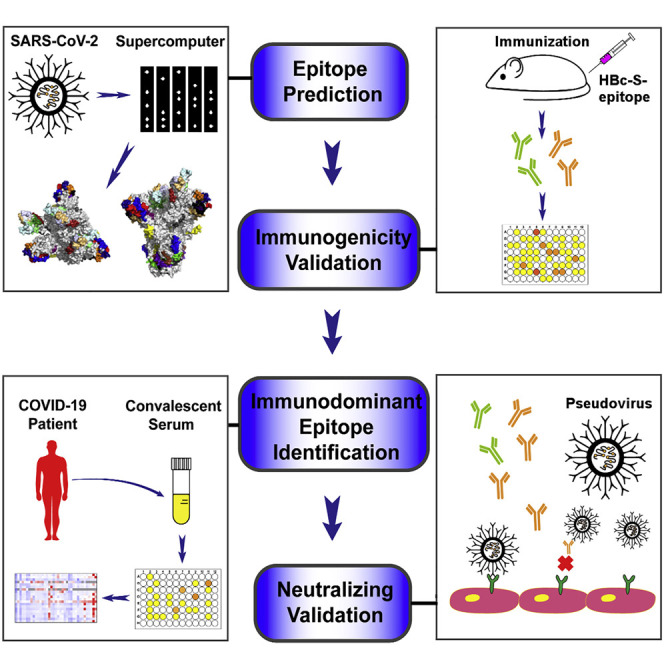
Abstract
Although vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are under development, the antigen epitopes on the virus and their immunogenicity are poorly understood. Here, we simulate the 3D structures and predict the B cell epitopes on the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins of SARS-CoV-2 using structure-based approaches and validate epitope immunogenicity by immunizing mice. Almost all 33 predicted epitopes effectively induce antibody production, six of these are immunodominant epitopes in individuals, and 23 are conserved within SARS-CoV-2, SARS-CoV, and bat coronavirus RaTG13. We find that the immunodominant epitopes of individuals with domestic (China) SARS-CoV-2 are different from those of individuals with imported (Europe) SARS-CoV-2, which may be caused by mutations on the S (G614D) and N proteins. Importantly, we find several epitopes on the S protein that elicit neutralizing antibodies against D614 and G614 SARS-CoV-2, which can contribute to vaccine design against coronaviruses.
Keywords: COVID-19, SARS-CoV-2, vaccine, immunodominant epitope, neutralizing epitope
Graphical Abstract
Highlights
-
•
B cell epitopes of SARS-CoV-2 are obtained using structure-based approaches
-
•
The predicted epitopes effectively induce robust antibody responses
-
•
D614 and G614 SARS-CoV-2 display different immunodominant epitopes
-
•
Epitopes on S protein elicit D614 and/or G614 SARS-CoV-2-neutralizing antibodies
Lu et al. predict and validate B cell epitopes on the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins of SARS-CoV-2 using structure-based approaches. The immunodominant epitopes vary between D614 and G614 SARS-CoV-2. The epitopes on the S protein elicit neutralizing antibodies against D614 and G614 SARS-CoV-2.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has had an unprecedented effect on global health. SARS-CoV-2 shares 79.5% of genomic sequence identity with SARS-CoV, but it is more contagious than SARS-CoV (Lu et al., 2020; Zhou et al., 2020). Prophylactic vaccines are an important means to curb pandemics of infectious diseases. Accordingly, an effective and safe SARS-CoV-2 vaccine are needed urgently. Several SARS-CoV-2 vaccine candidates, based on a variety of technologies, are being tested in clinical trials (Chen et al., 2020a; Thanh Le et al., 2020). However, the epitopes on these vaccines and SARS-CoV-2 are not well studied, and it is still urgent to identify epitopes that can elicit neutralizing antibodies and determine the immunodominant epitopes in humans for improvement and design of novel vaccines.
Four major structural proteins—spike (S), envelope (E), membrane (M), and nucleocapsid (N)—play vital roles in entry and replication of the virus (Chen et al., 2020b). Several epitopes on the S protein have been reported with little information regarding immunogenicity and neutralization, but most epitopes on the M, E, and N proteins remain unknown (Baruah and Bose, 2020; Bhattacharya et al., 2020; Yuan et al., 2020). The accuracy of the predicted epitopes using in silico methods is unclear, and the immunogenicity of the obtained epitopes needs further experimental verification (Ahmed et al., 2020; Grifoni et al., 2020; Kiyotani et al., 2020). Epitope prediction methods based on the 3D structure of a protein can greatly improve the precision of antigen epitopes (Jespersen et al., 2017). Therefore, the reported 3D structures of the S and M proteins are conducive for epitope prediction (Jin et al., 2020; Lan et al., 2020; Walls et al., 2020; Wrapp et al., 2020). Although the structures of the E and N proteins are still unsolved, it is possible to model these protein structures based on their reported gene sequence using molecular simulation and then predict their epitopes (Lu et al., 2020). Increasing evidence shows that some linear epitopes, as sites of virus vulnerability, conserved regions, or key components of conformational epitopes, play important roles in induction of virus neutralization (Alphs et al., 2008; Sok and Burton, 2018; Xu et al., 2018). For example, a linear epitope of HIV induced a broad-spectrum protection effect and could be used to develop universal vaccines (Kong et al., 2019). By identifying conformational B cell epitopes with a higher degree of continuity and the appropriate linear window with key functional residues of discontinuous B cell epitopes centralized and randomized, we may find the key components of conformational epitopes.
