Abstract
The lethal novel coronavirus disease 2019 (COVID-19) pandemic is affecting the health of the global population severely, and a huge number of people may have to be screened in the future. There is a need for effective and reliable systems that perform automatic detection and mass screening of COVID-19 as a quick alternative diagnostic option to control its spread. A robust deep learning-based system is proposed to detect the COVID-19 using chest X-ray images. Infected patient's chest X-ray images reveal numerous opacities (denser, confluent, and more profuse) in comparison to healthy lungs images which are used by a deep learning algorithm to generate a model to facilitate an accurate diagnostics for multi-class classification (COVID vs. normal vs. bacterial pneumonia vs. viral pneumonia) and binary classification (COVID-19 vs. non-COVID). COVID-19 positive images have been used for training and model performance assessment from several hospitals of India and also from countries like Australia, Belgium, Canada, China, Egypt, Germany, Iran, Israel, Italy, Korea, Spain, Taiwan, USA, and Vietnam. The data were divided into training, validation and test sets. The average test accuracy of 97.11 ± 2.71% was achieved for multi-class (COVID vs. normal vs. pneumonia) and 99.81% for binary classification (COVID-19 vs. non-COVID). The proposed model performs rapid disease detection in 0.137 s per image in a system equipped with a GPU and can reduce the workload of radiologists by classifying thousands of images on a single click to generate a probabilistic report in real-time.
Keywords: Chest X-ray radiographs, Coronavirus, Deep learning, Image processing, Pneumonia
1. Introduction
An eruption of novel coronavirus disease or COVID-19 (previously known as 2019-nCoV) started in China in December 2019. As of 16th September 2020, more than 29.5 million cases have been reported in more than 188 countries, and it has arrond 1 million deaths [1]. COVID-19 is a disease caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) that can be severe in patients with comorbidities and has a fatality rate of 2% [2]. There is an urgent need to take an effective step for the containment of COVID-19 by performing screening tests on a suspected fellow so that the infected person can receive immediate care with more specific treatment and quarantine of the patient can be ensured to limit the spread of the virus.
The SARS-CoV-2 infection has a wide range of clinical manifestations ranging from asymptomatic infection and mild upper respiratory tract illness to severe viral pneumonia that may culminate in failure of the respiratory system and sometimes death [3]. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) tests are performed for the qualitative detection of nucleic acid from upper and lower respiratory tract specimens (i.e. nasal, lower respiratory tract aspirates, sputum, nasopharyngeal or oropharyngeal swabs, nasal aspirate) of infected person [4]. Performing RT-PCR testing for COVID-19 will most probably remain the main detection method. However, it is expensive, complicated, and time-consuming for countless patients with a lack of time. Because of the shortage of kits for RT-PCR and relatively high false-negative rate, alternative methods such as the examination of chest X-rays (CXR) can be used for screening. It may be noted that the detection of lung involvement may predict a potentially life-threatening outcome in patients with COVID-19 [5,6].
CXR is a non-invasive imaging method and X-rays of the chest are usually done in either anteroposterior (AP view) or Posterior anterior (PA view) of a suspected patient's chest to generate cross-sectional images [7]. These X-ray images are examined by expert radiologists to find abnormal features suggestive of COVID-19 based on extent and type of lesions. Imaging features of the X-ray image of coronavirus affected persons varies as these depend on the stage of infection. The spectrum of radiological findings varies from normal (18% of cases) to 'whiteout Lung'. The usual abnormality seen is bilateral peripheral sub-pleural ground-glass opacities (GGO) and consolidations. "Crazy-paving" pattern and reversed halo sign may be seen [5,6]. There may be a rapid progression in the extent of the lesion in 24–48 h to multilobar to total lung involvement in severe disease [8]. With an increase in the number of patients with COVID-19 disease, the medical community may have to depend on portable CXR images because of its extensive accessibility and reduced infection controlling issues which presently limit the sutilisation of computed tomography (CT) services. With an increase in patient numbers, the workload on radiologists for this diagnostic process is also increasing and lack of availability of radiologists in certain places is also a challenge. Thus, there is an urgent requirement of a device or system which identifies the disease with an acceptable level of accuracy, even without a radiologist's help to save time as well as to preserve the effort for the neediest in these time-constrained settings.
The main contribution of the work is to develop a deep learning-based system that can automatically identify the COVID-19 disease in CXR images. For this purpose, we collected so far the largest dataset of COVID-19 patients and examined several different architectures where the most accurate was identified. The used dataset contains CXR database of 659 COVID-19, 1660 healthy and 4265 non-COVID (viral and bacterial pneumonia) samples which were also extended by 300 abnormal samples. Those samples were collected from three local hospitals of India and other countries like China, Italy, Australia, Iran, Spain, Germany, Vietnam, Israel Belgium, Canada, USA, Egypt, Korea and Taiwan making the multiple country dataset comprised of the large variety that may train the model for high robustness. The dataset was split into training, validation (5-fold cross-validation) and test sets. Different data configuration were examined including binary classification (COVID-19 or non-COVID), three-class classification (COVID-19, pneumonia or non-COVID), and four-class classification model (COVID-19, normal, bacterial pneumonia, viral pneumonia). The results outperform the previous works in terms of accuracy, speed, and other parameters. CXR images are augmented and annotated to introduce more variation in the dataset and develop a robust Convolutional Neural Network (CNN) model. The proposed framework can be ported into single board computers for low-cost and portable screening framework.
The variety of the sample images was collected from various public data sources and globally from several countries and extended from data collected from several hospitals. Thanks to this, we expect to achieve high comprehensiveness of the train model and robustness among a variety of different imaging devices with various settings. We suppose we achieved higher acceptance among a wide range of countries. The image test takes approximately 137 milliseconds per image (system with NVIDIA GeForce GTX 1060 GPU) thus making the model suitable for the online screening of COVID-19 patients on any system equipped with a modern GPU device. The dataset was released online so anyone can benefit from the work or can also extend the work in future with other sources. The experiment is fully reproducible.
The rest of the paper is structured as follows. Section 2 discusses the related works in the field of diagnosis of COVID-19. Section 3 describes the datasets used in the experiment and how it was created. It also discusses clinical aspects of the problem and architectures used for detection of COVID-19 cases, including the methodology used for evaluation. Section 4 includes experiments and results for COVID-19 detection and comparison to other works. Finally, section 5 concludes the paper and discusses possibilities regarding future work.
2. Related work
Artificial Intelligence assisted image tests can help in rapid detection of COVID-19 and subsequently control the influence of the disease. Medical image analysis for disease classification is one of the highest priority research areas. With the help of expert radiologists and based on the aforementioned features of CXR images, a computer-aided diagnostic system can be generated to correctly interpret COVID-19 cases from the input X-ray image. Several studies have been conducted with this motivation to lower the over-burden on medical professionals and contribute towards the rapid screening COVID-19 sutilising artificial intelligence and machine learning.
Several artificial intelligence-based systems using deep learning [9] as a pre-screening test for COVID-19 detection using CXR images are discussed in Refs. [10,11]. Narin et al. [12] and Zhang et al. [13] used ResNet 50 and basic ResNet, respectively, as a base neural network to classify normal (healthy) and COVID-19 patients. A range of fine-tuned deep convolutional neural network (DCNN) based COVID-19 detection method proposed by Khalid et al. [14] for classification of CXR image into normal (healthy) and pneumonia. Khan et al. [15] proposed CoroNet for detection of COVID-19 in which CXR images are trained on Xception deep neural architecture for COVID-19 classification.
Wang et al. [16] used deep-learning-based architecture COVID-Net for CXR image classification, which achieved the accuracy of 93.3%. Fine-tuned SqueezeNet is proposed by Ucar and Korkmaz [17] for COVID-19 diagnosis with Bayesian soptimisation additive giving the accuracy of 98.3% on CXR images. Ghosal and Tucker [18] used CXR images for computer-aided diagnostic of COVID-19 and normal (healthy) patients. The CNN model is trained with 70 COVID-19 and other images of normal subjects and claimed 92% accuracy with that dataset. Vaid et al. [19] used a public dataset of 181 COVID-19 images, 364 healthy images to detect COVID-19 using deep transfer learning. Their model achieved the accuracy of 96.3% and loss of 0.151. Brunese et al. [20] proposed a COVID-19 detection method using deep learning on the dataset of 250 COVID-19 CXR images. Shi et al. implemented random forest classifier-based screening system to differentiate between COVID-19 patients and community-acquired pneumonia with 87.9% accuracy [21]. Khobahi et al. proposed a novel semi-supervised deep neural network architecture that can distinguish between healthy, non-COVID pneumonia, and COVID-19 infection based on the CXR manifestation of these classes utilising very few numbers of parameters [22]. It comprised of Task-Based Feature Extraction Network (TFEN), and COVID-19 Identification Network (CIN). Ozturk et al. used 17 convolutional layers where each layer has different filters using DarkNet as a feature extractor layer [23]. Abbas et al. proposed a Decompose, Transfer, and Compose (DeTraC) method for COVID-19 classification where chest computes tomography (CT) dataset is trained with pre-trained ResNet model [24].
