SUMMARY
Intracellular vesicle fusion is catalyzed by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). Vesicle-anchored v-SNAREs pair with target membrane-associated t-SNAREs to form trans-SNARE complexes, releasing free energy to drive membrane fusion. However, trans-SNARE complexes are unable to assemble efficiently unless activated by Sec1/Munc18 (SM) proteins. Here, we demonstrate that SNAREs become fully active when the v-SNARE is split into two fragments, eliminating the requirement of SM protein activation. Mechanistically, v-SNARE splitting accelerates the zippering of trans-SNARE complexes, mimicking the stimulatory function of SM proteins. Thus, SNAREs possess the full potential to drive efficient membrane fusion but are suppressed by a conformational constraint. This constraint is removed by SM protein activation or v-SNARE splitting. We suggest that ancestral SNAREs originally evolved to be fully active in the absence of SM proteins. Later, a conformational constraint coevolved with SM proteins to achieve the vesicle fusion specificity demanded by complex endomembrane systems.
Graphical Abstract
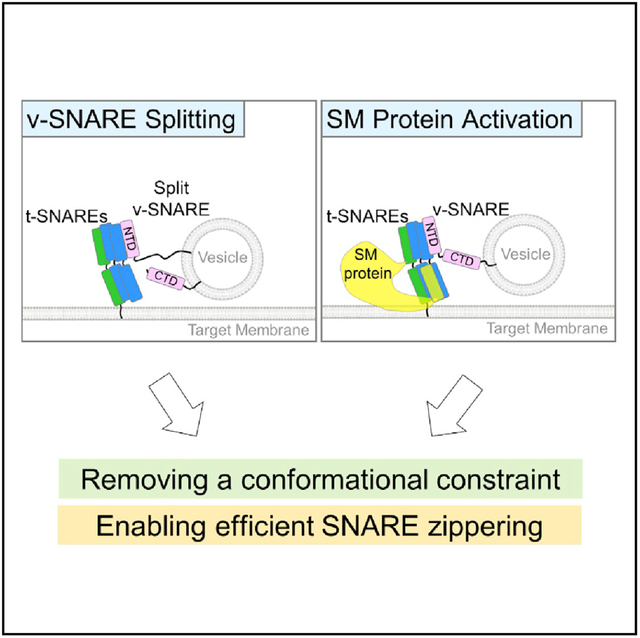
In Brief
SNAREs are unable to drive efficient membrane fusion unless activated by Sec1/Munc18 (SM) proteins. In this work, Liu et al. demonstrate that v-SNARE splitting mimics SM protein activation and unleashes the full membrane fusion potential of SNAREs.
INTRODUCTION
Cargo transport between membrane-bound organelles requires the fusion of cargo-carrying vesicles with target membranes. Vesicle fusion is catalyzed by a class of membrane-bound proteins known as soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (Rizo and Südhof, 2012; Südhof and Rothman, 2009). The vesicle fusion reaction is initiated when vesicle-anchored SNAREs (v-SNAREs) pair with target membrane-associated SNAREs (t-SNAREs) to form trans-SNARE complexes between the two membrane bilayers (Chapman, 2008; Ellena et al., 2009; Reese et al., 2005; Söllner et al., 1993; Weber et al., 1998). A fully assembled SNARE complex consists of a parallel, four-helix, coiled-coil bundle held together by 15 hydrophobic layers of interacting side chains (numbered −7 to −1 and +1 to +8), and a hydrophilic 0 layer (Stein et al., 2009; Sutton et al., 1998). One helix of the bundle is contributed by the v-SNARE, whereas three helices are from t-SNAREs (Stein et al., 2009; Sutton et al., 1998; Wickner, 2010).
The SNARE bundle assembles in distinct stages in the membrane fusion reaction. The N-terminal domains (NTDs, −7 to −1 layers) of SNAREs pair first, restructuring the t-SNAREs and setting the stage for the subsequent zippering of the C-terminal domains (CTDs, +1 to +8 layers) (Li et al., 2014; Zhang et al., 2016). Free energy released by CTD zippering is used to overcome the energy barrier of membrane merging (Gao et al., 2012; Li et al., 2014; Pobbati et al., 2006). Despite powering the membrane fusion reaction, trans-SNARE complexes are unable to assemble efficiently unless activated by Sec1/Munc18 (SM) proteins (Baker et al., 2015; Jiao et al., 2018; Kasula et al., 2016; Ma et al., 2013; Shen et al., 2007). Soluble factors of 60–70 kDa, SM proteins recognize their cognate pairs of v- and t-SNAREs and promote their assembly into energy-releasing trans-SNARE complexes (Dulubova et al., 2007; Garcia et al., 1994; Hata et al., 1993; Lobingier et al., 2014; Ma et al., 2015; Novick and Schekman, 1979; Pevsner et al., 1994).
In this work, we unexpectedly discovered that SNAREs become fully active when the v-SNARE is split into two fragments, eliminating the requirement of SM protein activation. Split SNARE-driven fusion is kinetically similar to the SM protein-activated fusion reaction and is highly sensitive to point mutations that abolish vesicle fusion in vivo. We observed that v-SNARE splitting accelerates the zippering of trans-SNARE complexes, mimicking the stimulatory function of SM proteins. However, split SNARE-driven fusion lacks the specificity observed in SM protein-activated fusion reactions. These data demonstrate that SNAREs possess the full potential to drive efficient membrane fusion but are suppressed by a conformational constraint. The constraint can be removed by binding to a cognate SM protein or by splitting the v-SNARE.
RESULTS AND DISCUSSION
The energy released by the SNARE complex is comparable to that from other membrane fusion proteins such as viral fusion proteins (Jiao et al., 2015). However, viral fusion proteins are self-sufficient engines that do not require activation by other proteins (Earp et al., 2005; Harrison, 2008). Thus, we posit that SNAREs are energetically competent in driving membrane fusion but are kinetically impeded by an inherent constraint that could be experimentally removed. To test this possibility, we engineered SNARE variants and examined whether they could drive efficient membrane fusion without requiring activation by a SM protein. A SNARE variant we engineered was a split v-SNARE, in which VAMP2/synaptobrevin, a v-SNARE involved in synaptic exocytosis, was severed at the zero layer. The detached NTD and CTD fragments were reconstituted into the same liposomes (Figures 1A and 1B). Although the NTD and CTD fragments were previously characterized in biochemical studies (Li et al., 2014; Melia et al., 2002; Yu et al., 2018), it was unclear whether they are capable of driving biologically relevant membrane fusion when detached and how their activities are linked to SM proteins.
Figure 1. Split SNAREs Drive Efficient Membrane Fusion without Requiring Activation by a SM Protein.