The immunogenicity, immunodominance, and especially neutralization of the epitopes are crucial for development of effective SARS-CoV-2 vaccines. Although the epitope immunodominance landscape of the S protein has been mapped (Zhang et al., 2020), mutation of virus proteins might alter the antigenicity of the virus and possibly affect human immune responses to the epitopes, making it a central challenge for vaccine development. Phylogenetic analysis showed that SARS-CoV-2 mutates with a mutation rate around 1.8 × 10−3 substitutions per site per year (Li et al., 2020). Within all identified mutations of the S protein, further investigation of the 614th amino acid is needed. G614 in the S1 protein of SARS-CoV-2, found in almost all individuals with COVID-19 outside of China, causes a higher fatality rate and might be more easily spread than D614, which is mainly found in China (Becerra-Flores and Cardozo, 2020; Korber et al., 2020). The 614th amino acid is located on the surface of the S protein protomer, and G614 destabilizes the conformation of the viral spike and facilitates binding of the S protein to ACE2 on human host cells (Becerra-Flores and Cardozo, 2020). However, little is known about how G614 influences human immune responses to SARS-CoV-2. In fact, mutations not only on the S protein but also on the E, M, and N proteins might affect human immune responses to the virus. The limited neutralizing effect by a vaccine using the S protein as the only antigen suggests that epitopes on the E, M, and N proteins might also be important for SARS-CoV-2 vaccine design, and it is necessary to understand how mutations affect human immune responses to the virus (van Doremalen et al., 2020).
In this study, we predicted and synthesized the B cell epitopes on the surface of the S, M, E, and N proteins of SARS-CoV-2, prepared 37 vaccines based on hepatitis B core protein (HBc) virus-like particles (VLPs) using the SpyCatcher/SpyTag system, validated the immunogenicity of the epitopes by immunizing mice, and identified epitopes that could elicit neutralizing antibodies. We also determined immunodominant epitopes on SARS-CoV-2 by mapping the epitopes with sera from COVID-19 convalescent individuals and analyzed the relevance of epitope immunodominance and the mutations on SARS-CoV-2 proteins.
Results
Prediction of SARS-CoV-2 B cell epitopes and preparation of HBc-S VLPs displayed with the epitopes
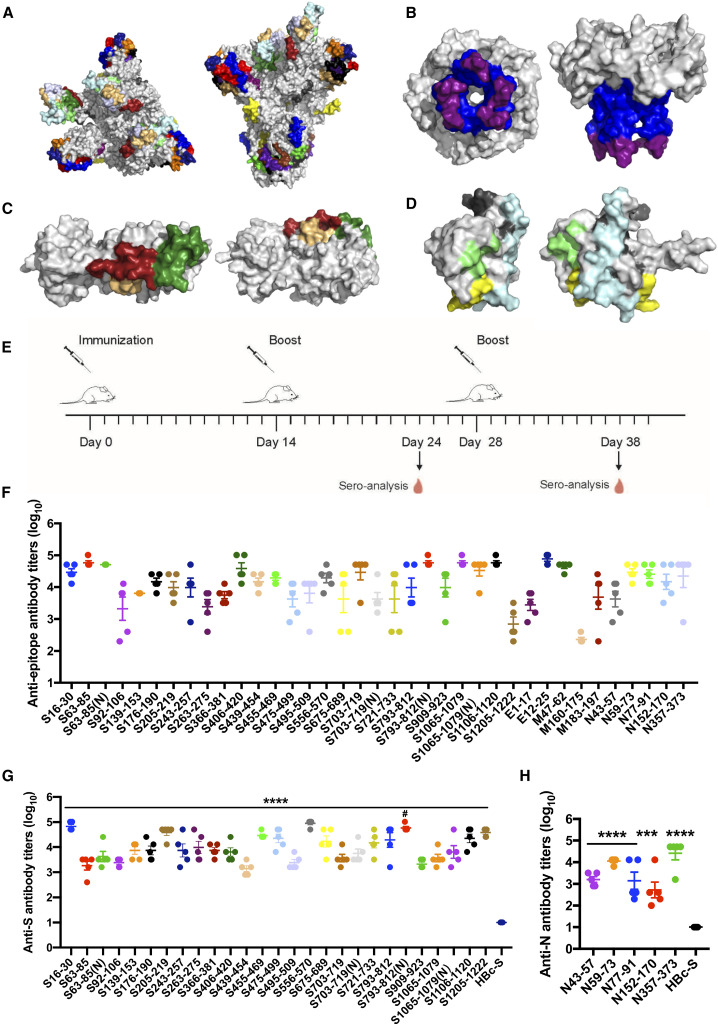
To predict antigen epitopes on SARS-CoV-2, we used a high-performance computer to simulate the 3D structures of the S, M, E, N proteins and then used computational simulation calculations to obtain preliminary antigen epitope information based on epitope surface accessibility. Spatial structure information of the S, M, E, and N protein structure models was obtained (Figures 1 A–1D); the structures of S and M proteins were consistent with the reported structures, and the structures of the E and N proteins were obtained for the first time (Wrapp et al., 2020; Yan et al., 2020). A total of 33 B cell epitopes were predicted on the basis of protein structures, and during the selection process, priority was given to select sequences with high homology within the SARS-CoV and RaTG13 coronavirus strains (Table S1; Data S1). Within these epitopes, four contained glycosylation sites (Watanabe et al., 2020; Wrapp et al., 2020), and the corresponding GlcNAc glycosylated epitopes were synthesized (Table S1); 13 from the S protein, two from the E protein, three from the M protein, and five from the N protein are conserved with more than 80% homology among SARS-CoV-2, SARS-CoV, and bat coronavirus RaTG13, respectively (Table S1). All epitopes were exposed on the surface of the virus proteins (Figures 1A–1D) and had a high antigenicity score, indicating their potential for initiating immune responses. Therefore, they were considered to be promising epitope candidates against B cells for vaccine preparation.
Figure 1.
