Abstract
Introduction
SARS-CoV-2 a new virus of the zoonotic coronavirus family causes the disease COVID-19, which has become a global pandemic. One of the ways for prevention of COVID-19 is by disabling its spike protein which results in inhibiting its binding with angiotensin-converting enzyme 2 (ACE-2). The other alternative is to inhibit its replication once inside the body. The aim of this study was to explore the literature to identify whether there were any Ayurvedic remedies which contained ingredients which demonstrated this dual effect.
Methods
In silico studies were carried out to find the structures of the targets i.e. spike protein of the virus and its main protease (Mpro). Databases were searched to identify the composition of Ayurvedic decoctions used for respiratory ailments.
Results
We have found that two components out of 26 active ingredients of Ayurvedic decoctions are strong binders for spike protein as well as corresponding Mpro (3CL protease) which plays an essential role in mediating viral replication and transcription, making it an attractive antiviral drug target. Out of 26 components of Ayurvedic herbal decoction used for influenza, one compound was found to be most active. It is a well-known antioxidant, antinflammatory and hepatoprotective molecule.
Conclusion
The resultant compound could act as a repurposed drug or like other methoxyphenols, could be a good lead molecule for a potent drug for COVID-19.
Keywords: SARS-CoV-2, SARS-CoV-2 Spike protein, SARS-CoV-2 main protease, COVID-19, Decoction, Ayurveda, Herbal medicine
1. Introduction
SARS-CoV2, the new coronavirus that causes the COVID-19 disease is causing havoc globally [1]. The analysis of the genetic template of spike proteins, that the virus uses to attach and penetrate human and animal cells has shown that these have a receptor-binding domain (RBD), a hook grips onto host cells, and the cleavage site, a molecular bottle opener allows the virus to crack open and enter host cells [2].These constitute two subunits of the virus spike protein, S1 and S2 [3].
The RBD portion of the SARS-CoV-2 spike proteins effectively targets angiotensin-converting enzyme 2 (ACE2), a receptor present throughout the human body [2], normally involved in regulating blood pressure. Once inside, the virus hijacks the host cell's machinery, making myriad copies of itself and invading new cells.
The“spikes” that SARS-CoV-2 uses to establish infections on human cells are about 4 times stronger than those on the SARS – that caused an epidemic in 2002 and resulted in hundreds of deaths [4].The coronavirus particles that are inhaled via the nose or mouth have a high probability of attaching to cells in the upper respiratory tract [5], implying that relatively only a few viruses are required to infect and rapidly replicate.
This virus hijacks living cells to replicate and spread. When the virus finds a suitable cell, it injects a strand of RNA that contains the entire coronavirus genome.The genome of the new coronavirus is less than 30,000 “letters” long. (The human genome is over 3 billion.) Scientists have identified genes for as many as 29 proteins, which carry out a range of jobs from making copies of the coronavirus to suppress the body's immune responses. The microRNAs are present in all human beings hsa-miR-27b [6] is one of the six antiviral microRNAs that can potentially bind to virus genes and affect its activity.
The gene for the spike protein in SARS-CoV-2 has an insertion of 12 genetic letters: ccucggcgggca. This mutation may help the spikes bind tightly to human cells [7] — a crucial step in its evolution from a virus that infected bats and other species. A number of scientific teams are now designing vaccines that could prevent the spikes from attaching to human cells [8].
The virus spike's transmembrane-glycoprotein,a homotrimer [9] is responsible for the entry of viral RNA into the host cell. Its S1 subunit attaches to ACE2 receptors of the host cell which stimulates formation of TMPRSS2 (Transmembrane protease Serin-2) [10]. Further, protease helps fusion of S2 subunit to the cell membrane of the host. Hence targeting ACE2 binding S1 subunit or protease binding S2 subunit would prevent the entry of viral RNA inside the host cell. Main protease of coronavirus is crucial for processing quality proteins which are translated by coronavirus RNA [11]. Therefore targeting them would stop further spread of infection. If a molecular structure can bind to the sites on the virus more frequently and strongly than the ACE-2 receptor, it will obstruct the virus from binding to the cells, making it a reliable drug to treat COVID-19.The same sites can be used to develop vaccines to prevent future infections.
Main proteases (Mpro) of SARS-CoV-2, a cysteine protease, helps virus in proteolytic processing of polyproteins. Mpro targets Glutamine residue and cleaves the polypeptide after it. Enzymes like RdRp or nsp13 which are essential replication enzymes of the virus cannot become functional without prior proteolytic release. This makes Mpro a very important enzyme in viral replication cycle and thereby its inhibition can stop production of viral particles [12].
The objective of our study was to block the attachment of the spike protein's S1 and S2 subunits to ACE2 receptors or protease which helps viruses fuse with cell membrane and release its RNA into the host cell. We also focused on targeting coronavirus main protease (Mpro) that processes replicase polyproteins and is pivotal for gene expression and replication.
We selected 26 active compounds from 4 essential herbal components of an Ayurvedic preparation known as “Kadha” or decoction, a beverage given to treat common cold and flu problems [13]. Our studies have shown that out of 26, three showed significant results while one, moupinamide, was found to interact strongly with the RBD site of Spike protein as well as the corresponding protease. Therefore, the molecule which is already known for its therapeutic values can be a good candidate for not only blocking the spike protein for binding with ACE2 but also for growth of the virus once inside cells by interfering with the formation of viral proteins.
2. Materials and methods
2.1. Ligand library collection
In Ayurveda herbs and spices are recommended for the treatment of respiratory ailments, often in the form of drinks. On this basis, 26 active compounds from selected plants were selected, namely Zingiber officinale (Ginger) [12], Glycyrrhiza glabra (Liquorice) [13], Piper nigrum (Black pepper) [14] and Ocimum basilicum (Sweet Basil) [15], These plants have exhibited curing and immune boosting properties and parts of these plants such as fruits, seeds, stem or leaves are used to prepare remedial beverages. The Ligand library collection of compounds was based on collection of active phytochemicals from the plants having anti-inflammatory, anti-viral, anti-microbial and immune boosting properties. A ligand library of the 26 active compounds from the above plants was downloaded from PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The pubchem IDs of the compounds obtained are given in Table 1 .
