Abstract
SARS-CoV-2 infection has become a worldwide pandemic and is spreading rapidly to people across the globe. To combat the situation, vaccine design is the essential solution. Mutation in the virus genome plays an important role in limiting the working life of a vaccine. In this study, we have identified several mutated clusters in the structural proteins of the virus through our novel 2D Polar plot and characterization descriptor. We have also studied several biochemical properties of the proteins to explore the dynamics of evolution of these mutations. This study would be helpful to understand further new mutations in the virus and would facilitate the process of designing a sustainable vaccine against the deadly virus.
Keywords: SARS-CoV-2, COVID-19, Mutation, Structural protein, GRANCH, Hotspot, Statistical data, Graph theory
1. Introduction
SARS-CoV-2 is the newest member of Coronaviridae family. After the COVID-19 (SARS-COV-2) infection broke out suddenly in Wuhan, China, it had spread across more than 200 countries worldwide affecting 70,476,836 people and causing 1,599,922 deaths as of 15th December 2020 [1]. The World Health Organization (WHO) declared this as a public health emergency of international concern (PHEIC) on 30th January 2020 and a pandemic on 11th March.
Coronaviruses can cause both mild and severe infections in human. Human coronaviruses OC43, HKU1, 229E and NL63 cause mild to moderate seasonal common colds in adults and children [2]. On the other hand, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), Severe Acute respiratory Syndrome Coronavirus (SARS-CoV) and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) can be fatal sometimes. The SARS-CoV-2 is likely to have jumped the species barrier like most of the other coronaviruses [3], and both bats and pangolins can be the possible hosts for this virus [4].
The Reproductive Number (R0) of SARS-CoV-2 is approximately 3.28 [5] which is relatively high and hence makes the virus extremely contagious. It spreads through respiratory droplets and contact routes. More recent studies show airborne transmission of COVID-19 too [6] increasing its capability to spread in communities. The symptoms of SARS-CoV-2 infection mainly include fever, cough, shortness of breath, sore throat, fatigue and so on [7,8]. In some cases, gastrointestinal malfunctions are also observed [9].
As of now, Pfizer and BioNTech's mRNA vaccine BNT162b2 is being administered in US, UK and Canada. Moderna's mRNA-1273, Russia's Sputnik V and AstraZeneca's AZD1222 have also shown tremendous success in their Phase 3 trials and are being considered for use at an emergency basis. [10]. . Generally, the potential vaccine candidates comprise of either inactivated or live attenuated or subunit viruses, or DNA or RNA vaccines. Mutation becomes a very important factor in determining the sustainability of a vaccine. High mutation rate of a virus or its proteins sometimes makes the vaccine less effective after a period of time. In another study, our lab has also proposed epitope-based peptide vaccine candidates [12].
In this article we analyzed the mutations in SARS-CoV-2 structural proteins and Orf1ab polyprotein to understand the regions in its genome where mutation is playing an important role so that it may help us to understand the dynamics of its evolution and guide us in designing a sustainable vaccine. In the course of doing so, we computed around one hundred thousand sequences and analyzed them through a sequence descriptor to cluster the similar strains or proteins. Later on, in each cluster we identified the point of mutation and for single point mutation we studied its biochemical properties through bioinformatic tools. Eventually, a detailed study of the origin of mutations for each protein and its growth with time is analyzed through temporal graphs to understand the dynamics of its spread. Furthermore, we also identified the hotspot regions for mutation which can be essential to locate mutations. This study helps us to identify the significant mutations in the proteins of SARS-CoV-2 to understand their pattern of growth and suggest future studies on these proteins.
2. The cell biology of the virus
SARS-CoV-2 has some genetic similarity with MERS and SARS. Although there are some resemblances, a detailed study of the proteins helps us understand their differences in pathogenicity [13]. In this section, a general study of the proteins in corona virus and their functions are described.
Coronavirus is a circular or pleomorphic, enveloped virus hosting a positive-sense single stranded RNA [13]. It has the largest genome compared to other RNA viruses giving it an opportunity to house a variety of genes [14]. A typical coronavirus genome consists of a 5′-cap, 3′-poly-adenylated (A) tail, at least six open reading frames (orfs), and both 3′ and 5′-Untranslated Regions (UTR). SARS-CoV-2 genome codes for 4 structural proteins: Spike glycoprotein (S), Envelope protein (E) Membrane protein (M) and Nucleocapsid protein (N) in 5′ to 3′ order. The Spike glycoprotein (S) helps the virus to attach with the host cell receptors and assists in viral entry. The small, hydrophobic Envelope protein (E) plays a vital role in virion assembly, virus exit, host-stress response, ion channel activity and host protein interactions [15]. The type III transmembrane glycoprotein, Membrane protein (M) plays a critical role during the virus budding process and the Nucleocapsid protein (N) helps in genomic RNA binding, capsid formation and host cell-cycle disruption [16].
A −1 frameshift between Orf1a and Orf1b leads to the production of two polyproteins (pp1a and pp1ab) which are processed by viral-encoded chymotrypsin-like protease (3CLpro) or main protease (Mpro) and papain-like proteases into 16 nsps (Non-structural proteins) [17,18]. The nsps play a very important role in viral pathogenicity. For example, nsp3 blocks the innate immune responses of the host cells and enhance cytokine production [19]; nsp16 negatively regulates the host innate immune system to promote viral proliferation [20] and so on. Some nsps also acts as cofactor of others to activate them and amplify their functions. On the other hand, functions of nsp11 and nsp2 are still not clear enough [21].
The SARS-CoV-2 S protein is cleaved by host proteases into S1 and S2 domains which are required for host receptor recognition and membrane fusion respectively [22]. The S1 harbors a Receptor Binding Domain (RBD) that efficiently recognizes human Angiotensin Converting Enzyme 2 (ACE2) as its receptor. Cell surface proteases like Transmembrane protease, serine 2 (TMPRSS2) and lysosomal proteases also helps in the virus’ entry [23,24].
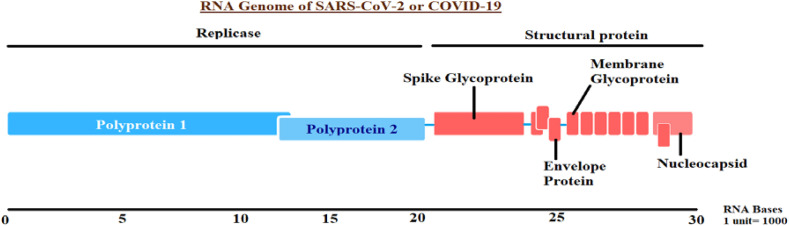
A schematic diagram of the genome of a typical SARS-CoV-2 is given in Fig. 1 below.
Fig. 1.
Genome of a typical SARS-CoV-2 The sequential arrangement of proteins over SARS-CoV-2 genome is shown in this picture.
A good analysis of mutations going on in both the structural and non-structural proteins in a virus is necessary to examine the origin and evolution of the virus, to decipher the functions of its proteins that are yet unknown, to understand the stability of the proteins and most importantly, for designing sustainable therapeutics. For example, recent studies on various proteins for conserved sequences have identified that besides the S-protein, the N-protein may also be a good target for drug design [25]. So, investigating mutations in the proteins becomes crucial for us to win this battle against COVID-19.
3. Methodology
3.1. 2D polar plot
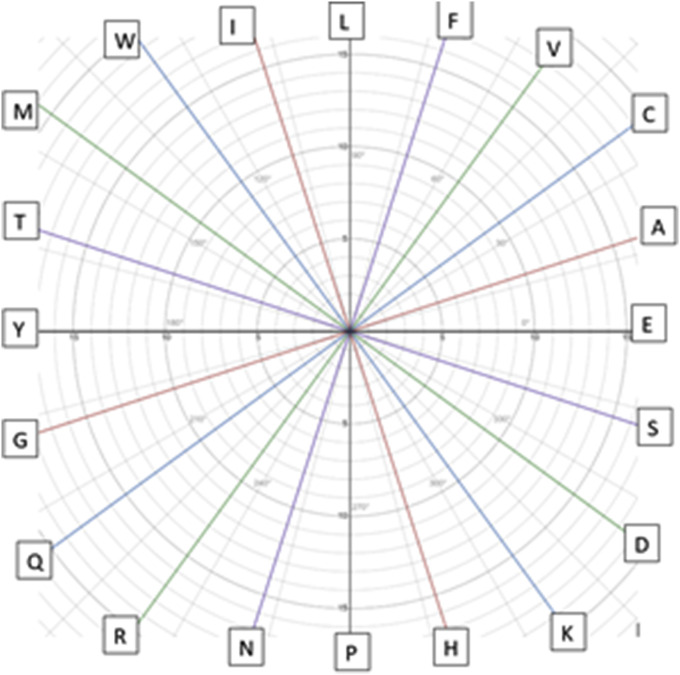
2D polar plot is an algorithm to represent amino acid sequences in 2-dimensional polar coordinate system. In this method an angle (with respect to positive axis) is assigned to each amino acid with respect to their biochemical properties. We considered the hydrophilicity index at pH 7B to assign the angles to amino acids [26]. The assigned angles are all equi-intervaled. While mapping the graph, we read the sequence starting from the origin and for each amino acid unit distance is moved to respective direction and the origin of the coordinate system is shifted to the later point. In this way a graph is drawn. A graph drawn from a sequence of length can be described mathematically as in equations (1), (2), (3)) [26].
| (1) |
| (2) |
| (3) |
In equation (1), represents the 2D polar graph which is a tuple consisting of two sets, namely (set of vertices) and (set of edges). is defined in equation (2). Starting from origin, it contains a vertex for each amino acid in the sequence. The vertex for an amino acid is determined by moving a unit distance in its angular direction from the coordinate of the last amino acid, so, and components of the angle are respectively added in and coordinate of the last vertex. In equation (3), the elements of (edge) set is defined as a tuple of two coordinate points which belong to set. So, the edge is present between those two coordinate points. All the constraints are described below equation (3).
