Abstract
mRNA vaccines have evolved from being a mere curiosity to emerging as COVID-19 vaccine front-runners. Recent advancements in the field of RNA technology, vaccinology, and nanotechnology have generated interest in delivering safe and effective mRNA therapeutics. In this review, we discuss design and self-assembly of mRNA vaccines. Self-assembly, a spontaneous organization of individual molecules, allows for design of nanoparticles with customizable properties. We highlight the materials commonly utilized to deliver mRNA, their physicochemical characteristics, and other relevant considerations, such as mRNA optimization, routes of administration, cellular fate, and immune activation, that are important for successful mRNA vaccination. We also examine the COVID-19 mRNA vaccines currently in clinical trials. mRNA vaccines are ready for the clinic, showing tremendous promise in the COVID-19 vaccine race, and have pushed the boundaries of gene therapy.
Keywords: mRNA delivery, Gene delivery, Lipid nanoparticles, Self-assembly, Immunization, COVID-19
Acronyms: ACE2, Angiotensin-converting enzyme 2; APC, Antigen-presenting cell; ApoE, Apolipoprotein E; ARCA, “Anti-reverse" cap analog; BCR, B-cell receptor; COVID-19, Coronavirus Disease 2019; Cryo-(T)EM, Cryogenic (transmission) electron microscopy; DC, Dendritic cell; DGTS, 1,2-Dipalmitoyl-sn-glycero-3-O-4'-(N,N,N-trimethyl)-homoserine; DLin-MC3-DMA, MC3, (6Z,9Z,28Z,31Z)-Heptatriacont-6,9,28,31-tetraene-19-yl 4-(dimethylamino)butanoate; DMG-PEG-2000, 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000; DODMA, 1,2-Dioleyloxy-3-dimethylaminopropane; DOGS, N1,N1'-(8-(dioctadecylamino)-5,8-dioxooctane-1,4-diyl)bis(propane-1,3-diaminium); DOPE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine; DOTAP, 1,2-Dioleoyl-3-trimethylammonium-propane; DOTMA, 1,2-Di-O-octadecenyl-3-trimethylammonium propane; DSPC, 1,2-Distearoyl-sn-glycero-3-phosphocholine; EUA, Emergency Use Authorization; GMP, Good manufacturing practice; GC, Germinal center; HIV, Human immunodeficiency virus; HPLC, High-performance liquid chromatography; ID, Intradermal; IgA, IgG, Immunoglobulin A, G; IL, Ionizable lipid; IM, Intramuscular; IV, Intravenous; IVT, In vitro transcription; LN, Lymph node; LNP, Lipid nanoparticle; MHC, Major histocompatibility complex; mRNA, Messenger RNA; PBAE, Poly(beta-amino esters); pDNA, Plasmid DNA; PEG, Poly(ethylene glycol); PEI, Poly(ethylene imine); PET-CT, Positron emission tomography- computed tomography; PLGA, Poly(lactic-co-glycolic acid); RBD, Receptor Binding Domain; RSV, Respiratory syncytial virus; SAR, Structure-activity relationship; saRNA, Self-amplifying RNA; SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2; SAXS, Small-angle X-ray scattering; SC, Subcutaneous; siRNA, Small interfering RNA; TLR, Toll-like receptor; TH1, Type 1 T helper cell; UTR, Untranslated region
Graphical abstract
1. Introduction
Recent advances in biotechnology have revolutionized the field of medicine, offering pioneering solutions for clinically unmet needs. In search of the “silver bullet” – a universal solution to treat an illness in any patient regardless of their condition – the focus of the research community has shifted towards nucleic acids. Over the last three decades, in vitro transcription (IVT) has become a widespread technique to design and prepare messenger RNA (mRNA), a type of nucleic acid capable of encoding virtually any protein [1]. Successful implementation of mRNA therapeutics would prevent or treat any disease characterized by a deficit of one or several key proteins. The range of such diseases is expansive and includes cancers, as well as genetic and infectious diseases [[1], [2], [3], [4], [5], [6]]. Since 2019, the field's spotlight has turned towards the prevention of infectious diseases with mRNA vaccines due to the emergence of SARS-CoV-2.
mRNA vaccines have a unique feature of promoting the evanescent expression of antigen (typically days). The expression of the exogenous antigen is controlled by the lifetime of encoding mRNA, which is regulated by innate degradation pathways. While this transient nature of protein expression requires repeated administration for the treatment of genetic diseases and cancers, it is extremely beneficial for vaccines, where prime or prime-boost vaccination is sufficient to develop highly specific adaptive immunity without any exposure to the contagion. The triumph of mRNA vaccines, however, is held back by several factors. First, mRNA is a relatively fragile macromolecule susceptible to physical, chemical and enzymatic degradation [2]. This delicate nature of mRNA imposes a limitation on its storage and the administration of the treatment. Second, precise identification of antigen is crucial in altering the course of the disease. Inappropriate antigen sequences may cause undesired activation of the immune system [3]. Last, naked IVT mRNA has limited capability to pass the physiological barriers of the body and enter the target cells. As a result, the treatment with naked mRNA may require frequent administration and/or high doses, necessitating substantial investment in biopharmaceutical development and high patient compliance. However, these challenges may be circumvented by carefully designing the target mRNA sequences and the delivery vectors that would assist mRNA in entering the cells.
Self-assembly is an important concept in material sciences, which is widely explored in the design and development of nanomaterials [7]. The term ‘self-assembly’ refers to a spontaneous arrangement of individual molecules into a supramolecular assembly, driven by non-covalent interactions [8]. This approach is extremely attractive in the development of “smart” materials since the properties of individual building blocks may fine-tune the properties of the constructs, and the preparation may be done by simple mixing of the components. Additionally, self-assembly relies purely on the physical interactions between the building blocks and therefore can be applied to small molecules and biomacromolecules alike. The vast majority of the non-viral vectors for mRNA delivery are currently prepared through self-assembly, which includes a large fraction of mRNA vaccines.
In this review, we highlight the recent advances in the design of mRNA vaccines and the implementation of self-assembly in their preparation. We will cover several relevant topics – (1) design and properties of mRNA; (2) immunological activity of mRNA vaccines; (3) importance of the administration routes; (4) mechanisms of delivery to the cytoplasm; (5) design of the materials and their self-assembly; (6) desired physicochemical properties for optimal delivery; and (7) the clinical utility of mRNA vaccines. This interdisciplinary review may provide guidance to researchers seeking to answer fundamental questions in the field of mRNA vaccines.
2. mRNA vaccines
mRNA vaccines can be categorized into two types: (1) non-replicating and (2) self-amplifying (also known as self-replicating and replicon) mRNA vaccines. While both types of vaccines share a common structure in mRNA constructs, self-amplifying RNA vaccines contain additional sequences in the coding region for RNA replication.
2.1. Advantages of mRNA vaccines
The term ‘mRNA vaccine’ refers to the type of vaccines that delivers an antigen protein in the form of mRNA. Since the proof-of-concept report on mRNA vaccines in the 1990s [9,10], these vaccines have been thought to offer more advantages over the conventional ones: subunit, recombinant, live-attenuated, and inactivated vaccines [2].
The first important advantage of mRNA vaccines is the safety profile. In contrast to live-attenuated and inactivated vaccines, mRNA vaccines exclude any concern associated with endotoxin and infection. Unlike DNA vaccines and viral vector-based vaccines, mRNA vaccines do not pose the risk of genomic integration and insertional mutagenesis because they do not require nuclear entry for their activity [1]. The transient nature of mRNA activity is advantageous to avoid an incessant expression of the respective protein, yielding better temporal control of their activity.
The second benefit of the mRNA vaccine is efficacy. Since mRNA vaccines deliver essential antigens, they promote the development of adaptive immunity more specific to the antigen and minimize adverse effects compared to whole-cell vaccines. In addition, the pharmacology of mRNA vaccines offers the development of cellular immune responses and antibody responses, which can be useful to treat diseases that require cell-mediated immunity, such as cancer.
The third advantage of mRNA vaccines lies in their production. The synthesis of clinical-grade mRNA is based on well-standardized in-vitro-transcription (IVT) processes. The manufacturing process uses a DNA template and various enzymes in a cell-free system. This process is relatively robust and scalable when compared with the culture-based production of DNA- or protein-based vaccines. Additionally, less optimization is required for synthesizing new antigen sequences of similar size. This flexibility in mRNA manufacturing is exceptionally beneficial to produce mRNA vaccines against rapidly spreading infectious agents. One exemplar of the rapid mRNA vaccine development is the case in which the National Institute of Allergy and Infectious Diseases of the United States and Moderna produced clinical-grade mRNA vaccines for SARS-CoV-2 within 27 days after the sequence of the virus spike protein was released [11,12]. It enabled the human clinical trials phase I, II, and III to be initiated at unprecedented speed: within 66 days, 140 days, and 199 days after the viral sequence was determined, respectively (NCT04283461, NCT04405076, NCT04470427), resulting in the emergency use authorization (EUA) of their mRNA vaccine in the United States within a year [13].
While promising, there is still room for improvement in the realm of mRNA vaccines. Anionic macromolecules such as mRNA struggle to enter the cells because of the electrostatic repulsion from the cell membrane, and mRNA is prone to degradation due to abundant ribonucleases (RNases) in the extracellular space [2]. Additionally, extracellular mRNA endogenously acts as a ligand to pattern recognition receptors, such as toll-like receptors (TLRs) [14,15] and RIG-I-like receptors (RLRs) [16,17], which can cause adverse effects. In these regards, IVT mRNA is typically packaged in delivery vectors that can mask undesirable side-effects and facilitate cellular entry despite several reports of naked mRNA being effective in vivo [18,19] and ex vivo [20]. Geall and colleagues demonstrated that packaging mRNA in lipid-based nanoparticles (LNPs) protects mRNA cargos from RNases [21]. They also showed that vaccination of 0.1 μg mRNA encapsulated in LNPs enhanced in vivo transfection 10-fold greater than vaccination of 1.0 μg naked mRNA, indicating the benefits of using delivery vectors for mRNA delivery [21].
2.2. Structural elements of mRNA
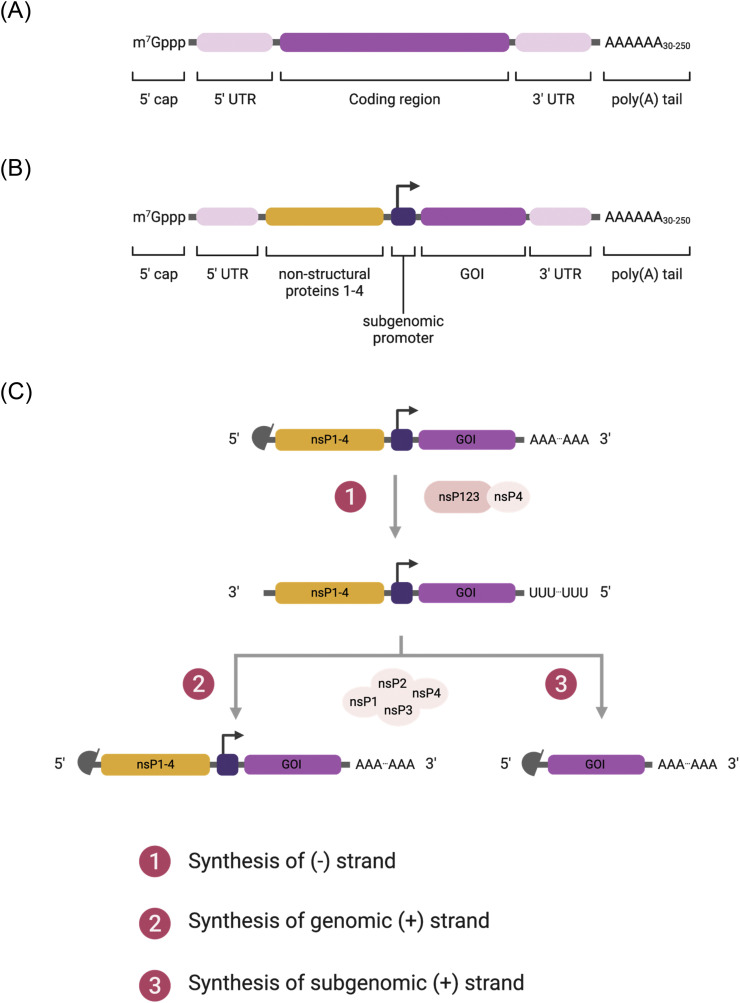
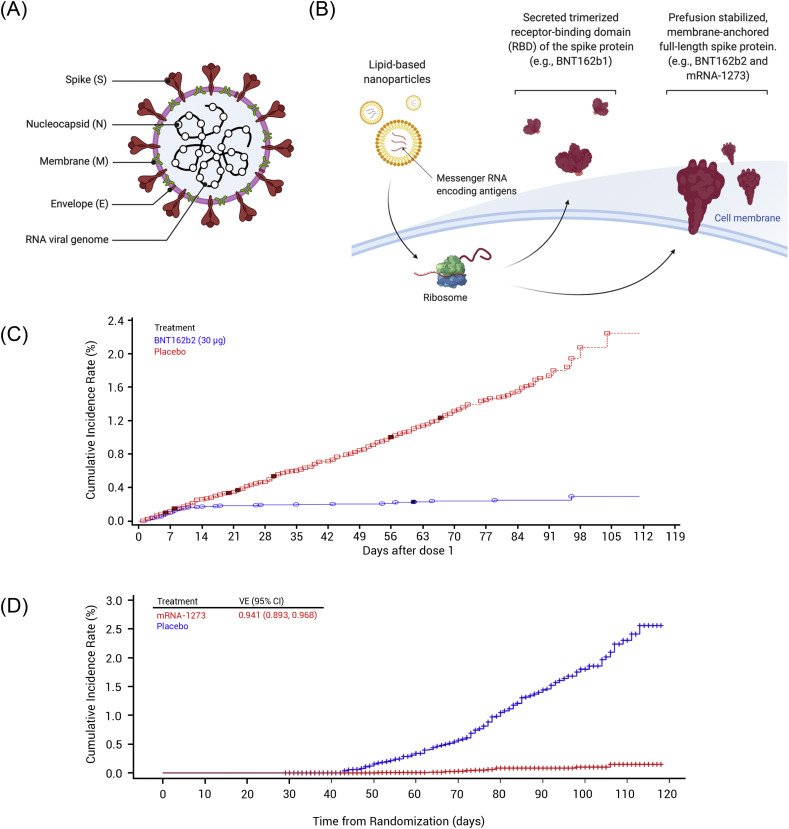
The structural elements of IVT mRNA are similar to those of natural mRNA, consisting of a 5′ cap, 5′ untranslated region (UTR), coding region, 3′ UTR, and a poly(A) tail (Fig. 1A). In this section, we will discuss the structural components of IVT mRNA and recent advances in RNA technology.
Fig. 1.
(A, B) Schematic diagrams of in-vitro-transcribed mRNA: (A) non-replicating mRNA and (B) self-amplifying RNA. (C) Replication of self-amplifying RNA inside cells. (1) After translation, non-structural proteins 1-4 (nsP1-4) are produced, forming the early replication complex that synthesizes (-) strand of mRNA transcript. (2, 3) The late replication complex produces (2) genomic (+) strand and (3) subgenomic (+) strand. Genomic transcript continues further replications and subgenomic transcript initiates antigen production.
2.2.1. 5′ cap
Natural mRNA in the eukaryotic cytoplasm contains the 7-methylguanosine (m7G) moiety at the 5′ end, followed by a triphosphate linkage to the first nucleotide (m7GpppN). This protective structure is called a 5′ cap and it regulates pre-mRNA splicing and nuclear export, protects RNA from exonuclease cleavage, and initiates mRNA translation [22]. The 5′ cap is also involved in the recognition of non-self RNA from self RNA by the innate immune system [23]. The m7GpppN is known as a cap 0 structure, which is an essential invariant for efficient translation of mRNA in eukaryotes. Early IVT methods to produce the cap 0 mRNA resulted in the inclusion of the cap analog either in the correct or reverse direction. Namely, when m7GpppG cap analog was used, the RNA polymerase produced m7GpppGN or Gpppm7GN. The development of an "anti-reverse" cap analog (ARCA, m2 7,3'-OGpppG) solved this directional issue [24]. Additional methylation of the 3′-OH in m7G prevents the ARCA from being incorporated in the reverse direction. However, co-transcriptional reactions with ARCA requires an excess of ARCA : GTP, which results in producing approximately 20% of the uncapped mRNA post-transcription [25]. Moreover, the use of ARCA introduces an additional methyl group to the mRNA, which is not present in native mRNA. An alternative for synthesizing the cap 0 mRNA is via a post-transcriptional enzymatic reaction using the Vaccinia virus capping complex.
While IVT mRNA with the cap 0 structure is sufficient to initiate mRNA translation, in mammalian cells, the mRNA undergoes further post-transcriptional modifications like 2′-O-methylation at position 2’ of ribose ring of the first nucleotide (cap 1, m7GpppN1m), and sometimes of the second nucleotide (cap 2, m7GpppN1mN2m) as well. The 2′-O-methylation in mRNA not only enhances the translation efficiency of IVT mRNA [26], but also prevents activation of endosomal and cytosolic sensors, such as RIG-I and MDA5, which are defensive mechanisms against viral RNA [23]. Therefore, 2′-O-methylation is highly desirable to improve protein production from IVT mRNA transfection and prevent undesirable immune responses. 2′-O-methylation can be achieved by using 2'-O-methyltransferase, an enzyme that catalyzes the transfer of a methyl group from S-adenosyl methionine to the IVT mRNA with cap 0 structure, yielding S-adenosyl-L-homocysteine and the IVT mRNA with cap 1 structure.
Another way to produce the cap 1 mRNA is by using trinucleotide cap analogs in co-transcriptional reactions. Ishikawa et al. used m7GpppAG analogs to cap IVT mRNA as the use of these analogs in the IVT reaction allowed mRNA to have the m7G moiety at 5′ end with no reverse capped products [27]. Furthermore, they produced various IVT mRNA with the first transcribed nucleotide as A, Am, m6A, or m6Am, and reported that m7Gpppm6AmG cap resulted in the highest luciferase expression in in vitro transfection experiments. Recently, Sikorski et al. further compared the effects of the first transcribed nucleotide in IVT mRNA [28]. They varied the first nucleotide among A, m6A, G, C, and U with or without 2′-O-methylation. They found that lipofectamine-delivered mRNA carrying A, Am, m6Am as the first nucleotide produced higher luciferase expression whereas the mRNA carrying G or Gm produced lower luciferase expression. Notably, the mRNA translation in the JAWSII cells, a murine dendritic cell (DC) line, was significantly affected by various caps, showing an 8-fold difference between m6A and m6Am. These findings may suggest the importance of the first nucleotide and the 5′ cap for mRNA vaccines targeting DCs.
2.2.2. 5′ and 3′ UTRs
UTRs are not translated into protein but are involved in regulating mRNA expression. 5′ and 3′ UTRs are located upstream and downstream of the coding region, respectively. They contain various regulatory sequences associated with the stability of mRNA, recognition of mRNA by the ribosomes, interaction with the components in translational machinery, and mRNA secondary structures [29]. In mRNA therapy, the inclusion of cis-regulatory sequences in the UTRs can improve the translation and half-life of mRNA. Many UTR sequences that promote mRNA translation have been identified from naturally occurring sequences. For instance, the UTR sequences derived from alpha- and beta-globins have been widely used to design IVT mRNA constructs [[30], [31], [32]].
Rather than searching the endogenous genes, UTR sequences can be designed de novo. Trepotec et al. designed a 14-nt-long 5′ UTR sequence that produces a comparable level of expression compared to human alpha-globin 5′ UTR [33]. Zeng et al. designed de novo 5′ UTR sequences based on the length and guanine-cytosine content as well as predicted resistance of resulting mRNA to miRNA for the development of mRNA vaccines [34]. Recent advances in big data and machine learning have enabled predicting and designing regulatory sequences in silico [35]. For example, Seelig and colleagues used deep learning to build a predictive model of the relationship between human 5′ UTR sequences to mRNA translation based on 280,000 random UTRs [36]. Their predictive model could explain 93% of the variability in the mean ribosome loads observed in the held-out data. These machine learning models may assist in the design of novel UTR sequences for mRNA therapeutics.
2.2.3. Poly(A) tail
mRNA holds a polyadenylated region at its 3′ end which is called as poly(A) tail. It is an essential determinant of the lifespan of mRNA molecules. Poly(A) tails of the natural mRNA molecules in mammalian cells have a length of ~ 250 nucleotides (nt), and they gradually shorten from 3′ to 5′ throughout the lifespan of mRNA in the cytosol [37]. Since the tail size affects the decay of mRNA by regulating 3′ exonucleolytic degradation, the incorporation of poly(A) tails at approximately 100 nt is desirable in the production of mRNA therapeutics. There are mainly two methods in the polyadenylation of IVT mRNA: (1) the enzymatic addition to the capped mRNA using recombinant poly(A) polymerases, and (2) poly(T) sequence embedded in the plasmid templates. Although poly(A) polymerase-mediated tailing works well for small-scale production, this method is limited by inconsistent length of poly(A) tail and it adds an extra step in synthesis, which is disadvantageous to large-scale production. On the other hand, embedding poly(T) sequences in the plasmid template enables one-step synthesis of poly(A) tails during the IVT reaction since the template for tailing is included in the plasmid DNA. Furthermore, it generates poly(A) tails with a definite length, which augments the reproducibility of mRNA therapy. A difficulty associated with this method is that the circular plasmids containing poly(A:T) regions tend to be unstable to clone and prone to shortening during the DNA propagation in E. coli [38]. To overcome the difficulties in cloning, Arbuthnot et al. proposed the repeated restriction digestion with type IIS enzymes, ligation, and propagation to extend the homopolymeric sequence up to 100 bp in circular plasmids [39]. Similarly, Grier et al. proposed linear plasmids as a template for mRNA synthesis to circumvent the limitations [40].
2.2.4. Modified nucleotides
Natural RNA molecules are synthesized based on four basic nucleotides (ATP, CTP, GTP, and UTP). During mRNA maturation, some nucleotides in mRNA are post-transcriptionally modified. These naturally-occurring modified nucleotides, such as pseudouridine and 5-methylcytidine, can be used in the synthesis of IVT mRNA [41]. Use of these modified nucleotides can avoid the recognition of IVT mRNA by the innate immune system, avoiding undesirable immune responses [15,42]. Karikó and coworkers pioneered modified nucleotides for producing IVT mRNA, reporting that substitution of uridine to pseudouridine in IVT mRNA not only reduces the innate immune responses against mRNA, such as TLRs [15] and Protein kinase R [42], but also increases protein translation [43]. 5-methylcytidine, 2-thiouridine, and N 1-methylpseudouridine were explored as alternatives to the basic nucleotides, and combinations of the modified nucleotides showed superior efficacy compared to the unmodified counterparts [[44], [45], [46]]. However, the counterexamples have been reported as well. Thess et al. showed that unmodified mRNA showed higher protein production and mild cytokine induction when compared to pseudouridine-substituted mRNA in mice [47]. Kauffman et al. demonstrated that pseudouridine substitution of mRNA did not change the in vivo protein expression and the levels of various cytokines as compared to unmodified mRNA when administered intravenously via LNPs [48]. More recently, Vaidyanathan et al. reported a comparative study about the influence of modified nucleotides on Cas9 mRNA activity, interleukins, and TNF-α levels [49]. They did not find a correlation between the incorporation of modified nucleotides and Cas9 mRNA activity. Although the modified nucleotides moderately decreased immune responses compared to unmodified nucleotides, the levels of cytokines were more significantly affected by HPLC purification. Taken together, the effects of modified nucleotides seem inconsistent and may depend on the experimental settings, such as cell type, administration route, and delivery vector. Nonetheless, considering that there are at least 143 natural nucleotide modifications, it is hard to argue that modified nucleotides are important determinants in regulating the RNA activity in the cells [50]. At the same time, more evidence suggests that aberrant presence or absence of RNA modifications could cause diseases in humans, which underpins the need for a thorough evaluation of the benefits of modified nucleotides [51].
