–78. | 305 | 33.89 | 54 |
| 76 | Gomez, JA | Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–54. | 301 | 43 | 52 |
| 77 | Kornienko, AE | Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC biology. 2013;11:59. | 290 | 41.43 | 75 |
| 78 | Zhang, B | Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell reports. 2012;2(1):111–23. | 288 | 36 | 53 |
| 79 | Khaitan, D | Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71(11):3852–62. | 286 | 31.78 | 37 |
| 80 | Han, P | Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514(7520):102–6. | 281 | 46.83 | 75 |
| 81 | Yang, F | Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. The FEBS journal. 2012;279(17):3159–65. | 278 | 34.75 | 50 |
| 82 | Qureshi, IA | Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain research. 2010;1338:20–35. | 278 | 27.8 | 39 |
| 83 | Hirata, H | Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75(7):1322–31. | 275 | 55 | 82 |
| 84 | Du, Z | Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nature structural & molecular biology. 2013;20(7):908–13. | 275 | 39.29 | 55 |
| 85 | Tian, D | Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. | 273 | 27.3 | 20 |
| 86 | St Laurent, G | St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends in genetics: TIG. 2015;31(5):239–51. | 272 | 54.4 | 73 |
| 87 | Ng, SY | Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. The EMBO journal. 2012;31(3):522–33. | 268 | 33.5 | 33 |
| 88 | Qi, P | Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26(2):155–65. | 267 | 38.14 | 60 |
| 89 | Volders, PJ | Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41(Database issue):D246-51. | 267 | 38.14 | 43 |
| 90 | Liang, WC | Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–25. | 264 | 52.8 | 79 |
| 91 | Martens-Uzunova, ES | Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. European urology. 2014;65(6):1140–51. | 264 | 44 | 79 |
| 92 | Chakravarty, D | Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S The estrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nature communications. 2014;5:5383. | 262 | 43.67 | 74 |
| 93 | Pasmant, E | Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25(2):444–8. | 258 | 28.67 | 36 |
| 94 | Zhang, EB | Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell death & disease. 2014;5:e1243. | 256 | 42.67 | 53 |
| 95 | Xiang, JF | Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell research. 2014;24(5):513–31. | 254 | 42.33 | 57 |
| 96 | Wang, K | Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circulation research. 2014;114(9):1377–88. | 254 | 42.33 | 59 |
| 97 | Xie, C | Xie C, Yuan J, Li H, Li M, Zhao G, Bu D NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42(Database issue):D98-103. | 253 | 42.17 | 39 |
| 98 | Li, CH | Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. The international journal of biochemistry & cell biology. 2013;45(8):1895–910. | 253 | 36.14 | 41 |
| 99 | Kumarswamy, R | Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circulation research. 2014;114(10):1569–75. | 249 | 41.5 | 53 |
| 100 | Kapusta, A | Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS genetics. 2013;9(4):e1003470. | 249 | 35.57 | 41 |
3.2. Year
The top 100 most-cited papers were published between 2007 and 2016. The vast majority of these papers (n = 30) was published in 2013 and 2011was the second peak with 21 papers. During this period, the number of articles was usually higher than the number of reviews, except for the years 2009 and 2016 (Figure 2a). From 2009 to 2015, the total citations of papers were on the decline, as well as the total citations of articles and reviews (Figure 2b).
Figure 2.
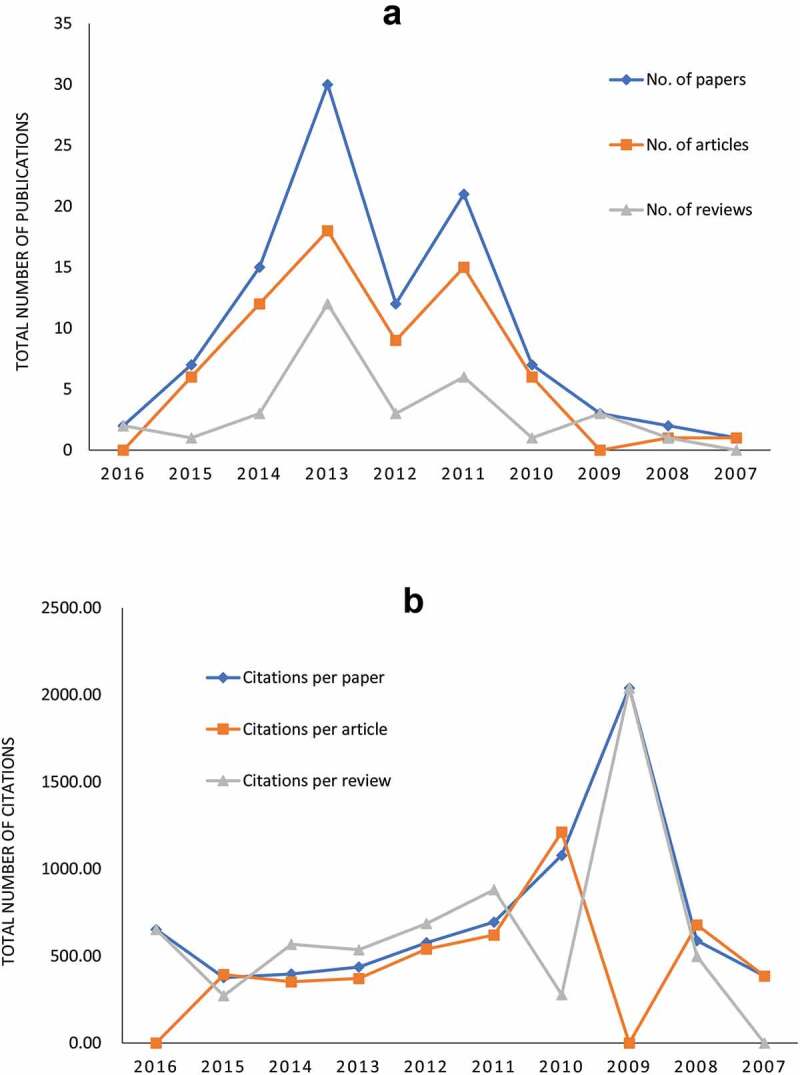
Number of publications and citations among different types of articles according to publication year. (a) Number of annual publications on lncRNAs research from 2007 to 2016. (b) Number of annual citations on lncRNAs research from 2007 to 2016
We executed a two-time point analysis to compare papers before and after 2011 (Table 2). Publications after 2011 approximately twice that of publications before 2011. However, the total number of citations per paper before 2011 was significantly higher than that after 2011 (P < .001), and the citation difference between these two periods was obvious in original articles (P < .001). The USA and China contributed more papers after 2011 than before this year, but the effect of time on the country constituent ratio was not significant (P = .096). Among the top 100 papers related to lncRNAs, 82% were open access and 92% were highly cited.
