Abstract
Coronavirus disease 2019 (COVID-19) exhibits variable symptom severity ranging from asymptomatic to life-threatening, yet the relationship between severity and the humoral immune response is poorly understood. We examined antibody responses in 113 COVID-19 patients and found that severe cases resulting in intubation or death exhibited increased inflammatory markers, lymphopenia, pro-inflammatory cytokines, and high anti-receptor binding domain (RBD) antibody levels. Although anti-RBD immunoglobulin G (IgG) levels generally correlated with neutralization titer, quantitation of neutralization potency revealed that high potency was a predictor of survival. In addition to neutralization of wild-type SARS-CoV-2, patient sera were also able to neutralize the recently emerged SARS-CoV-2 mutant D614G, suggesting cross-protection from reinfection by either strain. However, SARS-CoV-2 sera generally lacked cross-neutralization to a highly homologous pre-emergent bat coronavirus, WIV1-CoV, which has not yet crossed the species barrier. These results highlight the importance of neutralizing humoral immunity on disease progression and the need to develop broadly protective interventions to prevent future coronavirus pandemics.
Keywords: COVID-19, SARS-CoV-2, ELISA, RBD, spike, neutralizing antibodies, disease severity, pro-inflammatory cytokines, D614G, WIV1-CoV
Graphical abstract
Highlights
-
•
Severe COVID-19 associates with higher antibody production and neutralization titers
-
•
Neutralization potency of anti-RBD antibodies predicts disease severity and survival
-
•
Immunomodulatory COVID-19-directed therapies modulate antibody responses
-
•
COVID-19 sera neutralize D614 and G614 variants, but not pre-emergent WIV1-CoV
Garcia-Beltran et al. show that the development of more potent neutralizing antibodies during SARS-CoV-2 infection predicts COVID-19 survival. Protective antibody responses exhibit potent neutralization against the currently circulating SARS-CoV-2 D614G spike variant but lack significant activity against pre-emergent WIV1-CoV spike, suggesting that convalescent patients are likely to remain susceptible to future pandemics.
Introduction
Coronavirus disease 2019 (COVID-19), caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), exhibits significant variability in the severity of presentation. The impact of this variability on the development of protective immune responses and the role of antibodies in disease progression is unclear. Given the ongoing development of treatment regimens for mild and severe cases of COVID-19, there is limited understanding of the impact these investigational therapies have on immune responses against SARS-CoV-2.
Non-human primates (NHP) exposed to SARS-CoV-2 develop potent antibody responses and are largely immune to reinfection (Deng et al., 2020; Chandrashekar et al., 2020). Similarly, animal models testing candidate vaccine approaches have demonstrated that protection against SARS-CoV-2 challenge is positively correlated with the development of high titers of neutralizing antibodies (Mercado et al., 2020; Yu et al., 2020). Importantly, passive transfer of convalescent sera prevents infection in otherwise naive animals, highlighting the crucial role of antibodies in mediating protection against viral infection (Rogers et al., 2020; Hassan et al., 2020).
In contrast, the role of antibodies on the clearance of established SARS-CoV-2 infection and clinical outcomes is less clear. Ordinarily, infections with viruses require cell-mediated immunity for viral clearance. Antibodies mediate functions such as antibody-dependent cellular cytotoxicity (ADCC) and phagocytosis (ADCP) via innate immune cells such as natural killer (NK) cells and macrophages. Yet, the need for antibodies in the clearance of SARS-CoV-2 infection has been challenged by two recent cases of patients with X-linked agammaglobulinemia who acquired and survived SARS-CoV-2 infection without requiring oxygen or intensive care (Soresina et al., 2020). Some studies even propose the possibility of a pathogenic role of antibodies in primary infection via antibody-dependent enhancement (ADE) and augmentation of inflammation (Liu et al., 2019), although it is believed that this is insufficient to explain the prevalence of severe cases of SARS-CoV-2 infection (Arvin et al., 2020). As such, a beneficial, neutral, or harmful role of antibodies in active coronavirus infection remains controversial.
Numerous clinical studies testing a variety of COVID-19 therapies are ongoing, and thus far, suppression of the immune response with corticosteroids has emerged as a standard treatment regimen to limit COVID-19 disease severity (Siemieniuk et al., 2020a; Horby et al., 2020). Remdesivir, a nucleotide analog active against SARS-CoV-2, has shown modest benefit in severe COVID-19 cases by improving time to recovery (Beigel et al., 2020; Wang et al., 2020b). Hydroxychloroquine was initially tested in patients based on in vitro studies (Chen et al., 2020c; Wang et al., 2020b), but subsequent meta-analyses and randomized controlled trials have demonstrated no benefit in preventing or treating COVID-19 (Boulware et al., 2020; Tang et al., 2020; Ullah et al., 2020). Morbidity and mortality due to COVID-19 is largely a consequence of adult respiratory distress syndrome (ARDS) caused by a combination of both hyperinflammatory and hypercoagulable states (Domingo et al., 2020), and thus suppression of these will be key to improving outcomes, as evidenced by use of corticosteroids and current trials employing tocilizumab, an anti-IL-6 receptor antibody used to treat cytokine release syndrome (Guaraldi et al., 2020). However, the consequences of these and other current interventions on the development of humoral immunity are not known.
Recent studies have demonstrated the emergence of SARS-CoV-2 variants containing amino acid substitutions in the viral spike protein, raising concerns for potential resistance to neutralization. One mutation, D614G, has rapidly become the predominant transmitted variant by outcompeting wild-type infections (Korber et al., 2020; Lemieux et al., 2020). While it has been suggested that this mutant results in a more fit virus (Plante et al., 2020), the serological consequences of this change are unclear. Additionally, recent studies in bats have described a novel coronavirus (WIV1-CoV) with high homology to SARS-CoV-2 that uses the same ACE2 receptor for cell entry (Menachery et al., 2016). It has been postulated that this virus may present a similar pandemic risk if it were to spread from bats to humans. However, the consequences of prior SARS-CoV-2 seroconversion on neutralization of related pre-emergent coronaviruses like WIV1-CoV has not been described.
In this study, we characterized humoral immune responses and clinical outcomes in 113 SARS-CoV-2-infected patients of varying severity who received a range of treatments, as well as 1,257 pre-pandemic individuals. Our COVID-19 patient cohort contained a wide range of outcomes, including non-hospitalized, hospitalized, intubated, deceased, and immunosuppressed individuals. We assessed inflammatory markers, multiple cytokines, lymphocyte counts, and demographic variables such as age and sex. A quantitative ELISA that measures immunoglobulin G (IgG), immunoglobulin M (IgM), and immunoglobulin A (IgA) antibodies to the receptor binding domain (RBD) and spike protein of SARS-CoV-2 and a high-throughput neutralization assay using lentiviral vectors pseudotyped with SARS-CoV-2 and WIV1-CoV were developed to assess neutralization potency and cross-neutralizing responses. Remarkably, we find that anti-RBD antibody levels, neutralization titer, and neutralization potency associated with disease severity and predicted survival, but largely lacked cross-neutralizing activity to pre-emergent WIV1-CoV.
Taken together, our results highlight the impact of an effective humoral immune response on COVID-19, as quantified by a neutralization potency index, and describe both the cytokines associated with neutralization potency and the influence of current experimental therapies on antibody development. The limited cross-neutralizing potential of antibodies from SARS-CoV-2-infected patients highlights the need to focus future effort on the development of broadly protective interventions to mitigate future coronavirus pandemics.
Results
Spectrum of clinical severity of SARS-CoV-2 infection
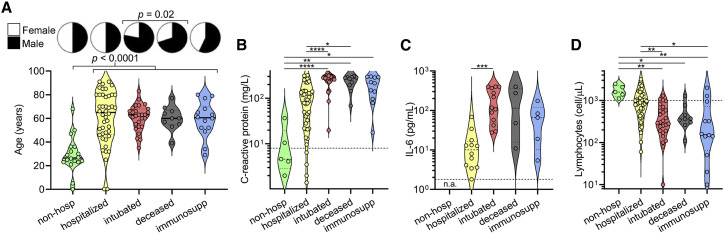
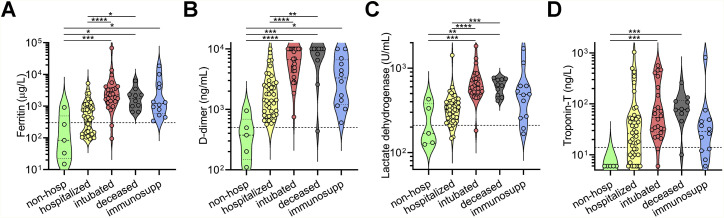
A cross-sectional cohort of 113 COVID-19 cases confirmed by SARS-CoV-2 nasopharyngeal PCR was studied and followed for at least 3 months. The cohort was divided into the following five groups based on disease severity, outcomes, and pre-existing health status: (1) “non-hospitalized,” which were never admitted to the hospital due to COVID-19; (2) “hospitalized,” which were admitted for at least 1 day but were never intubated and were eventually discharged, (3) “intubated,” which were intubated for at least 1 day but were subsequently extubated and discharged; (4) “deceased,” which passed away due to COVID-19 after sample collection; and (5) “immunosuppressed” due to medications or underlying medical conditions, which included some non-hospitalized, hospitalized, and intubated patients (but none deceased) (Table S1). When compared to non-hospitalized individuals, all cases of COVID-19 resulting in hospital admission were significantly older in age (median age 63 versus 28, p < 0.0001), and there was a significant enrichment for males in severe cases resulting in intubation and/or death (74% versus 51% males, p = 0.02) (Figure 1 A), consistent with prior studies (Chen et al., 2020a; Meng et al., 2020). Laboratory data showed that clinical severity correlated with markers of inflammation, namely, peak serum levels of C-reactive protein (Figure 1B), ferritin (Figure S1 A), D-dimer (Figure S1B), lactate dehydrogenase (Figure S1C), and interleukin (IL)-6 (Figure 1C), as well as lymphopenia (Figure 1D), as has been previously shown (Wang, 2020; Zhou et al., 2020b; Wynants et al., 2020; Chen et al., 2020b). Interestingly, COVID-19 severity was also associated with peak serum levels of troponin-T (Figure S1D), a marker of myocardial damage and/or ischemia that may reflect cardiac injury, as has been previously described (Tersalvi et al., 2020). Altogether, our cohort contained a wide range of clinical presentations of SARS-CoV-2 infection with our analyses confirming previously described associations.
Figure 1.
Clinical severity of SARS-CoV-2 infection is influenced by patient characteristics and coupled to clinical laboratory data
(A) A cross-sectional cohort of COVID-19 patients (n = 113) was divided into groups of varying clinical severity, i.e., non-hospitalized (n = 18), hospitalized (n = 45), intubated (n = 27), deceased (n = 10), and immunosuppressed (n = 13), and analyzed for age and sex. Median age was 28 years in patients who were never hospitalized (n = 20; includes 2 immunosuppressed) and 63 years in patients admitted to the hospital (n = 93), with a t test yielding p < 0.0001. Fisher’s exact test on males who were intubated or deceased (n = 31 males of 42 total; includes 5 immunosuppressed) versus not (n = 36 males of 71 total) demonstrated a significant enrichment (p = 0.02).
(B–D) Peak levels of C-reactive protein (B) and IL-6 (C) as well as lymphocyte count nadir (D) are presented in violin plots. In (C), none of the non-hospitalized patients had serum IL-6 levels measured (n.a., not assessed). For (B) and (C), clinical laboratory-defined cut-offs of the upper limit of normal are indicated with a dotted line; for (D), the dotted line represents the lower limit of normal. For each parameter, a non-parametric ANOVA was performed; statistical significance is indicated as follows: ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05.
Figure S1.
Clinical laboratory data from COVID-19 patients, related to Figure 1
(A−D) Peak serum levels of ferritin (A), D-dimer (B), lactate dehydrogenase (C), and troponin-T (D) documented for each COVID-19 patient in the indicated cohorts are shown as violin plots. Clinical laboratory-defined cut-offs of the upper limit of normal are indicated with a dotted line. For each parameter, a non-parametric ANOVA was performed; statistical significance is indicated as follows: ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05.