Panwar et al. proposed a deep learning neural network based model nCOVnet for COVID-19 detection using CXR images where VGG-16 model is trained to perform image classification for two different classes, i.e. COVID-19 and healthy [25]. Sarker et al. proposed an approach using the Densenet-121 for effective detection COVID-19 patients and make use of another deep-learning model CheXNet which was already trained on radiological dataset [26]. Accuracies of 96.49% and 93.71% were obtained for binary and 3-class classifications, respectively. The feasibility of decision-tree classifier is also investigated in the detection of COVID-19 from CXR images [27]. Three binary decision tree are trained by deep learning model and each tree is given to the classifier. The first decision tree is for normal-abnormal classification, second for tuberculosis, and third for COVID-19, which obtained the accuracy of 98%, 80%, and 95%, respectively. Grad-CAM based colour-visualisation approach is also employed for a more visual interpretation of CXR images through deep learning models. It took around 2 s time to process a single CXR image [28].
Table 1 summarises similar existing and reported methods for screening of COVID-19 using CXR images (Acc. = accuracy, Sens. = sensitivity, Spec. = specificity, Pre. = precision). For all previous works dataset for COVID-19 patients is too small or limited in size to make the training model robust.
Table 1.
Comparison with state-of-the-art methods.
| Work | Dataset | Methodology | Classification | Time (in seconds) | Acc. (%) | Sens. (%) | Spec. (%) | Pre. (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|---|---|
| [13] | 70 COVID-19, 1008 Pneumonia | ResNet-18 | Binary class | – | – | 96 | 70.65 | – | – |
| [29] | 455 COVID-19, 2109 Non-COVID Images | MobileNet V2 | Binary class | – | 99.18 | 97.36 | 99.42 | – | – |
| [31] | 224 Covid-19, 504 Normal, 400 Bacteria Pneumonia, 314 Viral Pneumonia | MobileNet | Binary class | – | 96.78 | 98.66 | 96.46 | – | – |
| [33] | 250 COVID-19, 3520 Normal, 2753 Other Pulmonary Diseases | VGG-16 | Binary class | 2.5 | 97 | 87 | 94 | – | – |
| [34] | 305 COVID-19, 1888 Normal, 3085 Bacterial Pneumonia, 1798 Viral Pneumonia | Stacked Multi-Resolution CovXNet | Binary class | – | 97.4 | 97.8 | 94.7 | 96.3 | 97.1 |
| [23] | 127 COVID-19, 500 Normal, 500 Pneumonia Images | DarkCovidNet (CNN) | Binary class | <1 s | 98.08 | 95.13 | 95.3 | 98.03 | 96.51 |
| 3-class | <1 s | 87.02 | 85.35 | 92.18 | 89.96 | 87.37 | |||
| [30] | 231 COVID-19, 1583 Normal, 2780 Bacterial Pneumonia, 1493 Viral Pneumonia | Inception ResNetV2 | 3-class | 0.1599 | 92.18 | 92.11 | 96.06 | 92.38 | 92.07 |
| [32] | 180 COVID-19, 8851 Normal, 6054 Pneumonia | Concatenation of Xception and ResNet50V2 | 3-class | – | 91.4 | – | – | – | – |
| [16] | 266 COVID-19, 8066 Normal, 5538 Pneumonia | COVID-Net | 3-class | – | 93.3 | 91 | – | – | – |
| [15] | 284 COVID-19, 310 Normal, 330 Bacterial Pneumonia, 327 Viral Pneumonia Images | CoroNet | Binary class | – | 99 | 99.3 | 98.6 | 98.3 | 98.5 |
| 3-class | 95 | 96.9 | 97.5 | 95 | 95.6 | ||||
| 4-class | 89.6 | 89.92 | 96.4 | 90 | 89.8 |
Thus, deep learning-based computer-aided screening tool is needed to be developed, which is economical as well as more efficient and accurate for deploying in real-world situations. The dataset collected from multiple places and multiple conditions to train the deep learning model can be helpful to develop a diagnostic system which will be less prone to errors with universal acceptance and such dataset can contribute to further research in this field. Data augmentation can also further enhance the performance of the training model and give more impactful results. Further, the screening systems should be developed in the way to diagnose the CXR of the person and classify that image according to the probability of the diagnosed disease. It creates a need for a user-friendly diagnosis system where there is no need for trained human resources. The whole framework should be standalone and for practical reasons, there is often required to be independent on internet connectivity. The system should work fast to reduce workload and give results much faster than human experts. Unlike many existing works [[25], [26], [27], [28], [29], [30]] that only consider a classification task on COVID-19 and non-COVID classes, the trained deep-learning network on comprehensive dataset belonging to various countries used in proposed work can extract the best region in the X-ray images to be further fed into the succeeding classifier network.
3. Materials and methodology
The experiment conducted in this research consists of data collection, data analysis, model architecture, and consequent experimental results evaluated in terms of various performance parameters. These parts are described in the following sections.
3.1. Dataset—chest X-ray images
Experiments are performed on the dataset collected from different sources for binary classification (COVID-19 vs. non-COVID) and multi-classification of diseases. The following classes are subjected to classification: COVID-19, Normal (Healthy), Bacterial Pneumonia, Viral Pneumonia, Abnormal, and Non-COVID. Non-COVID represents the combination of normal (healthy), bacterial pneumonia, viral pneumonia and abnormal categories of images. Abnormal cases are those cases which do not belong to COVID-19, normal and pneumonia category of CXR images. As abnormal images are limited in number in its category, so these are considered only for binary (COVID-19 vs. non-COVID) classification and not considered for multi-classification in 3 and 4 classes.
For this study, datasets of CXR images are taken from two publicly available databases which were supplemented by data collected from different hospitals in India. Indeed, the public database and collected X-ray images bring diversity and richness in the classification and performance assessment phase:
-
a)
Dataset A is from the open-source repository [35] that has 237 COVID-19 CXR images from various parts of the world like Australia, Belgium, Canada, China, Egypt, Germany, Iran, Israel, Italy, Korea, Spain, Taiwan, USA and Vietnam on (12th May, 2020). This open repository contains a database of chest images of COVID-19, acute respiratory distress syndrome (ARDS), severe acute respiratory syndrome (SARS) 1, SARS 2, Middle East respiratory syndrome (MERS) patients.
-
b)
Dataset B consists of chest X-ray images of pneumonia infected and normal (healthy) people of 5848 images from open source repository [36]. It is a combination of 1583 normal images, 2772 bacterial pneumonia images, and 1493 viral pneumonia CXR images.
-
c)
Dataset C1 is collected by the authors from 3 different hospitals from Uttar Pradesh and Rajasthan, India.
-
•
188 images (28 COVID-19, 83 abnormal, 77 healthy images were collected from King George's Medical University (K.G.M.U.), Lucknow, Uttar Pradesh, India.
-
•
68 images of COVID-19 patients were collected from Uttar Pradesh University of Medical Sciences (U.P.U.M.S.), Saifai, Etawah, Uttar Pradesh, India.
-
•
543 X-ray images (326 COVID-19, 217 abnormal) from Government Medical College, Kota, Rajasthan, India.
The statistics of the number of images in the datasets are given in Table 2 .
Table 2.