Table 2.
Two-time point analyses comparing journals’ top-cited articles before and after 2011
| Variables | Before 2011 | After 2011 | P value |
|---|---|---|---|
| Year | 2011(2007–2011) | 2013(2012–2016) | |
| No. of papers | 34 | 66 | 0.002 |
| Citations WOS per paper | 877.41(656.03) | 453.05(282.69) | 0.000 |
| Citations WOS per paper per year | 90.04(63.35) | 69.39(42.08) | 0.230 |
| Citations 2018 per paper | 137(119.77) | 101.32(69.08) | 0.476 |
| Country | |||
| USA | 18(36.73%) | 31(63.27%) | 0.096 |
| PEOPLES R CHINA | 4(16.67%) | 20(83.33%) | |
| Others | 12(44.44%) | 15(55.56%) | |
| Document type | |||
| No. of article | 23(33.82%) | 45(66.18%) | 0.957 |
| No. of review | 11(34.38%) | 21(65.63%) | |
| Citations per article | 767.30(568.65) | 402.84(277.93) | 0.000 |
| Citations per review | 1107.64(758.80) | 560.62(262.02) | 0.061 |
| Open access | 29(35.37%) | 53(64.63%) | 0.538 |
| Highly cited | 29(31.52%) | 63(68.48%) | 0.086 |
| Average IF (2018) | 22.049(2.929–43.704) | 19.154(3.144–43.704) | 0.421 |
3.3. Country and institution
The 100 top-cited papers originated from 15 countries; the USA and China were the most productive in this regard (Figure 3a). Nearly half of the papers published (n = 49) were from the USA, and around one-quarter (n = 24) was from China. Australia ranked third with 6 papers, followed by England and Germany, each contributing 4 papers to the list. Additionally, GDP and number of papers were positively correlated (r = 0.966, P < .001). Figure 3b shows the list of institutions, all the top three most-prolific institutions were rooted in the USA.
Figure 3.
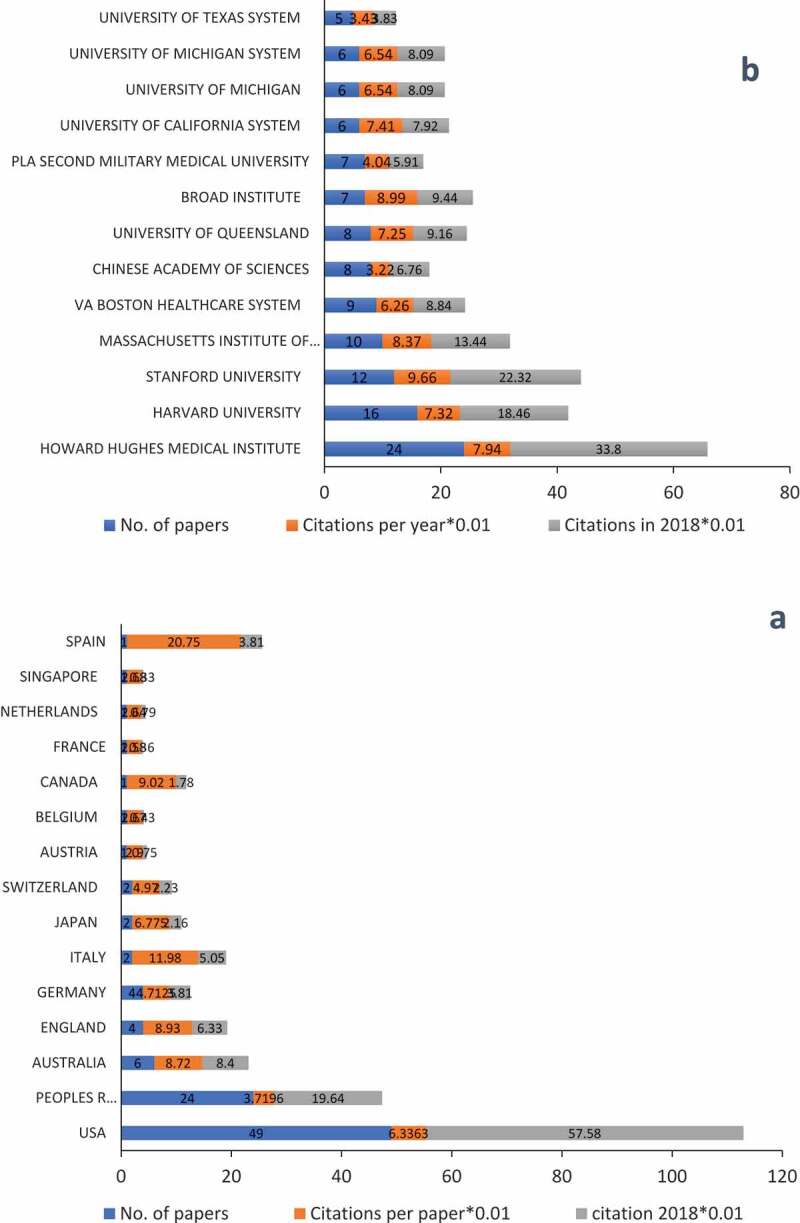
Countries and institutions of the top 100 most-cited papers. (a) Countries of region of the top 100 list. (b) Institutions with at least four papers in the top 100 most-cited papers
3.4. Journal
Exactly 47 academic journals contributed to the 100 top-cited papers, which were predominantly published in Cell (n = 12) and followed by Nature (n = 9), Nucleic Acids Research (n = 7), Molecular Cell (n = 6), and Science (n = 6). Table 3 presents journals with at least three publications.
Table 3.