Quantitative SARS-CoV-2 receptor binding domain and spike IgG, IgM, and IgA ELISAs
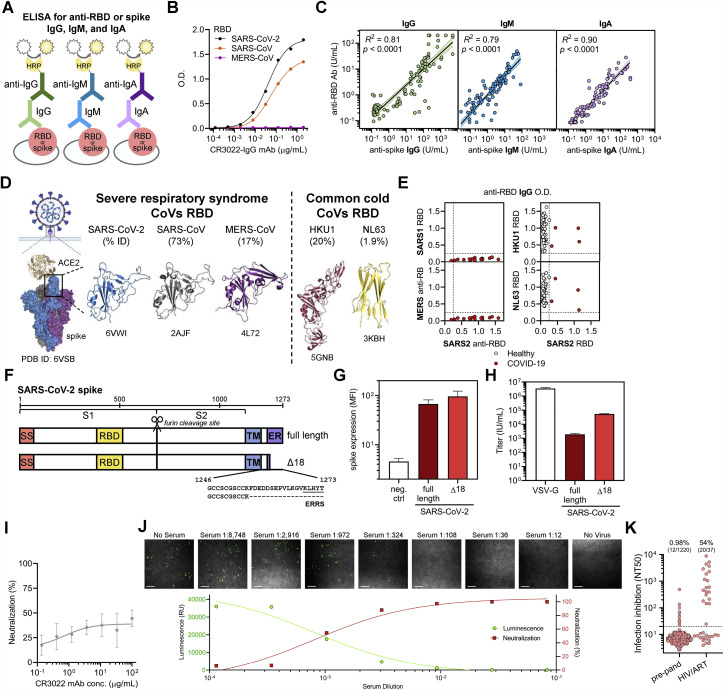
ELISAs that quantitatively measured IgG, IgM, and IgA antibodies that target either the RBD or full-length spike protein of SARS-CoV-2 were developed to characterize humoral immune responses (Figure S2 A), similar to what we have previously described (Roy et al., 2020; Iyer et al., 2020). Quantitation for both assays was achieved using a standard curve consisting of purified IgG, IgM, and IgA isotype of a monoclonal antibody, CR3022 (Figure 2 A), that cross-reacts to bind both SARS-CoV and SARS-CoV-2 RBD (Figure S2B) (ter Meulen et al., 2006; Tian et al., 2020).
Figure S2.
Cross-reactivity of anti-CoV antibody responses and high-throughput SARS-CoV-2 pseudovirus neutralization assay, related to Figure 2
(A) A schematic of the quantitative indirect ELISA that measures IgG, IgM, and IgA antibodies to the receptor binding domain (RBD) or spike for SARS-CoV-2 is shown.
(B) Reactivity of the anti-SARS-CoV and -CoV-2-specific monoclonal antibody (CR3022 mAb) toward SARS-CoV-2, SARS-CoV, and MERS-CoV RBD was measured.
(C) Log-log regression analyses were performed to compare anti-RBD versus anti-spike antibody levels for IgG (left panel), IgM (middle panel), and IgA (right panel). Pearson correlations were calculated and R2 and p values are indicated.
(D) Published crystal structures of the ACE2:prefusion-stabilized SARS-CoV-2 spike (PDB ID: 6VSB) as well as the RBD of SARS-CoV-2 (PDB ID 6VWI), SARS-CoV (PDB ID 2AJF), MERS-CoV (PDB ID: 4L72), HKU1 (PDB ID 5GNB), and NL63 (PDB ID 3KBH) are presented, with the sequence homology to SARS-CoV-2 RBD indicated.
(E) Cross-reactivity of anti-RBD IgG from SARS-CoV-2-infected patient sera (n = 15) toward the RBD of SARS-CoV (top left) and MERS-CoV (bottom left), as well as the reactivity anti-RBD IgG from the sera of healthy blood donors (n = 43) and COVID-19 patients (n = 4) toward the RBD of two common cold coronaviruses, HKU1 (top right) and NL63 (bottom right), was measured using a modified anti-RBD IgG ELISA and optical density as a readout.
(F) A schematic of the full-length and truncated (Δ18) construct of SARS-CoV-2 spike used to pseudotype lentivirus is shown; ERRS denotes ER retention signal.
(G) Expression of the indicated spike constructs was measured on the surface of 293T cells via flow cytometry; mean and standard deviation are shown.
(H) Pseudovirus titers of the indicated spike constructs were quantified in 293T-ACE2 cells; mean and standard deviation are shown.
(I) Lack of neutralizing ability of CR3022 mAb was confirmed in pseudovirus neutralization assay; mean and standard deviation of neutralization (%) at each dilution is shown.
(J) Confocal microscopy of each well of a serum dilution series using a representative COVID-19 patient sample taken 60 - 72 h after assay setup demonstrated the correlation between luciferase activity and transduced (ZsGreen+) target cells. Neutralization percentage at each dilution was calculated by measuring luciferase activity (luminescence) and normalizing to control well with no serum. Scale bar equals 200 μm.
(K) False positive NT50 values were observed in individuals taking antiretroviral medications (n = 20 out of 37 individuals), while a large cohort of pre-pandemic individuals for which antiretroviral use was largely screened out showed a very low rate of infection inhibition (n = 12 out of 1,220).
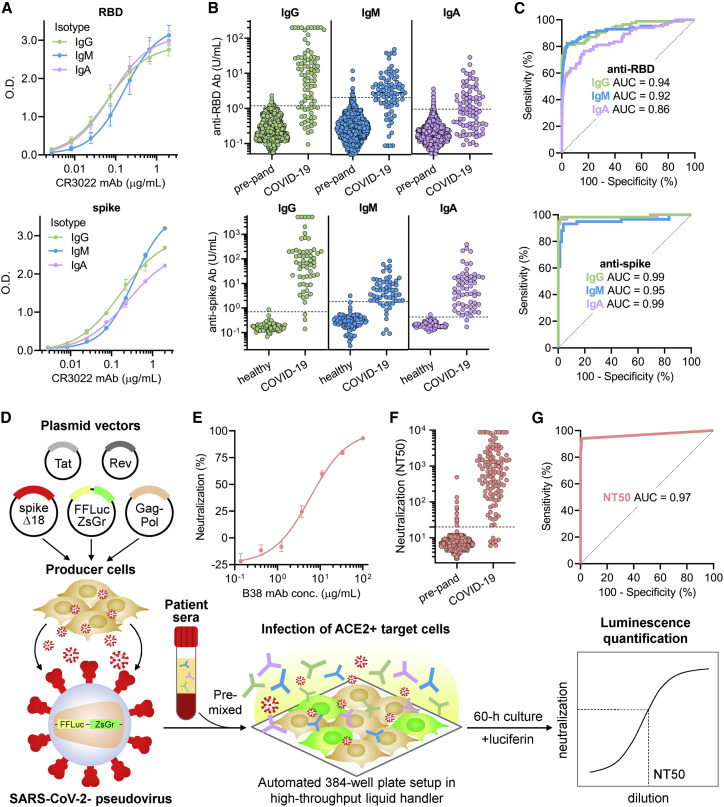
Figure 2.
Quantitative SARS-CoV-2 receptor binding domain and spike ELISA and high-throughput SARS-CoV-2 pseudovirus neutralization assay reveal highly variable IgG, IgM, and IgA responses and neutralization potency after SARS-CoV-2 infection
(A) For quantitation of anti-RBD (upper panel) and anti-spike (lower panel) IgG, IgM, and IgA antibodies, a standard curve consisting of a SARS-CoV-2 RBD-binding monoclonal antibody, CR3022, in IgG, IgM, and IgA isotypes was used. Error bars indicate standard deviation.
(B) Anti-RBD (upper panel) and anti-spike (lower panel) IgG, IgM, and IgA antibodies were measured in both negative controls (n = 1,257 pre-pandemic samples for anti-RBD; n = 78 healthy blood donors for anti-spike antibodies) and COVID-19 patient samples (n = 85 for anti-RBD; n = 59 for anti-spike antibodies). Dotted lines indicate the threshold of seropositivity that achieved a specificity of >99% for anti-RBD antibodies and >98% for anti-spike antibodies on ROC analyses.
(C) ROC analyses for anti-RBD (upper panel) and anti-spike (lower panel) IgG, IgM, and IgA antibodies were done to assess how seropositivity predicted COVID-19 status. Area under the curve (AUC) is indicated for each antibody target and isotype.
(D) A schematic of the high-throughput SARS-CoV-2 pseudovirus neutralization assay is shown.
(E) Validation of the neutralization assay using a recently discovered anti-RBD neutralizing monoclonal antibody, B38, was performed (IC50 = 6 μg/mL). Error bars indicate standard deviation.
(F) Neutralization titers that achieved 50% neutralization (NT50) were calculated for pre-pandemic samples (n = 1,220, individuals on antiretroviral therapy excluded) and samples from COVID-19 patients >14 days after symptom onset (n = 118).
(G) An ROC analysis demonstrated an AUC of 0.97, with an NT50 cut-off of 20 achieving a sensitivity of 94% and specificity of >99%.
See also Figure S2.
We determined the sensitivity and specificity of anti-RBD and anti-spike ELISAs by assessing antibody levels in a cohort of SARS-CoV-2-infected patient serum samples collected between 14 to 42 days after symptom onset (n = 85 for anti-RBD and n = 59 for anti-spike antibodies) in order to maximize seropositivity for IgG, IgM, and IgA. As controls, we also assessed antibody levels in 1,257 pre-pandemic serum samples (that included individuals with positive serology results for other infectious diseases) and 78 healthy blood donors (Figure 2B). Anti-RBD and anti-spike IgG, IgM, and IgA levels were measured for each sample by interpolation from the standard curve, and unsurprisingly, anti-RBD and anti-spike antibody levels correlated strongly to each other for each isotype (Figure S2C). A receiver operating curve (ROC) analysis was used to determine optimal cut-offs that distinguished SARS-CoV-2-infected patients from controls (Figure 2C) with high sensitivity and specificity (see STAR methods).
Although non-RBD anti-spike antibodies can cross-react between different coronaviruses (Secchi et al., 2020; Chan et al., 2005; Ju et al., 2020), we assessed for cross-reactivity of anti-RBD IgG in sera of SARS-CoV-2 seropositive individuals by modifying our ELISA to detect IgG antibodies against the RBD of SARS-CoV and MERS-CoV. Interestingly, no cross-reactivity was seen to SARS-CoV RBD despite 73% homology, nor to MERS-CoV, which has only 17% homology (Figures S2D and S2E, left), indicating that anti-RBD IgG antibodies induced during SARS-CoV-2 infection generally do not cross-react to recognize the RBD of other coronaviruses that cause severe respiratory syndrome. Additional experiments measuring IgG antibodies against the RBD of two common cold coronaviruses—NL63, which has 20% homology to SARS-CoV-2 RBD, and HKU1, which has 1.9% homology (Figure S2D)—showed a seroprevalence of >95% (Figure S2E), as has been shown in previously published studies (Gorse et al., 2010), with no correlation between the anti-RBD IgG antibody levels of NL63 or HKU1 with SARS-CoV-2 (Figure S2E). This indicates that anti-RBD IgG antibodies to common cold coronaviruses usually do not cross-react to recognize SARS-CoV-2 RBD; however, there may be rare individuals with anti-RBD IgG antibodies that exhibit low-level cross-reactivity, as seen in a small minority of individuals (<1%) in our pre-pandemic cohort (Figure 2B). Overall, these data suggest that natural infection with coronavirus results in anti-RBD antibodies with limited cross-reactivity.
High-throughput SARS-CoV-2 pseudovirus neutralization assay
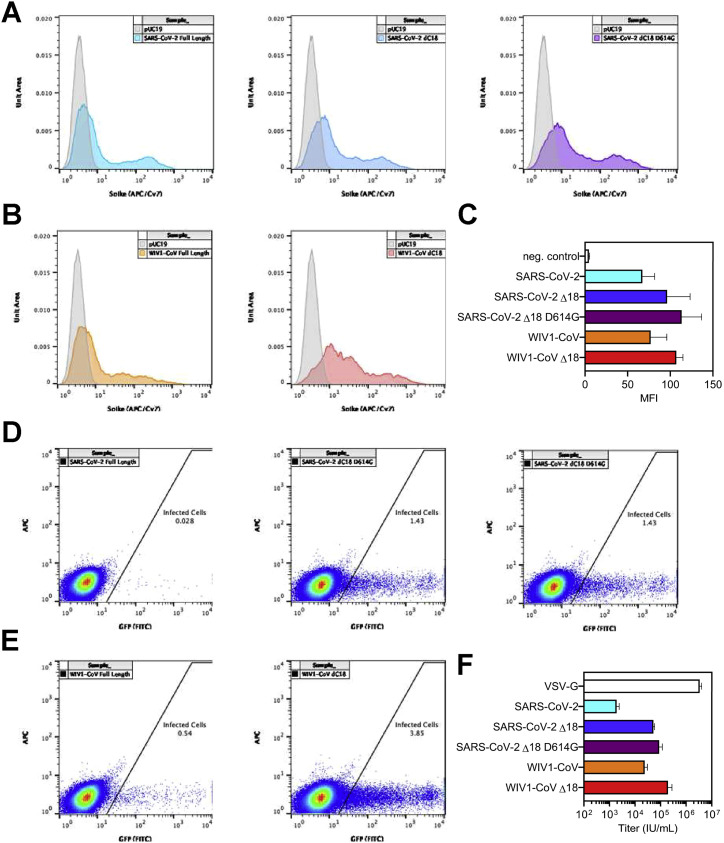
Previous studies have demonstrated the potential to pseudotype retroviral vectors with coronavirus spike proteins for pseudovirus neutralization assays (Moore et al., 2004) and have shown excellent correlation with results from live virus neutralization assays in the case of SARS-CoV-2 (Wang et al., 2020a; Ju et al., 2020; Pinto et al., 2020; Yang et al., 2020). However, pseudoviruses bearing SARS-CoV-2 spike produced by these methods yield low titers (Nie et al., 2020), hampering large-scale testing of neutralization. Recently, a forward genetics approach identified an efficiently replicating vesicular stomatitis virus (VSV) variant encoding SARS-CoV-2 spike containing a truncated form lacking the C-terminal 21 amino acids (Case et al., 2020). Interestingly, previous studies also showed a role of the cytoplasmic tail of SARS-CoV in altering surface expression and fusogenic potential (Corver et al., 2009). To determine whether analogous truncations might improve SARS-CoV-2 pseudovirus production, we examined the cell-surface expression of truncated forms of SARS-CoV-2 spike and found that removal of 18 amino acids from the C terminus (Δ18) resulted in significantly greater cell-surface expression and higher titers of pseudovirus (Figures S2F–S2H). This truncation removed a putative ER-retention signal (McBride et al., 2007; Ujike et al., 2016; Lontok et al., 2004) while retaining cysteine-rich domains that are highly conserved among coronaviruses. Using these spike modifications, we developed a CoV pseudovirus neutralization assay compatible with high-throughput liquid handling instrumentation in 384-well plate format using our previously published lentiviral vector system expressing both luminescent and fluorescent marker transgenes (Figure 2D) (Crawford et al., 2020).
