SUMMARY
Piwi proteins are a subfamily of Argonaute proteins that maintain germ cells in eukaryotes. However, the role of their human homologs in cancer stem cells, and more broadly in cancer, is poorly understood. Here, we report that Piwi-like family members are overexpressed in glioblastoma (GBM), with Piwil1 (Hiwi) most frequently overexpressed (88%). Piwil1 is enriched in glioma stem-like cells (GSCs) to maintain self-renewal. Silencing Piwil1 in GSCs leads to global changes in gene expression resulting in cell-cycle arrest, senescence, or apoptosis. Piwil1 knockdown increases expression of the transcriptional co-regulator BTG2 and the E3-ubiquitin ligase FBXW7, leading to reduced c-Myc expression, as well as loss of expression of stem cell factors Olig2 and Nestin. Piwil1 regulates mRNA stability of BTG2, FBXW7, and CDKN1B. In animal models of GBM, Piwil1 knockdown suppresses tumor growth and promotes mouse survival. These findings support a role of Piwil1 in GSC maintenance and glioblastoma progression.
Graphical Abstract
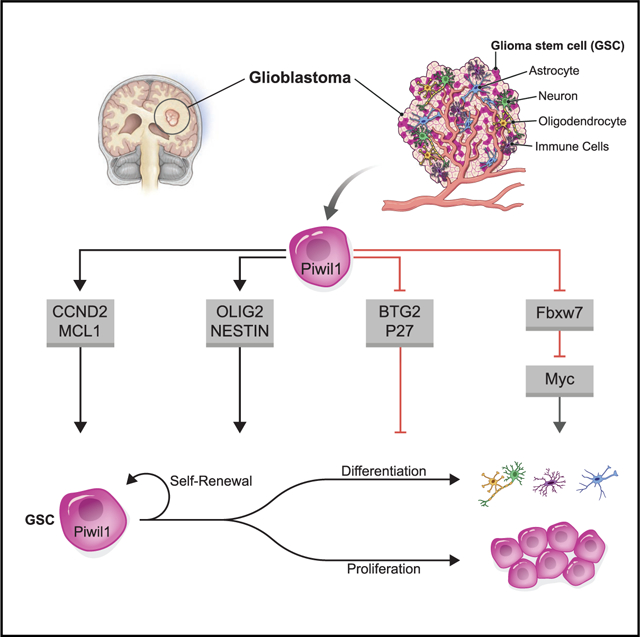
In Brief
Huang et al. find that Piwil1 protein is overexpressed in glioblastoma and glioma stem cells (GSCs). Piwil1 maintains GSC self-renewal and survival by regulating gene expression. Targeting Piwil1 extends survival in mouse models of glioblastoma. Piwil1 represents a therapeutic vulnerability.
INTRODUCTION
Glioblastoma (GBM) is an aggressive primary, malignant brain cancer. Despite maximal therapy, the cancer inevitably recurs. Glioma stem cells (GSCs) are a subset of cells that are highly resistant to conventional treatment. GSCs have the ability of self-renewal and differentiate and are implicated in tumor progression and recurrence (Bao et al., 2006; Chen et al., 2012; Eramo et al., 2006; Lathia et al., 2015). Understanding the molecular mechanisms that govern GSC maintenance is pivotal in the development of novel therapeutics.
Piwi proteins belong to a subfamily of Argonaute proteins that bind small RNAs. Piwi proteins are expressed highly in germ cells where they help to maintain genome stability by re-pressing transposons. Piwi was first discovered as an essential gene that guides asymmetric division of germline stem cells in Drosophila (Lin and Spradling, 1997). Piwi was found to be expressed in the germline stem cell niche to regulate bone morphogenic protein (BMP) signaling to control stem cell self-renewal and differentiation (Szakmary et al., 2005). Later, Piwi was found to control gene expression in germline stem cells by interacting with the PRC2 complex and preventing it from binding to chromatin (Peng et al., 2016). The most well-studied function of Piwi family proteins is their processing of and binding to piwi-interacting RNAs (piRNAs) to silence transposable elements to maintain genome integrity in germ cells (Iwasaki et al., 2015; Ozata et al., 2019). Since their discovery, Piwi proteins and piRNAs have been implicated in regulating gene expression outside of the germline (Peng and Lin, 2013; Rojas-Ríos and Simonelig, 2018; Ross et al., 2014).
The Piwi proteins are highly conserved across species. In humans, there are 4 Piwi proteins termed Piwi-like (Piwil) proteins, Piwil1 (Hiwi), Piwil2 (Hili), Piwil3 (Hiwi3), and Piwil4 (Hiwi2). Similar to their Drosophila homologs, these Piwi-like proteins regulate a broad range of activities including RNA stability, protein translation, gene transcription, chromatin dynamics, and stability. Whether the Piwi-like proteins play a role in somatic stem or progenitor cell maintenance is controversial. The expression of Piwi-like proteins was initially found to be elevated in hematopoietic stem cells, suggesting a role in stem cell maintenance (Nolde et al., 2013; Sharma et al., 2001). However, deletion of the three Piwi proteins in mice revealed normal hematopoiesis (Nolde et al., 2013). Thus, the role of Piwi-like proteins in normal stem or progenitor cell maintenance is yet to be determined.
Accumulating evidence suggests that Piwi proteins are dysregulated in cancer and may serve as cancer testis antigens (CT antigens) (Han et al., 2017; Litwin et al., 2017; Liu et al., 2019). The expression of Piwil1 through Piwil4 proteins were reported to be elevated in a variety of cancers including pancreatic cancer, endometrial, breast, and sarcoma (Chen et al., 2015; Han et al., 2017; Li et al., 2020; Litwin et al., 2017; Qu et al., 2015; Siddiqi et al., 2012; Taubert et al., 2007; Wang et al., 2016; Ye et al., 2010). In many of these cancers, overexpression of Piwi-like genes is correlated with aggressive tumor phenotype and poor patient survival (Han et al., 2017; Litwin et al., 2017). The Piwi-like proteins are thought to drive cancer progression by deregulating the cell cycle or altering cell motility. For example, in breast cancer cell lines, Piwil1 overexpression increases CDK4, CDK6, and CDK8 levels (Cao et al., 2016), and in sarcoma cell lines, Piwil1 regulates the cyclin-dependent kinase inhibitors (CDKI) p21 and p27 (Siddiqi et al., 2012). In pancreatic cancer, Piwil1 facilitates metastasis by reducing cell-cell adhesion (Li et al., 2020). Piwil2 is implicated in regulating the proliferation and survival of breast cancer stem cells through increased CyclinD1 and BCL-XL expression (Lee et al., 2010), and in non-small cell lung cancer, Piwil2 overexpression increases CDK2 and cyclin A (Qu et al., 2015). However, the functional consequences of Piwi-like proteins in cancer progression remains poorly understood. Considering the important role of Piwi proteins in germline stem cell maintenance, we interrogated their role in GSCs and more broadly in GBM.
RESULTS
Piwil1 Is Overexpressed in Glioblastoma
We first explored the expression of Piwil1 through Piwil4 in GBM using RNA sequencing (RNA-seq) data from the TCGA via cBioPortal. Expression data for the Piwi-like genes were available in 160 GBM specimens. The Piwi-like genes were overexpressed in 24/160 (16.9%) GBM patient specimens (Figure S1A). Piwil1 mRNA was most frequently overexpressed (10%) compared to Piwil2 (1.3%), Piwil3 (2.6%), or Piwil4 (3%) mRNA (Figure S1A). We next stained human GBM and normal brain for expression of Piwil1 by immunohistochemical analysis. In a panel of 25 human GBM specimens, 88% were positive of Piwil1 proteins with 64% showing strong staining. A subset of cells in perivascular areas showed strong positive staining for Piwil1 (Figures 1A and S1B). In contrast, in 6 non-neoplastic brain samples, there was no evidence of Piwil1 staining (Figure S1B). We further performed immunofluorescence staining in frozen human GBM specimens for Piwil1 with the stem cell marker Sox2 and found co-expression of Piwil1 and Sox2 in these samples (Figure 1B). Together, these data suggest that Piwil1 is overexpressed in a subset of GBM cells.
Figure 1. Piwil1 Is Frequently Overexpressed in GBM.
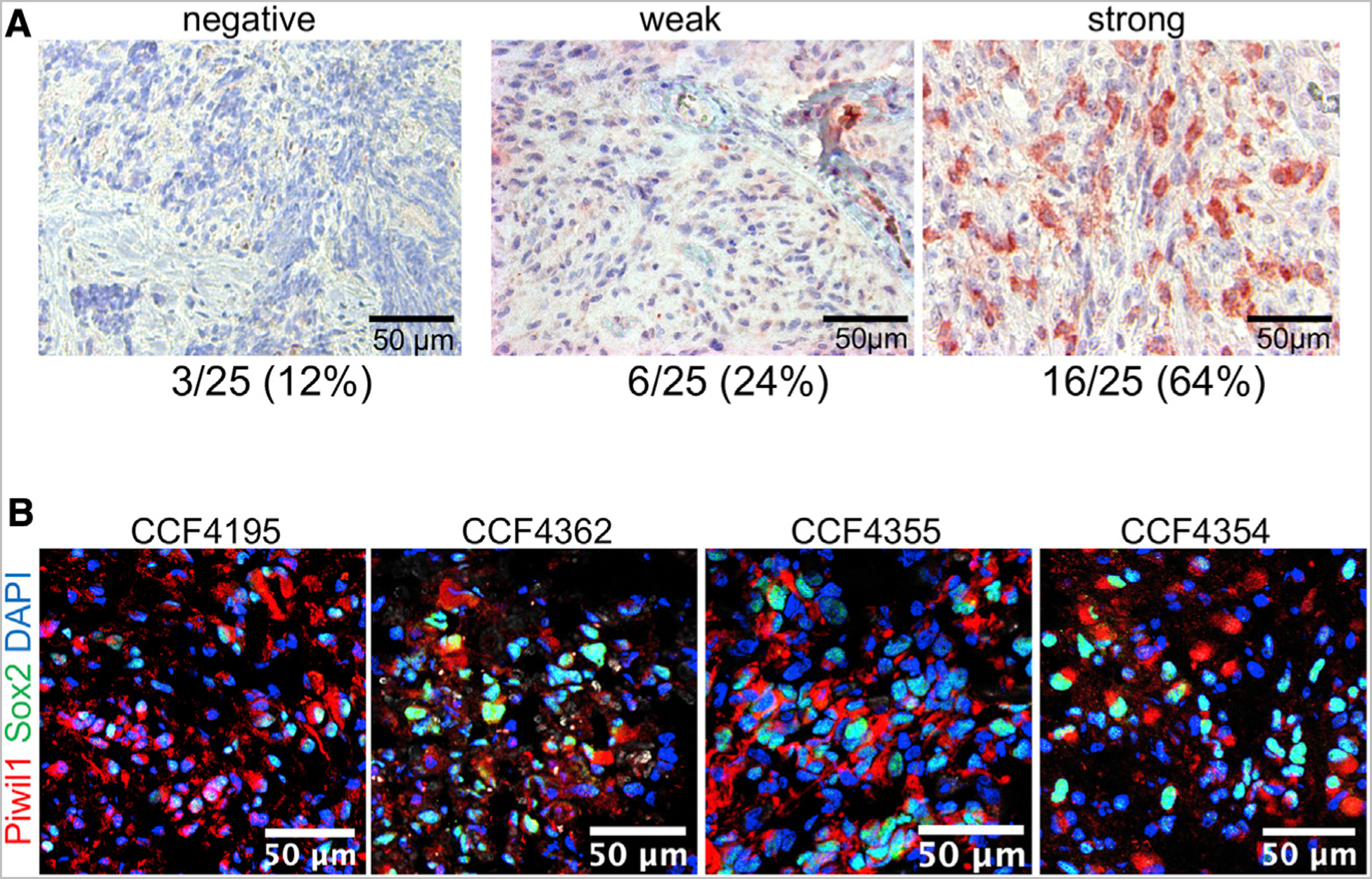
(A) Immunohistochemical staining of Piwil1 in human GBM.