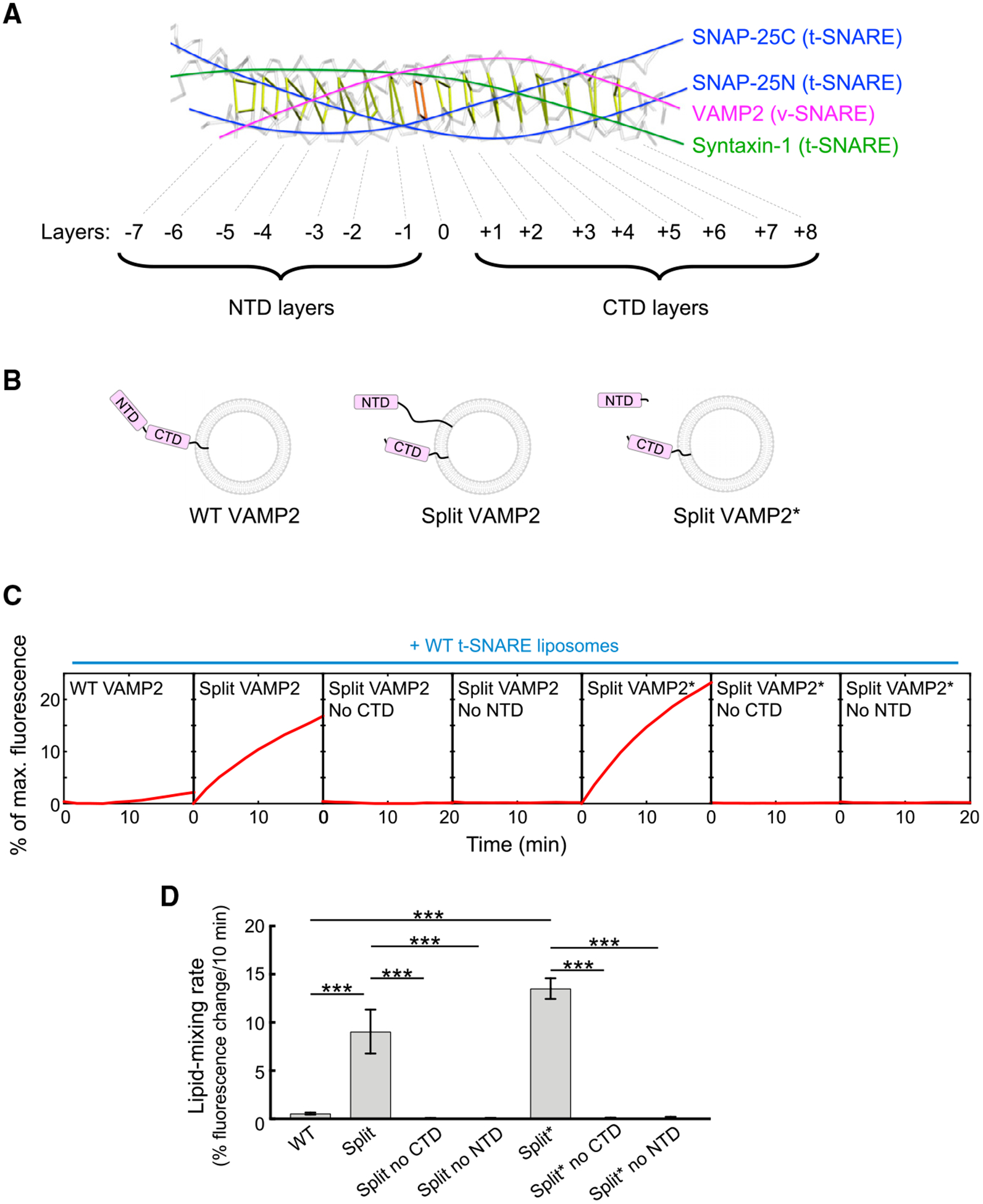
(A) Backbone view of the synaptic SNARE complex with individual layers of the SNARE motifs indicated (PDB: 1SFC).
(B) Diagrams illustrating two types of split v-SNAREs. In one split v-SNARE, the NTD and CTD of the v-SNARE VAMP2 were detached and anchored to the same liposomes. The NTD was connected to the transmembrane domain of VAMP2 through a TolA helix unrelated to SNAREs. In another split v-SNARE (marked with an asterisk), the CTD of VAMP2 is anchored to liposomes, whereas the NTD was added as a soluble fragment.
(C) Liposomes harboring WT or split VAMP2 (shown in B) were directed to fuse with liposomes containing WT t-SNAREs (syntaxin-1 and SNAP-25). The kinetics of the fusion reactions was measured using a fluorescence resonance energy transfer (FRET)-based lipid-mixing assay.
(D) Initial lipid-mixing rates of the fusion reactions shown in (C). Data are presented as mean ± SD (n = 3). The p values were calculated using Student’s t test. ***p < 0.001.
In the absence of a SM protein, wild-type (WT) SNAREs zippered inefficiently, driving a near-background level of liposome fusion (Figures 1C and 1D; Shen et al., 2007; Yu et al., 2015, 2018). Strikingly, split VAMP2 paired with WT t-SNAREs (syntaxin-1 and SNAP-25) and drove a highly efficient liposome fusion reaction (Figures 1C and 1D). Split SNARE-driven fusion was more than an order of magnitude faster than the WT SNARE-mediated fusion reaction (Figures 1C and 1D) and was kinetically similar to the SM protein-activated fusion reaction (Figures 2A and 2B). Omission of either NTD or CTD abolished split SNARE-driven liposome fusion (Figures 1C and 1D), consistent with the requirement of both domains in vesicle fusion (Gao et al., 2012; Li et al., 2014; Yu et al., 2018). We tested another split v-SNARE pair, in which the CTD of VAMP2 was anchored to liposomes yet the NTD was added as a soluble fragment (Figure 1B). We observed that this split VAMP2 also drove an efficient level of liposome fusion when paired with t-SNAREs (Figures 1C and 1D). These results demonstrate that when the NTD and CTD of the v-SNARE are physically detached, SNAREs are capable of driving efficient liposome fusion without requiring activation by a SM protein.
Figure 2. Split SNARE-Driven Membrane Fusion Mimics the SM Protein-Activated Fusion Reaction.