To validate our assay, the potency of a neutralizing monoclonal antibody, B38, and a non-neutralizing monoclonal antibody, CR3022, both of which target SARS-CoV-2 RBD with known IC50 values, was determined. This yielded IC50 values of ∼6 μg/mL for B38 and undetectable (>100 μg/mL) for CR3022, which were in agreement with previous reports (Wu et al., 2020; Tian et al., 2020) (Figures 2E and S2I). In addition, we found that luciferase activity was directly proportional to the number of infected cells, providing flexibility in assay readout (Figure S2J). To determine the performance of our assay on human sera, we measured the neutralization potency of human sera from 1,220 pre-pandemic individuals and 118 COVID-19 patient samples >14 days after symptom onset. The dilution titer that achieved 50% neutralization (NT50) was calculated for each specimen, and receiver operating characteristic curve (ROC) analysis was performed, revealing that an NT50 threshold of 1:20 achieves a sensitivity of 94% and specificity of >99% in identifying COVID-19 patients (Figures 2F and 2G). Importantly, we excluded individuals receiving antiretroviral therapy because their sera exhibited potent inhibition of pseudovirus infection (Figure S2K; STAR methods). Overall, we found median titers of 1:664 in COVID-19 patients, with potency ranging from <1:12 to >1:8,748. Comparatively, live virus neutralization titers of 1:40 for influenza are considered to indicate protective immunity (Hannoun et al., 2004; Plotkin, 2010). In the case of influenza, prior studies have demonstrated excellent correlations between live virus neutralization assays and pseudovirus neutralization assays (Du et al., 2010). However, thresholds for SARS-CoV-2 neutralization titers that confer protection from infection as measured by either live or pseudovirus assays have yet to be determined. Altogether, we established a highly accurate high-throughput SARS-CoV-2 pseudovirus neutralization assay that can be used to quantify the neutralization potency of humoral immune responses directed to SARS-CoV-2 spike protein.
Relationship between neutralizing humoral immunity in SARS-CoV-2 infection and clinical severity
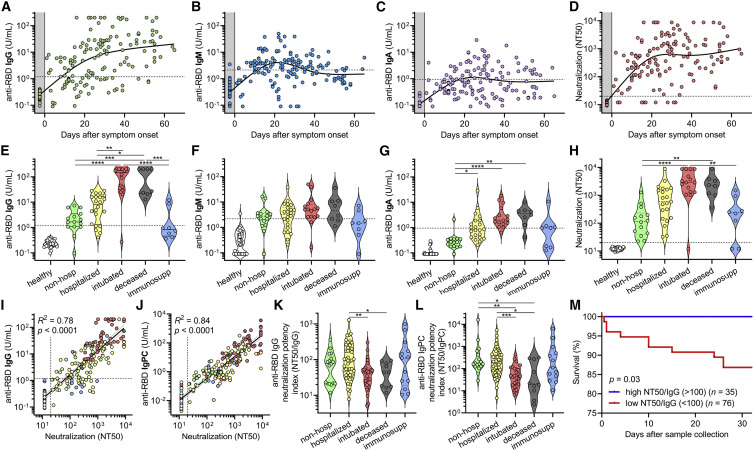
We proceeded to analyze antibody responses in our cohort of COVID-19 patients as well as a negative control cohort of 37 healthy blood donors sampled during the pandemic, and found that in contrast to the typical kinetics of antibody responses in viral infections (i.e., IgM before class-switched IgG and IgA), serum IgG antibodies appeared almost simultaneously with or sometimes even before serum IgM and IgA antibodies after symptom onset (Figures 3 A–3C and S3 A–S3C). Interestingly, IgG antibodies appeared to be sustained in the time frame analyzed (up to 72 days), whereas IgM and IgA decreased after ∼42 days. Neutralization titers were similarly sustained over time (Figure 3D).
Figure 3.
SARS-CoV-2 antibody levels and neutralization potency predict clinical severity and survival
(A–C) Levels of anti-RBD IgG (A), IgM (B), and IgA (C) were plotted over days after symptom onset for COVID-19 cases where this date was known (n = 98 patients, n = 147 samples total). Healthy blood donors (n = 37) are included as a negative control within the gray region. The dotted lines indicate the cut-offs for anti-RBD IgG, IgM, and IgA seropositivity.
(D) Titers that achieve 50% neutralization (NT50) were plotted over days after symptom onset for patient samples described in (A)–(C).
(E–H) COVID-19 patient samples were selected for collection between 14 and 42 days after symptom onset (earliest time point for each patient, n = 85), and for each cohort of non-hospitalized, hospitalized, intubated, deceased, and immunosuppressed individuals, anti-RBD IgG (E), IgM (F), IgA (G), and neutralization (NT50) (H) were plotted. Healthy blood donors (n = 37) are also included as negative controls for comparison. Non-parametric multivariate ANOVA was performed for each (excluding healthy blood donors); statistical significance is indicated as follows: ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05.
(I and J) Log-log regression analyses were performed on neutralization versus anti-RBD IgG (I) and anti-RBD IgPC (J), which is a principal component generated from multivariate analysis of anti-RBD IgG, IgM, and IgA levels. For (I) and (J), the severity cohort is indicated as follows: healthy (white), non-hospitalized (green), hospitalized (yellow), intubated (red), deceased (gray), and immunosuppressed (blue). Pearson correlations were performed and R2 and p values are indicated.
(K and L) Anti-RBD IgG neutralization potency index (NT50/IgG) (K) and anti-RBD IgPC neutralization potency index (NT50/IgPC) (L) was calculated for all 111 COVID-19 patients (at earliest time point) and plotted by cohort. A non-parametric multivariate ANOVA was performed; unadjusted p values are indicated as follows: ∗∗p < 0.01, ∗p < 0.05.
(M) Survival analysis of COVID-19 patients classified as having a high (≥100) (n = 35) or low (< 100) (n = 76) neutralization potency index (NT50/IgG) was performed using Kaplan-Meier method and revealed an increased risk of death in individuals with low neutralization potency (p = 0.03).
See also Figure S3.
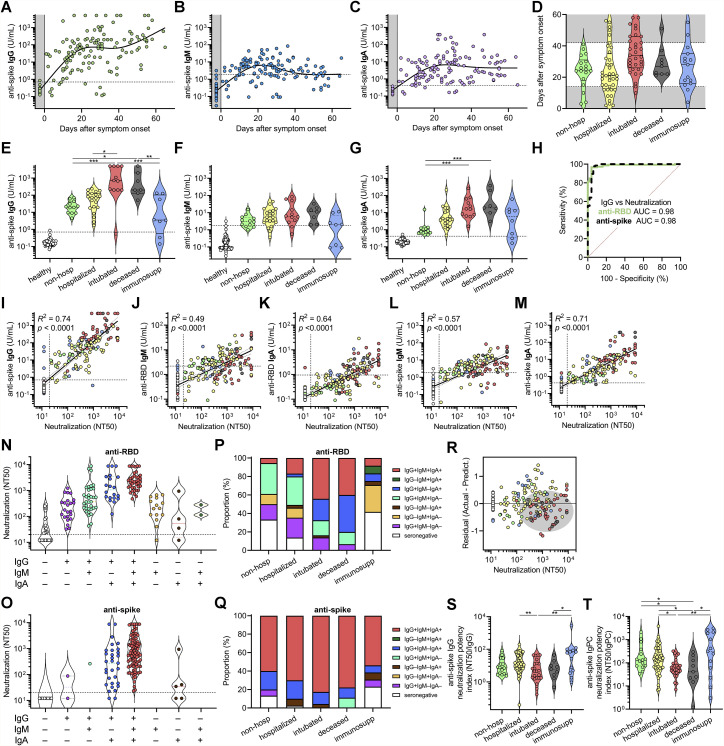
Figure S3.
Correlates between clinical outcomes and humoral immune responses against SARS-CoV-2, related to Figure 3
(A−C) Anti-spike IgG (A), IgM (B), and IgA (C) levels were plotted over days after symptom onset for confirmed COVID-19 cases for which date of symptom onset was known (n = 87 patients, n = 133 samples total). Healthy blood donors (n = 37) are included as a negative control within the gray region. The dotted lines indicate the cut-offs for anti-spike IgG, IgM, and IgA seropositivity.
(D) Standardization of cohorts by days after symptom onset to samples collected between 14 and 42 days was done to mitigate sampling biases and balance out representation from each cohort indicated.
(E−G) COVID-19 patient samples were selected for collection between 14 and 42 days after symptom onset (earliest time point for each patient), and for each cohort of non-hospitalized, hospitalized, intubated, deceased, and immunosuppressed individuals, anti-spike IgG (E), IgM (F), and IgA (G) was plotted (n = 54 total). An additional cohort of healthy blood donors (n = 78) is also included as negative controls for comparison. Non-parametric multivariate ANOVA was performed for each (excluding healthy blood donors); statistical significance is indicated as follows: ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05.
(H) ROC curve analysis of anti-RBD and anti-spike IgG for the prediction of neutralization was performed; AUC is indicated.
(I−M) Log-log regression analyses were performed on neutralization versus anti-spike IgG (I), anti-RBD IgM (J), anti-RBD IgA (K), anti-spike IgM (L), and anti-spike IgA (M). Severity cohort is indicated as follows: healthy (white), non-hospitalized (green), hospitalized (yellow), intubated (red), deceased (gray), and immunosuppressed (blue). Pearson correlations were performed and R2 and p values are indicated.
(N and O) Neutralization (NT50) of COVID-19 patient samples were grouped by serostatus as determined by anti-RBD antibodies (N); n = 165) and anti-spike antibodies (O); n = 148).
(P and Q) Proportion of COVID-19 patients of each indicated anti-RBD (P); n = 165) and anti-spike (Q); n = 148) serostatus group is presented for each severity cohort.
(R) A residual plot for neutralization titer versus anti-RBD IgG was generated from the log-log correlation. The gray ellipse indicates a cluster of samples from deceased (gray) patients.
(S and T) Anti-spike IgG neutralization potency index (NT50/IgG) (S) and anti-spike IgPC neutralization potency index (NT50/IgPC) (T) was calculated for all 100 patients (at earliest time point) and plotted by cohort. A non-parametric multivariate ANOVA was performed; unadjusted p values are indicated as follows: ∗∗p < 0.01, ∗p < 0.05.