Journals contributed ≥3 papers in the top 100 most cited list
| Rank | Journal | Country | No. of papers | Citations per paper | Citations WOS | IF (2017) | IF (5 year) | JCR category | JCR partition |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CELL | USA | 12 | 792.21 | 9515 | 36.216 | 36.430 | Biochemistry & Molecular Biology; Cell Biology | Q1,Q1 |
| 2 | NATURE | ENGLAND | 9 | 818.89 | 7370 | 43.07 | 45.819 | Multidisciplinary Sciences | Q1 |
| 3 | NUCLEIC ACIDS RESEARCH | ENGLAND | 7 | 345.14 | 2416 | 11.147 | 10.727 | Biochemistry & Molecular Biology | Q1 |
| 4 | MOLECULAR CELL | USA | 6 | 678.17 | 4064 | 14.548 | 15.090 | Biochemistry & Molecular Biology; Cell Biology | Q1,Q1 |
| 5 | SCIENCE | USA | 5 | 728 | 3640 | 41.037 | 43.644 | Multidisciplinary Sciences | Q1 |
| 6 | GENOME RESEARCH | USA | 4 | 848.75 | 3395 | 9.944 | 11.638 | Biochemistry & Molecular Biology; Biotechnology & Applied Microbiology; Genetics & Heredity | Q1,Q1,Q1 |
| 7 | CANCER CELL | USA | 3 | 565.67 | 1697 | 23.916 | 26.809 | Oncology; Cell Biology | Q1,Q1 |
| 8 | CANCER RESEARCH | USA | 3 | 450.67 | 1352 | 8.378 | 9.062 | Oncology | Q1 |
| 9 | CIRCULATION RESEARCH | USA | 3 | 297.33 | 892 | 15.862 | 14.552 | Cardiac & Cardiovascular Systems; Hematology; Peripheral Vascular Disease | Q1,Q1,Q1 |
| 10 | NATURE REVIEWS GENETICS | ENGLAND | 3 | 1469.67 | 4409 | 43.704 | 42.812 | Genetics & Heredity | Q1 |
| 11 | ONCOGENE | ENGLAND | 3 | 490.67 | 1472 | 6.634 | 6.429 | Biochemistry & Molecular Biology; Oncology; Cell Biology; Genetics & Heredity | Q1,Q1,Q1,Q1 |
The IF for journals within the top 100 cited papers ranged from 2.929 to 43.704. (Figure 4). Within the top 100 list, paper counts (r = 0.519, P < .001) and citation (r = 0.363, P < .001) counts were significantly related to IF.
Figure 4.
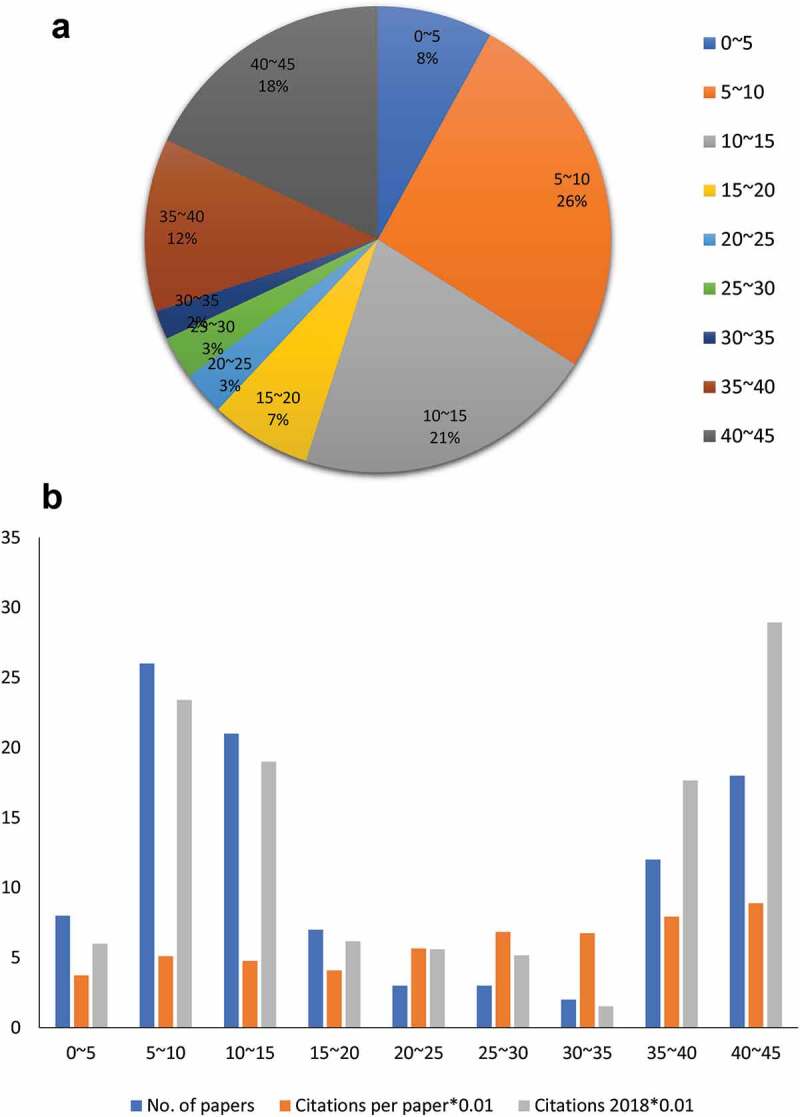
Impact factor of the top 100 most-cited papers
3.5. Author
The top 100 papers on lncRNAs research were drafted by 24 authors. Table 4 illustrates the most productive writers, i.e., those who authored at least three papers. Chang HY published the greatest number of papers (n = 11), and Mattick JS was the next biggest contributors owned 8 papers. Rinn JL and Chinnaiyan AM were tied for the third place with six papers.
Table 4.