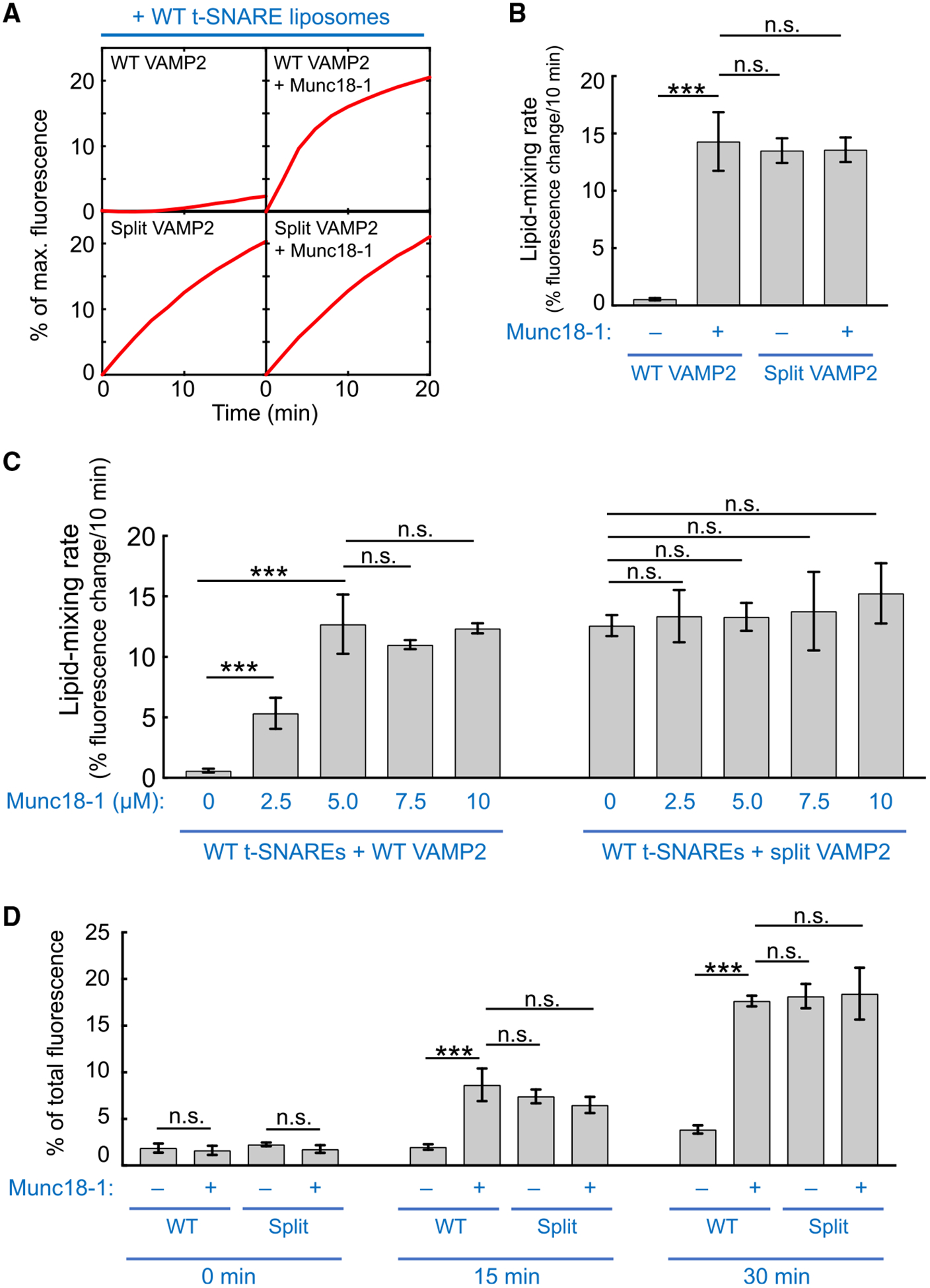
(A) Liposomes harboring WT or split VAMP2 (depicted in Figure 1B, right) were directed to fuse with liposomes containing WT t-SNAREs (syntaxin-1 and SNAP-25) in the absence or presence of 5 μM Munc18–1. The kinetics of the fusion reactions was measured using a FRET-based lipid-mixing assay. In this work, Munc18–1 was added at the same molar concentration (5 μM) as SNAREs to reflect their 1:1 binding stoichiometry (Dulubova et al., 2007; Shen et al., 2007; Yu et al., 2013). Higher concentrations of Munc18–1 did not further increase the rate of the liposome fusion reaction driven by WT SNAREs (Figure S1).
(B) Initial lipid-mixing rates of the fusion reactions shown in (A).
(C) Dose dependence of Munc18–1 in liposome fusion reactions mediated by WT or split VAMP2. Liposomes harboring WT or split VAMP2 (depicted in Figure 1B, right) were directed to fuse with liposomes containing WT t-SNAREs (syntaxin-1 and SNAP-25) in the absence or presence of Munc18–1 at the indicated concentrations. The kinetics of the fusion reactions was measured using a FRET-based lipid-mixing assay.
(D) Liposomes harboring WT or split VAMP2 (depicted in Figure 1B, right) were incubated with WT t-SNARE liposomes containing syntaxin-1 and SNAP-25 at 4°C to assemble trans-SNARE complexes between membrane bilayers. Relative amounts of assembled trans-SNARE complexes are presented as percentages of maximum rhodamine fluorescence.
In (B)–(D), data are presented as mean ± SD (n = 3). The p values were calculated using Student’s t test. n.s., p > 0.05. ***p < 0.001. See also Figure S1.
Next, we sought to determine the molecular mechanism by which v-SNARE splitting accelerates membrane fusion. The kinetics of split SNARE-driven fusion was comparable to that of SM protein-activated fusion reaction, in which the cognate SM protein Munc18–1 was added to WT SNAREs (Figures 2A–2C). Addition of Munc18–1 to the split SNARE-driven fusion reaction did not further increase the fusion rate (Figures 2A–2C), suggesting that SM protein and v-SNARE splitting promote membrane fusion through a similar mechanism. In a liposome coflotation assay, VAMP2 CTD bound to t-SNAREs and the interaction remained intact in the presence of VAMP2 NTD (Figure S1). Thus, the CTD of split VAMP2 interacts with the t-SNARE CTD, its native binding partner (Sutton et al., 1998), rather than recognizing t-SNAREs through a different binding mode. In a trans-SNARE assembly assay, which monitors the zippering of both NTDs and CTDs (Yu et al., 2019), WT SNAREs assembled inefficiently between membrane bilayers but were strongly stimulated by Munc18–1 (Figure 2D). v-SNARE splitting accelerated trans-SNARE assembly similarly, because Munc18–1 and the assembly reaction was not enhanced by Munc18–1 (Figure 2D). These data agree with the liposome fusion results (Figures 2A and 2B) and indicate that v-SNARE splitting permits efficient membrane fusion by augmenting trans-SNARE zippering, similar to the stimulatory function of SM proteins. Altogether, these findings suggest that split SNARE-driven fusion mimics the SM protein-activated fusion reaction.