To assess the humoral immune response among the pre-defined cohorts of varying disease severity, we focused on patients for whom samples were collected between 14 and 42 days after symptom onset. This time frame was chosen to prevent biases resulting from time of sampling post-infection (Figure S3D). We found that severely ill patients that were intubated or passed away due to COVID-19 had the highest levels of IgG and IgA antibodies targeting RBD and spike, but no significant differences were seen for IgM (Figures 3E–3G and S3E–S3G). These individuals also had the highest neutralization titers (Figure 3H). In contrast, individuals that were not hospitalized had the lowest IgG and IgA levels and neutralization titers. Unsurprisingly, immunosuppressed individuals—none of whom passed away—had significantly blunted IgG, IgA, and neutralizing responses. Upon analyzing IgG antibody seropositivity and neutralization titer, we found that both anti-RBD and anti-spike IgG were excellent predictors of neutralization (Figure S3H). However, although anti-spike IgG seropositivity was more sensitive at predicting neutralization (98% versus 78%), anti-RBD IgG was more specific (100% versus 92%). Indeed, of all the individual antibodies measured, anti-RBD IgG levels correlated the most with neutralization (R2 = 0.78) (Figure 3I). Anti-spike IgG also exhibited a strong, but slightly weaker, correlation with neutralization (Figure S3I), in line with prior studies demonstrating that RBD is the main target of neutralizing antibodies (He et al., 2005; Rogers et al., 2020). Anti-RBD IgM and IgA and anti-spike IgM and IgA also exhibited positive, but weaker, correlations with neutralization titer (Figures S3J–S3M). Triple positivity for anti-RBD and anti-spike IgG, IgM, and IgA antibodies was enriched in severely ill patients and was associated with the highest neutralization titers (Figures S3N–S3Q). However, anti-RBD IgM and IgA alone were capable of neutralization in serum samples where anti-RBD IgG could not be detected, indicating that anti-RBD IgM and IgA also contribute to neutralization (Figures S3N and S3O). Consequently, to better assess total anti-RBD antibody contribution to neutralization, we performed a multivariate analysis of anti-RBD IgG, IgM, and IgA levels in each patient and generated a principal component consisting of the sum of the weighted concentrations for each isotype, which we denoted anti-RBD “IgPC.” This total antibody composite variable exhibited an even tighter correlation with neutralization (R2 = 0.84) (Figure 3J), highlighting the importance of all antibody isotypes to neutralization.
Although anti-RBD IgG levels correlated with neutralization by regression analysis, there was variability that appeared to segregate by our pre-defined severity cohorts (Figure 3I). To better visualize this, we plotted residuals of each neutralization titer subtracted from its predicted titer based on the regression (Figure S3R). This revealed that samples from severely ill patients were biased toward lower-than-predicted neutralization titers, suggesting that they harbored higher levels of anti-RBD IgG antibodies that did not contribute to neutralization. Consequently, we calculated an anti-RBD IgG neutralization potency index (NT50/IgG) for each patient and found that intubated or subsequently deceased patients had a significantly lower index (Figure 3K). In addition, when using the composite variable IgPC to calculate neutralization potency index (NT50/IgPC), the differences in neutralization potency among intubated and deceased patients were even more pronounced (Figure 3L). However, when using anti-spike IgG and a similarly calculated anti-spike IgPC, differences in neutralization potency index were attenuated (Figures S3S and S3T), which may be due to the incorporation of a large fraction of non-neutralizing, non-RBD antibodies that are measured by anti-spike ELISAs.
To determine if anti-RBD IgG neutralization potency was predictive of outcomes, patients were classified as having neutralization potency indices that were “high” (≥100) or “low” (<100) and assessed for risk of death in the following days. Remarkably, there was a significant risk of death in the days following sample collection in the low index group (87% 30-day survival, n = 76), and there were no deaths in the high index group (100% 30-day survival, n = 35) (p = 0.03) (Figure 3M). This finding was true across our entire cohort of 111 COVID-19 patients for which we could calculate neutralization potency (including non-hospitalized and immunosuppressed individuals) and remained significant even when using a Cox proportional hazards model that accounted for age, sex, preferred language, and days between symptom onset and sample collection (p = 0.004). A similar analysis assessing anti-RBD IgPC neutralization potency across the full range of values in our cohort also yielded similar results (p = 0.005), with a risk ratio of 3.7 (i.e., for every 10-fold decrease in NT50/IgPC index, there is a 3.7-fold increased risk in mortality).
These results suggest that neutralization potency index may help risk stratify patients irrespective of where they are in their disease course. Altogether, severity of SARS-CoV-2 infection significantly correlates with higher anti-RBD antibody levels, but suboptimal neutralization potency is a significant predictor of mortality.
Correlation of neutralization potency to serum cytokine signatures
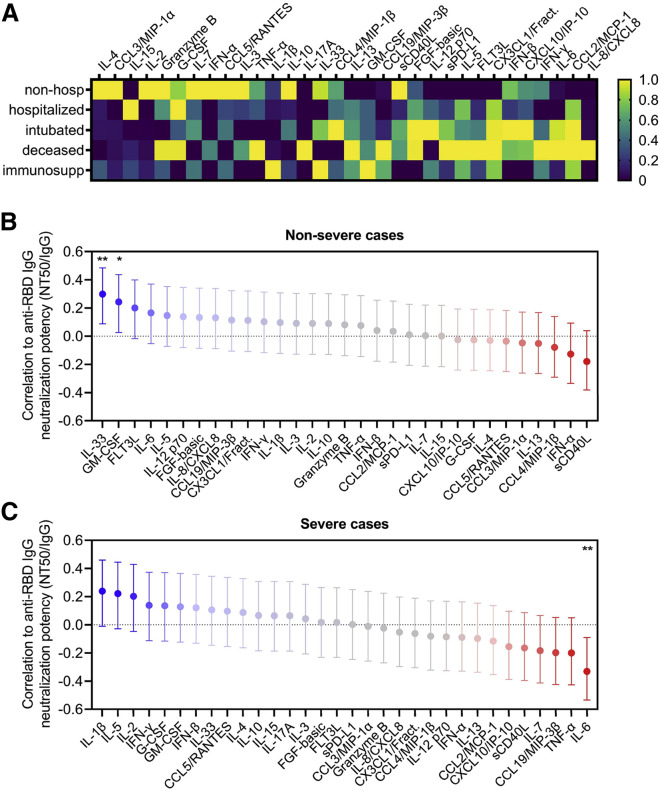
To explore the immunological implications of differences in neutralization potency, we quantified the level of 32 different cytokines in the serum of our COVID-19 patient cohort. We found that certain cytokines and chemokines were enriched in severe cases of COVID-19 resulting in intubation or death, including IL-6, IL-8, interferon (IFN)-γ, tumor necrosis factor alpha (TNF-α), CCL2, CXCL10, and sPD-L1. Interestingly, a separate set of factors were enriched in milder cases, namely, IFN-α, IL-4, IL-2, IL-15, IL-7, CCL3, CCL5, and granzyme B (an effector enzyme released by cytotoxic lymphocytes) (Figure 4 A).
Figure 4.
Neutralization potency correlates with distinct serum cytokine signatures in severe versus non-severe cases of COVID-19
(A) Serum cytokines were measured in COVID-19 patients that were non-hospitalized (n = 15), hospitalized (n = 38), intubated (n = 23), deceased (n = 9), and immunosuppressed (n = 13), and the average cytokine level for each cohort was calculated and presented as a heatmap. Color scales are normalized to each cytokine (column).
(B and C) A multivariate analysis was performed to calculate pairwise correlations between anti-RBD IgG neutralization potency index (NT50/IgG) and serum cytokine levels in non-severe (n = 61; upper panel) and severe cases of COVID-19 (n = 37; lower panel). Severe cases were defined as ones requiring intubation or resulting in death, and non-severe cases were all others (without accounting for immunosuppression status). Error bars indicate 95% confidence intervals and unadjusted p values are indicated as follows: ∗∗p < 0.01, ∗p = 0.05.
To determine the relationship between these cytokines and neutralization potency, we performed a multivariate analysis with pairwise correlations in COVID-19 patients categorized as either non-severe, consisting of non-hospitalized and hospitalized patients, or severe, consisting of those who were intubated or deceased (Figure 4B). These analyses revealed that in non-severe cases, GM-CSF and IL-33, which has been implicated in generating high-quality antibody responses (Sarkar et al., 2019), significantly correlated with increased neutralization potency. In contrast, sCD40L and IFN-α were inversely correlated with neutralization potency, although these were not statistically significant in our cohort.
Interestingly, severe cases of COVID-19 exhibited a different cytokine signature. Although positive trends between neutralization potency and IL-1β, IL-5, or IL-2 were observed, there was a significant correlation between IL-6 and decreased neutralization potency (Figure 4C). This was in stark contrast to a non-significant but weakly positive correlation between neutralization potency and IL-6 in non-severe cases. This suggests the possibility that although IL-6 is known to have a beneficial effect in the development of humoral immunity, it may be detrimental to neutralizing antibody responses if dysregulated. Interestingly, a recent report has suggested that soluble IL-6 receptor produced by dendritic cells is necessary for IL-6-induced class-switching (Yousif et al., 2020). Regardless, whether the observed cytokine signatures drive the production of neutralizing antibodies or if they are a consequence of antibody-driven cytokine dysregulation—as might be seen via antibody-dependent enhancement (ADE)—has yet to be determined.
The influence of pre-existing medical conditions and COVID-19 therapies on humoral immune responses to SARS-CoV-2
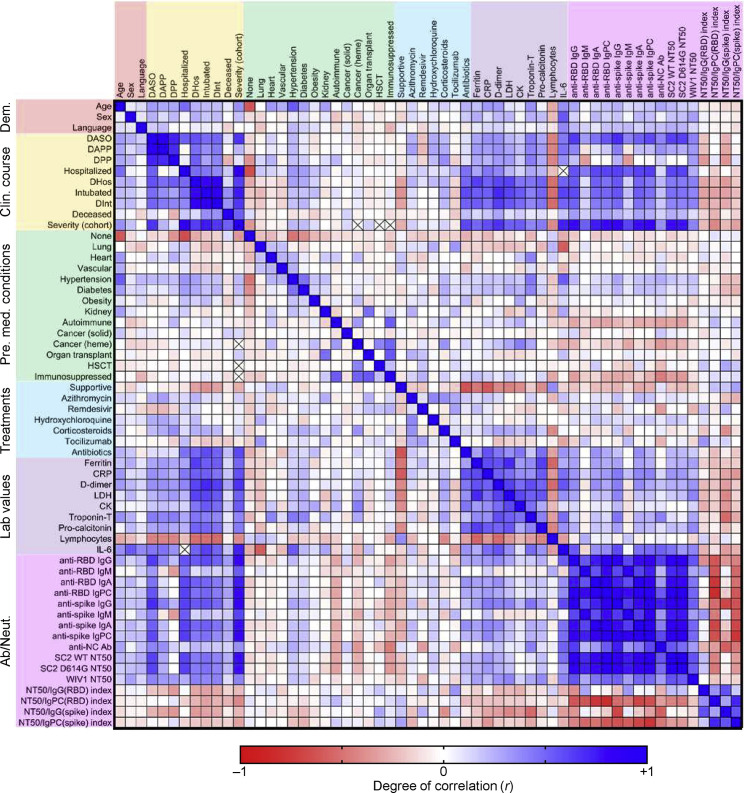
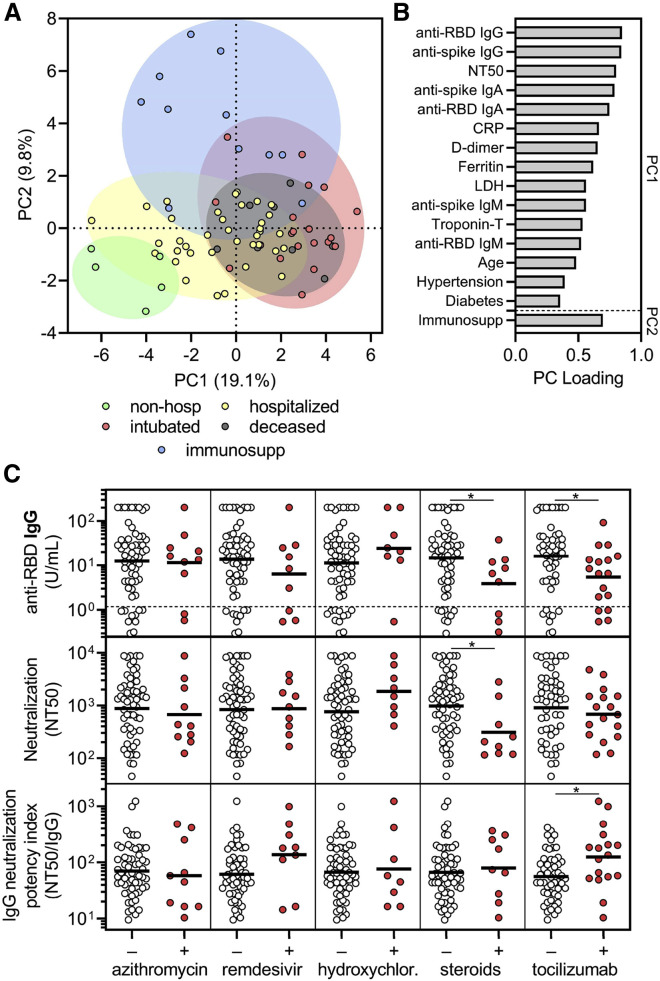
To explore the influence of pre-existing medical conditions and COVID-19 therapies on humoral immune responses to SARS-CoV-2, we performed multivariate analysis of all available demographic, clinical, laboratory, and experimental data (Figure S4 ). With the exception of immunosuppressed individuals, which had significantly decreased antibody and neutralizing responses, our cohort was not large enough to conclusively detect the effects of particular pre-existing medical conditions on the overall humoral immune response. However, a principle components analysis (PCA) that included demographic data, pre-existing medical conditions, laboratory data, treatments received, anti-RBD and anti-spike antibody levels, and neutralization titers, but not clinical outcomes, demonstrated clustering of patients by the severity cohorts (Figure 5 A). Principal components were mainly influenced by inflammatory markers, anti-RBD antibody levels, and neutralization titers, but a contribution from age and pre-existing medical conditions such as hypertension and diabetes was observed (Figure 5B).