Authors with at least three papers in the top 100 most-cited list
| Author | No. of papers | Citations per paper | Citations WOS | Citation in 2018 | Position on author list |
||
|---|---|---|---|---|---|---|---|
| First-author | Correspondent author | Other | |||||
| CHANG HY | 11 | 1087.27 | 11960 | 2287 | 0 | 9 | 2 |
| MATTICK JS | 8 | 724.5 | 5796 | 916 | 0 | 5 | 3 |
| DINGER ME | 6 | 808.33 | 4850 | 751 | 1 | 1 | 4 |
| RINN JL | 6 | 994.5 | 5967 | 867 | 0 | 0 | 6 |
| CHINNAIYAN AM | 4 | 658 | 2632 | 591 | 0 | 4 | 0 |
| GUTTMAN M | 4 | 668.75 | 2675 | 365 | 2* | 3 | 1 |
| LANDER ES | 4 | 668.75 | 2675 | 365 | 0 | 1 | 3 |
| LEE JT | 4 | 490.5 | 1962 | 351 | 2* | 4 | 0 |
| MERCER TR | 4 | 1120.25 | 4481 | 708 | 3 | 0 | 1 |
| PRENSNER JR | 4 | 658 | 2632 | 591 | 3 | 0 | 1 |
| YANG F | 4 | 434.5 | 1738 | 350 | 3 | 0 | 1 |
| CAO XH | 3 | 579.33 | 1738 | 411 | 0 | 0 | 3 |
| DHANASEKARAN SM | 3 | 579.33 | 1738 | 411 | 0 | 0 | 3 |
| GARBER M | 3 | 788 | 2364 | 304 | 0 | 0 | 3 |
| IYER MK | 3 | 579.33 | 1738 | 411 | 1 | 0 | 2 |
| REGEV A | 3 | 788 | 2364 | 304 | 0 | 0 | 3 |
| SUN M | 3 | 416.33 | 1249 | 300 | 0 | 0 | 3 |
| SUN SH | 3 | 486.67 | 1460 | 300 | 0 | 2 | 1 |
| WANG F | 3 | 486.67 | 1460 | 300 | 0 | 1 | 2 |
| WANG KC | 3 | 1844.67 | 5534 | 913 | 2 | 0 | 1 |
| WANG Y | 3 | 306.67 | 920 | 218 | 1 | 0 | 2 |
| ZHANG L | 3 | 331 | 993 | 182 | 0 | 0 | 3 |
| ZHANG Y | 3 | 353 | 1059 | 298 | 1 | 0 | 2 |
| ZHOU WP | 3 | 486.67 | 1460 | 300 | 0 | 0 | 3 |
3.6. Study field
In Table 5, the top 100 papers in lncRNAs were classified into various study fields based on WOS categories. The majority of the papers (n = 42) were categorized into “Biochemistry Molecular Biology,” and 38 papers were classified into “Cell Biology.” Considerable studies were also conducted in other fields, such as “Genetics Heredity,” “Oncology,” and “Science Technology Other Topics.”
Table 5.
Research fields of the top 100 most-cited list
| Rank | Research field | No. of papers | Citations per paper | Citations WOS |
|---|---|---|---|---|
| 1 | BIOCHEMISTRY MOLECULAR BIOLOGY | 42 | 605.38 | 25426 |
| 2 | CELL BIOLOGY | 38 | 609.39 | 23157 |
| 3 | GENETICS HEREDITY | 18 | 753.33 | 13560 |
| 4 | ONCOLOGY | 18 | 507.83 | 9141 |
| 5 | SCIENCE TECHNOLOGY OTHER TOPICS | 16 | 746.94 | 11951 |
| 6 | BIOTECHNOLOGY APPLIED MICROBIOLOGY | 6 | 790.67 | 4744 |
| 7 | BIOPHYSICS | 3 | 421 | 1263 |
| 8 | CARDIOVASCULAR SYSTEM CARDIOLOGY | 3 | 297.33 | 892 |
| 9 | HEMATOLOGY | 3 | 297.33 | 892 |
| 10 | LIFE SCIENCES BIOMEDICINE OTHER TOPICS | 3 | 301 | 903 |
| 11 | DEVELOPMENTAL BIOLOGY | 2 | 792.5 | 1585 |
| 12 | GASTROENTEROLOGY HEPATOLOGY | 1 | 420 | 420 |
| 13 | NEUROSCIENCES NEUROLOGY | 1 | 278 | 278 |
| 14 | PATHOLOGY | 1 | 267 | 267 |
| 15 | RESEARCH EXPERIMENTAL MEDICINE | 1 | 364 | 364 |
| 16 | SURGERY | 1 | 459 | 459 |
| 17 | UROLOGY NEPHROLOGY | 1 | 264 | 264 |
3.7. Keyword
The top 100 papers covered a wide range of keywords (Table 6). The terms “Gene-Expression,” “Expression,” “Chromatin,” “Gene,” and “Reveals” were the five most frequently used key words in the documents analyzed.
Table 6.
Key words with at least five papers in the top 100 most-cited list
| Key word | No. of papers | Key word | No. of papers |
|---|---|---|---|
| GENE-EXPRESSION | 27 | LONGNONCODINGRNA | 7 |
| EXPRESSION | 26 | PLURIPOTENCY | 7 |
| CHROMATIN | 23 | PROSTATE-SPECIFICGENE | 7 |
| GENE | 22 | BREAST-CANCER | 6 |
| REVEALS | 20 | EVOLUTION | 6 |
| TRANSCRIPTION | 18 | GENOME-WIDEASSOCIATION | 6 |
| CELLS | 15 | INTERGENICTRANSCRIPTION | 6 |
| GENOME | 15 | METHYLATION | 6 |
| DIFFERENTIATION | 13 | REPRESSION | 6 |
| IDENTIFICATION | 12 | X-CHROMOSOME | 6 |
| MESSENGER-RNA | 12 | ANTISENSERNA | 5 |
| EMBRYONICSTEM-CELLS | 11 | CELL-PROLIFERATION | 5 |
| TUMOR-SUPPRESSORGENE | 9 | DATABASE | 5 |
| CARCINOMA | 8 | GROWTH | 5 |
| HUMANGENOME | 8 | HISTONEH3 | 5 |
| IN-VIVO | 8 | METASTASIS | 5 |
| LONGNONCODINGRNAS | 8 | POOR-PROGNOSIS | 5 |
| X-CHROMOSOMEINACTIVATION | 8 | PROSTATE-CANCER | 5 |
| XISTRNA | 8 | STEM-CELLS | 5 |
| CANCER | 7 | UP-REGULATION | 5 |
| COMPLEXES | 7 |
3.8. Type of document
In terms of the type of document, original articles comprised 68% of the most-cited papers, and the remaining 34% were reviews. Total citations (P = .016), annual citations (P = .016), and citation 2018 (P = .001) significantly differed among the different document types.