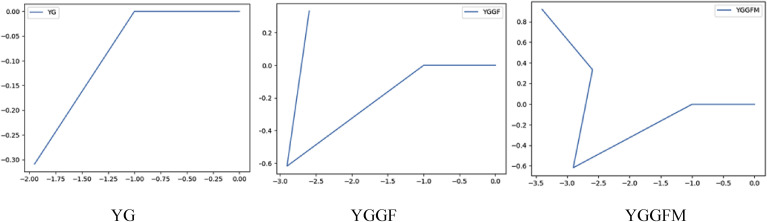
For an example, we take a small sequence of pentapeptide such as Met-enkephalin (YGGFM) and try to draw the graph from the theory explained above. According to the assignment of angles to amino acids, Y, G, F and M obtain angles respectively [26]. So, the first coordinate in the graph moving from will be , then the next one will be and in this way it will go on drawing the graph and the final coordinate will be . The step by step drawing of the graph is shown below.
3.2. characterization
characterization is an Alignment Free Sequence Descriptor (AFSD) used for characterizing the protein sequences where a numerical value is assigned to similar sequences of amino acids, which is found to be characterizing property of that sequence. Two dissimilar sequences differ by their values. In this method we draw the 2-D polar graph of the sequence and assume a unit mass to be at rest in all the vertices except at the origin. Now we calculate the centre for mass of that mass distribution. The distance of the centre of mass from the origin is defined as the value of that sequence. The mathematical definition of value of a sequence of length with a set of vertices () can be defined as in equations (4), (5)) [26].
| (4) |
| (5) |
In equation (4), represents a vertex from the set and mathematical equation for calculating is given in equation (5). Together, 2D Polar plot and characterization completes the GRANCH (Graphical Representation and Numerical Characterization) technique for specifying a protein sequence [27].
For an example, we take the previously described penta-peptide of Met-enkephalin (YGGFM). The five coordinates of the graph stand as follows.
The and . So, .
algorithm is an alignment free sequence descriptor used to visually represent amino acid sequences through graph and mathematically characterize them. In this method, the concepts of graph theory have been used to plot the sequences. On the other hand, methods like multiple alignment, as the name says, use the concept of aligning strings through dynamic programming or any other method of computer science.
In our method we assigned angles to each amino acid which lead the sequence of amino acids to different directions. Although, the angle assignment is not random. The arrangement has been specifically done in decreasing index of hydrophobicity. The more is the value of , the more hydrophobic this amino acid is. Hence, the graph having higher gradient is hydrophobic than with the lower gradient one. Mathematically it can be represented as follows.
Suppose, there are two sequences which are drawn by 2D Polar plot.
So, First Sequence contains more hydrophobic amino acids than the second sequence.
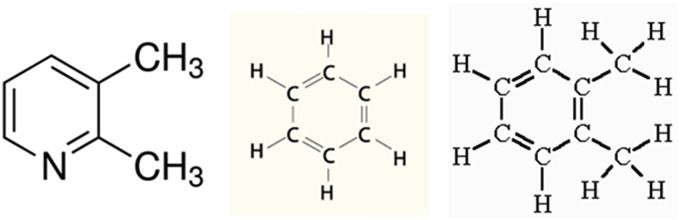
Indeed, this gives another hand of advantage over other traditional methods. In characterization the graph doesn't just give the sequence any random shape, rather it signifies about the type of the structural units. This hypothesis may be further illustrated to the point of finding surface exposure from graphs. Whereas, methods like multiple alignment doesn't use any biochemical properties to quantify the rational interpretation in visualization. is different from those, in using these concepts rather than developing a system completely based on pattern recognition. The graph itself carries information beyond general coordinate points and graph theory. Alignment-free methods use global descriptors whereas alignment-based methods use local aspects. To give a simple example, suppose we want to compare two chemicals, say benzene and Ortho xylene (see Fig. 3 ).
Fig. 3.
Organic Compounds. From the left, 2,3 - dimethyl pyridine, Benzene, Ortho-Xylene.
We can either superimpose the 6 aromatic carbons one on top of another and look at the difference between the two structures to make sense of their observed property like toxicity. Alternately, we can experimentally determine or calculate various properties of the two substances and use those or orthogonal descriptors like PCs derived from those to predict toxicity. In QSAR the method called CoMFA (comparative molecular field analysis) uses the alignment-based methods to compute intermolecular similarity. On the other hand, our group at the University of Minnesota used the second method to compute intermolecular similarity of molecules from their Euclidean distance in the n-dimensional PC space derived from the calculated properties. In our CCADD papers on Zika and SARS we used the same approach to characterize the viral sequences using PCs derived from a large number of alignment-free methods. One serious problem with the alignment-based methods in chemistry is that when the general structural form is similar, but the specific contents are different such methods cannot work.
For example, if we want to align the above pyridine derivative on the Ortho xylene structure, in one position we have C in one molecule and N in the other which are chemically quite different. But if we used alignment-free PC based method we can still calculate their intermolecular similarity. Analogously, if quite different biological sequences have similar properties, alignment-based methods may have difficulty, but alignment-free methods like our approach may still be able to characterize them.
3.3. Distribution graph with
is defined as the difference of values of two sequences. Since in this article our goal is to identify the strains that have mutated from the initial Wuhan strain, we define for two sequences as,
| (6) |
Distribution graph is a graph having the values in axis and the frequency of that value in axis. So, a point in the graph means, there are such sequences in the total set, whose value is . This graph will help us to know the presence of any mutation and to understand its spread. There can be many point mutations in a sequence, with some having lower frequency (less value), that is, there are very few sequences which have that particular value and hence those are considered insignificant. While mutated sequences having value with higher frequency, i.e. a large value indicates that it is important for further study. We know that a significant mutation which helps the virus sustain adverse conditions, should be more frequent in nature than others due to natural selection. Studies regarding the mutations in SARS-CoV-2 have been previously done on a smaller scale [28].
3.4. Data retrieval
Full GenBank and FASTA data files of sequences of various proteins of SARS-CoV-2 have been retrieved from National Centre for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) [29].Overall 103,245 sequences were retrieved and analyzed. They included Full Genome sequences (9199), Spike Glycoprotein sequences (9525), Nucleocapsid sequences (9581), Membrane Glycoprotein sequences (9486), Envelope Protein sequences (9571) and the open-reading frames (55,883).
3.5. Sequence alignment
The sequences obtained from NCBI database were aligned with the help of MEGA X software [30].
3.6. Protein stability calculation
The change in stability of the protein on single point mutations was calculated by online based software, iMutant 2.0 [31].
3.7. Proteolytic site prediction
We used PROSPER (Protease Specificity Prediction Server) [32] for in silico identification of proteolytic sites in the spike protein.
3.8. Temporal graph
In another study, we computed a temporal graph, where for each value we computed its presence in the collected sample set with respect to time in days. The graph reveals about the origin of a strain and its growth helping one to estimate its probable fate. A point on the graph refers to that, samples of a particular strain were collected on the day .
3.9. Hotspot regions
Hotspot regions on a genome refer to the zones where mutations are most probable. As in our study we are analyzing a large number of sequences, the most occurring changes can be easily identified. The result is shown in the form of bar graph, where a bar with centre at refer to the amino acid position and height refer to the number of changes occurred.
3.10. Computational work
All the necessary Computational Works have been performed through Python 3.8 Programming Language (https://www.python.org/) and GNU Octave 5.2 Programming Language (https://www.gnu.org/software/octave/). We used Google Collab (https://colab.research.google.com/) as Cloud Host Kernel and also used Jupiter Notebook (https://jupyter.org/) for local running.
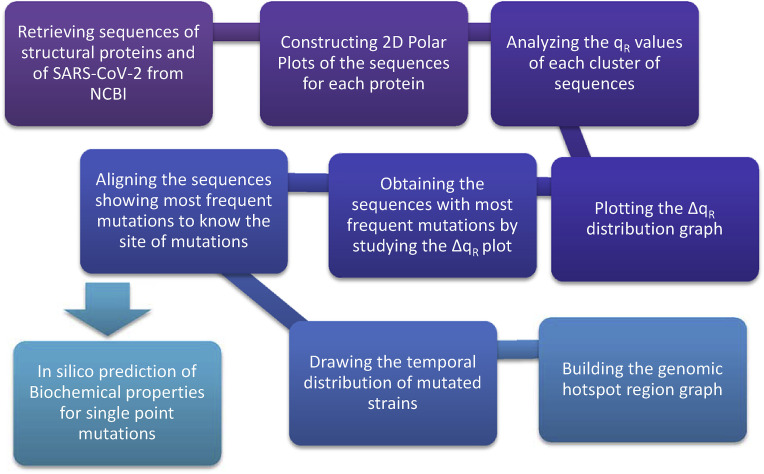
A schematic flow chart of our work is given below.