Predication and validation of epitopes on SARS-CoV-2
(A–D) Molecular simulated structures and predicted epitopes of major proteins of SARS-CoV-2. Shown are top and side views of 3D structures (gray) and the predicted epitopes (colored) of the spike (S) protein (A), envelope (E) protein (B), membrane (M) protein (C), and nucleocapsid (N) protein (D).
(E–H) Epitope-conjugated HBc-S VLPs induce high antibody titers against epitope peptides and SARS-CoV-2 proteins.
(E) Schematic of the immunization design.
(F–H) 96-well plates were coated with peptides (F) and S (G) and N (H) proteins.
Data are shown as mean ± SEM (compared with the HBc-S control; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; one-way ANOVA followed by Dunnett’s test; compared with non-glycosylated epitope; #p < 0.05; Student’s t test).
The predicted epitope peptides of the S, M, E, and N proteins were synthesized and conjugated onto the surface of HBc-SpyCatcher (HBc-S) VLPs via SpyCatcher/SpyTag isopeptide formation, forming epitope peptides displaying HBc-S VLPs called HBc-S-P VLPs. SDS-PAGE confirmed that the epitope peptides were conjugated successfully onto HBc-S VLPs, as shown by HBc monomers with higher molecular weight (Figure S1A). Our transmission electron microscopy (TEM) results showed that all HBc-S-P self-assembled into morphologically correct VLPs (Figure S1B). We further assessed the hydrodynamic diameter of HBc-S-P VLPs by dynamic light scattering (DLS), and the results showed that all HBc-S-P VLPs were relatively uniform (polydispersity index ([PDI] < 0.25) with a diameter around 40 nm (Figure S1C; Table S2), consistent with previous reports (Ji et al., 2020).
The epitopes elicit highly specific antibody responses
To assess immunogenicity, the HBc-S-P VLPs were administered subcutaneously to BALB/c mice three times, and the serum antibody titers were assayed by ELISA (Figure 1E). Epitopes S455–469, S556–570, E12–25, M47–62, N59–73, and N357–373 elicited robust antibody responses against peptides and/or the S and N proteins in mice 10 days after the second injection (≥1,000; Figure S2). All predicted epitopes boosted antibodies response against the corresponding epitope peptides, and the antibody titer reached at least 1,000 after the third immunization, except for M160–170, whose antibody titer was only 230 (Figure 1F). Accordingly, epitopes S16–30, S205–219, S455–469, S475–499, S556–570, S721–733, S793–812, S1106–1120, S1205–1222, N59–73, and N353–373 on the S and N proteins also induced robust antibodies with titers greater than 10,000 against the S and N proteins, respectively (Figures 1G and 1H). These results demonstrated that almost all predicted epitopes on the S, M, E, and N proteins elicited immune responses with high levels of antibodies, suggesting that these epitopes have good immunogenicity. The GlcNAc glycosylated epitopes also elicited sufficient amounts of antibodies toward the corresponding epitope peptides and S protein, and only S793–812(N) induced more antibodies than the non-glycosylated epitope (Figures 1F and 1G).
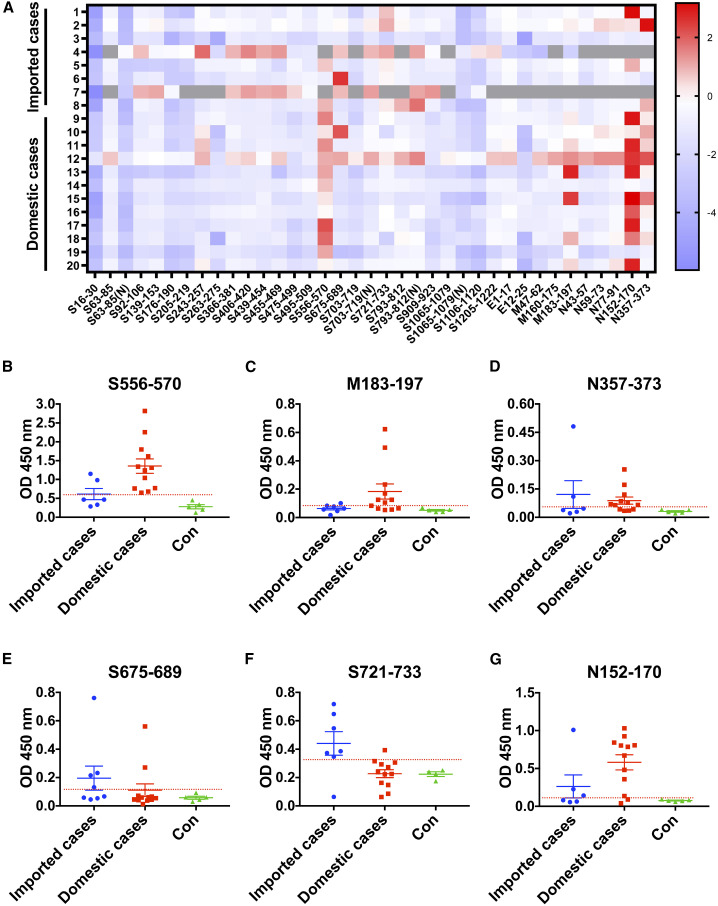
Imported and domestic COVID-19 cases have different immunodominant epitopes
To investigate the spectrum of antibodies in individuals with COVID-19, we detected binding of early convalescent sera of eight individuals with imported (Europe) COVID-19 who were infected with SARS-CoV-2 in early April, 2020 and 12 individuals with domestic (China) COVID-19 who were infected in early February, 2020 to various epitopes (Table 1 ). The mean value plus three times the standard deviation in healthy volunteers was used as the cutoff value to define positive reactions, and an epitope showing an average positive rate of 50% or more among individuals was considered an immunodominant epitope (Figures 2 A and S3). Our results showed that S556–570, M183–197, and N357–373 were immunodominant in individuals with domestic COVID-19 (Figures 2B–2D), whereas S675–689 and S721–733 were immunodominant in imported COVID-19 groups (Figures 2E and 2F). Only N152–170 was immunodominant in both groups (Figure 2G). Notably, S556–570, N152–170, and S721–733 reacted with the sera of almost all individuals (≥80%) with domestic or imported COVID-19 (Figures 2B, 2F, and 2G). These results indicated that imported and domestic COVID-19 had different immunodominant epitopes.