2.3. Self-amplifying RNA vaccines
Self-amplifying RNA (saRNA) vaccines utilize RNA-dependent RNA polymerases (RDRP) derived from RNA viruses, mostly alphaviruses, to amplify the delivered RNA, thus increasing the production of antigen proteins. In addition to the normal components of mRNA, the saRNA contains large open reading frames (ORF) that encode the components for RDRP, consisting of non-structural proteins 1-4 (nsP1-4), and gene of interest (antigen sequence) following a subgenomic promoter (Fig. 1B). The nsP1, 2, 3, and 4 encode the proteins responsible for mRNA capping, NTPase/Helicase/protease, macrodomain, and RDRP, respectively. A recent review by Abu Bakar and Ng summarizes the details of each component [52].
Once saRNA is delivered to the cytosol, it goes through the endogenous translation machinery in ribosomes. The translation of the nsP series leads to the production of precursor polyproteins, forming the early replication complex that synthesizes the negative strand of RNA. Further cleavages of polyproteins yield the late replication complex that synthesizes the positive strand of genomic and subgenomic RNA by using the negative strand as a template. As a result, one copy of the saRNA produces multiple copies of RNA transcripts (Fig. 1C), overcoming the limited cytosolic delivery of in vivo mRNA therapeutics.
Antigen expression of saRNA increases gradually initially and lasts for an extended period of time [53], which prolongs the stimulation of antigen-presenting cells (APCs). In 2012, Geall and coworkers demonstrated that LNP-delivered saRNA elicited adaptive immune responses against the respiratory syncytial virus (RSV) after intramuscular injection to mice [21]. The administration of the non-viral saRNA vaccine led to potent cellular and humoral responses comparable to the viral counterpart. Further preclinical studies demonstrated the potentials of saRNA vaccines against influenza [54], HIV [55], and Zika virus [56]. Recently, Shattock and colleagues demonstrated the efficacy of saRNA-LNP vaccines against SARS-CoV-2 [57], which is currently in phase I clinical trial in the UK (ISRCTN17072692).
One shortcoming associated with the saRNA vaccines is the large and complicated saRNA sequence. The length of the nsP1-4 sequence is around 7 kilobases, which often makes the full length of saRNA vaccines more than 9 kilobases. Recent efforts to reduce the size of saRNA constructs include splitting the sequences of the nsP series and gene of interest in separate RNA molecules, referred to as a trans-amplification or trans-replicating system [58]. This trans-replicating system can be favorable due to its potential advantages in terms of versatility, safety, and ease of large-scale production. Beissert et al. compared the translation efficiencies of self-amplifying and trans-amplifying RNA vaccines and found that mRNA translation of the trans-amplifying RNA vaccine was as efficient as the saRNA vaccine, warranting further exploration of trans-amplifying RNA vaccines [59]. Despite the relatively compact size of trans-amplifying RNA systems, the mRNA length of the replicase is still longer than 7 kilobases, which demands delivery vectors with a large loading capacity for successful RNA delivery.
3. Mechanism of action of mRNA vaccines
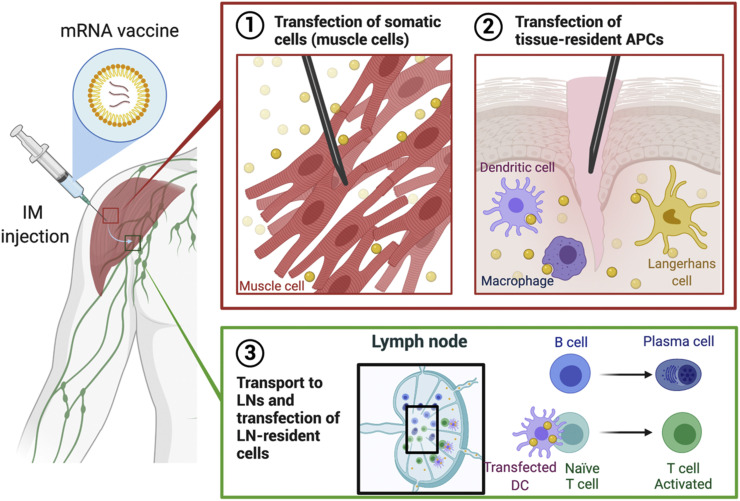
mRNA vaccination leads to adaptive immunity via several possible pathways: (1) transfection of somatic cells, such as muscle cells and epidermal cells, (2) transfection of tissue-resident immune cells at the injection sites, and (3) transfection of immune cells in the secondary lymphoid tissues, including lymph nodes (LNs) and the spleen (Fig. 2 ). mRNA vaccines administered by parental routes, such as intradermal, intramuscular, and subcutaneous injections, can transfect non-immune cells near the injection sites [60,61]. Non-immune cell transfection leads to the production of antigen that is subsequently degraded in the proteasomes. Following degradation, epitopes derived from the antigen can form a complex with major histocompatibility complex (MHC) class I for the antigen presentation to cytotoxic T cells expressing CD8, leading to the establishment of cellular immunity to the antigen. Transfection of myocytes is also known to activate bone-marrow-derived DCs, followed by CD8+ T cell priming [62]. Although the exact mechanism is unknown, it is thought that the antigen is transferred from the myocytes to DCs [62].
Fig. 2.
Modes of action of intramuscularly administered mRNA vaccines. (1, 2) mRNA vaccines can transfect (1) muscle cells as well as (2) tissue-resident APCs near the injection site. (3) mRNA vaccines can flow into proximal lymph nodes (LNs) and transfect LN-resident cells, resulting in activation of T and B cell development.
mRNA vaccines can also transfect tissue-resident immune cells, mostly APCs, such as DCs and macrophages [63]. Parenteral administration of mRNA vaccines can trigger local immune responses at injection sites [2,64]. This recruits immune cells to the area, facilitating the transfection of tissue-resident immune cells, such as DCs and macrophages [63,65]. As in the first case, mRNA transfection to these cells results in antigen presentation on MHC class I, which gives rise to the maturation of CD8+ T cells (Fig. 3 ) [2,66]. Moreover, the APCs can process the antigen through MHC class II pathway so mRNA transfection of the APCs can lead to the activation of T helper cells expressing CD4 (Fig. 3) [2]. Otherwise, the mRNA vaccine is drained by lymph and transported to the neighboring LNs by the lymphatic system [63,66]. LNs are the sites where various immune cells, including monocytes and naïve T and B cells, reside, and the antigens located in these secondary lymphoid organs initiate adaptive immune responses. In the LNs, mRNA vaccines transfect LN-resident cells, such as APCs and endothelial cells [63,66,67]. Transfection of these cells can initiate the priming of not only T cells but also B cells.
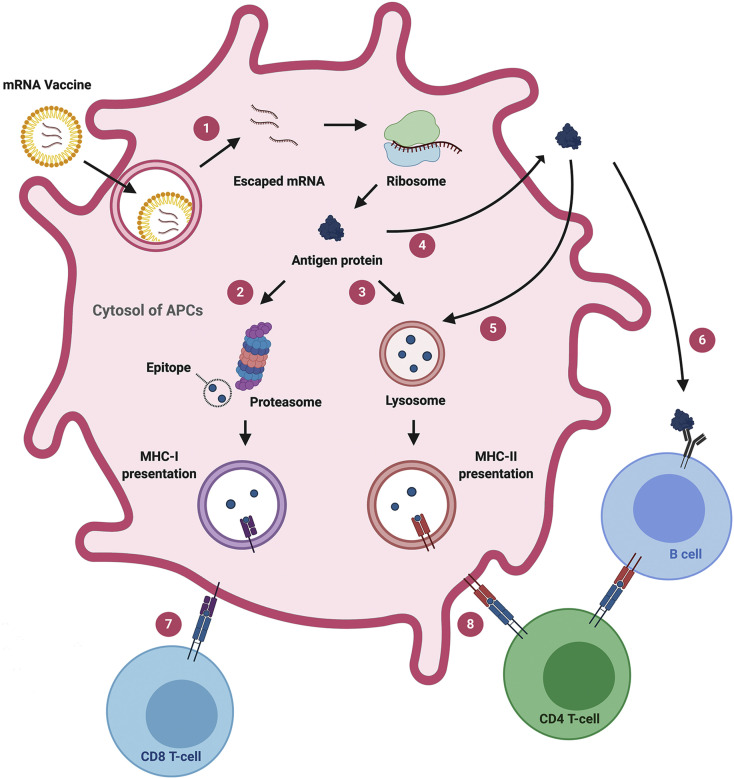
Fig. 3.
Mechanism of adaptive immune responses induced by mRNA vaccines. (1) Endosomal escape of mRNA to the cytosol after endocytosis-mediated internalization. (2) Antigen protein translated from exogenous mRNA is degraded into fragments in proteasome, followed by MHC-I presentation. (3) Antigen protein can undergo lysosomal degradation via various mechanisms, such as autophagy and signal peptide, followed by MHC-II presentation. (4) Antigen protein can be destined to extracellularly express in secreted or membrane-anchored form. (5) Extracellularly expressed antigen can be taken up again by APCs, directed to lysosomal degradation. (6) Instead, the extracellular antigen can be recognized by B cell receptor on B cells, leading to B cell maturation. (7) MHC-I presents the epitope to CD8+ T cells whereas (8) MHC-II presents the epitope to CD4+ T cells.
To elicit adaptive immunity against the antigen, mRNA vaccines must be translated to antigen by the endogenous translational machinery (Fig. 3). Because the translated antigen is originally located in the cytosol, it is preferentially considered an endogenous antigen by the cells. It means that endogenous antigens produced by mRNA vaccines are readily presented on MHC class I molecules, priming CD8+ T cells [2]. However, priming CD4+ T cells is also essential to build potent and robust cellular immunity [68]. Furthermore, considering that the roles of T helper cells in B cell priming is substantial [69], the ability of mRNA vaccines to prime CD4+ T cells is highly desirable to develop humoral immunity as well. As briefly mentioned earlier, mRNA vaccination leads to the activation of both CD8+ T cells and CD4+ T cells, suggesting that transfected APCs have alternative pathways to process endogenous antigens to MHC class II presentation [68]. One example of presenting endogenous antigens on MHC class II is via autophagy that is associated with lysosomal degradation of cytosolic antigens [70]. It is also known that the presentation of endogenous antigen on MHC class II can be mediated by heat shock protein 90 and transporter associated with antigen processing (TAP) complex as non-autophagic pathways [71]. Antigen presentation of endogenous antigen on MHC class II can be further potentiated by engineering mRNA sequence (Fig. 3). By encoding signal peptides that translocate proteins to the specific intracellular compartment, antigen presentation on MHC class II can be enhanced [72]. For example, Bonehill et al. inserted the targeting sequence of the invariant chain before the antigen sequence to program the translated protein to be presented on MHC class II [73,74]. Other signal peptides, such as MHC class I [75,76] and II [77,78], and lysosome-associated membrane protein-1 (CD107a) [68,79], can be incorporated in the mRNA coding region to prime antigen-specific CD4+ and CD8+ T cells. Alternatively, an mRNA sequence can be designed to express the antigens extracellularly (Fig. 3), either in secreted forms [11,80,81] or transmembrane form [11,81,82]. For instance, Richner et al. investigated mRNA vaccines against the Zika virus by encoding M-E proteins of the virus which generates virus-like subviral particles in mice [80]. Tiwari et al. engineered mRNA to encode glycosylphosphatidylinositol membrane anchor sequence from the decay-accelerating factor in order to express the protein of interest with anchored to the plasma membrane [83]. Corbett et al. designed the mRNA sequence to encode the prefusion-stabilized spike protein of SARS-CoV-2 as a transmembrane-anchored protein [11]. They also compared the efficacies of two mRNA vaccines against MERS - one encoding secreted form of the MERS spike protein and the other encoding transmembrane-anchored MERS spike protein - and showed that the latter elicited more potent pseudovirus neutralizing antibody responses. Nonetheless, it is difficult to generalize that transmembrane form is more efficacious than secreted form. Therefore, mRNA constructs for antigen expression - either secreted or membrane-anchored forms – can be tailored for individual virus strains to maximize the safety and efficacy of the vaccines. Extracellularly expressed antigens can be recognized by APCs and effectively elicit CD4+ and CD8+ T cell responses by MHC class II presentation and cross-presentation, respectively. Notably, the mRNA-derived extracellular antigens also help to develop antigen-specific humoral responses [84,85].
Antibody responses are a crucial mechanism of vaccines to neutralize foreign antigens. In the case of mRNA vaccines, proper activation of B cells is still pivotal to induce and maintain antibody production. B cell activation is initiated by encountering intact antigens in the extracellular space with the B cell receptor (BCR) (Fig. 3). Since naïve B cells reside in LNs, B cell activation requires the transport of antigens to the LNs for an encounter. Soluble antigens reach the draining LNs from the periphery; however, small proteins (less than 10 nm in size) tend to enter the blood capillaries because blood flow rates are 100-500 fold higher than lymph flow rates, limiting the antigen availability in the LNs [86]. APCs such as monocytes and migratory DCs carry the antigens to the LNs and provide them to naïve B cells; however, it is relatively inefficient compared to delivering antigens directly to the LNs [87]. Therefore, mRNA vaccines that can target the cells within LNs could improve antibody responses by increasing the local concentration of antigens in the LNs [87]. While soluble antigens in the LNs can be recognized by B cells directly, they can be captured by other cells for B cell activation, leading to more efficient B cell recognition of antigens [88]. Depending on whether the antigen is opsonized or not, subcapsular sinus macrophages or DCs can capture the antigens and present on their membranes for B cell activation [87,89].
The BCR-bound antigen is internalized by endocytosis, digested, and presented on MHC class II. Afterward, B cells can differentiate into short-lived plasma cells which rapidly secrete low-affinity antibodies or enter germinal centers (GCs) where they undergo somatic hypermutation and affinity-maturation. The activated B cells entering GCs present the antigen on MHC class II to T helper cells and receive co-stimulatory signals at the T cell/B cell borders. Subsequently, they undergo somatic hypermutation in the dark zone of the GCs, proliferating, and honing the specificity of BCR against the antigen. Then, GC B cells leave for the light zone where they undergo affinity maturation by interactions with follicular DCs. Follicular DCs that received the antigens from B cells and DCs store the antigen in their non-degradative compartments and present the antigens for long-term periods to B cells, helping their affinity maturation. A cyclic cascade of affinity maturation enables the selection of B cells having high-affinity BCRs and clearance of B cells having low-affinity BCR by apoptosis. The B cell populations with high BCR affinity exit the GCs as long-lived plasma cells or memory B cells [69,90]. The establishment of these two cells by vaccination is a key for producing antibodies with high affinity and securing long-term memory against the antigen. Upon secondary exposure to specific antigens, memory B cells reactivate the antibody production, enabling the rapid antibody-mediated immune responses [91]. Long-lived plasma cells mainly residing in the bone marrow provide long-lasting protective humoral immunity, for years and perhaps even decades [[91], [92], [93]]. Laczkó et al. recently demonstrated that a single dose of mRNA vaccines against SARS-CoV-2 generates antigen-specific memory B cells and long-lived plasma cells, supporting the capability of mRNA vaccines to develop long-lasting humoral immunity [94].
Consequently, ideal mRNA vaccines not only lead to the activation of CD8+ and CD4+ T cells, but also provide the antigens to the LNs for activating B cells and developing antibody responses against the pathogen. mRNA constructs can be engineered to elicit more potent T cell priming by encoding signal peptides or extracellular forms of antigens. Furthermore, considering the importance of GC reaction in the antibody responses, one way to design potent mRNA vaccines could be to target LNs with efficient delivery vectors [95].
4. Routes of administration
Administration routes for vaccination can have an impact on the efficacy of mRNA vaccines. The modality of vaccine administration affects the accessibility of immune cells for priming and the extent of consequential effects (local or systemic immune responses) after vaccination. Despite the evident influence of administration routes, the fundamental understanding of the impact of the administration route on the spatiotemporal distribution of mRNA vaccines remains limitedly understood.
4.1. Intramuscular, subcutaneous, and intradermal administration
Parenteral administration is the prevalent route for vaccination. The majority of vaccines are administered via invasive procedures, such as intramuscular (IM), subcutaneous (SC), and intradermal (ID) injections [96]. These administration routes require less training to perform and are considered safer from potential risk of infection than intravenous (IV) routes [97]. Skin and muscle tissues are attractive sites for vaccination due to the presence of large numbers of APCs, such as Langerhans cells and DCs, present near or in the tissues. Additionally, administration to these tissues provides the vaccines with entry to the lymphatic vessels, enhancing the vaccine delivery to the LNs. Therefore, mRNA vaccines administered by IM, SC, and ID injections can elicit strong immune responses by transfecting tissue-resident and LN-resident cells. Recently, Santangelo and colleagues visualized early trafficking of the mRNA vaccines labeled with dual radionuclide-NIR probes after IM injection to cynomolgus macaques using PET-CT and infrared imaging [63]. mRNA vaccines traveled to the LNs that drain from the injection sites; however, they rarely reached distal LNs. This result corroborates previous reports with protein-based vaccines [98,99]. Also, mRNA transfection was observed primarily in monocytes and to a less extent in DCs, B cells, and CD4+ T cells in the injected muscle tissues and the draining LNs, which is in accord with previous findings [65]. It suggests that mRNA vaccination may involve the monocyte transfection as a major mechanism, although it could be due to characteristics of the mRNA vector that was used in the study. Even though SC injection is considered more immunogenic than IM injection due to the proximity of the injection sites to the dermis that contains a large number of tissue-resident immune cells [100], multiple independent studies with protein-based vaccines showed no significant difference in antibody titers between IM and SC injections [99,101]. Nonetheless, injection methods for vaccination were shown to affect biodistribution [99] and GC reaction [101] of the vaccines in non-human primates. In line with the results of protein-based vaccines, a comparative study with mRNA vaccines in mice showed no significant difference in antibody production between IM and SC injections [102]. ID administration has been considered a promising route for vaccination because of the abundance of immune cells in the dermis; however, it is less frequently used than SC and IM injections due to the technical difficulty in administration. Nevertheless, it appears evident that mRNA vaccines administered by ID injection could result in more robust humoral responses [102]. Loré and coworkers showed that ID injection of mRNA vaccines encapsulated in LNPs led to the rapid and extended expression of B cell responses in non-human primates, suggesting that ID injection of mRNA vaccines is likely beneficial to provide rapid development of adaptive immunity to influenza virus [65,84].
4.2. Intravenous administration
Intravenous (IV) injection of mRNA therapeutics can lead to the highest production of the encoded protein in the body. Generally, mRNA therapeutics that are injected intravenously target the liver, efficiently transfecting hepatocytes, Kupffer cells, and endothelial cells [60]. However, in vaccination, IV injection is less favored because of the wide biodistribution of mRNA vaccines through the circulatory system, potentially leading to systemic side effects. Nonetheless, it is achievable to develop adaptive immune responses from IV-injected mRNA vaccines by targeting the spleen, another secondary lymphoid organ, as a site of transfection [103,104]. Kranz et al. demonstrated that the lipoplexes containing cancer antigen mRNA transfected the spleen after IV injection, leading to antitumor immunity in mice [105]. Of note, they showed that the surface charge of mRNA-lipoplexes altered the biodistribution and transfection, ranging from liver, spleen, to lungs. In addition, they demonstrated that IV injection of mRNA-lipoplex vaccines elicited more robust CD8+ T cell priming than SC injection. Clinical evaluation of this systemic mRNA approach is ongoing in the pursuit to treat melanoma and triple-negative breast cancer (NCT02410733 and NCT02316457). In spite of the encouraging results, further optimization is needed in IV administration of mRNA vaccines to enhance target-specific mRNA delivery and to minimize systemic side effects [106].
Alternatively, IV injection of mRNA therapeutics can be utilized to produce antibodies and proteins from the liver for neutralizing the target pathogen or cancer cells [[107], [108], [109]]. This strategy stems from the concept of mRNA-based protein replacement therapies which use the liver as a biological factory to produce immunologic proteins. Rybakova et al. engineered mRNA sequences to encode the heavy and light chains of Trastuzumab, a monoclonal antibody targeting the HER2 receptor on cancer cells and demonstrated that IV injection of the mRNA-LNP led to the in vivo production of Trastuzumab from the mouse liver [108]. We also reported that mRNA-LNP could be used to produce the soluble Angiotensin-Converting Enzyme 2 (ACE2) in the liver that neutralizes SARS-CoV-2 virus by binding to the spike proteins [109]. These results suggest that systemic administration of mRNA therapeutics provides an alternative method of monoclonal antibody-based immunotherapy.
4.3. Intranasal administration
Another approach for vaccination is mucosal vaccine delivery, targeting non-gastrointestinal mucosal lymphoid tissues and lymph nodes. mRNA vaccines administered to the mucosal layers, such as the nasal and pulmonary mucosa, travel to the draining mucosal LNs via lymphatic systems under the epithelium [86,110,111]. Particularly, prophylactic mRNA therapeutics to prevent respiratory diseases, such as influenza [112] and RSV [83], are preferred to be delivered via intranasal (IN) administration [113] because mucosal vaccination can offer pathogen-specific antibodies to be secreted to the mucus where the antibodies can neutralize the pathogens at the early stage of infection [114]. Additionally, mucosal delivery of mRNA vaccines can cause the secretion of immunoglobulin A (IgA). IgA typically exists in its dimeric form in the mucus and acts in the front line of defense against infection by blocking and neutralizing bacterial toxins and viruses [115]. For example, Li et al. reported that IN-administered PEI-based polymeric nanoparticles delivered HIV gp120 mRNA to nasal epithelium, resulting in the production of antigen-specific IgG and IgA in mice [113].
One aspect to consider for IN administration is the mucus penetration of delivery vectors. Mucus protects the epithelial cells by removing foreign materials, which also serves to get rid of the nanoparticles administered intranasally. Given that viscoelastic and adhesive mucus can block mRNA vaccines from transfecting cells, the development of mucus-penetrating vectors could benefit the effective mRNA vaccination after IN administration. It is known that the optimization of physicochemical properties of nanoparticles improves mucus penetration [116,117]. The strategies to make nanoparticles mucus-penetrating or muco-inert are reviewed in detail elsewhere [116,117]. Lastly, most evaluations of mRNA vaccines after IN administration are limited to rodent models, urging a further investigation of the advantages of IN administration in larger animals.