Figure S4.
Multivariate analysis of demographic data, clinical course, pre-existing medical conditions, treatments, laboratory data, and humoral immune response in COVID-19 patients, related to Figure 5
A multi-variate analysis of all available data including age, sex, language, hospital course and events, pre-existing medical conditions, treatments received, clinical laboratory data, and antibody and neutralization data was performed, with Pearson coefficients (r) ranging from −1 (red) to 0 (white) to +1 (blue). The presence of an ‘x’ indicates that there were insufficient data to correlate the variables in question. The following abbreviations were used: DASO, days after symptom onset; DAPP, days after PCR positivity; DPP, days PCR positive (total number of days between first PCR positive results and last PCR positive result that was followed by one negative result); DHos, days hospitalized; HSCT, hematopoietic stem cell transplant; CRP, C-reactive protein; LDH, lactate dehydrogenase; CK, creatine kinase; anti-RBD, anti-receptor binding domain; IgPC, total antibody principal component (IgG, IgM, and IgA); anti-NC Ab, anti-nucleocapsid antibody (as measured by the commercially available Roche SARS-CoV-2 total antibody chemiluminescent assay); SC2, SARS-CoV-2.
Figure 5.
Corticosteroid and tocilizumab therapy decrease humoral immune responses to SARS-CoV-2
(A and B) Principal components analysis was performed using the following variables: age, sex language, pre-existing medical conditions, treatments received, clinical laboratory data (ferritin, CRP, D-dimer, LDH, troponin-T, and lymphocyte nadir), anti-RBD and anti-spike antibody levels, and neutralization titers. The severity cohort of each patient is indicated by color. Patients with missing data were excluded. Loading of principle components (PC) is shown in (B).
(C) Sub-analyses of anti-RBD IgG levels (upper panel), neutralization titer (middle panel), and neutralization potency index (NT50/IgG) (lower panel) were performed on COVID-19 patients that were in the hospital for at least 3 days to (n = 69) and were performed on the last collected specimen to show the effect of azithromycin (n = 10 out of 69 received), remdesivir (n = 9 out of 69), hydroxychloroquine (n = 8 out of 69), corticosteroids (n = 9 out of 69), and tocilizumab (n = 17 out of 69; treated as part of a trial with 2:1 randomization to placebo). Several patients received more than one treatment regimen and thus were included in more than one treatment category. A t test was performed for each comparison of patients who received (+) versus did not receive (–) the indicated treatment; ∗ indicates unadjusted p < 0.05.
See also Figure S4.
To assess the effect of different treatments on the humoral immune response, we performed a retrospective analysis in patients that were in the hospital for at least 3 days and received one or more or none of the COVID-19-directed therapies (n = 69). COVID-19-directed treatment regimens included azithromycin, remdesivir, hydroxychloroquine, corticosteroids, and tocilizumab. Individuals in the tocilizumab-treated cohort included 16 patients enrolled in a blinded clinical trial with 2:1 tocilizumab-to-placebo randomization. We compared anti-RBD IgG levels, neutralization titers, and neutralization potency indices in individuals that received or did not receive a given treatment, and found that azithromycin, remdesivir, and hydroxychloroquine—for which there was concern of attenuating antibody responses (de Miranda Santos and Costa, 2020)—did not significantly impact these parameters (Figure 5C). However, use of corticosteroids and tocilizumab significantly decreased anti-RBD IgG concentration, and in the case of corticosteroids, neutralization titer as well (Figure 5C). Corticosteroids are a general immunosuppressant known to decrease antibody production, whereas IL-6 signaling is important in several aspects of antibody responses (Kopf et al., 1998). Interestingly, tocilizumab-treated patients had a significant increase in the neutralization potency index stemming from the larger effect on anti-RBD IgG as compared to neutralization (Figure 5C). Paired with our previous data that showed a negative correlation between neutralization potency and IL-6 levels, this result raises new questions regarding the role of IL-6 signaling in the production of non-neutralizing versus neutralizing antibodies and how these might become decoupled. However, it is important to note that these analyses are retrospective and that many patients received more than one COVID-19-directed treatment, which could have resulted in confounders including selection bias (i.e., more ill patients were more likely to receive therapy) and interactions between different treatment regimens. Indeed, standard least-squares analysis that adjusted for covariates such as age, severity, and multiple treatments resulted in a loss of significance in this relatively small and heterogeneous cohort of patients. Regardless, our findings suggest that immunomodulatory therapies, some of which have shown clinical efficacy or are actively being studied, may influence humoral immune responses in SARS-CoV-2-infected patients, although prospective or randomized control trials will be necessary to more definitively assess this.
Cross-neutralization of SARS-CoV-2-infected patients to emerging coronaviruses
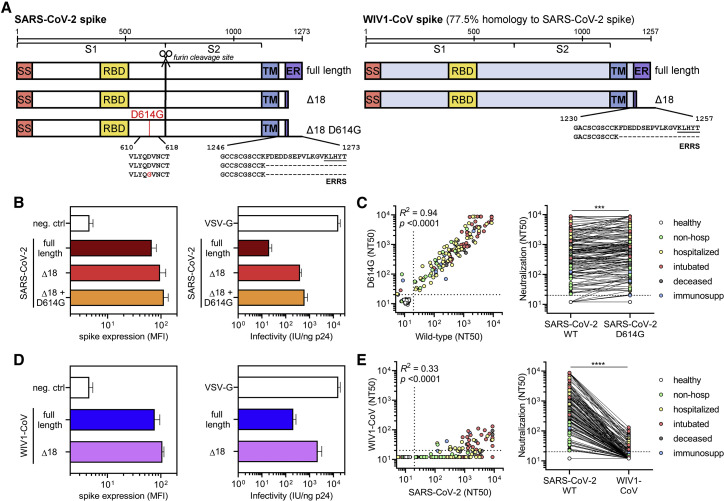
The recent emergence of a mutation in the SARS-CoV-2 spike protein (D614G) has raised concerns for the potential for convalescent patients to become reinfected. Recent studies have demonstrated that infection with live SARS-CoV-2 harboring the D614G spike variant yielded higher virus titers in respiratory cultures and increased transmissibility in hamster models (Plante et al., 2020; Zhou et al., 2020a; Hou et al., 2020). It has also been suggested that D614G spike may exist in a more open conformation (Yurkovetskiy et al., 2020) that does not impact antibody neutralization (Korber et al., 2020; Plante et al., 2020). To determine the impact of this variant on the neutralization potency of patients previously infected with SARS-CoV-2, we introduced the D614G mutation into the SARS-CoV-2 Δ18 spike (Figure 6 A). When characterizing this new construct, we found that both surface expression and infectivity were further increased relative to that of the wild-type SARS-CoV-2 Δ18 spike (Figures 6B, S5 A, S5C, S5D, and S5F), in line with previous studies (Korber et al., 2020). We tested this new pseudovirus in our cohort of 163 COVID-19 patient samples and found a very small, but statistically significant increase in neutralizing titers (Figure 6C), an effect of unknown clinical significance that was seen in prior studies (Korber et al., 2020; Plante et al., 2020). This indicates that individuals that have been infected with either D614 wild-type or G614 mutant SARS-CoV-2 will have cross-neutralization to the opposite strain, both of which are circulating in Boston, Massachusetts (Lemieux et al., 2020) and were likely represented in our study cohort.
Figure 6.
SARS-CoV-2-infected patient sera cross-neutralizes both wild-type and D614G mutant SARS-CoV-2 spike but not the highly homologous pre-emergent bat coronavirus WIV1-CoV
(A) A schematic of the SARS-CoV-2 and WIV1-CoV spike proteins, including full-length, truncated (Δ18), and mutant (D614G) forms is shown; ERRS denotes putative ER retention signal.
(B) Expression of full-length, Δ18, and Δ18 D614G SARS-CoV-2 spike constructs in 293T cells in comparison to empty vector (neg. ctrl) was measured by flow cytometry (left panel). Infectivity of lentivirus, which was defined as the infectious units divided by the quantity of p24 in lentiviral supernatant, was also measured and compared to VSV-G-pseudotyped lentivirus (right panel). Error bars indicate standard deviation.
(C) Cross-neutralization of serum samples from COVID-19 patients that were non-hospitalized (green, n = 16), hospitalized (yellow, n = 67), intubated (red, n = 43), deceased (gray, n = 15), or immunosuppressed (blue, n = 21) and healthy blood donors (n = 35) was measured for wild-type versus D614G mutant SARS-CoV-2 Δ18 spike pseudovirus. For the left panel, Pearson correlations were performed and R2 and p values are indicated. For the right panel, a paired non-parametric t test was performed; ∗∗∗p < 0.001.
(D) Similar to (B), expression and infectivity of full-length and Δ18 WIV1-CoV spike was measured. Error bars indicate standard deviation.
(E) Similar to (C), cross-neutralization of serum samples from COVID-19 patients was measured for wild-type SARS-CoV-2 versus WIV1-CoV pseudovirus. ∗∗∗∗p < 0.0001.
See also Figure S5.
Figure S5.
Characterization of CoV spike expression vectors, related to Figure 6
(A) Surface level expression of SARS-CoV-2 spike protein following transfection of 293T cells. Several constructs of spike were tested: codon-optimized full-length spike from SARS-CoV-2, a truncated version with 18 amino acids deleted from the cytoplasmic tail (Δ18), and a truncated version that also includes a D614G mutation. Expression was measured via flow cytometry by staining with B38 antibody at a concentration of 10 μg/mL followed by staining with an anti-human IgG antibody conjugated to AF647 at 2 μg/mL.
(B) Surface level expression of full-length and truncated (Δ18) WIV1-CoV spike proteins were also measured following transfection of 293T cells via flow cytometry. Expression was measured via flow cytometry by staining with CR3022 antibody at a concentration of 10 μg/mL followed by staining with an anti-human IgG antibody conjugated to AF647 at 2 μg/mL.
(C) Summary of spike expression data are shown with mean and standard deviation; MFI, median fluorescence intensity.
(D and E) Titers of lentivirus pseudotyped with the (D) SARS-CoV-2 or (E) WIV1-CoV spike proteins were measured by transducing ACE2-expressing 293T cells with 100 μL of lentivirus supernatant.
(F) Transduction with 10-fold serial dilutions and subsequent assessment of ZsGreen expression by flow cytometry was performed to calculate pseudovirus titer (U/mL) for each construct indicated. Summary data are presented with mean and standard deviation.
The emergence of SARS-CoV, MERS-CoV, and now SARS-CoV-2 within the last 2 decades has demonstrated the ability of zoonotic coronaviruses to cross the species barrier and pose pandemic threats. This has prompted microbiologists and epidemiologists to seek out and characterize zoonotic coronaviruses that have the potential to cross into humans. Recent studies in bats have identified a novel coronavirus, Wuhan Institute of Virology 1 coronavirus (WIV1-CoV), which, like SARS-CoV-2 and SARS-CoV, has a spike that uses ACE2 receptor for cell entry and bears high sequence homology to both SARS-CoV (92%) and SARS-CoV-2 (77%). We generated WIV1-CoV pseudovirus using an analogous spike truncation (Δ18) (Figure 6A), which resulted in high expression of WIV1-CoV spike on producer cells as well as infectivity and titer (Figures 6D, S5B, S5C, S5E, and S5F). These results suggest that this C-terminal truncation can serve as a general approach for modifying coronavirus spike proteins for efficient pseudovirus production. Interestingly, WIV1-CoV spike could be detected at the cell surface by the SARS-CoV and -CoV-2-specific monoclonal antibody CR3022 (Figure S5B). Using WIV1-CoV pseudovirus, we found that sera from SARS-CoV-2-infected individuals showed a lack of cross-neutralization except for relatively low-level neutralization in a few individuals with very high SARS-CoV-2 neutralization titers (Figure 6E). This indicates that humoral immunity raised against one coronavirus generally exhibits limited cross-neutralizing immunity to even highly related coronavirus strains.
