Abstract
Utilising diverse molecular platforms has formed a solid foundation in New Zealand’s COVID-19 response. We evaluated multiple extraction and PCR assays for the detection of SARS-CoV-2.
We included 65 positive samples which were run on the Panther Fusion using a laboratory developed test (LDT, E gene target). Where viral RNA was extracted by MagNA Pure (MP) 96 extraction platform or EpMotion 5075/Geneaid extraction kit, SARS-CoV-2 detection was performed on Light Cycler (LC) 480 using a LDT (E gene) or 3 commercial assays; Certest Viasure (Orf1ab, N genes) GenePro (E, RdRp genes) and A* Star Fortitude (proprietary target).
Median Cts on LC 480 LDT for specimens (n = 9) extracted on MP 96 (26.6) were lower than on EpMotion (31.6) whereas median Cts for specimens (n = 10) extracted on the Panther Fusion LDT (23.1) were comparable with MP 96 /LC480 LDT (23.6).
Specimens tested on Panther Fusion LDT (n = 28), extracted by MP 96, and amplified using commercial assays showed good concordance with a few exceptions; lower median Ct values were seen for 2 targets on GenePro (16.9, 21.5) and Viasure (19.5, 21.1) than for the Panther Fusion LDT (24.2) and A* Star Fortitude (25.6).
Specimens tested on MP 96 (n = 18) had comparable results using commercial assays, with lower median Cts for Viasure (22.2, 23.7) compared with the LC 480 LDT (24.7), GenePro (24.7,25.7) and A*Fortitude (25.1) assays.
The study provides an early assessment of the performance characteristics of 3 extraction methods for viral RNA and 5 PCR assays for the detection of SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, Viral RNA extraction, RT-PCR, New Zealand
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of Coronavirus disease 2019 (COVID-19), an outbreak of respiratory illness that originated in Wuhan, China in December 2019 and has infected over 9 million people globally to date (Johns Hopkins University, 2020). Symptoms of SARS-CoV-2 include fever, cough, shortness of breath, coryzal symptoms and anosmia (Harapan et al., 2020). Molecular detection of the SARS-CoV-2 RNA by real time reverse transcription polymerase chain reaction (RT-PCR) is considered the diagnostic benchmark for the diagnosis of acute COVID infection (Anon., 2020).
The New Zealand government’s COVID-19 elimination strategy requires testing individuals who meet a broad suspect case definition, which from 1 April 2020 included anyone with mild symptoms of an acute respiratory tract infection (NZ Government, 2020). The broad criteria for testing have contributed to high testing rates per population in NZ, peaking, to date, in excess of 7000 tests per day (1.4 tests per 1000 population per day). The inherent challenges of processing daily high specimen numbers with a short turn-around-time (TAT) are exacerbated by the unprecedented global demand for molecular reagents, and deterioration of international transport links which occurred over a matter of days from March 2020. As a solution, in NZ, multiple high-throughput extraction and PCR platforms were engaged simultaneously for SARS-CoV-2 RNA extraction and RT-PCR to circumvent these constraints and to achieve optimal TAT from sample collection to result delivery in order to support diagnosis and public health activities for individuals tested in hospital and the community (Verrall, 2020).
We present evaluations of the different RNA extraction strategies, commercial and laboratory developed tests (LDT) for RT-PCR undertaken at Auckland Hospital laboratory, LabPLUS, during the pandemic. This evaluation included 65 samples from which SARS-CoV-2 viral RNA was initially detected by Light Cycler (LC) 480 LDT or the Panther Fusion LDT (Fig 2). The clinical specimens and the extracted RNA were stored at 4 °C and the evaluation was performed within a week unless otherwise noted. The samples included 58 patient samples in Universal Transport Media (Fort Richard Laboratories, New Zealand), all of which were upper respiratory tract swabs and 7 external quality assurance program (QAP) samples [3 PCR positive from WHO CDC QAP and 4 PCR positive from RCPA QAP]. The information about matrix of the QAP specimens was not available at the time. The volume provided in the QAP specimens allowed for only one extraction, hence MP 96 extraction and RT-PCR amplification by the LC 480 LDT or the commercial PCR kits.