Table 1.
Pubchem IDs of selected active phytochemicals.
| S.No | Plants Name | Ligand Pubchem ID |
|---|---|---|
| 1 | Zingiberofficinale (Ginger) | 31,211, 12,689, 51,976, 92,776, 637,566, 643,779, 969,516, 5,317,270, 11,141,699, 22,321,203 |
| 2 | Glycyrrhiza glabra (Liquorice) | 5901, 44,654, 5,318,998, 135,413,551, 37,542, 14,982 |
| 3 | Piper nigrum (Black pepper) | 5,280,537, 9,938,436, 131,752,909, 117,443, 10,131,321 |
| 4 | Ocimumbasilicum (Sweet Basil) | 70,678,558, 17,868, 6654, 442,359, 5,317,844 |
2.2. Protein selection and preparation
The SARS-COV-2 main protease (Mpro) with PDBID:6Y84 [16] and spike protein with PDBID:6LXT [17] having resolution of 1.39 Å and 2.90 Å, respectively, were selected and retrieved from RCBS.
Both the proteins were prepared by using Glide v8.3, Schrodinger, LLc, New York, NY, 2019–2 Protein preparation wizard that adds protons, fixes bond orders, optimizes protonation states and hydrogen bond networks and performs minimization under restraints [18]. As the co-crystallized ligands in most of the proteins show better interaction to the protein with conserved water molecules within 5 Å, hence we used the same analogy and kept the highly conserved water molecule in the 5 Å and prepared the protein. Since no natural ligand was present with the protein defining any active site, hence using “Site-mapping” from glide we found binding sites in both the proteins. Site mapping finds the binding site with presence of hydrophobic groups, ligand – hydrogen bond donors, acceptors or metal binding functionality. Binding site selection was done on the basis of the site with the highest site score and volume.
2.2.1. Grid generation
Grid generation allows determining the position, size, shape and properties of the active site.To generate grid, in the Receptor grid generation panel of Glide, we uploaded the prepared protein and selected the residue of the most suited site map (SiteMap1) with maximum Site Score and volume and generated the grid using OPLS2005 force field. For bounding and enclosing boxes, we preferred the default size.
2.2.2. Virtual screening of compounds for ADMET, ligand preparation and molecular docking
Using the Virtual Screening Workflow from Glide module of Schrodinger, we performed the ADMET screening of the ligand library checking for the criteria like QPLogHERG (range below-5), QPLogS (range −6 to −0.5), CIQPLogS (range −6.5 to −0.5), QPPMDCK (range <25 for poor and >500 for better), PISA (range 0 to 450), FOSA (range 0 to 750), FISA (range 7 to 330), SASA (range 300 to 1000), QPPCaco range (<25 for poor and >500 for better), Mol. (MW) (range 130 to 725). Obtained ligands were already prepared as minimized energy 3D molecular structures with expanded ionization states, stereoisomers and ring confirmations for expanded chemical and structural diversity.
2.2.3. Docking
Virtual Screening Workflow of Glide also performs Protein-Ligand docking with accuracy levels-high throughput virtual screening (HTVS), standard precision (SP), and extra precision (XP) [19]. The docking results were analysed comparing the Glide Emodel and Glide Score as the Glide uses both the scoring function to select between protein ligand complexes of the given ligand and to provide a ranked order to compounds to separate compounds that bind strongly in comparison to others [20,21].
2.2.4. Relative binding affinity estimation using MM-GBSA
In order to find the binding free energy from Protein-ligand complex, we approached Prime- MM- GBSA, to add more validation to our docking results. It uses local optimization features in Prime to minimize the docked poses and calculates energy of complex using OPLS-AA force field and generalized-Born surface area (GBSA) continuum solvent model. The outcome is strongly related to the experimental results,but it did not include corrections for entropic changes [22], [23].
Prime/MM-GBSA follows the following equation to obtain Binding free energy:
2.3. Molecular dynamics simulation study
Molecular dynamics is an approach of computational simulation to analyze the physical movement of atoms and molecules that were allowed to interact with each other in a fixed period of time and in a given virtual environment. Desmond based MD simulation was used to evaluate the stability of protein-ligand complex. Protein-ligand contact bar graph, RMSD and simulation event analysis. It is used to study the stability and molecular movement of molecules of protein and ligand in a developed system throughout simulation run time.
2.3.1. System building
System building was done by system builder Desmond, Schrodinger, LLC, New York, NY, 2019–1. TIP3P was chosen as a solvent model that is a water model in orthorhombic shape of box, where the dimension of box was 10 × 10 × 10 Å and angle 90 × 90 × 90°. 0.15 M NaCl was added in a prepared box for salt addition to neutralize and stabilize the system after recalculation of ions and salt addition which made the complete real environment to run the MD simulation.
2.3.2. Molecular dynamics
After system building of protein ligand complex MD simulation was executed. Here all the parameters were set like recording interval (ps), energy, trajectory, NTP. This had been chosen as given default value. Check point interval of simulation was set as default value (240.06 ps). MD simulation was performed using Desmond, Schrodinger, LLC, New York, NY, 2019–1.
3. Results
The study was planned to find a multi targeting naturally occurring active compound that targets and inhibits the SARS COV-2 main protease Mpro) (6Y84) and SARS COV-2 spike protein (6LXT) simultaneously.