The SM protein-activated fusion reaction is highly sensitive to point mutations in the CTD layers of the v-SNARE, because these mutations reduce energy output and zippering cooperativity of SNAREs (Jiao et al., 2018; Walter et al., 2010; Yu et al., 2015, 2018). By contrast, a nonbiological SNARE zippering pathway (e.g., the basal fusion without a SM protein) is insensitive to these layer mutations (Yu et al., 2015, 2018). Here, we tested four layer mutations in VAMP2 CTD known to abolish synaptic exocytosis in the cell (Figure 3A; Walter et al., 2010; Yu et al., 2015, 2018). We observed that split SNARE-driven fusion was abrogated when any of the layer mutations was introduced (Figures 3B and 3C), similar to the effects of the mutations on SM protein-activated fusion in vitro and vesicle fusion in vivo (Figure 3D). These data suggest that split SNARE-driven fusion proceeds through the same route as the biologically relevant SM protein-activated fusion reaction.
Figure 3. Split SNARE-Driven Fusion Is Highly Sensitive to Layer Mutations that Impair Vesicle Fusion In Vivo.
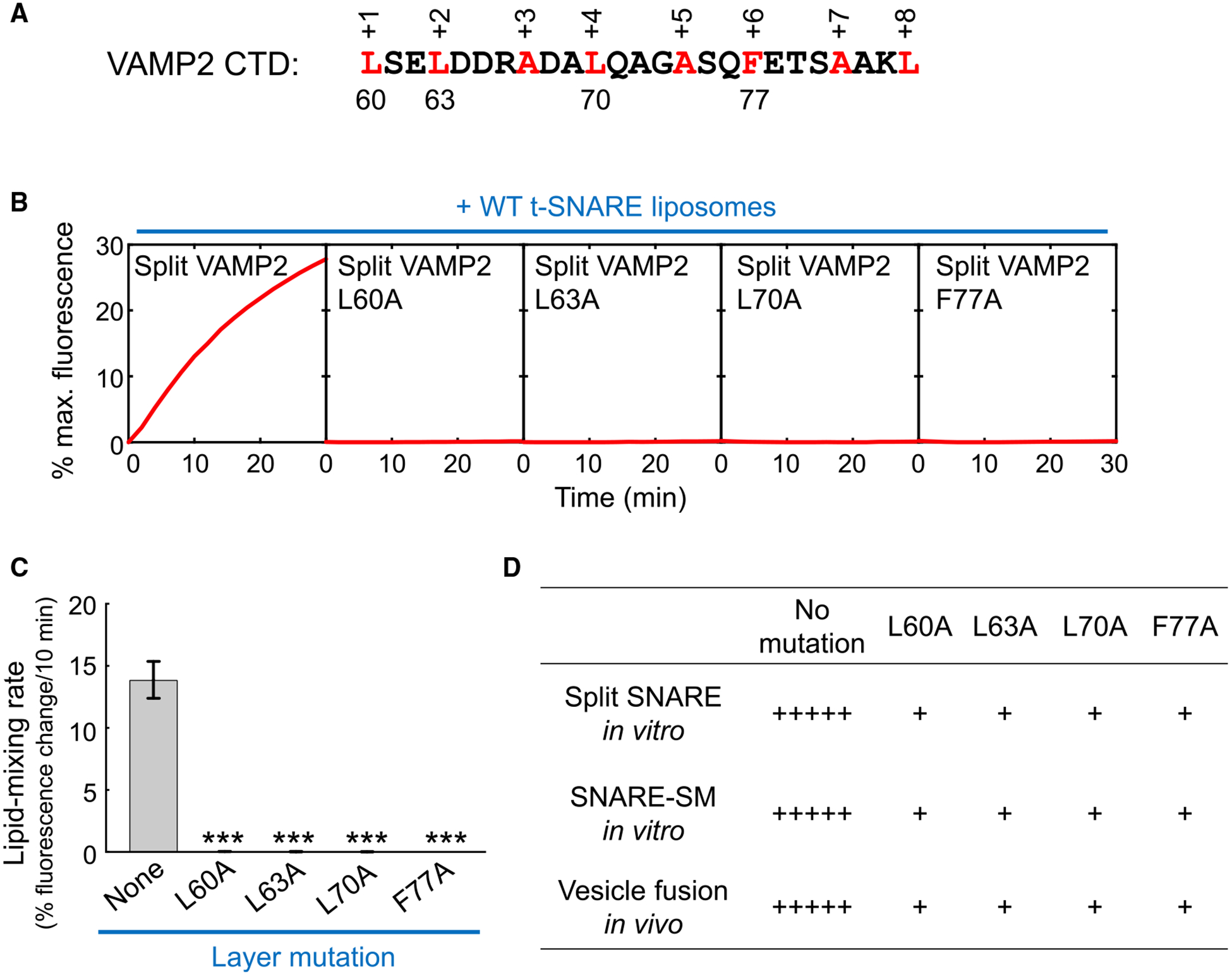
(A) Sequence of VAMP2 CTDs with layer residues numbered and highlighted.
(B) Liposomes harboring split VAMP2 (shown in Figure 1B, right, with or without layer mutations) were directed to fuse with liposomes containing WT t-SNAREs (syntaxin-1 and SNAP-25). The kinetics of the fusion reactions was measured using a FRET-based lipid-mixing assay.
(C) Initial lipid-mixing rates of the fusion reactions shown in (B). Data are presented as mean ± SD (n = 3). The point mutation data are compared with the no mutation data. The p values were calculated using Student’s t test. ***p < 0.001.
(D) Correlation of the effects of VAMP2 layer mutations on split SNARE-driven liposome fusion, SNARE-SM-mediated liposome fusion, and in vivo vesicle fusion. In vivo data are based on published genetic studies (Walter et al., 2010; Yu et al., 2015). +++++, WT levels of in vitro liposome fusion or in vivo vesicle fusion; +, <20% of WT levels of fusion.
Intracellular vesicle fusion is exquisitely specific such that a vesicle only fuses with its destined organelle (Jahn and Scheller, 2006; Südhof and Rothman, 2009). However, SNAREs alone are insufficient to achieve fusion specificity, because they possess similar hydrophobic layers (Brandhorst et al., 2006; Shen et al., 2007). SM proteins play a key role in determining vesicle fusion specificity by selectively recognizing and activating cognate SNARE pairs. VAMP8, a v-SNARE involved in endosomal/lysosomal vesicle fusion (Jahn and Scheller, 2006), exhibits no sequence similarity to VAMP2 except layer residues (Figure 4A). VAMP8 was able to pair with synaptic exocytic t-SNAREs and drove a minimal level of liposome fusion (Figures 4B and 4C). However, this noncognate SNARE pair was not activated by Munc18–1 (Figures 4B and 4C). A VAMP2-VAMP8 chimera, in which the NTD of VAMP2 was substituted with that of VAMP8, fully supported Munc18–1 activation (Figure S2), suggesting that the CTD of the v-SNARE plays a key role in determining the specificity of SM protein activation. Split VAMP8, by contrast, drove a highly efficient level of liposome fusion when paired with synaptic exocytic t-SNAREs (without Munc18–1), comparable to the kinetics of split VAMP2-mediated fusion (Figures 4B and 4C). These data suggest that split SNARE-driven fusion lacks the specificity of SM protein-activated fusion reactions, consistent with the nonselective nature of SNARE pairing.