(B) Immunofluorescence staining of Piwil1 (red) and Sox2 (green) in human GBM. Nuclei were counterstained with DAPI (blue).
See also Figure S1.
Piwil1 Is Expressed in Glioma Stem Cells
Because Piwil1 was first discovered as a stem cell factor in germline cells and co-stained with stem cell markers in GBM specimens, we assessed its expression in GSCs and their counterpart non-GSCs. Using four different GSC and non-GSC pairs, we found that Piwil1 is expressed at higher levels in GSCs than non-GSCs (Figure 2A). We then co-stained Piwil1 with stem cell markers Sox2, Olig2, and Nestin in human GBM tissues and found strong concordance in staining between Piwil1 and the stem cell markers (Figures 1B and 2B). More than 90% of Piwil1-positive cells were also Sox2-positive, whereas ~40% were Olig2-positive (Figure 2C). Our immunohistochemical staining studies suggest that Piwil1-positive cells can localize around blood vessels (Figures 1A and S1B). Notably, GSCs are known to reside in a perivascular niche that provides nutrients to support them (Calabrese et al., 2007). We next assessed Piwil1 expression with Sox2 and the endothelial cell marker CD31. Consistent with our immunohistochemical staining, Piwil1 was co-expressed with Sox2 in tumor cells around blood vessels (Figure 2D). Piwil1 is highly expressed in blood vessel-rich regions, with ~50% of cells within 100 μm to blood vessel are Piwil1+ (Figures 2E and S2). Together, these data suggest that Piwil1 is preferentially expressed in GSCs, particularly those in perivascular areas.
Figure 2. Piwil1 Is Preferentially Expressed in GSCs.
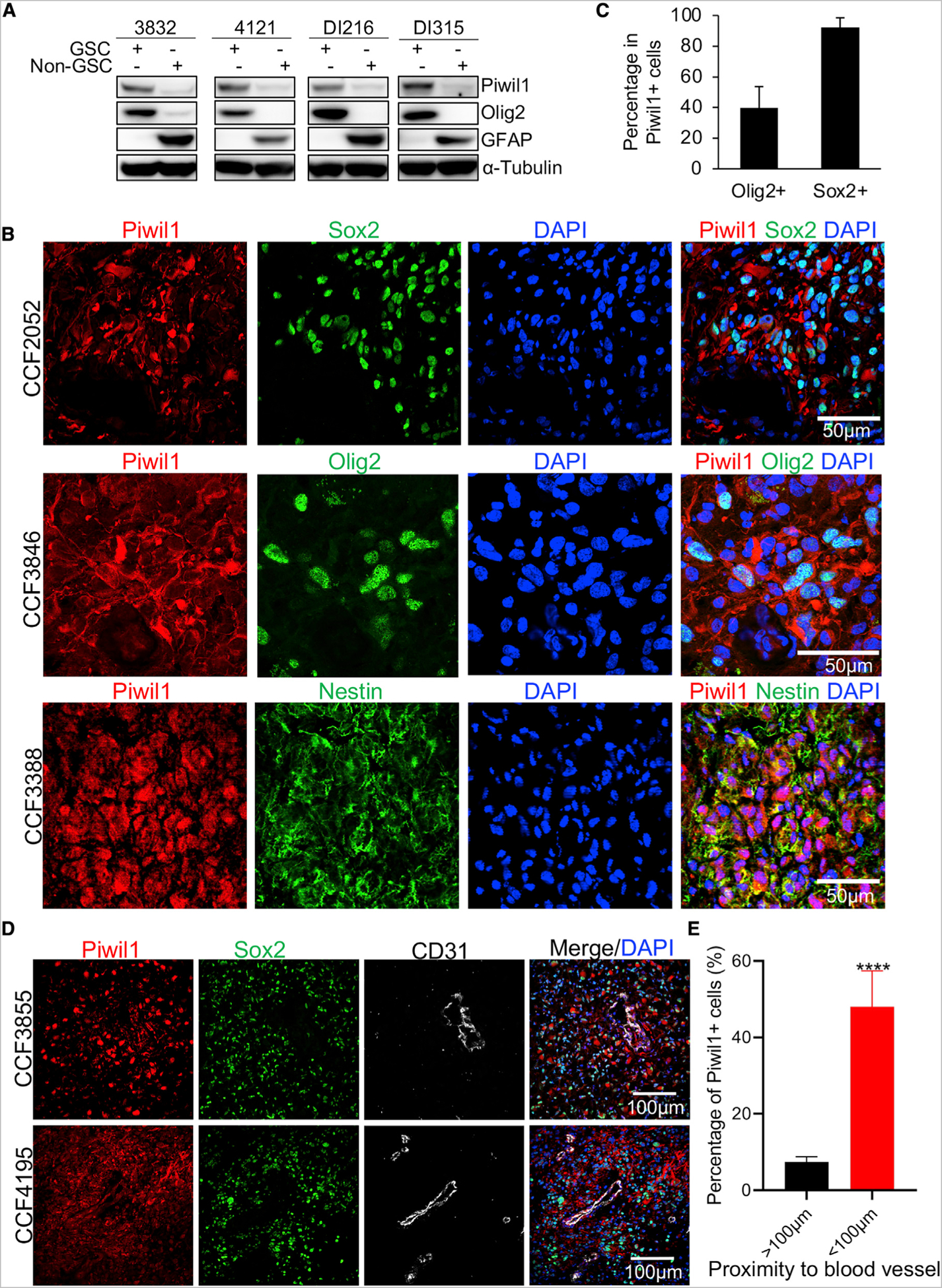
(A) Western blots of Piwil1, stem cell maker Olig2, and astrocyte marker GFAP in four paired GSCs and non-GSCs.
(B) Immunofluorescence imaging of Piwil1 (red) and stem cell markers (green) Sox2 (top), Olig2 (middle), and Nestin (bottom) in human GBM specimens. Nuclei were counterstained with DAPI (blue).
(C) Quantification of percentages of Sox2+ (n = 9 high-powered fields) and Olig2+ (n = 9 high-powered fields) cells in Piwil1+ cells.
(D) Immunofluorescence imaging of Piwil1 (red), Sox2 (green), and endothelial cell marker CD31 (gray) in human GBM specimens. Nuclei were counterstained with DAPI (blue).
(E) Quantification of Piwil1+ cells percentage within and beyond 100 μm distance from blood vessels (n = 15 high-powered fields).
****p < 0.0001. Data are represented as mean ± SD. See also Figure S2.
Piwil1 Is Required for GSC Survival and Self-Renewal
We then explored Piwil1 expression in GSCs derived from different patients. Consistent with its expression in GBM tissue specimens (Figure 1A), Piwil1 protein levels varied in 6 GSC lines we tested, with the highest levels in lines 4121 and 3832 and lower levels in lines 387 and 3359 (Figure S3A). To explore the function of Piwil1 in GSCs, we investigated the effect of Piwil1 disruption on proliferation and self-renewal ability. We silenced Piwil1 using two different short hairpin RNAs (shRNAs) or non-targeting control shRNA (shNT) (Figure 3A). Reducing Piwil1 expression significantly decreased cell viability in two different GSC populations (Figure 3A). In addition, Piwil1 knockdown decreased tumorsphere number and size (Figure 3B). Furthermore, Piwil1 knockdown (KD) in GSCs reduced proliferation, as determined by 5-ethynyl-2′-deoxyuridine (EdU) incorporation (Figure 3C). Silencing Piwil1 induced apoptosis, as indicated by increased levels of cleaved caspase 3 and cleaved PARP (Figure 3D). On extreme limiting dilution assay, silencing Piwil1 also decreased the frequency of sphere-forming cells (Figures 3E and 3F). To assess the function of Piwil1 in cells in which it is expressed at lower levels, we also knocked down Piwil1 in lines 387 and 3359. Piwil1 knockdown reduced cell viability and tumorsphere formation (Figures S3B and S3C), suggesting that even low levels of Piwil1 expression are biologically significant. Together, these studies indicate that Piwil1 regulates GSC survival and self-renewal.
Figure 3. Piwil1 Is Required for GSC Maintenance and Survival.