Table 1.
Characteristics of Individuals with COVID-19
| Group | Case | Sex | Age | Country | S mutation | N mutation |
|---|---|---|---|---|---|---|
| Imported cases | individual 1 | M | 47 | Denmark | NT | NT |
| individual 2 | M | 19 | Russia | G614 | R203G204 | |
| individual 3 | M | 23 | United Kingdom | D614 | R203G204 | |
| individual 4 | F | 17 | United Kingdom | NT | NT | |
| individual 5 | M | 19 | United Kingdom | G614 | K203R204 | |
| individual 6 | F | 49 | Russia | G614 | K203R204 | |
| individual 7 | M | 50 | Russia | G614 | K203R204 | |
| individual 8 | F | 21 | United Kingdom | G614 | G189R203G204 | |
| Domestic cases | individual 9 | M | 42 | China | D614 | R203G204 |
| individual 10 | M | 72 | China | D614 | R203G204S344 | |
| individual 11 | M | 56 | China | NT | NT | |
| individual 12 | M | 66 | China | D614 | R203G204 | |
| individual 13 | F | 40 | China | D614 | R203G204 | |
| individual 14 | F | 34 | China | NT | NT | |
| individual 15 | F | 60 | China | D614 | R203G204 | |
| individual 16 | F | 44 | China | NT | NT | |
| individual 17 | M | 67 | China | D614 | R203G204 | |
| individual 18 | F | 67 | China | D614 | R203G204 | |
| individual 19 | F | 10 | China | NT | NT | |
| individual 20 | F | 51 | China | NT | NT |
NT, not tested.
Figure 2.
Imported and domestic COVID-19 have different immunodominant epitopes
(A) The landscape of adjusted epitope-specific antibody levels in early convalescent sera of individuals with imported and domestic COVID-19. Gray indicates not tested.
(B–G) Immunodominant epitopes binding with the antibodies in early convalescent sera from individuals with imported and domestic COVID-19. Data are shown as mean ± SEM. The cutoff lines were based on the mean value plus 3 SD in 4–5 healthy persons.
To elucidate the possible cause of the difference in immunodominant epitopes, we sequenced the S and N genes of SARS-CoV-2 from individuals with imported and domestic COVID-19, respectively. We found that the gene sequences of imported and domestic strains contained G614 or D614 in the S protein and K203R204/G189R203G204/R203G204/R203G204S344 in the N protein, respectively (Table 1), which may result in different immunodominant epitopes of different virus sub-strains.
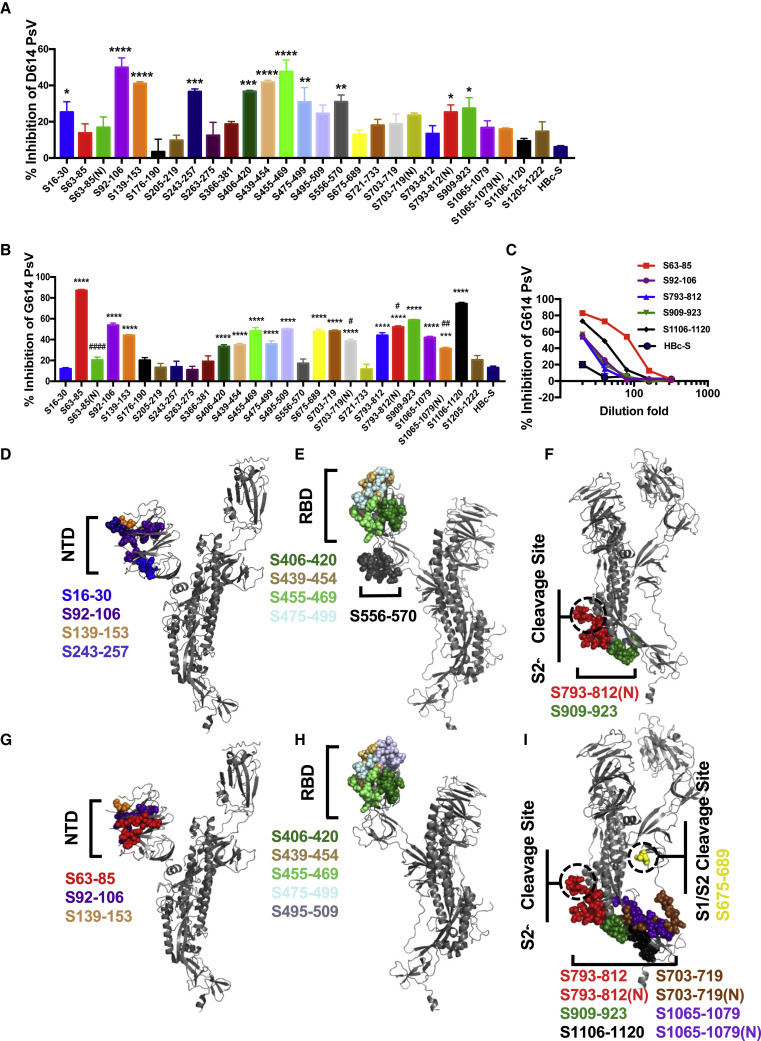
The predicted epitopes induce neutralization antibody production
The SARS-CoV-2 pseudovirus neutralization assay is a well-accepted method to detect the ability of a vaccine to inhibit SARS-CoV-2 infection (Ni et al., 2020; Wang et al., 2020). To assess neutralization antibodies induced by S protein epitopes, we incubated the immunization sera with D614 or G614 SARS-CoV-2 pseudoviruses, and then the mixture was added to ACE2-293T cells that stably expressed ACE2. The results showed that immunized sera of the S92–106, S139–153, S439–454, and S455–469 epitopes inhibited SARS-CoV-2 pseudovirus infection compared with the HBc-S control (p < 0.0001), with inhibition rates around 40%–50% (Figure 3 A). The sera of S16–30, S243–257, S406–420, S475–499, S556–570, S793–812(N), and S909–923 inhibited SARS-CoV-2 infection with an inhibition rate of 20%–40% (Figure 3A), indicating that these 11 epitopes induced neutralization antibody production. To detect the effect of epitope immunization on the neutralizing responses of G614 SARS-CoV-2, we incubated the epitope-immunized sera with the G614 SARS-CoV-2 pseudoviruses. The results showed that sera of epitopes inhibiting D614 SARS-CoV-2 also inhibited G614 SARS-CoV-2 infection, except for S16–30, S243–257, and S556–570 (Figure 3B). However, the immunized sera of epitopes S63–85, S495–509, S675–689, S703–719, S793–812, S1065–1079, S1065–1079(N), and S1106–1120 only inhibited G614 SARS-CoV-2 pseudovirus infection. Interestingly, compared with the non-glycosylation epitope, S63–85(N), S703–719(N), and S1065–1079(N) induced