4. Discussion
In this study, we conducted bibliometric analysis to identify the top 100 papers with highest citations in the field of lncRNAs research over the past two decades. We now summarize several features of these papers to gain insights into the history and prospects of this specialty.
4.1. Characteristics of the top 100 papers
4.1.1. Citations
The citation count, a reliable objective indicator of the quality and impact of a paper, varies across different subspecialties and depends on the size of the scientific community. Papers with high numbers of citations are usually called “citation classic;” hence, new researchers in a particular field could read these papers first before conducting further studies.41 The number of citations of the top 100 papers in our study on lncRNAs varied from 249 to 2,828, which is higher than that of other subjects.44–46 This finding indicates that lncRNAs research is a major concern in the medical and health fields. In comparison with a bibliometric study on lncRNAs published in 2018, in which the top 100 papers were cited 36,033 times, the total number of citations of the top 100 papers in our research (59,733 times) was much higher.41
To prevent bias wherein older papers are likely to receive more citations due to their longer citable period,47 we calculated annual citations to evaluate the relative impact of a paper. Papers with a large number of total citations but low number of annual citations are historically important in a certain period. By contrast, papers with high numbers of total and annual citations may be related to current studies and should be regarded as a true medical classics or landmarks in lncRNAs research. Papers published in the last 2 years have not accumulated enough citations to be included in the top 100 list. Hence, we tabulated the top 20 most-cited papers from 2017 to 2018 to show the “rising stars” in the lncRNAs field (Table 7).
Table 7.
The top 20 most-cited papers on long non-coding RNAs from 2017 to 2018
| Rank | Article | Citation WOS | Citations per year | Citations in 2018 |
|---|---|---|---|---|
| 1 | Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172(3):393–407. | 200 | 100 | 78 |
| 2 | Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J An atlas of human long non-coding RNAs with accurate 5 ‘ ends. Nature. 2017;543(7644):199-+. | 198 | 66 | 93 |
| 3 | Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77(15):3965–81. | 186 | 62 | 98 |
| 4 | Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017;18(4):558–76. | 160 | 53.33 | 63 |
| 5 | Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355(6320):10. | 154 | 51.33 | 75 |
| 6 | Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:13. | 116 | 38.67 | 46 |
| 7 | Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16:13. | 108 | 36 | 51 |
| 8 | Gupta SC, Tripathi YN. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–67. | 104 | 34.67 | 45 |
| 9 | Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F Genome-scale activation screen identifies a lncRNA locus regulating a gene neighborhood. Nature. 2017;548(7667):343-+. | 85 | 28.33 | 45 |
| 10 | Liu BY, Ye BQ, Yang LL, Zhu XX, Huang GL, Zhu PP Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499–508. | 85 | 28.33 | 55 |
| 11 | Cui Y, Zhang FM, Zhu CK, Geng L, Tian TD, Liu HM. Upregulated lncRNA SNHG1 contributes to progression of nonsmall cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8(11):17785–94. | 84 | 28 | 42 |
| 12 | Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–72. | 79 | 26.33 | 41 |
| 13 | Liu D, Li YW, Luo G, Xiao XY, Tao D, Wu XC LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2*. Cancer Lett. 2017;388:281–91. | 79 | 26.33 | 38 |
| 14 | Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–7. | 78 | 26 | 40 |
| 15 | Zhang EB, Han L, Yin DD, He XZ, Hong LZ, Si XX H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45(6):3086–101. | 77 | 25.67 | 39 |
| 16 | Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143(6):991–1004. | 75 | 25 | 36 |
| 17 | Xiong H, Ni Z, He J, Jiang S, Li X, He J LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–40. | 74 | 24.67 | 42 |
| 18 | Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J Biol Chem. 2017;292(1):82–99. | 73 | 24.33 | 38 |
| 19 | Lin CR, Yang LQ. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018;28(4):287–301. | 70 | 35 | 30 |
| 20 | Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77(5):1155–67. | 70 | 23.33 | 32 |
In comparison with annual citations, the citations in 2018 of most papers are much higher, which reflects that the top 100 papers gained sustained attention from scientists and may have potential academic importance in the future. The paper ranked first was published in Nature in 2010 and written by Gupta, RA, et al., reported that lncRNAs enhances the regulation of cancer expression10 and gained the highest number of annual citations. Interestingly, this paper also gained the most number of citations in bibliometric research published in 2018, with 1,281 total citations and 227.62 annual citations, but its overall number of citations in our study was twice more that in 2018 (2,828 times).41
4.1.2. Year
The top 100 papers were published from 2007 to 2016, although our search spanned the period from 1998 to 2018. Prior to 2007, only one paper entitled “Clusters of internally primed transcripts reveal novel long-noncoding RNAs” was searched. This paper was published by the journal PLOS GENETICS and gained 133 citations. Exactly 4,737 papers on lncRNAs were published from 2017 to 2018, but they were not cited enough to be included in the list of top 100 papers.
Previous bibliometric study reported that lncRNAs have been continuously studied since 2006 and would reach the inflection point of publication growth rate in 2021.41 The results of our study are consistent with those of previous research. The paper with the earliest publication in the top 100 list was published in 2007, and publication counts increased with fluctuations from 2007 to 2016. The peak citations of scientific papers are usually obtained approximately 10 years after publication.48 Hence, the papers included in our study could be expected to attract more attention from researchers in following years.
In our study, the most influential papers on lncRNAs were published in 2011–2014, which is a little different from a former study that the most boom years was 2009–2012.42 A possible explanation for this difference is that our study was conducted 2 years after this previous work, which allowed some papers more time to gather citations.
Two-time point analyses demonstrated that more papers were published but fewer citations were made per paper after 2011 than before 2011. This result is in line with the bibliometric analysis results on lncRNAs research from 2007 to 2016, in which the publication year was divided into a slowly increasing phase (2007–2011) and a sharply growing phase (2012–2016).42 The reason why papers published before 2011 were more frequently cited than those after 2011 might because the former have a longer citation period and academic importance in the lncRNAs field.