In summary, the selection of the administration route for mRNA vaccination can influence the safety and efficacy of mRNA vaccines. Hence, the appropriate route of administration for mRNA vaccination can be determined with consideration of disease and its pathogenesis. Moreover, understanding of biological characteristics in each delivery route will promote the rational design of delivery vectors for mRNA vaccines.
5. Intracellular delivery of mRNA
For efficacious vaccination, mRNA vaccines must reach the cytosol where they can produce antigens, triggering adaptive immunity against the antigens. However, as briefly mentioned before, the mRNA molecule is a large polyanion due to the presence of phosphate groups in its backbone, obstructing its direct entry to the cytosol. For this reason, nanoscale biomaterials are of a vast interest for the delivery of mRNA vaccines due to their capability to smuggle mRNA payload into the cell in a safe and efficient manner.
Cellular entry of the nanosized mRNA carriers is predominantly mediated by endocytosis, an interconnected network of trafficking pathways of the cell. Endocytosis of nanocarriers is extensively discussed in the field of nanomedicine [[118], [119], [120], [121]]. Endocytosis is initiated by dynamic non-covalent interactions between the plasma membrane and the nanocarrier in proximity to the cells. It is known that the physicochemical properties of nanocarriers, such as size, shape, surface charge, and surface composition, affect the defined pathways of endocytosis - macropinocytosis, caveolin-mediated, clathrin-mediated, clathrin, and caveolin-independent [122,123]. After endocytosis, internalized nanocarriers are transported to the early endosomes, followed by maturation to multi-vesicular late endosomes. The late endosomes further progress to lysosomes, where most nanocarriers are degraded by digestive enzymes. Otherwise, the internalized nanocarriers can be eliminated by recycling pathways during endo-lysosomal trafficking. Approximately 70% of the internalized LNPs encapsulating siRNA are cleared by exocytosis via recycling pathways, limiting cytosolic delivery of genetic cargos [124]. While recycling of mRNA-LNPs is likely similar based on their size and composition, the endocytic recycling of mRNA-loaded LNPs has not been reported to the date. Only a modicum of the internalized nanocarriers is retrieved from the recycling and degradation, and ultimately reach the cytosol. Numerically, Gilleron et al. demonstrated that only 2% of siRNA-loaded MC3 LNPs escape to the cytosol within a limited time window when they reside in the early- and late-endosomes [125]. Recently, Sabnis et al. reported that MC3- and lipid 5-based LNPs encapsulating mRNA exhibited 2.5% and 15% endosomal escape efficiencies, respectively [126]. Despite myriads of endeavors to decipher the mechanism of endosomal escape, mechanistic understanding remains limited.
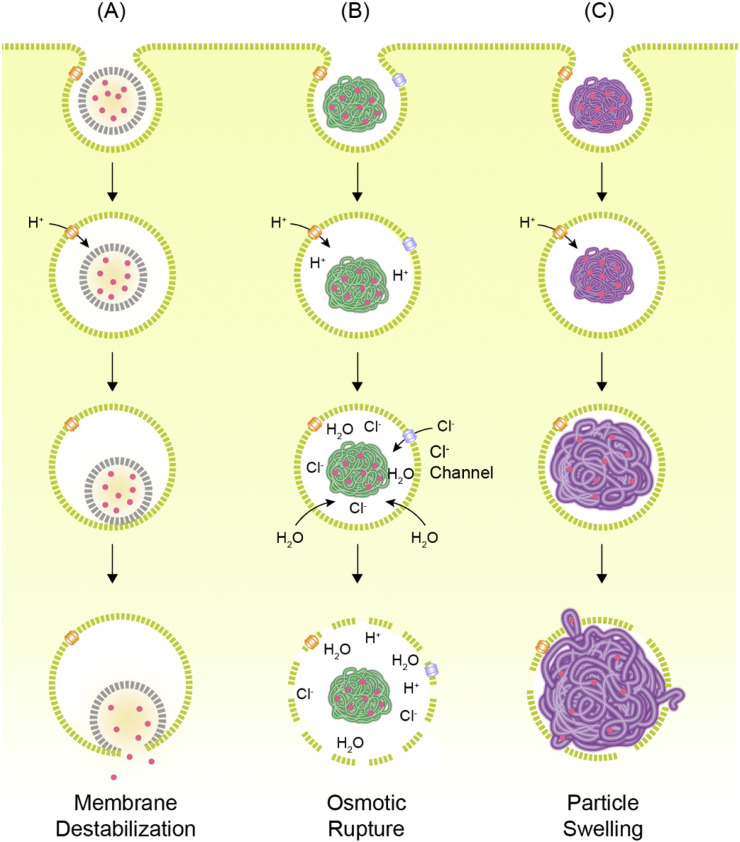
The most conspicuous characteristic of endo-lysosomal pathway is the gradient change of pH. For this reason, the primary approach to facilitate the endosomal escape of the nanocarriers has been to make them responsive to the acidification of endosomes. Among various strategies to induce the endosomal escape of the nanocarriers, three scenarios are broadly accepted: (1) destabilization of the endosomal membrane, (2) osmotic rupture of the endosomes by the “proton-sponge” effect, and (3) endosome rupture by particle swelling (Fig. 4 ). Generally speaking, lipid-based materials are considered to have a higher propensity for destabilizing the membrane, and polymeric materials are thought to disrupt the endosomes by osmotic or swelling-mediated ruptures [121]. Regardless of the material and the mechanism, the efficiency of endosomal escape remains extremely low, which calls for the implementation of novel methods to capture the intricacies of endosomal escape.
Fig. 4.
Hypothetical mechanisms of endosomal escape of nanocarriers. (A) Nanocarriers can induce destabilization of endosomal membrane for cytosolic release of genetic cargos. (B) Nanocarriers, particularly polyplexes, can scavenge protons and become cationic in acidic lumens of endosome compartments, resulting in the inflow of more protons and counter ions. This osmotic gradient induces influx of water to the endosomes, causing endosome rupture. (C) Nanocarriers swell in acidic pH due to the electrostatic repulsion and physically rupture the endosome. Reproduced from [121] with permission.
One way to pinpoint the endosome rupture is by tracking β-galactoside-binding lectins (galectins) that rapidly conglomerate around destabilized endocytic compartments [127]. Ruptured endosomes expose the luminal glycans to the cytosol, followed by binding of cytosolic galectins to the glycans. Recently, this phenomenon was exploited to indicate the occurrence of nanocarrier-mediated endosomal escape [128,129]. Reporter cell lines expressing the galectin-8 fused with fluorescent proteins exhibited the punctate fluorescent spots in response to various nanocarriers, visualizing their endosomal escape [128,130,131]. Our group also reported that endosomal escape of mRNA-loaded LNPs can be visualized as galectin recruitment in the reporter cells expressing galectin-8 fused with GFP and the galectin-8 recruitment was colocalized with Rab7 late-endosome marker, suggesting that mRNA-LNP escape from late endosomes to the cytosol [132]. Such tools can be extremely useful for high-throughput screening of a large number of nanocarriers, facilitating the discovery of potent vectors that are capable of triggering endosomal escape more efficiently.
6. Materials for mRNA delivery
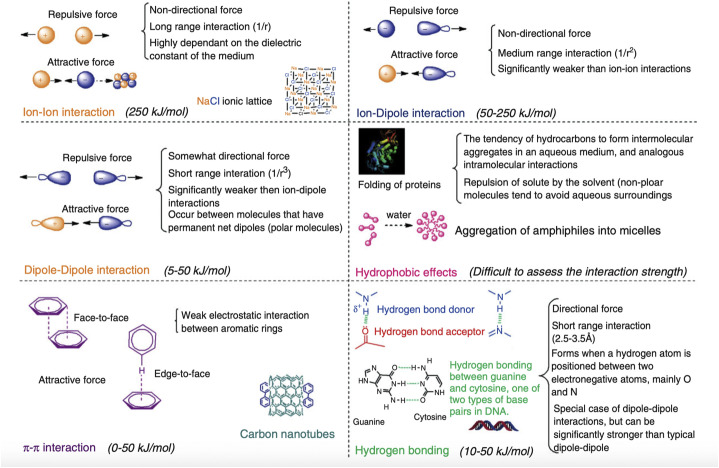
Self-assembly of “smart” materials is a highly sought-after approach in many areas of materials science - simply mix the components, and intermolecular interactions will assemble these components into the desired structure with the desired properties. This spontaneous organization is typically governed by non-covalent intermolecular interactions such as electrostatic, hydrophobic, van der Waals, and π-effects (Fig. 5 ) [[133], [134], [135]]. The use of self-assembly has found applications in numerous fields of nanoscience [7], including nanomedicine. In gene delivery, building blocks such as small molecules or polymers interact with nucleic acids and self-assemble into ordered structures.
Fig. 5.
Non-covalent interactions in supramolecular chemistry. Electrostatic interactions determine the encapsulation of mRNA and endosomal escape while hydrophobic interactions likely affect the formation and long-term stability of the delivery vector. The roles of other interactions in self-assembly of mRNA vaccines are not yet understood. Adapted with permission from [135].
Self-assembly has been primarily favored in the design of mRNA delivery vectors due to its versatility. However, the main challenge of self-assembly is determining the necessary properties of the building blocks that will translate into the desired properties of the self-assembled gene delivery vectors. These properties are (1) protection of nucleic acids from nucleases, (2) controlled release of nucleic acid, (3) cell and tissue selectivity, (4) high delivery yield, (5) minimal toxicity, and (6) stability, especially in long-term storage. Despite significant efforts devoted to the design of the gene delivery vehicles, the independent effect of building block properties on efficient cellular uptake remains mostly unclear [136].
To date, self-assembled nanoarchitectures loaded with mRNA yield higher delivery efficiency than naked mRNA vaccination and allow for a much broader variety of administration routes, expanding the therapeutic potential of mRNA nanomedicines [137]. Here, we will focus on the discussion of lipid-based materials due to their advances towards clinical development [12,[138], [139], [140]], and polymer-based materials due to their recent successes in delivering mRNA to the lung, an important yet hard-to-reach target. A reader interested in a more detailed discussion of polymer, peptide, and hybrid materials may direct their attention to seminal and recent reviews on the matter [[140], [141], [142], [143], [144], [145]].
6.1. Lipid-based systems
LNPs are currently the leading non-viral delivery vector for gene therapy [146]. As LNP-mediated siRNA therapeutic Onpattro® (patisiran) advanced towards clinical trials and subsequent approval [138], it was only natural that the mRNA delivery field adopted LNP technology. However, the knowledge acquired through siRNA-LNP development does not readily translate to mRNA-LNP delivery [147]. This observation prompted extensive research about the structure-activity relationship (SAR) of novel lipid materials tailored for efficient mRNA delivery [4,139,[148], [149], [150]]. While the question remains just what parameters must be customized precisely to maximize the delivery of a particular nucleic acid payload, it is clear that the structure of LNPs is different depending on the nucleic acid payload [147,151,152] and is determined by the initial stages of self-assembly [153,154].
The self-assembly of LNPs is driven by an ongoing competition between kinetics and thermodynamics, making them highly fluid and dynamic systems. Historically, lipid vesicles were commonly prepared through rehydration of lipid thin films and subsequent extrusion to achieve a homogenous size distribution [[155], [156], [157], [158]]; the formation of lipid vesicles, in this case, is likely driven by thermodynamics due to the poor solubility of lipids in aqueous media. As a result, these lipid vesicles yielded poor encapsulation of nucleic acid cargo [159]. Nowadays, LNPs are prepared through rapid mixing, often facilitated by microfluidic devices [153,154,160,161]. A particularly popular microfluidic preparation method is ethanol dilution, referring to the rapid condensation of lipids into nanodroplets when their ethanol solution is added to the excess of aqueous media [159]. The resulting LNPs may be considered a kinetic product and typically yield considerable encapsulation of nucleic acid.
Generally, LNPs are thought to be formed through a cascade of merging smaller lipid vesicles [153] similar to a classical LaMer model (nucleation - growth - ripening). The initial formation of the LNP seeds is directed by electrostatic interactions between anionic nucleic acid and positively charged lipids. In the case of siRNA-loaded LNPs, the seeds are proposed to have a liposome-like structure, where siRNA is sandwiched between the layers of charged ionizable lipids, and the growth results in neutral ionizable lipids filling up the core of the nanoparticle [153]. Kulkarni et al. showed, in a mechanistic study using a FRET-based fusion assay, that the LNP growth relies more on pH neutralization and migration of neutral, unbound ionizable lipid towards the LNP core regardless of the payload (mRNA, minicircular DNA, or pDNA) [162]. Based on their study, the internal structure of resulting LNPs, in general, may be lamellar or even “onion-like.” Indeed, the onion-like structures have been observed for mRNA-LNPs, although there is no indication that mRNA is captured between lipid layers [[163], [164], [165]]. On the other hand, mRNA-LNPs might form an inverted-hexagonal inner structure [166]. Yanez Arteta et al. showed that mixtures of ionizable lipid, cholesterol, and firefly luciferase mRNA form different structures depending on the solvent environment, assembling into inverted lipid tubules in conditions similar to those of a traditional ethanol dilution method [166]. This observation would indicate that LNP growth might start from inverted micelle-like seed structures, and the rest of the lipids aggregates to those purely based on hydrophobic interactions. Regardless of the seed structure, the hydrophobic interactions appear to be the dominant driving force of the LNP growth, and electrostatic interactions guide the seed formation and the stability of the final assembly. The differences in the seed and final structures may arise from the size of the payload (e.g., siRNA is 20-30 nucleotides vs mRNA is ca. 1000 or more nucleotides) or their flexibility (siRNA is a rigid duplex vs mRNA is a flexible single strand). Additionally, various mRNA sequences may form local secondary structures depending on the nucleotide sequence [167,168], which might interfere with RNA-lipid complexation. To the date, the effect of the nature of the nucleic acid payload on the self-assembly of LNPs and the factors for consequential optimization of lipid formulations to accommodate mRNA remain elusive and require further investigation.
Interestingly, the nucleic acid does not have to be embedded in the core of the LNP to be protected from nuclease degradation [169]. Previously, Brito et al. reported that cationic nanoemulsions could protect saRNA without losing functionality when saRNA was added to a preformed nanoemulsion [55]. The cryoEM micrographs of these nanoemulsions indicated the formation of homogenous structures of various sizes, which may indicate either that RNA is located inside the droplets or wrapped around the charged surface. This finding might suggest that any mechanism of self-assembly resulting in the formation of mRNA lipoplexes protects against RNase degradation, possibly by selectively interfering with RNase recognition machinery without affecting translation.
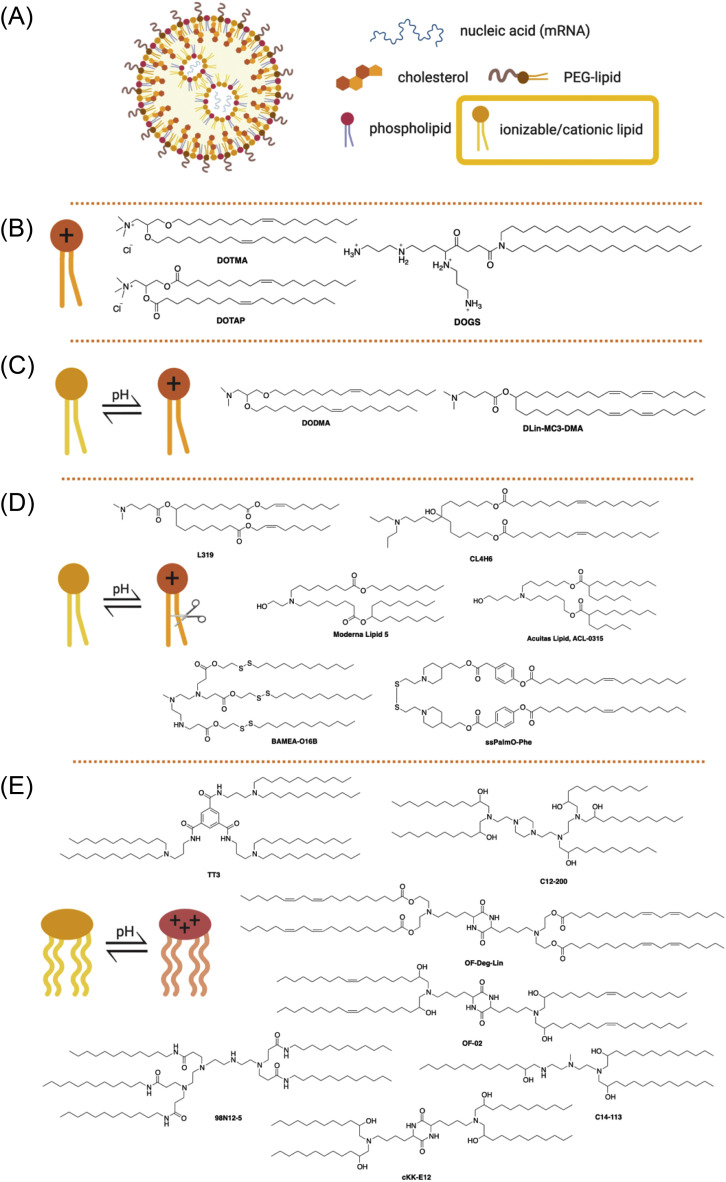
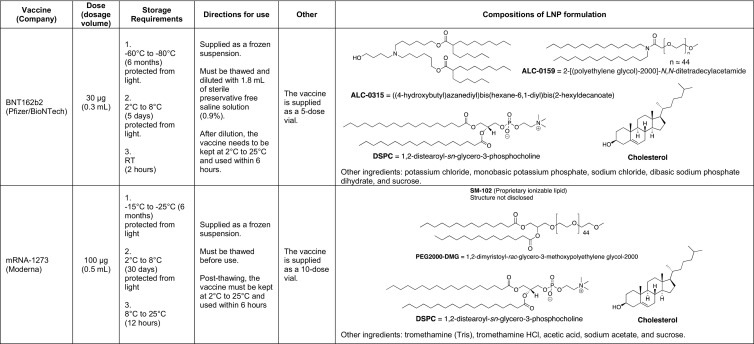
LNPs are arguably the most sophisticated mRNA delivery to the date, highly variable in structure and quantities of comprising lipids. The current clinical standard of LNPs (for siRNA delivery) – the first clinically approved formulation containing DLin-MC3-DMA, DSPC, cholesterol, and DMG-PEG-2000 [138] – comprises of nearly 60,000 lipid molecules per particle (Fig. 6A). Each lipid in the formulation serves a separate function, which will be discussed below. The chemical structure of lipids defines their molecular geometry, which is crucial in lipid packing, formation of nanostructures, and biological activity. As lipid-based materials for mRNA delivery have rapidly evolved in the last decades, the complexity of their structures along with considerations of synthetic limitations for vaccine manufacturing warrants a detailed discussion.
Fig. 6.
(A) General structure of lipid nanoparticles. Ionizable or cationic lipids (yellow box) are the main component responsible for the encapsulation of nucleic acid and intracellular delivery. These lipids may be divided into groups shown below. (B) Structures of first-generation cationic lipids DOTMA, DOTAP, and DOGS; (C) Structures of ionizable lipids DODMA and DLin-MC3-DMA; (D) Structures of selected ester-based and disulfide-based biodegradable ionizable lipids; (E) Structures of selected ionizable lipidoids.
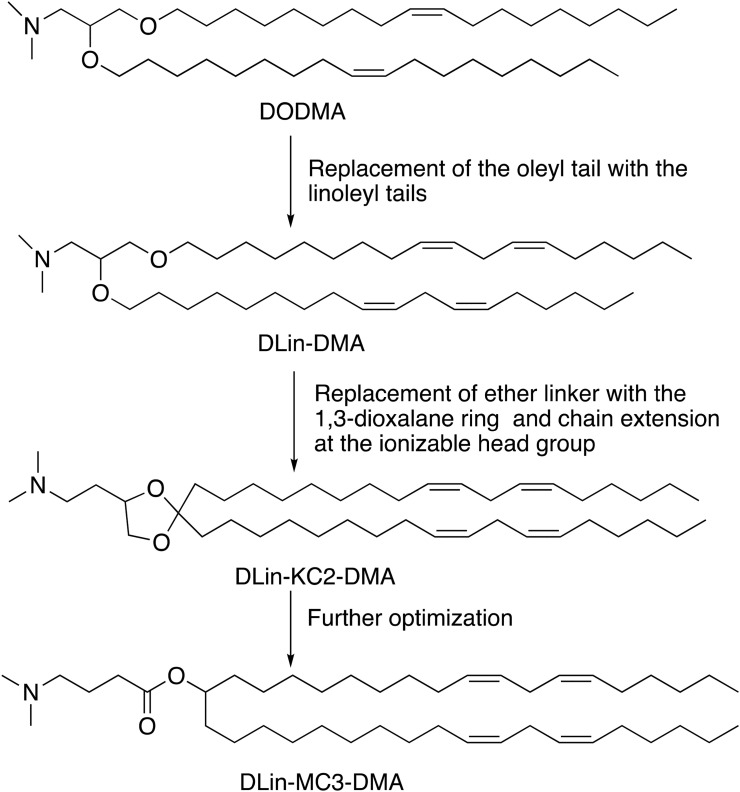
Cationic lipids are the first generation of lipids developed for mRNA delivery. They contain either a quaternary nitrogen atom giving them a permanently positive charge or a primary amine imparting a positive charge at or below physiological pH. The positive charge enables these lipids to form ionic interactions with the negatively charged mRNA forming a lipid complex, often referred to as lipoplex [170]. Examples of cationic lipids include DOTMA [171], DOTAP [172], and DOGS [173] (Fig. 6B). Early attempts of gene delivery used formulations comprised entirely of cationic lipids. While the early cationic lipids showed promising gene delivery in vitro, they suffered from limited in vivo efficacy and cytotoxicity issues arising potentially due to their permanent charge and non-biodegradable properties [3]. The cytotoxicity could be reduced by the incorporation of structural lipids such as DOPE. Lipofectin, a mixture of DOTMA and DOPE, is one of the first LNPs formulation that has been proved successful in the in vivo translation of mRNA [60]. Kranz et al. used Lipofectin RNA-lipoplex to target the DCs for cancer immunotherapy in mice [105], and in humans for the treatment of advanced melanoma (NCT02410733). Ionizable lipids (ILs), the second generation of cationic lipids, also referred to as pH-dependent lipids, have virtually no charge at the physiological pH but become positively charged at acidic pH. The pH-sensitive properties of ILs prevent their metabolism by the reticuloendothelial system (RES), thereby improving their half-life [174]. ILs are also less likely to cause immune activation or interactions with serum proteins, improving their safety profile [3]. DODMA was one of the first ILs evaluated for gene delivery (Fig. 6C and 7 ) [175]. Replacing the oleyl tails of DODMA with linoleyl tails led to DLinDMA [175,176] which elicited broad, potent, and protective immunity against the respiratory syncytial virus (RSV) in vivo [21]. Systematic SAR on the DLinDMA structure led to the identification of DLin-KC2-DMA, which was further optimized leading to the discovery of DLin-MC3-DMA, a key component of the first siRNA drug Onpattro® (Fig. 6C and 7) [[177], [178], [179], [180]]. MC3 was consequently shown to be effective in delivering mRNA as well [166,[181], [182], [183], [184], [185], [186], [187], [188]]. MC3 has a long plasma half-life of 72 hours, which limits its chronic administration [126,189]. Therefore, the next generation of lipids employed biodegradable functional groups that can facilitate clearance. One way to add biodegradability is the inclusion of ester moieties. The ester motifs are easy to incorporate, chemically stable, bio-cleavable, and their biodegradation kinetics can be effectively controlled [[189], [190], [191]]. Some examples of ester-based biodegradable lipids include L319 [179,189], YSK12-C4 [192], CL4H6 [193], and the Moderna proprietary lipid series [126,194], and Acuitas proprietary lipid series [195] (Fig. 6D). These biodegradable ester-based ILs often were found to be more potent in gene delivery as compared to MC3. For example, L319 exhibited better gene silencing when compared to MC3 in mice [189]. Similarly, Moderna lipid 5 was found to be 3-fold more potent [126] and Acuitas lipid, ACL-0315 (the IL used for Pfizer/BioNTech COVID-19 vaccine) was approximately 6-fold more potent than MC3 in delivering firefly luciferase mRNA to animals [195]. An alternate way of introducing biodegradability involves the addition of disulfide bonds into the backbone. Disulfide bonds are bio-reduced by glutathione (GSH) or other disulfide-reductases present in the cell [196]. Examples of ILs containing bio-reducible disulfide linkage includes BAMEA-O16B [197] and ssPalmO-Phe lipid (Fig. 6D) [198].