Discussion
Traditionally, cellular immunity is responsible for clearing an established viral infection, whereas humoral immune responses play a more critical role in preventing future infection. Here, we found that severely ill COVID-19 patients had the highest levels of anti-RBD and anti-spike antibodies, which is in agreement with previous studies (Shrock et al., 2020; Secchi et al., 2020). To further characterize this antibody response, we measured neutralization titers and developed a neutralization potency index derived from our quantitative readouts (NT50/IgG) to assess the quality of anti-RBD IgG antibodies irrespective of the quantity produced. Remarkably, anti-RBD IgG neutralization potency was significantly diminished in severely ill patients, and survival analysis demonstrated that an index of ≥100 was predictive of 100% 30-day survival, whereas <100 was associated with 87% 30-day survival in our limited cohort of 111 COVID-19 patients. Further analyses using a total antibody composite variable (IgPC) revealed even more significant differences in neutralization potency among severe cases of COVID-19, highlighting the importance of accounting for all antibody isotypes when assessing the neutralization response. Thus, this anti-RBD antibody neutralization potency index may be a useful metric for physicians seeking to risk-stratify COVID-19 patients.
Despite the clear correlation between COVID-19 severity and development of humoral immunity, the cause-effect relationship between these two is unclear. One possibility is that severe disease caused by hyperinflammation and/or uncontrolled viral replication induces overproduction of antibodies that serve as a “biomarker” of severity. This is supported by our finding that the most severely affected patients, which had the highest anti-RBD and anti-spike antibody levels, also had the highest levels of inflammatory markers and pro-inflammatory cytokine signatures. In support of this possibility, a recent study suggests a pathogenic role of immune activation and exuberant antibody production from extrafollicular B cells in critically ill patients (Woodruff et al., 2020). Indeed, of all the COVID-19 treatment regimens being used and tested, dampening of the immune response with corticosteroids has proven to have one of the greatest benefits in improving outcomes and survival (Siemieniuk et al., 2020a), and we find that corticosteroids decrease both anti-RBD IgG levels and neutralization titers. However, another possibility is that high levels of antibodies with low neutralization potency worsen disease severity, possibly via ADE. This is supported by our finding that increased pro-inflammatory cytokines signatures, mainly IL-6, correlated to low neutralization potency in severely ill patients, and raises concerns over the use of convalescent plasma as a treatment strategy. One exception, however, may be in immunosuppressed individuals, which generally have sub-optimal antibody levels and neutralization titers. Further studies in animal models of COVID-19 testing passive transfer of low-potency index sera may help resolve this controversy.
A multitude of vaccines are presently being evaluated for SARS-CoV-2 prevention, including inactivated virus (Gao et al., 2020), spike antigen (Jackson et al., 2020; Keech et al., 2020), and RBD antigen (Mulligan et al., 2020; Dai et al., 2020). Each vaccine will likely result in humoral immunity with different ratios of neutralizing and non-neutralizing antibodies. Given our results, it will be important to assess the potency index of each candidate to determine those with maximal potential. Interestingly, one study showed that vaccination of mice with RBD generated potently neutralizing antibodies without ADE. This was postulated to be due to the lack of immunodominant non-neutralizing epitopes present on the remainder of the spike protein (Quinlan et al., 2020).
The diverse and atypical kinetics of antibody production—in particular, early rise of IgG and in some cases IgA—suggests the possibility of a contribution from class-switched (IgG+ or IgA+) memory B cells early in the humoral immune response rather than solely from the naive (IgM+) B cell pool, as has been recently postulated (Song et al., 2020). Regardless, our results support a role for IgM and IgA antibodies in contributing to SARS-CoV-2 neutralization, despite their transient nature in serum. IgG responses and neutralization, on the other hand, were sustained in the time frame analyzed (72 days), but several studies have emerged that question the longevity of these responses, which has yet to be determined. It is tempting to speculate that severely afflicted individuals may have more enduring immunity than mild cases. The differences in humoral response induction may stem from a combination of factors, including host permissibility to viral replication and a rapid response from innate immune effector cells and cytotoxic T cells, some of which have been postulated to arise from cross-reactive memory cells to other coronaviruses (Grifoni et al., 2020).
Although the mutation rate of coronaviruses is very low when compared to other viruses such as influenza or HIV, certain mutations in the spike protein of SARS-CoV-2 have emerged in the setting of the rapidly spreading pandemic. We found that one such mutation, D614G, which has now spread and become a dominant strain worldwide, does not affect the neutralizing ability of patient sera, reducing concerns for re-infection. Still, prior coronavirus epidemics (e.g., SARS-CoV, MERS-CoV, and now SARS-CoV-2) have occurred due to zoonotic coronaviruses crossing the species barrier, indicating an ongoing threat of future pandemics even in the face of effective vaccines to current viruses. One pre-emergent bat coronavirus, WIV1-CoV, is highly homologous to SARS-CoV and SARS-CoV-2 and can infect ACE2-expressing human cells (Menachery et al., 2016). Our data demonstrate that sera from SARS-CoV-2-infected patients exhibit very limited cross-neutralization of WIV1-CoV, except for rare individuals with relatively low-level neutralization of WIV1-CoV, suggesting that generation of broadly neutralizing antibodies is indeed possible, as has been previously described (Wec et al., 2020).
In summary, the development of potently neutralizing humoral immunity against SARS-CoV-2 appears to increase survival and may protect against re-infection with other circulating strains of SARS-CoV-2. However, it is generally unlikely to provide protection against subsequent coronavirus pandemics. As such, future efforts should focus on the development of broadly active therapies and prevention modalities that generate potently neutralizing antibodies with activity across different coronavirus strains.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CR3022-IgG1 | Obtained from the lab of Dr. Aaron Schmidt | IEDB Cat# CR3022, RRID:AB_2848080 |
| CR3022-IgM | Obtained from MassBio | N/A |
| CR3022-IgA1 | Obtained from MassBio | N/A |
| Anti-human IgG-HRP | Bethyl | Cat# A80-104P; RRID:AB_67064 |
| Anti-human IgA-HRP | Bethyl | Cat# A80-102P, RRID:AB_67047 |
| Anti-human IgM-HRP | Bethyl | Cat# A80-100P, RRID:AB_67082 |
| VRC07 | Produced in this study | N/A |
| B38 | Obtained from the lab of Dr. Aaron Schmidt | N/A |
| Anti-human IgG (H+L) AF647 | Thermo Fisher | Cat# A-21445; RRID:AB_2535862 |
| Mouse anti-HIV-1 IgG mAb Leidos Kit | Leidos | Lot #: PP292-01 |
| Rabbit anti-HIV-1 (MN) p24 Ledios Kit | Leidos | Lot #: SP1321C |
| Goat anti-rabbit IgG H+L HRP Conjugated | Bethyl | Cat# A120-201P; RRID:AB_67265 |
| Polyclonal human sera | This study | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 pseudotyped pHAGE-CMV-Luc2-IRES-ZsGreen-W lentivirus | This study | N/A |
| SARS-CoV-2 ΔC18 pseudotyped pHAGE-CMV-Luc2-IRES-ZsGreen-W lentivirus | This study | N/A |
| SARS-CoV-2 ΔC18 D614G pseudotyped pHAGE-CMV-Luc2-IRES-ZsGreen-W lentivirus | This study | N/A |
| WIV1-CoV pseudotyped pHAGE-CMV-Luc2-IRES-ZsGreen-W lentivirus | This study | N/A |
| WIV1-CoV ΔC18 pseudotyped pHAGE-CMV-Luc2-IRES-ZsGreen-W lentivirus | This study | N/A |
| VSV-G pseudotyped pHAGE-CMV-Luc2-IRES-ZsGreen-W lentivirus | This study | N/A |
| DH5α Zymo-Competent E. coli | Zymo | Cat# T3009 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 receptor binding domain protein | Obtained from the lab of Dr. Aaron Schmidt | N/A |
| SARS-CoV-2 spike protein | Obtained from the lab of Dr. Aaron Schmidt | N/A |
| Phosphate buffered saline (PBS) | Corning | Cat# 21-031-CV |
| Dulbecco’s modified eagle medium (DMEM) | Corning | Cat# 10-013-CV |
| Fetal bovine serum (FBS) | VWR | Cat# 89510-186 |
| Penicillin/streptomycin | Corning | Cat# 30-002-CI |
| TMB 2-component microwell Peroxidase substrate kit | Seracare | Cat# 50-76-00 |
| Carbonate-bicarbonate buffer | Sigma | Cat# C3041-50CAP |
| 10% bovine serum albumin (BSA) | Seracare | Cat# 5140-0006 |
| Tween-20 | Fisher | Cat# BP-337-100 |
| Sulfuric Acid | VWR | Cat# BDH3068 |
| Tris buffer (pH 8.0) | Amresco | Cat# 0497-5KG |
| Hydrochloric acid | BDH | Cat# BDH3030-2.5LPC |
| Sodium chloride | VWR | Cat# BDH9286-2.5KG |
| Polyethylenimine 25K MW, linear | Polysciences Inc | Cat# 23966 |
| Puromycin | Sigma | Cat# P8833-10MG |
| ATP | Sigma | Cat# A2383-5G |
| Magnesium chloride | BDH | Cat# BDH9244-500G |
| Magnesium sulfate | BDH | Cat# BDH9246-500G |
| Dithiothreitol (DTT) | VWR | Cat# 97061-338 |
| D-luciferin | Gold Bio | Cat# LUCK-2G |
| EDTA | Sigma | Cat# 03690-100ML |
| Triton X-100 | Fisher | Cat# BP151-500 |
| Polybrene | Sigma | Cat# TR-1003-G |
| Critical commercial assays | ||
| Human Magnetic Luminex Assays – Human XL Cytokine Discovery Premixed Kit | R&D | Cat#FCSTM18-32 |
| Experimental models: cell lines | ||
| HEK293T/17 Cells | ATCC | Cat# CRL-11268 |
| 293T/ACE2.MF | Obtained from the lab of Dr. Michael Farzan | N/A |
| Recombinant DNA | ||
| pHAGE-CMV-Luc2-IRES-ZsGreen-W (backbone) | This study | Addgene Cat# 164432 |
| pHDM-SARS-CoV-2 (spike) | This study | Addgene Cat# 164433 |
| pHDM-SARS-CoV-2 Δ18 (spike) | This study | Addgene Cat# 164434 |
| pTwist-SARS-CoV-2 (spike) | This study | Addgene Cat# 164435 |
| pTwist-SARS-CoV-2 Δ18 (spike) | This study | Addgene Cat# 164436 |
| pTwist-SARS-CoV-2 Δ18 D614G (spike) | This study | Addgene Cat# 164437 |
| pTwist-WIV1-CoV (spike) | This study | Addgene Cat# 164438 |
| pTwist-WIV1-CoV Δ18 (spike) | This study | Addgene Cat# 164439 |
| pHDM-VSV-G (spike) | This study | Addgene Cat# 164440 |
| pHDM-Hgpm2 (Gag-Pol) | This study | Addgene Cat# 164441 |
| pHDM-Tat1b (helper) | This study | Addgene Cat# 164442 |
| pRC-CMV-Rev1b (helper) | This study | Addgene Cat# 164443 |
| pUC19 (empty) | Norrander et al., 1983 | Joachim Messing: Addgene Cat# 50005; RRID:Addgene_50005 |
| Software and algorithms | ||
| GraphPad Prism 8.4.3 | Graphpad Software | https://www.graphpad.com:443/scientific-software/prism/; RRID:SCR_002798 |
| Geneious Prime 2020 | Geneious | https://www.geneious.com/; RRID:SCR_010519 |
| JMP Pro 15 | SAS Institute | https://www.jmp.com/en_us/software/data-analysis-software.html?utm_source=software&utm_medium=redirect; RRID:SCR_014242 |
| FlowJo 10 | FlowJo | https://www.flowjo.com; RRID:SCR_008520 |
| Fluent Control | Tecan | https://lifesciences.tecan.com/fluent-laboratory-automation-workstation?p=tab-4 |
| R v4.0.2 | Open source software | https://cran.r-project.org/bin/windows/base/; RRID:SCR_001905 |
| CellCapTure | Stratedigm | https://stratedigm.com/cellcapture/ |
| Other | ||
| Nunc-Immuno/MaxiSorp 96-well plates, clear, flat-bottom, nonsterile | Fisher | Cat#446612 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Alejandro Balazs (abalazs@mgh.harvard.edu).
Materials availability
Plasmids generated in this study will be available through Addgene. Recombinant proteins and antibodies are available from their respective sources.
