Highlights
-
•
We proposed a novel (L, 2) transfer feature learning (L2TFL) approach.
-
•
L2TFL can elucidate the optimal layers to be removed prior to selection.
-
•
We developed a novel selection algorithm of pretrained network for fusion approach.
-
•
SAPNF can determine the best two pretrained models for fusion.
-
•
We introduced a deep CCT fusion discriminant correlation analysis fusion method.
Keywords: Chest CT, COVID-19, Deep fusion, transfer learning, pretrained model, Discriminant correlation analysis, Micro-averaged F1
Abstract
Aim
: COVID-19 is a disease caused by a new strain of coronavirus. Up to 18th October 2020, worldwide there have been 39.6 million confirmed cases resulting in more than 1.1 million deaths. To improve diagnosis, we aimed to design and develop a novel advanced AI system for COVID-19 classification based on chest CT (CCT) images.
Methods
: Our dataset from local hospitals consisted of 284 COVID-19 images, 281 community-acquired pneumonia images, 293 secondary pulmonary tuberculosis images; and 306 healthy control images. We first used pretrained models (PTMs) to learn features, and proposed a novel (L, 2) transfer feature learning algorithm to extract features, with a hyperparameter of number of layers to be removed (NLR, symbolized as L). Second, we proposed a selection algorithm of pretrained network for fusion to determine the best two models characterized by PTM and NLR. Third, deep CCT fusion by discriminant correlation analysis was proposed to help fuse the two features from the two models. Micro-averaged (MA) F1 score was used as the measuring indicator. The final determined model was named CCSHNet.
Results
: On the test set, CCSHNet achieved sensitivities of four classes of 95.61%, 96.25%, 98.30%, and 97.86%, respectively. The precision values of four classes were 97.32%, 96.42%, 96.99%, and 97.38%, respectively. The F1 scores of four classes were 96.46%, 96.33%, 97.64%, and 97.62%, respectively. The MA F1 score was 97.04%. In addition, CCSHNet outperformed 12 state-of-the-art COVID-19 detection methods.
Conclusions
: CCSHNet is effective in detecting COVID-19 and other lung infectious diseases using first-line clinical imaging and can therefore assist radiologists in making accurate diagnoses based on CCTs.
1. Introduction
COVID-19 (coronavirus disease 2019) was declared as a Public Health Emergency of International Concern on 30/Jan/2020, and a worldwide pandemic on 11/March/2020. Up to 18/Oct/2020, globally there have been 39.6 million confirmed cases and more than 1.1 million deaths (including US 222.5k Brazil 153.6k, India 114.0k, Mexico 86.0k, UK 43.5k, etc.) [1].
Two prevailing diagnostic methods are available for COVID-19 detection. One is viral testing by nasopharyngeal swabs [2] to test the existence of viral RNA fragments using real-time reverse-transcriptase PCR (rRT-PCR) and the other is imaging methods such as chest X-ray (CXR) [3] and chest computed tomography (CCT) [4]. Compared to viral testing, CCT can avoid the problem of sample contamination. For example, the swab can touch contaminated surfaces or gloves, samples can be cross-contaminated, etc. It was reported that in March 2020, due to the problem of reagent contamination, the US Center for Disease Control and Prevention (CDC) withdrew testing kits [5]. As an alternative, CCT scans can help to detect hazy, patchy, “ground glass” white spots in the lung, a tell-tale sign of COVID-19 infection, which can provide a more accurate result than viral tests. Furthermore, previous studies have shown that CCT can detect 97% of COVID-19 infections; whereas viral testing only detected 52% of patients with COVID-19 infection [6].
There are currently two imaging modalities that are used to detect COVID-19 infection. CXR is the most widely used diagnostic X-ray examination in medical practice, producing images of the blood vessels, airways, lungs, heart, bones of the spine, and chest. On the other hand, CCT uses computer-processed combinations of numerous X-ray images taken at different angles to produce a cross-sectional image of the region being scanned and to examine abnormalities. CCT is able to detect very small nodules in the lung compared to CXR [7]. In addition, CCT has advantages over CXR since it generates high-quality, detailed images by taking a 360-degree image of the chest and its internal organs. Moreover, CXR provides a 2D image that contains less information; whereas CCT provides 3D volumetric data that can highlight additional spatial features and abnormalities.
For diagnosis of COVID-19, CXR is sub-optimal since important abnormalities are undetectable due to the normal black appearance of the lung. However, CCT can clearly show a combination of multifocal peripheral lung changes of ground-glass opacity (GGO) [8] and/or consolidation [9], which indicate infection with COVID-19. Hence, in this study we used CCT to aid diagnosis of COVID-19 infection. Nevertheless, manual labeling by radiologists is tedious and time-consuming, while being affected by inter- and/or intra-expert factors (e.g., emotion, tiredness, lethargy, etc.). Further, diagnostic throughputs of radiologists are not comparable with digital methods and early symptoms are more difficult to measure and hence can potentially be missed by experts.
Improved diagnostic systems using image processing and machine learning can potentially benefit patients, experts, radiologists, consultants, and hospitals. Currently, most AI methods can differentiate COVID-19 infection in images from healthy subjects and/or community acquired pneumonia (CAP). Deep learning (DL) approaches are an emerging new type of machine learning, which consists of stacks of convolution layers and fully connected layers (FCLs).
For example, Li and Liu [10] employed wavelet packet Tsallis entropy as a feature descriptor, and used a real-coded biogeography-based optimization (RCBO) approach as a classifier. Lu [11] employed bat algorithm to optimize extreme learning machine. Their method was called ELM-BA. Jiang [12] proposed a six-level convolutional neural network (6L-CNN) towards therapy and rehabilitation, while improving performance by replacing the traditional rectified linear unit with leaky rectified linear unit. Guo and Du [13] used ResNet-18 (RN-18) to classify thyroid ultrasound standard plane (TUSP), achieving a classification accuracy of 83.88%. Their experiment verified the effectiveness of RN-18. Fulton, et al. [14] utilized ResNet-50 to classify Alzheimer's disease (RN-50-AD) with and without imagery. The authors stated that ResNet-50 models might help identify AD patients prior to provider review. Although these previous five studies did not analyze COVID-19 positive patients, their algorithms can be easily transferred to the multi-class classification task of COVID-19 diagnosis in this study.
Numerous cutting-edge AI methods have been proposed to diagnose COVID-19 using either CXR or CCT. For CXR, Loey, et al. [15] employed generative adversarial network (GAN) to produce new simulated images showing that the combination of GAN and GoogleNet (GAN-GN) is optimal for two-class classification than AlexNet and ResNet-18. Togacar, et al. [16] utilized SqueezeNet and MobileNetV2 to obtain image descriptors. The authors chose social mimic optimization (SMO) as a feature selection tool. The obtained features were then combined and passed into support vector machines. Cohen, et al. [17] employed a sizable non-COVID-19 CXR set to improve extracted features from images of CXRs from COVID-19 patients and predicted two scores: (i) lung opacity score; and (ii) geographic extent score. Their method could gage severity of COVID-19. The method (termed COVID severity score or CSS) achieved a mean absolute error (MAE) of 1.14 on geographic extent score, and a MAE of 0.78 on lung opacity score. Tabik, et al. [18] built COVIDGR-1.0, a homogeneous and balanced database that includes all levels of severity, and presented a novel COVID-SDNet in order to classify COVID-19 based on CXR images.
For CCT, Ni, et al. [19] proposed NiNet, utilizing both 3D U-Net and MVP-Net on more than 90 COVID-19 patients in CCT scanning, for the aim of (i) pulmonary lobe segmentation, (ii) lesion segmentation, and (iii) lesion detection. The authors found the deep learning algorithm could assist radiologists to make quicker diagnosis (all p values are less than 0.01%) with first-class performances. Ko, et al. [20] presented a straigtforward 2D fast-track deep learning system for single CCT image termed FCONet (fast-track COVID-19 classification network). They analyzed 4 pretrained models: ResNet-50, VGG16, Xception, and Inception-V3, finding that ResNet-50 performed the best when classifying COVID-19 positive patients. They used two augmentation methods: zoom and image rotation, while proposing extra layers consisting of a flatten layer, a FCL (32 neurons), and a FCL (3 neurons). The final FCL has 3 neurons since their task is to classify 3 categories: COVID-19, other pneumonia, and non-pneumonia. As validation, the authors tested the FCONet approaches on an external set from embedded low-quality CCT images of COVID-19 patients. Li, et al. [21] developed COVNet, choosing ResNet50 as the backbone network. In their study, the deep representations were merged by a max-pooling procedure, with the obtained feature map being passed into a FCL to produce the probability score of three categories: (i) COVID-19 infection, (ii) CA), and (iii) non-pneumonia. Wang, et al. [22] proposed DeCovNet, a weakly-supervised DL framework via three-dimensional CT data for (i) lesion localization; and (ii) COVID-19 classification. The lung region was firstly segmented via a pre-trained UNet. Next, the segmented 3D lung region was passed to a three-dimensional deep network to predict the probability of COVID-19. When using a probability threshold of 0.5, the DeCovNet yielded an accuracy of 90.1%, a negative predictive value of 98.2%, and a positive predictive value of 84.0%. Satapathy, et al. [23] proposed a seven-layer CNN by stochastic pooling. Their method achieved a specificity of 93.63%, a sensitivity of 94.44%, and an accuracy of 94.03%. Wu [24] combined wavelet Renyi entropy with a three-segment biogeography-based optimization (TSBO). Their proposed TSBO can optimize weights, biases, and order of Renyi entropy at the same time.
The inspiration for this study was to improve detection of COVID-19 infection in CCT images by developing a novel method to fuse the features from two neural network models. The main contributions of this paper are that: (i) We proposed a novel (L, 2) transfer feature learning (L2TFL) approach to elucidate the optimal layers to be removed prior to selection by testing various pretrained networks with various settings. (ii) We developed a novel selection algorithm of pretrained network for fusion (SAPNF) approach that can determine the best two pretrained models and proved it gives better performance than the proposed greedy selection algorithm for fusion (GSAF). (iii) We introduced a deep CCT fusion discriminant correlation analysis (DCFDCA) fusion method that gives better performance than traditional addition and concatenation fusion methods; and (iv) we improved performance over current methods by implementing multiple-way data augmentation.
The structure of the paper is organized as below. Section 2 introduces the dataset, imaging protocol, slice selection method, ground-truth labeling, and preprocessing of the images. Section 3 describes every component of the proposed AI model for COVID-19 detection, and Section 4 presents the experimental results and discussions. Finally, Section 5 concludes the paper.
2. Dataset and preprocessing
Table 10 in Appendix.A and Table 11 in Appendix.B list the abbreviation and variable meanings exercised for easy reading.
2.1. Slice selection
Four types of CCT were used in this study: (i) COVID-19 positive; (ii) community-acquired pneumonia (CAP); (iii) second pulmonary tuberculosis (SPT); (iv) healthy control (HC). The three diseased classes were chosen since they are all infectious diseases of the chest regions. Our aim was to develop an AI system that can automatically predict the four categories.
For each subject, slices were chosen and a slice level selection (SLS) method was employed: For the three diseased groups, the slice displaying the largest number of lesions and size was chosen. For healthy subjects, any slice of the 3D image was randomly chosen.