fewer neutralizing antibodies to the G614 pseudovirus, whereas those of S793-812(N) increased (Figure 3B). We then 2-fold serially diluted sera with an inhibition rate of more than 50% and determined the neutralizing antibody titers induced by these epitopes. S63–85 induced the highest neutralizing effect with an antibody titer at 1:80 (Figure 3C). The structural analysis showed that most of these neutralizing epitopes to D614 and G614 SARS-CoV-2 were in or near the N-terminal domain (NTD), receptor-binding domain (RBD), or S2′ cleavage site of the S protein and were spatially clustered (Figures 3D–3I), except S675–689, which was in or near the S1/S2 cleavage site at the interface of the S1 and S2 subunits of the S protein (Figure 3I).
Figure 3.
Antibodies induced by epitopes of the S protein inhibit SARS-CoV-2 pseudovirus infection
(A and B) Neutralizing potency of mice sera after the third immunization with each vaccine was measured with D614 (A) or G614 (B) SARS-CoV-2 pseudoviruses (PsVs). Data are shown as mean ± SEM (compared with the HBc-S control;∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; compared with non-glycosylated epitope; #p < 0.05; ##p < 0.01; ####p < 0.0001).
(C) 2-Fold serial dilution neutralizing assay against G614 SARS-CoV-2 pseudoviruses. Data are shown as mean ± SEM.
(D–I) Spatial positions of D614 (D–F) and G614 pseudovirus (G–I) neutralizing epitopes (colored), respectively, in or near the N-terminal domain (NTD; D and G), receptor-binding domain (RBD; E and H), and S2′ cleavage site (F and I) of the S protein (gray).
Discussion
In this study, we used a high-performance computer to simulate the 3D structures of major proteins on SARS-CoV-2 and predicted 33 surface area epitopes using the modeled protein structures, which proved to be efficient and accurate by mouse immunization and pseudovirus neutralization assay. Of the 33 identified epitopes, 24 were conserved with more than 80% homology, and 18 shared more than 90% homology between SARS-CoV-2, SARS-CoV, and bat coronavirus RaTG13 (Table S1), suggesting that these epitopes could be used for designing broad-spectrum betacoronavirus vaccines.
Some surface area epitopes of SARS-CoV-2 were determined to be immunodominant in the present study. Consistent with a previous report, S556–570 was an immunodominant epitope was able to elicit neutralizing antibodies (Figure 3A; Poh et al., 2020). S675–689 and S721–733 were immunodominant epitopes in imported strains but not in domestic strains, which may result from antigenic drift by the 614th amino acid variance on the S protein of SARS-CoV-2 between imported and domestic cases. In most imported cases, glycine was in the 614th position of the S protein, which possibly made the S675–689 and S721–733 regions more accessible by specifically destabilized the “up” state of the viral spike via unpacking with T859 in the adjacent helical stalk (Becerra-Flores and Cardozo, 2020). Moreover, S556–570 was no longer an immunodominant and neutralizing epitope in the G614 strain, and the antibodies induced by S675–689 inhibited G614 but not D614 pseudovirus entry into ACE2-expressing 293T cells, suggesting that antigenic drift was caused by the D614G mutation. Two epitopes, N152–170 and N357–373, are highly conserved among SARS-CoV-2, SARS-CoV, and bat coronavirus RaTG13. Consistent with previous reports, these two epitopes were immunodominant sites (Guo et al., 2004). Importantly, they bound to neutralizing antibodies in recovered SARS-CoV individuals (Guo et al., 2004; Shichijo et al., 2004). M183–197 is another immunodominant epitope in domestic cases. Because this epitope is located in the S4 subsite of the active center of the M protein, it is possible for it to elicit neutralizing antibodies that inhibit the protease function of the M protein (Dai et al., 2020; Yang et al., 2005).