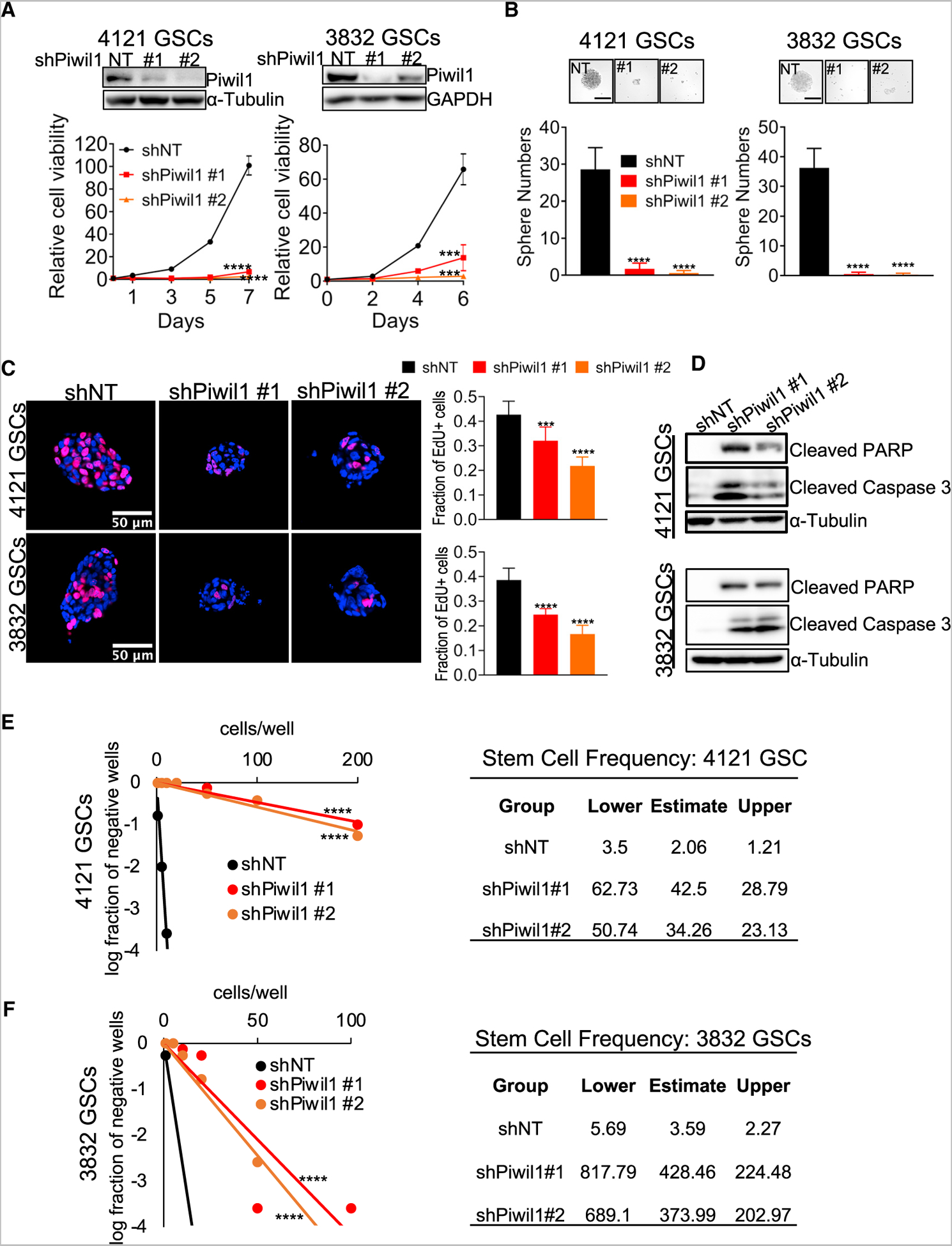
(A) Top panels: immunoblots show Piwil1 knockdown efficiency. Bottom panels: cell viability of 4121 and 3832 GSCs after Piwil1 knockdown (n = 4 replicates for each group).
(B) Top panels: representative images of tumorspheres in control shNT and shPiwil1 #1 and #2 GSCs. Scale bar, 200 μm. Bottom panels: quantification of tumor sphere numbers after Piwil1 knockdown (n = 12 replicates each group).
(C) Left panels: immunofluorescence imaging of EdU incorporation in 4121 and 3832 GSC tumorspheres after Piwil1 knockdown. Right panel: quantification of the fraction of EdU+ cells (n > 1,000 cells for each group).
(D) Western blot of cleaved PARP and cleaved caspase 3 proteins in GSCs after Piwil1 knockdown.
(E and F) in vitro limiting dilution assays of 4121 (E) and 3832 (F) GSCs after Piwil1 knockdown. Tables on the right show the estimated stem cell frequencies in control shNT and shPiwil1 knockdown groups.
***p < 0.001; ****p < 0.0001. Data are represented as mean ± SD. See also Figures S3 and S4.
Piwil1 was reported to be involved in metastasis in certain cancers (Li et al., 2020; Zhao et al., 2015a). We examined whether Piwil1 regulated the migration and invasion of GSCs. We performed in vitro transwell migration and wound healing assay in 4121 GSCs. Piwil1 knockdown had no significant effect on migration (Figures S4A and S4B). We then stained human nuclear antigen (HNA) to label human cells in a patient-derived xenograft model of GBM (Figure S4C). We found that tumor cells in both shNT and shPiwil1 GSC-derived xenograft models aggressively invaded mouse brains. These data indicated that Piwil1 is not required for GSC migration and invasion.
Piwil1 Gene Regulatory Network
To examine how Piwil1 affects GSC viability and self-renewal, we performed microarray analysis from GSCs transduced with two Piwil1 shRNAs (shPiwil1) and shNT. A volcano plot showed that Piwil1 knockdown globally affected mRNA expression (Figure 4A). Gene ontology and network analyses of differentially expressed genes revealed that Piwil1 regulated cell cycle, cellular differentiation, DNA damage response, and metabolism (Figures 4B and 4C).
Figure 4. Molecular Pathways Regulated by Piwil1.
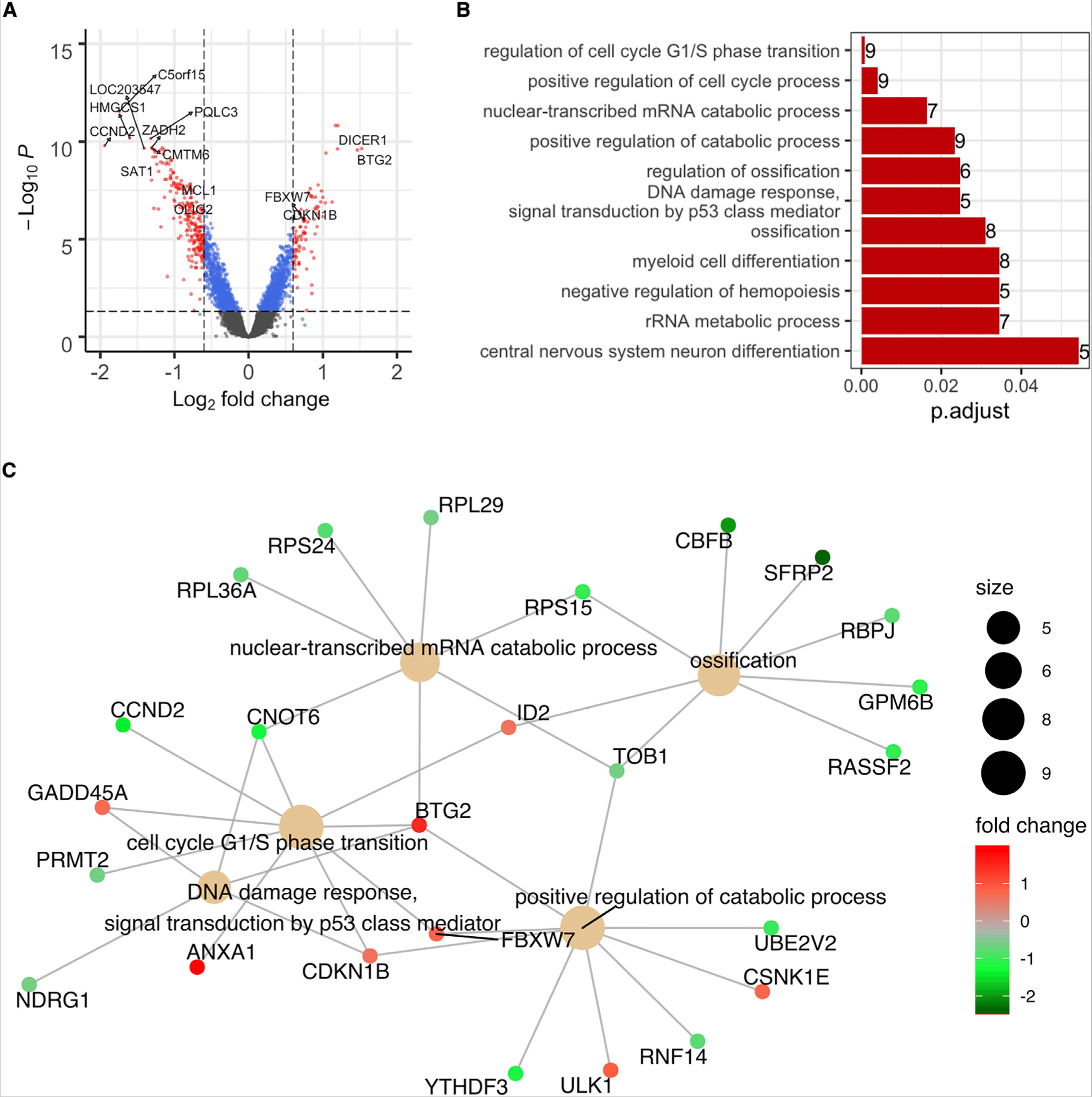
(A) Volcano plot of differentially expressed genes in 3832 GSCs after Piwil1 knockdown.
(B) Gene Ontology analysis of differentially expressed genes in 3832 GSCs after Piwil1 knockdown. Bar plot shows top altered pathways and the numbers at the right end of each bar show the numbers of genes affected in each pathway.
(C) Network map of top differentially expressed genes and pathways in 3832 GSCs after Piwil1 knockdown. The size of the circle denotes gene numbers in each pathway. Fold change in gene expression is shown.
The top gene dysregulated by Piwil1 silencing was B cell translocation gene 2 (BTG2, also known as PC3 or Tis21), a tumor suppressor gene that is downregulated in many cancers (Figures 4A and 4C) (Mao et al., 2015; Yuniati et al., 2019). BTG2 is a member of the BTG/TOB gene family and regulates cellular differentiation, proliferation, DNA damage response, apoptosis, RNA processing, and stability (Karve and Rosen, 2012; Mauxion et al., 2009; Stupfler et al., 2016). Using two different GSC populations, we validated that Piwil1 knockdown strongly increased BTG2 expression at both mRNA and protein levels (Figures 5A and 5B).
Figure 5. Piwil1 Regulates Cell Cycle, Senescence, and Stem Cell Marker Expression.