4. Results
4.1. Spike glycoprotein
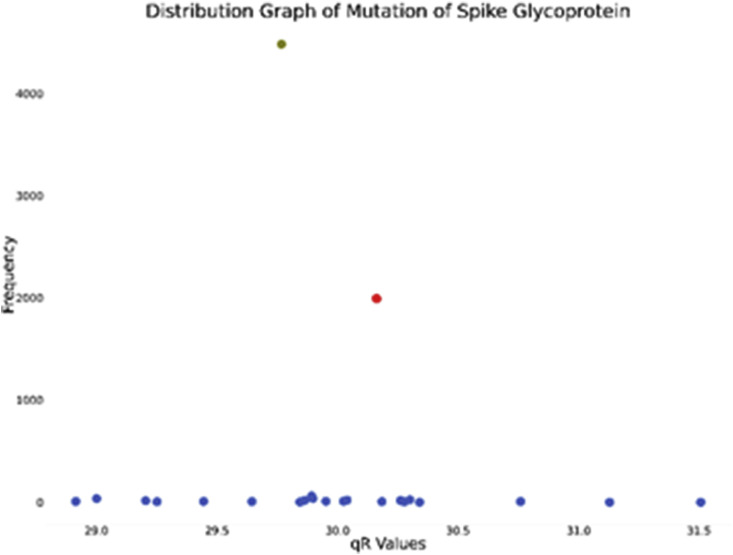
The Spike glycoprotein helps the virus to attach with ACE2 and TMPRSS2. It is of length 1273 amino acids. We collected 9525 Spike glycoprotein sequences out of which 7455 sequences were complete with no missing or unknown amino acids. The initial Wuhan sequence (YP_009724390) had value 30.16105055. We plotted the distribution graph to find out the significant mutations.
A perusal of Fig. 4 indicates that there is only one such significant mutation. We further proceeded to identify that mutation and find its properties.
Fig. 4.
Distribution graph of qR values of SARS-CoV-2 Spike Glycoprotein (red dot and green dot signifies wild protein and mutated protein respectively).
It is evident that the mutated protein is more prevalent than the wild one (see Table 1). To detect the mutation, we aligned the sequences in MEGA-X whose results are given in Table 2 .
Table 1.
Frequency and accession id of Spike glycoprotein mutated clusters.
| Accession Id | value | Frequency |
|---|---|---|
| YP_009724390 | 30.16105055 | 1993 |
| QLI46289 | 29.76636445 | 4482 |
Table 2.
Mutational change in spike glycoprotein clusters.
| Accession Id | Amino Acid Site | Amino Acid in Wild strain | Mutated Amino Acid |
|---|---|---|---|
| QLI46289 (D614G) | 614 | D | G |
Thermodynamics plays an important part in protein stability, protein folding and its activities inside host cell [33]. Eventually, the change in the free energy of a protein, the ΔΔG value plays quite a significant role in determining the change in stability of a protein on mutation. We calculated the ΔΔG value for the mutation at position 614 of the spike protein where ΔΔG = ΔGmutated – ΔGwild. The pH was taken 7.40 as it is the average pH of healthy lungs [34]. The obtained result is given in Table 3 .
Table 3.
ΔΔG value calculation.
| pH | Temperature | ΔΔG |
|---|---|---|
| 7.40 | 298.0K | −0.94 kcal/mol |
We also analyzed the protein sequence for protease reactions which may guide us to understand the reason for the widespread of the mutated spike protein. The results are as given in Table 4 .
Table 4.
Analysis of protease cleavage sites in SARS-CoV2 spike proteins at the vicinity of the mutated amino acid (the bar indicates the site of proteolysis).
| Protein | Enzyme | Site | Segments |
|---|---|---|---|
| Wild | Cathepsin G | 612 | AVLY|QGVN |
| D614G | Cathepsin G | 612 | AVLY|QGVN |
| Elastase 2 | 615 | YQGV|NCTE |
So we can see that due to the mutation at site 614 of the spike protein, a novel Elastase 2 or Neutrophil Elastase protease site has developed in between sites 615 and 616.
4.1.1. Temporal study
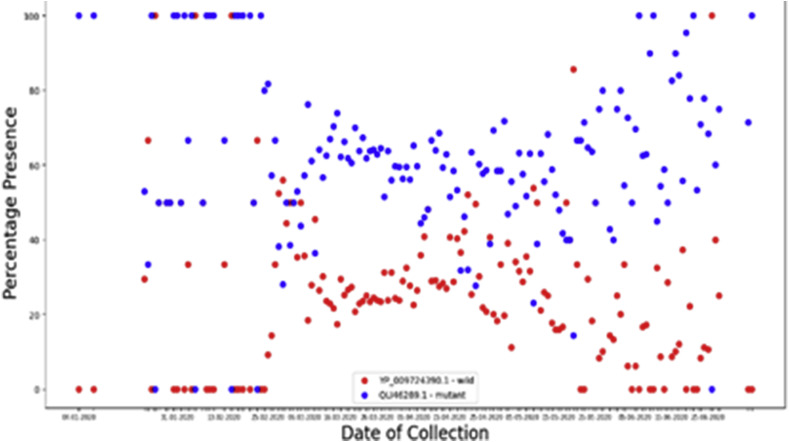
We plotted the percentage-presence of the mutated and wild protein w.r.t. time, which is shown in Fig. 5 .
Fig. 5.
Comparison of Evolution of Mutated D614G Spike Glycoproteins with wild strain spike glycoprotein with respect to time (wild strain in red and mutated strain in blue).
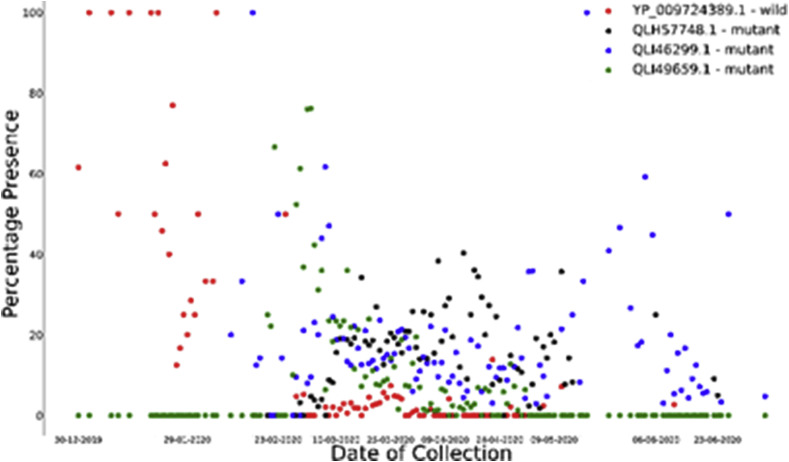
We see that with time, the two proteins evolved simultaneously. The D614G strain evolved quite soon after the first outbreak in China. The first D614G was collected on 4th January 2020 from Thailand. Thus, comparing the graph, we can infer that the point mutation at site 614 is more favorable for the virus. The graph indicates about the possibility of high infectivity of this protein over the wild one. Also, the graph looks quite symmetric.
Along with Temporal study, we also performed demographic analysis of Spike Protein. This analysis enabled us to interpret the transmission of D614G strain in various countries and its dominance over wild strain. The result of this study is shown in Table 5 .
Table 5.
Demographic analysis of spread of D614G Spike protein and first date of collection.
| Sl. No. | Country | % Wild | % D614G | First Appearance of D614G |
|---|---|---|---|---|
| 1 | USA | 27.32% | 58.99% | 20.02.2020 |
| 2 | India | 23.78% | 59.59% | 11.03.2020 |
| 3 | China | 2.63% | 23.68% | 22.01.2020 |
| 4 | Russia | 25% | 62.5% | 18.03.2020 |
| 5 | Italy | 27.27% | 54.54% | 01.03.2020 |
| 6 | France | 14.63% | 76.82% | March 2020 |
We have gone through the demographic study of SARS-CoV-2 D614G strain. It shows that most of the highly infected countries encountered a very early exposure to this mutated strain. Indeed, which resulted into a higher percentage of the same. The results also ensure that D614G is not limited to a particular geographical region or country or continent. It has spread all over the world with sheer dominance over the wild protein. The table confirms a very early detection of D614G in China, although the very first one is not from China. The first D614G strain was collected from Thailand on 4th January 2020, with accession ID QJX59860.
4.1.2. Genome hotspot region
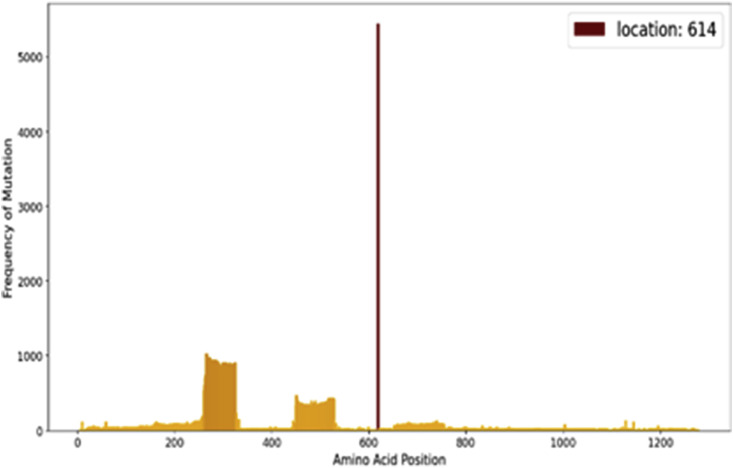
We analyzed the sequences to search for regions inthe gene which can be hotspots for mutation. In this graph, a bar on location refers to amino acid position and its height refers to the number of mutational changes at that site. The graph is show in Fig. 6 .
Fig. 6.
Mutational hotspots in SARS-CoV-2 spike glycoprotein.
Although some low frequent mutations are observed around sites 300 and 500 which lie in the spike Receptor Binding Domain (RBD), most of the sequences showed mutation at site 614. Indeed, the SARS-CoV-2 spike glycoprotein is changing mostly in this location of its protein.