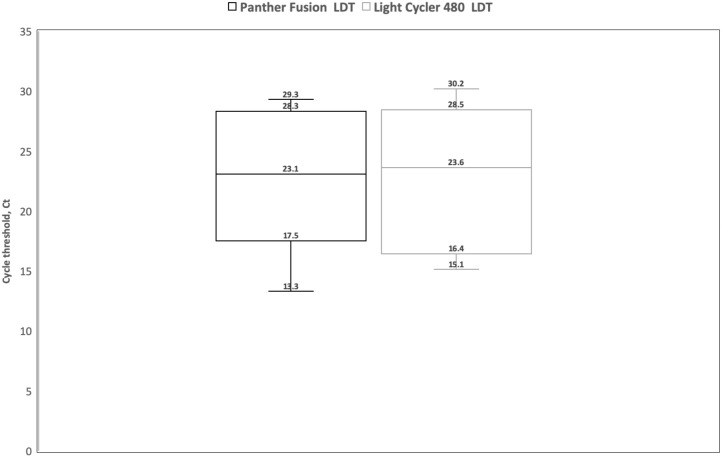
Fig. 2.
below shows box plots comparing the cycle threshold, Ct using Panther Fusion LDT (extraction and amplification) versus MagNAPure 96 extraction/Light Cycler 480 LDT [n = 10]. Three out of the 10 specimens were tested on the EpMotion were stored at 4 °C for longer than 1 week. Ten specimens processed on the MP 96/LC 480 using the LDT with a median Ct of 23.6 (IQR 17.5–27.6) had comparable results on the Panther Fusion LDT with a median Ct of 23.1 (IQR 18.4–27.9) for the same samples.
The MagNA Pure (MP) 96 semi-automated extraction platform (Roche Molecular Diagnostics, Pleasanton, CA, US) was used for viral RNA extraction. The EpMotion 5075 (Eppendorf, Hamburg, Germany), liquid handling workstation was programmed for high-throughput viral RNA extraction using viral DNA/RNA kit from Geneaid (Geneaid Biotech, Taiwan), as part of the diversification strategy to address the bottle-neck at the RNA extraction step (Fig. 1 ). Table 1 provides the details of the viral RNA extraction and PCR cycling parameters with five different platforms for detection of SARS-CoV-2 virus in clinical specimens. Briefly, for both MP 96 RNA and EpMotion 5075/ Geneaid viral DNA/RNA kit extraction, 200 μl of respiratory specimens (oropharyngeal or nasopharyngeal swabs in universal transport medium) were pre-treated with 100 μl MagNA Pure 96 External Lysis Buffer (Roche) followed by incubation for 10 min at 72 °C. Following further addition of 2 μl RNA internal extraction control (Bioline, London, UK) and 5 μl carrier RNA (Qiagen), the specimens underwent high-throughput automated extraction.
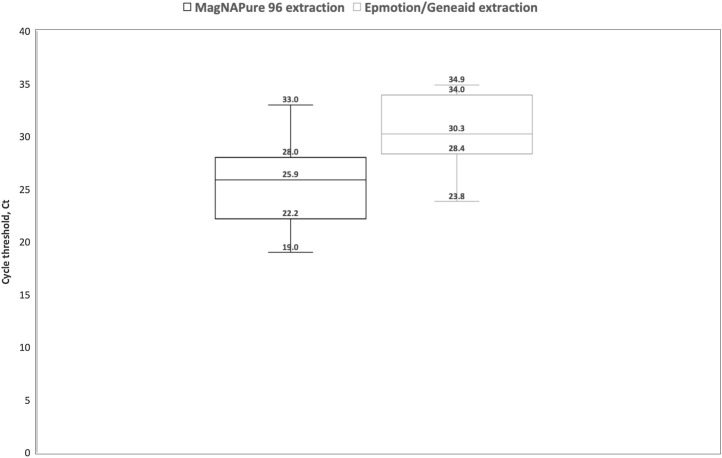
Fig. 1.
below shows box plots comparing the cycle threshold, Ct using the LDT Light Cycler 480 [n = 9] when specimens were extracted using MagNAPure 96 extraction versus when specimens were extracted using EpMotion 5075/Geneaid Viral RNA extraction kit. Nine specimens extracted on MP 96 extraction with a median Ct of 26.6 (interquartile range, IQR 23.3-28) had a higher median Ct of 31.6 (IQR 29.1–33.8) when extracted on the EpMotion 5075/Geneaid Viral RNA extraction kit and amplified using the LDT on the LC 480.