Fig. 7.
Rational development of Dlin-MC3-DMA (MC3) from DODMA.
Hybrid molecules derived from ionizable lipids and dendrimers are referred to as lipidoids. These third-generation cationic lipids are usually synthesized by straightforward chemistry, involving a limited number of steps, thus enabling a high-throughput turn around [199]. Akinc et al. reported the first example of lipidoid 98N12-5 [200], which was further modified, leading to the discovery of superior analogs C12-200 [201] and C14-113 (showed selectivity towards the heart) [201,202]. In subsequent studies, Dong et al. discovered cKK-E12 as a potent siRNA delivery agent [203]. cKK-E12 LNPs have been utilized for various therapeutic purposes such as cancer immunotherapy [66,108] and genome editing [204]. Modifications around the lipid tail of cKK-E12 resulted in lipids OF-02 [205] and OF-Deg-Lin. The latter was surprisingly selective towards the spleen (about 85%) and able to transfect B cells, highlighting that small structural variations can lead to tissue selectivity [206]. In another study, Li et al. reported TT3 as a potent delivery agent for various mRNA molecules encoding Factor IX [207], CRISPR/Cas9 [208], and more recently viral antigens against SARS-CoV-2 [34]. The structures of the lipidoids are enlisted in Fig. 6E.
LNPs typically also contain structural lipids (usually two – a phospholipid and cholesterol) and a PEG-lipid (Fig. 6A). The primary purpose of these lipids is to provide particle stability, control over size, and blood compatibility [209]; however, their quantities in the formulation and chemical properties may need adjustments for efficient mRNA delivery from the standard MC3 formulation [207,210]. The adjustments may also be warranted depending on the route of administration. For example, we investigated whether replacing the phospholipid DSPC with naturally-occurring glycerol derivatives could change mRNA delivery efficiency and found that introducing DGTS in place of DSPC improves delivery to the liver via IV injection but diminishes the efficacy of nebulized LNPs for IN delivery of mRNA [188]. Although it is not yet clear how to harness the power of these changes, structural lipids are likely located at the periphery of mRNA-LNPs [166], and it is a known fact that lateral organization and heterogeneity of lipid membranes play an essential role in cellular functions [211]. Specifically, the molecular geometry may change rigidity, curvature strain, and deformability of LNP lipid membranes [[212], [213], [214], [215]]. For example, Patel et al. explored the impact of varying the structure of cholesterol derivatives in LNPs on cellular uptake and trafficking and found up to 25-fold improvement in delivery efficiency [216], which may have been caused by morphological changes in mRNA-LNP internal and external structure [163,216]. Additionally, simulations of mRNA-LNP endosomal escape suggest that lateral diffusion and the tail protrusion of the peripheral lipids into the endosomal membrane may be key types of lipid dynamics responsible for achieving endosomal fusion [165]. The outermost lipid, PEG-lipid, might have other corrections to the properties of the LNP membrane.
PEG-lipid, typically the least abundant lipid in the LNP formulation, affects the properties of LNPs in several ways. First, the amount of incorporated PEG-lipid dictates the particle size [217], although excessive PEGylation hinders transfection [218]. Second, PEG-lipids prevent destabilization and aggregation [217,219]. Third, PEG-lipids prevent rapid uptake of LNPs, and reduce opsonization by serum proteins and reticuloendothelial clearance, improving LNP circulation lifetime [220]. Finally, the hydrophilic coating of PEG may be beneficial in navigating viscous media such as lung mucus [116,221]. The extent of these properties depends on the chemical structure of PEG-lipid, which contains a hydrophilic part (PEG) and a hydrophobic part (lipid anchor). Both may affect the LNP size, lipid membrane permeability, and immune response [66,222,223]. Unlike other components of LNP, PEG-lipid is designed to eventually disassociate and shed the PEG, controlling the kinetics of cellular uptake. The PEG-shedding is necessary to prevent the potential induction of PEG-specific antibodies that cause rapid systemic clearance of subsequent doses of PEGylated nanomaterials via accelerated blood clearance (ABC) phenomenon [224]. The severity of ABC may also be suppressed by adjusting the PEG-lipid structure, affecting the kinetics of shedding and the recognition of the chains [224,225]. These findings indicate that the structure and the amount of PEG-lipid must be adjusted carefully to preserve the “stealth” effect.
The long-term storage of mRNA-LNP vaccines is a significant yet underexplored part of the LNP lifecycle. Since self-assembled constructs are held together by weak intermolecular interactions, they may undergo structural changes to advance towards equilibrium in prolonged storage. Little is yet known what changes are inflicted on self-assembled materials for mRNA delivery in storage; however, a recent report by Zhao et al. dedicated to the long-term storage conditions of mRNA-loaded lipid materials indicates that lipid-like nanoparticles (mRNA-LLNs) stored in aqueous solution undergo size changes and lose their efficacy in vivo [226]. In comparison, freeze-dried mRNA-LLNs maintained their efficacy when lyophilized with 5% cryoprotectant solution, and changed the preferential organ uptake from the liver to spleen when lyophilized with 20% cryoprotectant solution. These findings support the hypothesis that self-assembled materials are dynamic systems sensitive to the environment, and highlight the need to further investigate the impact of long-term storage of mRNA vaccines.
In conclusion, there is a virtually endless parameter space that can be modified in order to achieve a highly efficient, nontoxic, and tissue, organ, or cell-selective LNP formulation. Although there are some structural criteria (e.g., biodegradable groups, pKa of ionizable group etc.) that may be used in the design of next-generation lipids for mRNA delivery, any changes on the atomic level may require additional optimization and drastically improve (or worsen) the mRNA delivery efficiency. The field of mRNA delivery would greatly benefit from in-depth SAR and morphological investigations to establish the guidelines for controlled and targeted delivery of mRNA via LNPs.
6.2. Polymer-based systems
Polyplexes, or polymer-based nucleic acid carriers, are another popular type of mRNA delivery system. Generally, the forces driving the self-assembly of mRNA polyplexes are the same as in the case of lipid-based carriers – mainly, electrostatic attraction (between the polymer carrying a positive charge, and negatively charged nucleic acid), hydrophobic interactions between polymer chains, and van der Waals forces between polymer, solvent, and nucleic acid cargo. However, polymeric formulations rely on different parameters than lipid-based ones. Polymers can include different functionalities in the same molecule and have different length and arrangement of monomer units [141]. Therefore, polyplexes typically consist of only one polymer that may attain several functionalities simultaneously. Nonetheless, polymeric materials, just like LNPs, require optimization of polymer composition depending on the payload [227], which has largely been empirical to the date.
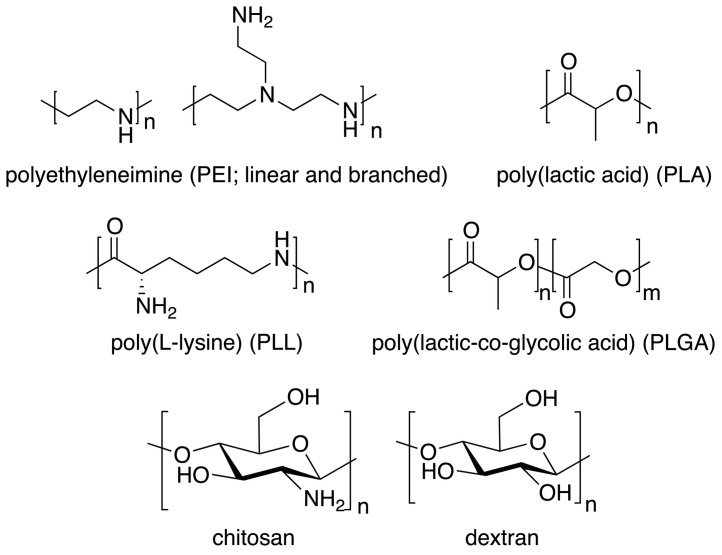
Polymeric gene delivery systems have initially struggled with cytotoxicity, partially due to their cationic charge [141,228]. Early attempts for gene delivery have been largely based on three types of synthetic polymers – poly(ethylene imine) (PEI), poly(L-lysine) (PLL), and poly(amidoamine) (PAMAM) (Fig. 8 ); however, only PEI has been widely used for mRNA vaccine delivery [150]. Even though optimized PEI structures offer high gene transfection efficiency, its high cationic charge density still creates toxicity issues [111]. Decorating these cationic polymers with PEG-chains and targeting moieties alleviated cytotoxicity and greatly improved their delivery in vitro and in vivo [141,[228], [229], [230]]. Another concern with regards to polymer structure is biodegradability, which made the design of polymeric mRNA vectors move towards polyesters (e.g., poly(beta-amino esters) (PBAE), poly(amine-co-esters) (PACE), poly(lactic acid) (PLA), poly(lactic-co-glycolic acid) (PLGA), and poly(2-dimethylaminoethyl methacrylate) (PDMAEMA)), natural polymers (e.g., chitosan and dextran) derivatives (Fig. 8), and other biodegradable structures [4,229,[231], [232], [233], [234]]. For example, Yan et al. reported that systemically administered polyester-based nanoparticles delivered mRNA to the lungs preferentially [235], and Fornaguera et al. demonstrated that oligopeptide end-modified PBAE nanocarriers transfect the splenic APCs after IV injection, highlighting their potential as mRNA vaccines[236].
Fig. 8.
Selected structures of natural and synthetic polymers used for mRNA complexation.
Polyplexes with mRNA form more rigid (compared to lipid-based) supramolecular structures with highly entangled polymeric chains due to higher molecular weight and slower mobility of polymer chains [145]. The superior stability of polyplexes compared to lipid-based materials makes them extremely attractive in the exploration of alternative delivery routes. mRNA polyplexes show great promise for mucosal vaccination [[237], [238], [239]] because their structure recovers after aerosolization. For example, Patel et al. reported successful delivery of mRNA, encapsulated by hyperbranched PBAE, to the lung epithelium via nebulized mist [240], and Li et al. explored intranasal delivery of mRNA via cyclodextrin-PEI complexes for HIV treatment [111,113]. Additionally, the stability of polyplexes can be further enhanced by crosslinking [145]. Various approaches for the stabilization of block copolymer nanostructures via crosslinking are summed up in a recent review [241]. The excessive stability of polyplexes, however, can become a disadvantage, compromising or restricting the release of the genetic cargo into the intracellular space [242].
Polymers can also be utilized to build scaffolds for mRNA vaccination. Bryers and colleagues demonstrated that porous templated scaffolds consisting of poly (2-hydroxyethyl methacrylate) [243] or chitosan-alginate [244] can load and release mRNA nanoparticles in a sustained manner at the site of implant/injection, resulting in the production of antigen-specific antibody in mice. In particular, scaffold-based mRNA delivery could be exploited to develop mRNA vaccines in the form of microneedles for less invasive and patient-compliant vaccination [114]. In spite of these promising features of polymeric mRNA vaccines, relatively low gene transfer efficiency and innate heterogeneity of polymers limit their clinical translation as well as industrial-scale production. In conclusion, mRNA delivery using polymers is an extremely promising and underexplored space that might provide distinctive benefits that lipid-based vectors cannot offer.
7. Physicochemical characteristics affecting mRNA vaccine delivery
The nanoscale and molecular structures of all potential gene carriers determine their efficiency. To date, the exact structural motifs of both the building blocks and the self-assemblies needed for the design of a superior mRNA-based gene delivery vehicle remain unclear. There are, however, some delivery system guidelines that are consistent regardless of the cargo and encapsulating material. Here, we will attempt to summarize the relevant information with regards to gene delivery vehicle design, discussing examples for mRNA vaccines whenever applicable.
7.1. Size
Size of a delivery vehicle is perhaps the most well-known parameter that affects the delivery efficiency. The size has a significant contribution to many functional parameters determining both the efficiency and the selectivity of a delivery vector – to name a few, biodistribution, immunogenicity, internalization, degradation, clearance, and cellular uptake [245]. As defined in vaccinology, subunit vaccines that are < 10 nm in size poorly reach the LNs because they are flushed by blood flow rather than being transported by lymph flow. In contrast, vaccines that are too large (> 200 nm) struggle to enter fluid flow across the interstitium due to diminished diffusion and convection [86]. In the context of mRNA vaccines to be administered to the interstitial space (for example, by IM, SC, or ID injection), the size of < 200 nm is generally desired as the nanoparticles of this size tend to be transported to the lymphatic system – in comparison, larger particles are retained at the injection site [86,137]. Overall, the size requirements highly depend on the administration route, experimental condition, and target organ. Irvine, Aung, and Silva recently reported that antigen delivery to DCs in LNs is strongly associated with the size of vaccines, with smaller particles (< 50 nm in diameter) showing higher delivery irrespective of the composition of the particles [87]. Similarly, Cabral et al. reported that enhanced permeation of 30 nm polymeric micelles to the BxPC3 tumors implanted on mouse flanks [246]. Chen et al. reported that gene silencing in the liver was most potent for 45 nm siRNA-LNPs when delivered via SC injection, and for 80 nm LNPs when delivered by IV injection to mice [247,248]. The size preference may also vary depending on the cells – e.g., 50 nm mRNA-carrying particles were superior to 100 nm ones in transfection of human embryonic kidney 293 cells and human adipocytes, but showed no significant difference in human hepatocytes [5]. Moreover, the route of administration may impose additional size restrictions depending on the delivery device – e.g., 1-5 μm microparticles or microdroplets containing nanoparticles may be better suited for delivery to the lung due to the aerodynamics of the pulmonary deposition [249].
Although the size of mRNA delivery vector is initially determined by the composing materials, it is subject to change depending on their fate. For example, nebulization of mRNA polyplexes for lung delivery leads to increases in both size and polydispersity of the polyplexes [240]; exposure of lipid-based nanoparticles to biological medium causes formation of a biomacromolecular corona, increasing size by ca. 20% [165]; lyophilization of lipid-based nanoparticles for long-term storage may cause up to 4-fold increase in particle size depending on storage conditions [226]. However, it may not be possible to extract the effect of the size on transfection independently of other properties – formation of protein corona on nanoparticle surface can determine the uptake pathway [165], and lyophilization may change not only the incidence of particle aggregation but also the reorganization of lipid components. Overall, more studies of size-dependent mRNA delivery are needed to fully reveal the impact of this criterion.
7.2. Charge
Charge plays a key role in the biodistribution and efficacy of mRNA vaccines, as the inherent dense negative charge of mRNA imposes an additional challenge to the packaging and delivery. Negatively charged carbohydrate barriers - such as glycocalyx and mucins in the interstitium, mucus, and at the surface of the plasma membranes – influence the trafficking of nanocarriers in the in vivo milieu. These barriers form an anionic gel-like matrix of glycolipids, proteoglycans, oligosaccharides, and glycoproteins [86,121,148,250]. As a result, the transport of positively charged vectors may be impeded – for example, cationic nanoparticles struggle to reach LNs due to limited convection across the interstitium despite their affinity for APCs. In contrast, anionic nanoparticles are diffusible in the interstitium and show better retention in LNs [86]; but cannot readily enter intracellular space due to the negative charge of the cellular membrane. Therefore, the charge of the delivery vector plays an instrumental role in dictating the transport of mRNA vaccines across the biological barriers.
A positively charged material like cationic liposomes can fully envelop anionic mRNA [251]. As a result, such mRNA nanocarriers possess positive surface charge – the cationic material should be abundant for the electrostatic complexation with mRNA molecules – which can lead to association with negatively charged cell surfaces such as cell and endosomal membranes, thus releasing the cargo [252]. Although cationic delivery vectors have various successes with transfection, they show cytotoxicity because of inadvertent associations with negatively charged biomacromolecules and off-target cells (e.g., albumin and red blood cells in the bloodstream) [228]. For these reasons, the RNA delivery vectors have evolved to have a relatively neutral surface charge.
Even though a minimal surface charge of the delivery vector is preferred to avoid off-target interactions, the positive charge is still required to breach cellular and endosomal membranes. Therefore, a delivery system must be responsive to changes in the biological environment. Fortunately, biological systems have variations in pH that can be exploited for the purpose of pH-responsive release of nucleic acids. For example, in endolysosomal pathways, lumen pH becomes gradually acidic (in a range of 6.8 to 4.5) [253], and thus a delivery vesicle can be engineered in such a way that it only acquires a positive charge upon entering the endosome. As mentioned above, a positive charge on the delivery vehicle is crucial for cellular uptake and endosomal escape of packaged mRNA; however, this charge should not be persistent. Using fluorescent assays or titration systems, one could determine the pKa, which is a measure of the material’s ability to attain a positive charge. If the pH of the environment is more acidic than material pKa, the material will become positively charged at that pH. This acidification process may be the key to controlled delivery to certain cells and subcellular compartments that may have a slightly acidic pH (for example, tumor microenvironment and endosomes). However, it is not entirely clear what pKa is needed for optimal gene delivery. For example, Jayaraman et al. prepared a library of ionizable lipids for in vivo siRNA delivery and found that the ideal pKa range was 6.2 - 6.5 for gene silencing [179]. However, even in this pKa range, the potency of gene silencing varied from 4 to 33-fold, indicating that pKa is not the only factor that influences activity. The desired pKa of LNPs may also vary depending on the administration route – Hassett et al. reported pKa of 6.6 - 6.9 as optimal for IM delivery of mRNA-LNP, which differed from the previous report of optimal pKa of 6.2 - 6.6 for IV delivery of similar mRNA-LNP [126,254]. Moreover, pKa determination for polymers is not trivial. Polymers carry significantly more ionizable groups per molecule than ionizable lipids, which confers a buffering effect and can cause swelling of polyelectrolytes [141]. The flexibility of polyelectrolyte chains may also influence how these large cationic molecules interact with the negatively charged mRNA cargo [230]. Thus, different materials may exploit different routes of cellular uptake depending on their electrostatic interactions with both the cargo and the environment.
Interestingly, incorporation of charged moieties in the core of the nanoparticles delivering the genetic payload may direct organ-specific uptake. Cheng et al. reported that incorporation of permanently charged lipids resulted in variations in organ delivery when LNPs were administered to mice intravenously – positively charged lipids lead to the preferential targeting of the mouse lungs while negatively charged lipids targeted the spleen [255]. In the absence of these charged lipids, LNPs preferentially targeted the liver. Notably, the LNPs possessed no substantial surface charge as determined via zeta-potential measurements, indicating that the charge was fully contained internally. This important discovery emphasizes the role of the charge in the cellular uptake pathways and should encourage further investigation.
7.3. Shape and structure
Shape and internal structure are both important structural parameters that can affect interactions with biological environment, influencing cellular uptake [136,245,256,257]. For example, both shape and internal structure may affect the rigidity of the surface of delivery vectors, changing the nature of the interaction with cellular and endosomal membranes. Although manufacturing of shape-specific materials is common in nanotechnology, this aspect of gene delivery vehicle design has been underexplored due to the technological challenges – many common X-ray and electron-based techniques are destructive to delicate samples and may misrepresent the internal structure. Fortunately, recent advances in cryogenic transmission electron microscopy (cryo-TEM) and small-angle X-ray scattering (SAXS) have made the probing of the “soft” nanoparticles structure more accessible, yielding some exciting results in the field of gene delivery [[258], [259], [260], [261], [262], [263]]. However, characterization of mRNA delivery vehicles with cryo-TEM became more common than SAXS. The reason for this is partly because the analysis of the nanomaterial structure can be done in both quantitative and qualitative manner, while SAXS provides information about the nanostructure of the bulk aggregates without direct visualization. Consequentially, cryo-TEM may allow for a more comprehensive analysis of nanomaterial shape and structure, although application of this analysis in the field of mRNA delivery is still in its infancy. For example, Kulkarni et al. utilized cryo-TEM to study morphological differences of LNPs depending on cargo, composition, and preparation method [162]; Eygeris et al. performed a quantitative analysis of LNPs containing cholesterol analogs [163]; Sun et al. used both TEM and cryo-TEM to confirm the structure of the PLGA-based lipopolyplexes [214]. Evidently, polymorphic particle shape, heterogeneity of the core, and multilamellar architecture may be beneficial for the cellular uptake and release of mRNA as suggested by the in vitro studies [163] although the exact mechanisms explaining these findings and their translation to in vivo environment remain obscured. The shape and the internal structure of mRNA delivery vehicles should be further investigated as these parameters may provide unique insights into the deformation of biological membranes and, inevitably, the therapeutic efficiency [214].
7.4. Surface composition
Delivery efficiency and biodistribution of mRNA vectors are affected by the surface composition of vectors. A prominent example of surface modification is the incorporation of PEG-chains. PEGylation of nanoparticles changes the trafficking of nanocarriers, extends half-life of delivery vectors in vivo by preventing interactions with serum proteins and phagocytes, and enhances the solubility and stability of delivery vectors by increasing hydrophilicity [264]. While PEG moiety plays a key role in biodistribution and half-life of nanocarriers, it hinders efficient uptake of nanoparticles by cells because of steric hindrance and inhibited interactions with the plasma membrane [218]. Therefore, PEG-chains are designed to detach in the serum to alleviate the steric hindrance and allow the nanocarriers to interact with ApoE and to subsequently enter the cells by ApoE-mediated endocytosis [218,265,266].
As with ApoE-mediated nanoparticle uptake, there is interest in biomolecular surface modification that may facilitate nanoparticle-mediated gene transfer. A recent report by Schöttler et al. suggests that the “stealth” effect of PEG-coating may be due to the formation of a biomolecular corona via attractive interactions [267]. Although the formation of the protein corona is a well-known fact [268], studies considering and exploiting this aspect in the context of mRNA gene therapy have been limited [165]. This circumstance could explain why there has been a weak correlation between in vitro and in vivo studies of LNPs since biomolecular corona can cause particle agglomeration, premature release of LNP content, or lipid dissociation depending on the content of the corona [269,270].