The resolutions of all images were . In total, we enrolled 521 subjects, and generated 1164 slice images using the SLS method, viz., 284 COVID-19 images, 281 CAP images, 293 SPT images; and 306 HC images. Image collection is challenging since it is expensive and labor-intensive, as well as requiring expert curation. Table 1 lists the demographics of the four-category subject cohort.
Table 1.
Subjects and images of four categories.
| Category | Patients (n) | CCT Images (n) |
|---|---|---|
| COVID-19 | 125 | 284 |
| CAP | 123 | 281 |
| SPT | 134 | 293 |
| HC | 139 | 306 |
(n = number).
2.2. Ground-truth labelling
Three radiologists (Two juniors: and , and one senior: ) were assigned to curate all the images. Suppose means one CCT scan, means the labeling of each individual expert, and the final labeling of the CCT scan is obtained by
| (1) |
Where denotes the labeling of all radiologists, viz.,
| (2) |
MV denotes majority voting. The above equation means the situation of disagreement between the analyses of two junior radiologists , we need to consult a senior radiologist to reach a consensus.
2.3. Preprocessing
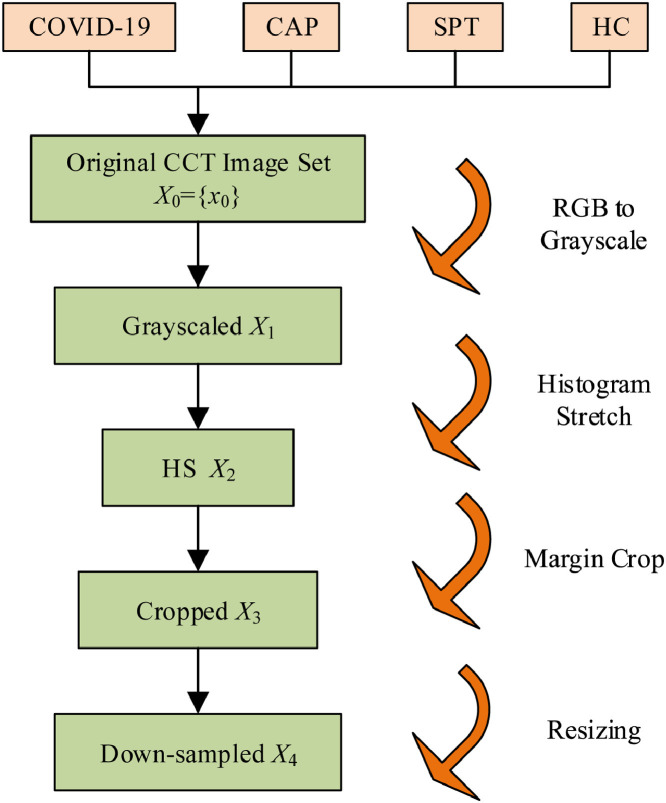
Preprocessing has already shown its success in medical image analysis [25, 26]. The original dataset contained slice images . The size of each image was . Fig. 1 shows the pipeline for preprocessing of our dataset.
Fig. 1.
Illustration of preprocessing. (CAP: community-acquired pneumonia; SPT: secondary pulmonary tuberculosis; HC: healthy control; CCT: chest CT; HS: histogram stretching).
First, the color CCT images from four classes were converted into grayscale by retaining the luminance channel, and yielding the grayscale data set :
| (3) |
where means the grayscale operation. Note that
Second, the histogram stretching (HS) [27, 28] was utilized to increase the contrast of all images. Take the i th image as an example, its minimum and maximum grayscale values and were calculated as:
| (4) |
here (w, h, c) means the index of width, height, and channel directions along image , respectively. The new histogram stretched image set was calculated as:
| (5) |
Third, cropping was carried out to remove the checkup bed at the bottom area, and to remove the texts at the margin areas. The cropped dataset is obtained as
| (6) |
where represents crop operation. Parameter means pixels to be cropped in unit of pixel from four directions. The subscript is the initial letter of top, bottom, left, and right, respectively. After this step, the resolution of each image .
Fourth, we down-sampled each image to a size of , obtaining the resized image set as
| (7) |
where represents the downsampling (DS) function, in which b is a down-sampled image of the raw image a.
After the preprocessing procedure, each image was approximately 1.64% (explained below) of its original storage or size. The compression ratio (CR) rates of i th image of the final stage to the raw stage was measured by two variables: the storage CR and size CR
| (8) |
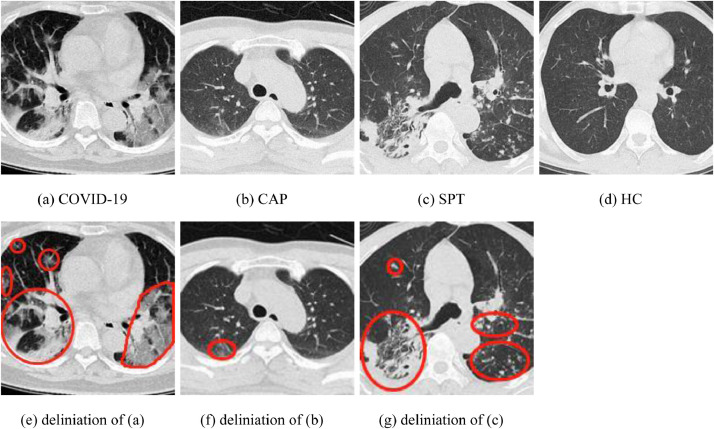
We can have , and . Hence, we can obtain , . Hence, it proves the importance of preprocessing. Furthermore, Fig. 2 displays four samples from the preprocessed set . The top row presents the preprocessed images, and the bottom row the delineated results in red curves. Overall, the advantages of preprocessing is three-fold: (i) Compression ratio helps to minimize the storage size; (ii) Histogram stretching helps to normalize the contrast of all samples; (iii) Cropping removes irrelevant contents from CCT images, so the AI model will focus on the lung region. Table 2 compares the storage and size of every image at each preprocessing step.
Fig. 2.
Samples of X4. (CAP: community-acquired pneumonia; SPT: secondary pulmonary tuberculosis; HC: healthy control).
Table 2.
Storage and size per preprocessing step.
| Preprocessing Step | Variable | W | H | C | Storage* | Size* |
|---|---|---|---|---|---|---|
| Raw | 1024 | 1024 | 3 | 12,582,912 | ||
| Grayscale | 1024 | 1024 | 1 | 4194,304 | ||
| HS | 1024 | 1024 | 1 | 4194,304 | ||
| Crop | 724 | 724 | 1 | 2096,704 | ||
| DS | 227 | 227 | 1 | 206,116 |
* Storage and size are measured per image.
3. Methodology
The motivation of our algorithm was to use pretrained models to generate features from CCT images, and fuse those features using the discriminant correlation analysis (DCA) method. Section 3.1 introduces what transfer learning is. Section 3.2 briefs several state-of-the-art pretrained models and proposes a novel (L, 2) transfer feature learning (L2TFL) algorithm, to answer the question of how to extract features using pretrained networks. Section 3.3 determines how to choose the optimal two pretrained models, and proposes a novel selection algorithm of pretrained networks for fusion (SAPNF). Section 3.4 details how to fuse, and introduces the DCA technology. Section 3.5 presents a novel data augmentation method to further improve the performance. Section 3.6 presents the experimental setup and measures. Section 3.7 summarizes and gives the pseudocode of the proposed algorithms.
3.1. Transfer learning
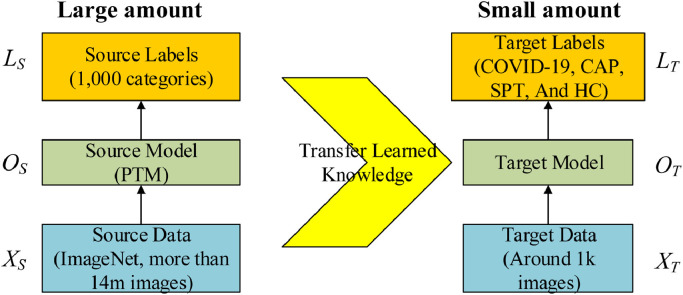
The basic ideas of transfer learning (TL) are utilizing a complicated and successfully pre-trained model (PTM) [29], taught from a sizable amount of source data, viz., (1000 categories from ImageNet), and then “transfer” the learnt knowledge [30] to the relatively simple task (4 categories of COVID-19, CAP, SPT and HC in this study) with a small quantity of data.
Mathematically, suppose the source data is representing ImageNet, the source label the 1000-category labeling, and means the source objective-predictive function (i.e., the classifier), we have the source domain knowledge as a triple variable of
| (9) |
Now we have the triple target: target data represents the training set, presents the 4-class labeling (COVID-19, CAP, SPT, or HC), and represents the classifier to be established.
| (10) |
Using TL, the classifier to be created can be written as . Without using transfer learning, the classifier is written as .
| (11) |
Then we can say is expected to be much closer to the ideal classifier than the classifier using only the target domain , viz. suppose we have a large number of samples and its labels .
| (12) |
Where is an error function measuring the two inputs and .
In practice, three elements are vital to help transfer learning improve its performance than building and training a network [31] from scratch: (i) Successful PTM can help the user remove hyper-parameter tuning; (ii) The initial layers in PTM can be thought of as feature descriptors, which extract low-level features, e.g., tints, edges, blobs, shades, and textures; (iii) The target model may only need to re-train the last several layers of the pre-trained model, since we believe the last several layers carry out the complex identification tasks. The basic idea of transfer learning is shown in Fig. 3 .
Fig. 3.
Idea of transfer learning. (PTM: pretrained mode; CAP: community-acquired pneumonia; SPT: secondary pulmonary tuberculosis; HC: healthy control).
3.2. Novelty 1: (L, 2) transfer feature learning
As shown in Table 3 , pretrained models were tested in this study: AlexNet, DenseNet201, ResNet50, ResNet101, VGG16, and VGG19. Traditional transfer learning usually modifies the neuron number of the last fully connected layer. Then the user may choose to retrain the whole network (The weights of reserved layers may be initialized by either pretrained models or re-initialization) or only retrains the modified layer.
Table 3.
Candidate pretrained models.
| PTM | PTM Symbol | Parameters (millions) | Input Size |
|---|---|---|---|
| AlexNet | 61.0 | 227×227 | |
| DenseNet201 | 20.0 | 224×224 | |
| ResNet50 | 25.6 | 224×224 | |
| ResNet101 | 44.6 | 224×224 | |
| VGG16 | 138 | 224×224 | |
| VGG19 | 144 | 224×224 |
In this study, we proposed a new (L, 2) transfer feature learning algorithm (abbreviated as L2TFL). The motivation for L2TFL is two-fold: (i) We make , the number of layers to be removed (NLR), adaptive, and the value of was optimized to improve performance. (ii) We chose to add two newly fully connected layers due to the arbitrary width case of universal approximation theorem.
For ease of understanding, the pseudocode of proposed L2TFL algorithm is presented in Algorithm 1, where is a parameter and its value was optimized.
Step 1. Read the PTM network in Table 3, and store it into variable , suppose its number of learnable layers is .
- Step 2. Remove the last NLR l-learnable layers from and get ,
(13)
where means remove layer function, and parameter means the number of last layers to be removed. If there are shortcuts with their outputs located within the last learnable layers, those shortcuts must be removed.