Table 1.
Viral RNA extraction and PCR cycling parameters with five different platforms for detection of SARS-CoV-2 virus in clinical specimens.
| Panther Fusion LDT |
Light Cycler 480 LDT |
Certest Viasure |
GenePro |
A*Star Fortitude |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Extraction platform | Integrated | MagNAPure 96 (Roche) OR EpMotion 5075/Geneaid viral DNA/RNA extraction kit | ||||||||||||||
| Extraction | Specimen volume | 500μL | 200 μLa | 200 μLa | 200 μLa | 200 μLa | ||||||||||
| Lysis buffer | 710 μL (provided) | 100 μLa | 100 μLa | 100 μLa | 100 μLa | |||||||||||
| Heat-inactivation | NA | 72 °C for 10 min | 72 °C for 10 min | 72 °C for 10 min | 72 °C for 10 min | |||||||||||
| RNA extraction control | Included | 2 μL | 2 μL | 2 μL | 2 μL | |||||||||||
| Carrier RNA | Included | 5 μL | 5 μL | 5 μL | 5 μL | |||||||||||
| Elution volume | 50 μL | 100 μL | 100 μL | 100 μL | 100 μL | |||||||||||
| PCR | Volume of extracted RNA | 5 μL | 10 μL | 5 μL | 5 μL | 2.5 μL | ||||||||||
| Final master mix volume | 20 μL | 15 μL | 15 μL | 20 μL | 22.5 μL | |||||||||||
| Temp. | Time | Cycles | Temp. | Time | Cycles | Temp. | Time | Cycles | Temp. | Time | Cycles | Temp. | Time | Cycles | ||
| PCR conditions | Reverse transcription | 50 °C | 8′5″ | 1 | 50 °C | 15ꞌ | 1 | 45°C | 15′ | 1 | 55°C | 15′ | 1 | 48°C | 15′ | 1 |
| Initial activation | 95 °C | 2′ | 1 | 95 °C | 2′ | 1 | 95 °C | 2′ | 1 | 94 °C | 3′ | 1 | 95 °C | 2′30″ | 1 | |
| Denaturation | 95 °C | 5″ | 44 | 95 °C | 5″ | 45 | 95 °C | 10″ | 45 | 94 °C | 15″ | 45 | 95 °C | 20″ | 45 | |
| Annealing/Extension | 58 °C | 7″ | 44 | 60 °C | 30″ | 45 | 60 °C | 50″ | 45 | 58 °C | 30″ | 45 | 59°C | 42″ | 45 | |
| Additional Optic On step | 60 °C | 14″ | 72 °C | 30″ | 45 | |||||||||||
| Real-time PCR platforms used | Panther Fusion LDT | Light Cycler 480 | Viasure | ABI 7500 | ABI 7500 | |||||||||||
NA = Not applicable.
After mixing 200 μL of specimen and 100 μL lysis buffer, 200 μL used for the following steps.
We compared the different extraction methods and PCR kits targeting different genes by analysis of the median cycle threshold (Ct) values, the delta Ct (difference between Ct values) and the interquartile ranges (IQR).
Amplification of the Sarbeco E gene of SARS-CoV-2 using a LDT (Corman et al., 2020) was performed on the LC 480 (Roche Molecular Diagnostics, Pleasanton, CA, US). Commercial PCR kits obtained from alternate supply chains for SARS-CoV-2 RT-PCR tests were also evaluated. Viasure SARS-CoV-2 Real Time PCR Detection Kit (Certest Biotec SL, Spain) targets a conserved region of ORF1ab and N genes for SARS-CoV-2 developed based on the sequences published by the Chinese Centre for Disease Control and Prevention and comes in a multiplexed single tube format where the primer/probes and master mixes are lyophilised in PCR tubes. The PCR with Viasure kits were evaluated on the Viasure PCR machine. The two other commercial kits evaluated were A*STAR Fortitude Kit 2.0 (MiRXES, Singapore) with a proprietary gene target, and GenePro PCR kit (Gencurix, Korea) kit which targets the E-gene and the RdRp genes in two single-plexed reaction. Both were run on the ABI 7500 real-time PCR [Applied Biosystems]. We analysed of the median cycle threshold (Ct) values, the delta Ct (difference between Ct values) and the interquartile ranges (IQR) for comparison.