Chest X-ray images in different datasets.
| Dataset | Non-COVID |
Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 |
Normal (healthy) |
Viral Pneumonia |
Bacterial Pneumonia |
Abnormal (used only for Binary classification) |
|||||||
| Training & validation | Test | Training & validation | Test | Training & validation | Test | Training & validation | Test | Training & validation | Test | ||
| Dataset A | 237 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 237 |
| Dataset B | 0 | 0 | 1000 | 583 | 1000 | 493 | 1000 | 1772 | 0 | 0 | 5848 |
| Dataset C | 228 | 194 | 77 | 0 | 0 | 0 | 0 | 0 | 230 | 70 | 799 |
| Total | 465 | 194 | 1077 | 583 | 1000 | 493 | 1000 | 1772 | 230 | 70 | 6884 |
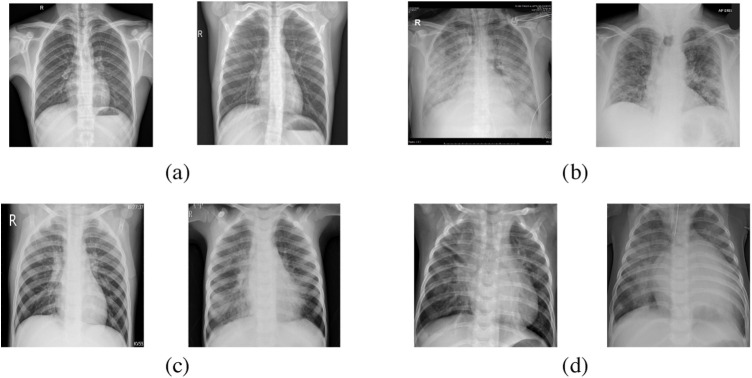
CXR images from all the databases are divided into training, validation, and test sets. Test dataset contains 3112 samples for multi-class classification in 4 classes (194 COVID vs. 583 normal vs. 1772 bacterial pneumonia vs. 493 viral pneumonia cases) whereas 3042 samples for binary classification and COVID-19 detection (194 positives and 2848 non-COVID samples). Sample CXR images of COVID-19, healthy, viral pneumonia, bacterial pneumonia is shown in Fig. 1 .
Fig. 1.
Chest X-ray example images: (a) healthy person; (b) COVID-19 patient; (c) viral pneumonia patient; (d) bacterial pneumonia patient.
CXR images of COVID-19 contain patches and opacities which look similar to viral pneumonia images. At the initial stage of the COVID-19 infection, images do not show any kind of abnormalities. But with the increase of viruses, the images gradually become unilateral. The lower zone and the mid-zone of the lung started transforming into patchy and get smudged.
3.2. Clinical perspective of X-ray images for COVID-19 detection
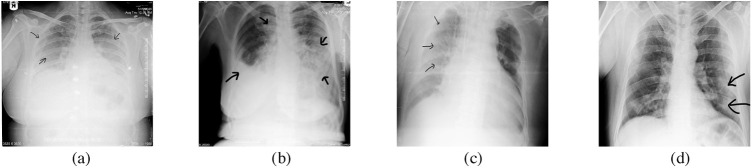
Bilateral and peripheral opacities (areas of hazy opacity) are the common characteristic features of COVID-19 affected patients X-ray report [37] with consolidations of the lungs (compressible lung tissue filled with fluid instead of air). The presence of air space opacities in more than one lobe is unlikely bacterial pneumonia since bacterial pneumonia is likely to be unilateral and involves a single lobe [38]. Other significant signs for COVID-19 are consolidation, peripheral, and diffused air space opacities. Initially, the researcher of COVID-19 found the air-space disease likely to have a lower lung distribution and is most commonly bilateral and peripheral [39]. These kinds of peripheral lung opacities also have characteristics to be confluent, either patchy or, multifocal, and can be easily srecognised on CXR images. Diffused lung opacities in COVID-19 patients have a similar pattern of CXR as other prevalent inflammatory or infectious processes such as in ARDS. Some other rare findings in COVID-19 affected patients are pneumothorax, lung cavitation, and pleural effusion (water in pleural spaces of the lung) which were mostly found at the later stage of the disease [40]. Some of the COVID-19 CXR features are depicted in Fig. 2 .
Fig. 2.
Chest X-ray images of COVID-19 infected patients: (a) diffuse ill-defined hazy opacities (black arrows); (b) diffuse lung disease and right pleural effusion (black arrows); (c) subtle ill-defined hazy opacities in right side (black arrows); (d) patchy peripheral left mid to lower lung opacities (black arrows).
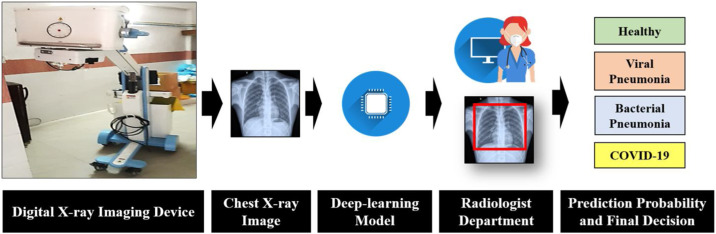
The conceptual schematic diagram of the proposed work is given in Fig. 3 . Once the model is trained using a deep learning algorithm it can be sutilised for rapid screening in health care centres. Mobile van-based screening can be performed in hot spot areas and public places. For pre-screening, a digital X-ray machine is required to generate CXR image which will be taken as an input to the deep learning model.
Fig. 3.
Conceptual schematic representation of the proposed COVID-19 screening framework.
Thereafter, the image can be tested on any computing device which contains the proposed model. The model can classify the image in 0.137 s. It can classify thousands of images on a single click and generate a report.
3.3. Methodology
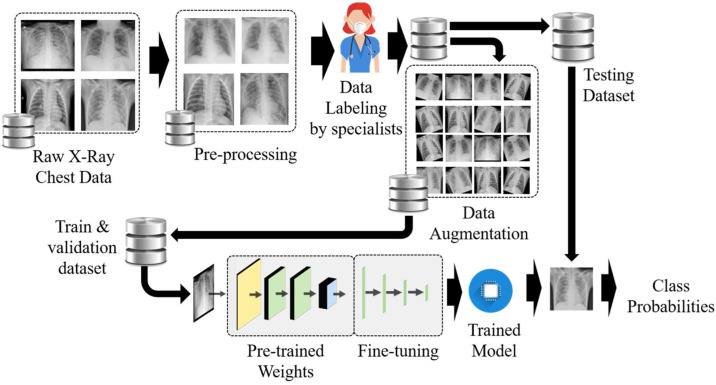
Deep learning (DL) is a part of machine learning which is sutilised to solve complex problems with the state-of-the-art performance on computer vision and image processing [41]. DL methods are widely used for medical imaging giving a high performance in segmentation, classification, and detection tasks including breast cancer detection, tumour detection and skin cancer detection [42]. The block diagram for dataset preparation, training and analysing using the deep learning model in the proposed work is depicted in Fig. 4 .
Fig. 4.
Methodology of training and testing of the deep learning based COVID-19 detection algorithm.
All the collected images from various sources of different countries were merged into one large dataset. Most of the collected samples were in Digital Imaging and Communications in Medicine (DICOM) format with extension ".dcm". All digital X-ray files were converted into one common image format. The samples were then pre-processed where CXR images were cropped to remove redundant portions and resized to fit better to dimensions of used artificial neural networks. Augmentation was carried out, which increases the dataset, gives robustness to the trained model and mitigates the occurrence of overfitting problems. Rotation, shear, scaling, flips, and shifts are few of the augmentation techniques which were used to prepare the model to increase the efficiency of test images in a different orientation. Dataset was labelled in different classes as per the opinion of the medical experts which are annotated and scategorised accordingly. CXR images are annotated manually for proper training and bounding boxes made around the targeted area. The respective information about the labels and area is saved. After dividing the dataset of CXR in training, test, and validation set, deep learning models are trained for multiple iterations with the prepared dataset. The validation set is used for tuning the parameters and for escaping overfitting of the training model. After sufficient training, the model adjusts its weights and the final trained model is tested on the new set of CXR images of various categories which were analysed for performance evaluation of the trained model.
For the image recognition and classification tasks, various architectures of convolutional neural networks or CNNs have proven their accuracy and are used widely. CNNs are commonly composed of multiple building blocks of layers consisting of convolution layer, activation layers, pooling layers, and fully connected (FC) layers which are designed to learn spatial hierarchies of features automatically and adaptively through backpropagation to perform vision task. The convolutional layer is an important part of the deep learning neural network, which extracts the features from the input images. Input images are convolved with a filter or kernel to generate a convolved feature matrix using different strides. After convolution, the output is passed through an activation function (ReLU, Tanh, or Sigmoid). The activation layer is used to increase non-linearity without effecting its receptive field. Convolution layers are interleaved with pooling layers that are used to decrease the spatial size of the convolved feature matrix. It looks for a larger area of the input image matrix and takes aggregate information (maximum, average, and sum). FC layer is a dense layer which is the final learning phase of CNN architecture performing classification tasks.