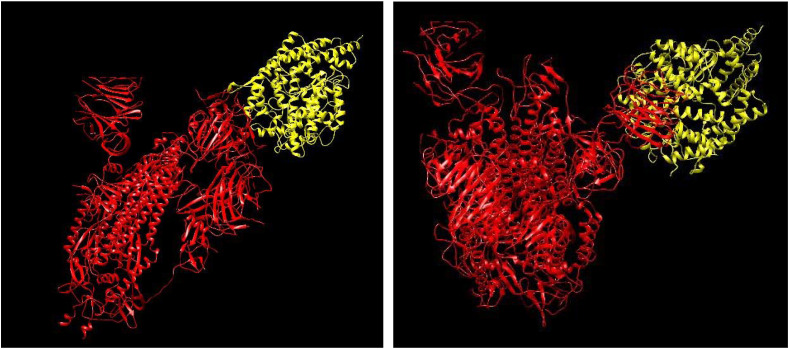
We have included the docking results to make a comparative study of the interaction energy between the host proteins and the SARS-CoV-2 viral proteins. That is to check whether the mutation helps the protein to bind more efficiently and hence becomes potentially more infectious. In this regard, the spike glycoprotein becomes the most important viral protein as it is involved in direct contact with the host ACE-2 receptor which helps the virus to enter the human cells. We searched for the 3-dimensional structures for both the wild and mutated variety of the spike glycoprotein in the protein data bank. Though we found the 3-dimensional structure for wild spike protein, unfortunately the complete structure of the mutated D614G spike protein was unavailable. The only structure available lacked the receptor binding domain (PDB ID: 6XS6). Considering the fact that the receptor binding domain in the spike protein plays the key role in the binding of human ACE-2 and viral spike protein, we are afraid that docking the ACE-2 with an incomplete spike protein will not give us the correct results. Thus, the ultimate goal of comparison remains unfulfilled due to the lack of protein structures in the database. Although, there is incompleteness in the pdb files, we performed the blind docking and the results with the binding energy are shown in the table. The atomic energies and docking structure are given in Table 6 and Fig. 7 respectively.
Table 6.
Result of blind docking of wild and mutated spike protein with ACE2.
| Sl. No. | Receptor Protein | Ligand Protein | Atomic Contact Energy (ACE) | Global Energy |
|---|---|---|---|---|
| 1 | ACE2 | Wild Spike Protein | 165.83 | 0.17 |
| 2 | ACE2 | Mutant Spike Protein (D614G) | 110.81 | −2.79 |
Fig. 7.
Docking Image of Spike Protein (in Red) with ACE2 (in yellow). Left image is of Wild Protein Docking and Right Image is o mutated protein Docking.
4.2. Nucleocapsid phosphoprotein
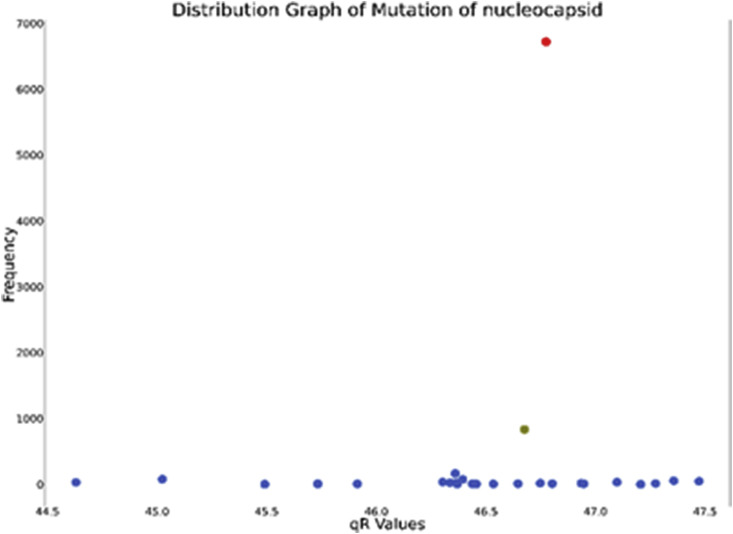
Nucleocapsid which forms the core of the nucleocapsid Phosphoprotein helps in genomic RNA binding and capsid formation. There were 9581 sequences of nucleocapsid phosphoprotein out of which 8689 sequences were complete. The initial Wuhan protein (YP_009724397) had value 46.77401554. The distribution graph is shown in Fig. 8 .
Fig. 8.
Distribution graph of qR values of Nucleocapsid Phosphoprotein (wild strain is shown in red and mutated strain in green).
Although the only notable mutated cluster shows lower frequency, we still considered to highlight it because of its trends that we found from the time span graph.
The notable clusters were identified (see Table 7). Furthermore, we identified the mutations of these two proteins.
Table 7.
Frequency and accession id of nucleocapsid phosphoprotein mutated clusters.
| Accession ID | value | Frequency |
|---|---|---|
| YP_009724397 | 46.77401554 | 6719 |
| QLI46309 | 46.67568301 | 833 |
Here we see, two consecutive locations have changed by mutation.
4.2.1. Temporal study
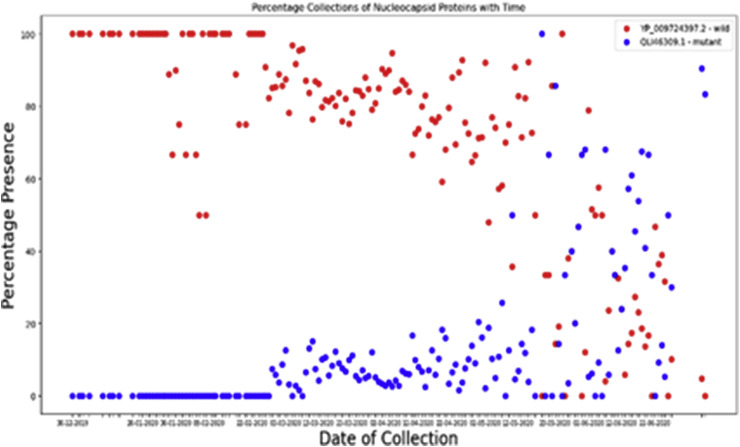
We see in Fig. 9 that, initially the wild protein was prevalent. But from the beginning of March 2020, the mutated protein has started to grow. After an initial lag phase, the growth has now become exponential. Studying the mutated protein growth curves from other cases, we infer this as anindication of future growth .
Fig. 9.
Comparison of evolution of Mutated Nucleocapsid Phosphoprotein and Wild Nucleocapsid Phosphoprotein with respect to time (wild strain in red and mutated strain in green).
4.2.2. Genome hotspot region
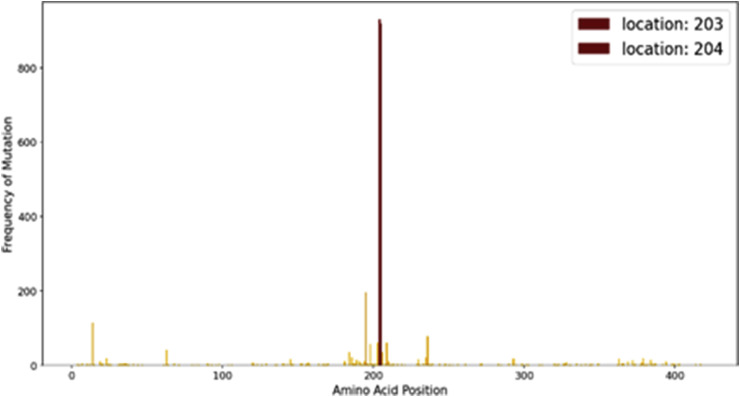
The mutational hotspot graph given here shows that the region around site 200 is more prone to mutations in the gene. This region falls in the core nucleocapsid protein and hence might be responsible for some novel traits in the virus which can be further analyzed in wet lab experiments. (refer to Fig. 10 )
Fig. 10.
Mutational hotspots in SARS-CoV-2 nucleocapsid phosphoprotein.
4.3. Envelope protein
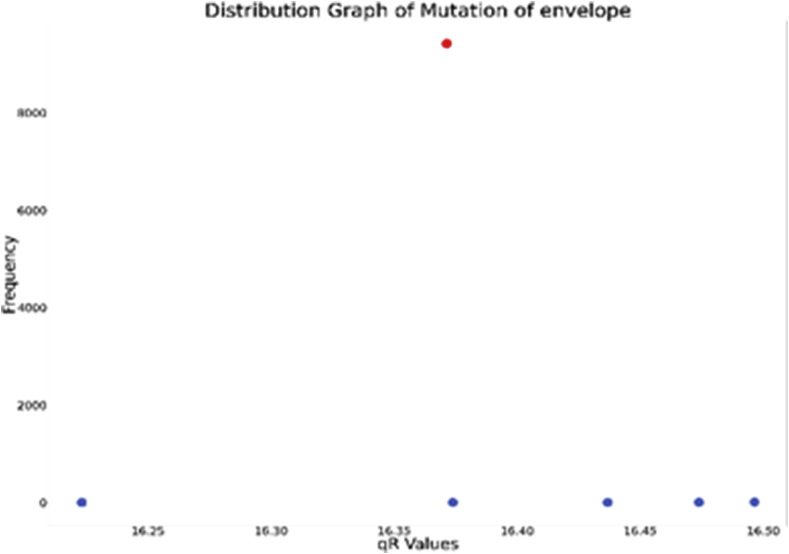
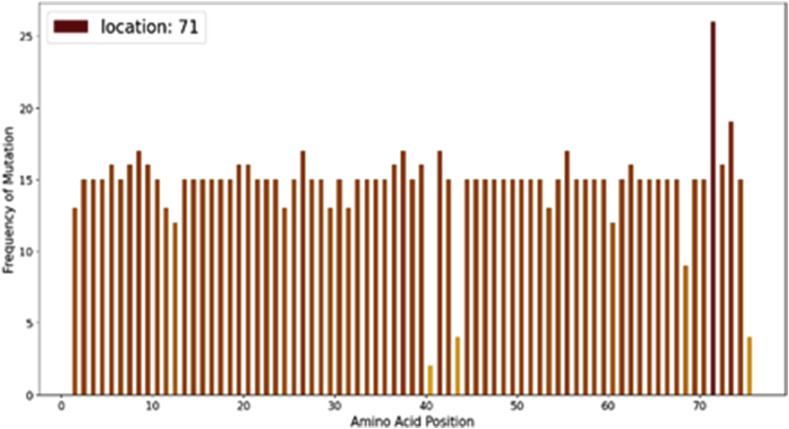
The envelope protein is a hydrophobic protein formed by 75 amino acids. We collected 9571 sequences of Envelope protein out of which 9481 were complete. The initial protein obtained from Wuhan (YP_009724392) has value 16.37137772.