The Panther Fusion platform (Hologic, San Diego, CA) which incorporates the RNA extraction and RT-PCR steps was also employed for testing. Amplification of the Sarbeco E gene of SARS-CoV-2 using the same LDT described above (Corman et al., 2020) was performed using the open access channel of the Panther Fusion platform (Hologic Inc.)
Twenty eight specimens initially tested on the Panther Fusion, extracted by MP 96, and run with 3 commercial PCR kits, Certest Viasure, GenePro, and A* Star Fortitude showed good concordance with a few exceptions (Table 2 ); lower median Ct values were seen for GenePro (E gene 19.9, IQR 16.7–26.9 and RdRp gene 21.5, IQR 16.7–26.9) and Viasure (Orf1ab gene 19.5, IQR 14.9–23.7 and N gene 21.1, IQR 18–25.1) than for the Panther Fusion LDT (24.2, IQR 20.1–30.1) and A* Star Fortitude (25.6, IQR 19.1–27.7). One specimen that gave a Ct of 42 on Panther and a Ct of 37 using A* Star Fortitude was not detected in GenePro or the Certest Viasure kit. Additionally, one other specimen was detected on all platforms except the Orf1ab gene on the Certest Viasure kit and both targets on GenePro kit. E gene of the GenePro was also not detected in 4 additional specimens.
Table 2.
Four-way same-sample cycle threshold (Ct) comparison on Panther Fusion LDT, Certest, GenePro, and A* Star Fortitude [n = 28].
| Panther LDT | GenePro |
Certest |
A*STAR | |||
|---|---|---|---|---|---|---|
| Specimen # | E gene | E gene | RdRp gene | Orf1ab gene | N gene | Proprietary |
| 1 | 34.2 | 27.0 | 27.2 | 24 | 24.6 | 26.5 |
| 2 | 27.3 | 26.6 | 27.0 | 23.2 | 23.2 | 26.6 |
| 3 | 30.2 | ND | 30.0 | 27.3 | 27.4 | 30.3 |
| 4 | 16.7 | 17.3 | 16.8 | 13.6 | 16 | 16.4 |
| 5 | 18.4 | 18.8 | 18.6 | 15 | 16.4 | 17.9 |
| 6 | 28.3 | 27.3 | 26.9 | 23.6 | 25.6 | 27.4 |
| 7 | 42.8 | ND | ND | ND | ND | 37.0 |
| 8 | 25.66 | 26.8 | 27.2 | 23.5 | 24.2 | 26.7 |
| 9 | 34.7 | ND | ND | 30.1 | 29.8 | 34.3 |
| 10 | 24.1 | 22.9 | 23.0 | 19.6 | 21.1 | 25.6 |
| 11 | 24.2 | 23.0 | 22.9 | 19.4 | 20.8 | 22.1 |
| 12 | 22 | 20.1 | 20.4 | 16.5 | 18.3 | 19.3 |
| 13 | 19.89 | 20.1 | 20.0 | 16.2 | 18.2 | 18.6 |
| 14 | 27.9 | 27.5 | 27.6 | 24.1 | 24.9 | 27.0 |
| 15 | 31.2 | ND | 32.1 | 28.1 | 29 | 30.6 |
| 16 | 18.5 | 16.4 | 17.4 | 13.9 | 14.6 | 17.7 |
| 17 | 27.1 | 25.8 | 24.6 | 22.9 | 24.7 | 27.0 |
| 18 | 20.4 | 15.6 | 16.4 | 14.4 | 16.5 | 18.4 |
| 19 | 19.7 | 20.7 | 6.3 | 15.7 | 18.2 | 20.3 |
| 20 | 23.9 | 14.4 | 13.8 | 11.9 | 13.4 | 15.6 |
| 21 | 19.9 | 20.9 | 19.5 | 16.0 | 18.9 | 19.8 |
| 22 | 36.6 | ND | ND | ND | 28.7 | 32.6 |
| 23 | 23.9 | 23.7 | 22.6 | 17.9 | 20.1 | 23.1 |
| 24 | 32.5 | ND | 28.7 | 27.1 | 27.5 | 31.9 |
| 25 | 15.2 | 12.2 | 12.2 | 14.7 | 17.3 | 14.2 |
| 26 | 30.1 | ND | 26.6 | 29.2 | 31.1 | 28.7 |
| 27 | 20.2 | 19.7 | 18.7 | 19.7 | 21.5 | 19.8 |
| 28 | 23.4 | 27.0 | 24.8 | 20.7 | 21.1 | 25.7 |
| Median | 24.2 | 19.9 | 21.5 | 19.5 | 21.1 | 25.6 |
| IQR range | 20.1, 30.1 | 9.2, 24.3 | 16.7, 26.9 | 14.9, 23.7 | 18, 25.1 | 19.1, 27.7 |
ND = Not detected.