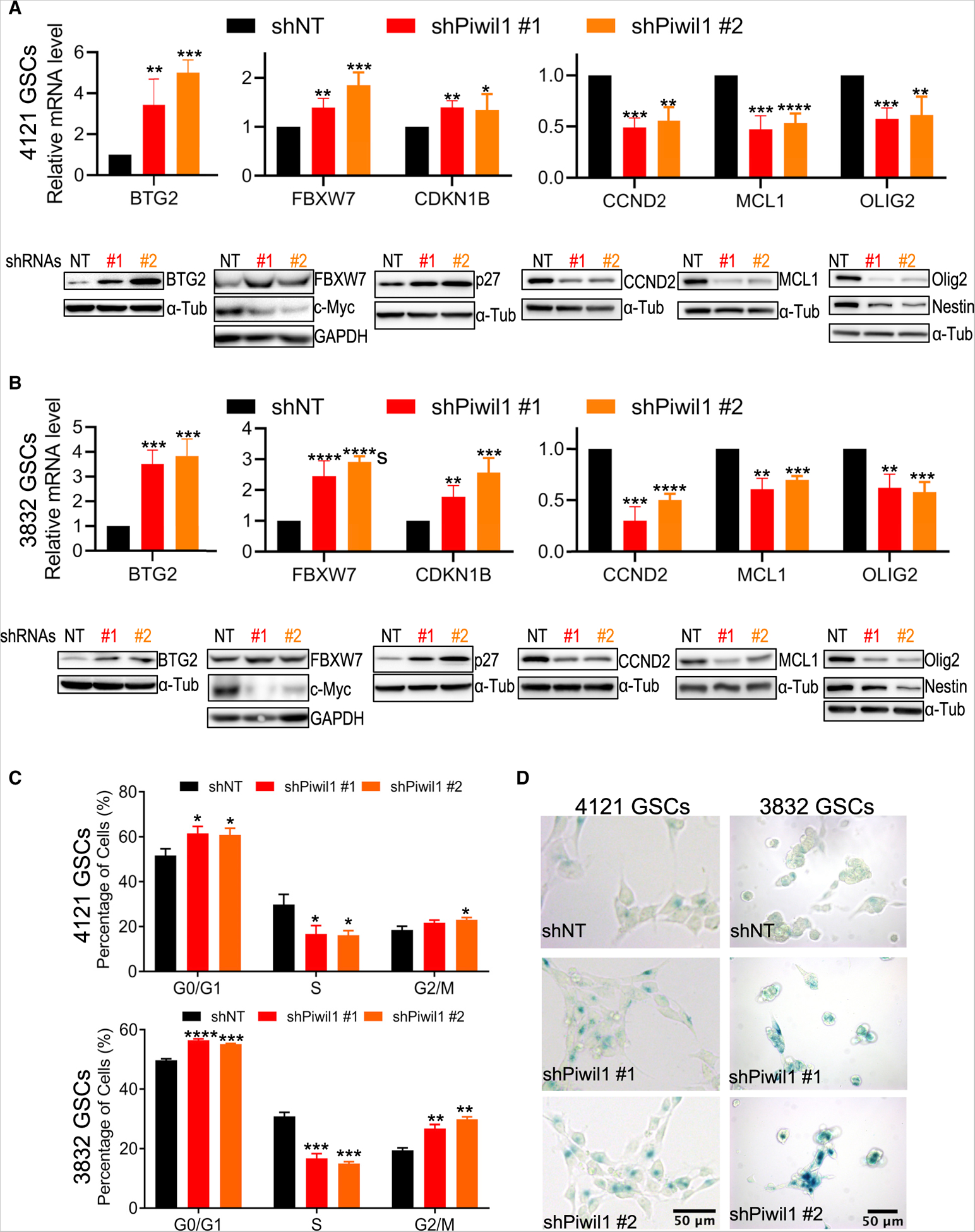
(A and B) qRT-PCR (upper panel, n = 3 replicates for each group) and immunoblot (lower panel) of BTG2, FBXW7, CDKN1B, CCND2, MCL1, and OLIG2 in 4121 (A) and 3832 (B) GSCs after Piwil1 knockdown.
(C) Cell-cycle analysis showing percentages of cells in each phase of the cell cycle in GSCs after Piwil1 knockdown (n = 3 replicates for each group).
(D) β-galactosidase staining of 4121 and 3832 GSCs on Matrigel after Piwil1 knockdown.
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Data are represented as mean ± SD. See also Figure S5.
Our gene expression analyses also revealed increased expression of the tumor suppressor FBXW7 after Piwil1 silencing (Figures 4A and 4C). We confirmed these results in GSCs and found upregulation of FBXW7 at both mRNA and protein levels in GSCs after Piwil1 knockdown (Figures 5A and 5B). FBXW7 is a component of the SCF (SKP1-CUL1-F-box) E3 ubiquitin-protein ligase complex that regulates the stability of many proteins including MYC, cyclin-E (CCNE1 or CCNE2), and the NOTCH1 intracellular domain (NICD) (Yeh et al., 2018). In accordance with increased expression of FBXW7, Piwil1 knockdown reduced c-Myc protein levels (Figure 5A and 5B). Expression of cyclin-E, NICD1, and Notch targets Hes-1/Hey-1 remained unchanged (data not shown).
Interestingly, the neural stem and progenitor cell marker Olig2 was also one of the top downregulated genes in GSCs in which Piwil1 was silenced (Figure 4A). We validated that both mRNA and protein levels of Olig2 were reduced in GSCs after Piwil1 knockdown (Figures 5A and 5B). Consistent with a loss of self-renewal capacity (Figures 3B, 3E, and 3F), Piwil1 knockdown also reduced expression of the stem cell marker Nestin (Figures 5A and 5B). These data suggest that Piwil1 may play an important role in regulating the expression of stem cell programs that maintain GSC self-renewal.
Piwil1 Regulates Cell-Cycle Progression
Gene ontology analysis of differentially expressed genes in Piwil1 knockdown showed significant changes in cell cycle (Figures 4B and 4C). We found altered expression of two key cell-cycle regulators, CDKN1B and CCND2, after Piwil1 knockdown. Silencing Piwil1 increased expression of CDKN1B at both the mRNA and protein levels (Figures 5A and 5B). CDKN1B encodes a cyclin-dependent kinase inhibitor p27 that regulates G1 progression by inhibiting the activity of cyclin D2-CDK4, cyclin E-CDK2, and cyclin A-CDK2 complexes (Bencivenga et al., 2017). Silencing Piwil1 also reduced expression of cyclin D2 (CCND2) (Figures 5A and 5B), which regulates G1/S transition (Sherr and Roberts, 2004). We next analyzed cell cycle by flow cytometry. Consistent with changes in the expression of these cell-cycle regulators, Piwil1 knockdown reduced the proportion of cells in S phase and increased those in G0/G1 and G2/M phases (Figure 5C).
Piwil1 Regulates Apoptosis and Senescence
Our microarray data also suggested that Piwil1 alters expression of MCL1 to regulate apoptosis and senescence (Bolesta et al., 2012; Wu et al., 2019). We validated that Piwil1 knockdown reduced expression of MCL1 at both the mRNA and protein levels (Figures 5A and 5B). Silencing Piwil1 induced apoptosis (Figure 3D) and facilitated senescence as seen by strong lysosomal β-galactosidase activity (Figure 5D). Taken together, these data suggest that Piwil1 modulates cell cycle, apoptosis, and senescence by coordinating the expression of multiple genes.
To determine which of these genes are causally related to Piwil1 function in GSCs, we performed rescue experiments. We overexpressed CCND2, Olig2, or MCL1 or knocked down BTG2, FBXW7, and CDKN1B in Piwil1 knockdown GSCs and measured cell viability. We found that MCL1 and Olig2 overexpression can partially but significantly rescue the Piwil1 knockdown phenotype (Figure S5). This suggested that Piwil1 regulated apoptosis, senescence, and stemness.
Piwil1 Regulates mRNA Stability and Binds to Specific mRNAs
Steady-state gene expression changes reflect alterations in transcription or mRNA stability. We assessed the possibility that the gene expression changes are due to alteration of transcription or mRNA stability by treating GSCs with actinomycin D, which inhibits global transcription, in GSCs expressing control shRNA or two different Piwil1 shRNAs (Figures 6A and 6B). Two GSC lines were used for these experiments. We then measured levels of transcripts over time. We found that Piwil1 knockdown increased the stability of BTG2, FBXW7, and CDKN1B mRNAs. The mRNA decay rates of MCL1, Olig2 (Figures 6A and 6B), and CCND2 (not shown), were not significantly changed, suggesting that Piwil1 knockdown decreased their transcription rather than regulated their mRNA stability.
Figure 6. Piwil1 Regulates mRNA Stability and Binds to Specific mRNAs.