4.1.3. Country and institution
In terms of countries, the USA and China were the two largest contributors (73% of all publications) to the top 100 list. More papers were published in these countries after 2011 than before 2011; indeed, publications from China increased by fivefold over this time (Table 2). The USA dominated the top 100 list and can be regarded as the leading nation in lncRNAs research.42 The dominance of this country in other clinical disciplines, such as miRNAs,33 DNA repair,35 and cancer,39 has been noted, thus reflecting the enormous influence of the USA in the medical field.44 China presented a remarkable increase in both amount and quality of publications in the lncRNAs field. In one bibliometric study, China was the most productive country with 2,462 papers published (63.47%);41 in another bibliometric study, increasing numbers of papers from China were published in top academic journals with an IF higher than 10.7
Many scientists speculate that scientific activities are tightly connected to the social and economic issues of a country.35,49,50 Countries with high GDP may allot substantial investments in scientific investigation and foster a large sum of senior researchers.49,51,52 Similarly, the publication count is strongly related to a nation’s GDP. However, financial support partly explains the strong dominance of the USA in lncRNAs research. Furthermore, USA authors are more likely to cite local papers than foreign ones, and their papers are easier to publish in American journals than foreign papers.43
Although the institution rankings usually resemble country rankings, some important distinctions can be found in our study. Figure 3b reports that 13 institutions owned no less than 4 papers, including 10 from the USA and 2 from China. This result is quite different from a previous bibliometric study on lncRNAs (including all papers), in which 9 of the top 10 most prolific institutions were from China.41,42 Different types of bibliometric research depict the institutions’ advantages in different aspects. Scholars could find productive or influential institutions in the lncRNAs area for academic cooperation according to their needs.
4.1.4. Journal
Journals with high IFs, such as Cell, Nature, and Science, published the majority of the top-cited papers. This result is quite different from that of a previous bibliometric study on lncRNAs (including all papers), in which only 18.86% of all publications appeared in journals with an IF higher than 3.000.42 The IF value of a journal may be an effective predictor of citations. Our study supports the theory that paper counts and citation counts are positively related to the IF of the journals. In previous bibliometric studies, the journal Oncotarget published the largest number of papers in lncRNAs research.41,42 However, this journal contributed only one paper to the list of top 100 papers.
Moreover, the most-cited lncRNAs papers are nearly almost published in journals from USA and England. Considering that these prestigious journals have a higher rank and wider influence in their area to attract audience and citations, successful scientists may prefer to submit their high-quality works to these journals, which, in turn, maintains the latter’s high IF.53,54 For beginners in the lncRNAs field, choosing to read papers in these journals would provide them with a quick track to understand the fundamentals and follow developing trends.55
4.1.5. Author
In comparison with a bibliometric study that included all lncRNAs papers, nearly none of the most prolific authors in our study were included in their most productive list.42 Hence, authors should strive to conduct quality of research while working on increasing their number of works. Previous study found lncRNAs papers written by Gupta RA, Derrien T, Tsai MC, and Guttman M had been cited mostly by 2017,41,42 these authors also appear in our study whose paper citations ranked top 10. Gupta, who owned the top-cited paper in the lncRNAs field, was the most influential author in 2017.41 He remained the top author in our study, and citations of his paper increased from 1,821 to 2,828 within 2 years.
4.1.6. Study hotpots
The majority of included papers in our study fall into the fields of “Biochemistry Molecular Biology” and “Cell Biology,” thus stressing the critical role of lncRNAs in cancer transition and cell proliferation. We summarized the keywords of the top 100 papers and found “Gene-Expression,” “Expression,” “Chromatin,” and “Gene” to be the most frequently used keywords. This finding is similar to the results of a bibliometric analysis by Miao Y et al. in which “dosage compensation,” “in vivo,” “genome-wide association,” and “xist RNA” were the most common keywords.42
Previous lncRNAs bibliometrics studies found that “TNM stage,” “epithelial mesenchymal transition (EMT),” and “cell apoptosis” were the latest research areas up to 2017.41 Moreover, interest in lncRNA-related studies gradually shifted from “characteristics” to “application” during 2013–2017.41 Our study also supports this development trend because the function, human atlas and human disease may be promising future hotpots in the lncRNAs field (Table 7). Understanding the main areas and hotpots of lncRNAs papers is critical for editorial boards and scientific organizations when choosing and judging future research work; such knowledge may also help young researchers publish articles more effectively.56
4.1.7. Type of document
In our initial search, a total of 7,754 papers on lncRNAs were identified from 1998 to 2018, including 6,815 (87.89%) papers published as articles and 939 (12.11%) papers classified as reviews. In comparison with the initial search, the proportion of reviews (34%) is higher in our list of the top 100 most-cited papers. Moreover, reviews received significantly higher total and annual numbers of citations than articles. Hence, although original articles account for a large part of the citation classics, reviews have also attracted considerable attention among researchers.
4.2. Citation bias
Notwithstanding that citation count offers a valuable quantitative evaluation to determine academic importance, inherent methodologic limitations of the bibliometric study must be considered.57
First, the citation count for an article increases over time. Thus, earlier publications are potentially cited more frequently, regardless of their actual impact, whereas the importance of recent works may be underestimated due to the insufficient time to accumulate citation rates.43,58
Second, specific landmark publications are rarely cited due to the issue of “obliteration by incorporation.” This phenomenon occurs when the information provided by classics becomes part of the current body of knowledge in the field and embedded in the daily practice of clinicians.59–61
Another limitation is “orientated-citing bias,” which involves various types of conscious or unconscious incomplete citation biases. For instance, a researcher may prefer to cite works written by himself, a powerful person, colleagues, or friends, or select references from certain journals in which he prepared to submit work or journals with high IF; he may also avoid citing contradictory studies or competitors’ papers.44,56,61
Finally, other factors, such as open access, language preference, and the inherent design of the Science Citation Index,62 may also affect the citation count.
4.3. Strengths and limitations
A strength of our research is that the inherent time bias of bibliometrics was fully considered. We conducted a two-time analyze before and after 2011 and summarized the top 20 most-frequently cited papers from 2017 to 2018. Then, we listed the authors (e.g., first author, corresponding author), journal (e.g., originating country, IF, JCR category, and JCR partition), and most common keywords in detail. Finally, we performed statistical analyses to determine the underlying factors that may be related to citation counts.
Our study has several limitations. First, the search strategy may have missed some papers without the search words in their titles. Second, the language of the papers was restricted to English; thus, studies written in other languages may have been omitted. Third, only the WOS was searched to collect data; other databases such as Scopus and Google Scholar were not analyzed.