Incorporation of carbohydrates (such as mannose) on the nanocarriers can also influence the trafficking to the LNs and uptake to immune cells [271,272]. Tokatlian et al. demonstrated that tri-mannosylation of the HIV nanoparticle immunogens leads to strong GC reactions because of the facilitated accumulation in LNs with a complement-, mannose-binding lectin-mediated, immunogen glycan-dependent manner [271]. Exploitation of innate immune recognition of glycans can be used for mRNA vaccination as well. It was reported that mannosylated LNPs encapsulating mRNA encoding hemagglutinin of H3N2 influenza virus exhibited enhanced inhibition in hemagglutination assay as compared to the plain LNPs [112].
7.5. Methods of characterization
The self-assembly of the mRNA delivery vehicles has been extensively studied by a large variety of physicochemical methods. Nowadays, almost every paper related to the field of nucleic acid delivery utilizes three characterization methods - dynamic light scattering (DLS; to determine size and polydispersity of a self-assembled material), luminescence spectroscopy (e.g., fluorescent and luminescent assays to characterize encapsulation efficiency, transfection etc.), and transmission electron microscopy (TEM; to visualize the morphology and internal structure). More specialized methods include, but not limited to electron tomography (a variation of electron microscopy where the sample is tilted at different angles to recreate a 3D structure) [273], small-angle neutron or X-ray scattering (SANS and SAXS, respectively; to characterize the internal structure) [166,274], differential scanning calorimetry (DSC; transformation of phases in response to heat) [275], nuclear magnetic resonance spectroscopy (NMR; a variety of structural parameters) [276], and liquid chromatography (especially high-performance variety, or HPLC; composition or degradation of material)[197].
While the aforementioned techniques are extremely useful in gathering a large amount of information about material properties, they all have limitations in time – setup and acquisition requires minutes, if not hours. Therefore, these methods likely capture properties of late-stage, stable self-assemblies, and miss entirely the very event of formation of the delivery vector. Studying the early stages of self-assembly requires an extremely fresh sample and a technique capable of nanoscale acquisition both in spatial and temporal dimensions. For this reason, computational approaches are becoming increasingly more popular for understanding the early structural developments of self-assembly, which may provide insights into the cellular uptake and payload release [165,277]. Latest advances in molecular dynamics simulations of biomolecule self-assembly are highlighted in a recent perspective [278]. Despite the breakthroughs in the fields of computer science and molecular dynamics, significantly improving accuracy and processing speed of simulation, computational findings so far remain empirical and it is yet unclear whether the simulations could ever replace experiments or dictate material design. One exciting approach to supplement theoretical exploration in silico with much needed experimental data is utilizing microfluidics, which have become a useful method not only for nanomaterial synthesis but also for the in-situ analysis of the nanomaterial formation [279]. Even though co-flow microfluidics have been useful in studying the incorporation of DNA into lipid vesicles [280], this approach to the date has not been implemented for mRNA-loaded systems.
In conjunction with various techniques to characterize material properties, there are new techniques for in vivo trafficking of mRNA carriers, enabling more accurate evaluation of various carriers. One of the recently emerging tools is intravital microscopy. Lymphatic trafficking of nanovaccines labeled with fluorophores can be tracked at a subcellular level in real-time with high resolution [281]. Furthermore, it can be performed in a non-invasive manner, allowing for the spatiotemporal dynamics of nanocarriers for an extended period of time [272]. Positron emission tomography (PET) is another way to monitor nanocarriers in vivo. PET offers quantitative and translational detection of nanocarriers at the outstanding resolution [121]. A recent example of PET-mediated trafficking of mRNA vaccines revealed the spatiotemporal trafficking of the vaccines in non-human primates [63]. While intravital microscopy and PET imaging provide real-time in vivo distribution of nanocarriers, visualization of RNA molecules in tissues at nanoscale can be achieved by in situ hybridization. Although single molecule fluorescent in situ hybridization (smFISH) enables the delivered mRNA molecules to be detectable and quantifiable in fixed cells [126], it is difficult to use for tissue samples in clinical settings due to the low signal-to-noise ratio [282]. Non-specific interactions of the smFISH probes in tissue samples generally result in high background fluorescence, limiting the sensitivity and specificity necessary to detect low abundance RNA molecules accurately [282]. RNAScope® utilizes target-specific double Z probes, preamplifiers, and fluorophore-conjugated amplifiers in order to increase the signal-to-noise ratio. It was recently used to visualize synthetic mRNA delivered by LNPs to mouse liver at wide field-of-view [283]. With deep, multiplexed imaging and image processing, it can be used to reveal in-depth trafficking of administered mRNA vaccines in animals.
In conclusion, experimental developments in the characterization of nanomedicines would be extremely useful to understand the nanomaterial design criteria. Self-assembly (and disassembly) of mRNA-nanomaterial, spatiotemporal distribution of mRNA vectors, and pharmacokinetic and pharmacodynamic profiles of mRNA vaccines are all critical areas that urge further investigation, and the understanding of these phenomena can consequentially lead to the establishment of superior mRNA vaccines.
8. Clinical utilities of mRNA vaccines for COVID-19
As mentioned before, mRNA vaccines have gained significant interest in the last decade, with the current COVID-19 pandemic further fueling their development for mass use. The status of mRNA vaccines has recently been reviewed [2,159,284]. In this section, we focus on the development of clinically investigated mRNA vaccines against Coronavirus Disease 2019 (COVID-19) pandemic (Table 1 ). COVID-19 is caused by a beta-coronavirus SARS-CoV-2, a single-stranded positive sense RNA virus. SARS-CoV-2 consists of an RNA genome, four structural proteins [nucleocapsid (N), membrane (M), envelope (E) proteins, and spike (S) protein], and sixteen non-structural proteins (nsp1-16) (Fig. 9A). The notorious “crown”-like spike (S) protein consists of S1 and S2 subunits. The receptor-binding domain (RBD) within the S1 subunit binds to human ACE2 receptors, and S2 subunit subsequently catalyzes the fusion between the viral envelope and the host cell membrane, allowing the virus to ultimately enter the host cells. Following the cellular entry, the viral genomic RNA undergoes translation, replication, and transcription into essential viral components. In the final steps, the virus is assembled, and packed in the host cells, and the viral particles are released [[285], [286], [287]]. Due to its indispensable functions and high conservation, the spike protein serves as a primary target for therapeutic interventions [288].
Table 1.
List of mRNA vaccines against COVID-19 in clinical trials (till December 2020).
| Vaccine Name | Target antigen | Delivery Vehicle | Delivery Route | Sponsor | Phase | Results or Comments | EUA | Reference |
|---|---|---|---|---|---|---|---|---|
| BNT162b2 | Membrane-anchored prefusion-stabilized spike protein of SARS-CoV-2 | LNP | IM | BioNTech/ Pfizer |
III | In the phase I/II trials the vaccine was found to be safe with transient side effects. It was given in the prime-booster regimen. After the booster dose, it produced robust level of antibodies comparable to or better than convalescent plasma. In an ongoing phase III trials consisting of more than 40,000 participants, it showed an overall efficacy of 95% with an acceptable safety profile. |
UK, Bahrain, Canada, US, Mexico, Kuwait, Singapore, Jordan, Oman, Costa Rica, Panama, Chile, and Saudi Arabia | [81,298,300,301] NCT04368728 |
| mRNA-1273 | Membrane-anchored prefusion-stabilized spike protein of SARS-CoV-2 | LNP | IM | Moderna | III | In the phase I clinical trials, two doses of mRNA-1273 were administered by IM injection. It was found to be safe and produced good antibody responses after the second dose. Interim analysis of the phase III clinical trials involving approximately 30,000 participants showed that the vaccine was safe with 94.1% efficacy. |
US | [12,304,305] NCT04283461 NCT04405076 NCT04649151 NCT04470427 |
| ARCoV | RBD of the SARS-CoV-2 spike protein | LNP | IM | Academy of Military Medical Science/ Sushou Abogen Biosciences/ Walvax Biotechnology |
Ib | Two doses of either 100 μg or 1000 μg (2 weeks apart) led to antibody responses in Cynomolgus Macaques. ARCoV is reported to be stable at 25 °C for at least one week. |
[306] ChiCTR2000034112 ChiCTR2000039212 |
|
| BNT162b1 | Trimerized RBD of the SARS-CoV-2 spike protein | LNP | IM | BioNTech/ Pfizer |
II | In the phase I/II clinical trials, two doses of 10 μg, 30 μg, or 100 μg (3 weeks apart) were tolerated. After the second dose, it produced robust antibody responses and T cell responses. Based on the better tolerability of BNT162b2, particularly in older adults, Pfizer/BioNTech advanced BNT162b2 in the phase-II/III trials. |
[81,296,297] NCT04368728 |
|
| COVAC1 (LNP-nCoVsaRNA) | Prefusion-stabilized spike protein of SARS-CoV-2 and an alphavirus replicase | LNP | IM | Imperial College London/ Morning Side Ventures |
I/II | COVAC1 is a self-amplifying vaccine. In mice two injections of 0.01 μg, 0.1 μg, 1 μg, or 10 μg of the vaccine were given 1-month apart. Found a strong Th1-biased cell response at 6 weeks after the initial vaccination. |
[57] ISRCTN17072692 |
|
| LUNAR® -COV19 (ARCT-021) | Prefusion-stabilized spike protein of SARS-CoV-2 and an alphavirus replicase | LNP | IM | Arcturus Therapeutics/ Duke NUS |
I/II | Uses proprietary self-transcribing and replicating RNA (STARR™) platform. In mice, 60 days after the administration of a single dose (2 μg or 10 μg) of the vaccine elicited robust CD8+ cell and CD4+ TH1 cell response. |
[307] | |
| CVnCoV | Prefusion-stabilized spike protein of SARS-CoV-2 | LNP | IM | CureVac | IIa | In the phase I clinical trials, two doses were given in 4 weeks interval by IM injection. The vaccine was found to be safe with dose-dependent mild to moderate side effects. The antibody response after the second dose was similar to the convalescent sera. | [308] NCT04515147, NCT04449276 |
|
| ChulaCoV19 mRNA vaccine | Virus-specific antigen | LNP | IM | Chulalongkorn University | I | NCT04566276 |
Fig. 9.
(A) Structure of SARS-CoV-2 virus depicting a RNA genome and structural proteins [nucleocapsid (N), membrane (M), envelope (E) proteins, and spike (S) protein], (non-structural proteins are not shown). (B) Pfizer/BioNTech (BNT162b2) and Moderna (mRNA-1273) COVID-19 vaccines both utilize LNP platform and carry mRNA encoding the prefusion-stabilized, membrane-anchored, full-length spike protein of SARS-CoV-2. BNT162b1, another vaccine from Pfizer/BioNTech, is a LNP-formulated mRNA vaccine encoding the secreted trimerized RBD of the SARS-CoV-2 spike protein. (C) Cumulative incidence curves for the first COVID-19 occurrence after dose 1 of BNT162b2 vaccine. Reprinted from [301]. (D) Cumulative incidence curves for the first COVID-19 occurrence after randomization (same as date of dose 1) of mRNA-1273 vaccine. Reprinted from [305].
The COVID-19 pandemic, which likely began in late 2019, is responsible for more than 77 million infections and 1.69 million casualties worldwide as of December 21, 2020 [289]. Within the United States, there are more than 18 million cases with more than 320,000 deaths [289]. The symptoms of the disease can range from cough and mild fever to respiratory and systemic failure [[290], [291], [292]]. The countermeasures for containment of this contagion involve wearing face masks, proper handwashing, large-scale testing, social distancing, and avoiding in-person interactions, especially in large groups [293]. However, these efforts cannot be relied upon in the long term, and thus a safe and effective vaccine is urgently needed to end the misery caused by the pandemic. Scientific communities have come together and worked at a lightning speed to decode the virus components in order to develop vaccines against COVID-19 [294]. According to the WHO, there are 166 candidates in preclinical development and 56 vaccines are currently in clinical trials as of December 2020 [295]. Out of the 56 clinical candidates, 7 candidates are mRNA-based, which are discussed below with special emphasis on 2 vaccines (BNT162b and mRNA-1273) that have received EUA in the United States and other countries.
Researchers at Pfizer and BioNTech co-developed mRNA vaccine candidates BNT162b1 (NCT04368728) and BNT162b2 (NCT04368728). BNT162b1 mRNA encodes for the trimerized RBD of the SARS-CoV-2 spike protein held together by the T4 fibritin domain to enhance immunogenicity [296]. On the other hand, BNT162b2 mRNA encodes the prefusion-stabilized spike protein with two key proline substitutions (see Fig. 9B) [81]. Both candidates are formulated with LNPs and nucleotide-modified with uridine being replaced with 1-methyl-3’-pseudouridine. Phase I/II trials of the vaccines were conducted in young adults (18–55 years) and older adults (65-85 years) [81,296,297]. BNT162b1 and BNT162b2 were well-tolerated in phase I studies when multiple doses were administered in a prime-boost regimen (3 weeks apart). Local and systemic side effects were mild to moderate, transient, and dose-dependent for both vaccines with BNT162b2 exhibiting less reactogenicity than BNT162b1 [81]. Both vaccines produced high levels of binding antibodies and 50% neutralization titers, which were higher when compared with convalescent plasma. Antibody response was enhanced within 7 days of the booster dose, indicating a beneficial effect of the booster dose [81]. Both vaccines also induced robust CD4+ type 1 helper T cell (TH1) and CD8+ T cell responses [81,298], alleviating the concerns associated with vaccine-associated enhanced respiratory disease (VAERD) [299]. While both vaccines were effective and well-tolerated, a 30 μg dose of BNT162b2 was advanced to the phase II/III trials (NCT04368728) due to its milder systemic reactogenicity, especially in older adults [81]. Recently, Pfizer and BioNTech announced promising results of the ongoing phase III trials [300,301]. Approximately 43,000 individuals, who were 16 years or older and who were healthy or had stable chronic medical conditions were randomized to receive either two 30 μg doses of the vaccine or saline placebo (3 weeks apart). Although the study was intended to evaluate the efficacy based on a two-dose regimen, BNT162b2 was shown to provide a partially protective effect of immunization as early as 12 days after the first dose [300]. Between the first dose and the second dose, the vaccine efficacy was approximately 52% (39 cases in the BNT162b2 group vs. 82 cases in the placebo group, Fig. 9C). In the efficacy studies to evaluate the protective effects of the vaccine against confirmed COVID-19 diagnoses with onset at least seven days after the booster dose, BNT162b2 exhibited 95.0% efficacy in the participants with no history of prior SARS-CoV-2 infection (specifically, 8 cases of infections were noted in the vaccine group while the placebo group had 162 cases) [300], and 94.6% efficacy when the participants with prior infection of COVID-19 were included (9 cases of infection in the vaccine group and 169 cases in the placebo group) [300]. Similar to the earlier trial results [81], BNT162b2 was well tolerated with mild to moderate side effects [300]. The most common side effects were pain at the injection site, fatigue, headache, muscle pain, chills, joint pain, and fever, which were more common after the booster injection [300]. Severe adverse events occurred in only about 0.6% of the vaccine recipients, with similar frequency noted in the placebo group (0.5%) [300]. The promising results of phase III trial suggested BNT162b2 to be a safe and effective vaccine against COVID-19. Based on the positive interim results, Pfizer/BioNTech received EUA for BNT162b2 (also known as Tozinameran) in several countries including the UK [302] and the US [303].
mRNA-1273, another LNP-based mRNA vaccine, is being developed by a joint collaboration between Moderna and Vaccine Research Center (VRC, part of NIH). mRNA-1273 encodes for the prefusion-stabilized SARS-CoV-2 spike protein with two key proline substitutions, which is similar to the BNT162b2 [11]. After successful pre-clinical testing [11], the mRNA-1273 vaccine went through two phase I clinical trials - one in younger healthy adults (age of 18-55) [12] and the other in older adults (age 55+) (NCT04283461) [304]. In young adults, 45 healthy individuals were divided into three groups (15 participants each) and were given 25, 100, and 250 μg doses of mRNA-1273 via IM injections in a prime-boost regimen (4 weeks apart) [12]. Akin to BNT162b2, mRNA-1273 was well-tolerated with dose-dependent and transient side effects (pain, fever, fatigue, chills, headache, and myalgia) which were more prevalent after the booster dose [12]. In participants who received 25 μg and 100 μg does of mRNA-1273, levels of binding antibodies and neutralization titers were lesser than the convalescent sera, 15 days after the first dose. However, after the booster dose, the levels of binding antibodies and neutralization titer became similar or higher than the convalescent sera, clearly indicating the benefits of the prime-booster regimen. mRNA-1273 elicited robust CD4+ type 1 T helper cell (TH1) responses at both 25 μg and 100 μg doses although a lower CD8+ T cell response was noted [12]. The safety and efficacy profiles in older adults (age 55+) were fairly similar to those in younger adults [304]. Recently, the US FDA released the interim results from the ongoing phase III trial (NCT04470427) of mRNA-1273 involving approximately 30,400 individuals. mRNA-1273 was administered by IM injection in two doses at 100 μg each, 4 weeks apart [305]. mRNA-1273 exhibited 94.1% of vaccine efficacy (Fig. 9D): 11 cases of COVID-19 were noted in the vaccine group and 185 cases of COVID-19 in the placebo group [305]. Of note, none of the mRNA-1273 recipient developed severe COVID-19 (0 in the vaccine group vs 30 in the placebo group) [305]. The data also suggest that mRNA-1273 reduced asymptomatic infection by 63% after the first dose [305]. The reported side effects were mild to moderate and included pain, fatigue, headache, muscle pain, joint pain, and chills that typically occurred after the second dose [305]. These encouraging results led to mRNA-1273 being the second vaccine with EUA in the US [13].
BNT162b2 and mRNA-1273 share many similarities. Both utilize LNPs to deliver mRNA encoding the prefusion-stabilized spike protein of the SARS-CoV-2 (see Fig. 9B) with uridine substitution and showed similar efficacy (ca. 95%) in ongoing phase III trials with the two-dose regimen (prime-booster) [301,305]. However, their mRNA doses, dosing intervals, formulation characteristics, and storage conditions are different as highlighted in Table 2 [301,305]. While the BNT162b2 requires a 30 μg dose, mRNA-1273 requires a 100 μg dose for each vaccination. BNT162b2 requires thawing followed by dilution with saline before administration whereas mRNA-1273 could be injected as is after thawing [301,305]. Their formulations also use different ILs, PEG-lipids, and buffers [301,305]. In terms of storage, BNT162b2 requires ultra-cold storage (-60 °C to -80 °C) while mRNA-1273 requires less stringent conditions (-15 °C to -25 °C) [301,305]. This difference in the storage conditions may make mRNA-1273 a potentially better option for hospitals, clinics, and pharmacies without an ultra-cold freezer. The exact reason for the disparity in storage conditions remains largely unknown and it may be due to the differences in the secondary structure of mRNA or lipids used in the formulations. Nevertheless, the optimization of the mRNA vaccine composition for less strict storage conditions represents a pharmaceutical challenge that requires further investigation.
Table 2.
Dosing profile, storage requirements, directions, and formulation composition of BNT162b2 and mRNA-1273.
The following five mRNA vaccines are also in the clinical trials; however, due to the lack of clinical data, we discuss them briefly. ARCoV is in development by a collaboration between Academy of Military Medical Sciences, Suzhou Abogen Biosciences, and Walvax Biotechnology [306]. It encodes the RBD of SARS-CoV-2 in mRNA and is formulated in LNPs [306]. In the preclinical studies with cynomolgus monkeys, ARCoV was administered twice by IM injection at 100 μg or 1000 μg, 2 weeks apart [306]. The vaccinated monkeys developed strong binding antibody responses and neutralization titers at 100 μg administration, which were comparable to the convalescent sera. In the high dose (1000 μg), ARCoV produced stronger humoral responses. It also elicited CD4+ TH1 cell responses in Cynomolgus Macaques [306]. ARCoV is supplied as a thermostable liquid formulation that can be stored at room temperature for at least 1 week and is currently being evaluated in phase I clinical trials in China (ChiCTR2000034112).
COVAC1, a saRNA vaccine, is being developed by the Imperial College of London [57]. The saRNA vaccine encodes the prefusion stabilized SARS-CoV-2 spike protein and an alphavirus replicase responsible for replicating RNA in the cytoplasm [57]. In mice, the vaccine was administered twice, one month apart, at doses of 0.01, 0.1, 1, or 10 μg. 6 weeks after the initial vaccination, there was a dose-dependent increase in the binding antibody titer with values ranging from 105 - 106 ng/mL, approximately 100-1000 fold more than in the convalescent sera [57]. Similar results were obtained in the pseudovirus neutralization assay with ID50 value ranging from 103 - 105, which is up to 100-fold higher as compared to the levels of convalescent sera. COVAC1 induced TH1-biased responses that lessen the risks of VAERD [299]. The vaccine is currently undergoing phase I clinical trials in the UK (ISRCTN17072692).
ARCT-021 (LUNAR-COV19) is being developed by Arcturus Therapeutics and Duke-NUS Medical Center [307]. LUNAR-COV19 is a single dose, LNP-based vaccine that utilizes the proprietary self-transcribing and replicating RNA (STARR™) platform. This saRNA vaccine encodes alphavirus replicase and the full length, unmodified, prefusion spike protein. 60 days after the IM injection of either 2 μg or 10 μg doses to the human ACE2 transgenic mice, LUNAR-COV19 produced significantly higher titers of binding antibody compared to the non-replicating counterpart. A similar trend was found in the neutralization titer assay where LUNAR-COV19 outperformed the non-replicating mRNA vaccine. Furthermore, LUNAR-COV19 produced strong CD4+, CD8+, and TH1-responses and protected the mice when challenged with a lethal dose of SARS-CoV-2. LUNAR-COV19 is currently in phase I clinical trials (NCT04480957).
CVnCoV is being developed by CureVac. CVnCoV is an mRNA vaccine encoding the prefusion full-length spike protein and formulated with LNPs by their proprietary formulation (RNActive®). In the phase-I clinical trials (NCT04515147, NCT04449276), two doses of 2 μg to 12 μg each, were administered by an IM injection, 4 weeks apart [308]. In healthy adults (age of 18-60) with no history of prior SARS-CoV-2 infection, CVnCoV was well tolerated with mild to moderate side effects [308]. Local reactions and systemic adverse events displayed dose-dependent increases in frequency and severity, and no vaccine-related serious adverse events were observed [308]. CVnCoV elicited dose-dependent antibody responses, and the neutralizing antibody titers after the second 12 μg dose were similar to those observed in the convalescent sera [308].
Lastly, a mRNA vaccine being developed by the Chulalongkorn University in Thailand is in the phase I clinical trial (NCT04566276); however, no preliminary data is reported yet.
9. Conclusions and Future aspects
The current COVID-19 crisis has led to unprecedented investments in the manufacturing of mRNA-LNPs. Billions of doses are being made to be administered to the human population. This is a seismic shift that was hard to imagine just a year ago. If in the future an LNP platform is found to traverse thick mucus of respiratory challenged patients (such as with cystic fibrosis) or can target extrahepatic tissues, the steps to manufacture billions of doses are in place to supply a multitude of patients suffering from rare genetic disorders (>100,000 patients). The modularity of LNPs for delivering genome editing components means that a successful technology could be quickly scaled and effectively wipe out a rare disease with a one-time treatment.