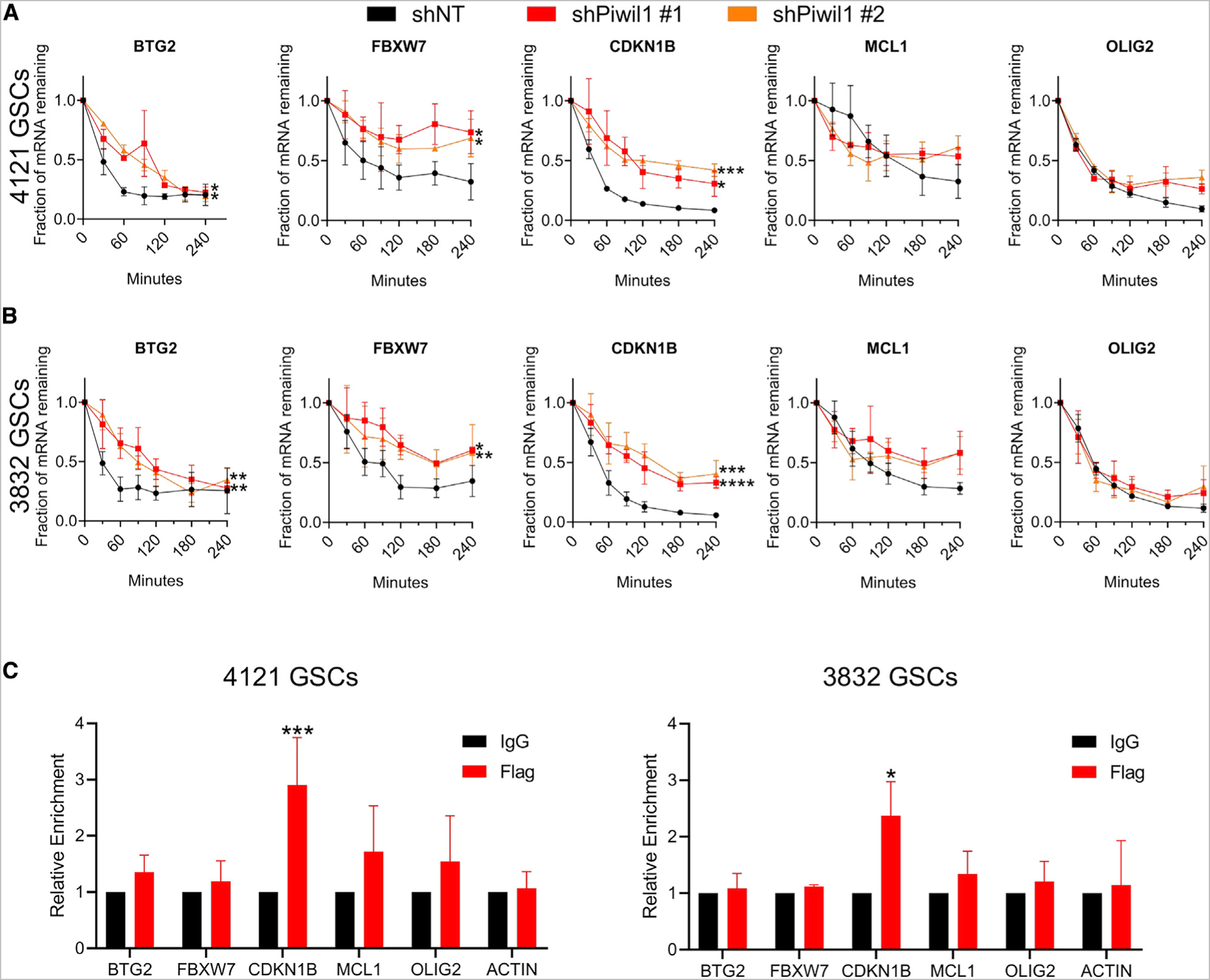
(A and B) qRT-PCR showing the fraction of mRNA remaining at different time points after actinomycin D treatment in 4121 (A) and 3832 (B) GSCs (n = 3 replicates for each group).
(C) qRT-PCR of mRNAs in Flag-Piwil1 UV-RNA immunoprecipitation in 4121 and 3832 GSCs (n = 3 replicates for each group).
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Data are represented as mean ± SD.
Given that Piwil1 is localized in the cytoplasm, and PIWI family proteins can bind mRNAs (Iwasaki et al., 2015; Ozata et al., 2019; Ross et al., 2014), we hypothesized that Piwil1 might bind to certain mRNAs to regulate their stability. To test whether Piwil1 binds to these mRNAs directly, we performed RNA immunoprecipitation (RIP) using anti-Flag antibody in GSCs expressing Flag-Piwil1 following UV crosslinking. UV crosslinks proteins and nucleic acids that directly interact. We found that only CDKN1B mRNAs were significantly enriched in Flag-Piwil1 RIP sample compared to immunoglobulin G (IgG) control (Figure 6C). These data suggest that Piwil1 directly binds to CDKN1B mRNA to regulate its stability. Piwil1 did not appear to bind to BTG2 or FBXW7 mRNAs. It is likely that Piwil1 regulates their stability indirectly through other factors.
Piwil1 Knockdown Reduces Tumor Growth and Improves Animal Survival
To examine the functional consequences of Piwil1 knockdown in vivo, we established orthotopic xenografts derived from shNT or shPiwil1-expressing GSCs (shNT mice or shPiwil1 mice, respectively). In the two GSC lines tested, shNT cells established intracranial tumors efficiently. In contrast, shPiwil1 mice showed only small tumors or in some animals no evidence of tumors at the pre-determined, 6-month post-injection endpoint (data not shown). Consistent with the reduction in tumor growth, shPiwil1 mice showed greatly increased animal survival compared to control shNT mice (Figure 7A).
Figure 7. Piwil1 Knockdown Reduces Tumor Growth and Improves Animal Survival.
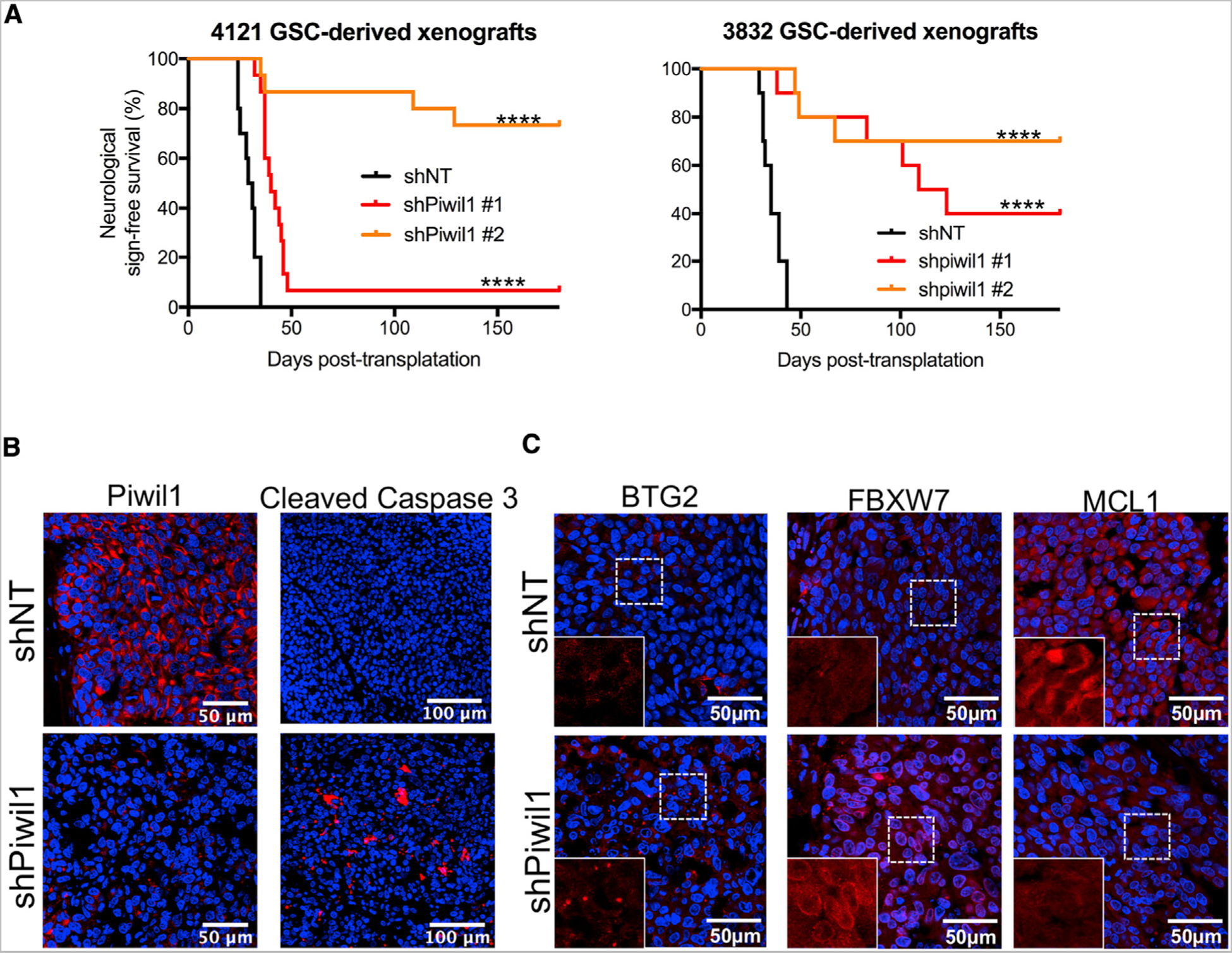
(A) Kaplan-Meier survival curve of mice implanted with 4121 or 3832 GSCs transduced with shNT or shPiwil1 shRNAs (n = 12 mice for each group).
(B) Immunofluorescence staining of Piwil1 and cleaved caspase 3 in shNT and shPiwil1 mice. Representative data from 3832 GSC-derived xenografts are shown. Nuclei were counterstained with DAPI (blue).
(C) Immunofluorescence staining of BTG2, FBXW7, and MCL1 in shNT and shPiwil1 mice. Representative data from 3832 GSC-derived xenografts are shown. Nuclei were counterstained with DAPI (blue).
****p < 0.0001.
Further analysis of tumors from these mice revealed higher levels of apoptosis in shPiwil1 mice, as indicated by increased cleaved Caspase 3 staining within the tumor (Figure 7B). Additionally, in accordance with our in vitro findings, tumors from shPiwil1 mice showed increased levels of BTG2 and FBXW7 staining and decreased MCL1 expression (Figure 7C). Together, these data reveal a critical role of Piwil1 in GSC-mediated tumor progression.
DISCUSSION
The Piwi proteins help maintain stemness in germline stem cells. In GBM, we report that all four human Piwi-like proteins (Piwil1 through Piwil4) are dysregulated. Of these Piwil-like genes, we found that Piwil1 mRNA is most frequently overexpressed in GBM. Piwil1 protein was overexpressed in 88% of GBM. Our data suggest that Piwil1 maintains GSC self-renewal and regulates their tumorigenic potential. Piwil1 is preferentially expressed in Sox2-, Olig2-, and Nestin-positive GSCs, particularly those in the perivascular niche. Piwil1 knockdown reduced GSC self-renewal and extended mouse survival in GSC-derived models of GBM.
We have found that Piwil1 coordinates the expression of multiple genes involved in cellular differentiation and cell-cycle progression. Piwil1 knockdown impaired expression of the stem cell transcription factor Olig2, and Olig2 overexpression was able to rescue the Piwil1 knockdown phenotype. These data suggest that Piwil1 may help maintain stemness at least in part through Olig2. Silencing Piwil1 also reduced expression of BTG2, the loss of which promotes tumorigenesis in leukemia, medulloblastoma, and oligodendroglioma by regulating differentiation (Appolloni et al., 2012; Dolezal et al., 2017; Kuiper et al., 2007; Mullighan et al., 2007; Presutti et al., 2018). In mouse models of GBM, we found that tumors in which Piwil1 was silenced exhibited increased BTG2 expression. These data suggest that Piwil1 is a key factor in maintaining the undifferentiated state of GSCs.