3.1. Ligand selection and screening
Ligand library collection of 26 active chemicals compounds from herbal components of an Ayurvedic preparation known as “Kadha” or decoction, screened for ADMET. After screening of 26 compounds, specifically the one with antiviral and anti-inflammatory property, using the QikProp tool of Glide, we found 5 active compounds (PubChem CID 5,280,537: Moupinamide, 5,318,998: Licochalcone A, 10,131,321: Coumaperine, 969,516: Curcumin and 22,321,203: 6-Dehydrogingerdione) that fulfilled all the screening parameters as shown in Table 2 .
Table 2.
QikProp result of ADMET properties of selected compounds.
| Ligand IDADMET criteria | 5,280,537 | 5,318,998 | 10,131,321 | 969,516 | 22,321,203 |
|---|---|---|---|---|---|
| Mol MW | 313.35 | 338.40 | 257.33 | 368.38 | 290.36 |
| Dipole | 5.035 | 2.56 | 5.547 | 3.64 | 4.83 |
| SASA | 634.02 | 653.93 | 562.96 | 708.92 | 638.81 |
| FOSA | 186.02 | 282.35 | 259.49 | 261.23 | 377.49 |
| FISA | 161.74 | 129.87 | 96.48 | 190.59 | 131.23 |
| PISA | 286.26 | 241.71 | 206.99 | 257.09 | 130.11 |
| QPlogS | −4.37 | −5.03 | −4.21 | −4.62 | −4.11 |
| CIQPlogS | −4.27 | −5.03 | −3.34 | −4.61 | −3.31 |
| QPlogHERG | −6.16 | −5.74 | −5.32 | −6.35 | −5.47 |
| QPPCaco | 289.83 | 581.21 | 1205.09 | 154.35 | 564.27 |
| QPPMDCK | 129.72 | 275.19 | 605.23 | 65.65 | 266.52 |
| Rule Of Five | 0 | 0 | 0 | 0 | 0 |
3.2. Docking
Docking was performed to find the compound with maximum affinity to both the target proteins: COVID-19 Mpro and Spike protein. Both the docking score results showed maximum interaction of 6Y84 and 6LXT with Moupinamide (Tables 3 and 4 ). The Glide Score of 6Y84 with Moupinamide(5,280,537) was 5.066 and the Glide Score of 6LXT with Moupinamide(5,280,537) was 5.962. The compound Moupinamide, in comparison to other ligands demonstrated great affinity to both the proteins simultaneously.
Table 3.
Docking results of main protease Mproprotein (PDBID: 6Y84) with selected compounds.
| Protein: 6Y84 | |||||
|---|---|---|---|---|---|
| S No. | Title | Docking Score | Glide gscore | Glide Emodel | Glide Energy |
| 1 | 5,280,537 | −4.997 | −5.066 | −45.684 | −38.576 |
| 2 | 5,318,998 | −4.981 | −5.052 | −43.648 | −36.996 |
| 3 | 10,131,321 | −4.631 | −4.632 | −38.415 | −32.058 |
| 4 | 969,516 | −3.259 | −4.677 | −40.258 | −35.528 |
| 5 | 22,321,203 | −3.149 | −3.552 | −38.959 | −35.976 |
Table 4.
Docking results of spike protein(PDBID: 6LXT) with selected compounds.
| Protein: 6LXT | |||||
|---|---|---|---|---|---|
| S No. | Title | Docking score | Glide gscore | Glide Emodel | Glide energy |
| 1 | 5,280,537 | −5.893 | −5.962 | −55.307 | −43.897 |
| 2 | 5,318,998 | −5.63 | −5.701 | −45.867 | −36.75 |
| 3 | 10,131,321 | −5.32 | −5.321 | −44.368 | −33.673 |
| 4 | 969,516 | −5.261 | −5.484 | −61.698 | −47.61 |
| 5 | 22,321,203 | −3.146 | −3.151 | −44.202 | −38.294 |
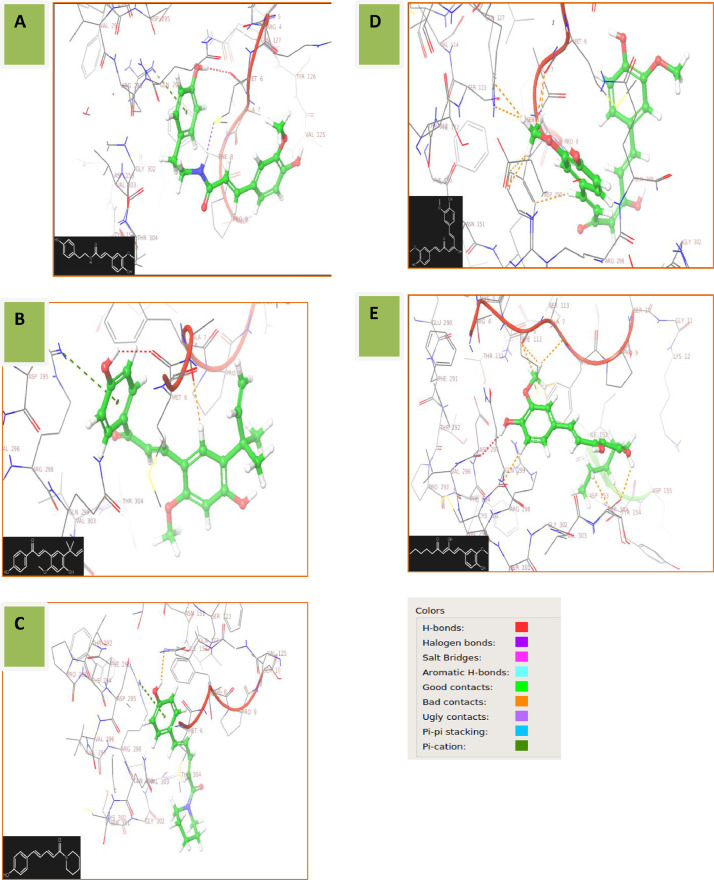
The ligand-interaction diagram obtained from the docking results of SARS COV-2 Main Protease(6Y84) and Moupinamide(5,280,537) reveals that the binding affinity in the cavity formed by the residues 4–10(ARG-4, LYS-5, MET-6, ALA-7, PHE-8, PRO-9, SER-10), 125–127 (VAL-125, GLN-127), and 295–304 (ASP-295, ARG-298, GLN-299, GLY-302, VAL-303, THR-304)from A-chain, is mainly due to the presence of hydroxyl (-OH) group that shows hydrogen bond with MET-6 residue and the benzene ring to which hydroxyl group is attached, showing pi-cation interaction with ARG-298 residue (Fig. 1 A). Protein-ligand(s) post docking interaction of Mpro with Licochalcone, Coumaperine, Curcumin and 6-Dehydrogingerdioneis shown in Fig. 1. B, C, D, and E.