3.3.1. Training model
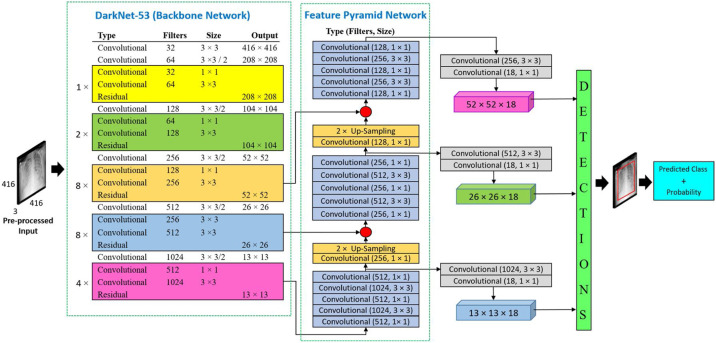
Architecture selection for backbone network plays an important role in feature extraction in object detection tasks. Stronger the backbone network, stronger the detection speed and accuracy of the detection result. DarkNet-53 is used as a backbone network which consists of 53 layers pre-trained on ImageNet [43]. Instead of random weights for sinitialisation of training of the model, pre-trained weights using transfer learning is used in the proposed work, which reduces the training time and makes more efficient training. The DarkNet-53 network composed of 3 × 3 and 1 × 1 filters with shortcut connections. To perform detection tasks, 53 additional layers are merged with DarkNet layers resulting in a total of 106 network layers. The considered YOLO-v3 based-architecture [44] for the proposed work with processed data to train with different CXR images of various classes, is shown in Fig. 5 .
Fig. 5.
Architecture of deep learning model for COVID-19 detection with processed dataset.
This neural network architecture provides high speed of detection and desired precision. Due to the multiscale search, it can detect large or as well as smaller objects. DarkNet-53 reaches the highest measured floating-point operations per second, resulting in higher sutilisation of GPU by the structure of the network, which offers higher performance.
DarkNet-53 uses residual networks for better feature extraction. Detection takes place at three different scales like feature pyramid network (FPN) [45], which is done by down-sampling the input image dimensions by 32, 16, and 8. FPN is used to extract both spatial and semantic information-rich feature maps from a given CXR image of dimensions 416 × 416. The last layer of the DarkNet-53 network generates the first feature map and additional convolution operation of 1 × 1, 3 × 3, 1 × 1, 3 × 3, 1 × 1, and 1 × 1 are applied to generate the second feature map which is up-sampled by a factor of two. Then the up-sampled feature map from the last step is concatenated with a feature map generated from the fourth residual layer of DarkNet-53 backbone network. Same 5 layers of convolution layer with a different number of filters to generate the third feature map from the second feature map which later up-sampled by a factor of a two and concatenated with the third residual layer of DarkNet-53. Detection at three different scales makes the deep learning model detect the smallest objects. The last layer of the training network performs a bounding box and class prediction for an input CXR image.
For training the proposed model with CXR images in a computationally efficient manner, an advanced form of ReLU activation function, leaky ReLU (LReLU) is used. LReLU activation function saves the value of gradients from getting saturated in case of constant negative bias alike in ReLU. Instead of pruning the negative part to completely zero (as ReLU does), the negative part is multiplied by , which is a small constant value and non-zero number, usually taken as 0.01. The output of the LReLU activation function used in the trained model can be represented as:
| (1) |
In the proposed methodology, max-pooling is sutilised together with convolutional layers for extraction of sharp features such as edges from input CXRs. In max-pooling, the maximum value from the rectified feature map is selected. For a CNN architecture, where is the pooling size and is pooling function, the output feature on jth local receptive for ith pooling layer is:
| (2) |
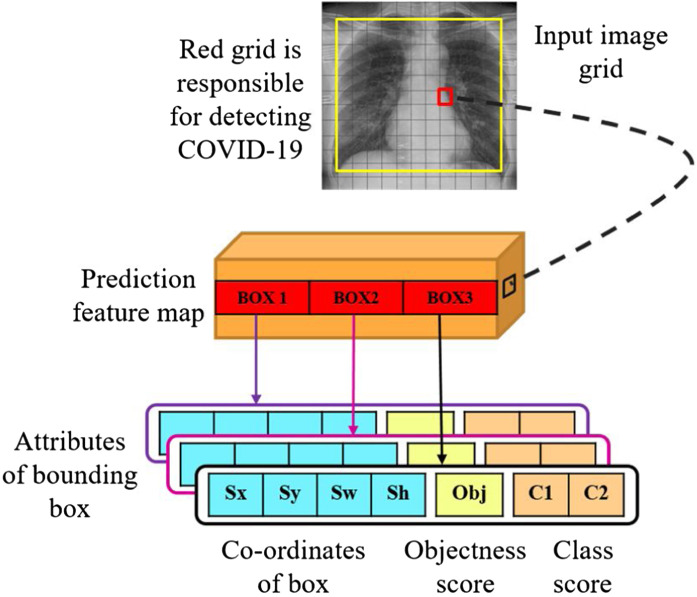
Binary cross-entropy loss is used during training for the class predictions of input CXR images. The input CXR images are divided into N × N grids. In the proposed work, predictions were made on three different scales as shown in Fig. 8. Thus, an input CXR image of 416 × 416 dimension is divided into grids of 13 × 13, 26 × 26 and 52 × 52 for the respective stride values of 32, 16, and 8. Detection of the targeted object for input CXR image in the proposed methodology is shown in Fig. 6 . The cell which contains the centre of ground truth box is responsible for predicting the trained object class of CXR. The red grid cell in Fig. 6 is depicting the center of ground truth which responsible for detecting COVID-19 related features.
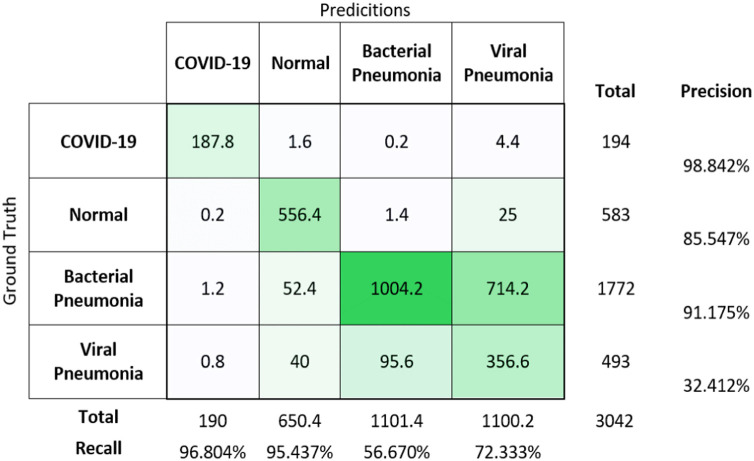
Fig. 8.
Confusion matrix for multi-classification on the test dataset.
Fig. 6.
Detection task through the trained model in an image.
The grid cells are responsible for detecting the objects if the centres of the objects lie in those grid cells. The grid cells predict bounding boxes and determine the confidence score associated with those boxes. The confidence score describes the confidence of the model that the object lies in the box and the accuracy of the box is predicted. Each grid in the input CXR image predicts B number of bounding boxes with confidence scores, as well as C class conditional probabilities. B represents the number of bounding boxes predicted per single grid cell, which is 3 for each detection at a different scale. All three boxes will be used for the estimation of the parameters of predicting bounding box. Confidence score formula is given in Eq. (3):
| (3) |
where represents the common value in between predicted and reference bounding box and if the target is in the grids, otherwise it would be 0.
Attributes of the bounding box contain coordinate points of the bounding box, objectness score, and target classes (COVID-19 and non-COVID). Objectness score is defined as the likelihood of containing the targeted object in a given bounding box. Objectness score is calculated by logistic regression for each bounding box and it should be one if the ground truth object has more overlapping of bounding box prior as compared to others. The best bounding box is selected out of multiple bounding boxes with the help of non-maximum suppression (NMS). It suppresses less likely bounding box and keeps the best bounding box. NMS considers objectness score and intersection over union (IoU) parameters of the bounding box where IoU is the ratio between the area of overlap and area of the union of the predicted bounding box and true bounding box. NMS chooses the box with the highest score for multiple iterations and eliminates higher overlapping bounding boxes after the computation of overlap with other boxes. The deep neural network computes four coordinate points for each bounding box, Sx, Sy, Sw, Sh. Then, corresponding predictions for respective x-coordinate, y-coordinate, width and height of bounding box represented by Bx, By, Bw, and Bh, respectively calculated as:
| Bx = σ(Sx) + COx | (4) |
| By = σ(Sy) + COy | (5) |
| Bw = BpweSw | (6) |
| Bh = BpheSh | (7) |
where the cell is offset from the top left corner of the image by (COx, COy). Bpw and Bph are bounding box width and height prior, respectively. For training, the dataset sum of squared error loss is used. Logistic regression is used for predicting each class score and threshold for multiple labels for multi-labels prediction on CXR images. Objects which has higher class score value than the defined threshold value are assigned to the respective bounding box. In the testing stage, images will pass through the same neural network and the same operations will be executed on the test image. It will calculate the score which will be projected to the fully connected layer. The score obtained during the testing phase is compared with responses got during the training, then it will make a decision in favour of that class which was giving similar response during the training of CXR images of different categories. Then, corresponding predictions for respective x-coordinate, y-coordinate, width and height of bounding box represented by Bx, By, Bw, and Bh, respectively.