The Piwi-like proteins have also been implicated in cell-cycle regulation and proliferation. Piwi-like proteins were identified in breast cancers to guide cell-cycle progression by regulating the expression of cyclin D1, CDK4, CDK6, and CDK8 (Cao et al., 2016). In GSCs, we found that Piwil1 regulates G1/S progression. This is in part mediated by transcriptional regulation of CDKN1B, CCND2, and FBXW7, and its target substrate, c-Myc. These data suggest that Piwi-like proteins may regulate different cell-cycle components in different cancer types. Piwil1 silencing also augmented MCL1 expression and prompted cells to enter senescence. Thus, Piwil1 coordinates the expression of several key regulators to modulate cell-cycle progression, apoptosis, senescence, proliferation, and stemness. Our rescue experiments showed that among the genes regulated by Piwil1, MCL1 and Olig2 overexpression could partially rescue the Piwil1 knockdown phenotype. Therefore, Piwil1 regulates apoptosis, senescence, and stemness pathways. Piwil1 is involved in various functions through different downstream targets that coordinately promote GSC proliferation and maintenance.
Piwi-like proteins regulate mRNA decay in Drosophila embryo, mouse testis, and in the planarian Schmidtea mediterranea (Ramat and Simonelig, 2020). In GSCs, we found that Piwil1 regulates the stability of a subset of mRNAs. Piwil1 directly bound to CDKN1B mRNA to regulate its stability. Piwil1 may regulate BTG2 and FBXW7 mRNA stability indirectly. Further work to identify Piwil1 protein partners and its associated mRNAs globally is required to illuminate the details of Piwil1 functions.
mRNA decay regulation by Piwi-like proteins is reported to be dependent on piRNAs (Ramat and Simonelig, 2020). However, whether Piwil1 regulates gene expression through piRNAs in GSCs is not clear. Although small RNAs in the size range of piRNAs have been found in normal mouse tissues including brain and in some human cancers, their association with endogenous Piwi-like proteins or function in a piRNA/Piwi complex has yet to be demonstrated. Interestingly, Viljetic et al. (2017) found that Piwil1 knockout in mice resulted in profound changes in neocortical projections yet had no effect on piRNA-like small RNAs in mouse brain. This suggests that Piwil1 may function independently of piRNAs in the brain. In pancreatic and colon cancer cell lines that robustly expressed Piwil1, there was no evidence that Piwil1 bound to piRNA-like small RNAs (Genzor et al., 2019; Li et al., 2020). Furthermore, loss of Piwil1 had no effect on gene expression or transposon transcript abundance (Genzor et al., 2019). A recent study showed Piwil1 drives pancreatic cancer metastasis as a co-activator for anaphase promoting complex/cyclosome in a piRNA-independent way (Li et al., 2020). These data support a role for Piwil1 in cancer progression independent of piRNAs. Further research is needed to define the role, if any, of piRNAs in cancer.
The Piwi-like proteins may serve as cancer testis (CT) antigens. CT antigens are characterized by their expression in the reproductive system, predominantly in germ cell precursors with minimal if any expression in other normal tissues, and their ectopic expression in some cancers. Due to their restricted expression, CT antigens including Piwil1 are promising targets for cancer treatments. In GBM, silencing Piwil1 impaired self-renewal of GSCs and triggered senescence or apoptosis. This led to reduced tumor growth and prolonged mouse survival in two different GSC-derived xenograft models of GBM. In mouse embryos, Piwil1 was detected in some areas of the brain and may play a role in newborn neuron formation and neocorticogenesis. However, Piwil1 knockout mice are viable and do not exhibit overt neurologic deficits (Viljetic et al., 2017; Zhao et al., 2015a). Therefore, targeting Piwil1 in GBM patients may improve tumor control with minimal neurologic side effects.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jennifer S. Yu (yuj2@ccf.org)
Materials Availability
All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Microarray data generated in this study are available in GEO database under the accession number GEO: GSE160437
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committee at Cleveland Clinic. Mice used in these studies were 4–6-week old, NSG mice (from The Jackson Laboratory) and were maintained in the pathogen-free barrier animal facility at Cleveland Clinic Lerner Research Institute, which is accredited by the AAALAC (American Association for Accreditation of Laboratory Animal Care). A maximum of 5 mice per cage were allowed. Both male and female mice were used and littermates of the same sex were randomly assigned to experimental groups.
Human Subjects
GBM surgical specimens were collected for this study in accordance with a Cleveland Clinic Institutional Review Board-approved protocol. The sex of patients from whom tumor specimens were assessed are as follows: CCF4195, female; CCF4362, male; CCF4355, male; CCF4354, male; CCF2052, female. Age and sex of all other patients from whom tumor specimens were assessed are not available.
Cell Lines
293T used for virus production, were maintained as adherent cultures in DMEM supplemented with 10% fetal bovine serum, 1 mM L-Glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. Glioma stem cell lines 4121, 3832, 387 and 3359 were maintained as suspension cultures in Neurobasal-A medium with B27 supplement, 20 ng/ml EGF and 20 ng/ml FGF, 1 mM L-Glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin). All cells were kept in a humidified 5% CO2 incubator at 37°C. GSCs were from GBM patients of the following age and sex: 4121, 53 year old male (recurrent GBM), 3832, 75 year old female; 387, 69 year old, sex not available; 3359, 35 year old male.
METHOD DETAILS
Cells, Tissues, and Cell Culture
GBM surgical specimens and non-neoplastic brain tissues were collected for this study in accordance with a Cleveland Clinic Institutional Review Board-approved protocol. GSCs and non-GSCs were isolated and characterized from GBM surgical specimens or xenografts as previously described (Man et al., 2018). Briefly, tumors were disaggregated using the Papain Dissociation System (Worthington Biochemical) according to the manufacturer’s instructions. Isolated cells were recovered in stem cell medium (Neurobasal-A medium with B27 supplement, 20 ng/ml EGF and 20 ng/ml FGF, 1 mM L-Glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin) for at least 6 hours and then sorted by magnetic cell sorting for GSCs using the surface marker CD133 (Miltenyi Biotec.). CD133+ GSCs were cultured in stem cell medium as described above. The cancer stem cell phenotype of GSCs was confirmed by functional assays of self-renewal (serial neurosphere passage), stem cell marker expression, differentiation induction, and tumor propagation as previously described (Man et al., 2018). Non-GSCs were derived from GSCs that underwent a differentiation protocol using DMEM supplemented with 10% fetal bovine serum (FBS). Non-GSCs exhibited gain of differentiation markers including GFAP and loss of stem cell marker OLIG2 (Figure 2A).
Immunofluorescence Staining, Immunohistochemistry
Human glioblastoma (n = 25) and non-neoplastic brain tissues (n = 6) were obtained under an approved Cleveland Clinic IRB. Immunofluorescent staining of cells and tissues sections was performed as previously described (Man et al., 2014, 2018). Briefly, cells or tissue sections were fixed in 4% paraformaldehyde for 15 min at room temperature. After washing with PBS three times, samples were permeabilized with 0.3% Triton X-100 in PBS for 10 min, followed by three additional PBS washes. Samples were then blocked with 5% normal donkey serum for 60 min at room temperature and then incubated with primary antibodies overnight at 4°C followed by the appropriate secondary fluorescently labeled antibodies (Invitrogen) for one hour at room temperature. Nuclei were counterstained with DAPI. Images were acquired using a SP-5 confocal microscope. The following antibodies were used: Piwil1 (Abcam #ab12337), Sox2 (Millipore #MAB4423), OLIG2 (Millipore, #MABN50), CD31 (Abcam # ab187377), BTG2 (Santa Cruz #sc-517187), FBXW7 (Abcam #ab109617), MCL1 (Santa Cruz #sc-819). Immunohistochemical staining of paraffin embedded tissue sections were performed with Mouse and Rabbit Specific HRP/AEC IHC Detection Kit (Abcam # ab236467). The Piwil1 antibody used was from Abcam #ab12337. Images were processed with ImageJ (Schneider et al., 2012).
β-Galactosidase Staining
β-Galactosidase staining was performed using Senescence β-Galactosidase Staining Kit (Cell signaling #9860) following the instruction. Briefly, cells transduced with the indicated shRNAs were seeded on Matrigel and fixed with 1X Fixative Solution for 15 min at room temperature after rinsed with PBS. Cells were then incubated with β-Galactosidase Staining Solution at pH 6.0 at 37°C for 24 hr. Images were then captured under bright-field microscopy.
Immunoblot
Immunoblotting was performed as previously described (Man et al., 2014, 2018). Briefly, cells were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors. Protein samples were resolved by SDS-PAGE and transferred onto PVDF membranes. Blots were incubated with primary antibodies overnight at 4°C followed by HRP-conjugated species-specific secondary antibodies. The images were captured with the ChemiDoc Touch Imaging System (Bio-Rad) and analyzed with Image Lab software (Bio-Rad). The following antibodies were used: cleaved Caspase 3 (Cell Signaling #9664), cleaved PARP (Cell Signaling #5625), BTG2 (Santa Cruz #SC-517187), FBXW7 (Abcam #ab109617), Cyclin D2 (Novus #NBP2-14460), p27 (Cell Signaling #2552), MCL1 (Santa Cruz #sc-819), Olig2 (Millipore, #MABN50), Nestin (BD Biosciences #611658).
DNA Constructs and Lentiviral Transfection
Lentivirus DNA clones of control and Piwil1 shRNAs were purchased from Sigma-Aldrich. Flag-Piwil1 was generated by PCR from cDNA and cloned into the pCDH-MCS-T2A-Puro-MSCV lentiviral vector (System Biosciences), CCND2, MCL1 and OLIG2 were generated by PCR from cDNA and cloned into pCDH-EF1α-MCS-IRES-neo lentiviral vector (System Biosciences).
Lentivirus were produced in 293T cells. Briefly, lentivirus plasmids were transfected with packaging plasmid psPAX2 and envelope plasmid VSV-G (at a ratio of 1:1:1) into 293T cells. After 12 hours, the media were changed to neural basal media and incubated for another 48 hours. The collected mediums were passed through 0.45 μm filters and stored at −80°C.
Cell Viability Assays
For cell viability assays, 1000 cells transduced with shRNAs were seeded in 96-well plate. Cell titers were measured at the indicated days using the Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega). All data were performed in triplicates and normalized to day 0 and presented as mean ± standard deviation.