Step 3. Add 2 new fully connected layers
| (14) |
where means add fully-connected layer function, and the constant 2 means the number of fully-connected layers to be appended to . Here the number of learnable layers of network can be calculated as . The first layer FCL layer has nodes, and the second FCL layer has nodes.
Step 4. Keep the learning rate [32] of all the transfer layers zero, in order to freeze those layers
| (15) |
Where means the leaning rate, and means the layers from to in network , in total layers are considered in .
Step 5. Let the last two added new fully connected layers be retrainable, i.e., set their learning rate as 1
| (16) |
Step 6. Retrain the whole network using our four-class data and get the trained network .
| (17) |
where is some dataset, and means the retrain function.
Step 7. Using to generate learnt features
| (18) |
where is the activation function, means to extract the activation functions from network at the -th layer, means features learnt from network by removing learnable layers.
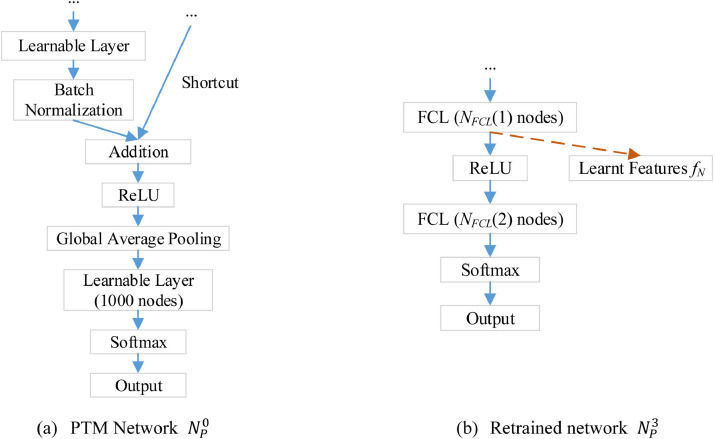
Take ResNet18 as an example, Fig. 4 shows the diagram of our L2TFL algorithm, where . Fig. 4(a) shows the part of ResNet18 with the last two learnable layers. Fig. 4(b) shows the structure of using our L2TFL, by which the last two learnable layers of ResNet18 were replaced by two newly added FCL layers with number of nodes of and , respectively.
Fig. 4.
A simplistic example of L2TFL algorithm for ResNet18 (Here NLR L = 2). (ReLU: rectified linear unit; FCL: fully-connected layer; L2TFL: (L, 2) transfer feature learning; NLR: number of layers to be removed).
To search the optimal value of NLR , we set a range of , where we searched the optimal NLR value from this range for each PTM. is the maximum removable layer (MRL). Note that SqueezeNet [33] and GoogleNet [34] were not considered since their structure contains parallel branches and were not appropriate in our L2TFL algorithm.
3.3. Novelty 2: selection algorithm of pretrained networks for fusion
Previously, we discussed how to extract features from PTMs. Now the question is how to select the two pretrained models? The naive idea is to use greedy selection algorithm for fusion (GSAF), i.e., select the best two pretrained models, and extract their features, and fuse those two features.
Suppose there is a dataset will be split into a training set , a validation set , and a test set , resulting in . The GSAF uses to create a performance rank list , choose the best two PTMs from that list, and fuse their corresponding features.
The procedure of GSAF is briefly described as: For a given -th PTM , we use L2TFL via data and removing layers to obtain
| (19) |
where is our proposed L2TFL operation. Subsequently, an empty one-hidden layer neural network (OHNN) [35] was created for validation. The initial and trained one-hidden neural network are symbolized as and , respectively. The input of is , and the number of hidden neurons is symbolized to . Performance indicator was calculated by comparing the output of over validation set , viz.,
| (20) |
with its ground truth labels , so is calculated as
| (21) |
Where is the measuring indicator function. It can be accuracy or sensitivity or specificity or any other measuring indicators. The is gathered overall all possible hyperparametric combination, so we get indicator vector
| (22) |
The indicator vector is used to compare all the possible models and all possible removable layers, and we obtain the rank list by
| (23) |
Where is the sort function in descending way, and means the indicator by learnt features from k-th PTM with removing NLR learnable layers. Now and means the index of the top two best models by GSAF method, as shown in Table Algorithm 2.
Nevertheless, this greedy selection algorithm cannot ensure the fused feature can obtain the best performance. For example, if the two best models are all focusing on one region, their fusion does not help improve the performance.
Hence, we proposed a novel selection algorithm of pretrained networks for fusion (SAPNF) to help choose the best two pretrained models that can specifically improve the performance of the fused features. The difference between SAPNF and GSAF is the former will investigate a larger search space that covers both PTM candidates to be fused, while the latter only searches a smaller space which contains only one PTM candidate. The pseudocode of SAPNF is presented in Algorithm 3.
Mathematically, we retrained two models (with hyperparameters as PTM and NLR) in SAPNF. Hence, Eq (19) was updated as
| (24) |
where the subscript in and means the index of candidate model. Note we should guarantee
| (25) |
Which helped ensure the two candidate models are not the same one.
Now we can generate two features and from two different models and , respectively. We used some fusion operation to generate a fused feature as
| (26) |
Where is the deep fusion function, which will be discussed in next Section.
The indicator vector is updated as
| (27) |
Similarly, we obtain the rank list by
| (28) |
3.4. Novelty 3: deep CCT fusion by discriminant correlation analysis
Feature-level fusion (FLF) aims to combine discriminative multiple features, while decision-level fusion (DLF) combines multiple decision answers. Commonly, DLF is simpler than FLF, but FLF outperforms DLF [36, 37]. In this study we chose feature-level fusion. In our future research, we will also consider some advanced fusion rules, such as score-level fusion, DLF, and hybrid fusion methods [38].
We have discussed how to carry out transfer feature learning and how to select pretrained models. Now we need to answer the question of how to fuse those extracted features. There are two commonly used FLF methods. Based on having two -dimension features from two PTMs, the features were generated by our L2TFL method, and the selection of PTM was by SAPNF method.
Assume the two features are symbolized as with length and with length , the fused feature is symbolized as . Serial fusion (SF) [39] concatenates the two features into one single feature
| (29) |
where represents the SF operation. The length of equals .
Parallel fusion (PF) [40] combines and into one complex vector
| (30) |
where represents the PF operation, and the imaginary unit.
Sun, et al. [41] proposed a canonical correlation analysis (CCA), which finds optimal linear combination of and which have maximum correlation with each other. Suppose , , where means the number of trained features. First, we can define two covariance matrixes and as
| (31) |
where is the cross-covariance operation. Also, we can define the covariance matrix as
| (32) |
We have .
The overall covariance matrix can be computed as
| (33) |
The aim of CCA is to seek the best linear projection
| (34) |
where and are transformation matrices of CCA. The aim is to find the optimal that maximizes the pair-wise correlation over the two feature sets:
| (35) |
Where means the pair-wise correlation, defined as
| (36) |
The detailed derivation and solution can be found in [41]. For the optimal weights , we have , and . Hence, the combination of the transformed features is carried out by either concatenation or summation as:
| (37) |
where and represent the concatenation and summation of CCA features, respectively.
CCA has two issues: (i) The number of samples is less than the number of features in many real world scenarios: , which makes the covariance matrices non-invertible and singular. (ii) CCA neglects the class structure information. To solve these two issues, Haghighat, et al. [42] presented a discriminant correlation analysis (DCA) approach. DCA has been proven to offer improved performance than recent fusion approaches.
In this study, we used DCA to fuse features from CCT images, and we named it as deep CCT fusion by discriminant correlation analysis (DCFDCA). Similar to CCA, suppose , where means the number of trained features. The columns of the data matrix can be segmented into classes, suppose columns belong to the i th class, we have
| (38) |
Let denotes feature extracted from i th image of j-th category via model , and and denotes the mean of over i th class and the whole set, respectively. We can get
| (39) |
Thus, the between-class scatter (BCS) matrix is defined as
| (40) |
where is defined as
| (41) |
Note the number of features is greater than the number of classes in this study, i.e., , so a method [43] is chosen here to calculate the covariance matrix of . The most significant eigenvectors of can be economically attained by mapping the eigenvectors of . Hence, it is ncessary to acquire the eigenvectros of this covraicne matrix . Assue the classes were well-separated, is a diagonal matrix as
| (42) |
where denotes the matrix of orthogonal eigenvectors, the diagonal matrix of real and non-negtive eigenvalue in decreasing order.
Assue entails the first eigenvectors from , so corrresponds to the largest non-zero eigenvalues in . We can deduce folowing equation as
| (43) |
Therefore, the most significant eigenvectors of are acquired by the mapping as
| (44) |
Assume is the transformation which uses and reduces the data's dimensionality from to , we have
| (45) |
and
| (46) |
where denotes the projection of in a temporary space, in which the BCS matrix of the 1st feature set to be fused is and the classes are all separated. Notice that
| (47) |
where is the rank function.
Similarly, to the second feature set , we can find a transform matrix , which employes the BCS matrix for the second feature sets to be fused and reduces the dimensionality of from to as
| (48) |
| (49) |
The updated and are now non-square orthonormal matrices. Note that , nevertheless, the matrices and are strictly diagonally dominant matrices (DDMs), namely, if denotes the entry of a DDM, then . In our study, the diagonal entries are near to 1 and the off-diagonal entries are near to zero.
So far, we have transformed and , i.e., we have finished the unitization of BCS matrices. The next step is to transform the features in one set to have nonzero correlation with their cognate features in the other set.
Mathematically, the between-set covariance (BSC) matrix of the transformed features set need to be diagonalized. The singular value decomposition (SVD) approach is utilized at this step.
| (50) |
Remember that and are of rank and is non-degenerate. We can deduce is a diagonal matrix, of which the main diagonal elements are non-zero. Assume
| (51) |
We have
| (52) |
which unitizes the BSC matrix . Finally, the DCA-transformed features can be written as
| (53) |
Where and are the final transformation matrices of DCA for and , respectively. Similarly, the combination of the transformed DCA features is done by either concatenation or summation as:
| (54) |
where and represent the concatenation and summation of DCA features, respectively. In this study, was chosen, since (i) summation procedure features in lower number of dimensions, and (ii) the summation and concatenation have similar results reported in [42]. In addition, feature fusion can help improve the performance compared to using a single PTM model (See Sections 4.3 and 4.4).