Fig. 1.
COVID-19 Mpro-ligand post docking interaction. – A (Mpro-5,280,537), B (Mpro-5,318,998), C (Mpro-10,131,321), D (Mpro-969,516) and E (Mpro-22,321,203).
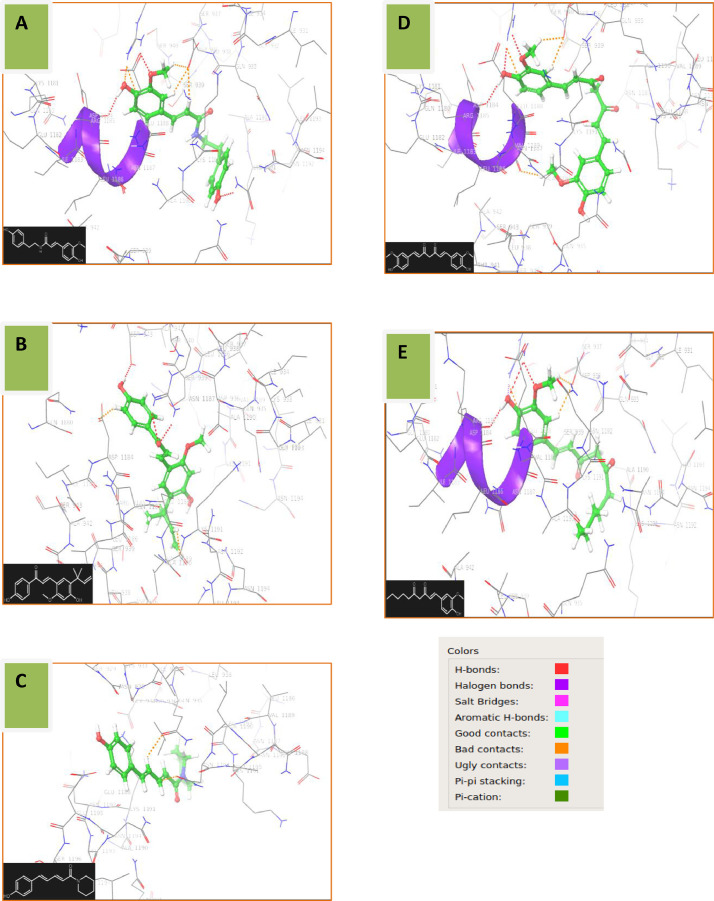
Ligand Interaction diagram of SARS COV-2 spike protein(6LXT) and Moupinamide(5,280,537) reveals that the binding affinity in the cavity formed by the residues 1184–1194 of B (ASP-1184, ARG-1185, ASN-1187, GLU-1188, ALA-1190, LYS-1191, ASN-1192, ASN-1194), E (ARG-1185) and F chains (ASN-1187, ALA-1190, LYS-1191, ASN-1194) and 935–939 of F chain (GLN-935, ASP-936, SER-939) is mainly due to the presence of both the hydroxyl (-OH) groups which shows hydrogen bond with ASN-1194 and ASP-1184 residues of B-chain each and the ether group shows pi-cation interaction with ARG-1185 residue of E-chain (Fig. 2 A). Protein-ligand(s) post docking interaction of spike protein with Licochalcone, Coumaperine, Curcumin and 6-Dehydrogingerdione is shown in Fig. 2. B, C, D, and E.
Fig. 2.
Spike protein-ligand post docking interaction. – A (Mpro-5,280,537), B (Mpro-5,318,998), C (Mpro-10,131,321), D (Mpro-969,516) and E (Mpro-22,321,203).
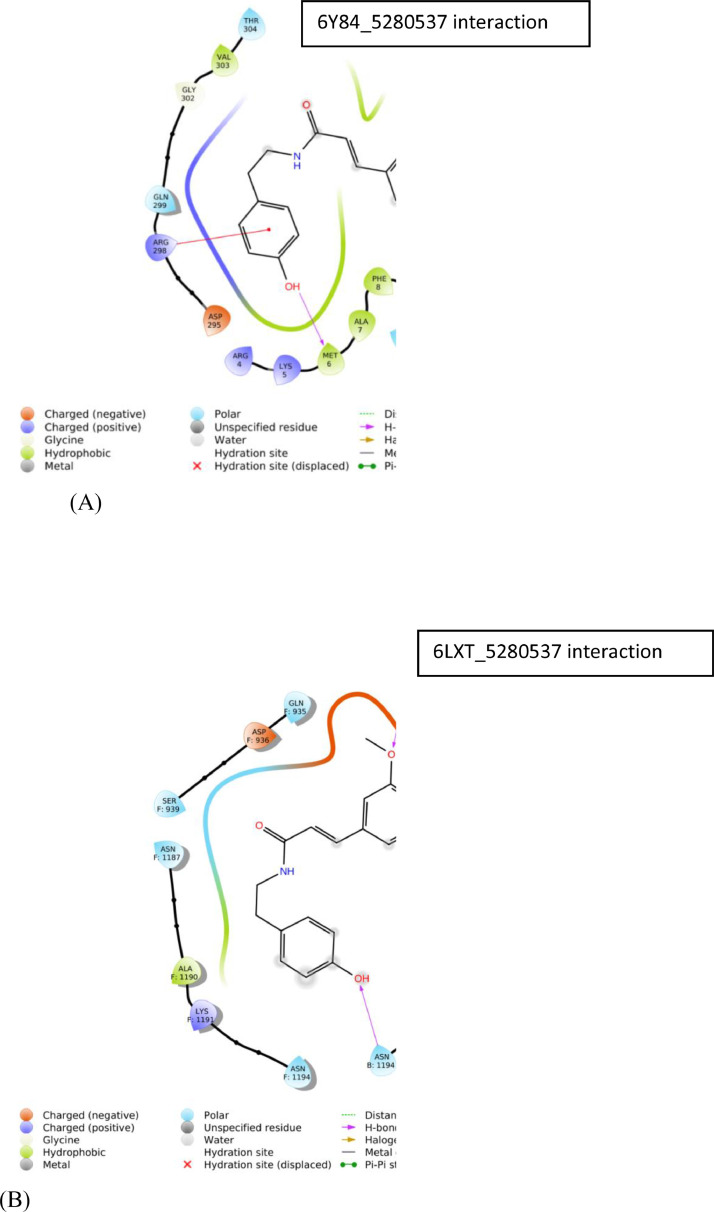
The detailed analysis of residues and binding interactions of Moupinamide and COVID-19 MPRO (6Y84) and Spike protein (6LXT) at the binding pocket is shown in Fig. 3 (A) and 3(B).