The trained deep-learning network on the comprehensive dataset can extract the best region in the X-ray images to be further fed into the succeeding classifier network. As the predictive framework works on detection rather than the only classification task, it can sometimes predict more than one class in a given CXR image. For an input CXR image for testing the trained model, single or multiple detection results can be predicted based on the values of threshold probability (0.5 in the proposed work), the best one is chosen as output to make screening processes rapid for a large number of testing samples. When CXR image is tested with the trained model, it predicts the class according to the detection probability percentage. In most of the cases, the detection percentage is greater than 80%. In few images having some artefacts, it can detect the multiple numbers of class, but the prediction probability is greater than 80% for the highest and nearly 50% for the others. In those cases, the best one is chosen as output to make screening processes rapid for a large number of testing samples with the threshold value of 50%. Otherwise in general scenarios, it may be due to some improper CXR images and if the results are not clear or percentage probability of detection is close, the subject can be tested again for better diagnostic results. Suppose for an input CXR image sample, the network is predicting it is in viral pneumonia category with 83% probability as well as 61% for Healthy category. Thus, only the highest predicting results are considered, which make the testing rapid in the time of COVID-19 pandemic. Otherwise, in general scenarios, it may be due to some improper CXR image and if the results are not clear, the subject can be tested again for better diagnostic results.
4. Experimental results and discussion
All training and testing of the deep learning models is done on Python language based framework in a system with Intel Xenon processor equipped with graphics processing unit (GPU). Considering the memory limitations of the server, the batch size of the training model is taken as sixteen. Other factors such as momentum to accelerate network training, an initial learning rate to affect the speed at which the algorithm reaches the optimal weights, weight decay to sregularise the training model are given in Table 3 , with complete software and hardware specifications for training different CXR images.
Table 3.
Parameters for training a deep learing model.
| Name | Parameters |
|---|---|
| Development Environment | Anaconda, Jupyter Notebook, Tensorflow, Keras, OpenCV |
| Processor | Intel Xenon Gold 5218 CPU @ 2.30 GHz, 2.29 GHz |
| Installed RAM | 64 GB |
| Operating System | Windows 10, 64 bit |
| Graphics | NVIDIA, Quadro P600 |
| Graphics Memory | 24 GB |
| Programming Language | Python |
| Input | Image Dataset |
| Input dimension | 416 × 416 |
| Batch Size | 16 |
| Decay | 0.0001 |
| Initial Learning Rate | 0.001 (will reduced to 10−2 times after every 50,000 steps) |
| Momentum | 0.9 |
| Epochs | 250 |
| Optimisation algorithm | Stochastic Gradient Descent (SGD) |
Training and validation sets were kept constant while test dataset was kept updating with new CXR images during the experiments. Apart from keeping images in the test set, the remaining dataset from all the sources is divided into training and validation set, which is further cross-validated. The deep learning model is trained with CXR images of different categories for several iterations until the loss gets saturated. Generated trained models are analysed with multiple images in test dataset to get overall performance.
Performance metrics used for the calculation of the experiment are:
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
where true positive (TP) are those cases where the model correctly predicts the positive labelled image, false positive (FP) is the case where the model predicts as COVID-19 although the image is labelled as non-COVID-19. True negative (TN) is when the model correctly predicts negative image and false-negative (FN) is the case where the model incorrectly predicts a positive labelled negative image. The confusion matrix is used for measuring the performance of the machine learning classification problem. It is a combined representation of ground truth and predicted classes. First, image data is analysed without augmentation and later the proposed methodology is tested with the application of augmentation techniques.
4.1. Binary classification
The combined database of datasets A, B, and C are used for performing the binary classification, i.e. COVID-19 or non-COVID, which contains 465 images (237 images from dataset A and 228 from dataset C) of COVID-19 and 3307 CXR images (3000 images from dataset B and 307 from dataset C) belonging to non-COVID cases.
Confidence interval (CI) is used to analyse the results, which give more information than point estimates. It measures the degree of certainty and uncertainty in a sampling method and gives the range of values which likely to contain the unknown parameter. 95% confidence interval is the most commonly used criteria for such estimations. The confidence intervals are calculated in terms of the mean value and standard deviation for different folds of cross-validation as in Eq. (13):
| (13) |
where is the mean, is standard deviation and N is the sample size. The constant z = 1.96 is confidence level value for 95% confidence interval.
To better examine the classification model generated by a deep learning algorithm, 5-fold cross-validation is performed. The complete CXR image dataset is divided into five different parts and trained for five iterations. The model is trained with the four-fifth part and validated with remaining one-fifth part of CXR image dataset. After having 5-fold cross-validation, overall performance evaluation of detected outputs in binary classification is achieved as in Table 4 . The values of TP, TN, FP and FN are averaged for 5-fold cross-validation and that mean value is used to calculate those parameters. The standard deviation of all the obtained results after different fold of cross-validation is calculated. Results are also interpreted in terms of the confidence interval (95%). Accuracy in terms of confidence interval (95%) is achieved as 99.61 ± 0.17%, which shows very less false positives and false negatives cases.
Table 4.
5-fold cross validation result for 2-class classification: COVID-19 vs. non-covid on dataset (A + B + C).
| Parameter | Accuracy | Sensitivity | Specificity | Precision | F1-score |
|---|---|---|---|---|---|
| Standard deviation | ±0.19 | ±2.08 | ±0.11 | ±0.77 | ±0.81 |
| Overall results (95% CI) | 99.61 ± 0.17 | 98.57 ± 1.83 | 99.76 ± 0.10 | 98.30 ± 0.68 | 98.42 ± 0.71 |
Tests are performed on new 3112 number of test CXR images belonging to different classes for each of the 5 models received after cross validation. Results of the binary classification on test images are shown in Table 5 . The values of TP, TN, FP, and FN are averaged and other parameters values are calculated. Overall results are represented in terms of the confidence interval of 95%.
Table 5.
Averaged test result after cross validation for 2-class classification: COVID-19 vs. non-covid on dataset (A + B + C).
| COVID-19 | Non-COVID | TP | TN | FP | FN | Accuracy | Sensitivity | Specificity | Precision | F1-Score |
|---|---|---|---|---|---|---|---|---|---|---|
| 194 | 2918 | 188.8 | 2916.8 | 1.2 | 5.2 | 99.79 | 97.32 | 99.96 | 99.37 | 98.33 |
| Standard deviation | ±0.12 | ±2.04 | ±0.02 | ±0.22 | ±1.00 | |||||
| Overall results (95% CI) | 99.79 ± 0.10 | 97.32 ± 1.79 | 99.96 ± 0.02 | 99.37 ± 0.20 | 98.32 ± 0.88 | |||||
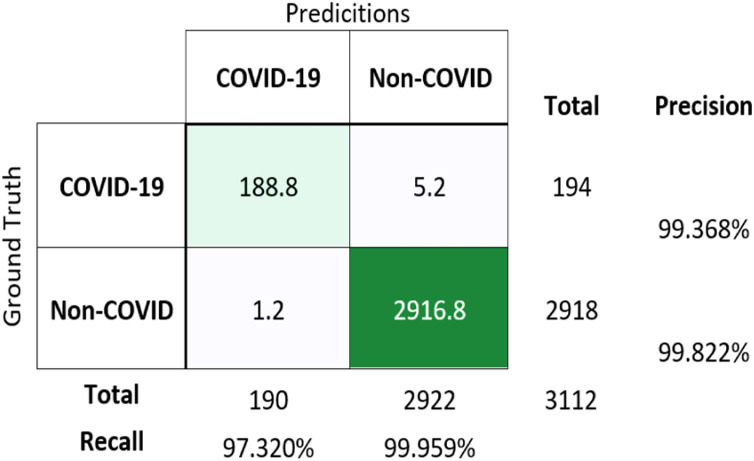
The confusion matrix for binary classification using averaged values of results obtained from 5 different weights of the trained model after cross-validation, is shown in Fig. 7 .