3.5. Data augmentation
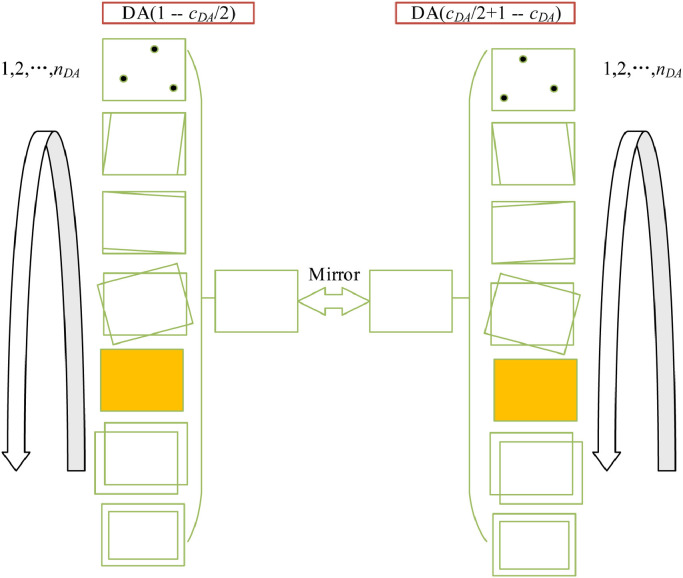
Multiple-way data augmentation (MDA) technology [44] was used in this study. The disparity of MDA to conventional DA is that MDA utilizes a large number of different data augmentation methods. There are two types [45] of MDA, offline and online. Offline means editing and storing data on the disk, and online means on-the-fly augmentation. In this study, we chose to use offline multiple-way data augmentation, as shown in Fig. 6 . Usually, online data augmentation is mainly applied when the dataset is large. The transformations happen in mini-batches and then, the transformed data is fed into the model to improve the generalization of the model. However, we have a small dataset in this study. Therefore, we chose offline data augmentation as a preprocessing step to expand the dataset.
Fig. 6.
Diagram of proposed offline MDA technology. (DA: data augmentation; MDA: multiple-way DA).
Suppose the number of different DA techniques used is , and there is one training image , where means the training set. Assume each offline MDA technique will generate images, so for each image, we will generate new images. Over the entire training image set , we perform the subsequent seven DA methods:
(i) noise injection
The -mean -variance Gaussian noises were added to all training images to produce new noised images.
| (55) |
where means the noise injection function.
(ii) horizontal shear (HS) transform
New images were made by HS transform
| (56) |
Where denotes the HS transform function. HS factors does not include the value of .
(iii) vertical shear (VS) transform
| (57) |
where means VS transform function, which operates similarly as ST transform. The VS factor has the same value of HS factor .
(iv) rotation
(i) Rotation angle vector skips the value of 0.
| (58) |
where means rotation operation.
(v) gamma correction (GC)
The factor of GC skips the value of 1.
| (59) |
Where means GC operation.
(vi) random translation (RT)
Every image in the training set is translated times with random vertical shift and random horizontal shift . The values of and are in the range of , and obey uniform distribution .
| (60) |
where is the maximum shift range. Hence, we have
| (61) |
(vii) scaling
All training images are scaled with scaling factor , skipping .
| (62) |
where is the scaling operation.
(ix) mirror
All the above results are mirrored:
| (63) |
where represents the mirror function.
(x) concatenation
All the -way results are concatenated as
| (64) |
where means concatenation operation, is the collection of generated MDA images with original image . is the set of all augmented images. is the size of the augmented dataset. the data augmentation factor (DAF), representing the ratio of size of augmented training set to the size of original training set. is calculated as
| (65) |
We can calculate . Therefore, the MDA is a function making the enhanced training set times as large as the original training set .
| (66) |
3.6. Experiment setup and measures
Two types of measures were performed in our experiment. One is for validation to choose the best PTMs, and the other is on the test set to relate the unbiased performances so as to compare with state-of-the-art approaches. the whole preprocessed dataset is split into a non-test set , and a test set , i.e., . Roughly, the non-test set comprises 80% of the whole dataset, and the test set the remaining 20%. So we have
| (67) |
where means the number of samples in the non-test set in k-th class and the number of samples of the test set in k-th class. Hence, , and .
For the validation phase, a runs of 10-fold cross validation [46] was run to obtain the validation performance. The ideal confusion matrix combining all runs of 10 folds is
| (68) |
In the test phase, we ran our selected best models with times, each run with various initial seeds, the ideal confusion matrix is
| (69) |
For realistic runs, suppose is the run index, and each run we will generate either validation confusion matrix [47] or test confusion matrix . After summarizing all runs, we can obtain the summation of validation confusion matrix as
| (70) |
And the summation of test confusion matrix as
| (71) |
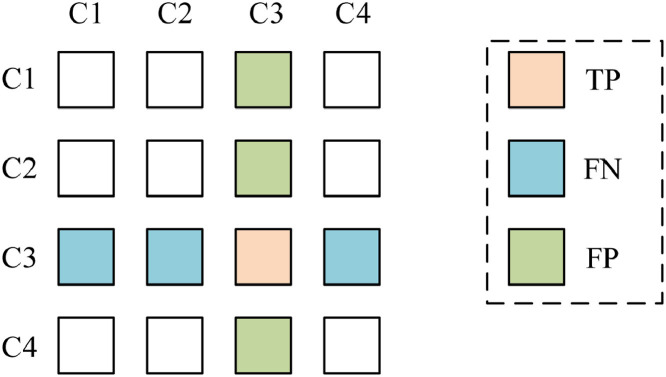
For each class , we set the that class label as “positive”, and all other three classes are “negative”. Fig. 7 shows a schematic of a multiple-class confusion matrix, where we focus on the 3rd class. Hence, the element on the 3rd row and 3rd column is TP, the summation of the remaining entries on the 3rd row is FN, and the summation of the remaining entries in the 3rd column is FP. So, we can define this measure per class as
| (72) |
| (73) |
| (74) |
Fig. 7.
Confusion matrix of multiple class conditions.
Measures can also be given at an overall level. One is called macro-level, which computes the metric independently for each class and takes the average that gives equal weight to each class (treating all classes equally) [48]. In contrast, the other is micro-level, weighting all samples equally [49]. In this multiple classification research, we prefer the micro-averaged (MA) F1 as the dataset is slightly unbalanced. The MA F1 [50] () is defined below as the main indicator in the validation phase.
| (75) |
where and are micro-averaged precision and micro-averaged sensitivity, defined as
| (76) |
| (77) |
We used in this study, since its values equals and .
3.7. Pseudocode of CCSHNet
Algorithm 4 lists the pseudocode of proposed AI model, named CCSHNet, which is an acronym of the four categories analyzed in this study: COVID-19, CAP, SPT, and HC. The proposed algorithm and experiment setup consisted of five phases: Phase I shows the preprocessing. Phase II shows runs of ten-folds CV on the non-test set. Phase III shows the PTM selection. Phase IV presents CCSHNet model creation, and Phase V reports the test performance of the CCSHNet model.
Gradient-weighted Class Activation Mapping (Grad-CAM) [51] was used to give an explainable heat map. It utilizes the gradient of the classification score in terms of the convolutional features regulated by the AI model to help users comprehend which regions of the input image are the most vital for AI model to make decisions.
4. Experiments, results, and discussions
4.1. Hyperparameter values
Table 4 itemizes the hyperparameter setting. The image size using slice level section was obtained as . The size of each raw image was . The crop values along four directions were all set to 150 (We tested larger values and found some important chest regions are removed). The number of PTM candidates was set to 6. The number of the first FCL was set to 512, and the number of the second FCL was set to 4, which corresponds to the number of classes in this task. The maximum removable layer was set 3, so we searched the best at the range of . The number of hidden neurons in OHNN was set to 10.
Table 4.
Hyperparameter Setting.
| Parameter | Value |
|---|---|
| 1024 | |
| 1024 | |
| 3 | |
| 150 | |
| 150 | |
| 150 | |
| 150 | |
| 6 | |
| 512 | |
| 4 | |
| 3 | |
| 10 | |
| 14 | |
| 30 | |
| 0 | |
| 0.01 | |
|
, . |
|
|
, . |
|
|
, . |
|
| 20 | |
|
, . |
|
| 422 | |
| 10 | |
| 10 |
For the offline MDA technique, the number of different DA techniques was adjusted to 14. The number of generated images by each offline MDA technique was 30. The mean and variance of Gaussian noise injected were 0 and 0.01, respectively. The HS factor ranged from −0.15 to +0.15, excluding the value of 0. The RO factor ranged from −30 to 30 excluding the value of 0. The GC factor ranged from 0.4 to 1.6 skipping the value of 1. The maximum shift range was set to 20. The SC factor ranged from 0.7 to 1.3 excluding the value of 1. Data augmentation factor was calculated as 422. The number of runs over validation and test sets were all set to 10.
Table 5 itemizes the training, validation, and test set for each category. For the non-test set, 10-fold cross validation was used for validation, with 9 folds being for training and the remaining fold for validation, which repeated 10 times, so all the non-test set was used in the validation set. The above 10-fold cross validation repeat runs, and thus generated a summation of validation confusion matrix . For the test set, runs generated a summation of test confusion matrix .
Table 5.
Training, validation, and test set.
| Non-test (9 folds for training and 1-fold for validation) |
Test | Total | |
|---|---|---|---|
| COVID-19 | |||
| CAP | |||
| SPT | |||
| HC |
4.2. Illustration of multiple data augmentation
Fig. 8 displays the MDA results, where the hyperparameters can be found in Section 4.1. The raw image is Fig. 2(a), which generates 421 new images (1 mirror image, 210 new images obtained from the original image, and 210 new images obtained from the mirrored original image). Fig. 8(a-g) shows the noise injection, HS transform, VS transform, rotation, GC, RT, and scaling results, respectively.
Fig. 8.
Results of proposed MDA.
4.3. Top three models of the validation set
On the validation set, we found the best three models using GSAF were: (i) , i.e., DenseNet201 with NLR of 1; (ii) DenseNet201 with NLR of 2; and (iii) ResNet1–1 with NLR of 1. Those top best three models found by GSAF are listed in Table 6 . For the best model (DenseNet201 with NLR of 1), we observed the sensitivity of the four classes were 94.63%, 93.16%, 98.12%, and 99.18%, the precision of the four classes were 96.45%, 96.68%, 96.15%, and 96.16%, the F1 score of the four classes were 95.53%, 94.88%, 97.12%, and 97.65%. The MA F1 score was 96.35%. For the other two best models, their values were 96.06%, and 95.83%, respectively.
Table 6.
Top best three models on validation set.
| Model | Class | Sen (%) | Prc (%) | F1 (%) |
|---|---|---|---|---|
| DensetNet201 (NLR=1) |
C1 | 94.63 | 96.45 | 95.53 |
| C2 | 93.16 | 96.68 | 94.88 | |
| C3 | 98.12 | 96.15 | 97.12 | |
| C4 | 99.18 | 96.16 | 97.65 | |
| MA | 96.35 | |||
| DensetNet201 (NLR=2) |
C1 | 94.93 | 97.07 | 95.99 |
| C2 | 93.91 | 95.57 | 94.73 | |
| C3 | 97.26 | 94.09 | 95.65 | |
| C4 | 97.92 | 97.52 | 97.72 | |
| MA | 96.06 | |||
| ResNet101 (NLR=1) |
C1 | 96.91 | 96.57 | 96.74 |
| C2 | 96.22 | 94.45 | 95.33 | |
| C3 | 94.44 | 95.75 | 95.09 | |
| C4 | 95.79 | 96.50 | 96.14 | |
| MA | 95.83 |
(MA: micro-averaged; Sen: Sensitivity; Prc: Precision).
4.4. GSAF against SAPNF
Using the greedy version GSAF to select the two models, we chose the best two models as DenseNet201 with NLR of 1, and DenseNet201 with NLR of 2. Conversely, using the non-greedy algorithm SAPNF showed the two best models to be fused were DenseNet201 with NLR of 1 and ResNet101 with NLR of 1. The comparative results are presented in Table 7 .