We see in Fig. 11 that no significant mutations are observable. Although some point mutations exist, it has significantly less frequency.
Fig. 11.
Distribution graph of qR values of Envelope protein.
4.3.1. Genome hotspot region
Plotting the hotspot graph we ensure the possibilities of mutation at various locations.
We see in Fig. 12 that the range of mutation at various sites is very less, only about 0.26% of the total no of sequences hence these mutations are not deemed to be highly important for further study .
Fig. 12.
Mutational hotspots in SARS-CoV-2 envelope protein.
4.4. Membrane glycoprotein
It is a SARS-CoV-2 structural protein of length 222. We collected 9486 sequences out of which 9231 were complete. The protein collected from Wuhan (YP_009724393) had value 31.53950918.
Here also we see some point mutations but with very less frequency to be considered as a significant one.
4.4.1. Genome hotspot region
Fig. 14 clearly depicts that some locations in the protein have alterations in amino acid, but it merely covers 0.27% of the total number of sequences collected.
Fig. 14.
Mutational hotspots in SARS-CoV-2 membrane glycoprotein.
4.5. Orf 1 ab
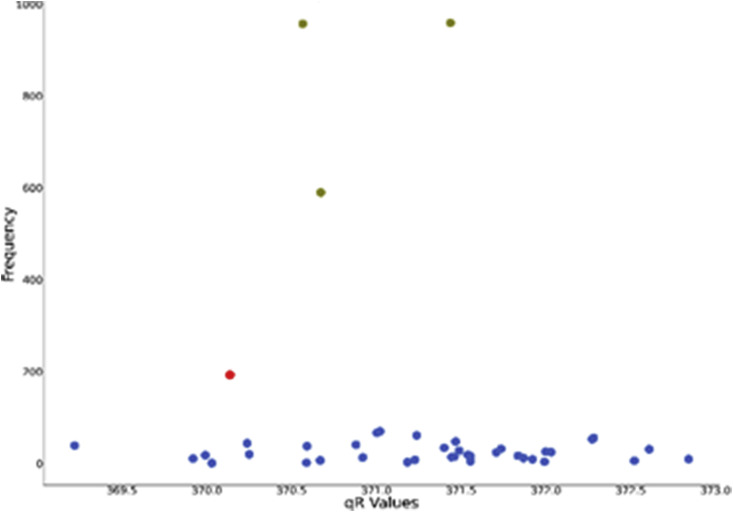
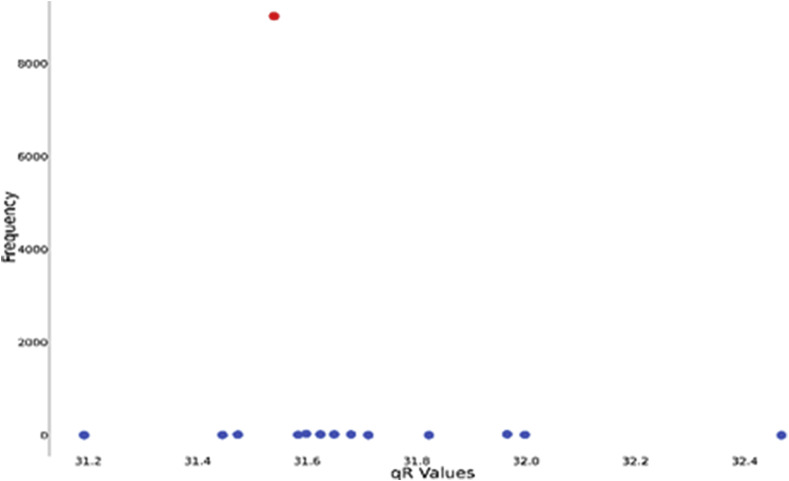
A total of 9199 sequences were retrieved from NCBI Database out of which 6394 sequences were complete. We computed value of all of them and plotted the distribution graph. A point in the distribution graph represents that there are such sequences which have value of . The initial Wuhan strain, (YP_009724389.1) had value 370.1371472 and is represented with red point in Fig. 2. A sequence having a mutation or a change in amino acid from wild strain is expected to have a different value from wild strain. The mutations that help the virus sustain adverse conditions are expected to be present in large numbers due to natural selection. So, our goal through this graph is to identify large mutated clusters having high values.
Fig. 2.
Arrangement of Amino Acid in various angles. Visual Organisation of 2D Polar Plot Algorithm.
Observing Fig. 15 , we can identify the three points with olive colour having much higher frequency than the wild strain. Clearly, those mutations have helped the virus sustain adverse conditions, resulting in having much more abundance in society. The frequencies and accession ids of one of such sequences are given in Table 9 .
Fig. 15.
Distribution graph of qR values of Orf 1 ab.
Table 9.
Frequency and accession id of full genome mutated clusters.
| Accession ID | Value | Frequency |
|---|---|---|
| YP_009724389 | 370.1371472 | 189 |
| QLH57748 | 371.4376792 | 959 |
| QLI46299 | 370.5669977 | 957 |
| QLI49659 | 370.6726746 | 590 |
These three sequences were aligned in Mega-X and the mutations are identified which is given in Table 10 .
Table 10.
Mutational change in full genome clusters.
4.5.1. Temporal study
To understand the evolution of the strains we plotted them against time, starting from the initial collection date, 23rd December 2019. The graph is shown in Fig. 16 .
Fig. 16.
Comparison of evolution of Mutated orf 1 ab polyproteins and wild orf1ab polyprotein with respect to time (wild strain in red and other are mutated ones).
From Fig. 13, we can find out how the wild protein (in red) which was more widespread at the beginning eventually becomes less predominant and now is almost on the verge of extinction. The third strain (in green) has reached its peak and is less dominant now. But the visual observations seem to indicate that the second strain (in blue) is very much prevalent right now and growing. The first strain (in black) seems to be fluctuating. Observing the graph, we can see that in between March and April 2020 the presence of all the protein strains have been fluctuating so much as if to filter out the more effective one. So, in this time span, it seems that the orf1ab genome had undergone several recombination to achieve viability.
Fig. 13.
Distribution graph of qR values of Membrane Glycoprotein.
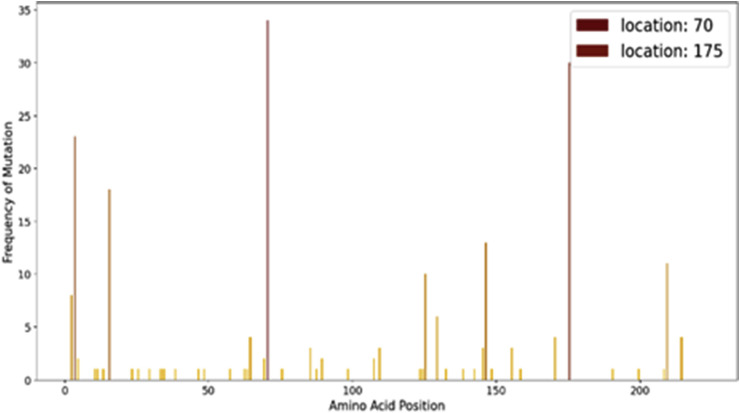
4.5.2. Genome hotspot region
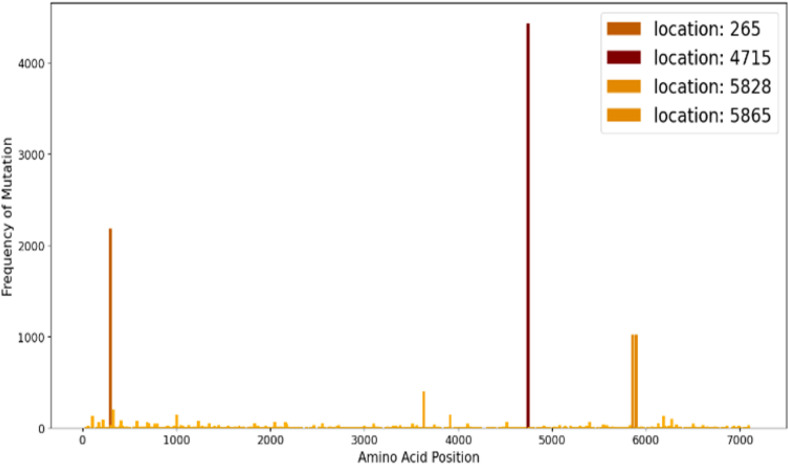
In another study we computed the frequency of mutation in various sites across the genome to identify the hotspot regions. Fig. 17 , depicts it clearly about the most mutatively active sites in the full genome..
Fig. 17.