Certainly, these multicomponent systems are not perfect. Synthesis of ionizable lipids alone is a multi-step process that requires several building blocks, and the complexity of lipids formulations discussed above makes these supramolecular assemblies rather intricate. Additionally, LNPs currently require strict storage conditions, limiting their distribution to locations where cold storage is scarce or unavailable. In the nearest future, lyophilization or other pharmaceutical processing methods may help to resolve this problem and even enable nasal, oral, or respiratory administration. In comparison, the synthesis of polymers is often a one-step process requiring one or several monomers; however, polymer products have inherent variability in the molecular weight and arrangement of monomers in the chain that increases batch-to-batch variability. Therefore, even though polymer-based systems have lagged as compared to LNPs, these technologies must not be discounted, especially for extrahepatic or localized delivery. Moreover, polymeric materials may be another solution to the distribution challenge of mRNA vaccines. Lastly, the self-assembly of mRNA and the encapsulating materials may require extensive optimization depending on the payload. The mixing of the components must be done in a highly controlled and reproducible manner, most often achieved with microfluidic mixers. Recent advances in the microfluidic technology allow for scale-up with the precision that is required for GMP facilities.
In conclusion, the COVID-19 pandemic led to the rapid deployment of mRNA vaccines. These technologies have been developed through years of painstaking work by scientists in academia and industry. Although not enough time has yet passed to evaluate the long-term effects of mRNA vaccines, the phase III clinical studies of Pfizer/BioNTech and Moderna vaccines revealed ca. 95% efficacy and favorable safety profiles. The COVID-19 pandemic has paved the way to scale up and resolved distribution barriers which otherwise would have taken decades. RNA therapeutics and nanomedicine as a field will never remain the same and are ready for their time to shine. The Age of Enlightenment of RNA nanotherapeutics is nearing its end, and the field is ready for the transition to a full-scale industrial revolution.
Conflicts of Interest
There are no conflicts to declare.
Acknowledgments
Acknowledgement
This project was supported through funding from the National Heart Lung and Blood Institute (N.H.L.B.I) 5R01HL146736 (G.S), National Eye Institute 1R21EY031066 (G.S), and Cystic Fibrosis Foundation SAHAY19XX0 (G.S). We thank Antony Jozic and Madeleine Landry for the review of the manuscript. Some figures were created using BioRender.com.
References
- 1.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 2.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granot Y., Peer D. Delivering the right message: Challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—An innate immune system standpoint. Interact. Nanoparticles Immune Syst. Advant. Disadv. 2017;34:68–77. doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Yu C. Emerging concepts of nanobiotechnology in mRNA delivery. Angew. Chem. Int. Ed. 2020 doi: 10.1002/anie.202003545. [DOI] [PubMed] [Google Scholar]
- 6.Mi P., Miyata K., Kataoka K., Cabral H. Clinical translation of self-assembled cancer nanomedicines. Adv. Ther. 2020 doi: 10.1002/adtp.202000159. [DOI] [Google Scholar]
- 7.Whitesides G.M., Grzybowski B.A. Self-assembly at all scales. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 8.Bishop K.J.M., Wilmer C.E., Soh S., Grzybowski B.A. Nanoscale forces and their uses in self-assembly. Small. 2009;5:1600–1630. doi: 10.1002/smll.200900358. [DOI] [PubMed] [Google Scholar]
- 9.Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.-G., Lévy J.-P., Meulien P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 10.Conry R.M., LoBuglio A.F., Wright M., Sumerel L., Pike M.J., Johanning F., Benjamin R., Lu D., Curiel D.T. Characterization of a messenger rna polynucleotide vaccine vector. Cancer Res. 1995;55:1397. [PubMed] [Google Scholar]
- 11.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A., Fritch E.J., Martinez D.R., Bock K.W., Minai M., Nagata B.M., Hutchinson G.B., Wu K., Henry C., Bahi K., Garcia-Dominguez D., Ma L., Renzi I., Kong W.-P., Schmidt S.D., Wang L., Zhang Y., Phung E., Chang L.A., Loomis R.J., Altaras N.E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M.K., Shi W., Leung K., Yang E.S., West A., Gully K.L., Stevens L.J., Wang N., Wrapp D., Doria-Rose N.A., Stewart-Jones G., Bennett H., Alvarado G.S., Nason M.C., Ruckwardt T.J., McLellan J.S., Denison M.R., Chappell J.D., Moore I.N., Morabito K.M., Mascola J.R., Baric R.S., Carfi A., Graham B.S. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ledford H. Moderna COVID vaccine becomes second to get US authorization. Nature. 2020 doi: 10.1038/d41586-020-03593-7. d41586-020-03593–7. [DOI] [PubMed] [Google Scholar]
- 14.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 15.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.-K., Schlee M., Endres S., Hartmann G. 5’-triphosphate RNA Is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 17.Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., Sousa C. Reis E. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 18.Weide B., Carralot J.-P., Reese A., Scheel B., Eigentler T.K., Hoerr I., Rammensee H.-G., Garbe C., Pascolo S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 19.Diken M., Kreiter S., Selmi A., Britten C.M., Huber C., Türeci Ö., Sahin U. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18:702–708. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- 20.Sahin U., Derhovanessian E., Miller M., Kloke B.-P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B., Omokoko T., Vormehr M., Albrecht C., Paruzynski A., Kuhn A.N., Buck J., Heesch S., Schreeb K.H., Müller F., Ortseifer I., Vogler I., Godehardt E., Attig S., Rae R., Breitkreuz A., Tolliver C., Suchan M., Martic G., Hohberger A., Sorn P., Diekmann J., Ciesla J., Waksmann O., Brück A.-K., Witt M., Zillgen M., Rothermel A., Kasemann B., Langer D., Bolte S., Diken M., Kreiter S., Nemecek R., Gebhardt C., Grabbe S., Höller C., Utikal J., Huber C., Loquai C., Türeci Ö. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 21.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T., O’Hagan D.T., Singh M., Mason P.W., Valiante N.M., Dormitzer P.R., Barnett S.W., Rappuoli R., Ulmer J.B., Mandl C.W. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramanathan A., Robb G.B., Chan S.-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Jr., Shi P.-Y., Diamond M.S. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3’-O-methyl)GpppG and 7-methyl (3’-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 25.Grudzien-Nogalska E., Stepinski J., Jemielity J., Zuberek J., Stolarski R., Rhoads R.E., Darzynkiewicz E. Methods Enzymol. Elsevier; 2007. Synthesis of anti-reverse cap analogs (ARCAs) and their applications in mRNA translation and stability; pp. 203–227. [DOI] [PubMed] [Google Scholar]
- 26.Cowling V.H. Regulation of mRNA cap methylation. Biochem. J. 2010;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa M., Murai R., Hagiwara H., Hoshino T., Suyama K. Preparation of eukaryotic mRNA having differently methylated adenosine at the 5’-terminus and the effect of the methyl group in translation. Nucleic Acids Symp. Ser. 2009;53:129–130. doi: 10.1093/nass/nrp065. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski P.J., Warminski M., Kubacka D., Ratajczak T., Nowis D., Kowalska J., Jemielity J. The identity and methylation status of the first transcribed nucleotide in eukaryotic mRNA 5′ cap modulates protein expression in living cells. Nucleic Acids Res. 2020;48:1607–1626. doi: 10.1093/nar/gkaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee S., Pal J.K. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 30.Carralot J.-P., Probst J., Hoerr I., Scheel B., Teufel R., Jung G., Rammensee H.-G., Pascolo S. Polarization of immunity induced by direct injection of naked sequence-stabilized mRNA vaccines. Cell. Mol. Life Sci. 2004;61:2418–2424. doi: 10.1007/s00018-004-4255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babendure J.R. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrom E., Huber M., Aneja M., Dohmen C., Emrich D., Geiger J., Hasenpusch G., Herrmann-Janson A., Kretzschmann V., Mykhailyk O., Pasewald T., Oak P., Hilgendorff A., Wohlleber D., Hoymann H.-G., Schaudien D., Plank C., Rudolph C., Kubisch-Dohmen R. Translation of angiotensin-converting enzyme 2 upon liver- and lung-targeted delivery of optimized chemically modified mRNA. Mol. Ther. Nucleic Acids. 2017;7:350–365. doi: 10.1016/j.omtn.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trepotec Z., Aneja M.K., Geiger J., Hasenpusch G., Plank C., Rudolph C. Maximizing the translational yield of mRNA therapeutics by minimizing 5′-UTRs. Tissue Eng. A. 2019;25:69–79. doi: 10.1089/ten.tea.2017.0485. [DOI] [PubMed] [Google Scholar]
- 34.Zeng C., Hou X., Yan J., Zhang C., Li W., Zhao W., Du S., Dong Y. Leveraging mRNA sequences and nanoparticles to deliver SARS-CoV-2 antigens in vivo. Adv. Mater. 2020;32 doi: 10.1002/adma.202004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Jongh R.P.H., van Dijk A.D.J., Julsing M.K., Schaap P.J., de Ridder D. Designing eukaryotic gene expression regulation using machine learning. Trends Biotechnol. 2020;38:191–201. doi: 10.1016/j.tibtech.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Sample P.J., Wang B., Reid D.W., Presnyak V., McFadyen I.J., Morris D.R., Seelig G. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019;37:803–809. doi: 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckmann C.R., Rammelt C., Wahle E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 38.Godiska R., Mead D., Dhodda V., Wu C., Hochstein R., Karsi A., Usdin K., Entezam A., Ravin N. Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli. Nucleic Acids Res. 2010;38:e88. doi: 10.1093/nar/gkp1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arbuthnot P., Ely A., Bloom K. A convenient method to generate and maintain poly(A)-encoding DNA sequences required for in vitro transcription of mRNA. BioTechniques. 2019;66:37–39. doi: 10.2144/btn-2018-0120. [DOI] [PubMed] [Google Scholar]
- 40.Grier A.E., Burleigh S., Sahni J., Clough C.A., Cardot V., Choe D.C., Krutein M.C., Rawlings D.J., Jensen M.C., Scharenberg A.M., Jacoby K. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded Poly(A) sequences. Mol. Ther. - Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karikó K., Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 42.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine Into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warren L., Manos P.D., Ahfeldt T., Loh Y.-H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A., Daley G.Q., Brack A.S., Collins J.J., Cowan C., Schlaeger T.M., Rossi D.J. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 46.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Controlled Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 47.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaidyanathan S., Azizian K.T., Haque A.K.M.A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. - Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCown P.J., Ruszkowska A., Kunkler C.N., Breger K., Hulewicz J.P., Wang M.C., Springer N.A., Brown J.A. Naturally occurring modified ribonucleosides. WIREs RNA. 2020;11 doi: 10.1002/wrna.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu A.-M., Jian C., Yu A.H., Tu M.-J. RNA therapy: are we using the right molecules? Pharmacol. Ther. 2019;196:91–104. doi: 10.1016/j.pharmthera.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abu Bakar F., Ng L. Nonstructural proteins of alphavirus—potential targets for drug development. Viruses. 2018;10:71. doi: 10.3390/v10020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pepini T., Pulichino A.-M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J., Yu D., Ulmer J.B., Iavarone C. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hekele A., Bertholet S., Archer J., Gibson D.G., Palladino G., Brito L.A., Otten G.R., Brazzoli M., Buccato S., Bonci A., Casini D., Maione D., Qi Z.-Q., Gill J.E., Caiazza N.C., Urano J., Hubby B., Gao G.F., Shu Y., De Gregorio E., Mandl C.W., Mason P.W., Settembre E.C., Ulmer J.B., Craig Venter J., Dormitzer P.R., Rappuoli R., Geall A.J. Rapidly produced SAM ® vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013;2:1–7. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W., Dey A.K., Lilja A., Valiante N.M., Mason P.W., Mandl C.W., Barnett S.W., Dormitzer P.R., Ulmer J.B., Singh M., O’Hagan D.T., Geall A.J. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luisi K., Morabito K.M., Burgomaster K.E., Sharma M., Kong W.-P., Foreman B.M., Patel S., Fisher B., Aleshnick M.A., Laliberte J., Wallace M., Ruckwardt T.J., Gordon D.N., Linton C., Ruggiero N., Cohen J.L., Johnson R., Aggarwal K., Ko S.-Y., Yang E.S., Pelc R.S., Dowd K.A., O’Hagan D., Ulmer J., Mossman S., Sambor A., Lepine E., Mascola J.R., Pierson T.C., Graham B.S., Yu D. Development of a potent Zika virus vaccine using self-amplifying messenger RNA. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blakney A.K., McKay P.F., Shattock R.J. Structural components for amplification of positive and negative strand VEEV splitzicons. Front. Mol. Biosci. 2018;5:71. doi: 10.3389/fmolb.2018.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beissert T., Perkovic M., Vogel A., Erbar S., Walzer K.C., Hempel T., Brill S., Haefner E., Becker R., Türeci Ö., Sahin U. A Trans-amplifying RNA Vaccine Strategy for Induction of Potent Protective Immunity. Mol. Ther. 2020;28:119–128. doi: 10.1016/j.ymthe.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Controlled Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.-G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 62.Lazzaro S., Giovani C., Mangiavacchi S., Magini D., Maione D., Baudner B., Geall A.J., De Gregorio E., D’Oro U., Buonsanti C. CD8 T-cell priming upon mRNA vaccination is restricted to bone-marrow-derived antigen-presenting cells and may involve antigen transfer from myocytes. Immunology. 2015;146:312–326. doi: 10.1111/imm.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindsay K.E., Bhosle S.M., Zurla C., Beyersdorf J., Rogers K.A., Vanover D., Xiao P., Araínga M., Shirreff L.M., Pitard B., Baumhof P., Villinger F., Santangelo P.J. Visualization of early events in mRNA vaccine delivery in non-human primates via PET–CT and near-infrared imaging. Nat. Biomed. Eng. 2019;3:371–380. doi: 10.1038/s41551-019-0378-3. [DOI] [PubMed] [Google Scholar]
- 64.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., Fotin-Mleczek M., Hoerr I., Clemens R., von Sonnenburg F. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 65.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., John S., Hassett K., Yuzhakov O., Bahl K., Brito L.A., Salter H., Ciaramella G., Loré K. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Firdessa-Fite R., Creusot R.J. Nanoparticles versus dendritic cells as vehicles to deliver mRNA encoding multiple epitopes for immunotherapy. Mol. Ther. - Methods Clin. Dev. 2020;16:50–62. doi: 10.1016/j.omtm.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Nuffel A.M., Benteyn D., Wilgenhof S., Pierret L., Corthals J., Heirman C., van der Bruggen P., Coulie P.G., Neyns B., Thielemans K., Bonehill A. Dendritic cells loaded with mRNA encoding full-length tumor antigens prime CD4+ and CD8+ T cells in melanoma patients. Mol. Ther. 2012;20:1063–1074. doi: 10.1038/mt.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Silva N.S., Klein U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Münz C. Antigen processing for MHC class II presentation via autophagy. Front. Immunol. 2012;3 doi: 10.3389/fimmu.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leung C.S.K. Endogenous antigen presentation of MHC class II epitopes through non-autophagic pathways. Front. Immunol. 2015;6 doi: 10.3389/fimmu.2015.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lizée G., Basha G., Jefferies W.A. Tails of wonder: endocytic-sorting motifs key for exogenous antigen presentation. Trends Immunol. 2005;26:141–149. doi: 10.1016/j.it.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 73.Bonehill A., Heirman C., Tuyaerts S., Michiels A., Zhang Y., van der Bruggen P., Thielemans K. Efficient presentation of known HLA class II-restricted MAGE-A3 epitopes by dendritic cells electroporated with messenger RNA encoding an invariant chain with genetic exchange of class II-associated invariant chain peptide. Cancer Res. 2003;63:5587–5594. [PubMed] [Google Scholar]
- 74.Bonehill A., Heirman C., Tuyaerts S., Michiels A., Breckpot K., Brasseur F., Zhang Y., van der Bruggen P., Thielemans K. Messenger RNA-electroporated dendritic cells presenting MAGE-A3 simultaneously in HLA Class I and Class II molecules. J. Immunol. 2004;172:6649–6657. doi: 10.4049/jimmunol.172.11.6649. [DOI] [PubMed] [Google Scholar]
- 75.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci Ö., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 76.Kreiter S., Konrad T., Sester M., Huber C., Türeci Ö., Sahin U. Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA. Cancer Immunol. Immunother. 2007;56:1577–1587. doi: 10.1007/s00262-007-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kreiter S., Selmi A., Diken M., Sebastian M., Osterloh P., Schild H., Huber C., Türeci Ö., Sahin U. Increased antigen presentation efficiency by coupling antigens to MHC Class I trafficking signals. J. Immunol. 2008;180:309–318. doi: 10.4049/jimmunol.180.1.309. [DOI] [PubMed] [Google Scholar]
- 78.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., Wagner W., Granados A., Greenhouse J., Walker M., Willis E., Yu J.-S., McGee C.E., Sempowski G.D., Mui B.L., Tam Y.K., Huang Y.-J., Vanlandingham D., Holmes V.M., Balachandran H., Sahu S., Lifton M., Higgs S., Hensley S.E., Madden T.D., Hope M.J., Karikó K., Santra S., Graham B.S., Lewis M.G., Pierson T.C., Haynes B.F., Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su Z., Dannull J., Yang B.K., Dahm P., Coleman D., Yancey D., Sichi S., Niedzwiecki D., Boczkowski D., Gilboa E., Vieweg J. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8 + and CD4 + T cell responses in patients with metastatic prostate cancer. J. Immunol. 2005;174:3798–3807. doi: 10.4049/jimmunol.174.6.3798. [DOI] [PubMed] [Google Scholar]
- 80.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against zika virus infection. Cell. 2017;168:1114–1125. doi: 10.1016/j.cell.2017.02.017. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Şahin U., Gruber W.C. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Espeseth A.S., Cejas P.J., Citron M.P., Wang D., DiStefano D.J., Callahan C., Donnell G.O., Galli J.D., Swoyer R., Touch S., Wen Z., Antonello J., Zhang L., Flynn J.A., Cox K.S., Freed D.C., Vora K.A., Bahl K., Latham A.H., Smith J.S., Gindy M.E., Ciaramella G., Hazuda D., Shaw C.A., Bett A.J. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. Npj Vaccines. 2020;5:16. doi: 10.1038/s41541-020-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tiwari P.M., Vanover D., Lindsay K.E., Bawage S.S., Kirschman J.L., Bhosle S., Lifland A.W., Zurla C., Santangelo P.J. Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat. Commun. 2018;9:3999. doi: 10.1038/s41467-018-06508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J., Brito L.A., Salter H., Ciaramella G., Loré K. Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., LaBranche C.C., Montefiori D.C., Lobby J.L., Saunders K.O., Liao H.-X., Korber B.T., Sutherland L.L., Scearce R.M., Hraber P.T., Tombácz I., Muramatsu H., Ni H., Balikov D.A., Li C., Mui B.L., Tam Y.K., Krammer F., Karikó K., Polacino P., Eisenlohr L.C., Madden T.D., Hope M.J., Lewis M.G., Lee K.K., Hu L., Hensley S.E., Cancro M.P., Haynes B.F., Weissman D. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trevaskis N.L., Kaminskas L.M., Porter C.J.H. From sewer to saviour — targeting the lymphatic system to promote drug exposure and activity. Nat. Rev. Drug Discov. 2015;14:781–803. doi: 10.1038/nrd4608. [DOI] [PubMed] [Google Scholar]
- 87.Irvine D.J., Aung A., Silva M. Controlling timing and location in vaccines. Adv. Drug Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.06.019. S0169409X2030065X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carrasco Y.R., Batista F.D. B cell recognition of membrane-bound antigen: an exquisite way of sensing ligands. Curr. Opin. Immunol. 2006;18:286–291. doi: 10.1016/j.coi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 89.Gordon S., Plüddemann A., Mukhopadhyay S. Sinusoidal immunity: macrophages at the lymphohematopoietic interface. Cold Spring Harb. Perspect. Biol. 2014;7 doi: 10.1101/cshperspect.a016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heesters B.A., van der Poel C.E., Das A., Carroll M.C. Antigen presentation to B cells. Trends Immunol. 2016;37:844–854. doi: 10.1016/j.it.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 91.Ionescu L., Urschel S. Memory B cells and long-lived plasma cells. Transplantation. 2019;103:890–898. doi: 10.1097/TP.0000000000002594. [DOI] [PubMed] [Google Scholar]
- 92.Lightman S.M., Utley A., Lee K.P. Survival of long-lived plasma cells (LLPC): piecing together the puzzle. Front. Immunol. 2019;10:965. doi: 10.3389/fimmu.2019.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bortnick A., Allman D. What is and what should always have been: long-lived plasma cells induced by T cell-independent antigens. J. Immunol. 2013;190:5913–5918. doi: 10.4049/jimmunol.1300161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., Ojha A., Zhang L., Mu Z., Parks R., Manzoni T.B., Roper B., Strohmeier S., Tombácz I., Arwood L., Nachbagauer R., Karikó K., Greenhouse J., Pessaint L., Porto M., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Lin P.J.C., Tam Y.K., Sempowski G.D., Farzan M., Choe H., Saunders K.O., Haynes B.F., Andersen H., Eisenlohr L.C., Weissman D., Krammer F., Bates P., Allman D., Locci M., Pardi N. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020 doi: 10.1016/j.immuni.2020.07.019. S1074761320303265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schudel A., Francis D.M., Thomas S.N. Material design for lymph node drug delivery. Nat. Rev. Mater. 2019;4:415–428. doi: 10.1038/s41578-019-0110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang L., Wang W., Wang S. Effect of vaccine administration modality on immunogenicity and efficacy. Expert Rev. Vaccines. 2015;14:1509–1523. doi: 10.1586/14760584.2015.1081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jones G.B., Collins D.S., Harrison M.W., Thyagarajapuram N.R., Wright J.M. Subcutaneous drug delivery: An evolving enterprise. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf9166. [DOI] [PubMed] [Google Scholar]
- 98.Liang F., Lindgren G., Sandgren K.J., Thompson E.A., Francica J.R., Seubert A., De Gregorio E., Barnett S., O’Hagan D.T., Sullivan N.J., Koup R.A., Seder R.A., Loré K. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal2094. [DOI] [PubMed] [Google Scholar]
- 99.Ols S., Yang L., Thompson E.A., Pushparaj P., Tran K., Liang F., Lin A., Eriksson B., Karlsson Hedestam G.B., Wyatt R.T., Loré K., et al. Cell Rep. 2020;30:3964–3971. doi: 10.1016/j.celrep.2020.02.111. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Malissen B., Tamoutounour S., Henri S. The origins and functions of dendritic cells and macrophages in the skin. Nat. Rev. Immunol. 2014;14:417–428. doi: 10.1038/nri3683. [DOI] [PubMed] [Google Scholar]