Table 7.
GSAF against SAPNF on validation set.
| Selection Approach |
Selected Model |
Class | Sen (%) | Prc (%) | F1 (%) |
|---|---|---|---|---|---|
| GSAF | DenseNet201 (NLR =1) & DenseNet201 (NLR =2) |
C1 | 94.80 | 96.58 | 95.68 |
| C2 | 93.42 | 96.59 | 94.98 | ||
| C3 | 97.90 | 96.22 | 97.05 | ||
| C4 | 99.06 | 96.11 | 97.56 | ||
| MA | 96.37 | ||||
| SAPNF | DenseNet201 (NLR=1) & ResNet101 (NLR=1) |
C1 | 96.43 | 98.07 | 97.24 |
| C2 | 95.95 | 97.03 | 96.49 | ||
| C3 | 97.64 | 96.82 | 97.23 | ||
| C4 | 98.53 | 96.83 | 97.67 | ||
| MA | 97.18 |
(MA: micro-averaged; Sen: Sensitivity; Prc: Precision).
There are two findings we can observe from comparing Table 7 with Table 6. (i) First, fusion can give improved performance than individual models alone. The MA F1 score of the best single model was 96.35%, while the two fused model gave improved performance, with GSAF of 96.37%, and SAPNF of 97.18%. (ii) A non-greedy selection approach (SAPNF) can obtain better results than the greedy selection approach, GSAF. The reason is the two best models have similar advantages. For example, both DenseNet201 (NLR=1) and DenseNet201 (NLR=2) work optimally on the 3rd and 4th classes, so their fusion will not help to handle the weak spots (1st and 2nd classes). Nevertheless, the 3rd best model, i.e., RESNet101 (NLR = 1) shows exceptional classification ability on 1st and 2nd classes. Hence, fusing the 1st best model and 3rd best model is more logical, which is the core idea of our SAPNF.
4.5. Visual explanation of fusion
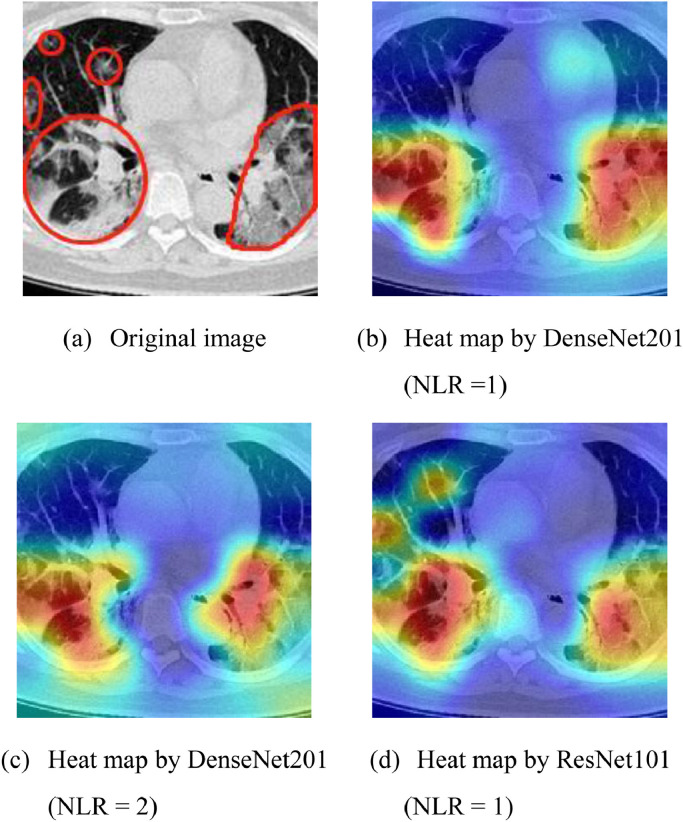
Grad-CAM [51] was used to illustrate why the fusion of Heat map by DenseNet201 (NLR =1) and Heat map by ResNet101 (NLR = 1) works the best among all possible fusion model combinations.
Fig. 9 displays the heat map results of a COVID-19 CCT slice by Grad-CAM over three models. Fig. 9(b, c, & d) presents the heat maps generated by DenseNet201 (NLR =1), DenseNet201 (NLR = 2), and ResNet101 (NLR = 1), respectively. We can observe that DenseNet201 networks with NLR equaling 1 & 2 capture the same GGO lesion on the bottom-half of the pictures (See Fig. 9b & c), so their fusion will not aid the other model. In contrast, ResNet101 (NLR=1) captures the top left GGO areas, which are neglected by the two DenseNet models. Thus, fusing DenseNet201 (NLR =1) and ResNet101 (NLR=1) is reasonable and has a solid visual explanation.
Fig. 9.
Grad-CAM result of a COVID-19 slice. (The “jet” pseudo-color was used. Red colors mean part and parcel areas for AI diagnosis, and blue colors less important areas for AI decision.).
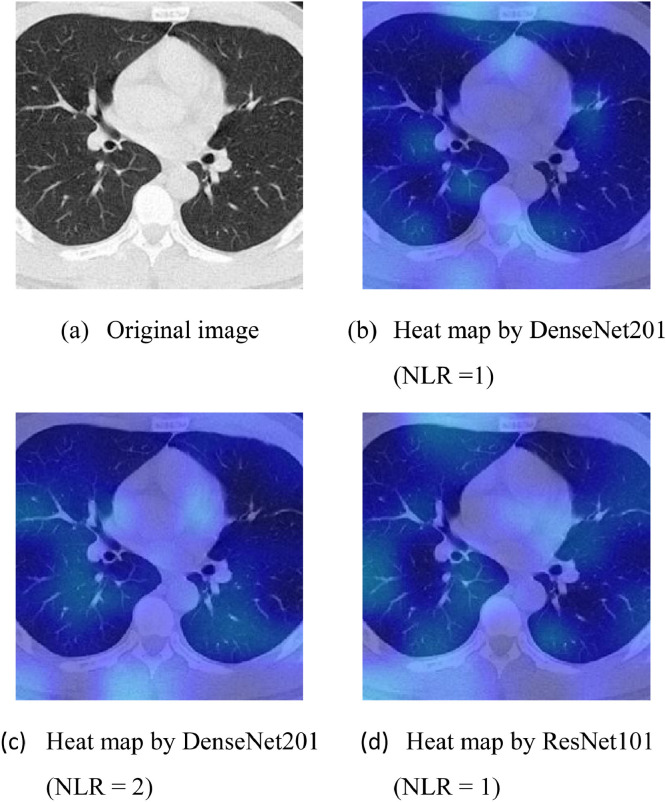
Fig. 10 displays the Grad-CAM heat map of a normal CCT slice using the top three models. Fig. 10(a) shows the original CCT image, and Fig. 10(b-d) gives the heat maps using DenseNet201 (NLR=1), DenseNet201 (NLR=2), and ResNet101 (NLR=1). All three AI models did not locate any strong indications of suspicious areas. Therefore, all three AI models classified this image as “normal”, which was subsequently confirmed by a radiologist.
Fig. 10.
Grad-CAM result on a normal case.
4.6. Performance of CCSHNet on the test set
After completing our previous experiments on the validation set, and selecting the optimal pretrained models and optimal NLR values, we ran our model CCSHNet, i.e., fusion of DenseNet201 (NLR =1) and ResNet101 (NLR=1) via DCA, on the test set and reported its performance. Test results are summarized in Table 8 . The sensitivities of four classes were 95.61%, 96.25%, 98.30%, and 97.86%, respectively. The precision values for the four classes were 97.32%, 96.42%, 96.99%, and 97.38%, respectively. The F1 scores of the four classes were 96.46%, 96.33%, 97.64%, and 97.62%, respectively. The MA F1 score of CCSHNet on test set was 97.04%, which is slight lower than the validation of 97.18% (See Table 7).
Table 8.
Performance of proposed CCSHNet on test set (%).
| Class | Sen (%) | Prc (%) | F1 (%) |
|---|---|---|---|
| C1 | 95.61 | 97.32 | 96.46 |
| C2 | 96.25 | 96.42 | 96.33 |
| C3 | 98.30 | 96.99 | 97.64 |
| C4 | 97.86 | 97.38 | 97.62 |
| MA | 97.04 |
(MA: micro-averaged; Sen: Sensitivity; Prc: Precision).
4.7. Comparison to state-of-the-art approaches
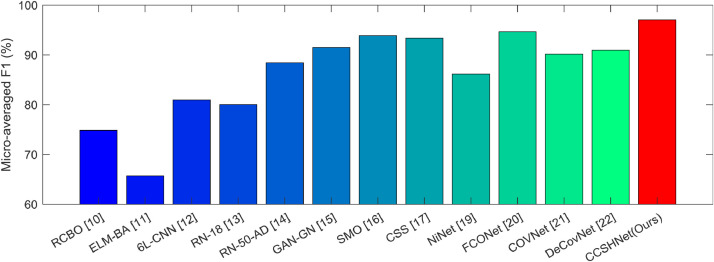
Proposed CCSHNet method was compared with 12 state-of-the-art approaches: RCBO [10], ELM-BA [11], 6L-CNN [12], RN-18 [13], RN-50-AD [14], GAN-GN [15], SMO [16], CSS [17], NiNet [19], FCONet [20], COVNet [21], and DeCovNet [22]. All these approaches were compared using our dataset. The comparison and their MA F1 plots are presented in Table 9 and Fig. 11 , respectively.
Table 9.
Comparison results of state-of-the-art methods.