In vitro Limiting Dilution Assay
GSCs transduced with shRNAs were digested into single cells and seeded into 96-well plates with gradient cell concentrations of 1, 5, 10, 20, 50, 100 and 200 cells per well and 12 wells for each concentration. After incubation for 7 days, wells without tumor spheres were counted. The stem cell frequency was estimated with the online extreme limiting dilution assay (ELDA) tool (Hu and Smyth, 2009) (http://bioinf.wehi.edu.au/software/elda/).
EdU Incorporation
EdU incorporation experiment was performed using Click-iT EdU Imaging Kits (Thermo Fisher) following the instruction. Briefly, GSC spheres or matrigel-attached cells were incubated with Edu (1:2000) for 3 hours. Then the cells were fixed in 3.7% formaldehyde in PBS. After wash with 3% BSA in PBS, cells were permeabilized in 0.5% Triton X-100 for 20 min followed by two washes. The cells were incubated with Click-iT reaction cocktail for 30 min. After wash, cells were counter-stained with Hoechst 33342 and mounted for microscopic imaging. For each condition, EdU incorporation numbers were counts in at least 200 cells.
In vitro Transwell Migration and Wound Healing Assays
For in vitro transwell migration assays, 10000 cells transduced with control shRNA (shNT) or shPiwil1 lentivirus were suspended with neural basal medium with 1/10 supplements and plated on top of the filter membrane in transwell inserts (Corning), both sides of which were coated with 1% Matrigel (Corning). The inserts were placed in a 24-well plate with neural basal medium with full supplements. After incubation for 24 hours, the inserts were removed from the plate. Cells and media inside the inserts were removed using cotton-tipped applicator. Then the inserts were put in 70% ethanol to allow cell fixation for 10 minutes and then stained with 0.2% crystal violet for 10 minutes and washed extensively with distilled water. Transwell membranes were allowed to dry. Cells were viewed using an inverted microscope and the number of cells in different fields of view were counted.
For the in vitro wound healing assay, 3 × 106 cells transduced with control shRNA (shNT) or shPiwil1 lentivirus were seeded on 2% Matrigel (Corning) in a 6-well plate. After overnight incubation, the cells reached 100% confluence. A 200 μL pipette tip was used to scratch the cells to make a wound. The width at 20 sites per wound was measured immediately and after 24 hours incubation. The migration distances were calculated.
RNA Isolation and Real-time PCR
Total RNA was extracted from cells using PureLink RNA Mini Kit (Thermo Fisher). cDNA was synthesized with Invitrogen SuperScript IV VILO Master Mix (Thermo Fisher). Real-time PCR was performed with PowerUp SYBR Green Master Mix (Thermo Fisher) in ABI7500 instrument. Data were analyzed from at least three independent experiments and shown as the mean ± SD.
For mRNA stability measurements, cells expressing shNT or shPiwil1 were treated with Actinomycin D (Thermo Fisher) and collected at different time points (0, 30, 60, 90,120,180,240 min). RNAs were isolated and relative mRNA levels were calculated by normalizing with ACTIN and 0 min time point after real-time PCR.
UV-RNA Immunoprecipitation
GSCs stably expressing Flag-Piwil1 were collected and washed with cold PBS, and then irradiate once with 800 mJ/cm2 at 254 nm on ice. After wash with cold PBS, cells were lysed in polysome lysis buffer (10 mM HEPES, pH7.0, 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0.5% NP-40, 2 mM DTT) with protease inhibitors and RNase inhibitor and passed through 27G needles. After centrifugation at 1000 g for 10 min, the supernatants were collected and added to RIP buffer (50 mM Tris-Cl, pH7.4, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate with protease inhibitors and RNase inhibitor). The lysates were incubated with anti-Flag antibody (Sigma) or control mouse IgG and then with Protein G Dynabeads (Thermo Fisher). The samples were washed extensively with RIP buffer and RIP buffer with 500 mM NaCl, then with 1 M urea. After washing, the beads were digested with protease K in PK buffer (50 mM Tris-Cl, pH 7.4, 1% SDS, 100 mM NaCl, 0.5% NP-40) at 55°C for 1 hour and then extracted with Phenol/Chloroform and precipitated in 70% ethanol at −80 °C. The pellets were dissolved in water and cDNA was synthesized to perform qPCR. The relative enrichment was normalized to IgG for each gene and at least three independent experiments were performed.
Cell Cycle Analysis
Cells were digested with Accutase solution (Thermo Fisher) into single cells and fixed in cold 70% ethonal at −20°C. After washed with PBS, the cells were stained with Propidium Iodide Solution (Thermo Fisher) for 30 min at room temperature and then analyzed at BD Biosciences LSR II instrument. Cell cycle data were analyzed with Modfit LT software and at least three independent experiments for each cell line were performed.
Orthotopic Mouse Xenograft Studies
Intracranial transplantation of GSCs to establish GBM xenografts was performed as described (Man et al., 2014). Briefly, 24 hours after infected with shNT or shPiwil1 lentivirus, 2 × 104 GSCs were implanted into the right frontal lobes of 4-week old NOD scid gamma (NSG) mice. Animals were monitored daily and euthanized when they displayed neurologic signs. Animals were necropsied and underwent cardiac perfusion with PBS. Brains were harvested and fixed with 4% paraformaldehyde for 48 hours, cryopreserved in 30% sucrose at 4°C and cryosectioned for immunostaining and histological analysis.
Microarray and Bioinformatics Analysis
Total RNA was extracted from GSCs transduced with the indicated shRNAs using PureLink RNA Mini Kit (Thermo Fisher) and microarray analysis using HumanHT-12 v4.0 Gene Expression BeadChip (Illumina). Four replicates were used for each sample. Differential gene expression analysis and pathway analysis were performed using publicly available platforms including the biostatistics program R with packages beadarray (Dunning et al., 2007), limma (Ritchie et al., 2015), clusterProfiler (Yu et al., 2012).
The TCGA GBM, provisional dataset (including 160 patients) in cBioPortal (https://www.cbioportal.org/) was used to analyze the expression of Piwil1 through Piwil4 genes. mRNA Expression z-Scores (RNA Seq V2 RSEM) were set to 1.
QUANTIFICATION AND STATISTICAL ANALYSIS
All the statistical analyses were performed at Graphpad Prism software. All grouped data are presented as mean ± SD unless otherwise specified. All in vitro experiments were repeated at least three times and values of n were reported in the figure legends. Differences in means between groups were analyzed using two-tailed unpaired Student’s t test. A probability value of 0.05 or less was considered significant. For mouse xenograft studies, 12 mice were used for each group and significance testing was done by log rank test.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-BTG2 | Santa Cruz | Cat#sc-517187 |
| Rabbit monoclonal anti-CCND2 | Novus Biologicals | Cat# NBP2-14460 |
| Mouse monoclonal anti-CD31 | Abcam | Cat#ab187377, RRID:AB_2756834; lot#GR317844-4 |
| Rabbit monoclonal anti-Cleaved-Caspase 3 | Cell Signaling Technology | Cat#9664, RRID: AB_10891784‘ |
| Rabbit monoclonal anti-Cleaved-PARP | Cell Signaling Technology | Cat#5625S, RRID: AB_10699459; lot#13 |
| Rabbit polyclonal anti-FBXW7 | Abcam | Cat#ab109617; RRID:AB_2687519; lot#GR190912-1 |
| Mouse monoclonal anti-Flag | Sigma-Aldrich | Cat#F1804, RRID: AB_262044 |
| Mouse monoclonal anti-GAPDH | Santa Cruz | Cat# sc-47724, RRID:AB_627678; lot#F2216 |
| Rabbit polyclonal anti-GFAP | Agilent | Cat#Z0334, RRID:AB_10013382; lot#00085136 |
| Mouse monoclonal Human Nuclei antibody | Millipore | Cat#MAB1281, RRID:AB_94090; lot#3129247 |
| Rabbit polyclonal anti-MCL1 | Santa Cruz | Cat# sc-819; RRID:AB_2144105 |
| Mouse monoclonal anti-Nestin | BD Biosciences | Cat# 611658, RRID:AB_399176; lot#13748 |
| Mouse monoclonal anti-Olig2 | Millipore | Cat# MABN50, RRID:AB_10807410; lot#3109744 |
| Rabbit polyclonal anti-p27 | Cell Signaling Technology | Cat# 2552, RRID:AB_10693314; lot#7 |
| Rabbit polyclonal anti-Piwil1 | Abcam | Cat#ab12337; RRID:AB_470241; lot#GR312421-1 |
| Mouse monoclonal anti-Sox2 | Millipore | Cat#MAB4423, RRID: AB_11213224; lot#3112625 |
| Mouse monoclonal anti-α-Tubulin | Sigma-Aldrich | Cat# T6199, RRID:AB_477583 |
| Biological Samples | ||
| GBM specimens | Cleveland Clinic Brain Tumor Bank | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| EGF | R&D | Cat#236-EG |
| FGF | R&D | Cat#4114-TC |
| Actinomycin D | Invitrogen | Cat#A7592 |
| Critical Commercial Assays | ||
| Cell Titer-Glo | Promega | Cat#G7572 |
| Mouse and Rabbit Specific HRP/AEC IHC Detection Kit | Abcam | Cat#236467 |
| Senescence β-Galactosidase Staining Kit | Cell Signaling Technology | Cat#9860S |
| Click-iT EdU Imaging Kits | Thermo Fisher | Cat#C10338 |
| Transwell polycarbonate membrane cell culture inserts | Corning | Cat#3422 |
| SuperScript IV VILO Master Mix | Thermo Fisher | Cat# 11756050 |
| PowerUp SYBR Green Master Mix | Thermo Fisher | Cat#A25742 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE160437 |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | Cat# CRL-3216, RRID:CVCL_0063 |
| 4121 | Gifts from Dr. Jeremy Rich | N/A |
| 3832 | Gifts from Dr. Jeremy Rich | N/A |
| 387 | Gifts from Dr. Jeremy Rich | N/A |
| 3359 | Gifts from Dr. Jeremy Rich | N/A |
| Experimental Models: Organisms/Strains | ||
| NSG mice | The Jackson Laboratory | Cat# 005557 |
| Oligonucleotides | ||
| qPCR primers, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pCDH-MCS-T2A-Puro-MSCV lentiviral vector | System Biosciences | Cat#CD522A-1 |
| pCDH-EF1 α-MCS-IRES-neo lentiviral vector | System Biosciences | Cat#CD532A-2 |
| pMSCV-Flag-Piwil1 | This paper | N/A |
| pEF1 α-Flag-MCL1 | This paper | N/A |
| pEF1 α-Flag-OLIG2 | This paper | N/A |
| pEF1 α-Flag-CCND2 | This paper | N/A |
| shRNA human pLKO.1 CTRL shRNA | Sigma-Aldrich | Cat# SHC002 |
| shRNA human pLKO.1 Piwil1 shRNA#1 | Sigma-Aldrich | TRCN0000417558 |
| shRNA human pLKO.1 Piwil1 shRNA#2 | Sigma-Aldrich | TRCN0000007878 |
| Software and Algorithms | ||
| GraphPad Prism 8 | GraphPad Software | RRID:SCR_002798; https://www.graphpad.com/ |
| ImageJ | Schneider et al., 2012 | RRID:SCR_003070; https://imagej.nih.gov/ij/ |
| ELDA | Hu and Smyth, 2009 | RRID:SCR_018933; http://bioinf.wehi.edu.au/software/elda/ |
| Modfit LT | Verify Software House | RRID:SCR_016106; http://www.vsh.com/products/mflt/ |
| R Bioconductor package: Beadarray 2.36.1 | Dunning et al., 2007 | RRID:SCR_001314; https://bioconductor.org/packages/release/bioc/html/beadarray.html |
| R Bioconductor package: clusterProfiler 3.14.3 | Yu et al., 2012 | RRID:SCR_016884; https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| R Bioconductor package: limma 3.42.2 | Ritchie et al., 2015 | RRID:SCR_010943; https://bioconductor.org/packages/release/bioc/html/limma.html |
Highlights.