Figure 4. Split SNARE-Driven Fusion Lacks Compartmental Specificity.
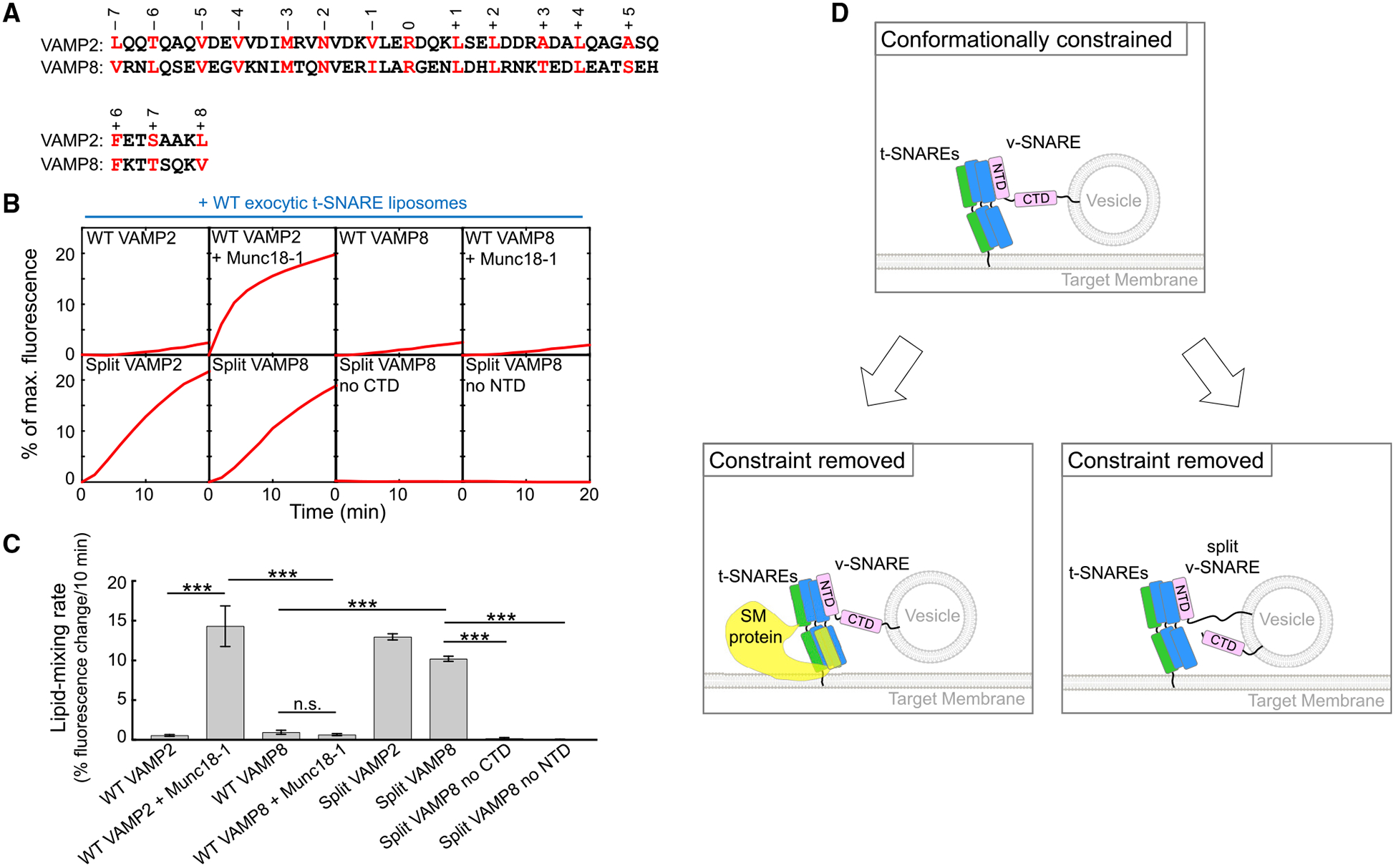
(A) Alignment of SNARE motifs of VAMP2 and VAMP8 with layer residues highlighted and numbered.
(B) Liposomes harboring the indicated v-SNAREs were directed to fuse with liposomes containing WT exocytic t-SNAREs (syntaxin-1 and SNAP-25) with or without 5 μM Munc18–1. The kinetics of the fusion reactions was measured by a FRET-based lipid-mixing assay.
(C) Initial lipid-mixing rates of the fusion reactions shown in (B). Data are presented as mean ± SD (n = 3). The p values were calculated using Student’s t test. n.s., p > 0.05. ***p < 0.001.
(D) Model illustrating the activation of the SNARE vesicle fusion machinery by a SM protein in a biological setting or by v-SNARE splitting in an engineered system. The CTDs of WT SNAREs are unable to zipper efficiently because of the presence of a conformational constraint. The SM protein uses its SLP to restructure t-SNARE CTDs, enabling the latter to properly zipper with the v-SNARE CTD. When the v-SNARE is split, the freed CTD is able to optimally zipper with t-SNARE CTDs, achieving the same effect as SM protein binding to t-SNAREs.
See also Figures S2–S4.
Finally, we characterized v-SNARE splitting in another vesicle fusion pathway: the exocytosis of the glucose transporter GLUT4 in adipocytes and muscles. When reconstituted into proteoliposomes, GLUT4 exocytic SNAREs—syntaxin-4, SNAP-23, and VAMP2—drove a minimal level of liposome fusion (Figure S3). However, splitting the v-SNARE strongly accelerated the fusion kinetics (Figure S3). The split SNARE-driven fusion was diminished when any of the four CTD layer mutations was introduced (Figure S3). These results are consistent with the data of synaptic exocytic SNAREs and suggest that SNARE activation by v-SNARE splitting represents a conserved feature of SNARE proteins.