| Method | Class | Sen (%) | Prc (%) | F1 (%) |
|---|---|---|---|---|
| RCBO [10] | C1 | 71.93 | 84.19 | 77.58 |
| C2 | 72.86 | 72.73 | 72.79 | |
| C3 | 73.56 | 76.41 | 74.96 | |
| C4 | 80.66 | 68.91 | 74.32 | |
| MA | 74.85 | |||
| ELM-BA [11] | C1 | 62.63 | 67.61 | 65.03 |
| C2 | 64.29 | 65.10 | 64.69 | |
| C3 | 71.86 | 66.77 | 69.22 | |
| C4 | 63.93 | 63.52 | 63.73 | |
| MA | 65.71 | |||
| 6L-CNN [12] | C1 | 72.46 | 83.94 | 77.78 |
| C2 | 78.93 | 77.82 | 78.37 | |
| C3 | 81.86 | 75.00 | 78.28 | |
| C4 | 89.84 | 87.54 | 88.67 | |
| MA | 80.94 | |||
| RN-18 [13] | C1 | 82.81 | 82.66 | 82.73 |
| C2 | 81.07 | 74.43 | 77.61 | |
| C3 | 74.24 | 76.98 | 75.58 | |
| C4 | 82.13 | 86.38 | 84.20 | |
| MA | 80.04 | |||
| RN-50-AD [14] | C1 | 87.72 | 85.03 | 86.36 |
| C2 | 87.68 | 91.26 | 89.44 | |
| C3 | 93.39 | 89.89 | 91.60 | |
| C4 | 84.92 | 87.65 | 86.26 | |
| MA | 88.41 | |||
| GAN-GN [15] | C1 | 91.75 | 89.86 | 90.80 |
| C2 | 92.86 | 91.87 | 92.36 | |
| C3 | 89.83 | 89.98 | 89.91 | |
| C4 | 91.64 | 94.27 | 92.93 | |
| MA | 91.50 | |||
| SMO [16] | C1 | 97.02 | 92.63 | 94.77 |
| C2 | 89.11 | 95.23 | 92.07 | |
| C3 | 94.92 | 94.92 | 94.92 | |
| C4 | 94.26 | 92.89 | 93.57 | |
| MA | 93.86 | |||
| CSS [17] | C1 | 94.04 | 92.25 | 93.14 |
| C2 | 93.75 | 95.11 | 94.42 | |
| C3 | 91.36 | 93.58 | 92.45 | |
| C4 | 94.43 | 92.75 | 93.58 | |
| MA | 93.39 | |||
| NiNet [19] | C1 | 87.89 | 91.59 | 89.70 |
| C2 | 80.89 | 85.47 | 83.12 | |
| C3 | 83.22 | 82.11 | 82.66 | |
| C4 | 92.30 | 85.95 | 89.01 | |
| MA | 86.18 | |||
| FCONet [20] | C1 | 92.28 | 95.64 | 93.93 |
| C2 | 96.79 | 94.43 | 95.59 | |
| C3 | 94.75 | 95.88 | 95.31 | |
| C4 | 94.92 | 92.94 | 93.92 | |
| MA | 94.68 | |||
| COVNet [21] | C1 | 89.82 | 86.63 | 88.20 |
| C2 | 89.82 | 92.63 | 91.21 | |
| C3 | 93.73 | 90.66 | 92.17 | |
| C4 | 87.38 | 90.96 | 89.13 | |
| MA | 90.17 | |||
| DeCovNet [22] | C1 | 91.05 | 90.58 | 90.81 |
| C2 | 93.75 | 90.99 | 92.35 | |
| C3 | 90.51 | 86.97 | 88.70 | |
| C4 | 88.69 | 95.58 | 92.01 | |
| MA | 90.94 | |||
| CCSHNet (Ours) |
C1 | 95.61 | 97.32 | 96.46 |
| C2 | 96.25 | 96.42 | 96.33 | |
| C3 | 98.30 | 96.99 | 97.64 | |
| C4 | 97.86 | 97.38 | 97.62 | |
| MA | 97.04 |
Fig. 11.
Comparison plot of MA F1 for our algorithm compared to 12 state-of-the-art methods.
The results in Table 9 and Fig. 11 demonstrate that our CCSHNet accomplished the best outcomes among all methods. The reason our CCSHNet obtains the best overall performance is that we have proposed several new algorithms to improve our fusion model: (L, 2) transfer feature learning (L2TFL), the selection algorithm of pretrained network for fusion (SAPNF), and deep CCT fusion discriminant correlation analysis (DCFDCA). The fusion framework demonstrates their effectiveness. Meanwhile, the proposed multiple-way data augmentation prevents our AI model from overfitting, thus increasing its performances.
Our method is unique in comparison to other strategies. The RCBO [10] used real-coded strategy in traditional biogeography-based optimization method; however, their method still needs to manually select the features, and they cannot validate their manually curated features to fit this four-class classification task. ELM-BA [11] used extreme learning classifier as the backbone, which employed random features (i.e., non-tuned random hidden nodes), so its performance may not be reliable. 6L-CNN [12] was proposed for fingerspelling classification during patients’ rehabilitation. It used leaky rectified linear unit to replace traditional rectified linear unit. Nevertheless, the structure itself is shallow (only six layers), thus may not handle the complicated internal mapping from CCT images to the four class labels. RN-18 [13] and RN-50-AD [14] used two variants of ResNet to classify thyroid ultrasound standard plane and Alzheimer's disease, respectively. The weights of the corresponding two networks were already adapted to their corresponding data, so retraining of the weights is required, which results in suboptimal performance. GAN-GN [15] combined generative adversarial network (GAN) and GoogleNet, but the image size and size of the dataset affects the generated images produced by GAN. SMO [16] used social mimic optimization for feature selection and fusion. Nevertheless, SMO's performance needs further verification. CSS [17] predicted COVID severity score in their model. We transferred the score prediction in their paper to COVID-19 recognition in this task. Those geographic extent score and lung opacity score may not have direct relation to our COVID-19 recognition, so this transfer is cross-field, which makes it more challenging. NiNet [19] combined 3D U-Net and MVP-Net. However, the 3D neural network needs more samples to train; otherwise it is susceptible to overfitting. FCONet [20] is a type of fast-track COVID-19 classification network. Again, the authors used ResNet50 and trained their models on three categories. In contrast our CCSHNet used deeper models and four categories of CCT images; hence, our model is more complicated and effective. COVNet [21] chose ResNet50, which has fewer layers than our proposed models (DenseNet201 and ResNet101). They trained their models with three categories; in contrast, our model was trained with four categories, which provides an additional class such as secondary pulmonary tuberculosis. DeCovNet [22] is a weakly-supervised DL method. Nevertheless, their model needs to train a UNet to extract lung regions, which requires more samples and more precise expert annotations.
5. Conclusions
This paper proposed a novel CCSHNet for COVID-19 detection in CCTs. Our model is based on the proposed DCFDCA algorithm of the selected two optimal models, of which we developed a SAPNF algorithm to optimally determine the best PTM and NLR. The feature learning procedures of the two models were achieved by the proposed L2TFL algorithm. Overall, our experiments showed our CCSHNet can achieve the best performance compared to 12 state-of-the-art approaches, and potentially aid radiologists in making more accurate, quicker diagnoses of COVID-19 using CCTs.
The impacts of our method in hospitals and society are promising. From the experimental results, our CCSHNet system can aid decision making when diagnosing lung-related diseases using CCTs. Furthermore, our CCSHNet can be improved by integration with other AI models developed by other teams from other universities/countries. In addition; our algorithm has the potential to be re-deployed to a new hospital's server, with little costs if using cloud-computing based techniques.
The shortcomings of our CCSHNet are three-fold: (i) It cannot handle heterogeneous data, such as the mixed data of CCT with CXR and patient history and other data. (ii) It has not yet been through a strict clinical verification. (iii) The dataset in this study is size-limited and category-limited.
The future work contains following aspects: (i) Expand the size of the dataset and test CCSHNet model on a larger and heterogeneous dataset. (ii) Try to use some advanced PTMs, particularly those trained from medical lung images. (iii) Try some advanced data preprocessing techniques to check whether the performance of our AI system can be improved. (iv) Our AI system can be embedded into other automated healthcare systems [52], [53], [54]. (v) IoT [55], [56], [57], [58] and communication technologies [59] can help make our AI system more powerful. (vi) Some advanced or hybrid fusion rules will be tested.
CRediT authorship contribution statement
Shui-Hua Wang: Conceptualization, Methodology, Software, Validation, Data curation, Writing - original draft, Investigation, Data curation. Deepak Ranjan Nayak: Formal analysis, Writing - original draft, Writing - review & editing. David S. Guttery: Writing - original draft, Writing - review & editing. Xin Zhang: Writing - original draft, Writing - review & editing. Yu-Dong Zhang: Resources, Formal analysis, Investigation, Data curation, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This paper is partially supported by British Heart Foundation Accelerator Award, UK; Royal Society International Exchanges Cost Share Award, UK (RP202G0230); Hope Foundation for Cancer Research, UK (RM60G0680); Medical Research Council Confidence in Concept Award, UK (MC_PC_17171).
Appendix A
Table 10.
Abbreviation List.
| Abbreviation | Full Name |
|---|---|
| (M)DA | (multiple-way) data augmentation |
| BCS | between-class scatter |
| BSC | between-set covariance |
| CAP | community-acquired pneumonia |
| CCA | canonical correlation analysis |
| CCT | chest computed tomography |
| DAF | data augmentation factor |
| DCA | discriminant correlation analysis |
| DCFDCA | deep CCT fusion by discriminant correlation analysis |
| DDM | diagonally dominant matrix |
| DL | deep learning |
| DLF | decision-level fusion |
| FCL | fully-connected layer |
| FLF | feature-level fusion |
| GSAF | greedy selection algorithm for fusion |
| HC | healthy control |
| ISP | incompatible size problem |
| L2TFL | (L,2) transfer feature learning |
| MRL | maximum removable layer |
| MV | Majority voting |
| NLR | number of layers to be removed |
| OHNN | one-hidden layer neural network |
| PF | parallel fusion |
| PTM | pre-trained model |
| SAPNF | selection algorithm of pretrained networks for fusion |
| SF | serial fusion |
| SPT | secondary pulmonary tuberculosis |
| SVD | singular value decomposition |
| TL | transfer learning |
Appendix B
Table 11.
Symbol List.
| Symbol | Meaning |
| raw dataset | |
| raw slice CCT image | |
| size of | |
| index of | |
| labeling | |
| radiologist | |
| class (viz., COVID, CAP, SPT, HC) | |
| grayscale operation | |
| minimum grayscale of an image | |
| maximum grayscale of an image | |
| crop operation | |
| crop values in unit of pixel from four directions | |
| preprocessed dataset | |
| preprocessed slice CCT image | |
| size of | |
| storage compression ratio | |
| size compression ratio | |
| data, labeling, and classifier of source domain | |
| data, labeling, and classifier of target domain | |
| error function | |
| learning rate | |
| number of PTMs | |
| a specified model among all six models. See Table 3. | |
| a pretrained model | |
| with last L layers removed | |
| with 2 new fully connected layers added | |
| retrained | |
| k-th PTM | |
| 1st model | |
| 2nd model | |
| number of last layers to be removed | |
| number of last layers to be removed at 1st model | |
| number of last layers to be removed at 2nd model | |
| number of learnable layers of | |
| number of learnable layers of | |
| maximum removable layer | |
| layers from to in network | |
| features learnt from network | |
| remove layer function | |
| add fully-connected layer function | |
| retrain function | |
| activation function | |
| node number of first added FCL layer | |
| node number of second added FCL layer | |
| training, validation, and test set of a dataset . | |
| number of hidden neurons | |
| initialized OHNN | |
| trained OHNN | |
| sort function in descending way | |
| rank list | |
| indicator by k-th PTM and removing layers | |
| indicator vector | |
| proposed L2TFL operation | |
| output on validation set | |
| output on test set | |
| measuring indicator function | |
| fused feature | |
| deep fusion function | |
| features to be fused from two models | |
| serial fusion operation | |
| parallel fusion operation | |
| length of feature | |
| number of trained features. | |
| cross-covariance operation | |
| transformation matrix of CCA for model 1 | |
| transformation matrix of CCA for model 1 | |
| transformed features by CCA | |
| concatenation of CCA features | |
| summation of CCA features | |
| feature extracted from i th image of j-th category via model | |
| between-class scatter matrix | |
| matrix of orthogonal eigenvectors | |
| diagonal matrix of real and non-negtive eigenvalue in decreasing order. | |
| rank function | |
| projection of where the BCS matrix is | |
| between-set covariance matrix of transformed feature sets | |
| transform matrix of DCA for model 1 | |
| transform matrix of DCA for model 2 | |
| transformed feature sets by DCA | |
| concatenation of DCA features | |
| summation of DCA features | |
| number of different DA techniques | |
| number of generated images by each offline MDA technique | |
| one training image | |
| training set | |
| validation set | |
| mean of Gaussian noise injected | |
| variance of Gaussian noise injected | |
| noise injection operation | |
| horizontal shift transform function | |
| vertical shift transform function | |
| Gamma correction operation | |
| image rotation operation | |
| random translation operation | |
| random horizontal shift | |
| random vertical shift | |
| maximum shift range | |
| uniform distribution | |
| image scaling operation | |
| mirror function | |
| concatenation operation | |
| collection of generated MDA images with original image | |
| set of all augmented images | |
| data augmentation factor | |
| non-test set of preprocessed dataset | |
| test set of preprocessed dataset | |
| number of samples of non-test set in k-th class | |
| number of samples of test set in k-th class. | |
| ideal confusion matrix over validation set | |
| ideal confusion matrix over test set | |
| number of runs on validation set | |
| number of runs on test set | |
| micro-averaged F1 | |
| micro-averaged precision | |
| micro-averaged sensitivity | |
| run index | |
| fold index |
Algorithm 1.