- 101.Havenar-Daughton C., Carnathan D.G., Boopathy A.V., Upadhyay A.A., Murrell B., Reiss S.M., Enemuo C.A., Gebru E.H., Choe Y., Dhadvai P., Viviano F., Kaushik K., Bhiman J.N., Briney B., Burton D.R., Bosinger S.E., Schief W.R., Irvine D.J., Silvestri G., Crotty S. Rapid germinal center and antibody responses in non-human primates after a single nanoparticle vaccine immunization. Cell Rep. 2019;29:1756–1766. doi: 10.1016/j.celrep.2019.10.008. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tan L., Zheng T., Li M., Zhong X., Tang Y., Qin M., Sun X. Optimization of an mRNA vaccine assisted with cyclodextrin-polyethyleneimine conjugates. Drug Deliv. Transl. Res. 2020;10:678–689. doi: 10.1007/s13346-020-00725-4. [DOI] [PubMed] [Google Scholar]
- 103.Mockey M., Bourseau E., Chandrashekhar V., Chaudhuri A., Lafosse S., Le Cam E., Quesniaux V.F.J., Ryffel B., Pichon C., Midoux P. mRNA-based cancer vaccine: prevention of B16 melanoma progression and metastasis by systemic injection of MART1 mRNA histidylated lipopolyplexes. Cancer Gene Ther. 2007;14:802–814. doi: 10.1038/sj.cgt.7701072. [DOI] [PubMed] [Google Scholar]
- 104.Perche F., Benvegnu T., Berchel M., Lebegue L., Pichon C., Jaffrès P.-A., Midoux P. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine Nanotechnol. Biol. Med. 2011;7:445–453. doi: 10.1016/j.nano.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 105.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Hüsemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Türeci Ö., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 106.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K., Shaheen F., Collman R.G., Karikó K., Danet-Desnoyers G.A., Madden T.D., Hope M.J., Weissman D. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8 doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rybakova Y., Kowalski P.S., Huang Y., Gonzalez J.T., Heartlein M.W., DeRosa F., Delcassian D., Anderson D.G. mRNA delivery for therapeutic Anti-HER2 antibody expression in vivo. Mol. Ther. 2019;27:1415–1423. doi: 10.1016/j.ymthe.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim J., Mukherjee A., Nelson D., Jozic A., Sahay G. Rapid generation of circulating and mucosal decoy ACE2 using mRNA nanotherapeutics for the potential treatment of SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.07.24.205583. 2020.07.24.205583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lindsay K.E., Vanover D., Thoresen M., King H., Xiao P., Badial P., Araínga M., Park S.B., Tiwari P.M., Peck H.E., Blanchard E.L., Feugang J.M., Olivier A.K., Zurla C., Villinger F., Woolums A.R., Santangelo P.J. Aerosol delivery of synthetic mRNA to vaginal mucosa leads to durable expression of broadly neutralizing antibodies against HIV. Mol. Ther. 2020;28:805–819. doi: 10.1016/j.ymthe.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li M., Li Y., Peng K., Wang Y., Gong T., Zhang Z., He Q., Sun X. Engineering intranasal mRNA vaccines to enhance lymph node trafficking and immune responses. Acta Biomater. 2017;64:237–248. doi: 10.1016/j.actbio.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 112.Zhuang X., Qi Y., Wang M., Yu N., Nan F., Zhang H., Tian M., Li C., Lu H., Jin N. mRNA vaccines encoding the HA protein of influenza A H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines. 2020;8:123. doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li M., Zhao M., Fu Y., Li Y., Gong T., Zhang Z., Sun X. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Controlled Release. 2016;228:9–19. doi: 10.1016/j.jconrel.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 114.Shakya A.K., Chowdhury M.Y.E., Tao W., Gill H.S. Mucosal vaccine delivery: Current state and a pediatric perspective. J. Controlled Release. 2016;240:394–413. doi: 10.1016/j.jconrel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Woof J.M., Kerr M.A. The function of immunoglobulin A in immunity. J. Pathol. 2006;208:270–282. doi: 10.1002/path.1877. [DOI] [PubMed] [Google Scholar]
- 116.Ensign L.M., Schneider C., Suk J.S., Cone R., Hanes J. Mucus penetrating nanoparticles: biophysical tool and method of drug and gene delivery. Adv. Mater. 2012;24:3887–3894. doi: 10.1002/adma.201201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khutoryanskiy V.V. Beyond PEGylation: alternative surface-modification of nanoparticles with mucus-inert biomaterials. Adv. Drug Deliv. Rev. 2018;124:140–149. doi: 10.1016/j.addr.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 118.Sahay G., Alakhova D.Y., Kabanov A.V. Endocytosis of nanomedicines. J. Controlled Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Akinc A., Battaglia G. Exploiting endocytosis for nanomedicines. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang S., Gao H., Bao G. Physical principles of nanoparticle cellular endocytosis. ACS Nano. 2015;9:8655–8671. doi: 10.1021/acsnano.5b03184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Patel S., Kim J., Herrera M., Mukherjee A., Kabanov A.V., Sahay G. Brief update on endocytosis of nanomedicines. Adv. Drug Deliv. Rev. 2019;144:90–111. doi: 10.1016/j.addr.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stewart M.P., Lorenz A., Dahlman J., Sahay G. Challenges in carrier-mediated intracellular delivery: moving beyond endosomal barriers. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2016;8:465–478. doi: 10.1002/wnan.1377. [DOI] [PubMed] [Google Scholar]
- 123.Kim J., Narayana A., Patel S., Sahay G. Advances in intracellular delivery through supramolecular self-assembly of oligonucleotides and peptides. Theranostics. 2019;9:3191–3212. doi: 10.7150/thno.33921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R., Buganim Y., Schroeder A., Langer R., Anderson D.G. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M., Epstein-Barash H., Zhang L., Koteliansky V., Fitzgerald K., Fava E., Bickle M., Kalaidzidis Y., Akinc A., Maier M., Zerial M. Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 126.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson Ö., Stanton M.G., Benenato K.E. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Johannes L., Jacob R., Leffler H. Galectins at a glance. J. Cell Sci. 2018;131 doi: 10.1242/jcs.208884. [DOI] [PubMed] [Google Scholar]
- 128.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., Manoharan M., Kirchhausen T., Lieberman J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Du Rietz H., Hedlund H., Wilhelmson S., Nordenfelt P., Wittrup A. Imaging small molecule-induced endosomal escape of siRNA. Nat. Commun. 2020;11:1809. doi: 10.1038/s41467-020-15300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kilchrist K.V., Dimobi S.C., Jackson M.A., Evans B.C., Werfel T.A., Dailing E.A., Bedingfield S.K., Kelly I.B., Duvall C.L. Gal8 visualization of endosome disruption predicts carrier-mediated biologic drug intracellular bioavailability. ACS Nano. 2019;13:1136–1152. doi: 10.1021/acsnano.8b05482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rui Y., Wilson D.R., Choi J., Varanasi M., Sanders K., Karlsson J., Lim M., Green J.J. Carboxylated branched poly(β-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aay3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Herrera M., Kim J., Eygeris Y., Jozic A., Sahay G. Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery. Bioengineering. 2020 doi: 10.1101/2020.12.02.407601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yáñez-Mó M., Siljander P.R.-M., Andreu Z., Bedina Zavec A., Borràs F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J., Colás E., Cordeiro-da Silva A., Fais S., Falcon-Perez J.M., Ghobrial I.M., Giebel B., Gimona M., Graner M., Gursel I., Gursel M., Heegaard N.H.H., Hendrix A., Kierulf P., Kokubun K., Kosanovic M., Kralj-Iglic V., Krämer-Albers E.-M., Laitinen S., Lässer C., Lener T., Ligeti E., Linē A., Lipps G., Llorente A., Lötvall J., Manček-Keber M., Marcilla A., Mittelbrunn M., Nazarenko I., Nolte E.N.M., Nyman T.A., O’Driscoll L., Olivan M., Oliveira C., Pállinger É., del Portillo H.A., Reventós J., Rigau M., Rohde E., Sammar M., Sánchez-Madrid F., Santarém N., Schallmoser K., Ostenfeld M. Stampe, Stoorvogel W., Stukelj R., Van der Grein S.G., Vasconcelos M. Helena, Wauben M.H.M., De Wever O. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4 doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Michelle Grandin H., Guillaume-Gentil O., Zambelli T., Mayer M., Houghtaling J., Palivan C.G., Textor M., Höök F. Bioinspired, nanoscale approaches in contemporary bioanalytics. Biointerphases. 2018;13 doi: 10.1116/1.5037582. review. [DOI] [PubMed] [Google Scholar]
- 135.Mendes A.C., Baran E.T., Reis R.L., Azevedo H.S. Self-assembly in nature: Using the principles of nature to create complex nanobiomaterials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013;5:582–612. doi: 10.1002/wnan.1238. [DOI] [PubMed] [Google Scholar]
- 136.Petros R.A., DeSimone J.M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 137.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28 doi: 10.1016/j.nantod.2019.100766. [DOI] [Google Scholar]
- 138.Rizk M., Tuzmen S. Update on the clinical utility of an RNA interference-based treatment: focus on Patisiran. Pharmacogenomics Pers. Med. 2017;10:267–278. doi: 10.2147/PGPM.S87945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 140.Weng Y., Li C., Yang T., Hu B., Zhang M., Guo S., Xiao H., Liang X.-J., Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40 doi: 10.1016/J.BIOTECHADV.2020.107534. [DOI] [PubMed] [Google Scholar]
- 141.Park T.G., Jeong J.H., Kim S.W. Current status of polymeric gene delivery systems. Gene Deliv. Tissue Eng. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 142.Hadinoto K., Sundaresan A., Cheow W.S. Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur. J. Pharm. Biopharm. 2013;85:427–443. doi: 10.1016/j.ejpb.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 143.Zhang R., Billingsley M.M., Mitchell M.J. Biomaterials for vaccine-based cancer immunotherapy. J. Controlled Release. 2018;292:256–276. doi: 10.1016/j.jconrel.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Peng L., Wagner E. Polymeric carriers for nucleic acid delivery: current designs and future directions. Biomacromolecules. 2019;20:3613–3626. doi: 10.1021/acs.biomac.9b00999. [DOI] [PubMed] [Google Scholar]
- 145.Iqbal S., Blenner M., Alexander-Bryant A., Larsen J. Polymersomes for therapeutic delivery of protein and nucleic acid macromolecules: from design to therapeutic applications. Biomacromolecules. 2020;21:1327–1350. doi: 10.1021/acs.biomac.9b01754. [DOI] [PubMed] [Google Scholar]
- 146.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/J.YMTHE.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hajj K.A., Ball R.L., Deluty S.B., Singh S.R., Strelkova D., Knapp C.M., Whitehead K.A. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small. 2019;15:1–7. doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 148.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2 doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 149.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: From concept to clinical reality. Genome Med. 2017;9:1–16. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zeng C., Zhang C., Walker P.G., Dong Y. Top. Microbiol. Immunol; Springer, Berlin, Heidelberg: 2020. Formulation and Delivery Technologies for mRNA Vaccines, in: Curr; pp. 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kulkarni J.A., Darjuan M.M., Mercer J.E., Chen S., van der Meel R., Thewalt J.L., Tam Y.Y.C., Cullis P.R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12:4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 152.Roces C.B., Lou G., Jain N., Abraham S., Thomas A., Halbert G.W., Perrie Y. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jahn A., Stavis S.M., Hong J.S., Vreeland W.N., Devoe D.L., Gaitan M. Microfluidic mixing and the formation of nanoscale lipid vesicles. ACS Nano. 2010;4:2077–2087. doi: 10.1021/nn901676x. [DOI] [PubMed] [Google Scholar]
- 154.Leung A.K.K., Tam Y.Y.C., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 155.MacDonald R.C., MacDonald R.I., Menco B.P.M., Takeshita K., Subbarao N.K., Hu L. Rong. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. BBA Biomembr. 1991;1061:297–303. doi: 10.1016/0005-2736(91)90295-J. [DOI] [PubMed] [Google Scholar]
- 156.Dittrich P.S., Heule M., Renaud P., Manz A. On-chip extrusion of lipid vesicles and tubes through microsized apertures. Lab. Chip. 2006;6:488–493. doi: 10.1039/b517670k. [DOI] [PubMed] [Google Scholar]
- 157.Hope M.J., Bally M.B., Webb G., Cullis P.R. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. BBA - Biomembr. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 158.Mayer L.D., Hope M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. BBA - Biomembr. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 159.Samaridou E., Heyes J., Lutwyche P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020;154–155:37–63. doi: 10.1016/j.addr.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 160.Kim H., Sung J., Chang Y., Alfeche A., Leal C. Microfluidics synthesis of gene silencing cubosomes. ACS Nano. 2018;12:9196–9205. doi: 10.1021/acsnano.8b03770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. - Nucleic Acids. 2012;1 doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kulkarni J.A., Witzigmann D., Leung J., van der Meel R., Zaifman J., Darjuan M.M., Grisch-Chan H.M., Thöny B., Tam Y.Y.C., Cullis P.R. Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale. 2019;11:9023–9031. doi: 10.1039/C9NR02004G. [DOI] [PubMed] [Google Scholar]
- 163.Eygeris Y., Patel S., Jozic A., Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hou X., Zhang X., Zhao W., Zeng C., Deng B., McComb D.W., Du S., Zhang C., Li W., Dong Y. Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis. Nat. Nanotechnol. 2020;15:41–46. doi: 10.1038/s41565-019-0600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Miao L., Lin J., Huang Y., Li L., Delcassian D., Ge Y., Shi Y., Anderson D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020;11:2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Peeri M., Tuller T. High-resolution modeling of the selection on local mRNA folding strength in coding sequences across the tree of life. Genome Biol. 2020;21:63. doi: 10.1186/s13059-020-01971-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mauger D.M., Cabral B.J., Presnyak V., Su S.V., Reid D.W., Goodman B., Link K., Khatwani N., Reynders J., Moore M.J., McFadyen I.J. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. U. S. A. 2019;116:24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wu Y., Phillips J.A., Liu H., Yang R., Tan W. Carbon nanotubes protect DNA strands during cellular delivery. ACS Nano. 2008;2:2023–2028. doi: 10.1021/nn800325a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rafael D., Andrade F., Arranja A., Luís S., Videira M. Lipoplexes and polyplexes: gene therapy. Encycl Biomed Polym Polym Biomater. 2015:4335–4347. [Google Scholar]
- 171.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. 1987;84:7413. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stamatatos L., Leventis R., Zuckermann M.J., Silvius J.R. Interactions of cationic lipid vesicles with negatively charged phospholipid vesicles and biological membranes. Biochemistry. 1988;27:3917–3925. doi: 10.1021/bi00411a005. [DOI] [PubMed] [Google Scholar]
- 173.Behr J.P., Demeneix B., Loeffler J.P., Perez-Mutul J. Efficient gene transfer into mammalian primary endocrine cells with lipopolyamine-coated DNA. Proc. Natl. Acad. Sci. 1989;86:6982. doi: 10.1073/pnas.86.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Controlled Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 176.Zimmermann T.S., Lee A.C.H., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., Harborth J., Heyes J.A., Jeffs L.B., John M., Judge A.D., Lam K., McClintock K., Nechev L.V., Palmer L.R., Racie T., Röhl I., Seiffert S., Shanmugam S., Sood V., Soutschek J., Toudjarska I., Wheat A.J., Yaworski E., Zedalis W., Koteliansky V., Manoharan M., Vornlocher H.-P., MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 177.Lin P.J.C., Tam Y.Y.C., Hafez I., Sandhu A., Chen S., Ciufolini M.A., Nabi I.R., Cullis P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomedicine Nanotechnol. Biol. Med. 2013;9:233–246. doi: 10.1016/j.nano.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 178.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W.Y., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., de Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 179.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., Mui B.L., Semple S.C., Tam Y.K., Ciufolini M., Witzigmann D., Kulkarni J.A., van der Meel R., Cullis P.R. The onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 181.Patel S., Ryals R.C., Weller K.K., Pennesi M.E., Sahay G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Controlled Release. 2019;303:91–100. doi: 10.1016/j.jconrel.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nabhan J.F., Wood K.M., Rao V.P., Morin J., Bhamidipaty S., LaBranche T.P., Gooch R.L., Bozal F., Bulawa C.E., Guild B.C. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. 2017;114 doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Robinson E., MacDonald K.D., Slaughter K., McKinney M., Patel S., Sun C., Sahay G. Lipid nanoparticle-delivered chemically modified mRNA restores chloride secretion in cystic fibrosis. Mol. Ther. 2018;26:2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Veiga N., Goldsmith M., Granot Y., Rosenblum D., Dammes N., Kedmi R., Ramishetti S., Peer D. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9:4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sedic M., Senn J.J., Lynn A., Laska M., Smith M., Platz S.J., Bolen J., Hoge S., Bulychev A., Jacquinet E., Bartlett V., Smith P.F. Safety evaluation of lipid nanoparticle–formulated modified mRNA in the Sprague-Dawley rat and cynomolgus monkey. Vet. Pathol. 2017;55:341–354. doi: 10.1177/0300985817738095. [DOI] [PubMed] [Google Scholar]
- 187.Zhang M., Sun J., Li M., Jin X. Modified mRNA-LNP Vaccines Confer Protection against Experimental DENV-2 Infection in Mice. Mol. Ther. - Methods Clin. Dev. 2020;18:702–712. doi: 10.1016/j.omtm.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kim J., Jozic A., Sahay G. Naturally derived membrane lipids impact nanoparticle-based messenger RNA delivery. Cell. Mol. Bioeng. 2020 doi: 10.1007/s12195-020-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., Zhang X., Wang Q., Panesar S., Hutabarat R., Carioto M., Hettinger J., Kandasamy P., Butler D., Rajeev K.G., Pang B., Charisse K., Fitzgerald K., Mui B.L., Du X., Cullis P., Madden T.D., Hope M.J., Manoharan M., Akinc A. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gilham D., Lehner R. Techniques to measure lipase and esterase activity in vitro. Mol. Cell Biol. Lipids. 2005;36:139–147. doi: 10.1016/j.ymeth.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 191.Wong H., Schotz M.C. The lipase gene family. J. Lipid Res. 2002;43:993–999. doi: 10.1194/jlr.R200007-JLR200. [DOI] [PubMed] [Google Scholar]
- 192.Sato Y., Hashiba K., Sasaki K., Maeki M., Tokeshi M., Harashima H. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J. Controlled Release. 2019;295:140–152. doi: 10.1016/j.jconrel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 193.Shobaki N., Sato Y., Suzuki Y., Okabe N., Harashima H. Manipulating the function of tumor-associated macrophages by siRNA-loaded lipid nanoparticles for cancer immunotherapy. J. Controlled Release. 2020;325:235–248. doi: 10.1016/J.JCONREL.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 194.Huang E.Y.-C., Tse S.-W., Iacovelli J., Mckinney K., Hopson K. Messenger ribonucleic acids for enhancing immune responses and methods of use thereof. 2018. https://patents.google.com/patent/WO2018081459A1/en WO2018081459A1. (accessed December 21, 2020)
- 195.Ansell S.M., Du X. Lipids and lipid nanoparticle formulations for delivery of nucleic acids, US10166298B2. 2019. https://patents.google.com/patent/US10166298B2/en?oq=US10166298B2
- 196.Tanaka H., Sakurai Y., Anindita J., Akita H. Development of lipid-like materials for RNA delivery based on intracellular environment-responsive membrane destabilization and spontaneous collapse. Adv. Drug Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 197.Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L., Xu Q., Wang M. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019;31 doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tanaka H., Takahashi T., Konishi M., Takata N., Gomi M., Shirane D., Miyama R., Hagiwara S., Yamasaki Y., Sakurai Y. Self-degradable lipid-like materials based on “hydrolysis accelerated by the intra-particle enrichment of reactant (HyPER)” for messenger RNA delivery. Adv. Funct. Mater. 2020;30 [Google Scholar]
- 199.Goldberg M. In: RNA Interf. Biol. Ther. Howard K.A., editor. Springer US; Boston, MA: 2013. Lipidoids: A Combinatorial Approach to siRNA Delivery; pp. 143–160. [DOI] [Google Scholar]
- 200.Akinc A., Zumbuehl A., Goldberg M., Leshchiner E.S., Busini V., Hossain N., Bacallado S.A., Nguyen D.N., Fuller J., Alvarez R., Borodovsky A., Borland T., Constien R., de Fougerolles A., Dorkin J.R., Narayanannair Jayaprakash K., Jayaraman M., John M., Koteliansky V., Manoharan M., Nechev L., Qin J., Racie T., Raitcheva D., Rajeev K.G., Sah D.W.Y., Soutschek J., Toudjarska I., Vornlocher H.-P., Zimmermann T.S., Langer R., Anderson D.G. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Love K.T., Mahon K.P., Levins C.G., Whitehead K.A., Querbes W., Dorkin J.R., Qin J., Cantley W., Qin L.L., Racie T., Frank-Kamenetsky M., Yip K.N., Alvarez R., Sah D.W.Y., de Fougerolles A., Fitzgerald K., Koteliansky V., Akinc A., Langer R., Anderson D.G. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. 2010;107:1864. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Turnbull I.C., Eltoukhy A.A., Fish K.M., Nonnenmacher M., Ishikawa K., Chen J., Hajjar R.J., Anderson D.G., Costa K.D. Myocardial delivery of lipidoid nanoparticle carrying modRNA induces rapid and transient expression. Mol. Ther. 2016;24:66–75. doi: 10.1038/mt.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., Bogorad R.L., Yin H., Chen Y., Vegas A.J., Alabi C.A., Sahay G., Olejnik K.T., Wang W., Schroeder A., Lytton-Jean A.K.R., Siegwart D.J., Akinc A., Barnes C., Barros S.A., Carioto M., Fitzgerald K., Hettinger J., Kumar V., Novobrantseva T.I., Qin J., Querbes W., Koteliansky V., Langer R., Anderson D.G. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. 2014;111:3955. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yin H., Song C.-Q., Suresh S., Wu Q., Walsh S., Rhym L.H., Mintzer E., Bolukbasi M.F., Zhu L.J., Kauffman K., Mou H., Oberholzer A., Ding J., Kwan S.-Y., Bogorad R.L., Zatsepin T., Koteliansky V., Wolfe S.A., Xue W., Langer R., Anderson D.G. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fenton O.S., Kauffman K.J., McClellan R.L., Appel E.A., Dorkin J.R., Tibbitt M.W., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F., Yang J.H., Oberli M.A., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater. 2017;29 doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 207.Li B., Luo X., Deng B., Wang J., McComb D.W., Shi Y., Gaensler K.M.L., Tan X., Dunn A.L., Kerlin B.A., Dong Y. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jiang C., Mei M., Li B., Zhu X., Zu W., Tian Y., Wang Q., Guo Y., Dong Y., Tan X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27:440–443. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/J.ADDR.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 210.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., Derosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 211.L. V Schäfer, D.H. De Jong, A. Holt, A.J. Rzepiela, A.H. De Vries, B. Poolman, J.A. Killian, S.J. Marrink, Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes, Proc. Natl. Acad. Sci. U. S. A. 108 (2011) 1343–1348. doi.10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed]