The antibodies induced by S406–420, S439–454, S455–469, and S475–499 but not S366–381 or S495–509 in the RBD region had a neutralizing effect on the D614 and G614 pseudoviruses, consistent with the interaction interface between the SARS-CoV-2 receptor-binding motif (RBM) and ACE2 (Seydoux et al., 2020; Shang et al., 2020), indicating that these epitopes are suitable for designing universal vaccines. Consistent with previous reports, the antibodies induced by epitope S366–381 did not have a neutralizing effect on entry of the pseudovirus (Wrapp et al., 2020; Yuan et al., 2020). Interestingly, not only antibodies targeting the interaction interface between the RBD and ACE2 but also antibodies binding with the NTD of the S protein, such as S16–30, S92–106, S139–153, and S243–257, had a neutralizing effect on the D614 strain. Of the neutralizing NTD epitopes, S92–106 and S139–153 also had a neutralizing effect on the G614 strain. Antibodies induced by S63–85, but not its glycosylated form, inhibited cell entry of the G614 pseudovirus rather than the D614 pseudovirus, and the epitopes S703–719(N) and S1064–1079(N) induced fewer neutralizing antibodies compared with the unglycosylated ones, indicating that native oligomannose and complex-type N-glycan might have a shield effect on the epitope and that mutation at the 614th position possibly affected exposure of the epitope by altering the pose of the glycan shield (Barnes et al., 2018). In contrast, the glycosylated epitope S793–812(N) had a more inhibitory effect than S793–812 on the G614 and D614 pseudoviruses, suggesting that glycosylation on the epitope might affect viral infectivity. Two conserved epitopes (S793–812(N) and S909–923) near the highly conserved S2′ protease cleavage site of the S protein also induced neutralizing antibodies on the D614 and G614 pseudoviruses, indicating that there may be mechanism that blocks cell entry of SARS-CoV-2. Notably, several identified neutralizing epitopes are consistent with the epitopes of some important neutralizing antibodies, such as S139–153 to antibody 4A8 (PDB: 7C2L), S406–420 to antibody C105 (PDB: 6XCN), and S16–30 to antibody P2B-2F6 (PDB: 7BWJ) (Barnes et al., 2020; Chi et al., 2020; Ju et al., 2020), suggesting that these epitopes might be the antibody-targeting sites. Importantly, a shift of the immunodominant and neutralizing epitope region from S556–S570 to S675–689 was observed upon D614G mutation. S675–689 is at the S1/S2 cleavage site located at the interface of the S1 and S2 subunits of the S protein, which is important for S protein-mediated virus-cell membrane fusion. Our results suggested that the S675–689 epitope was at the vulnerability site of SARS-CoV-2 and might be an ideal candidate and targeting site for vaccine development. Moreover, our results showed that the neutralizing epitopes are highly spatially clustered, indicating that conformational epitopes in the above regions may be used for designing an effective vaccine.
In conclusion, we successfully predicted SARS-CoV-2 epitopes based on the 3D structures of the S, M, N, and E proteins; validated their immunogenicity; characterized the homology of the epitopes among betacoronaviruses; and identified the neutralizing and immunodominant epitopes (Table S3). Our findings provide a wide neutralizing and immunodominant epitope spectrum for design of an effective and safe vaccine, differential diagnosis, and virus classification.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HRP-conjugated goat anti-mouse IgG | Abcam | Cat#ab6789; RRID: AB_955439 |
| HRP-conjugated goat anti-human IgG | Abcam | Cat#ab6858; RRID: AB_955433 |
| Chemicals, peptides, and recombinant proteins | ||
| BSA | Sigma-Aldrich Corporation | Cat#V900933 |
| Epitope peptides | GL Biochem | N/A |
| HBc-S | Ji et.al., 2020 | N/A |
| Imject Alum Adjuvant | Thermo Fisher | Cat#77161 |
| D614 SARS-CoV-2 pseudoviruses | PackGene Biotech | Cat#LV-nCov1 |
| G614 SARS-CoV-2 pseudoviruses | PackGene Biotech | Cat#LV-nCov1 |
| chromogenic substrate TMB | Thermo Fisher | Cat#34028 |
| SARS-CoV-2 Spike S1+S2 | Sino Biological Inc. | Cat#40589-V08B1 |
| SARS-CoV-2 nucleocapsid | Sino Biological Inc. | Cat#40588-V08B |
| Critical commercial assays | ||
| Bright-Glo™ Luciferase Assay System | Promega | Cat#E2620 |
| SARS-CoV-2 Nucleic Acid Extraction Kit | Daan Gene | Cat#DA0931 |
| Deposited data | ||
| Sequence data | This paper | GenBank: MW362746-MW362764, MW368449-MW368461 |
| Experimental models: cell lines | ||
| ACE2-239T cells | PackGene Biotech | Cat#nCov-3 |
| Experimental models: organisms/strains | ||
| C57BL/6 | Beijing HFK Bioscience CO., LTD | N/A |
| Software and algorithms | ||
| Gromacs v5.1 | GROMACS | http://www.gromacs.org/ |
| Discotope 2.0 | Immune epitope database and analysis resource (IEDB) | http://tools.iedb.org/discotope/ |
| ClustalW2 | EMBL’s European Bioinformatics Institute (EMBL-EBI) | https://www.ebi.ac.uk/Tools/msa/clustalw2/ |
| GraphPad Prism 7.0a | GraphPad Software | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rui-tian Liu (rtliu@ipe.ac.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any unique code. The sequence data of S and N gene of SARS-CoV-2 from COVID 19 patients is available at NCBI GenBank repository. The accession numbers for the S and N gene sequencing reported in this paper are GenBank: MW368449-MW368461 and GenBank: MW362746-MW362764.
Experimental model and subject details
Specimens from SARS-CoV-2 patients
Serum samples were collected from 20 early convalescent patients with COVID-19 which were confirmed by SARS-CoV-2 real-time reverse transcriptase–polymerase chain reaction (RT-PCR). 12 patients were infected in China and the other eight were imported cases from Europe. The median age of imported and domestic patients was 50.8 years (range, 10-72 years) and 30.6 years (range, 17-50 years), respectively. Patient information including age and sex was shown in Table 1. Biological samples from patients were randomly and blindly selected with no considerations for age or sex. This study was approved by the Institutional Review Board of the Shijiazhuang Fifth Hospital. Waiver of informed consent for collection of samples from infected individuals was granted by the institutional ethics committee. Nucleic acids from throat swab samples were extracted using SARS-CoV-2 Nucleic Acids Extraction Kit (Daan Gene, Zhongshan, China) according to the manufacturer’s instructions. The genes of S and N were reverse transcripted, amplified, and sequenced.