The split v-SNAREs we engineered offer key insights into the molecular mechanisms of SNAREs and SM proteins in vesicle fusion. Our findings demonstrate that SNAREs possess the full membrane fusion potential but are suppressed by an intrinsic conformational constraint (Figure 4D). The conformational constraint is created by the relative spatial organization of the v- and t-SNAREs, rather than by either of them alone, and is expected to require the presence of apposed membrane bilayers. We posit that the conformational constraint precludes optimal pairing of SNARE CTDs, resulting in incomplete CTD zippering and a concordant decrease in available energy to overcome the kinetic barrier for fusion (Figure 4D). When the v-SNARE is split, its freed CTD is able to align properly with t-SNARE CTDs to achieve full zippering (Figure 4D). In the cell, the conformational constraint is removed by a cognate SM protein that uses its SNARE-like peptide (SLP) to restructure t-SNARE CTDs (Yu et al., 2018), enabling the latter to zipper properly with the v-SNARE CTD (Figure 4D). This mechanism is distinct from v-SNARE splitting but achieves the same effect of relieving the conformational constraint. Overall, the role of the SM protein is to unleash the inherent fusion-driving potential of SNAREs, without directly contributing to the energetics of the membrane fusion reaction. The conformational constraint cannot be removed by simply inserting flexible residues between NTD and CTD of the v-SNARE (Figure S4), suggesting that SM proteins induce a large spatial rearrangement of SNAREs. The conformational constraint may also arise from off-pathway SNARE assemblies such as the 2:1 t-SNARE complex (containing an extra copy of syntaxin) and the anti-parallel trans-SNARE complex. We postulate that v-SNARE splitting diverts SNAREs from these nonfusogenic misassembled structures by altering the energetic landscape of SNARE interactions, mimicking the roles of SM proteins in guiding SNARE assembly (Baker et al., 2015; Jiao et al., 2018; Lai et al., 2017; Ma et al., 2015; Wang et al., 2019; Zhang et al., 2016). Further research will be needed to define the precise nature of the conformational constraint of SNAREs. We suggest that a powerful approach to address the question is single-molecule biophysical measurements using membrane-anchored proteins (Ma et al., 2017).
Split SNAREs are not known to regulate vesicle fusion in extant eukaryotes. However, SNAREs’ possession of a full membrane fusion potential raises the intriguing possibility that SNAREs originally evolved to be fully active without requiring activation by SM proteins. The ancestral v-SNARE might exhibit a split form similar to the one described in this work. Indeed, there is no physiochemical obstacle to anchor detached v-SNARE NTD and CTD to the same vesicles. Alternatively, the ancestral SNAREs might display another configuration free of a conformational constraint. Despite lacking pairing specificity, these constitutively active SNAREs were adequate in mediating vesicle fusion in a primordial endomembrane system (EMS), requiring no specificity in vesicle fusion (Klinger et al., 2016; Zaremba-Niedzwiedzka et al., 2017). For a complex EMS, however, it became critical to ensure compartmental specificity of vesicle fusion, which could not be achieved by SNAREs alone (Dacks and Field, 2007; Schlacht et al., 2014). The specificity issue was solved when a conformational constraint of SNAREs coevolved with SM proteins. As a result, SNAREs could not drive efficient fusion unless a cognate SNARE pair is recognized by a SM protein to remove the conformational constraint. Noncognate v- and t-SNARE can form initial interactions but are unable to progress to drive efficient fusion because of a lack of a cognate SM protein to relieve the constraint.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Jingshi Shen (jingshi.shen@colorado.edu).
Materials Availability
All the reagents generated in this study are available via material transfer agreement.
Data and Code Availability
This study did not generate unique code or dataset.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Microbial Strains
All the recombinant proteins in this study were expressed in E. Coli BL21 [B F− ompT hsdS(rB− mB−) dcm+ Tetr gal λ(DE3) endA Hte] at 37°C in a shaker incubator set at 220 rpm.
METHOD DETAILS
Protein Expression and Purification
Recombinant full-length (FL) v- and t-SNAREs were expressed in E. coli and purified using nickel affinity chromatography (Yu et al., 2019). The synaptic exocytic t-SNARE complex was composed of untagged rat syntaxin-1 and mouse SNAP-25 with an N-terminal His6 tag (plasmid TW34) (Weber et al., 2000). The GLUT4 exocytic t-SNARE complex was composed of untagged rat syntaxin-4 and mouse SNAP-23 with an N-terminal His6 tag. Recombinant mouse VAMP2 and VAMP8 proteins had no tags left after the His6-SUMO moiety was removed by proteolytic digestion (Shen et al., 2010; Yu et al., 2019). Recombinant untagged Munc18–1 was produced in E. coli using a procedure we previously established (Shen et al., 2007, 2015; Yu et al., 2013, 2015). The soluble fragments – VAMP2 NTD (residues 28–55) and VAMP8 NTD (residues 9–36) – were expressed and purified in the same way as Munc18–1. Membrane-bound fragments including VAMP2 CTD (residues 60–116), VAMP8 CTD (residues 41–101), and VAMP2-NTD-TolA (residues 60–84 of FL VAMP2 were replaced by a fragment from the bacterial TolA protein) were expressed and purified in a similar way as WT VAMP2. The sequence of the TolA helix is: GGSSIDAVMVDSGAVVEQYKRMQSQ. VAMP2 layer mutants were generated by site-directed mutagenesis and purified as their corresponding wild-type (WT) proteins.
Proteoliposome Preparation
All lipids used in this work were acquired from Avanti Polar Lipids. To prepare t-SNARE liposomes, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) and cholesterol were mixed in a molar ratio of 60:20:10:10. To prepare v-SNARE liposomes, POPC, POPE, POPS, cholesterol, (N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE), and N-(Lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (rhodamine-DPPE) were mixed at a molar ratio of 60:17:10:10:1.5:1.5. SNARE proteoliposomes were generated by detergent dilution and isolated on a Nycodenz density gradient flotation (Shen et al., 2010). Detergent was removed by overnight dialysis of the samples in Novagen dialysis tubes against the reconstitution buffer (25 mM HEPES [pH 7.4], 100 mM KCl, 10% glycerol, and 1 mM DTT). The protein: lipid ratio was 1:200 for v-SNARE liposomes and 1:500 for t-SNARE liposomes. Membrane-anchored SNARE fragments were reconstituted at the same density as FL v-SNAREs.