Eighteen specimens initially tested on MP 96/ LC 480 LDT (n = 18) had comparable results using the commercial assays (Table 3 ), with lower median Cts for Viasure (Orf1ab 22.2, IQR 20.2–26.9 and N gene 23.7, IQR 21.3–28.1) compared with the LC 480 LDT (24.7, IQR 23.4–28.2), GenePro (E gene 24.7, IQR 18.4–26.2, RdRp gene 25.7, IQR 22.4–27) and A*Fortitude (25.1, IQR 21.6–27.7) assays.
Table 3.
Four-way same-sample cycle threshold (Ct) comparison on LC 480 LDT, Certest, GenePro, and A* Star Fortitude [n = 18].
| LC 480 LDT | GenePro |
Viasure |
A*STAR | |||
|---|---|---|---|---|---|---|
| Specimen # | E gene | E gene | RdRp gene | Orf1ab gene | N gene | Proprietary |
| 29 | 28.2 | 26.1 | 26.7 | 22.6 | 24.1 | 25.9 |
| 30 | 28.2 | 25.2 | 23.7 | 21.3 | 22.3 | 24.9 |
| 31 | 17.6 | 17.5 | 19.0 | 13.3 | 15.8 | 16.3 |
| 32 | 23.2 | 24.4 | 26.1 | 19.8 | 21.4 | 23.0 |
| 33 | 24.7 | ND | ND | 27.3 | 28.4 | 30.7 |
| 34 | 20.4 | 21.1 | 22.6 | 16.5 | 18.8 | 19.9 |
| 35 | 27.0 | 28.7 | 27.9 | 21.9 | 23.2 | 25.3 |
| 36 | 30.0 | ND | ND | 27.2 | 28.1 | 30.5 |
| 37 | 30.0 | ND | ND | 28.7 | 27.4 | 31.6 |
| 38 | 24.0 | 24.6 | 25.7 | 20.4 | 21.1 | 23.7 |
| 39 | 28.0 | 26.3 | 27.9 | 22.5 | 23.0 | 25.2 |
| 40 | 23.7 | 25.0 | 27.0 | 22.9 | 29.0 | 22.4 |
| 41 | 29.8 | 30.0 | 31.1 | 26.6 | 33.0 | 26.3 |
| 42 | 23.0 | 24.8 | 26.9 | 21.5 | 28.0 | 20.8 |
| 43 | 29.5 | 28.2 | 28.1 | 27.0 | 27.3 | 27.3 |
| 44 | 23.4 | 21.5 | 22.5 | 20.1 | 21.3 | 21.3 |
| 45 | 23.8 | ND | 22.4 | ND | ND | ND |
| 46 | ND | 33.5 | ND | 32.5 | 33.7 | 33.7 |
| Median | 24.2 | 19.9 | 21.5 | 19.5 | 21.1 | 25.6 |
| IQR range | 20.1, 30.1 | 9.2, 24.3 | 16.7, 26.9 | 14.9, 23.7 | 18, 25.1 | 19.1, 27.7 |
ND = Not detected.