Mice
6-8 week-old BALB/c female mice were obtained from Beijing HFK Bioscience CO., LTD (Beijing, China) and maintained with access to food and water ad libitum in a colony room kept at 22 ± 2°C and 50 ± 5% humidity, under a 12:12 light/dark cycle. All animal experiments were performed in accordance with the China Public Health Service Guide for the Care and Use of Laboratory Animals. Experiments involving mice and protocols were approved by the Institutional Animal Care and Use Committee of Tsinghua University (AP#15-LRT1).
Method details
Epitope prediction
Homologous modeling and molecular dynamics simulation was used to predict the structure of S, M, N, E protein. The genome sequence of SARS-CoV-2 isolate Wuhan-Hu-1 was retrieved from the NCBI database under the accession number MN988669.1 and the protein sequences were acquired according to the annotation. The original pdb file of the S, M, N, E protein was established by homologous modeling using SWISS-MODEL (Waterhouse et al., 2018) according to the template structures of SARS-CoV spike glycoprotein (PDB: 6ACC), SARS-CoV main peptidase M(pro) (PDB: 2A5K), SARS-CoV envelope small membrane protein (PDB: 5X29) and SARS-CoV nucleocapsid protein (PDB: 1SSK), respectively. On the basis of the homologous modeled pdb file, added water, adjusted pH of chloride and sodium ions and ran molecular dynamics simulation program, obtaining the pdb file in the human body temperature (310K) state. We then calculated the full atomic structure of the protein for 1 μs using the molecular dynamics software GROMACS 5.1 on Sunway TaihuLight supercomputer and obtained the molecular orbital energy level and spatial structure information of the protein. In particular, the RBD region was referred to as the fragment from 347 to 520 amino acid of S protein. Structure-based conformational B cell epitope prediction was performed by using Discotope 2.0 (Kringelum et al., 2012) and −2.5 was used as a positivity cutoff. All the appropriate linear epitope windows were then selected by the following criteria: 1) solvate accessible regions with high surface probability; 2) regions with high antigenicity and flexibility; 3) the key functional residues of conformational B cell epitopes were centralized and with high degree of continuity in the window. The selected epitopes were then applied to homology analysis with the according sequences from SARS-CoV and RaTG13 via ClustalW (Thompson et al., 1994). N-glycosylated regions with homology >50% were selected for synthesis of N-glycosylated epitopes. Epitope homology was calculated by the following formula:
Preparation and characterization of HBc-S-peptide VLP vaccine
HBc-S was purified as described previously (Ji et al., 2020). Purified protein was concentrated and determined by BCA protein assay kit (Pierce, Rockford, IL, USA). The purity of the recombinant protein was analyzed by SDS-PAGE. The peptides of SpyTag-epitope were chemically synthesized by GL Biochem (Shanghai, China). For the preparation of epitope conjugated HBc-S VLP vaccine, HBc-S VLPs were incubated with 3-fold molar excess of epitope peptide for 3 h at room temperature in citrate reaction buffer (40 mM Na2HPO4, 200 mM sodium citrate, pH 6.2), and was then dialyzed with 100 kDa cut-off membrane to remove the unreacted epitope peptide.
Transmission electron microscopy (TEM) was used for the morphological examination of HBc-S-peptide VLP vaccine. 20 μl VLPs (0.1-0.3 mg/ml) were applied to 200 mesh copper grids for 5 min and negatively stained with 2% uranyl acetate for 1 min, and then blotted with filter paper and air-dried. VLPs were imaged in a Hitachi TEM system at 80 kV at 40,000 x magnification. To measure the hydrodynamic size of HBc-S-peptide VLP vaccine using dynamic light scattering (DLS), 9 μL of HBc-S or HBc-S-P VLPs at concentration of 0.1 mg/mL was loaded into a Uni tube and held at 2 min at room temperature. Each measurement was taken four times with 5 DLS acquisitions by an all-in-one stability platform Uncle (Unchained lab, USA).
Mice immunization
To evaluate the immunogenicity of the epitopes, female BALB/c mice (6-8 weeks) were subcutaneously vaccinated with HBc-S-P VLPs (containing 100 μg HBc-S) in citrate buffer (200 mM citrate acid, 40 mM NaH2PO4, pH 6.2, 100 μl) mixed with Alum (1:3 v/v) (ThermoFisher, Waltham, MA, USA). HBc-S was used as a control. Each group of mice (n = 5) received their first injection at day 0 and boosters at day 14 and 28. Serum samples were taken 10 days after each immunization.
Enzyme-linked immunosorbent assay (ELISA)
Serum antibodies specific for epitope peptides and SARS-CoV-2 proteins were detected by ELISA. 96-well plates (Dynex Technologies, Chantilly, VA) were coated with 0.5 μg peptides, 100 ng S or N protein per well at 4°C overnight, respectively, and then washed three times with PBS and blocked with 3% BSA (in 0.1% PBST) for 2 h at 37°C. After blocking, the plates were incubated with serial dilutions of the sera (100 μl/well, in two-fold dilution) for 2 h at 37°C. The bound serum antibodies were detected with HRP-conjugated goat anti-mouse IgG and chromogenic substrate TMB (ThermoFisher, Waltham, MA, USA). The cut-off for seropositivity was set as the mean value plus three standard deviations (3SD) in HBc-S control sera. The binding of the epitopes to the sera of COVID-19 infected patients were detected by ELISA using the same procedure as described above, 96-well plates were coated with 0.5 μg peptides and sera were diluted at 1:50. The cut-off lines were based on the mean value + 3SD in 4-5 healthy persons. All ELISA studies were performed at least twice.