Data and code availability
This study did not generate sequence data or code. Data generated in the current study (including ELISA, neutralization, and cytokine measurements) have not been deposited in a public repository but are available from the corresponding author upon request
Experimental model and subject details
Human subjects
Use of patient samples for the development and validation of SARS-CoV-2 diagnostic tests was approved by Partners Institutional Review Board (protocol 2020P000895). Serum samples from 113 patients diagnosed with COVID-19 (confirmed by at least one SARS-CoV-2 PCR-positive nasopharyngeal swab at Massachusetts General Hospital) were collected over course of several weeks, resulting in partially longitudinal, cross-sectional cohort consisting of 165 serum samples, with a prospective follow-up period of at least 3 months to assess clinical course and outcomes by manual chart review curated by at least two physicians. For each patient, the following information was obtained: age, sex, SARS-CoV-2 PCR results, date of symptom onset, hospitalization and discharge dates, intubation and extubation dates, and deceased date. Date of symptom onset was defined as the earliest date that at least one of the following COVID-19-related symptoms was reported as developing acutely and new from baseline: fever, chills, loss of smell or taste, body aches, rhinorrhea, nasal congestion, sore throat, cough, shortness of breath. If the date of symptom onset could not be determined with confidence, this information was excluded from the analysis. Patients were assessed for the presence of absence of the following pre-existing medical conditions: lung disease (e.g., asthma, COPD), heart disease (e.g., coronary artery disease, heart failure), vascular disease (e.g., peripheral vascular disease), hypertension, diabetes, obesity (BMI > 30), kidney disease, autoimmune disorder, solid organ cancer, chemotherapy for solid organ cancer, hematologic cancer, chemotherapy or immunotherapy for hematologic cancer, history of organ transplant, history of hematopoietic stem cell transplant, and pre-existing use of corticosteroids or other immunosuppressive medications. Based on these information, the cohort was divided into the following groups based on severity of disease and underlying health status: (i) non-hospitalized, consisting of individuals that were never admitted to the hospital and were sent home to quarantine; (ii) hospitalized, which included individuals that were hospitalized for at least one night but were never intubated and were eventually discharged; (iii) intubated, comprising hospitalized individuals that were intubated for at least one day but survived and were eventually discharged; (iv) deceased, for which we had obtained a specimen before they eventually passed away in the hospital; and (v) immunosuppressed, which consisted of people that were on immunosuppressive medication (including high-dose corticosteroid) and/or were afflicted by a clinically significant hematologic malignancy before being diagnosed with COVID-19. Laboratory data throughout admission were analyzed, and the maximum documented serum levels of ferritin, C-reactive protein, D-dimer, lactate dehydrogenase, troponin-T, and IL-6 were recorded for each patient, as well as the lowest absolute lymphocyte count documented (lymphocyte count nadir). In addition, use of the following treatments were documented: corticosteroids, hydroxychloroquine, azithromycin, atorvastatin, remdesivir, lopinavir/ritonavir, tocilizumab (part of treatment versus placebo trial, currently blinded), and anakinra. All information obtained from medical records was verified by at least two physicians. Pre-pandemic serum samples (n = 1,257) were obtained from the clinical laboratories at Massachusetts General Hospital (MGH). These samples were comprised of an unbiased cohort of individuals being tested for measles, mumps, and rubella titers (n = 1124), as well as a selected subset of 133 individuals with positive serology results for cytomegalovirus (n = 10), varicella-zoster virus (n = 25), hepatitis B virus (n = 25), hepatitis C virus (n = 24), HIV (n = 37), syphilis (n = 16), Toxoplasma (n = 1), and rheumatoid factor (n = 1). Specimens from anonymous pre-screened healthy blood donors (n = 78) were collected from the MGH Blood Donor Center.
Cell lines
HEK293T cells (ATCC) were cultured in DMEM (Corning) containing 10% fetal bovine serum (VWR), and penicillin/streptomycin (Corning) at 37°C and 5% CO2. 293T-ACE2 cells were a gift from Michael Farzan (Scripps Florida) and Nir Hacohen (Broad Institute) and were cultured under the same conditions as HEK293T cells. Confirmation of ACE2 expression in 293T-ACE2 cells was done via flow cytometry.
Method details
SARS-CoV-2 receptor binding domain and spike IgG, IgM, and IgA ELISA
To quantitatively detect IgG, IgM, and IgA antibodies to SARS-CoV-2 receptor binding domain (RBD) and spike protein, we developed an indirect ELISA using an anti-SARS-CoV and -CoV-2 monoclonal antibody (CR3022) with IgG1, IgM, and IgA1 isotypes (kindly provided by Galit Alter, Stephanie Fischinger, Caroline Atyeo, and Matt Slein in collaboration with Jeffrey Bernard at MassBiologics). SARS-CoV-2 RBD was designed based on GenBank sequence MN975262.1 and cloned into a pVRC vector containing HRV 3C-cleavable C-terminal 8xHis and SBP tags. Sequence confirmation was performed by Sanger sequencing from Genewiz. The SARS-CoV-2 spike plasmid was obtained from Dr. Jason McLellan at the University of Texas, Austin. It contained a C-terminal Foldon trimerization tag, as well as HRV 3C-cleavable C-terminal 6xHis and 2xStrep II tags. Transient transfections in Expi293F cells (ThermoFisher) were performed using Expifectamine reagents (ThermoFisher) according to the manufacturer’s protocol. At 5 to 7 days post-transfection, supernatants were subjected to centrifugation. Proteins were then purified using immobilized metal affinity chromatography (IMAC) with Cobalt-TALON resin (Takara). Eluent was concentrated and further purified using a Superdex 200 Increase 10/300 GL size exclusion column (GE Healthcare). 96-well Nunca MaxiSorp ELISA plates (ThermoFisher) were coated with purified RBD diluted in carbonate-bicarbonate buffer (Sigma) to a concentration of 1 μg/mL for IgG and IgA plates and 2 μg/mL for IgM plates for 1 h at room temperature. Plates for spike ELISAs were coated with purified spike protein diluted in carbonate-bicarbonate to 2 μg/mL for all antibody isotypes. Plates were washed with a wash buffer consisting of 50 mM Tris (pH 8.0) (Sigma), 140 mM NaCl (Sigma), and 0.05% Tween-20 (Sigma). Plates were incubated with a blocking buffer consisting of 1% BSA (Seracare), 50 mM Tris (pH 8.0), and 140 mM NaCl for 30 min at room temperature, and then washed. Serum samples were diluted 1:100 with a dilution buffer consisting of 1% BSA, 50 mM Tris (pH 8.0), 140 mM NaCl, and 0.05% Tween-20. A seven-point standard curve was created using each of the standards (i.e., CR3022-IgG1, CR3022-IgM, CR3022-IgA1) starting at 2 μg/mL by performing 1:3 serial dilutions with dilution buffer. Samples and standards were added to corresponding wells and incubated for 1 h at 37°C, followed by washing. Human antibody isotypes were detected with specific antibodies (Bethyl) diluted as indicated: anti-human IgG-HRP (1:25,000), anti-human IgM-HRP (1:20,000), and anti-human IgA-HRP (1:5,000). These were added to each plate and incubated for 30 min at room temperature. After washing, TMB substrate (Inova) was added to each well and incubated for ∼7 min (for IgG), ∼13 min (for IgM), and ∼10 min (for IgA), before stopping with 1 M H2SO4. Buffer compositions, reagent concentrations and incubation times and temperatures were optimized in separate experiments for each analyte to maximize signal-to-noise ratio. Optical density (O.D.) was measured at 450 nm with subtraction of the O.D. at 570 nm as a reference wavelength on a SpectraMax ABS microplate reader. Anti-RBD and anti-spike antibody levels were calculated by interpolating onto the standard curve and correcting for sample dilution; one unit per mL (U/mL) was defined as the equivalent reactivity seen by 1 μg/mL of CR3022. For anti-RBD ELISAs, cut-offs of 1.18 U/mL for anti-RBD IgG achieved a sensitivity of 73%, 2.14 U/mL for anti-RBD IgM achieved 66%, and 0.95 U/mL for anti-RBD IgA achieved 48%, with > 99% specificity for all three anti-RBD antibodies. For anti-spike ELISAs, cut-offs of 0.70 U/mL for anti-spike IgG achieved a sensitivity of 95%, 1.82 U/mL for anti-spike IgM achieved 80%, and 0.42 U/mL for anti-spike IgA achieved 97%, with > 98% specificity for all three anti-spike antibodies.
SARS-CoV-2 pseudovirus neutralization assay
To compare the neutralizing activity of patient sera against coronaviruses, we produced lentiviral particles, pseudotyped with different spike proteins, by transient transfection of 293T cells and titered the viral supernatants by flow cytometry on 293T-ACE2 cells (Moore et al., 2004). Virus production was also quantified by p24 ELISA on viral supernatants using the HIV-1 p24CA antigen capture assay (Leidos Biomedical Research, Inc). To increase throughput and consistency, assays and readouts were performed on a Fluent Automated Workstation (Tecan) using 384-well plates (Grenier). Following an initial 12-fold dilution, the liquid handler performed serial three-fold dilutions (ranging from 1:12 to 1:8,748) of each patient serum and/or purified antibody in 20 μL followed by addition of 20 μL of pseudovirus containing 125 infectious units and incubation for 1 h at room temperature. Finally, 10,000 293T-ACE2 (Moore et al., 2004) cells in 20 μL cell media containing 15 μg/mL polybrene were added to each well and incubated at 37°C for 60-72 h. Following transduction, cells were lysed using a modified form of a previously described assay buffer (Siebring-van Olst et al., 2013) containing a final concentration of 20 mM Tris-HCl, 100 μM EDTA, 1.07 mM MgCl2, 2.67-26.7 mM MgSO4, 17 mM dithiothreitol (DTT), 250 μM ATP, and 125-250 μM D-luciferin, 1% Triton-X and shaken for five minutes prior to quantitation of luciferase expression within 1h of buffer addition using a Spectramax L luminometer (Molecular Devices). Percent neutralization was determined by subtracting background luminescence measured in cell control wells (cells only) from sample wells and dividing by virus control wells (virus and cells only). Of note, repeated sera neutralization measurements in independent assays using 500, 250, and 125 infectious units of pseudovirus per well generated similar results (data not shown), indicating that the NT50 is not significantly influenced by pseudovirus titers. Data was analyzed using Graphpad Prism and NT50 values were calculated by taking the inverse of the 50% inhibitory concentration value for all samples with a neutralization value of 80% or higher at the highest concentration of serum or antibody. As a separate note for investigators using pseudovirus neutralization assays, we excluded pre-pandemic individuals taking antiretroviral therapy for human immunodeficiency virus infection or pre-exposure prophylaxis (n = 37 in the original cohort of 1,257) after finding that potent inhibition of pseudovirus infection occurred in a majority of these individuals (Figure S2K). We believe this was due to antiretroviral compounds in the patients’ sera inhibiting transduction with our lentivirus-based vector system, thus generating an artifact. Of note, undocumented antiretroviral use may explain a proportion of the false positives observed in the remaining specimens (n = 12 out of 1,220).
Flow cytometry
To quantify the pseudotyped lentiviral supernatants in terms of infectious units, we plated 400,000 of either 293T or 293T-ACE2 cells in 1 mL in a 12-well plate format (Corning). 24 h later, ten-fold serial dilutions of lentiviral transfection supernatant were made in 100 μL, which was then used to replace 100 μL of media on the plated cells. Cells were then incubated with lentivirus supernatant for 48 h at 37°C and then harvested with Trypsin-EDTA (Corning), resuspended in PBS supplemented with 2% FBS (PBS+), and measured on a Stratedigm S1300Exi Flow Cytometer. Samples were gated for ZsGreen expression. To compare the relative surface expression of pseudovirus spike protein, we plated 400,000 293T cells per well in 1 mL in a 12-well plate. 24 h later, we transfected each well with a lentiviral helper vector coding for different spike proteins. The cells were incubated for 48 h at 37°C and harvested into PBS containing 1% fetal bovine serum (Sigma) (called PBS+). Cells transfected with each vector were divided into 3 aliquots, stained with either PBS+, CR3022 SARS-CoV antibody (10 μg/mL in PBS+), or B38 SARS-CoV-2 antibody (10 μg/mL in PBS+) for 30 minutes at room temperature. Cells were then washed with 1 mL PBS+, spun at 1,150 x g, and stained with anti-human IgG-AF647 polyclonal antibody (Invitrogen) at 2 μg/mL in PBS+ for 30 minutes at room temperature. Cells were washed with 1 mL of PBS+, spun at 1,150 x g, resuspended in 150 μL of PBS+ and measured on a Stratedigm S1300Exi Flow Cytometer.
Confocal microscopy
60-72 hours after neutralization assay setup, each well in a serum dilution series within a 384-well plate was imaged using a FITC filter to detect cellular ZsGreen expression. Images were acquired using a 20X air objective on a Zeiss LSM510 instrument. Acquired images were analyzed using ImageJ to produce overlays.
Multiplexed serum cytokine measurements
Serum cytokines were measured using the Luminex technology-based Human XL Cytokine Discovery Kit (R&D) following the manufacturer’s instructions. The cytokines measured were: sPD-L1, CCL19/MIP-3β, CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CCL5/RANTES, sCD40L, CX3CL1/Fractalkine, CXCL10/IP-10, FGF-basic, FLT3L, G-CSF, GM-CSF, Granzyme B, IFN-α, IFN-β, IFN-γ, IL-10, IL-12 p70, IL-13, IL-15, IL-17A, IL-1β, IL-2, IL-3, IL-33, IL-4, IL-5, IL-6, IL-7, IL-8/CXCL8, and TNF-α. Samples were read in a flow cytometry-based FLEXMAP 3D System (Bio-Rad).