Liposome Fusion Assay
A standard liposome fusion reaction contained 5 μM t-SNAREs and 1.5 μM v-SNARE. NBD- and rhodamine-labeled v-SNARE liposomes were directed to fuse with unlabeled t-SNARE liposomes in the presence or absence of the indicated concentrations of Munc18–1. The macromolecular crowding agent Ficoll 70 (100 mg/mL) was included in all liposome fusion reactions to mimic the crowded cellular environment (Yu et al., 2015). In split v-SNARE fusion assays using membrane-anchored NTD, VAMP2-NTD-TolA and VAMP2 CTD were reconstituted together into liposomes. These liposomes were mixed with t-SNARE liposomes and loaded into a pre-warmed 96-well microplate to initiate fusion. In split v-SNARE fusion assays using soluble NTD, t-SNARE liposomes were first incubated with 5 μM soluble VAMP2 NTD peptide at 37°C for 30 min. Subsequently, the samples were mixed with VAMP2 CTD liposomes and loaded into a pre-warmed 96-well microplate to initiate fusion. All fusion reactions were conducted at 37°C. NBD fluorescence (excitation: 460 nm; emission: 538 nm) was measured every 2 min in a BioTek Synergy HT microplate reader. At the end of the reaction, 10 μL of 10% CHAPSO was added to each sample to obtain the values of maximum fluorescence. Fusion data were presented as the percentage of maximum fluorescence change. The initial fusion rate was calculated based on the average fusion rate within the first 10 min of a liposome fusion reaction. Full accounting of statistical significance was included for each dataset based on at least three independent experiments.
Liposome Co-flotation Assay
The cytosolic domains of t-SNAREs (syntaxin-1 and SNAP-25) were incubated with protein-free (PF) or VAMP2 CTD liposomes in the absence or presence of soluble VAMP2 NTD at 4°C with gentle agitation. After 1 h, an equal volume (150 μL) of 80% Nycodenz (w/v) in the reconstitution buffer was added and the mixture was transferred to 5 mm by 41 mm centrifuge tubes. The samples were overlaid with 200 μL each of 35% and 30% Nycodenz, and then with 20 μL reconstitution buffer on the top. The gradients were centrifuged for 4 h at 52,000 rpm in a Beckman SW55 rotor. Liposome samples were collected from the 0/30% Nycodenz interface (2 × 20 μL) and analyzed by SDS-PAGE.
Trans-SNARE Assembly Assay
WT t-SNARE liposomes containing syntaxin-1 and SNAP-25 were mixed with WT or split VAMP2 liposomes. After incubation at 4°C in the absence or presence of 5 μM Munc18–1, 20 μM VAMP2 CD (residues 1–95) was added to dissociate partially assembled trans-SNARE complexes in which CTDs had not fully zippered. Fully assembled trans-SNARE complexes, by contrast, were resistant to VAMP2 CD treatment. The t-SNARE liposomes and bound v-SNARE liposomes were pulled down using nickel Sepharose beads through binding to the His6 tag on SNAP-25. After washing three times with the reconstitution buffer, CHAPS was added to a final concentration of 1% to solubilize bead-bound liposomes. After centrifugation, rhodamine fluorescence in the supernatant was measured in a BioTek Synergy HT microplate reader. In a negative control reaction, v-SNARE liposomes were replaced with PF liposomes, allowing us to calculate background fluorescence. After subtraction of background fluorescence, the obtained rhodamine fluorescence reflected the relative amounts of assembled trans-SNARE complexes. The data were presented as percentage of total rhodamine fluorescence of input v-SNARE liposomes. All reactions were performed in the presence of 100 mg/mL Ficoll 70.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was calculated for each data point based on at least three independent experiments. Data were analyzed using the KaleidaGraph 3.6 software (Synergy) and are presented as means ± standard deviation.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Stains | ||
| BL21 Gold DE3 competent cells | Stratagene | Cat # 230132 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids | Cat # 850457C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), | Avanti Polar Lipids | Cat # 850757C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) | Avanti Polar Lipids | Cat # 840034C |
| Cholesterol | Avanti Polar Lipids | Cat # 700000P |
| N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE) | Avanti Polar Lipids | Cat # 810114C |
| N-(Lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (rhodamine-DPPE) | Avanti Polar Lipids | Cat # 810158C |
| Nycodenz | Axis-Shield | Cat # 1002424 |
| Protease inhibitor cocktail | Roche | Cat # 05056489001 |
| CHAPSO (3-((3-Cholamidopropyl) dimethylammonio)-2-hydroxy-1-propanesulfonate) | Soltec Ventures | Cat # 82473-24-3 |
| CHAPS (3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate) | Sigma-Aldrich | Cat # C3023 |
| OG (n-Octyl-β-D-glucopyranoside) | EMD Millipore | Cat # 494459 |
| Ficoll 70 | GE | Cat # 17-0310-10 |
| Recombinant DNA | ||
| pET28a | Novagen | Cat # 69864-3 |
| pET15b | Novagen | Cat # 69661-3 |
| pTW34 | (Weber et al., 2000) | N/A |
| pET-SUMO-Munc18–1 | Shen et al., 2007 | Cat # 135550 in Addgene |
| pET-SUMO-VAMP2 | Shen et al., 2007 | Cat # 135551 in Addgene |
| pET-syntaxin-4 | Yu et al., 2013 | N/A |
| pET-SNAP-23 | Yu et al., 2013 | N/A |
| pET-SUMO-VAMP8 | Yu et al., 2019 | Cat # 135553 in Addgene |
| Software and Algorithms | ||
| KaleidaGraph | Synergy | https://www.synergy.com/wordpress_650164087/kaleidagraph/ |
Highlights.