Fig. 3.
Protein-ligand interaction diagram, 3 (A) Mpro-5,280,537 interaction and 3(B) Spike Protein-5,280,537 interaction.
3.3. MMGBSA results
We followed the molecular mechanics,generalised born model and solvent accessibility method to elicit free energies from the protein ligand complex. Analysing the overall binding free energy generated for docking interactions of each protein, COVID-19 Main Protease and Spike protein, by observing receptor energy, ligand energy and complex energy we found that both the proteins show best interaction with the proposed druggable ligand Moupinamide, which is −69.38 kcal/mol for 6Y84 (Table 5 ) and −46.67 kcal/mol for 6LXT (Table 6 ).
Table 5.
Binding free energy analysis Prime-MMGBSA-result with Mproprotein (PDBID: 6Y84).
| S No | Title | MMGBSA dG Bind | MMGBSA dGBind Coulomb | Ligand Energy | Ligand Coulomb | Complex Energy | Complex Coulomb | Receptor Energy | Receptor Coulomb |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5,280,537 | −69.38 | −14 | −33.04 | −37.69 | −13,089.74 | −9968.35 | −12,987.33 | −9916.65 |
| 2 | 5,318,998 | −69.05 | −14.72 | 2.86 | −13.35 | −13,053.52 | −9944.73 | −12,987.33 | −9916.65 |
| 3 | 10,131,321 | −64.95 | −12.99 | 18.37 | −5.42 | −13,033.91 | −9935.06 | −12,987.33 | −9916.65 |
| 4 | 969,516 | −60.59 | −10.3 | −11.14 | −21.72 | −13,059.06 | −9948.67 | −12,987.33 | −9916.65 |
| 5 | 22,321,203 | −70.97 | −9.74 | 5.35 | 0.66 | −13,052.95 | −9925.72 | −12,987.33 | −9916.65 |
Table 6.
Binding free energy analysis Prime-MMGBSA-result with Spike protein (PDBID: 6LXT).
| S No | Title | MMGBSA dG Bind | MMGBSA dG Bind Coulomb | Ligand Energy | Ligand Coulomb | Complex Energy | Complex Coulomb | Receptor Energy | Receptor Coulomb | Energy |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5,280,537 | −46.67 | −24.6 | −29.026 | −32.369 | −32,644.8 | −23,164.1 | −32,569.1 | −23,107.1 | 13.937 |
| 2 | 5,318,998 | −49.85 | −13.46 | 0.111 | −15.822 | −32,618.9 | −23,136.4 | −32,569.1 | −23,107.1 | 27.325 |
| 3 | 10,131,321 | −39.65 | −0.94 | 15.936 | −4.577 | −32,592.8 | −23,112.6 | −32,569.1 | −23,107.1 | 31.379 |
| 4 | 969,516 | −44.09 | −16.19 | −26.501 | −29.778 | −32,639.7 | −23,153.1 | −32,569.1 | −23,107.1 | 23.761 |
| 5 | 22,321,203 | −42.37 | −18.71 | −19.618 | −15.043 | −32,631.1 | −23,140.9 | −32,569.1 | −23,107.1 | 13.811 |
3.4. MD simulation results
Finding promising docking hits and affinity between ligand and both the target proteins, we further performed MD simulation study to understand the dynamic behavior of protein-ligand molecular system. Results from the simulation run strongly support the docking outcomes showing stability of protein-ligand complexes.