- 212.Israelachvili J.N., Mitchell D.J., Ninham B.W. Theory of self-assembly of lipid bilayers and vesicles. BBA - Biomembr. 1977;470:185–201. doi: 10.1016/0005-2736(77)90099-2. [DOI] [PubMed] [Google Scholar]
- 213.Carnie S., Israelachvili J.N., Pailthorpe B.A. Lipid packing and transbilayer asymmetries of mixed lipid vesicles. Biochim. Biophys. Acta BBA - Biomembr. 1979;554:340–357. doi: 10.1016/0005-2736(79)90375-4. [DOI] [PubMed] [Google Scholar]
- 214.Sun J., Zhang L., Wang J., Feng Q., Liu D., Yin Q., Xu D., Wei Y., Ding B., Shi X., Jiang X. Tunable rigidity of (polymeric core)-(lipid shell) nanoparticles for regulated cellular uptake. Adv. Mater. 2015;27:1402–1407. doi: 10.1002/adma.201404788. [DOI] [PubMed] [Google Scholar]
- 215.Villamil Giraldo A.M., Kasson P.M. Bilayer-coated nanoparticles reveal how influenza viral entry depends on membrane deformability but not curvature. J. Phys. Chem. Lett. 2020:7190–7196. doi: 10.1021/acs.jpclett.0c01778. [DOI] [PubMed] [Google Scholar]
- 216.Patel S., Ashwanikumar N., Robinson E., Xia Y., Mihai C., Griffith J.P., Hou S., Esposito A.A., Ketova T., Welsher K., Joyal J.L., Almarsson Ö., Sahay G. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Heyes J., Hall K., Tailor V., Lenz R., MacLachlan I. Synthesis and characterization of novel poly(ethylene glycol)-lipid conjugates suitable for use in drug delivery. J. Controlled Release. 2006;112:280–290. doi: 10.1016/J.JCONREL.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 218.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y.C., Lin P.J., Chen S., Narayanannair J.K., Rajeev K.G., Manoharan M., Akinc A., Maier M.A., Cullis P., Madden T.D., Hope M.J. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. - Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Leung A.K.K., Tam Y.Y.C., Cullis P.R. Lipid nanoparticles for short interfering RNA delivery. Adv. Genet. 2014;88:71–110. doi: 10.1016/B978-0-12-800148-6.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jokerst J.V., Lobovkina T., Zare R.N., Gambhir S.S. Nanoparticle PEGylation for imaging and therapy. Nanomed. Lond. 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Xu Q., Ensign L.M., Boylan N.J., Schön A., Gong X., Yang J.-C., Lamb N.W., Cai S., Yu T., Freire E., Hanes J. Impact of surface polyethylene glycol (PEG) density on biodegradable nanoparticle transport in mucus ex vivo and distribution in vivo. ACS Nano. 2015;9:9217–9227. doi: 10.1021/acsnano.5b03876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hashizaki K., Taguchi H., Itoh C., Sakai H., Abe M., Saito Y., Ogawa N. Effects of poly(ethylene glycol) (PEG) concentration on the permeability of PEG-grafted liposomes. Chem. Pharm. Bull. (Tokyo). 2005;53:27–31. doi: 10.1248/cpb.53.27. [DOI] [PubMed] [Google Scholar]
- 223.Hashizaki K., Taguchi H., Itoh C., Sakai H., Abe M., Saito Y., Ogawa N. Effects of poly(ethylene glycol) (PEG) chain length of PEG-lipid on the permeability of liposomal bilayer membranes. Chem. Pharm. Bull. (Tokyo). 2003;51:815–820. doi: 10.1248/cpb.51.815. [DOI] [PubMed] [Google Scholar]
- 224.Abu Lila A.S., Kiwada H., Ishida T. The accelerated blood clearance (ABC) phenomenon: Clinical challenge and approaches to manage. J. Controlled Release. 2013;172:38–47. doi: 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 225.Suzuki T., Suzuki Y., Hihara T., Kubara K., Kondo K., Hyodo K., Yamazaki K., Ishida T., Ishihara H. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: Faster PEG shedding attenuates anti-PEG IgM production. Int. J. Pharm. 2020;588 doi: 10.1016/J.IJPHARM.2020.119792. [DOI] [PubMed] [Google Scholar]
- 226.Zhao P., Hou X., Yan J., Du S., Xue Y., Li W., Xiang G., Dong Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact. Mater. 2020;5:358–363. doi: 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Blakney A.K., Yilmaz G., McKay P.F., Becer C.R., Shattock R.J. One size does not fit all: the effect of chain length and charge density of poly(ethylene imine) based copolymers on delivery of pDNA, mRNA, and RepRNA polyplexes. Biomacromolecules. 2018;19:2870–2879. doi: 10.1021/acs.biomac.8b00429. [DOI] [PubMed] [Google Scholar]
- 228.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Controlled Release. 2006;114:100–109. doi: 10.1016/J.JCONREL.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 229.Wahane A., Waghmode A., Kapphahn A., Dhuri K., Gupta A., Bahal R. Role of lipid-based and polymer-based non-viral vectors in nucleic acid delivery for next-generation gene therapy. Molecules. 2020;25:2866. doi: 10.3390/molecules25122866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Miyazaki T., Uchida S., Nagatoishi S., Koji K., Hong T., Fukushima S., Tsumoto K., Ishihara K., Kataoka K., Cabral H. Polymeric nanocarriers with controlled chain flexibility boost mRNA delivery in vivo through enhanced structural fastening. Adv. Healthc. Mater. 2020;9 doi: 10.1002/adhm.202000538. [DOI] [PubMed] [Google Scholar]
- 231.Jiang Y., Lu Q., Wang Y., Xu E., Ho A., Singh P., Wang Y., Jiang Z., Yang F., Tietjen G.T., Cresswell P., Saltzman W.M. Quantitating endosomal escape of a library of polymers for mRNA delivery. Nano Lett. 2020;20:1117–1123. doi: 10.1021/acs.nanolett.9b04426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jiang Y., Gaudin A., Zhang J., Agarwal T., Song E., Kauffman A.C., Tietjen G.T., Wang Y., Jiang Z., Cheng C.J., Saltzman W.M. A “top-down” approach to actuate poly(amine-co-ester) terpolymers for potent and safe mRNA delivery. Biomaterials. 2018;176:122–130. doi: 10.1016/J.BIOMATERIALS.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Coolen A.-L., Lacroix C., Mercier-Gouy P., Delaune E., Monge C., Exposito J.-Y., Verrier B. Poly(lactic acid) nanoparticles and cell-penetrating peptide potentiate mRNA-based vaccine expression in dendritic cells triggering their activation. Biomaterials. 2019;195:23–37. doi: 10.1016/J.BIOMATERIALS.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 234.Gebhart C.L., Kabanov A.V. Evaluation of polyplexes as gene transfer agents. J. Controlled Release. 2001;73:401–416. doi: 10.1016/S0168-3659(01)00357-1. [DOI] [PubMed] [Google Scholar]
- 235.Yan Y., Xiong H., Zhang X., Cheng Q., Siegwart D.J. Systemic mRNA delivery to the lungs by functional polyester-based carriers. Biomacromolecules. 2017;18:4307–4315. doi: 10.1021/acs.biomac.7b01356. [DOI] [PubMed] [Google Scholar]
- 236.Fornaguera C., Guerra-Rebollo M., Ángel Lázaro M., Castells-Sala C., Meca-Cortés O., Ramos-Pérez V., Cascante A., Rubio N., Blanco J., Borrós S. mRNA delivery system for targeting antigen-presenting cells in vivo. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201800335. [DOI] [PubMed] [Google Scholar]
- 237.Sharma R., Agrawal U., Mody N., Vyas S.P. Polymer nanotechnology based approaches in mucosal vaccine delivery: challenges and opportunities. Biotechnol. Adv. 2015;33:64–79. doi: 10.1016/j.biotechadv.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 238.Vaghasiya K., Sharma A., Ray E., Adlakha S., Verma R.K. 2020. Methods to Characterize Nanoparticles for Mucosal Drug Delivery. AAPS/Springer. [DOI] [Google Scholar]
- 239.Lai S.K., Wang Y.Y., Hanes J. Mucus-penetrating nanoparticles for drug and gene delivery to mucosal tissues. Adv. Drug Deliv. Rev. 2009;61:158–171. doi: 10.1016/j.addr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Patel A.K., Kaczmarek J.C., Bose S., Kauffman K.J., Mir F., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 2019;31:1–7. doi: 10.1002/adma.201805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Deng Z., Liu S. Emerging trends in solution self-assembly of block copolymers. Polymer. 2020;207 doi: 10.1016/J.POLYMER.2020.122914. [DOI] [Google Scholar]
- 242.Wang Y., Ye M., Xie R., Gong S. Enhancing the in vitro and in vivo stabilities of polymeric nucleic acid delivery nanosystems. Bioconjug. Chem. 2019;30:325–337. doi: 10.1021/acs.bioconjchem.8b00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Chen R., Zhang H., Yan J., Bryers J.D. Scaffold-mediated delivery for non-viral mRNA vaccines. Gene Ther. 2018;25:556–567. doi: 10.1038/s41434-018-0040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yan J., Chen R., Zhang H., Bryers J.D. Injectable biodegradable chitosan-alginate 3D porous gel scaffold for mRNA vaccine delivery. Macromol. Biosci. 2019;19 doi: 10.1002/mabi.201800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mitragotri S., Lahann J. Physical approaches to biomaterial design. Nat. Mater. 2009;8:15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Cabral H., Matsumoto Y., Mizuno K., Chen Q., Murakami M., Kimura M., Terada Y., Kano M.R., Miyazono K., Uesaka M., Nishiyama N., Kataoka K. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 2011;6:815–823. doi: 10.1038/nnano.2011.166. [DOI] [PubMed] [Google Scholar]
- 247.Chen S., Tam Y.Y.C., Lin P.J.C., Leung A.K.K., Tam Y.K., Cullis P.R. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J. Controlled Release. 2014;196:106–112. doi: 10.1016/j.jconrel.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 248.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Controlled Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 249.Merkel O.M., Rubinstein I., Kissel T. siRNA delivery to the lung: what’s new? Adv. Drug Deliv. Rev. 2014;75:112–128. doi: 10.1016/j.addr.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Li L.D., Crouzier T., Sarkar A., Dunphy L., Han J., Ribbeck K. Spatial configuration and composition of charge modulates transport into a mucin hydrogel barrier. Biophys. J. 2013;105:1357–1365. doi: 10.1016/j.bpj.2013.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Malone R.W., Felgner P.L., Verma I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hafez I., Maurer N., Cullis P. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 253.Viricel W., Poirier S., Mbarek A., Derbali R.M., Mayer G., Leblond J. Cationic switchable lipids: PH-triggered molecular switch for siRNA delivery. Nanoscale. 2017;9:31–36. doi: 10.1039/c6nr06701h. [DOI] [PubMed] [Google Scholar]
- 254.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson Ö., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. - Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Barua S., Mitragotri S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: A review of current status and future prospects. Nano Today. 2014;9:223–243. doi: 10.1016/j.nantod.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Gratton S.E.A., Ropp P.A., Pohlhaus P.D., Luft J.C., Madden V.J., Napier M.E., DeSimone J.M. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Brosey C.A., Tainer J.A. Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr. Opin. Struct. Biol. 2019;58:197–213. doi: 10.1016/J.SBI.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Stewart P.L. Cryo-electron microscopy and cryo-electron tomography of nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017;9 doi: 10.1002/wnan.1417. [DOI] [PubMed] [Google Scholar]
- 260.Kuntsche J., Horst J.C., Bunjes H. Cryogenic transmission electron microscopy (cryo-TEM) for studying the morphology of colloidal drug delivery systems. Int. J. Pharm. 2011;417:120–137. doi: 10.1016/j.ijpharm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 261.Helvig S., Azmi I.D.M., Moghimi S.M., Yaghmur A. Recent advances in Cryo-TEM imaging of soft lipid nanoparticles. AIMS Biophys. 2015;2:116–130. doi: 10.3934/biophy.2015.2.116. [DOI] [Google Scholar]
- 262.Mulet X., Gong X., Waddington L.J., Drummond C.J. Observing self-assembled lipid nanoparticles building order and complexity through low-energy transformation processes. ACS Nano. 2009;3:2789–2797. doi: 10.1021/nn900671u. [DOI] [PubMed] [Google Scholar]
- 263.Barriga H.M.G., Holme M.N., Stevens M.M. Cubosomes: the next generation of smart lipid nanoparticles? Angew. Chem. Int. Ed. 2019;58:2958–2978. doi: 10.1002/anie.201804067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ambegia E., Ansell S., Cullis P., Heyes J., Palmer L., MacLachlan I. Stabilized plasmid–lipid particles containing PEG-diacylglycerols exhibit extended circulation lifetimes and tumor selective gene expression. Biochim. Biophys. Acta BBA - Biomembr. 2005;1669:155–163. doi: 10.1016/j.bbamem.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 266.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., Butler J.S., Qin L., Racie T., Sprague A., Fava E., Zeigerer A., Hope M.J., Zerial M., Sah D.W., Fitzgerald K., Tracy M.A., Manoharan M., Koteliansky V., de Fougerolles A., Maier M.A. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Schöttler S., Becker G., Winzen S., Steinbach T., Mohr K., Landfester K., Mailänder V., Wurm F.R. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016;11:372–377. doi: 10.1038/nnano.2015.330. [DOI] [PubMed] [Google Scholar]
- 268.Zhang Y., Wu J.L.Y., Lazarovits J., Chan W.C.W. An analysis of the binding function and structural organization of the protein corona. J. Am. Chem. Soc. 2020;142:8827–8836. doi: 10.1021/jacs.0c01853. [DOI] [PubMed] [Google Scholar]
- 269.Paunovska K., Sago C.D., Monaco C.M., Hudson W.H., Castro M.G., Rudoltz T.G., Kalathoor S., Vanover D.A., Santangelo P.J., Ahmed R., Bryksin A.V., Dahlman J.E. A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett. 2018;18:2148–2157. doi: 10.1021/acs.nanolett.8b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Francia V., Schiffelers R.M., Cullis P.R., Witzigmann D. The biomolecular corona of lipid nanoparticles for gene therapy. Bioconjug. Chem. 2020 doi: 10.1021/acs.bioconjchem.0c00366. [DOI] [PubMed] [Google Scholar]
- 271.Tokatlian T., Read B.J., Jones C.A., Kulp D.W., Menis S., Chang J.Y.H., Steichen J.M., Kumari S., Allen J.D., Dane E.L., Liguori A., Sangesland M., Lingwood D., Crispin M., Schief W.R., Irvine D.J. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2019;363:649–654. doi: 10.1126/science.aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Conniot J., Scomparin A., Peres C., Yeini E., Pozzi S., Matos A.I., Kleiner R., Moura L.I.F., Zupančič E., Viana A.S., Doron H., Gois P.M.P., Erez N., Jung S., Satchi-Fainaro R., Florindo H.F. Immunization with mannosylated nanovaccines and inhibition of the immune-suppressing microenvironment sensitizes melanoma to immune checkpoint modulators. Nat. Nanotechnol. 2019;14:891–901. doi: 10.1038/s41565-019-0512-0. [DOI] [PubMed] [Google Scholar]
- 273.Le Bihan O., Bonnafous P., Marak L., Bickel T., Trépout S., Mornet S., De Haas F., Talbot H., Taveau J.-C., Lambert O. Cryo-electron tomography of nanoparticle transmigration into liposome. J. Struct. Biol. 2009;168:419–425. doi: 10.1016/J.JSB.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 274.Siewert C.D., Haas H., Cornet V., Nogueira S.S., Nawroth T., Uebbing L., Ziller A., Al-Gousous J., Radulescu A., Schroer M.A., Blanchet C.E., Svergun D.I., Radsak M.P., Sahin U., Langguth P. Hybrid biopolymer and lipid nanoparticles with improved transfection efficacy for mRNA. Cells. 2020;9:1–19. doi: 10.3390/cells9092034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ziller A., Nogueira S.S., Hühn E., Funari S.S., Brezesinski G., Hartmann H., Sahin U., Haas H., Langguth P. Incorporation of mRNA in Lamellar Lipid Matrices for Parenteral Administration. Mol. Pharm. 2018;15:642–651. doi: 10.1021/acs.molpharmaceut.7b01022. [DOI] [PubMed] [Google Scholar]
- 276.Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced nmr spectroscopy. J. Phys. Chem. B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- 277.Tanaka H., Takahashi T., Konishi M., Takata N., Gomi M., Shirane D., Miyama R., Hagiwara S., Yamasaki Y., Sakurai Y., Ueda K., Higashi K., Moribe K., Shinsho E., Nishida R., Fukuzawa K., Yonemochi E., Okuwaki K., Mochizuki Y., Nakai Y., Tange K., Yoshioka H., Tamagawa S., Akita H. Self-degradable lipid-like materials based on “hydrolysis accelerated by the intra-particle enrichment of reactant (HyPER)” for messenger RNA delivery. Adv. Funct. Mater. 2020;34(34):1910575. doi: 10.1002/adfm.201910575. [DOI] [Google Scholar]
- 278.Sampath J., Alamdari S., Pfaendtner J. Closing the gap between modeling and experiments in the self-assembly of biomolecules at interfaces and in solution. Chem. Mater. 2020 doi: 10.1021/acs.chemmater.0c01891. acs.chemmater.0c01891. [DOI] [Google Scholar]
- 279.Seiffert S. Microfluidics and macromolecules: top-down analytics and bottom-up engineering of soft matter at small scales. Macromol. Chem. Phys. 2017;218 doi: 10.1002/macp.201600280. [DOI] [Google Scholar]
- 280.Otten A., Köster S., Struth B., Snigirev A., Pfohl T. Microfluidics of soft matter investigated by small-angle X-ray scattering. J. Synchrotron Radiat. 2005;12:745–750. doi: 10.1107/S0909049505013580. [DOI] [PubMed] [Google Scholar]
- 281.Cordeiro A.S., Crecente-Campo J., Bouzo B.L., González S.F., de la Fuente M., Alonso M.J. Engineering polymeric nanocapsules for an efficient drainage and biodistribution in the lymphatic system. J. Drug Target. 2019;27:646–658. doi: 10.1080/1061186X.2018.1561886. [DOI] [PubMed] [Google Scholar]
- 282.Wang F., Flanagan J., Su N., Wang L.-C., Bui S., Nielson A., Wu X., Vo H.-T., Ma X.-J., Luo Y. RNAscope. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Dammes N., Peer D. Special issue: advances in drug delivery systems paving the road for RNA therapeutics. Trends Pharmacol. Sci. 2020;41:755–775. doi: 10.1016/j.tips.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Poland G.A., Ovsyannikova I.G., Crooke S.N., Kennedy R.B. SARS-CoV-2 vaccine development: current status. Mayo Clin. Proc. 2020;95:2172–2188. doi: 10.1016/j.mayocp.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Tregoning J.S., Brown E.S., Cheeseman H.M., Flight K.E., Higham S.L., Lemm N.-M., Pierce B.F., Stirling D.C., Wang Z., Pollock K.M. Vaccines for COVID-19. Clin. Exp. Immunol. 2020 doi: 10.1111/cei.13517. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Samrat S.K., Tharappel A.M., Li Z., Li H. Prospect of SARS-CoV-2 spike protein: Potential role in vaccine and therapeutic development. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.CDC Coronavirus disease 2019 (COVID-19) Cent. Dis. Control Prev. 2020 https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/index.html (accessed October 24, 2020) [Google Scholar]
- 294.Batty C.J., Heise M.T., Bachelder E.M., Ainslie K.M. Vaccine formulations in clinical development for the prevention of severe acute respiratory syndrome coronavirus 2 infection. Adv. Drug Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.12.006. S0169409X20302775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.World Health Organization Draft landscape of COVID-19 candidate vaccines. 2020. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed December 21, 2020)
- 296.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Şahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 297.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., Brachtendorf S., Lörks V., Sikorski J., Hilker R., Becker D., Eller A.-K., Grützner J., Boesler C., Rosenbaum C., Kühnle M.-C., Luxemburger U., Kemmer-Brück A., Langer D., Bexon M., Bolte S., Karikó K., Palanche T., Fischer B., Schultz A., Shi P.-Y., Fontes-Garfias C., Perez J.L., Swanson K.A., Loschko J., Scully I.L., Cutler M., Kalina W., Kyratsous C.A., Cooper D., Dormitzer P.R., Jansen K.U., Türeci Ö. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 298.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., Pascal K., Maurus D., Brachtendorf S., Lörks V., Sikorski J., Koch P., Hilker R., Becker D., Eller A.-K., Grützner J., Tonigold M., Boesler C., Rosenbaum C., Heesen L., Kühnle M.-C., Poran A., Dong J.Z., Luxemburger U., Kemmer-Brück A., Langer D., Bexon M., Bolte S., Palanche T., Schultz A., Baumann S., Mahiny A.J., Boros G., Reinholz J., Szabó G.T., Karikó K., Shi P.-Y., Fontes-Garfias C., Perez J.L., Cutler M., Cooper D., Kyratsous C.A., Dormitzer P.R., Jansen K.U., Türeci Ö. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. Infectious Diseases (except HIV/AIDS) 2020 doi: 10.1101/2020.12.09.20245175. [DOI] [Google Scholar]
- 299.Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 300.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034577. NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Vaccines and Related Biological Products Advisory Committee December 10, 2020 Meeting Announcement. FDA; 2020. https://www.fda.gov/advisory-committees/advisory-committee-calendar/vaccines-and-related-biological-products-advisory-committee-december-10-2020-meeting-announcement [Google Scholar]
- 302.Ledford H., Cyranoski D., Van Noorden R. The UK has approved a COVID vaccine — here’s what scientists now want to know. Nature. 2020;588:205–206. doi: 10.1038/d41586-020-03441-8. [DOI] [PubMed] [Google Scholar]
- 303.Ledford H. US authorization of first COVID vaccine marks new phase in safety monitoring. Nature. 2020;588:377–378. doi: 10.1038/d41586-020-03542-4. [DOI] [PubMed] [Google Scholar]
- 304.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Vaccines and Related Biological Products Advisory Committee December 17, 2020 Meeting Announcement. FDA; 2020. https://www.fda.gov/advisory-committees/advisory-committee-calendar/vaccines-and-related-biological-products-advisory-committee-december-17-2020-meeting-announcement [Google Scholar]
- 306.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R., Guo Y., Sun S.-H., Fan H., Zu L., Chen Q., He Q., Cao T.-S., Huang X.-Y., Qiu H.-Y., Nie J.-H., Jiang Y., Yan H.-Y., Ye Q., Zhong X., Xue X.-L., Zha Z.-Y., Zhou D., Yang X., Wang Y.-C., Ying B., Qin C.-F., Thermostable A. mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283. doi: 10.1016/j.cell.2020.07.024. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.de Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., Yau C., Matsuda D., Allen E., Hartman P., Park J., Alayyoubi M., Bhaskaran H., Dukanovic A., Bao B., Clemente B., Vega J., Roberts S., Gonzalez J.A., Sablad M., Yelin R., Taylor W., Tachikawa K., Parker S., Karmali P., Davis J., Sullivan S.M., Hughes S.G., Chivukula P., Ooi E.E. A single dose of self-transcribing and replicating RNA based SARS-CoV-2 vaccine produces protective adaptive immunity In mice. BioRxiv. 2020 doi: 10.1101/2020.09.03.280446. 2020.09.03.280446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kremsner P., Mann P., Bosch J., Fendel R., Gabor J.J., Kreidenweiss A., Kroidl A., Leroux-Roels I., Leroux-Roels G., Schindler C., Schunk M., Velavan T.P., Fotin-Mleczek M., Müller S., Quintini G., Schönborn-Kellenberger O., Vahrenhorst D., Verstraeten T., Walz L., Wolz O.-O., Oostvogels L. Phase 1 assessment of the safety and immunogenicity of an mRNA- lipid nanoparticle vaccine candidate against SARS-CoV-2 in human volunteers. Infectious Diseases (except HIV/AIDS) 2020 doi: 10.1101/2020.11.09.20228551. [DOI] [Google Scholar]