Mutational Hotspots in SARS-CoV-2 orf 1 ab.
5. Discussions
In this paper we have used various approaches to analyze the sequences of different proteins, starting from identifying their mutations, to understand their growth and determining the mutation-hotspot regions of each protein. This has been a collective organization of discrete studies on those proteins from which we try to draw cumulative inferences about the mutations in SARS-CoV-2.
-
•
We performed the detailed stability check of the various point mutations described above. Through the web-based software i-Mutant [35], we calculated the values for each point mutation. A positive value () indicates decrease in stability while a negative indicates increase in stability. The table is given below. We computed each in and , which is optimum for respiratory tract. The results of i-Mutant study are shown in Table 11 .
Table 11.
ΔΔG values of all the mutations and their result in stability.
| Protein | Amino acid Position | Wild Amino Acid | Mutant Amino Acid | (Kcal/mol) | Stability |
|---|---|---|---|---|---|
| Spike | 614 | D | G | −1.76 | Increase |
| Nucleocapsid | 203 | R | K | −2.16 | Increase |
| 204 | G | R | +0.01 | Decrease | |
| Orf1ab | 265 | T | I | −1.68 | Increase |
| 4715 | P | L | −0.77 | Increase | |
| 5828 | P | L | +0.19 | Decrease | |
| 5865 | Y | C | +0.79 | Decrease |
In this regard, we computed the values for each point mutation at and , which is optimum for respiratory tract. But iMutant can only check the stability for mutations at a single amino acid, which is a valid case for spike glycoprotein and we had already mentioned about it in our paper. For others, mutations are happening at more than one position and iMutant is unable to compare their stability. Other software like DyanMut requires 3-dimensional structures of a protein which are either not available in database or are incomplete for the mutated SARS CoV −2 proteins. In this regard we think it is better not to incorporate the table of the stability changes that are happening due to mutations at several points in a protein to avoid inconsistency of bioinformatic and biochemical studies.
-
•
RNA viruses have lower replication accuracy than DNA viruses due to lack of proof-reading capabilities of RNA Polymerases. But as observed here, the mutation rate in COVID-19 is quite lower compared to other RNA viruses. The possible reason can be the size of the genome. As pointed out by Rafael Sanjuán et al., RNA viruses of family Coronaviridae which have the largest genome mutate slowly compared to other RNA viruses [36].
-
•
A significant mutation is observed in the Spike glycoprotein at position 614, where an aspartic acid (polar) is changed to a glycine (non-polar). The mutation is in fact destabilizing the native spike protein (having both the S1–S2 domains) which may eventually influence its cleavage. This mutation lies close to the S1–S2 junction of the spike protein. We found out that the point mutation has developed an additional cleavage site for elastase 2. From previous studies on coronavirus, it has been found that proteolysis at several points of the spike glycoprotein is essential for its entry inside the cell [37]. So, we can conclude from the data that the generation of a novel protease site at the vicinity of the S1–S2 junction has helped the virus enter the host cell more efficiently. This data indicate that the mutated protein may be increasing the potential of the virus to attach with host receptors and undergo cleavage. The temporal graph also revealed how the mutated protein gained dominance over the wild protein gradually with time. Indeed, this mutation is growing at a very fast rate and is obviously more infectious than the wild strain, which is in fact the reason of its higher presence in samples. Also, the genome hotspot analysis showed some less-frequent mutations around location 300 and 500. Besides the more-frequent missense mutation at 614, the ones that have occurred around positions 300 and 500 can also affect the receptor-spike attachment as it falls inside and near the receptor binding domain (RBD). As a conclusive remark from our studies on Spike glycoprotein, it indeed tells us about the significance of the mutations. A detailed study of effect of D614G mutation is described in the following articles [38,39].
-
•
In Nucleocapsid Phosphoprotein, two significant mutations have been observed successively at locations 203 and 204 which fall in the core nucleocapsid protein region. In those locations an Arginine (polar) is replaced by a Lysine (polar) and a Glycine (non-polar) is replaced by an Arginine (polar) respectively. It is conspicuous that the protein has accumulated a greater positive charge due to these mutations. Electrostatic interactions between the capsid proteins and the viral nucleic acid play a crucial role in viral biology. The positive charges in structural proteins, like the capsid protein plays a vital role in virion stability by neutralizing the negative charges in phosphate (PO4 2−) of viral nuclear material (RNA in case of SARS-CoV-2) [40]. The temporal graph reveals that the mutated protein is still in its growing phase. The mutational hotspot graph reveals that no other site in the nucleocapsid gene has been frequently mutated other than 203 and 204.
-
•
In case of Envelope and Membrane glycoprotein no such significant mutations are observed. Although temporal graphs show some mutations, but they have too low frequency to be considered important for further studies.
-
•
So, from observing the pattern in mutation of the structural proteins (Spike and Nucleocapsid) it is seen that the protein is gradually tending to accumulate more positive charges as compared to the wild strain. In spike protein, negatively charged aspartic acid is replaced by uncharged Glycine and similarly for nucleocapsid, where uncharged Glycine is mutated to positive charged Arginine. This is probably to attain more overall stability of the virion [40]. The combined data obtained from the genome hotspot analysis of all the structural proteins reveals that a directed mutagenesis has been occurring in them. This response can be probably due to selective pressure [41].
-
•
Mutations in orf 1 ab, which translates polyprotein 1 ab, give us an idea of mutational changes occurring in non-structural proteins (nsp1 – nsp16). The hotspot graph reveals about the four most active mutation-sites in the polyprotein; details of which are given in Table 8. Nsp2 is a non-structural protein which binds to the host cell Prohibitin 1 (PHB1) and its homolog Prohibitin 2(PHB 2) and disrupts the host cell-signaling pathway in SARS-CoV infection [42]; it might play similar roles in COVID-19 infection too and so an extensive study on this mutation becomes necessary. The mutations which are occurring in Helicase and RNA Dependent RNA Polymerase might affect the viral genome replication rate and its life cycle. Rdrp might be a target protein for therapeutics [43] and hence its mutations should be studied extensively.
Table 8.
Mutational change in nucleocapsid phosphoprotein clusters.
| Accession ID | Amino Acid Site | Amino Acid in Wild strain | Mutated Amino Acid |
|---|---|---|---|
| QLI46309 (L84S) | 203 | R | K |
| 204 | G | R |
6. Conclusion
We surveyed the genome of SARS-CoV-2 for mutations prevalent around the world. From our study we can conclude that mutations in the proteins of SARS-CoV-2 are slow yet steady. We observed how the wild and mutated spike proteins tussled with each other and ultimately the mutated protein became more widespread. From it we can conclude about the similar fate of nucleocapsid wild and mutated proteins. We further suggest wet lab studies which may reveal various important information regarding their properties and role in coronavirus life cycle.
A detailed documentation on the mutations in the viral enzymes, like RNA-Dependent RNA Polymerase [44], viral Main Protease (Mpro) [45] et cetera can also help researchers to identify potential drug candidates that can inhibit their functions. The tremendous importance of these enzymes in the life cycle of SARS-CoV-2 can make therapeutics targeted against them valuable to stop further spread of the virus.
The analysis of mutation in the SARS-CoV-2 will help us understand the genetics of coronaviruses. It can also be a path to understand the evolutionary linkage between RNA and DNA based organisms [46]. Thus, mutation, which plays one of the most important roles in progression of organisms and life itself, from simple to complex, becomes perhaps one of the most important fields to be studied in order to combat the virus and save millions of human lives worldwide.
A thorough study of the mutations that have occurred in various proteins encoded by the SARS-CoV-2 genome can also help researchers and medical personnel in designing suitable drugs and other therapeutics. Designing alternative vaccine strategies like peptide vaccines and mRNA vaccine can be boosted by this study as targeting the conserved regions of the proteins can only be done if one has sound knowledge regarding the mutation hot-spots. Computer-aided drug designing can also be improved with the help of this study. An advanced study correlating COVID-19 symptoms with subtle mutational changes can also be undertaken which will help us understand the virus better.
An appendix is given at the bottom to show the list of all mutations according to their position.
7. Data in brief
The data that we have used for our study has been deposited in GitHub Repository https://github.com/cire-org/Identification-and-Computational-Analysis-of-Mutations-in-SARS-CoV-2-.
Authors’ contribution
TD performed all the computational tasks and analysis regarding sequences and identified the different cluster of mutation. SC conducted various bioinformatic studies and performed the alignments to identify the points of mutation and interpreted their significance. SM assisted in the writeup and AN and SCB guided the overall concepts.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Declaration of competing interest
The Authors declare no conflict of interest.
Biographies

Tathagata Dey, is currently pursuing B. Tech in Computer Science and Engineering from West Bengal University of Technology, Kolkata and a full-time researcher at Centre for Interdisciplinary Research and Education, Kolkata, (CIRE). He is specialised in Data Science including Machine Learning and Artificial Deep Learning Models. He possesses research interests in the field of Bioinformatics and Computational Biology, Computer Science Algorithms, Theoretical Physics and Philosophy of Science. He is also a JBNSTS Senior Scholar of batch 2018. He passed his high school from Uttarpara Govt. High School in 2018.
E-mail: tathagata2403@gmail.com

Shreyans Chatterjee, currently pursuing BSc. Microbiology Hons. at St. Xavier's College (Autonomous), Kolkata is a full-time researcher at CIRE. He passed his 10th from Ramakrishna Mission Vidyalaya, Narendrapur in 2016 and his 12th from the same school in 2018 securing state rank 9th and 7th respectively. He currently works in bioinformatics. He is also a JBNSTS and an INSPIRE Scholar.