Fig. 7.
Confusion matrix for binary classification (COVID-19 vs non-COVID) on test dataset.
The testing results for new images gives an accuracy of 99.79 ± 0.10 %, which is signifying the robustness of the proposed model. Hence, this model can be sutilised for performing detection and classification of COVID-19 and non-COVID X-ray images.
4.2. Multi-class classification
The combined database, i.e. dataset A, B, and C, which contains 465 images of COVID-19, 1077 normal CXR images, 1000 bacterial pneumonia and 1000 viral pneumonia images are trained. In this set, CXR images of 77 normal classified people and 228 COVID-19 diagnosed patient data from local hospitals are also involved along with images of the dataset A and B for multi-class classification.
Overall performance evaluation after performing 5-fold cross-validation for detected outputs in multi-classification is given as in Table 6 . The accuracy for 95% confidence interval is achieved as 94.79 ± 3.81% whereas the results concerning only the COVID-19 patients the achieved accuracy is for 99.70 ± 0.23%, which shows significantly less false positives and false negatives cases.
Table 6.
5-fold cross validation for 4-class classification: normal vs. viral pneumonia vs. bacterial pneumonia vs. COVID-19.
| Parameters | Accuracy | Sensitivity | Specificity | Precision | F1-score |
|---|---|---|---|---|---|
| Standard deviation – 4 classes | ±3.89 | ±9.50 | ±2.52 | ±5.82 | ±6.69 |
| Overall results – 4 classes (95% CI) | 94.79 ± 3.81 | 90.82 ± 9.31 | 96.48 ± 2.47 | 91.35 ± 5.70 | 90.88 ± 6.56 |
| Standard deviation – COVID-19 | ±0.26 | ±1.72 | ±0.09 | ±0.61 | ±0.91 |
| Overall results for COVID-19 (95% CI) | 99.70 ± 0.23 | 98.14 ± 1.51 | 99.91 ± 0.08 | 99.42 ± 0.53 | 98.77 ± 0.80 |
The model made the decision based on the dataset division and search for features which can distinguish between the classes of objects used to train the model. The abnormal samples may have similarity with both COVID-19 and normal healthy CXR image samples because the abnormal samples are COVID-19 suspected samples which were declared negative after the conventional RT-PCR test. The abnormal CXR image sample removal in the multi-classification task help the detection task easier for the trained model and performance enhancement, as interpreted in the last two rows of Table 6.
Testing of trained models is done after different cross-validation folds to authenticate the performance. Trained model after each cross-validation has been tested on new test images and the mean values of different parameters for all 5 trained models are shown in Table 7 .
Table 7.
Averaged test result after cross validation for 4-class classification: normal vs. viral pneumonia vs. bacterial pneumonia vs. COVID-19 on dataset (A + B + C).
| Class | Samples of testing category | Samples of other classes | TP | TN | FP | FN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | 194 | 2848 | 187.8 | 2845.8 | 2.2 | 6.2 | 99.72 | 96.80 | 99.92 | 98.85 | 97.81 |
| Normal | 583 | 2459 | 556.4 | 2365 | 94 | 26.6 | 96.04 | 95.44 | 96.18 | 85.61 | 90.24 |
| Bacterial Pneumonia | 1772 | 1270 | 1004.2 | 1172.8 | 97.2 | 767.8 | 71.56 | 56.67 | 92.35 | 91.18 | 69.89 |
| Viral Pneumonia | 493 | 2549 | 356.6 | 1773.8 | 743.6 | 136.4 | 70.03 | 72.33 | 70.45 | 32.43 | 44.77 |
| Standard deviation – 4 classes | ±15.72 | ±19.35 | ±13.22 | ±30.22 | ±23.74 | ||||||
| Overall results – 4 classes (95% CI) | 84.34 ± 15.41 | 80.31 ± 18.96 | 89.72 ± 12.95 | 77.02 ± 29.61 | 75.68 ± 23.27 | ||||||
| Standard deviation – COVID-19 | ±0.07 | ±0.99 | ±0.06 | ±0.84 | ±0.54 | ||||||
| Overall results for COVID-19 (95% CI) | 99.72 ± 0.06 | 96.80 ± 0.87 | 99.92 ± 0.05 | 98.85 ± 0.74 | 97.81 ± 0.47 | ||||||
The confusion matrix for multi-classification using averaged values of results obtained from 5 different weights of the trained model is shown in Fig. 8. The overall accuracy for this classification is achieved as 69.2%.
If only the COVID-19 results are taken into consideration, classification accuracy is achieved as 99.72 ± 0.06%, the sensitivity of 96.80 ± 0.87%, specificity if 99.92 ± 0.05%, the precision value of 98.85 ± 0.74% and F1-score value of 97.81 ± 0.47%. While considering all four classes, the average accuracy is achieved as 84.34 ± 15.41% in terms of 95% CI. After analysing the testing results, the classification of bacterial pneumonia and viral pneumonia is giving lower classification results when compared with other classes, as shown in Table 4. Chaos occurs for a model to classify more precisely the CXR images of viral and bacterial pneumonia. So, the results after training a deep neural network for 4-class classification is represented in the form of 3-class classification where the results of bacterial and viral pneumonia are considered as a single class of pneumonia. After combining both pneumonia classes in one output class, results are significantly improved. Table 8 represents the test result after 5-fold cross-validation for performing three classifications.
Table 8.
Averaged cross validation test result for 3-class classification: normal vs. COVID-19 vs. pneumonia (viral pneumonia + bacterial pneumonia) on dataset (A + B + C).
| Class | Samples of testing category | Samples of other classes | TP | TN | FP | FN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | 194 | 2848 | 187.8 | 2845.8 | 2.2 | 6.2 | 99.72 | 96.80 | 99.92 | 98.84 | 97.81 |
| Normal | 583 | 2459 | 556.4 | 2365 | 94 | 26.6 | 96.04 | 95.44 | 96.18 | 85.55 | 90.22 |
| Pneumonia | 2265 | 777 | 2170.6 | 746 | 31 | 94.4 | 95.88 | 95.83 | 96.01 | 98.59 | 97.19 |
| Standard deviation | ±2.18 | ±0.70 | ±2.21 | ±7.57 | ±4.21 | ||||||
| Overall results (95% CI) | 97.21 ± 2.46 | 96.02 ± 0.80 | 97.37 ± 2.50 | 94.35 ± 8.57 | 95.08 ± 4.76 | ||||||
The testing results show the enhancement in the average classification accuracy for all classes from 84.34 ± 15.41% to 97.21 ± 2.46%, signifying high accuracy results in this representation. The values of other parameters are also substantially increased. Thus, this consideration can be used for 3 class classification for more surety in case of rapid large scale testing.
4.3. Dataset augmentation
After performing the augmentation on the training and validation set with rotation (15 degrees clockwise and anti-clockwise) and scaling (half and double) of images, the dataset of 3772 CXR images is increased to 18,860 images, 5 times that of the original dataset. After augmentation, CXR images are used for training which was tested on the test dataset of 3112 images for binary and 3042 images for multi-class classification. The results are shown in Table 9 , where average test accuracy of 99.81%, 88.25 ± 11.49%, and 97.11 ± 2.71% is achieved for binary(COVID-19 vs. non-COVID), 4-class (COVID-19 vs. Normal vs. Bacterial Pneumonia vs. Viral Pneumonia), and 3-class (COVID-19 vs. Normal vs. Pneumonia), respectively. The overall accuracy for 3-class and 4-class classification is achieved as 95.66% and 76.46%, respectively.
Table 9.