The delta Ct of <3.5 cycles between median Cts over the different assays indicates comparable performance though neither of the GenePro targets were detected in 3 clinical specimens that were detected by all other platforms, with LC 480 LDT Cts of 24.65, 30, and 30. Among the QAP specimens, 1 specimen was detected by LC480 LDT and GenePro RdRp gene only while a second QAP specimen was detected on all platforms but not by LC480 LDT and GenePro RdRp gene. The results of the QAP were not available at the time of submission of this article.
Rapid scale-up of SARS-CoV-2 RT-PCR testing capacity by utilising diverse platforms and assays has formed one of the pillars in New Zealand’s COVID-19 response. Accordingly we evaluated multiple extraction and PCR platforms and assays for the detection of SARS-CoV-2.
There were both logistical and technical advantages and disadvantages to the extraction platforms. The MP 96 reagents used for RNA extraction were initially in short supply due to high demands during this pandemic. An advantage of the EpMotion liquid handling workstation is that it does not require proprietary reagents and plasticware and hence the extraction kits could be sourced locally. A second advantage of the EpMotion liquid handling workstation is the flexibility to program and automate the required steps for an extraction run. We note the higher median Ct values in the EpMotion 5075 extraction with the Geneaid viral extraction kit, compared with the MP 96 platform, though the upper ranges were comparable, with a delta Ct <3 cycles. It is possible that the EpMotion with the Geneaid viral extraction kit could contribute to reduced detection of low viral loads and this warrants further study.
A*Fortitude kits supplied the reverse transcriptase reagent and the Taq polymerase in the kit and hence did not need to be sourced separately at the time of reagent scarcity. The mastermixes in the Viasure kits are supplied in lyophilised form in PCR tubes and only need reconstitution with a supplied rehydration buffer and addition of extracted RNA before running the PCR amplification step. These kits include two targets in a multiplexed reaction. GenePro kit also provided reverse transcriptase reagent and the Taq polymerase but unlike the other commercial kits lacked an internal control and had two single-plex targets which required additional wells to be used on the plate, limiting the number of samples per plate significantly. It also failed to detect higher number of positive samples than those detected on other platforms.
The advantages of commercial assays are offset by additional reagent cost, and under current circumstances, significant potential supply issues, where companies drip-feed supplies which are subject to postponement or cancellation without notice. To overcome these issues in NZ, the Ministry of Health sourced A*Fortitude kits direct from Singapore in bulk, which are stored and distributed centrally in NZ to overcome short-term supply issues.
The Panther LDT allows random access testing and offers a shorter turn-around-time to results compared with the non-integrated platforms, and in theory has the capacity to test >1000 samples in a 24 period, but some of the reagents and consumables such as the external lysis tubes are proprietary and remain in short supply in NZ, so the potential of this platform has been unfulfilled, and our machine often idles, awaiting the next shipment of reagents.
The study provides an early assessment of the performance characteristics of 3 extraction methods for viral RNA and 5 different PCR kits with different gene targets for the detection of SARS-CoV-2. During the current pandemic, the LDT and commercial assays examined here had acceptable performance criteria for testing such as each specimen well was positive for the internal control to check for PCR inhibition or no target signals detected in both the negative template control and extraction control or the positive template control lies within 2 cycles above or below the accepted Ct value. This evaluation equipped us with wider options for the ongoing testing of the SARS-CoV-2 specimens after the NZ alert levels are lifted and the testing numbers expected to increase.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The authors would like to acknowledge and thank Fahimeh Rahnama, Sally Roberts and Daniel Hunt for their support with this work.
References
- https://www.who.int/csr/sars/coronarecommendations/en/.
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.20000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapan H., Itoh N., Yufika A., Winardi W., Keam S., Te H., Megawati D., Hayati Z., Wagner A.L., Mudatsir M. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Publ. Health. 2020;13(5):667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins University . 2020. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University.https://coronavirus.jhu.edu/map.html [Google Scholar]
- NZ Government . 2020. Ministry of Health Website for Case Definition.https://www.health.govt.nz/our-work/diseases-and-conditions/covid-19-novel-coronavirus/covid-19-resources-health-professionals/case-definition-covid-19-infection [Google Scholar]
- Verrall A. 2020. Rapid Audit of Contact Tracing for Covid-19 in New Zealand.https://www.health.govt.nz/system/files/documents/publications/contact_tracing_report_verrallA.pdf [Google Scholar]