5. Conclusion
Our study identified papers responsible for the most significant developments in lncRNAs research. The number of citations in the top 100 most-cited papers varied from 249 to 2,828, and the publication years spanned from 2007 to 2016, with the year 2013 accounting for the most number of papers published. IF, GDP, and document type were strongly related to the citation count. The applications of lncRNAs in function, human atlas, and human disease may be future research hotpots.
Although the citation count does not accurately provide an evaluation of study quality, the top 100 most-cited papers offer investigators interesting insights into how lncRNAs research has evolved rapidly over the past decades, and help them choose targeted scientific issues to fill research gaps. Factors related to the citation count suggested researchers publishing researches in journals with high IF to gain higher influence, choosing countries with high GDP to cooperate or further education to receive more support. Furthermore, it could serve as a guide in policy making, R&D planning, and funding decision.
Funding Statement
This work was supported by the National Natural Science Foundation of China under Grant (number 81871844); Fok Ying-Tong Education Foundation of China under Grant (number 161092); the Shanghai Municipal Commission of Health and Family Planning under Grant (number 201840346); the Shanghai Key Lab of Human Performance (Shanghai University of Sport) under Grant (number 11DZ2261100); Shuguang Program supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission under Grant (number 18SG48).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Perkel JM. Visiting “noncodarnia”. Biotechniques. 2013;54:301, 303–304. PMID:23750541. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 2.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. PMID:24776770. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. PMID:22955620. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groux H, Huet S, Aubrit F, Tran HC, Boumsell L, Bernard A. A 19-kDa human erythrocyte molecule H19 is involved in rosettes, present on nucleated cells, and required for T cell activation. Comparison of the roles of H19 and LFA-3 molecules in T cell activation. J Immunol. 1989;142:3013–3020. PMID:2468708. [PubMed] [Google Scholar]
- 5.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. PMID:1985261. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 6.Briggs SF, Reijo Pera RA. X chromosome inactivation: recent advances and a look forward. Curr Opin Genet Dev. 2014;28:78–82. PMID:25461454. doi: 10.1016/j.gde.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing YH, Bai Z, Liu CX, Hu SB, Ruan M, Chen LL. Research progress of long noncoding RNA in China. IUBMB Life. 2016;68:887–893. PMID:27699981. doi: 10.1002/iub.1564. [DOI] [PubMed] [Google Scholar]
- 8.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. PMID:23498938. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. PMID:19188922. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. PMID:20393566. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skuratovskaia D, Vulf M, Komar A, Kirienkova E, Litvinova L. Promising directions in atherosclerosis treatment based on epigenetic regulation using MicroRNAs and long noncoding RNAs. Biomolecules. 2019;9. PMID:31212708. doi: 10.3390/biom9060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. PMID:24296535. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 13.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao G, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16(5):616–626. PMID:25525793. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. PMID:24105322. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Leary VB, Ovsepian SV, Carrascosa LG, Buske F, Radulovic V, Niyazi M, Moertl S, Trau M, Atkinson M, Anastasov N, et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015;11(3):474–485. PMID:25900080. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 16.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. PMID:21550244. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci Rep. 2014;4:6591. PMID:25307233. doi: 10.1038/srep06591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao XY, Lin JD. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40:586–596. PMID:26410599. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. PMID:19571179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. PMID:26430155. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. PMID:21925379. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Yan C, Zhang X, You Z. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017;18:558–576. PMID:27345524. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Sun Y, Guan N, Qu J, Huang Z-A, Zhu Z-X, Li J-Q. Computational models for lncRNA function prediction and functional similarity calculation. Brief Funct Genomics. 2019;18(1):58–82. PMID:30247501. doi: 10.1093/bfgp/ely031. [DOI] [PubMed] [Google Scholar]
- 24.Tang J, Yu Y, Yang W. Long noncoding RNA and its contribution to autism spectrum disorders. CNS Neurosci Ther. 2017;23:645–656. PMID:28635106. doi: 10.1111/cns.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Maegdefessel L. Non-coding RNA contribution to thoracic and abdominal aortic aneurysm disease development and progression. Front Physiol. 2017;8:429. PMID:28670289. doi: 10.3389/fphys.2017.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. PMID:22096659. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. PMID: 23827673. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. PMID:21489289. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. PMID:23243023. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. PMID:22289355. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. PMID:25842979. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23(9):1446–1461. PMID:23796952. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casey MC, Kerin MJ, Brown JA, Sweeney KJ. Evolution of a research field-a micro (RNA) example. PeerJ. 2015;3:e829. PMID:25802804. doi: 10.7717/peerj.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rousseau R. Library science: forgotten founder of bibliometrics. Nature. 2014;510:218. PMID:24919911. doi: 10.1038/510218e. [DOI] [PubMed] [Google Scholar]
- 35.Maisonobe M, Giglia-Mari G, Eckert D. DNA repair: a changing geography? (1964-2008). DNA Repair (Amst). 2013;12:466–471. PMID:23669398. doi: 10.1016/j.dnarep.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Lu X, Feng JB, Tian M, Liu QJ. Identification and validation of candidate radiation-responsive genes for human biodosimetr. Biomed Environ Sci. 2017;30:834–840. PMID:29216961. doi: 10.3967/bes2017.112. [DOI] [PubMed] [Google Scholar]