3.4.1. SARS COV-2 main protease (Mpro)
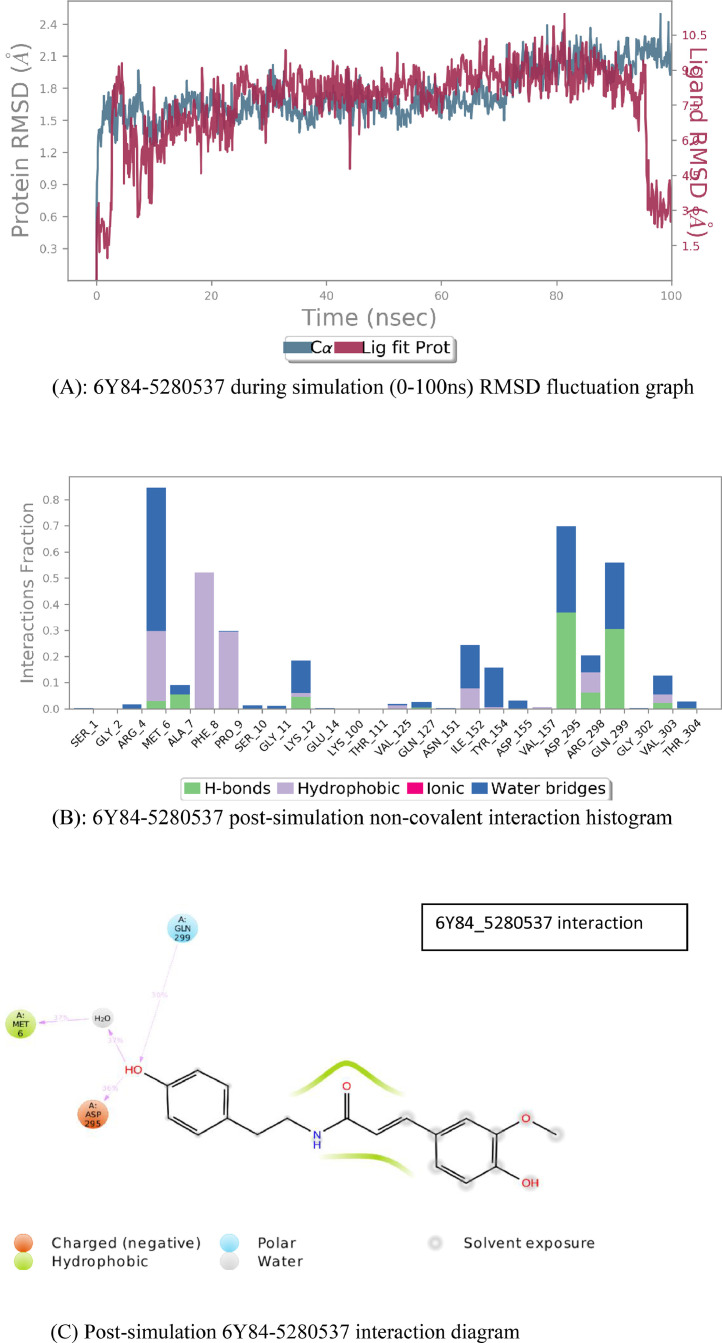
Root Mean Square Deviation graph (Fig. 4 (A)) depicts that while interacting with the ligand, fluctuations in the structural confirmation of protein attain stability within 20 ns and remained so upto 75nsec, then persists some fluctuation and again gets stable and continued for 100 ns, whereas, Moupinamide system remains stable for 20 to 90nsec. Changes in RMSD value of protein and ligand present the variation in conformation of protein and ligand as well as their stability. In the given complex of protease protein (PDB id: 6Y84) and ligand moupinamide (pubchem id: 5,280,537), the fluctuation of RMSD was not increased more than 2.5 Å all the time of simulation.
Fig. 4.
(A): 6Y84–5,280,537 during simulation (0–100 ns) RMSD fluctuation graph. (B): 6Y84–5,280,537 post-simulation non-covalent interaction histogram. (C) Post-simulation 6Y84–5,280,537 interaction diagrams.
From Simulation event analysis we found that the average RMSD of 6Y84 C-alpha is 1.91 with a fluctuation range of 0.00 to 2.7 for 0 to 100 ns, average RMSD of side chain of protein is 2.65 and average RMSD of ligand, Moupinamide throughout simulation run is 1.001 with a fluctuation range from 0.00 to 2.4. Visualizing the protein-ligand trajectory, the ligand appeared to be interacting with the same cavity of protein throughout the simulation run.
The Ligand interaction graph (Fig. 4(B)) depicts the fraction of interactions and bonds holding the ligand to the protein cavity. As the protein cavity is mainly formed of its terminal amino acids, present in it's a chain, i.e. from SER-1 to GLU-14 and from LYS-111 to THR-304, from SER-1 end, the major interactions behold Hydrophobic and water bridges, whilst a few contribution of H-bond.Towards THR-304 end, the major bond contributors are hydrogen bond and water bridges, with a few hydrophobic interactions. Graph results are parallely supported by simulation interaction diagrams where the specific interaction percent is clearly notable. Also, comparing the Simulation ligand interaction diagram (Fig. 4(C) with docking ligand interaction diagram we are fortunate enough to find the same interactions. Here in post simulation ligand interaction diagram also, the amide group of the ligand shows hydrophobic interaction at its surface and the hydroxyl group holds hydrogen bond with MET-6, polar bond with GLN-299 and ionic interaction with ASP-295.