Quantification and statistical analysis
Statistical and data analyses were performed using GraphPad Prism 8.4.3, JMP Pro 15.0.0 (SAS Institute), and R v4.0.2. Flow cytometry data was analyzed using FlowJo 10.6.2. Non-parametric multivariate ANOVAs were performed on the indicated figures where several cohorts were present; all p values were adjusted for multiple comparisons except when indicated. Statistical significance was defined as p < 0.05. Error bars throughout all figures represent one standard deviation unless otherwise specified. For survival, Kaplan-Meier method was used for survival analysis, and Cox proportional hazards models performed by both JMP Pro and R confirmed these findings after accounting for additional variables. When using R, the Cox proportional hazards model was performed using the coxph function from the survival package v3.2-7 (http://cran.rproject.org/package=survival) in R v4.0.2. The total anti-RBD antibody principal component (IgPC) described was generated using JMP Pro 15.0.0, and contained loadings of 0.90 for anti-RBD IgG, 0.80 for anti-RBD IgM, and 0.90 for anti-RBD IgA.
Acknowledgments
We wish to thank Nir Hacohen, PhD, and Jesse D. Bloom, PhD, for spike-expression plasmids, and Michael Farzan, PhD, for providing ACE2-expressing 293T cells. We also give thanks to Julio Silva, Daniel Claiborne, PhD, and Vivek Naranbhai, MD, PhD, for input on experiments and statistical analyses. This independent research was supported by the Gilead Sciences Research Scholars Program in HIV. K.L.C. is supported by Ruth L. Kirschstein National Research Service Award (NRSA) Individual Postdoctoral Fellowship (1F32AI143480). T.M.C. and B.M.H. were supported by the National Institute of General Medical Sciences (T32GM007753). J.F. was supported by the National Institute of General Medical Sciences (T32AI007245). A.G.S. was supported by NIH (R01 AI146779) and a Massachusetts Consortium on Pathogenesis Readiness (MassCPR) grant. J.A.B. has received research support from Zeus Scientific, bioMerieux, Immunetics, Alere, DiaSorin, the Bay Area Lyme Foundation (BALF), and the National Institute of Allergy and Infectious Diseases (NIAID) (1R21AI119457-01) for unrelated projects. A.J.I. is supported by the Lambertus Family Foundation. A.B.B. is supported by the National Institute on Drug Abuse (NIDA) Avenir New Innovator Award (DP2DA040254) and the MGH Transformative Scholars Program, as well as funding from the Charles H. Hood Foundation.
Author contributions
W.F.G.-B., E.C.L., M.G.A., D.Y., T.E.M., and A.B.B. designed the experiments. W.F.G.-B., E.C.L., M.G.A., D.Y., and K.L.C. carried out experiments and analyzed data. J.F., B.M.H., T.M.C., and A.G.S. provided key reagents and useful discussions and insights. D.L. offered suggestions for experiments and useful discussions. A.D.N. contributed to statistical analyses. A.J.I., J.L., J.A.B., and A.D. provided clinical samples. A.J.I., M.M., G.A., and R.C. contributed key inputs into experimental design. W.F.G.-B., E.C.L., and A.B.B. wrote the paper with contributions from all authors.
Declaration of interests
J.A.B. has served as a paid consultant to T2 Biosystems, DiaSorin, and Roche Diagnostics.
Published: December 15, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.12.015.
Supplemental information
Clinical data are shown for each pre-defined severity cohort.
References
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E. Remdesivir for the Treatment of Covid-19 - Preliminary Report. Reply. N. Engl. J. Med. 2020;383:994. doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., Skipper C.P., Nascene A.A., Nicol M.R., Abassi M. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N. Engl. J. Med. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Kafai N.M., Fox J.M., Smith B.K., Shrihari S., McCune B.T., Harvey I.B., Keeler S.P. Replication-Competent Vesicular Stomatitis Virus Vaccine Vector Protects against SARS-CoV-2-Mediated Pathogenesis in Mice. Cell Host Microbe. 2020;28:465–474.e4. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Cheng V.C.C., Woo P.C.Y., Lau S.K.P., Poon L.L.M., Guan Y., Seto W.H., Yuen K.Y., Peiris J.S.M. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol. 2005;12:1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hu J., Zhang Z., Jiang S., Han S., Yan D., Zhuang R., Hu B., Zhang Z. Efficacy of Hydroxychloroquine in Patients with COVID-19: Results of a Randomized Clinical Trial. medRxiv. 2020 doi: 10.1101/2020.03.22.20040758. [DOI] [Google Scholar]
- Corver J., Broer R., van Kasteren P., Spaan W. Mutagenesis of the transmembrane domain of the SARS coronavirus spike glycoprotein: refinement of the requirements for SARS coronavirus cell entry. Virol. J. 2009;6:230. doi: 10.1186/1743-422X-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:E513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M. A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo P., Mur I., Pomar V., Corominas H., Casademont J., de Benito N. The four horsemen of a viral Apocalypse: The pathogenesis of SARS-CoV-2 infection (COVID-19) EBioMedicine. 2020;58:102887. doi: 10.1016/j.ebiom.2020.102887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Zhang X., Liu Z., Yu H., Zheng B.-J., Zhou Y., Jiang S. Development of a safe and convenient neutralization assay for rapid screening of influenza HA-specific neutralizing monoclonal antibodies. Biochem. Biophys. Res. Commun. 2010;397:580–585. doi: 10.1016/j.bbrc.2010.05.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorse G.J., Patel G.B., Vitale J.N., O’Connor T.Z. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17:1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaraldi G., Meschiari M., Cozzi-Lepri A., Milic J., Tonelli R., Menozzi M., Franceschini E., Cuomo G., Orlando G., Borghi V. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoun C., Megas F., Piercy J. Immunogenicity and protective efficacy of influenza vaccination. Virus Res. 2004;103:133–138. doi: 10.1016/j.virusres.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhu Q., Liu S., Zhou Y., Yang B., Li J., Jiang S. Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology. 2005;334:74–82. doi: 10.1016/j.virol.2005.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., RECOVERY Collaborative Group Dexamethasone in Hospitalized Patients with Covid-19 - Preliminary Report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. Published online July 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;(November):eabe8499. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F. Dynamics and Significance of the Antibody Response to SARS-CoV-2 Infection. medRxiv. 2020 doi: 10.1101/2020.07.18.20155374. [DOI] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Herren S., Wiles M.V., Pepys M.B., Kosco-Vilbois M.H. Interleukin 6 influences germinal center development and antibody production via a contribution of C3 complement component. J. Exp. Med. 1998;188:1895–1906. doi: 10.1084/jem.188.10.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux J., Siddle K.J., Shaw B.M., Loreth C., Schaffner S., Gladden-Young A., Adams G., Fink T., Tomkins-Tinch C.H., Krasilnikova L.A. Phylogenetic Analysis of SARS-CoV-2 in the Boston Area Highlights the Role of Recurrent Importation and Superspreading Events. medRxiv. 2020 doi: 10.1101/2020.08.23.20178236. [DOI] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontok E., Corse E., Machamer C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Li J., Machamer C.E. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 2007;81:2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Wu P., Lu W., Liu K., Ma K., Huang L., Cai J., Zhang H., Qin Y., Sun H. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: A retrospective study of 168 severe patients. PLoS Pathog. 2020;16:e1008520. doi: 10.1371/journal.ppat.1008520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Structural and Functional Analysis of a Potent Sarbecovirus Neutralizing Antibody. bioRxiv. 2020 doi: 10.1101/2020.04.07.023903. [DOI] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. Published online October 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan B.D., Mou H., Zhang L., Guo Y., He W., Ojha A., Parcells M.S., Luo G., Li W., Zhong G. The SARS-CoV-2 Receptor-Binding Domain Elicits a Potent Neutralizing Response without Antibody-Dependent Enhancement. bioRxiv. 2020 doi: 10.1101/2020.04.10.036418. [DOI] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy V., Fischinger S., Atyeo C., Slein M., Loos C., Balazs A., Luedemann C., Astudillo M.G., Yang D., Wesemann D.R. SARS-CoV-2-specific ELISA development. J. Immunol. Methods. 2020;484–485:112832. doi: 10.1016/j.jim.2020.112832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Miranda Santos I.K.F., Costa C.H.N. Impact of Hydroxychloroquine on Antibody Responses to the SARS-CoV-2 Coronavirus. Front. Immunol. 2020;11:1739. doi: 10.3389/fimmu.2020.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Piepenbrink M.S., Basu M., Thakar J., Keefer M.C., Hessell A.J., Haigwood N.L., Kobie J.J. IL-33 enhances the kinetics and quality of the antibody response to a DNA and protein-based HIV-1 Env vaccine. Vaccine. 2019;37:2322–2330. doi: 10.1016/j.vaccine.2019.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchi M., Bazzigaluppi E., Brigatti C., Marzinotto I., Tresoldi C., Rovere-Querini P., Poli A., Castagna A., Scarlatti G., Zangrillo A. COVID-19 survival associates with the immunoglobulin response to the SARS-CoV-2 spike receptor binding domain. J. Clin. Invest. 2020;130:6366–6378. doi: 10.1172/JCI142804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., MGH COVID-19 Collection & Processing Team Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebring-van Olst E., Vermeulen C., de Menezes R.X., Howell M., Smit E.F., van Beusechem V.W. Affordable luciferase reporter assay for cell-based high-throughput screening. J. Biomol. Screen. 2013;18:453–461. doi: 10.1177/1087057112465184. [DOI] [PubMed] [Google Scholar]
- Siemieniuk R.A., Bartoszko J.J., Ge L., Zeraatkar D., Izcovich A., Kum E., Pardo-Hernandez H., Rochwerg B., Lamontagne F., Han M.A. Drug treatments for covid-19: living systematic review and network meta-analysis. BMJ. 2020;370:m2980. doi: 10.1136/bmj.m2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., He W.-T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S. Cross-Reactive Serum and Memory B Cell Responses to Spike Protein in SARS-CoV-2 and Endemic Coronavirus Infection. bioRxiv. 2020 doi: 10.1101/2020.09.22.308965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Focà E., Bezzi M., Baronio B., Giacomelli M., Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Cao Z., Han M., Wang Z., Chen J., Sun W., Wu Y., Xiao W., Liu S., Chen E. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369:m1849. doi: 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersalvi G., Vicenzi M., Calabretta D., Biasco L., Pedrazzini G., Winterton D. Elevated Troponin in Patients With Coronavirus Disease 2019: Possible Mechanisms. J. Card. Fail. 2020;26:470–475. doi: 10.1016/j.cardfail.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Huang C., Shirato K., Makino S., Taguchi F. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J. Gen. Virol. 2016;97:1853–1864. doi: 10.1099/jgv.0.000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah W., Abdullah H.M., Roomi S., Sattar Y., Almas T., Narayana Gowda S., Saeed R., Mukhtar M., Ahmad A., Oliver T. Safety and Efficacy of Hydroxychloroquine in COVID-19: A Systematic Review and Meta-Analysis. J. Clin. Med. Res. 2020;12:483–491. doi: 10.14740/jocmr4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020;50:332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., Bonten M.M.J., Damen J.A.A., Debray T.P.A., De Vos M. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Huang B., Ruhan A., Li W., Wang W., Deng Y., Tan W. Development and effectiveness of Pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosaf. Health. 2020 doi: 10.1016/j.bsheal.2020.08.004. Published online August 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousif A.S., Ronsard L., Shah P., Omatsu T., Sangesland M., Bracamonte Moreno T., Lam E.C., Vrbanac V.D., Balazs A.B., Reinecker H.-C., Lingwood D. The persistence of interleukin-6 is regulated by a blood buffer system derived from dendritic cells. Immunity. 2020 doi: 10.1016/j.immuni.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Tran T.N.T., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Portmann J., Halwe N.J. SARS-CoV-2 Spike D614G Variant Confers Enhanced Replication and Transmissibility. bioRxiv. 2020 doi: 10.1101/2020.10.27.357558. [DOI] [Google Scholar]
- Zhou Y., Yang Z., Guo Y., Geng S., Gao S., Ye S., Hu Y., Wang Y. A New Predictor of Disease Severity in Patients with COVID-19 in Wuhan, China. medRxiv. 2020 doi: 10.21203/rs.3.rs-29566/v1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical data are shown for each pre-defined severity cohort.
Data Availability Statement
This study did not generate sequence data or code. Data generated in the current study (including ELISA, neutralization, and cytokine measurements) have not been deposited in a public repository but are available from the corresponding author upon request