Proposed L2TFL algorithm.
| Step 1 Read one raw PTM network , |
| Step 2 Remove the last NLR l-learnable layers from and get , , |
| Step 3 Add two new fully connected layers, , |
| Step 4 Freeze early layers, , |
| Step 5 Let last two layers retrainable, , |
| Step 6 Retrain the whole network, and obtain the new network , |
| Step 7 Output learnt features . |
Algorithm 2.
Proposed GSAF for PTM selection.
| Step 1 Input: Training set and validation set |
| Step 2 for k = 1: (k is the index of PTM) |
| for (L is the index of NLR) |
| Step 2.1 PTM Retrain |
| Import -th PTM , |
| Use L2TFL via data and removing layers, |
| Obtain , |
| Step 2.2 Feature Extraction |
| Generate features from . |
| Step 2.3 Train OHNN |
| Initialize OHNN , |
| Train OHNN using input as , |
| Obtain , |
| Step 2.4 Obtain Indicator |
| Obtain performance indicator over validation set |
| end |
| end |
| Step 3 Generate and sort the indicator vector |
| Step 4 Obtain the rank list , |
| Step 5 Choose the top two best models (determine PTM and NLR): |
| and |
Algorithm 3.
Proposed SAPNF for PTM selection.
| Step 1 Input: Training set and validation set |
| Step 2 for ( is the index of PTM of 1st model) |
| for ( is the index of NLR of 1st model) |
| for ( is the index of PTM of 1st model) |
| for ( is the index of NLR of 1st model) |
| Step 2.1 1st model Retrain |
| Import -th PTM , |
| Use L2TFL via data and removing layers, |
| Obtain , |
| Step 2.2 2nd model Retrain |
| Import -th PTM , |
| Use L2TFL via data and removing layers, |
| Obtain |
| Step 2.3 Feature Extraction from two retrained PTMs |
| Generate features from , |
| Generate features from , |
| Step 2.4 Feature Fusion |
| Obtain |
| Step 2.5 Train OHNN |
| Initialize OHNN , |
| Train OHNN using input as , |
| Obtain , |
| Step 2.6 Obtain Indicator |
| Obtain performance indicator over validation set |
| end |
| end |
| end |
| end |
| Step 3 Generate and sort the indicator vector |
| Step 4 Obtain the rank list , |
| Step 5 Choose the top two best models (determine PTM and NLR) |
| and |
Algorithm 4.
Pseudocode of our CCSHNet algorithm.
| Input: Original Image Set and its ground truth label . |
| Phase I: Preprocessing |
| Grayscaling: . See Eq. (3). |
| HS: , See Eq. (5). |
| Crop: See Eq. (6). |
| Downsampling: See Eq. (7). |
| Phase II: Ten-folds CV on Non-test Set |
| Split into nontest set and test set: |
| for % is run index |
| for % is fold index |
| Step II.A Split into 10 folds |
| Split nontest set into 10 folds . |
| Step II.B Create Training and Validation set |
| Training Set: |
| Validation Set: |
| Step II.C MDA on training set |
| for |
| Training image: and its ground truth labels . |
| : i th training image in -th run |
| . See Eq. (66). |
| end |
| DA enhanced training set: . |
| Enhanced training set labels: . |
| Step II.D Model Selection by SAPNF, L2TFL, and DCFDCA. |
| See Algorithm 3, Algorithm 1, and Fig. 5. |
| Step II.E Validation confusion matrix at-th run and-th fold |
| Record , See Eq. (68) |
| End |
| Validation confusion matrix at -th run |
| end |
| Phase III: PTM and NLR Selection |
| Validation confusion matrix. See Eq. (70). |
| Indicator is chosen as micro-averaged F1. |
| Obtain . See Eq. (75) |
| Obtain the rank list . See Eq. (28). |
| Output the top two models, i.e., best PTM and NLR combinations. |
| Output and and the corresponding removed layers and |
| Phase IV: Create CCSHNet Model |
| Select the two optimal models and . |
| Feature learning by L2TFL with NLR and layers removed. |
| Deep CCT fusion by DCFDCA. |
| OHNN . |
| Phase V: Report the test performance of the CCSHNet model |
| Training set is , and its labels . |
| Test set is , and its labels . |
| for % is run index |
| We initialized a random seed at each run. |
| Trained CCSHNet Model |
| Prediction: ; |
| Test confusion matrix at -th run: . |
| Calculate Indicators. See Eq. (72)-(77). |
| End |
| Test confusion matrix: See Eq. (71). |
| Calculate indicators. |
| Output: The best model CCSHNet and its test performances. |
Fig. 5.
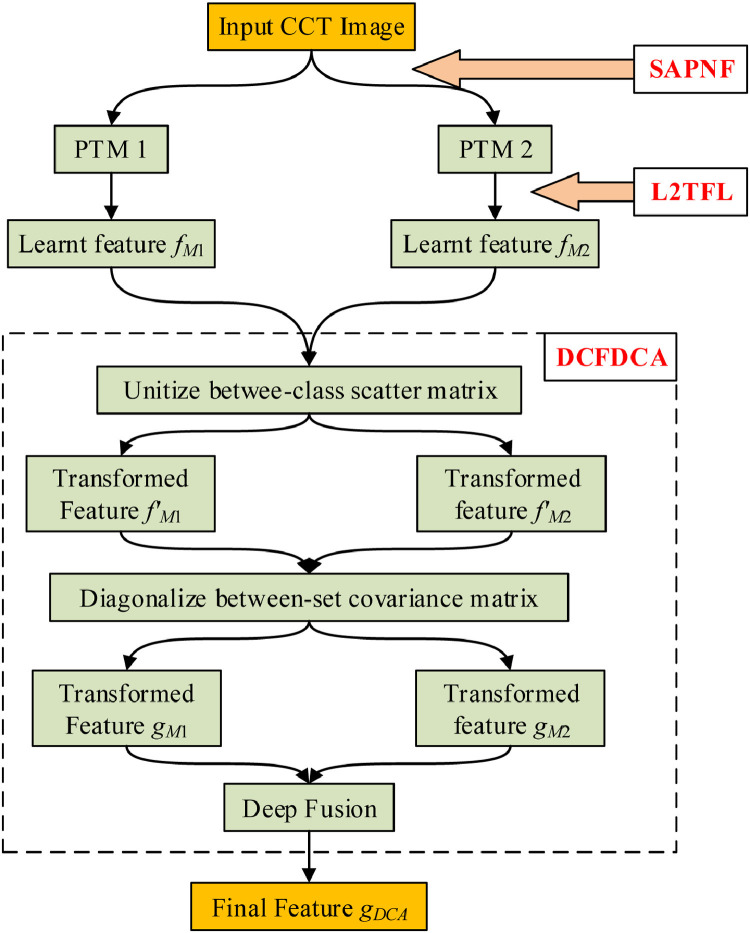
Diagram of our proposed fusion method, indicating the relation among SAPNF, L2TFL, and DCFDCA. (SAPNF: selection algorithm of pretrained networks for fusion; L2TFL: (L, 2) transfer feature learning; DCFDCA: deep CCT fusion by discriminant correlation analysis; CCT: chest CT; PTM: pretrained model).
References
- 1.COVID-19 CORONAVIRUS PANDEMIC, 2020. (12/Oct/2020). Available: https://www.worldometers.info/coronavirus.