E-mail: shreyansrkmv@gmail.com

Dr. Smarajit Manna, currently working as Student Advisor at Jagadis Bose National Science Talent Search, Kolkata, India, did his B.Sc and M.Sc in Physics. He received his Ph.D. degree from Jadavpur University, Kolkata and the title of his thesis was “A study of electrical properties of bilayer lipid membranes and the dynamics of ion channels incorporated in them”. His research interests include Statistical analysis of dynamical systems, Material Science and Bio-informatics. Dr. Manna has various research publications in these fields in national and international journals. He is one of the authors of two book chapters “Electrical noise in cells, membranes and neurons in: Understanding Complex Systems", Springer Berlin/ Heidelberg, 255–267, 2009 and “Computational Methodology for Peptide Vaccine Design for Zika Virus: A Bioinformatics Approach” Namrata Tomar (ed.), Immunoinformatics, Methods in Molecular Biology, vol. 2131, https://doi.org/10.1007/978-1-0716-0389-5_2, © Springer Science + Business Media, LLC, part of Springer Nature 2020.
Email: man_smarajit@yahoo.com

Dr Ashesh Nandy did his PhD in theoretical Physics and later switched to molecular biology. He was one of the pioneers in the new field of graphical representations of biomolecular sequences and study of sequence characteristics, sequence analysis, sequence homologies, molecular phylogeny, etc. His group also developed mathematical frameworks for quantitative assessment of the graphical plots and indexes of DNA/RNA/protein sequences for quantitative sequence comparison. He has edited books and journals and published widely in national and international journals, most recently on peptide vaccine designs.

Dr. Subhash C. Basak is currently an Adjunct Professor in the Department of Chemistry and Biochemistry, University of Minnesota Duluth, USA. He received his PhD in biochemistry in 1981 from the university of Calcutta, India. His current research interests involve discrete mathematical chemistry and its applications to chemoinformatics, bioinformatics, quantitative structure-activity relationship (QSAR), computational toxicology, mathematical quantification of DNA/ RNA sequences, mathematical proteomics, and computer aided vaccine design for emerging pathogens like Zika virus, COVID-19. Dr. Basak is the Editor-in-chief of the international journal Current Computer Aided Drug Design. He was involved in editing three books: 1) Statistical and Machine Learning Approaches for Network Analysis, Wiley, 2012; 2)Advances in Mathematical Chemistry and Applications, Volume 1 & 2, Elsevier & Bentham Science Publishers, 2015; 3) Zika virus: Basic biology, surveillance, vaccine design and anti-Zika drug discovery: Computer assisted strategies to combat the menace, Nova, 2019. Dr. Basak has authored more than 335 papers and book chapters (H index- 53).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.104166.
Appendix.
Here, in the following table, the position wise mutations are given. The information refers to that, the mutant amino acid is whose frequency of mutation is .
| Position | Wild Strain AA | Mutant AA wit Frequency | ||
|---|---|---|---|---|
| 2 | F | L, 2 | ||
| 5 | L | F, 65 | ||
| 7 | L | V, 1 | ||
| 12 | S | F, 3 | C, 1 | |
| 13 | S | I, 3 | ||
| 14 | Q | H, 10 | ||
| 17 | N | K, 1 | ||
| 18 | L | F, 3 | ||
| 21 | R | I, 1 | ||
| 22 | T | I, 5 | N, 2 | A, 1 |
| 25 | P | L, 2 | S, 2 | |
| 26 | P | L, 2 | S, 1 | |
| 27 | A | S, 1 | ||
| 28 | Y | H, 1 | ||
| 29 | T | I, 7 | ||
| 32 | F | L, 3 | ||
| 35 | G | V, 1 | ||
| 38 | Y | C, 1 | ||
| 49 | H | Y, 10 | ||
| 50 | S | L, 6 | ||
| 54 | L | F, 72 | ||
| 67 | A | V, 1 | ||
| 69 | H | Y, 2 | ||
| 71 | S | F, 2 | ||
| 72 | G | V, 1 | ||
| 75 | G | V, 3 | ||
| 76 | T | I, 4 | ||
| 78 | R | M, 10 | ||
| 86 | F | S, 1 | ||
| 88 | D | Y, 2 | A, 1 | |
| 95 | T | I, 8 | ||
| 96 | E | G, 1 | ||
| 97 | K | T, 1 | ||
| 98 | S | F, 3 | ||
| 102 | R | I, 1 | ||
| 111 | D | N, 1 | ||
| 127 | V | F, 1 | ||
| 132 | E | D, 1 | ||
| 138 | D | H, 20 | ||
| 142 | G | V, 1 | ||
| 145 | Y | H, 2 | ||
| 146 | H | Y, 21 | ||
| 148 | N | S, 1 | Y, 1 | |
| 151 | S | I, 1 | G, 1 | |
| 152 | W | L, 2 | ||
| 153 | M | I, 7 | ||
| 155 | S | I, 2 | ||
| 156 | E | D, 2 | ||
| 157 | F | L, 1 | ||
| 158 | R | S, 1 | ||
| 162 | S | I, 1 | ||
| 173 | Q | H, 1 | ||
| 176 | L | I, 1 | F, 1 | |
| 177 | M | I, 2 | ||
| 178 | D | N, 1 | ||
| 180 | E | K, 2 | ||
| 181 | G | A, 2 | ||
| 185 | N | K, 1 | ||
| 188 | N | K, 1 | D, 1 | |
| 190 | R | K, 1 | ||
| 197 | I | V, 2 | ||
| 203 | I | M, 3 | ||
| 211 | N | Y, 2 | ||
| 213 | V | L, 1 | ||
| 214 | R | L, 2 | ||
| 216 | L | F, 6 | ||
| 218 | Q | L, 1 | ||
| 220 | F | L, 9 | ||
| 221 | S | L, 22 | ||
| 222 | A | V, 1 | P, 1 | |
| 240 | T | I, 1 | ||
| 242 | L | F, 1 | ||
| 243 | A | V, 1 | ||
| 245 | H | R, 1 | ||
| 248 | Y | H, 1 | ||
| 252 | G | S, 1 | ||
| 253 | D | G, 17 | ||
| 254 | S | F, 2 | ||
| 255 | S | F, 3 | ||
| 258 | W | L, 4 | ||
| 261 | G | R, 1 | V, 1 | D, 4 |
| 262 | A | S, 4 | T, 4 | |
| 265 | Y | C, 1 | ||
| 267 | V | L, 1 | ||
| 273 | R | S, 2 | G, 1 | |
| 279 | Y | N, 1 | ||
| 288 | A | T, 1 | ||
| 289 | V | I, 1 | ||
| 301 | C | F, 1 | ||
| 307 | T | I, 2 | ||
| 308 | V | L, 8 | ||
| 309 | E | Q, 1 | ||
| 314 | Q | K, 1 | L, 1 | R, 1 |
| 315 | T | I, 1 | ||
| 321 | Q | L, 1 | ||
| 323 | T | I, 1 | ||
| 330 | P | S, 2 | ||
| 345 | T | S, 1 | ||
| 348 | A | S, 1 | ||
| 354 | N | K, 1 | ||
| 367 | V | F, 4 | ||
| 379 | C | F, 1 | ||
| 382 | V | L, 1 | E, 1 | |
| 384 | P | L, 2 | ||
| 393 | T | P, 1 | ||
| 403 | R | K, 8 | ||
| 408 | R | I, 2 | ||
| 441 | L | I, 1 | ||
| 453 | Y | F, 5 | ||
| 457 | R | K, 1 | ||
| 458 | K | Q, 1 | ||
| 471 | E | Q, 1 | ||
| 476 | G | S, 2 | ||
| 477 | S | N, 37 | G, 1 | |
| 479 | P | L, 1 | ||
| 483 | V | F, 1 | A, 13 | |
| 485 | G | R, 1 | ||
| 486 | F | L, 1 | ||
| 501 | N | Y, 13 | T, 1 | |
| 518 | L | I, 3 | ||
| 519 | H | Q, 1 | ||
| 520 | A | S, 4 | ||
| 522 | A | V, 1 | ||
| 547 | T | I, 4 | ||
| 553 | T | N, 1 | I, 2 | |
| 554 | E | D, 14 | ||
| 558 | K | R, 1 | ||
| 561 | P | L, 1 | ||
| 570 | A | V, 3 | S, 1 | |
| 572 | T | I, 13 | ||
| 574 | D | Y, 2 | ||
| 583 | E | D, 17 | ||
| 594 | G | S, 1 | ||
| 611 | L | F, 1 | ||
| 613 | Q | H, 1 | ||
| 614 | D | G, 4124 | ||
| 621 | P | S, 1 | ||
| 622 | V | F, 2 | A, 1 | |
| 623 | A | S, 1 | ||
| 626 | A | V, 1 | ||
| 640 | S | A, 1 | F, 2 | |
| 647 | A | S, 1 | ||
| 653 | A | V, 1 | ||
| 654 | E | Q, 1 | ||
| 655 | H | Y, 4 | ||
| 660 | Y | F, 1 | ||
| 672 | A | V, 1 | ||
| 675 | Q | R, 4 | K, 1 | H, 1 |
| 676 | T | I, 2 | ||
| 677 | Q | H, 14 | R, 1 | |
| 681 | P | L, 11 | ||
| 682 | R | Q, 2 | W, 2 | |
| 684 | A | V, 1 | S, 1 | T, 1 |
| 688 | A | V, 1 | ||
| 690 | Q | H, 3 | ||
| 691 | S | F, 1 | ||
| 698 | S | L, 3 | ||
| 701 | A | V, 1 | ||
| 704 | S | L, 3 | ||
| 706 | A | S, 2 | ||
| 708 | S | F, 1 | ||
| 724 | T | A, 1 | ||
| 731 | M | I, 2 | ||
| 732 | T | A, 1 | ||
| 740 | M | I, 1 | ||
| 745 | D | G, 1 | ||
| 751 | N | D, 1 | ||
| 765 | R | S, 1 | ||
| 769 | G | V, 1 | ||
| 778 | T | I, 1 | ||
| 783 | A | S, 3 | ||
| 789 | Y | D, 1 | ||
| 791 | T | I, 4 | ||
| 795 | K | Q, 1 | ||
| 808 | D | G, 1 | ||
| 809 | P | S, 1 | ||
| 812 | P | S, 3 | ||
| 827 | T | I, 1 | ||
| 829 | A | T, 37 | ||
| 832 | G | C, 1 | ||
| 836 | Q | P, 1 | L, 3 | |
| 838 | G | D, 8 | ||
| 839 | D | N, 1 | ||
| 845 | A | V, 2 | D, 8 | S, 7 |
| 846 | A | V, 2 | ||
| 854 | K | R, 1 | ||
| 859 | T | I, 8 | ||
| 879 | A | V, 1 | S, 3 | |
| 892 | A | S, 2 | V, 1 | |
| 922 | L | F, 2 | ||
| 924 | A | V, 1 | ||
| 931 | I | V, 2 | ||
| 936 | D | Y, 2 | ||
| 939 | S | F, 9 | Y, 1 | |
| 940 | S | F, 3 | ||
| 981 | L | F, 1 | ||
| 1002 | Q | E, 1 | ||
| 1020 | A | V, 2 | D, 1 | S, 1 |
| 1063 | L | F, 1 | ||
| 1078 | A | V, 2 | S, 2 | |
| 1079 | P | S, 1 | ||
| 1083 | H | Q, 2 | ||
| 1085 | G | R, 3 | ||
| 1091 | R | L, 1 | ||
| 1101 | H | Y, 4 | ||
| 1104 | V | L, 2 | ||