Test results for binary and multi-class classification after augmentation.
| Classification | Class | Samples of testing category | Samples of other categories | TP | TN | FP | FN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Binary (COVID-19 vs. Non-COVID) | COVID-19 | 194 | 2918 | 191 | 2915 | 3 | 3 | 99.81 | 98.45 | 99.90 | 98.45 | 98.45 |
| Multi-class (4-Class) | COVID-19 | 194 | 2848 | 189 | 2846 | 2 | 5 | 99.77 | 97.42 | 99.93 | 98.95 | 98.18 |
| Normal | 583 | 2459 | 565 | 2384 | 75 | 18 | 96.94 | 96.91 | 96.95 | 88.28 | 92.40 | |
| Bacterial Pneumonia | 1772 | 1270 | 1165 | 1210 | 60 | 607 | 78.07 | 65.74 | 95.28 | 95.10 | 77.74 | |
| Viral Pneumonia | 493 | 2549 | 407 | 1972 | 577 | 86 | 78.21 | 82.56 | 77.36 | 41.36 | 55.11 | |
| Standard deviation | ±11.73 | ±14.96 | ±10.20 | ±26.74 | ±19.20 | |||||||
| Average (class interval 95%) | 88.25 ± 11.49 | 85.66 ± 14.66 | 92.38 ± 9.99 | 80.92 ± 26.21 | 80.86 ± 18.82 | |||||||
| Multi-class (3-Class) | COVID-19 | 194 | 2848 | 191 | 2847 | 1 | 3 | 99.87 | 98.45 | 99.96 | 99.48 | 98.96 |
| Normal | 583 | 2459 | 555 | 2359 | 100 | 28 | 95.79 | 95.20 | 95.93 | 84.73 | 89.66 | |
| Pneumonia | 2265 | 777 | 2164 | 746 | 31 | 101 | 95.66 | 95.54 | 96.01 | 98.59 | 97.04 | |
| Standard deviation | ±2.39 | ±1.79 | ±2.30 | ±8.27 | ±4.91 | |||||||
| Average (class interval 95%) | 97.11 ± 2.71 | 96.40 ± 2.02 | 97.30 ± 2.61 | 94.27 ± 9.36 | 95.22 ± 5.56 | |||||||
The performance comparison table for all kind of classification with and without augmentation is given in Table 10 . The data augmentation enhanced the performance of the proposed methodology. Binary classification is giving the best output among others.
Table 10.
Comparision of the obtained test results using the proposed methodology.
| Augmentation | Classification | Sensitivity (%) | Specificity (%) | Precision (%) | F1-score (%) |
|---|---|---|---|---|---|
| Without augmentation | Binary (COVID-19 vs. Non-COVID) | 97.32 ± 1.79 | 99.96 ± 0.02 | 99.37 ± 0.20 | 98.32 ± 0.88 |
| Multi-class (4-classes) | 84.34 ± 15.41 | 80.31 ± 18.96 | 89.72 ± 12.95 | 77.02 ± 29.61 | |
| Multi-class (3-classes) | 96.02 ± 0.80 | 97.37 ± 2.50 | 94.35 ± 8.57 | 95.08 ± 4.76 | |
| With augmentation | Binary (COVID-19 vs. Non-COVID) | 98.45 | 99.90 | 98.45 | 98.45 |
| Multi-class (4-classes) | 85.66 ± 14.6 | 92.38 ± 9.99 | 80.92 ± 26.21 | 80.86 ± 18.82 | |
| Multi-class (3- classes) | 96.40 ± 2.02 | 97.30 ± 2.61 | 94.27 ± 9.36 | 95.22 ± 5.56 |
In the testing dataset which is collected, consists of only normal images without much rotational or scaling variation, so both augmentation and without augmentation accuracies are nearby (in binary classification it is more). The deep learning model trained on the augmented dataset will help to get better detection results in new CXR images with various images transformations.
All these generated models can be used in primary health care centres for performing the screening test on CXR images. It can also be sutilised at places where there is a lack of availability of expert radiologists or to validate expert’s opinion and can assist them to make an accurate diagnosis whenever there are more patients. The model can be sutilised as a real-time screening setup that has an average detection time of 0.137 s per image for detection and its classification from input CXR images with dimensions of 416 × 416 pixels in another system with 6 GB GPU (NVIDIA GeForce GTX 1060). Some of the detected CXR images are shown in Fig. 9 , where the bounding box is drawn around the detected portion with respective prediction probability.
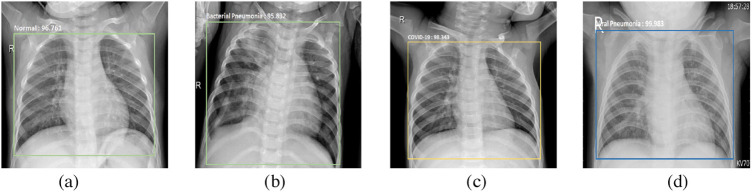
Fig. 9.
Prediction results of trained model on augmented dataset for multi-classification: (a) normal – 96.76%; (b) bacterial pneumonia – 95.83%; (c) COVID-19 – 98.34%; (d) viral pneumonia – 99.98%.
Different models are also compared with the collected dataset after augmentation as given in Table 11 where the proposed method achieved higher accuracy than other deep learning based methods.
Table 11.
Comparision of various deep learning models with the proposed methodology.
| Classification | Class | Accuracy (%) |
||||
|---|---|---|---|---|---|---|
| MobileNetV2 [46] | VGG-16 [47] | Faster-RCNN [48] | ResNet-50 [49] | Proposed method | ||
| Binary (COVID-19 vs. Non-COVID) | COVID-19 | 97.39 | 95.63 | 96.34 | 93.76 | 99.81 |
| Multi-class | COVID-19 | 94.12 | 91.16 | 94.74 | 90.76 | 99.87 |
| Normal | 86.52 | 86.26 | 89.41 | 84.91 | 95.79 | |
| Pneumonia | 88.13 | 88.26 | 89.41 | 83.10 | 95.66 | |
| Average (95% CI) | 89.52 ± 4.53 | 88.56 ± 2.788 | 91.1867 ± 3.482 | 86.257 ± 4.531 | 97.11 ± 2.71 | |
Table 12 compares different existing works for the diagnosis of COVID-19 detection using CXR images which gives a reference of some similar existing and reported methods. The proposed method achieved good accuracy with low time complexity for detection of COVID-19 using CXR images, which is encouraging. The comparison is questionable due to different datasets, but illustrative to a certain degree.
Table 12.
Comparision of the proposed methodology with state-of-the-art methods.
| Work | Classification | Time (in seconds) | Overall Acc. (%) | Sens. (%) | Spec. (%) | Pre. (%) | F1-Score (%) |
|---|---|---|---|---|---|---|---|
| MobileNet [31] | Binary class | – | 96.78 | 98.66 | 96.46 | – | – |
| Stacked Multi-Resolution CovXNet [34] | Binary class | – | 97.4 | 97.8 | 94.7 | 96.3 | 97.1 |
| DarkCovidNet (CNN) [23] | Binary class | <1 s | 98.08 | 95.13 | 95.3 | 98.03 | 96.51 |
| 3-class | <1 s | 87.02 | 85.35 | 92.18 | 89.96 | 87.37 | |
| CoroNet [15] | Binary class | 99 | 99.3 | 98.6 | 98.3 | 98.5 | |
| 3-class | 95 | 96.9 | 97.5 | 95 | 95.6 | ||
| 4-class | 89.6 | 89.92 | 96.4 | 90 | 89.8 | ||
| Proposed Approach | Binary class | 0.137 | 99.81 | 98.45 | 99.90 | 98.45 | 98.45 |
| 3-class | 95.66 | 96.40 | 97.30 | 94.27 | 95.22 | ||
| 4-class | 76.46 | 85.66 | 92.38 | 80.92 | 80.86 |
As multiple datasets are not available publicly, the collected dataset for proposed work can act as a remedy for more research in this domain. A large open-access dataset will be made available to the research community, which will help the researchers to try and develop different methods for better outcomes.
4.4. Misclassified images
On the analysis of the experimental results, it was found that most of the misclassified images were low-quality images or had some artefacts. Table 13 includes some of the images classified either as false-positives or false-negatives and has a clinical input given by radiologists as a possible reason for misclassification.
Table 13.
Clinical input by radiologist for misclassified images.
| Images | Ground truth | Prediction | Clinical input |
|---|---|---|---|
 |
COVID-19 | Normal | X-ray image of the pediatric patient has less filed of the lung than mediastinum, so the software learning algorithm picks up as normal (healthy). |
 |
COVID-19 | Normal | No explanation has to correlate with chest auscultation findings. |
 |
Normal | COVID-19 | X-ray image has an area of retro cardiac opacity and cardiac silhouettes deviation, so the software learning algorithm may have picked up as COVID-19. |
 |
Bacterial Pneumonia | COVID-19 | X-ray image has hilar lymph nodes and peripheral opacity, so the software learning algorithm may have picked up as COVID-19. |
 |
COVID-19 | Normal | No explanation has to correlate with chest auscultation findings |
In Table 9, three COVID-19 images were classified as normal, whereas one bacterial image is misclassified as COVID-19, and one normal image is classified as COVID-19. Clinically it was found that since the lungs of children are not fully developed, it is difficult to predict the diseases using their CXR image.