3.4.2. SARS COV-2 spike protein (6LXT)
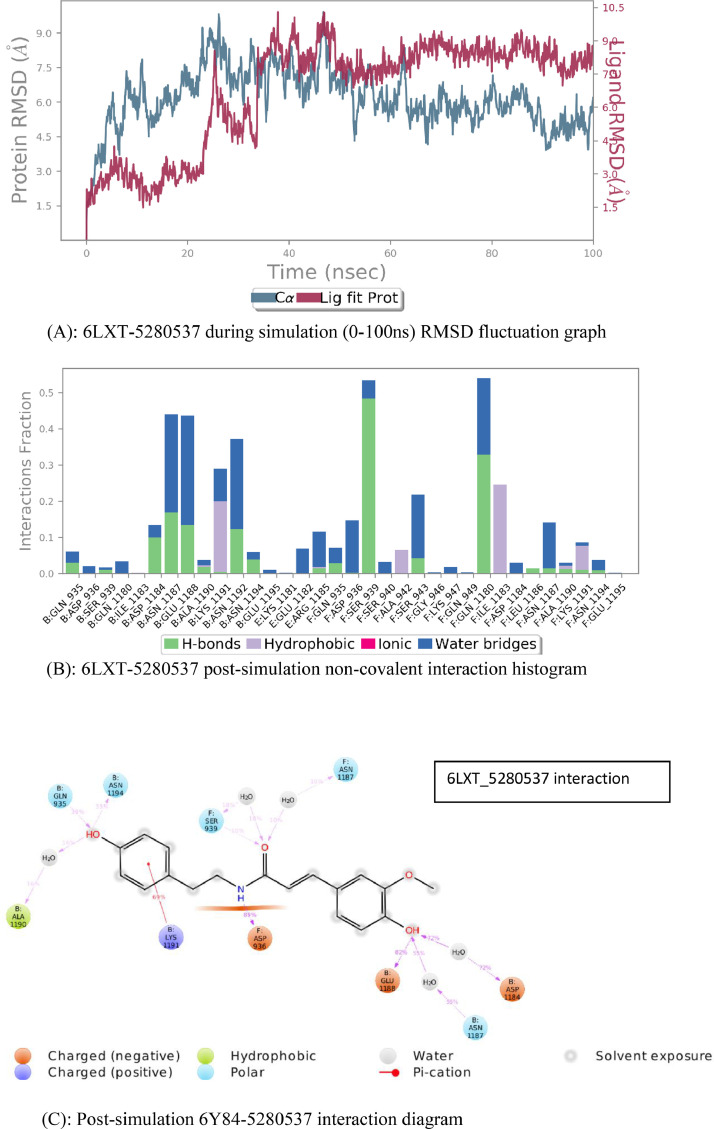
The Root Mean Square Deviation graph (Fig. 5 (A) of interaction of COVID-19 spike protein depicts that the protein and ligand remained bound together for more than half of the simulation run time. From 0 to 60 ns the interaction is quite significant. Though there is fluctuation in the initial simulation run, the stability is attained in the final half of the time period. Visualizing the Protein-Ligand trajectory, the ligand appeared to be interacting with the same cavity of protein throughout the simulation run.
Fig. 5.
(A): 6LXT-5,280,537 during simulation (0–100 ns) RMSD fluctuation graph. (B): 6LXT-5,280,537 post-simulation non-covalent interaction histogram. (C): Post-simulation 6Y84–5,280,537 interaction diagram.
The Simulation event analysis of the MD Simulation project of COVID-19 spike protein, depicts that the average RMSD of 6LXTC-alpha is 3.8 with a fluctuation range of 0.00 to 6.9 for 0 to 100 ns, average RMSD of side chain of protein is 4.5 and average RMSD of ligand, Moupinamide throughout simulation run is 1.12 with a fluctuation range from 0.00 to 2.8.
The Ligand interaction graph (Fig. 5(B) depicts interaction between SARS COV-2 spike protein (6LXT) and the selected ligand moupinamide(5,280,537). Result generated post-simulation clearly shows that the cavity formed by the chains B, E and F which are homotrimers, is from the same end. GLY-932 to GLN-935, ASN-1187 to GLU-1188 and ASN-1192 to ASN-1194 residues of B-chain, and GLN-935 to SER-939 of F-chain contributes all the hydrogen bond interaction between the taken protein and ligand. Other residues hold other interactions like hydrophobic interactions and water bridges. Involvement of these many cavity residues bonding with ligand depicts better interaction of ligand to protein. The exact sketch of the graph is visible in the ligand interaction diagram. The interaction diagram of SARS COV-2 spike protein(6LXT) with Moupinamide (Fig. 5(C) shows that ASP-936 holds hydrogen bond for 89 percent of the simulation run period, GLU-1188 holds hydrogen bond interaction for 82 percent of the simulation run period, LYS-1191 holds pi=cation interaction, providing complex more stability, for 69 percent of total simulation running period. Some water bridges also existed for more than 50 percent of the simulation run like ASP-1184 for 72percent and ASN-1187 for 56percent. Comparing this post-simulation interaction of taken ligand and protein to that of post-docking interaction diagram, we found that both results agree with another showing the very same residue binding and contribution in protein ligand interactions.
4. Discussion
Understanding the virus-receptor recognition mechanism responsible for COVID-19 infection, pathogenesis and host range provides direction for the development of therapy to combat and cure this worldwide pandemic. No effective treatment is available so far against Covid-19, although search for new therapy is proceeding actively. On the basis of virus- receptor mechanism two prominent therapeutic protein targets (spike and M-pro) were selected. Spike-protein present on the SARS-CoV-2 envelope is responsible for interaction with ACE2 receptor of host cells. The viral main proteinase (Mpro, also called 3CLpro), controls the activities of the coronavirus replication complex. As per WHO, approximately 80% of world population depends on medicinal plants for their primary healthcare source. For paradigm, the traditional medicine has been used from thousands of years in India, China, Africa, etc. Thus, knowledge based on the medical benefits of plants, has the potential to identify novel drug candidates. Active compounds of medicinal plants extracts are being considered as possible and promising approach for COVID-19 treatments and should be tested for efficacy and adverse side effects.
Molecular docking and simulation studies were performed over 26 active compounds from our 4 decoction ingredients to find a potential lead candidate for the treatment of SARS-CoV-2 infections. All the 26 active compounds docked with both the spike and M-pro and ranked on the basis of their dock score. Later, compounds which fulfilled the ADMET criteria were selected. As per docking results the Moupinamide proves to be the best ligand targeting both the therapeutic target proteins, followed by Licochalcone, Coumaperine, Curcumin and 6-Dehydrogingerdione. The rest of the 21 compounds docked with proteins but not fulfilled the ADMET criteria. These lead compounds can target S-RBD–ACE2 interface protein-protein interactions and stop the replication of viral RNA while inhibiting ACE2 can lead to such structural changes which could potentially inhibit the virus entry into host cell and thus, provide quick solution to control SARS-CoV-2.
MMGBSA further approved the better binding affinity of Moupinamide with both the target proteins.Running MD-Simulation for 100 ns showed the well stability of protein ligand complex and the work could be validated and continued further to wet lab validations. Also, comparing the Simulation ligand interaction with docking ligand interaction of Moupinamide with M-pro and spike protein, we are fortunate enough to find the same interactions. Comparing this post-simulation interaction of taken ligand and protein to that of post-docking interaction diagram, we found that both results agree with another showing the very same residue binding and contribution in protein ligand interactions.