| 1109 | F | L, 1 | ||
| 1118 | D | Y, 1 | ||
| 1120 | T | I, 1 | ||
| 1122 | V | L, 4 | ||
| 1124 | G | V, 14 | ||
| 1129 | V | A, 2 | ||
| 1136 | T | I, 2 | ||
| 1141 | L | F, 1 | ||
| 1143 | P | L, 1 | ||
| 1153 | D | Y, 1 | ||
| 1162 | P | S, 3 | L, 2 | |
| 1163 | D | G, 1 | ||
| 1176 | V | F, 1 | ||
| 1181 | K | R, 1 | ||
| 1187 | N | Y, 1 | K, 1 | |
| 1191 | K | N, 3 | ||
| 1195 | E | Q, 1 | ||
| 1201 | Q | K, 1 | ||
| 1203 | L | F, 2 | ||
| 1205 | K | N, 3 | ||
| 1219 | G | V, 5 | C, 1 | |
| 1228 | V | L, 2 | ||
| 1237 | M | T, 1 | ||
| 1243 | C | F, 2 | ||
| 1246 | G | S, 1 | ||
| 1250 | C | F, 2 | ||
| 1254 | C | F, 1 | ||
| 1260 | D | H, 1 | N, 4 | |
| 1263 | P | L, 14 | ||
| 1264 | V | L, 1 | ||
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.https://covid19.who.int/
- 2.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Yong-Zhen, Holmes Edward C. 2020. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam T.T., Jia N., Zhang Y. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Gayle Albert A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS Coronavirus. J. Trav. Med. 2020;27(2) doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Renyi, Li Yixin, Zhang Annie L., Wang Yuan, Molina Mario J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. Unit. States Am. Jun 2020;117(26):14857–14863. doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html
- 8.Grant, M. C., L. Geoghegan, M. Arbyn, Z. Mohammed, L. McGuinnes, E. L. Clarke, R. G. Wade, The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): A systematic review and meta-analysis of 148 studies from 9 countries, PloS One 15(6): e0234765. 10.1371/journal.pone.0234765. [DOI] [PMC free article] [PubMed]
- 9.Yang, L., L. Tu, Implications of gastrointestinal manifestations of COVID-19, The Lancet Gastroenterology and Hepatology, Volume 5, Issue 7, 629-630. [DOI] [PMC free article] [PubMed]
- 10.Folegatti P.M., Ewer K.J. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020 Aug 15;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas S., Chatterjee S., Dey T., Manna S., Nandy A., Basak S.C. International Conference on Multidisciplinary Sciences USINEWS-04. UMN; Duluth, USA: 2020. In silico approach for peptide vaccine design of CoVID-19, MOL2NET. [DOI] [Google Scholar]
- 13.Mousavizadeh Leila, Ghasemi Sorayya. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. PMCID: PMC7138183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patrick C.Y. Woo, Huang Yi, Sussana K.P. Lau, Yuen Kwok-Yung. 2010. Coronavirus Genomics and Bioinformatics Analysis. 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruch Travis R., Machamer Carolyn E. The coronavirus E protein: assembly and beyond. Viruses vol. 2012;4(3) doi: 10.3390/v4030363. 363–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surjit Milan, Sunil K. Lal. The nucleocapsid protein of the SARS coronavirus: structure, function and therapeutic potential. Molecular Biology of the SARS-Coronavirus. Jul. 2009;22:129–151. doi: 10.1007/978-3-642-03683-5_9. [DOI] [Google Scholar]
- 17.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(Pt 4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 19.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi P., Su Y., Li R., Liang Z., Dong S., Huang J. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 2019;265:57–66. doi: 10.1016/j.virusres.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Qihui, Zhang Yanfang, Wu Lili, Niu Sheng, Song Chunli, Zhang Zengyuan, Lu Guangwen, Qiao Chengpeng, Hu Yu, Yuen Kwok-Yung, Wang Qisheng, Zhou Huan, Yan Jinghua, Qi Jianxun. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. 0092–8674, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Scheiergens T.S., Herrler G., Wu N., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 16 April 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang Jian, Wan Yushun, Luo Chuming, Ye Gang, Geng Qibin, Auerbach Ashley, Li Fang. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. May 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta Noton K., Mazumdar Kaushiki, Gordy James T. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J. Virol. Jun 2020;94(13) doi: 10.1128/JVI.00647-20. e00647–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dey, T., S. Biswas, S. Chatterjee, S. Manna, A. Nandy, S. C. Basak, 2D Polar Co-ordinate Representation of Amino Acid Sequences With some applications to Ebola virus, SARS and SARS-CoV-2 (COVID-19), MOL2NET, International Conference on Multidisciplinary Sciences USINEWS-04, UMN, Duluth, USA, 10.3390/mol2net-06-06790. [DOI]
- 27.Nandy A. The GRANCH techniques for analysis of DNA, RNA and protein sequences. Advances in Mathematical Chemistry and Applications. 2015:96–124. doi: 10.1016/B978-1-68108-053-6.50005-3. [DOI] [Google Scholar]
- 28.Dey, T., S. Chatterjee, S. Manna, A. Nandy, S. C. Basak, New Computational Analysis to Identify the Mutational Changes in SARS-CoV-2, MOLNET, International Conference on Multidisciplinary Sciences, USINEWS-04, UMN, Duluth, USA, 10.3390/mol2net-06-06811. [DOI]
- 29.Wheeler D.L., Chappey C., Lash A.E., Leipe D.D., Madden T.L., Schuler G.D., Tatusova T.A., Rapp B.A. Database resources of the national center for Biotechnology information. Nucleic Acids Res. 2000 Jan 1;28(1):10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar Sudhir, Stecher Glen, Li Michael, Knyaz Christina, Tamura Koichiro. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33(Web Server issue):W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song J., Tan H., Perry A.J., Akutsu T., Webb G.I., Whisstock J.C., Pike R.N. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PloS One. 2012;7(11):e50300. doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gummadi S.N. What is the role of thermodynamics on protein stability? Biotechnol. Bioproc. Eng. 2003;8:9–18. doi: 10.1007/BF02932892. [DOI] [Google Scholar]
- 34.Giorno F. The pH level of Healthy Lungs. 2011. Livestrong. Oct 2012;27 [Google Scholar]
- 35.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjuán R., Domingo-Calap P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016;73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterjee S., Dey T., Manna S. Emergence of a pathogenic strain of COVID-19. Journal of Bioinformatics and Systems Biology. 2020;3 doi: 10.26502/jbsb.5107016. 081–091. [DOI] [Google Scholar]
- 39.Korber B., Fischer W.M. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020, Aug 20;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrillo P.J.P., Hervás M., Rodríguez-Huete A. Systematic analysis of biological roles of charged amino acid residues located throughout the structured inner wall of a virus capsid. Sci. Rep. 2018;8:9543. doi: 10.1038/s41598-018-27749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanjuán Rafael. Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2010;365(1548) doi: 10.1098/rstb.2010.0063. 1975–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornillez-Ty Cromwell T., Liao Lujian, Yates John R., III, Kuhn Peter, Buchmeier Michael J. Severe Acute respiratory Syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. Sep. 2009;83(19):10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aftab S.O., Ghouri M.Z., Masood M.U. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020;18:275. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan A., Khan M. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscipl. Sci. Comput. Life Sci. June 2020:1–14. doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan M.T., Ali A. Marine natural compounds as potent inhibitors against the main protease of SARS-CoV-2—a molecular dynamic study. J. Biomol. Struct. Dyn. 2020 Jun 1:1–11. doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith E.C., Sexton N.R., Denison M.R. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu Rev Virol. 2014;1(1):111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.