- 37.Gingras Y. The Revisiting “quiet debut” of the double helix: a bibliometric and methodological note on the “impact” of scientific publications. J Hist Biol. 2010;43:159–181. PMID:20503721. doi: 10.1007/s10739-009-9183-2. [DOI] [PubMed] [Google Scholar]
- 38.Lu K, Yu S, Sun D, Xing H, An J, Kong C, Yu M, Zhu Y. Scientometric analysis of SIRT6 studies. Med Sci Monit. 2018;24:8357–8371. PMID:30457131. doi: 10.12659/MSM.913644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabral BP, da Graca Derengowski Fonseca M, Mota FB. The recent landscape of cancer research worldwide: a bibliometric and network analysis. Oncotarget. 2018;9:30474–30484. PMID:30093962. doi: 10.18632/oncotarget.25730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Shi Y, Zhou K, Yu S, Cai W, Ying M. A bibliometric analysis of long non-coding RNA and chemotherapeutic resistance research. Oncotarget. 2019;10:3267–3275. PMID:31143372. doi: 10.18632/oncotarget.26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhai X, Zhao J, Wang Y, Wei X, Li G, Yang Y, Chen Z, Bai Y, Wang Q, Chen X, et al. Bibliometric analysis of global scientific research on lncRNA: a swiftly expanding trend. Biomed Res Int. 2018;2018:7625078. PMID:29992161. doi: 10.1155/2018/7625078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miao Y, Xu SY, Chen LS, Liang GY, Pu YP, Yin LH. Trends of long noncoding RNA research from 2007 to 2016: a bibliometric analysis. Oncotarget. 2017;8:83114–83127. PMID:29137328. doi: 10.18632/oncotarget.20851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baltussen A, Kindler CH. Citation classics in critical care medicine. Intensive Care Med. 2004;30:902–910. PMID:14985952. doi: 10.1007/s00134-004-2195-7. [DOI] [PubMed] [Google Scholar]
- 44.Kim ES, Yoon DY, Kim HJ, Jeon HJ, Lee JY, Cho B-M, Lee K. Citation classics in neurointerventional research: a bibliometric analysis of the 100 most cited articles. J Neurointerv Surg. 2017;9:508–511. PMID:27127230. doi: 10.1136/neurintsurg-2016-012399. [DOI] [PubMed] [Google Scholar]
- 45.Gu W, Yuan Y, Yang H, Qi G, Jin X, Yan J. A bibliometric analysis of the 100 most influential papers on COPD. Int J Chron Obstruct Pulmon Dis. 2015;10:667–676. PMID:25848243. doi: 10.2147/COPD.S74911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefaivre KA, Shadgan B, O’Brien PJ. 100 most cited articles in orthopaedic surgery. Clin Orthop Relat Res. 2011;469:1487–1497. PMID:20922583. doi: 10.1007/s11999-010-1604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pepe A, Kurtz MJ. A measure of total research impact independent of time and discipline. PLoS One. 2012;7:e46428. PMID:23144782. doi: 10.1371/journal.pone.0046428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu Y, Zhang C, Hu Z, Li S, NKonging Y, Shang Y, Bai C. The 100 most influential publications in asthma from 1960 to 2017: A bibliometric analysis. Respir Med. 2018;137:206–212. PMID:29605206. doi: 10.1016/j.rmed.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 49.Tao T, Zhao X, Lou J, Bo L, Wang F, Li J, Deng X. The top cited clinical research articles on sepsis: a bibliometric analysis. Crit Care. 2012;16(3):R110. PMID:22731930. doi: 10.1186/cc11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HJ, Yoon DY, Kim ES, Lee K, Bae JS, Lee JH. The 100 most-cited articles in neuroimaging: A bibliometric analysis. Neuroimage. 2016;139:149–156. PMID:27327516. doi: 10.1016/j.neuroimage.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 51.Oelrich B, Peters R, Jung K. A bibliometric evaluation of publications in urological journals among European Union countries between 2000-2005. Eur Urol. 2007;52:1238–1248. PMID:17673361. doi: 10.1016/j.eururo.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 52.Moon JY, Yun EJ, Yoon DY, Choi C, Seo Y, Cho Y, Lim K, Baek S, Hong S, Yoon S, et al. The 100 most-cited articles focused on ultrasound imaging: a bibliometric analysis. Ultraschall Med. 2017;38(3):311–317. PMID:28511228. doi: 10.1055/s-0042-120259. [DOI] [PubMed] [Google Scholar]
- 53.Garfield E. The history and meaning of the journal impact factor. Jama. 2006;295:90–93. PMID:16391221. doi: 10.1001/jama.295.1.90. [DOI] [PubMed] [Google Scholar]
- 54.Callaham M, Wears RL, Weber E. Journal prestige, publication bias, and other characteristics associated with citation of published studies in peer-reviewed journals. Jama. 2002;287:2847–2850. PMID:12038930. doi: 10.1001/jama.287.21.2847. [DOI] [PubMed] [Google Scholar]
- 55.Tregoning J. How will you judge me if not by impact factor? Nature. 2018;558:345. PMID:29921857. doi: 10.1038/d41586-018-05467-5. [DOI] [PubMed] [Google Scholar]
- 56.Loonen MP, Hage JJ, Kon M. Plastic surgery classics: characteristics of 50 top-cited articles in four plastic surgery journals since 1946. Plast Reconstr Surg. 2008;121:320e–327e. PMID:18453945. doi: 10.1097/PRS.0b013e31816b13a9. [DOI] [PubMed] [Google Scholar]
- 57.Chou CY, Chew SS, Patel DV, Ormonde SE, McGhee C. Publication and citation analysis of the Australian and New Zealand journal of ophthalmology and clinical and experimental ophthalmology over a 10-year period: the evolution of an ophthalmology journal. Clin Exp Ophthalmol. 2009;37:868–873. PMID:20092596. doi: 10.1111/j.1442-9071.2009.02191.x. [DOI] [PubMed] [Google Scholar]
- 58.Shadgan B, Roig M, Hajghanbari B, Reid WD. Top-cited articles in rehabilitation. Arch Phys Med Rehabil. 2010;91:806–815. PMID:20434622. doi: 10.1016/j.apmr.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Khan MS, Ullah W, Riaz IB, Bhulani N, Manning WJ, Tridandapani S, Khosa F. Top 100 cited articles in cardiovascular magnetic resonance: a bibliometric analysis. J Cardiovasc Magn Reson. 2016;18:87. PMID:27866473. doi: 10.1186/s12968-016-0303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seglen PO. Citation rates and journal impact factors are not suitable for evaluation of research. Acta Orthop Scand. 1998;69:224–229. PMID:9703393. doi: 10.3109/17453679809000920. [DOI] [PubMed] [Google Scholar]
- 61.Baltussen A, Kindler CH. Citation classics in anesthetic journals. Anesth Analg. 2004;98:443–451. PMID:14742385. doi: 10.1213/01.ane.0000096185.13474.0a. [DOI] [PubMed] [Google Scholar]
- 62.Ponce FA, Lozano AM. Highly cited works in neurosurgery. Part I: the 100 top-cited papers in neurosurgical journals. J Neurosurg. 2010;112:223–232. PMID:20078192. doi: 10.3171/2009.12.JNS091599. [DOI] [PubMed] [Google Scholar]