Moupinamide is a well-known antioxidant, anti-inflammatory and hepatoprotective molecule. It can act as a repurposed drug or like other methoxyphenols, can be a good lead molecule for finding a potent drug for COVID-19.
5. Limitations
Inferences drawn from this study are important because of the necessity to curb the havoc caused by COVID-19. These ligands described can be considered as potential active molecules to inhibit the function of both the Spike protein and Main Protease. However, these naturally occurring ligands, although safe for human consumption need further investigation and optimization in-vitro before being considered for clinical trials. They could be considered as potential lead molecules for designing potent drugs to combat COVID-19.
6. Conclusion
Ayurvedic decoction containing a mixture of 26 active components from 4 selected plants viz. Zingiber officinale (Ginger), Glycyrrhiza glabra (Liquorice), Piper nigrum(Black pepper) and Ocimum basilicum (Sweet Basil) have been shown to be effective for treatment of respiratory ailments.
We have concluded from our results that one of these components viz. Moupinamide (PubChem CID:5,280,537) present in Piper nigrum is the best fit since it binds strongly with both essential proteins of SARS-Cov-2, the SpikBeing e protein, which is responsible for the entry of virus into the host cell and the Main Protease, that plays very crucial role in viral replication. Being as well as an inflammation inhibitor and immune boosting compound, our multi-targeting molecule can be used as an inhibitor to hinder the activities of both proteins of COVID-19. This can also be used as a lead molecule for finding potent drugs for treatment of the disease.
Author contribution
Richa Shukla: Data curation, Investigation, Validation, Visualization, Methodology, Software, Writing - review & editing. Sangeeta Singh: Formal analysis, Investigation, Supervision, Writing - review & editing. Anirudh Singh: Methodology, Software. Krishna Misra: Conceptualization, Data curation, Supervision, Writing - original draft, Writ- ing - review & editing.
Financial support
None.
Declaration of Competing Interest
All related authors have asserted that there is no conflict of interest.
Acknowledgements
Authors are grateful to Director, Indian Institute of Information Technology, Allahabad, India for providing facilities for research. One of the authors (R.S.) is thankful to Ministry of Human Resource, New Delhi, India for providing her with research fellowship.
Data availability
Supplementary information or data can be obtained from the author on request.
References
- 1.Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S., et al. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020;49:717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.-.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. .e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai C.-.C., T.-P.Shih W.-C.Ko, Tang H.-.J., Hsueh P.-.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Ag. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardar R., Satish D., Birla S., Gupta D. Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. Bioinformatics. 2020 doi: 10.1101/2020.03.21.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W.-.H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 Vaccine Pipeline: an Overview. Curr. Trop. Med. Rep. 2020;7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalathiya U., Padariya M., Mayordomo M., Lisowska M., Nicholson J., Singh A., et al. Highly Conserved Homotrimer Cavity Formed by the SARS-CoV-2 Spike Glycoprotein: a Novel Binding Site. JCM. 2020;9:1473. doi: 10.3390/jcm9051473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. .e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., et al. Substrate specificity profiling of SARS-CoV-2 main protease enables design of activity-based probes for patient-sample imaging. Biochemistry. 2020 doi: 10.1101/2020.03.07.981928. [DOI] [Google Scholar]
- 12.A. Do, Perspective Chemistry and Biology of SARS-CoV-2, (2020) 1283–1295. DOI: 10.1016/j.chempr.2020.04.023. [DOI] [PMC free article] [PubMed]
- 13.Cavanagh D., Willis C. Ayurveda UK; 2004. Everyday Ayurveda. [Google Scholar]
- 14.Mousavi L., Salleh R.M., Murugaiyah V. Phytochemical and bioactive compounds identification of Ocimumtenuiflorum leaves of methanol extract and its fraction with an anti-diabetic potential. Int. J. Food Propert. 2018;21:2390–2399. doi: 10.1080/10942912.2018.1508161. [DOI] [Google Scholar]
- 15.Prasad S., Tyagi A.K. Ginger and its constituents: role in prevention and treatment of gastrointestinal cancer. Gastroenterol. Res. Pract. 2015;2015 doi: 10.1155/2015/142979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damle M. Glycyrrhiza glabra (Liquorice)-a potent medicinal herb. Int. J. Herb. Med. 2014;2(2):132–136. [Google Scholar]
- 17.Meghwal M., Goswami T.K. Chemical Composition, Nutritional, Medicinal And Functional Properties of Black Pepper: a Review. J. Nutr. Food Sci. 2012;1(2) [Google Scholar]
- 18.Owen C.D., Lukacik P., Strain-Damerell C.M., Douangamath A., Powell A.J., Fearon D., et al. COVID-19 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, SARS-CoV-2) RCSB Protein Data Bank (PDB) 2020:3–7. ID: 6Y84. [Google Scholar]
- 19.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion, inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenselink E.B., Louvel J., Forti A.F., van Veldhoven J.P.D., de Vries H., Mulder-Krieger T., et al. Predicting Binding Affinities for GPCR Ligands Using Free-Energy Perturbation. ACS Omega. 2016;1:293–304. doi: 10.1021/acsomega.6b00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra S., Singh S. Identification of Inhibitors against Metastasis Protein “Survivin:” In silico Discovery Using Virtual Screening and Molecular Docking Studies. Pharmacogn Mag. 2018;13:S742–S748. doi: 10.4103/pm.pm_178_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., et al. Glide: a New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava Mani, Harvinder Singh and Pradeep Kumar Naik. "Molecular modeling evaluation of the antimalarial activity of artemisinin analogues: molecular docking and rescoring using Prime/MM-GBSA Approach." (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary information or data can be obtained from the author on request.