A conformational constraint of SNAREs is removed by v-SNARE splitting
Split SNARE-driven fusion mimics the SM protein-activated fusion reaction
v-SNARE splitting enables efficient trans-SNARE zippering
Split SNARE-driven fusion lacks compartmental specificity
ACKNOWLEDGMENTS
We thank Drs. Yongli Zhang, Tom Perkins, Gus Lienhard, and Frederick Hughson for reagents or advice and Yan Ouyang for technical assistance. This work was supported by National Natural Science Foundation of China grants 91854117 and 31871425 (to H.Y.); a Natural Science Foundation of Jiangsu Province grant BK20200036 (to H.Y.); National Institutes of Health (NIH) grants GM126960 (to J.S.), DK124431 (to J.S.), and AG061829 (to J.S. and M.H.B.S.); and an American Diabetes Association Basic Science Award (to J.S.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108611.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, and Hughson FM (2015). A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst D, Zwilling D, Rizzoli SO, Lippert U, Lang T, and Jahn R (2006). Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl. Acad. Sci. USA 103, 2701–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman ER (2008). How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem 77, 615–641. [DOI] [PubMed] [Google Scholar]
- Dacks JB, and Field MC (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci 120, 2977–2985. [DOI] [PubMed] [Google Scholar]
- Dulubova I, Khvotchev M, Liu S, Huryeva I, Südhof TC, and Rizo J (2007). Munc18–1 binds directly to the neuronal SNARE complex. Proc. Natl. Acad. Sci. USA 104, 2697–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earp LJ, Delos SE, Park HE, and White JM (2005). The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol 285, 25–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellena JF, Liang B, Wiktor M, Stein A, Cafiso DS, Jahn R, and Tamm LK (2009). Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc. Natl. Acad. Sci. USA 106, 20306–20311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE, and Zhang Y (2012). Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337, 1340–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EP, Gatti E, Butler M, Burton J, and De Camilli P (1994). A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc. Natl. Acad. Sci. USA 91, 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC (2008). Viral membrane fusion. Nat. Struct. Mol. Biol 15, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, and Südhof TC (1993). Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366, 347–351. [DOI] [PubMed] [Google Scholar]
- Jahn R, and Scheller RH (2006). SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol 7, 631–643. [DOI] [PubMed] [Google Scholar]
- Jiao J, Rebane AA, Ma L, Gao Y, and Zhang Y (2015). Kinetically coupled folding of a single HIV-1 glycoprotein 41 complex in viral membrane fusion and inhibition. Proc. Natl. Acad. Sci. USA 112, E2855–E2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, He M, Port SA, Baker RW, Xu Y, Qu H, Xiong Y, Wang Y, Jin H, Eisemann TJ, et al. (2018). Munc18–1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife 7, e41771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasula R, Chai YJ, Bademosi AT, Harper CB, Gormal RS, Morrow IC, Hosy E, Collins BM, Choquet D, Papadopulos A, and Meunier FA (2016). The Munc18–1 domain 3a hinge-loop controls syntaxin-1A nano-domain assembly and engagement with the SNARE complex during secretory vesicle priming. J. Cell Biol 214, 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger CM, Ramirez-Macias I, Herman EK, Turkewitz AP, Field MC, and Dacks JB (2016). Resolving the homology-function relationship through comparative genomics of membrane-trafficking machinery and parasite cell biology. Mol. Biochem. Parasitol 209, 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Choi UB, Leitz J, Rhee HJ, Lee C, Altas B, Zhao M, Pfuetzner RA, Wang AL, Brose N, et al. (2017). Molecular Mechanisms of Synaptic Vesicle Priming by Munc13 and Munc18. Neuron 95, 591–607.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Kümmel D, Coleman J, Reinisch KM, Rothman JE, and Pincet F (2014). A half-zippered SNARE complex represents a functional intermediate in membrane fusion. J. Am. Chem. Soc 136, 3456–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier BT, Nickerson DP, Lo SY, and Merz AJ (2014). SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife 3, e02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Su L, Seven AB, Xu Y, and Rizo J (2013). Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Rebane AA, Yang G, Xi Z, Kang Y, Gao Y, and Zhang Y (2015). Munc18–1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife 4, e09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Cai Y, Li Y, Jiao J, Wu Z, O’Shaughnessy B, De Camilli P, Karatekin E, and Zhang Y (2017). Single-molecule force spectroscopy of protein-membrane interactions. eLife 6, e30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, Mahal LK, Sollner TH, and Rothman JE (2002). Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J. Cell Biol 158, 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, and Schekman R (1979). Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 76, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, and Scheller RH (1994). n-Sec1: a neural-specific syntaxin-binding protein. Proc. Natl. Acad. Sci. USA 91, 1445–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati AV, Stein A, and Fasshauer D (2006). N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science 313, 673–676. [DOI] [PubMed] [Google Scholar]
- Reese C, Heise F, and Mayer A (2005). Trans-SNARE pairing can precede a hemifusion intermediate in intracellular membrane fusion. Nature 436, 410–414. [DOI] [PubMed] [Google Scholar]
- Rizo J, and Südhof TC (2012). The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—guilty as charged? Annu. Rev. Cell Dev. Biol 28, 279–308. [DOI] [PubMed] [Google Scholar]
- Schlacht A, Herman EK, Klute MJ, Field MC, and Dacks JB (2014). Missing pieces of an ancient puzzle: evolution of the eukaryotic membrane-trafficking system. Cold Spring Harb. Perspect. Biol 6, a016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Tareste DC, Paumet F, Rothman JE, and Melia TJ (2007). Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell 128, 183–195. [DOI] [PubMed] [Google Scholar]
- Shen J, Rathore SS, Khandan L, and Rothman JE (2010). SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18–1 activation of membrane fusion. J. Cell Biol 190, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Rathore SS, Yu H, Gulbranson DR, Hua R, Zhang C, Schoppa NE, and Shen J (2015). The trans-SNARE-regulating function of Munc18–1 is essential to synaptic exocytosis. Nat. Commun 6, 8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, and Rothman JE (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. [DOI] [PubMed] [Google Scholar]
- Stein A, Weber G, Wahl MC, and Jahn R (2009). Helical extension of the neuronal SNARE complex into the membrane. Nature 460, 525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC, and Rothman JE (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, and Brunger AT (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Walter AM, Wiederhold K, Bruns D, Fasshauer D, and Sørensen JB (2010). Synaptobrevin N-terminally bound to syntaxin-SNAP-25 defines the primed vesicle state in regulated exocytosis. J. Cell Biol 188, 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li Y, Gong J, Ye S, Yang X, Zhang R, and Ma C (2019). Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly. Nat. Commun 10, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Parlati F, McNew JA, Johnston RJ, Westermann B, Sollner TH, and Rothman JE (2000). SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. J Cell Biol 149, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner TH, and Rothman JE (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Wickner W (2010). Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol 26, 115–136. [DOI] [PubMed] [Google Scholar]
- Yu H, Rathore SS, Lopez JA, Davis EM, James DE, Martin JL, and Shen J (2013). Comparative studies of Munc18c and Munc18–1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. USA 110, E3271–E3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Rathore SS, Shen C, Liu Y, Ouyang Y, Stowell MH, and Shen J (2015). Reconstituting Intracellular Vesicle Fusion Reactions: The Essential Role of Macromolecular Crowding. J. Am. Chem. Soc 137, 12873–12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Shen C, Liu Y, Menasche BL, Ouyang Y, Stowell MHB, and Shen J (2018). SNARE zippering requires activation by SNARE-like peptides in Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. USA 115, E8421–E8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Crisman L, Stowell MHB, and Shen J (2019). Functional Reconstitution of Intracellular Vesicle Fusion Using Purified SNAREs and Sec1/Munc18 (SM) Proteins. Methods Mol. Biol 1860, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, et al. (2017). Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rebane AA, Ma L, Li F, Jiao J, Qu H, Pincet F, Rothman JE, and Zhang Y (2016). Stability, folding dynamics, and long-range conformational transition of the synaptic t-SNARE complex. Proc. Natl. Acad. Sci. USA 113, E8031–E8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate unique code or dataset.