- 2.A. Azar, D.E. Wessell, J.R. Janus, and L.V. Simon. Fractured aluminum nasopharyngeal swab during drive-through testing for COVID-19: radiographic detection of a retained foreign body. Skeletal Radiol. [Article; Early Access]. 5 (2020). doi: 10.1007/s00256-020-03582-x. [DOI] [PMC free article] [PubMed]
- 3.de Barry O., Obadia I., Hajjam M.El, Carlier R.Y. Chest-X-ray is a mainstay for follow-up in critically ill patients with covid-19 induced. Eur. J. Radiol. 2020;129(2) doi: 10.1016/j.ejrad.2020.109075. Article ID: 109075, Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herpe G., Tasu J.P. Impact of the Prevalence on the Predictive Positive Value of Chest CT in the Diagnosis of Coronavirus Disease (COVID-19) Am. J. Roentgenol. 2020;215:W39. doi: 10.2214/AJR.20.23530. Sep. [DOI] [PubMed] [Google Scholar]
- 5.Willman D. The Washington Post [Internet]; 2020. Contamination At CDC Lab Delayed Rollout of Coronavirus Tests. [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: a Report of 1014 Cases,". Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imre A. A Typical Chest CT Appearance of a Case with Coronavirus Disease 2019 (COVID-19), Erciyes Med. J. Sep, 2020;42:346–347. [Google Scholar]
- 8.Flor N., Tonolini M. From ground-glass opacities to pulmonary emboli. A snapshot of the evolving role of a radiology unit facing the COVID-19 outbreak. Clin. Radiol. 2020;75:556–557. doi: 10.1016/j.crad.2020.04.009. Jul, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fry C.V., Cai X.J., Zhang Y., Wagner C.S. Consolidation in a crisis: patterns of international collaboration in early COVID-19 research. PLoS ONE. 2020;15:15. doi: 10.1371/journal.pone.0236307. Article ID: e0236307, Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li P., Liu G. Pathological Brain Detection via Wavelet Packet Tsallis Entropy and Real-Coded Biogeography-based Optimization. Fundam. Inform. 2017;151:275–291. [Google Scholar]
- 11.Lu S. A Pathological Brain Detection System based on Extreme Learning Machine Optimized by Bat Algorithm. CNS Neurol. Dis. - Drug Targets. 2017;16:23–29. doi: 10.2174/1871527315666161019153259. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X. Chinese Sign Language Fingerspelling Recognition via Six-Layer Convolutional Neural Network with Leaky Rectified Linear Units for Therapy and Rehabilitation. J. Med. Imaging Health Inform. 2019;9:2031–2038. [Google Scholar]
- 13.Guo M.H., Du Y.Z. Classification of Thyroid Ultrasound Standard Plane Images using ResNet-18 Networks. IEEE 13th International Conference on Anti-Counterfeiting, Security, and Identification; Xiamen, China; 2019. pp. 324–328. [Google Scholar]
- 14.Fulton L.V., Dolezel D., Harrop J., Yan Y., Fulton C.P. Classification of Alzheimer's Disease with and without Imagery Using Gradient Boosted Machines and ResNet-50. Brain. Sci. 2019;9:16. doi: 10.3390/brainsci9090212. Article ID: 212, Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loey M., Smarandache F., Khalifa N.E.M. Within the Lack of Chest COVID-19 X-ray Dataset: a Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry-Basel. 2020;12:19. Article ID: 651, Apr. [Google Scholar]
- 16.Togacar M., Ergen B., Comert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121:12. doi: 10.1016/j.compbiomed.2020.103805. Article ID: 103805, Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen J.P., Dao L., Morrison P., Roth K., Bengio Y., Shen B.Y. Predicting COVID-19 Pneumonia Severity on Chest X-ray With Deep Learning. Cureus. 2020;12:10. doi: 10.7759/cureus.9448. Article ID: e9448, Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabik S., Gómez-Ríos A., Martín-Rodríguez J., Sevillano-García I., Rey-Area M., Charte D. 2020. COVIDGR Dataset and COVID-SDNet Methodology For Predicting COVID-19 Based On Chest X-Ray Images. arXiv Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni Q.Q., Sun Z.Y., Qi L., Chen W., Yang Y., Wang L. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020:11. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko H., Chung H., Kang W.S., Kim K.W., Shin Y., Kang S.J. COVID-19 Pneumonia Diagnosis Using a Simple 2D Deep Learning Framework With a Single Chest CT Image: model Development and Validation. J. Med. Internet Res. 2020;22:13. doi: 10.2196/19569. Article ID: e19569, Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT: evaluation of the Diagnostic Accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X.G., Deng X.B., Fu Q., Zhou Q., Feng J.P., Ma H. A Weakly-Supervised Framework for COVID-19 Classification and Lesion Localization From Chest CT. IEEE Trans. Med. Imaging. Aug, 2020;39:2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 23.Satapathy S.C., Zhu L.Y., Górriz J.M. A seven-layer convolutional neural network for chest CT based COVID-19 diagnosis using stochastic pooling. IEEE Sens. J. 2020:1. doi: 10.1109/JSEN.2020.3025855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X. Diagnosis of COVID-19 by Wavelet Renyi Entropy and Three-Segment Biogeography-Based Optimization. Int. J. Comput. Intelligence Syst. 2020;13:1332–1344. 2020-09-17T09:29:20.000Z. [Google Scholar]
- 25.Chen Y. A Feature-Free 30-Disease Pathological Brain Detection System by Linear Regression Classifier. CNS Neurol. Dis. - Drug Targets. 2017;16:5–10. doi: 10.2174/1871527314666161124115531. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y. Wavelet energy entropy and linear regression classifier for detecting abnormal breasts. Multimed. Tools Appl. 2018;77:3813–3832. [Google Scholar]
- 27.Farhood H., Perry S., Cheng E., Kim J. Enhanced 3D Point Cloud from a Light Field Image. Remote Sens. (Basel) 2020;12 Article ID: 1125, Apr. [Google Scholar]
- 28.Debnath S., Talukdar F.A. Brain tumour segmentation using memory based learning method. Multimed. Tools. Appl. Aug, 2019;78:23689–23706. [Google Scholar]
- 29.Glatt R., Da Silva F.L., Bianchi R.A.D., Costa A.H.R. DECAF: deep Case-based Policy Inference for knowledge transfer in Reinforcement Learning. Expert Syst. Appl. 2020;156:13. Article ID: 113420, Oct, [Google Scholar]
- 30.Benbahria Z., Sebari I., Hajji H., Smiej M.F. Intelligent mapping of irrigated areas from landsat 8 images using transfer learning. Int. J. Eng. Geoscie. 2021;6:41–51. Feb, [Google Scholar]
- 31.Hundt A., Killeen B., Greene N., Wu H.T., Kwon H., Paxton C. Good Robot!": efficient Reinforcement Learning for Multi-Step Visual Tasks with Sim to Real Transfer. IEEE Robotics Automation Lett. 2020;5:6724–6731. Oct, [Google Scholar]
- 32.Gessert N., Bengs M., Wittig L., Dr?mann D., Keck T., Schlaefer A. Deep transfer learning methods for colon cancer classification in confocal laser microscopy images. Int. J. Comput. Assist. Radiol. Surg. 2019;14:1837–1845. doi: 10.1007/s11548-019-02004-1. Nov, [DOI] [PubMed] [Google Scholar]
- 33.Hassanpour M., Malek H. Learning Document Image Features With SqueezeNet Convolutional Neural Network. Int. J. Eng. Jul, 2020;33:1201–1207. [Google Scholar]
- 34.Hirano G., Nemoto M., Kimura Y., Kiyohara Y., Koga H., Yamazaki N. Automatic diagnosis of melanoma using hyperspectral data and GoogLeNet. Skin Res. Technol. [Article; Early Access]. 2020;7 doi: 10.1111/srt.12891. [DOI] [PubMed] [Google Scholar]
- 35.Venturi L., Bandeira A.S., Bruna J. Spurious Valleys in One-hidden-layer Neural Network Optimization Landscapes. J. Mach. Learn. Res. 2019;20:34. Article ID: 133. [Google Scholar]
- 36.Planet S., Iriondo I. Comparison between decision-level and feature-level fusion of acoustic and linguistic features for spontaneous emotion recognition. 7th Iberian Conference on Information Systems and Technologies (CISTI 2012; Madrid, Spain; 2012. pp. 1–6. [Google Scholar]
- 37.Gunatilaka A.H., Baertlein B.A. Feature-level and decision-level fusion of noncoincidently sampled sensors for land mine detection. IEEE Trans. Pattern Anal. Mach. Intell. 2001;23:577–589. [Google Scholar]
- 38.Grover J., Hanmandlu M. Hybrid fusion of score level and adaptive fuzzy decision level fusions for the finger-knuckle-print based authentication. Appl. Soft Comput. 2015;31:1–13. 2015/06/01/ [Google Scholar]
- 39.Liu C.J., Wechsler H. A shape- and texture-based enhanced fisher classifier for face recognition. IEEE Transactions on Image Processing. 2001;10:598–608. doi: 10.1109/83.913594. Apr. [DOI] [PubMed] [Google Scholar]
- 40.Yang J., Yang J.Y. Generalized K-L transform based combined feature extraction. Pattern Recognit. 2002;35:295–297. Jan. [Google Scholar]
- 41.Sun Q.S., Zeng S.G., Liu Y., Heng P.A., Xia D.S. A new method of feature fusion and its application in image recognition. Pattern Recognit. 2005;38:2437–2448. Dec. [Google Scholar]
- 42.Haghighat M., Abdel-Mottaleb M., Alhalabi W. Discriminant Correlation Analysis: real-Time Feature Level Fusion for Multimodal Biometric Recognition. IEEE Trans. Inform. Forensics Security. 2016;11:1984–1996. Sep. [Google Scholar]
- 43.Chaib S., Liu H., Gu Y.F., Yao H.X. Deep Feature Fusion for VHR Remote Sensing Scene Classification. IEEE Trans. Geosci. Remote Sens. Aug, 2017;55:4775–4784. [Google Scholar]
- 44.Wang S.-.H., Govindaraj V.V., Górriz J.M., Zhang X., Zhang Y.-.D. Information Fusion; 2020. Covid-19 Classification by FGCNet With Deep Feature Fusion from Graph Convolutional Network and Convolutional Neural Network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazzini D., Napoletano P., Piccoli F., Schettini R. A Novel Approach to Data Augmentation for Pavement Distress Segmentation. Comput. Industry. 2020;121 Article ID: 103225, Oct. [Google Scholar]
- 46.Duncan M.J., Eyre E.L.J., Cox V., Roscoe C.M.P., Faghy M.A., Tallis J. Cross-validation of Actigraph derived accelerometer cut-points for assessment of sedentary behaviour and physical activity in children aged 8-11 years. Acta Paediatr. Sep, 2020;109:1825–1830. doi: 10.1111/apa.15189. [DOI] [PubMed] [Google Scholar]
- 47.Hasnain M., Pasha M.F., Ghani I., Imran M., Alzahrani M.Y., Budiarto R. Evaluating Trust Prediction and Confusion Matrix Measures for Web Services Ranking. IEEE Access. 2020;8:90847–90861. [Google Scholar]
- 48.Pondenkandath V., Alberti M., Eichenberger N., Ingold R., Liwicki M. Cross-Depicted Historical Motif Categorization and Retrieval with Deep Learning. J. Imaging. 2020;6:20. doi: 10.3390/jimaging6070071. Article ID: 71, Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandes B., Gonzalez-Briones A., Novais P., Calafate M., Analide C., Neves J. An Adjective Selection Personality Assessment Method Using Gradient Boosting Machine Learning. Processes. 2020;8:24. Article ID: 618, May. [Google Scholar]
- 50.Krsnik I., Glavas G., Krsnik M., Miletic D., Stajduhar I. Automatic Annotation of Narrative Radiology Reports. Diagnostics. 2020;10:15. doi: 10.3390/diagnostics10040196. Article ID: 196Apr, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-CAM: visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020;128:336–359. 2020/02/01. [Google Scholar]
- 52.Zhang Y., Qian Y.F., Wu D., Hossain M.S., Ghoneim A., Chen M. Emotion-Aware Multimedia Systems Security. IEEE Trans. Multimedia. Mar, 2019;21:617–624. [Google Scholar]
- 53.Zhang Y., Gravina R., Lu H.M., Villari M., Fortino G. PEA: parallel electrocardiogram-based authentication for smart healthcare systems. J. Netw. Comput. Appl. Sep, 2018;117:10–16. [Google Scholar]
- 54.Zhang Y., Li Y., Wang R., Lu J., Ma X., Qiu M. PSAC: proactive Sequence-aware Content Caching via Deep Learning at the Network Edge. IEEE Trans. Netw. Scie. Eng. 2020:1. doi: 10.1109/TNSE.2020.2990963. [DOI] [Google Scholar]
- 55.Zhang Y., Wang R.R., Hossain M.S., Alhamid M.F., Guizani M. Heterogeneous Information Network-Based Content Caching in the Internet of Vehicles. IEEE Trans. Veh. Technol. Oct, 2019;68:10216–10226. [Google Scholar]
- 56.Zhang Y., Ma X., Zhang J., Hossain M.S., Muhammad G., Amin S.U. Edge Intelligence in the Cognitive Internet of Things: improving Sensitivity and Interactivity. IEEE Netw. 2019;33:58–64. May-Jun. [Google Scholar]
- 57.Zhang Y., Li Y., Wang R., Hossain M.S., Lu H. Multi-Aspect Aware Session-Based Recommendation for Intelligent Transportation Services. IEEE Trans. Intell. Transport. Syst. 2020:1–10. doi: 10.1109/TITS.2020.2990214. [DOI] [Google Scholar]
- 58.Zhang Y., Wen H., Qiu F., Wang Z., Abbas H. iBike: intelligent public bicycle services assisted by data analytics. Future Gen. Comput. Syst. 2019;95:187–197. 2019/06/01/ [Google Scholar]
- 59.Zhang Y., Hossain M.S., Ghoneim A., Guizani M. COCME: content-Oriented Caching on the Mobile Edge for Wireless Communications. IEEE Wireless Commun. 2019;26:26–31. [Google Scholar]