5. Conclusion and future work
The 2019 novel coronavirus (COVID-19) pandemic appeared in Wuhan, China in December 2019 and has become a serious public health problem worldwide. In the proposed work, a deep learning algorithm-based model is proposed for pre-screening of COVID-19 with CXR images. To make the system robust, it was trained with a dataset of chest X-ray images collected from local hospitals of India and also from countries like Australia, Belgium, Canada, China, Egypt, Germany, Iran, Israel, Italy, Korea, Spain, Taiwan, USA, and Vietnam. The database has been manually processed and trained with a deep convolutional neural network. In order to detect COVID-19 at an early stage, this study uses transfer learning methods. The performance of the convolutional neural network after 5-fold cross-validation was giving the average accuracy of 99.61 ± 0.17% for binary classification (is or is not COVID-19 disease) using 1132 CXR image samples and average accuracy of 94.79 ± 3.81% for multi-class classification of COVID-19, normal (healthy), bacterial pneumonia, and viral pneumonia using 1063 CXR image samples. The test accuracy for COVID-19 detection in the augmented dataset is achieved as 99.87% for 3112 CXR images samples for 3-class classification data (COVID-19, normal (healthy), and pneumonia) and 99.81% for binary (COVID-19 vs. non-COVID) classification of 3042 different CXR image samples in respective classes. Since the current scenario identification of the COVID-19 infected cases is the most important task, the experimental results of the proposed work support the COVID-19 identification with very high accuracy. For the future, the model can be trained with images of more diseases to make an automatic prediction for those diseases.
Authorship statement
It is certified that that the submitted work has not been published previously or submitted elsewhere for review and a copyright transfer. All persons who meet authorship criteria are listed as authors, and they have participated sufficiently in the work to take public responsibility for the content, including participation in the concept, design, analysis, writing, or revision of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this research paper.
Acknowledgements
We would like to acknowledge Department of Science and Technology (DST), Government of India, EU Interreg, niCE-life, CE1581 and MVCR VI04000039, Czech Rep. for partly funding this work.
Footnotes
COVID-19 local hospitals datasets: https://doi.org/10.17632/4n66brtp4j.1 (Mendeley database)
References
- 1.COVID-19 Dashboard by the Centre for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU)". ArcGIS. Johns Hopkins University. Retrieved 16th September 2020. (Web Link: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6).
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigues J.C., Hare S.S., Edey A., Devaraj A., Jacob J., Johnstone A. An update on COVID-19 for the radiologist—a British society of thoracic imaging statement. Clin Radiol. 2020;75(May (5)):323–325. doi: 10.1016/j.crad.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiting P., Singatullina N., Rosser J.H. Computed tomography of the chest: I. basic principles. Contin Educ Anaesth Crit Care Pain. 2015;15(6):299–304. [Google Scholar]
- 8.Li M., Lei P., Zeng B., Li Z., Yu P., Fan B. Coronavirus disease (COVID-19): spectrum of CT findings and temporal progression of the disease. Acad Radiol. 2020;27:603–608. doi: 10.1016/j.acra.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 10.Ulhaq A., Khan A., Gomes D., Paul M. Computer vision for COVID‐19 control: a survey. engrXiv. 2020:1–24. doi: 10.1109/ACCESS.2020.3027685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev Biomed Eng. 2020:1. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 12.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv. 2003 doi: 10.1007/s10044-021-00984-y. 2020. (Zonguldak Bulent Ecevit University, Turkey) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Xie Y., Li Y., Shen C., Xia Y. COVID-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv. 2003 2020. (China) [Google Scholar]
- 14.Khalid, et al. “Automated Methods for Detection and Classification Pneumonia based on X-Ray Images Using Deep Learning”. https://arxiv.org/abs/2003.14363v1.
- 15.Khan Asif Iqbal, Shah Junaid Latief, Bhat Mohammad Mudasir. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput Methods Programs Biomed. 2020 doi: 10.1016/j.cmpb.2020.105581. ISSN 0169-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Linda, Lin Zhong Qiu, Wong Alexander. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv. 2003 doi: 10.1038/s41598-020-76550-z. [eess.IV] 15 April 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ucar F., Korkmaz D. Covidiagnosis-net: deep bayes-squeezenet based diagnostic of the coronavirus disease 2019 (covid-19) from X-ray images. Med Hypotheses. 2020;109761 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghoshal B., Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv. 2003 2020. (Brunel University, London, United Kingdom) [Google Scholar]
- 19.Vaid S., Kalantar R., Bhandari M. Deep learning covid-19 detection bias: accuracy through artificial intelligence. Int Orthop. 2020:1. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus covid-19 detection from x-rays. Comput Methods Prog Biomed. 2020:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi F., Xia L., Shan F., Wu D., Wei Y., Yuan H. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. arXiv. 2003 doi: 10.1088/1361-6560/abe838. 2020. (China) [DOI] [PubMed] [Google Scholar]
- 22.Khobahi S., Agarwal C., Soltanalian M. CoroNet: a deep network architecture for semi-supervised task-based identification of COVID-19 from chest X-ray images. medRxiv. 2020;(January) doi: 10.1101/2020.04.14.20065722. 2020.04.14.20065722. (University of Illinois, Chicago) [DOI] [Google Scholar]
- 23.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2020;(September) doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of covid19 in X-rays using ncovnet. Chaos Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarker L., Islam M.M., Hannan T., Ahmed Z. COVID-DenseNet: a deep learning architecture to detect COVID-19 from chest radiology images. Preprints. 2020 doi: 10.20944/preprints202005.0151.v1. 2020050151. [DOI] [Google Scholar]
- 27.Yoo S.H., Geng H., Chiu T.L., Yu S.K., Cho D.C., Heo J. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front Med. 2020;7 doi: 10.3389/fmed.2020.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals. 2020;140(November) doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative covid-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J Med Biol Eng. 2020:1. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elasnaoui K., Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2020:1–22. doi: 10.1080/07391102.2020.1767212. no. just-accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahimzadeh M., Attar A. A modified deep convolutional neural network for detecting covid-19 and pneumonia from chest X-ray images based on the concatenation of xception and resnet50v2. Informn Med Unlocked. 2020 doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus covid-19 detection from X-rays. Comput Methods Prog Biomed. 2020 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmud T., Rahman M.A., Fattah S.A. Covxnet: a multidilation convolutional neural network for automatic covid-19 and other pneumonia detection from chest X-ray images with transferable multireceptive feature optimization. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen Joseph Paul, Morrison Paul, Dao Lan. COVID-19 image data collection. arXiv. 2003 https://github.com/ieee8023/COVID-chestxray-dataset 2020. [Google Scholar]
- 36.Chest X-Ray Images (Pneumonia). https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- 37.Kobayashi Y., Mitsudomi T. Management of ground-glass opacities: should all pulmonary lesions with ground-glass opacity be surgically resected? Transl Lung Cancer Res. 2013;2(5):354–363. doi: 10.3978/j.issn.2218-6751.2013.09.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vilar J., Domingo M.L., Soto C., Cogollos J. Radiology of bacterial pneumonia. Eur J Radiol. 2004 doi: 10.1016/j.ejrad.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Wong H.Y.F., Lam H.Y.S., Fong A.H.-T., Leung S.T., Chin T.W.-Y., Lo C.S.Y. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020;296:E72–E78. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. Am J Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 41.Alom M.Z., Taha T.M., Yakopcic C., Westberg S., Hasan S.M., Van Esesn B.C. The history began from AlexNet: a comprehensive survey on deep learning approaches. arXiv preprintarXiv. 2018 [Google Scholar]
- 42.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Krizhevsky A., Sutskever I., Hinton G. ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems; Lake Tahoe, NV, USA, 3 – 6 December; 2012. pp. 1097–1105. [Google Scholar]
- 44.Redmon J., Farhadi A. Yolov3: an incremental improvement. arXiv preprint arXiv. 2018 1804.02767. [Google Scholar]
- 45.Lin T.-Y., Dollar P., Girshick R., He K., Hariharan B., Belongie S. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Feature pyramid networks for object detection. [DOI] [Google Scholar]
- 46.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.C. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2018. MobileNetV2: inverted residuals and linear bottlenecks. [DOI] [Google Scholar]
- 47.Simonyan K., Zisserman A. 3rd International Conference on Learning Representations, ICLR 2015 – Conference Track Proceedings. 2015. Very deep convolutional networks for large-scale image recognition. [Google Scholar]
- 48.Ren S., He K., Girshick R., Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017 doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 49.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition. [DOI] [Google Scholar]