SARS-CoV-2 pseudovirus neutralization assay
Pooled mice sera collected at day 10 after the third immunization were diluted in DMEM supplemented with 10% fetal bovine serum, mixed with SARS-CoV-2 pseudoviruses (31600 TCID50 per well) and incubated at 37°C for 1 h. The mixture was then added to 1.5 × 104 ACE2-293T cells and the medium was replaced after 6 h. Firefly luciferase activity was measured 72 h post-infection using Bright-Glo Luciferase Assay System (Promega). All neutralization studies were performed at least twice. Three independently mixed replicates were measured for each experiment.
Quantification and statistical analysis
The data presented in this study were expressed as mean ± SEM. Data were analyzed by one-way (ANOVA), followed by multiple comparisons using Dunnett’s test within GraphPad Prism 7.0 software. Student’s t test was used to analyze the data of non-glycosylated and glycosylated epitopes. p < 0.05 was considered to be significant.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81971610, 81971073, and 81903531); the National Science and Technology Major Projects of New Drugs (2018ZX09733001-001-008); the Innovation Academy for Green Manufacture, Chinese Academy of Sciences (IAGM2020C29); and the Zhejiang University Special Scientific Research Fund for COVID-19 Prevention and Control (2020XGZX075).
Author contributions
R.-t.L. designed experiments and wrote the manuscript. S.L. and X.-x.X. designed experiments, performed research, analyzed data, and wrote the manuscript. C.-g.J. designed experiments. L.Z. (Lei Zhao), Y.-l.W., J.X., H.-x.G., and X.-h.S. collected blood samples and carried out experiments. B.W., J.Z., T.-r.Y., M.J., C.-p.L., D.-q.L., L.Z. (Lun Zhang), S.-j.H., X.-l.Y., G.-w.Y., X.-m.F., C.-w.K., and B.-x.K. performed research.
Declaration of interests
R.-t.L, S.L., and X.-x.X. have filed a provisional patent on epitopes for designing a coronavirus vaccine.
Published: January 26, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108666.
Supplemental information
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphs H.H., Gambhira R., Karanam B., Roberts J.N., Jagu S., Schiller J.T., Zeng W., Jackson D.C., Roden R.B. Protection against heterologous human papillomavirus challenge by a synthetic lipopeptide vaccine containing a broadly cross-neutralizing epitope of L2. Proc. Natl. Acad. Sci. USA. 2008;105:5850–5855. doi: 10.1073/pnas.0800868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Gristick H.B., Freund N.T., Escolano A., Lyubimov A.Y., Hartweger H., West A.P., Jr., Cohen A.E., Nussenzweig M.C., Bjorkman P.J. Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat. Commun. 2018;9:1251. doi: 10.1038/s41467-018-03632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020;74:e13525. doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S.S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 Vaccine Pipeline: an Overview. Curr. Trop. Med. Rep. 2020:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.P., Petric M., Campbell W., McGeer P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45(W1):W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji M., Xie X.X., Liu D.Q., Lu S., Zhang L.X., Huang Y.R., Liu R.T. Engineered hepatitis B core virus-like particle carrier for precise and personalized Alzheimer’s disease vaccine preparation via fixed-point coupling. Applied Materials Today. 2020;19:100575. [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kiyotani K., Toyoshima Y., Nemoto K., Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R., Duan H., Sheng Z., Xu K., Acharya P., Chen X., Cheng C., Dingens A.S., Gorman J., Sastry M., NISC Comparative Sequencing Program Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization. Cell. 2019;178:567–584.e19. doi: 10.1016/j.cell.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelum J.V., Lundegaard C., Lund O., Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012;8:e1002829. doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li X., Wang W., Zhao X., Zai J., Zhao Q., Li Y., Chaillon A. Transmission dynamics and evolutionary history of 2019-nCoV. J. Med. Virol. 2020;92:501–511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y.-P., Chee R.S.-L., Yeo N.K.-W., Lee W.-H., Leo Y.-S., Chen M.I.C. Potent neutralizing antibodies in the sera of convalescent COVID-19 patients are directed against conserved linear epitopes on the SARS-CoV-2 spike protein. bioRxiv. 2020 doi: 10.1101/2020.2003.2030.015461. [DOI] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichijo S., Keicho N., Long H.T., Quy T., Phi N.C., Ha L.D., Ban V.V., Itoyama S., Hu C.J., Komatsu N. Assessment of synthetic peptides of severe acute respiratory syndrome coronavirus recognized by long-lasting immunity. Tissue Antigens. 2004;64:600–607. doi: 10.1111/j.1399-0039.2004.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D., Burton D.R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018;19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.2005.2013.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Acharya P., Kong R., Cheng C., Chuang G.Y., Liu K., Louder M.K., O’Dell S., Rawi R., Sastry M. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 2018;24:857–867. doi: 10.1038/s41591-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.-z., Hu Y.-f., Chen L.-l., Tong Y.-g., Hu J.-c., Cai J.-p., Chan K.-H., Dou Y., Deng J., Gong H.-r. Mapping the Immunodominance Landscape of SARS-CoV-2 Spike Protein for the Design of Vaccines against COVID-19. bioRxiv. 2020 2020.2004.2023.056853. [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique code. The sequence data of S and N gene of SARS-CoV-2 from COVID 19 patients is available at NCBI GenBank repository. The accession numbers for the S and N gene sequencing reported in this paper are GenBank: MW368449-MW368461 and GenBank: MW362746-MW362764.