Piwil1 is overexpressed in glioblastoma and particularly in GSCs
Targeting Piwil1 impairs GSC maintenance
Piwil1 regulates mRNA stability in GSCs
Targeting Piwil1 decreases tumor growth and promotes mouse survival
ACKNOWLEDGMENTS
We thank Dr. Jeremy N. Rich for his generous gift of some of the GSCs used in this study. We thank the Burkhardt Brain Tumor and Neuro-Oncology Center brain tumor bank for glioblastoma specimens. We thank the Cleveland Clinic Center for Medical Art and Photography for assistance with the graphical abstract. The Graphical Abstract is ©2020 Cleveland Clinic Center for Medical Art & Photography. This work was funded by NIH/NINDS (R01NS094199 and R01NS092641), VeloSano, Case Comprehensive Cancer Center, and the Cleveland Clinic (to J.S.Y.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108522.
DECLARATION OF INTERESTS
E.J. is a co-founder of Bainom.
REFERENCES
- Appolloni I, Curreli S, Caviglia S, Barilari M, Gambini E, Pagano A, and Malatesta P (2012). Role of Btg2 in the progression of a PDGF-induced oligodendroglioma model. Int. J. Mol. Sci 13, 14667–14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, and Rich JN (2006). Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760. [DOI] [PubMed] [Google Scholar]
- Bencivenga D, Caldarelli I, Stampone E, Mancini FP, Balestrieri ML, Della Ragione F, and Borriello A (2017). p27Kip1 and human cancers: A reappraisal of a still enigmatic protein. Cancer Lett. 403, 354–365. [DOI] [PubMed] [Google Scholar]
- Bolesta E, Pfannenstiel LW, Demelash A, Lesniewski ML, Tobin M, Schlanger SE, Nallar SC, Papadimitriou JC, Kalvakolanu DV, and Gastman BR (2012). Inhibition of Mcl-1 promotes senescence in cancer cells: implications for preventing tumor growth and chemotherapy resistance. Mol. Cell. Biol 32, 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. (2007). A perivascular niche for brain tumor stem cells. Cancer Cell 11, 69–82. [DOI] [PubMed] [Google Scholar]
- Cao J, Xu G, Lan J, Huang Q, Tang Z, and Tian L (2016). High expression of piwi-like RNA-mediated gene silencing 1 is associated with poor prognosis via regulating transforming growth factor-β receptors and cyclin-dependent kinases in breast cancer. Mol. Med. Rep 13, 2829–2835. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu T-SS, McKay RM, Burns DK, Kernie SG, and Parada LF (2012). A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Che Q, Jiang F-Z, Wang H-H, Wang F-Y, Liao Y, Wan X-P, McLendon RE, Turner K, Perkinson K, and Rich J (2015). Piwil1 causes epigenetic alteration of PTEN gene via upregulation of DNA methyltransferase in type I endometrial cancer. Biochem. Biophys. Res. Commun 463, 876–880. [DOI] [PubMed] [Google Scholar]
- Dolezal E, Infantino S, Drepper F, Börsig T, Singh A, Wossning T, Fiala GJ, Minguet S, Warscheid B, Tarlinton DM, et al. (2017). The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nat. Immunol 18, 911–920. [DOI] [PubMed] [Google Scholar]
- Dunning MJ, Smith ML, Ritchie ME, and Tavaré S (2007). beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23, 2183–2184. [DOI] [PubMed] [Google Scholar]
- Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C, and De Maria R (2006). Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 13, 1238–1241. [DOI] [PubMed] [Google Scholar]
- Genzor P, Cordts SC, Bokil NV, and Haase AD (2019). Aberrant expression of select piRNA-pathway genes does not reactivate piRNA silencing in cancer cells. Proc. Natl. Acad. Sci. USA 116, 11111–11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y-N, Li Y, Xia S-Q, Zhang Y-Y, Zheng J-H, and Li W (2017). PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell. Physiol. Biochem 44, 1–20. [DOI] [PubMed] [Google Scholar]
- Hu Y, and Smyth GK (2009). ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78. [DOI] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, and Siomi H (2015). PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem 84, 405–433. [DOI] [PubMed] [Google Scholar]
- Karve TM, and Rosen EM (2012). B-cell translocation gene 2 (BTG2) stimulates cellular antioxidant defenses through the antioxidant transcription factor NFE2L2 in human mammary epithelial cells. J. Biol. Chem 287, 31503–31514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, and Hoogerbrugge PM (2007). High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia 21, 1258–1266. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, and Rich JN (2015). Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Jung C, Javadian-Elyaderani P, Schweyer S, Schütte D, Shoukier M, Karimi-Busheri F, Weinfeld M, Rasouli-Nia A, Hengstler JG, et al. (2010). Pathways of proliferation and antiapoptosis driven in breast cancer stem cells by stem cell protein piwil2. Cancer Res. 70, 4569–4579. [DOI] [PubMed] [Google Scholar]
- Li F, Yuan P, Rao M, Jin CH, Tang W, Rong YF, Hu YP, Zhang F, Wei T, Yin Q, et al. (2020). piRNA-independent function of PIWIL1 as a co-activator for anaphase promoting complex/cyclosome to drive pancreatic cancer metastasis. Nat. Cell Biol 22, 425–438. [DOI] [PubMed] [Google Scholar]
- Lin H, and Spradling AC (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463–2476. [DOI] [PubMed] [Google Scholar]
- Litwin M, Szczepańska-Buda A, Piotrowska A, Dzięgiel P, and Witkie-wicz W (2017). The meaning of PIWI proteins in cancer development. Oncol. Lett 13, 3354–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, Tao J, Li W, Yin X, and Xu W (2019). The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man J, Shoemake J, Zhou W, Fang X, Wu Q, Rizzo A, Prayson R, Bao S, Rich JN, and Yu JS (2014). Sema3C promotes the survival and tumor-igenicity of glioma stem cells through Rac1 activation. Cell Rep. 9, 1812–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man J, Yu X, Huang H, Zhou W, Xiang C, Huang H, Miele L, Liu Z, Bebek G, Bao S, and Yu JS (2018). Hypoxic Induction of Vasorin Regulates Notch1 Turnover to Maintain Glioma Stem-like Cells. Cell Stem Cell 22, 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Zhang Z, and Wang G (2015). BTG2: a rising star of tumor suppressors (review). Int. J. Oncol 46, 459–464. [DOI] [PubMed] [Google Scholar]
- Mauxion F, Chen CY, Séraphin B, and Shyu AB (2009). BTG/TOB factors impact deadenylases. Trends Biochem. Sci 34, 640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. (2007). Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–764. [DOI] [PubMed] [Google Scholar]
- Nolde MJ, Cheng EC, Guo S, and Lin H (2013). Piwi genes are dispensable for normal hematopoiesis in mice. PLoS ONE 8, e71950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, and Zamore PD (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet 20, 89–108. [DOI] [PubMed] [Google Scholar]
- Peng JC, and Lin H (2013). Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr. Opin. Cell Biol 25, 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Liu N, and Lin H (2016). Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Poly-comb group proteins. Nat. Genet 48, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presutti D, Ceccarelli M, Micheli L, Papoff G, Santini S, Samperna S, Lalli C, Zentilin L, Ruberti G, and Tirone F (2018). Tis21-gene therapy inhibits medulloblastoma growth in a murine allograft model. PLoS ONE 13, e0194206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Liu J, Zhong X, Li X, and Zhang Q (2015). PIWIL2 promotes progression of non-small cell lung cancer by inducing CDK2 and Cyclin A expression. J. Transl. Med 13, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramat A, and Simonelig M (2020). Functions of PIWI Proteins in Gene Regulation: New Arrows Added to the piRNA Quiver. Trends Genet. Published online September 17, 2020 10.1016/j.tig.2020.08.011. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Ríos P, and Simonelig M (2018). piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 145, dev161786. [DOI] [PubMed] [Google Scholar]
- Ross RJ, Weiner MM, and Lin H (2014). PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, and Hoffman R (2001). Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 97, 426–434. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, and Roberts JM (2004). Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18, 2699–2711. [DOI] [PubMed] [Google Scholar]
- Siddiqi S, Terry M, and Matushansky I (2012). Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS ONE 7, e33711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupfler B, Birck C, Séraphin B, and Mauxion F (2016). BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat. Commun 7, 10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A, Cox DN, Wang Z, and Lin H (2005). Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol 15, 171–178. [DOI] [PubMed] [Google Scholar]
- Taubert H, Greither T, Kaushal D, Würl P, Bache M, Bartel F, Kehlen A, Lautenschläger C, Harris L, Kraemer K, et al. (2007). Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 26, 1098–1100. [DOI] [PubMed] [Google Scholar]
- Viljetic B, Diao L, Liu J, Krsnik Z, Wijeratne SHR, Kristopovich R, Dutre-Clarke M, Kraushar ML, Song J, Xing J, et al. (2017). Multiple roles of PIWIL1 in mouse neocorticogenesis. bioRxiv. 10.1101/106070. [DOI] [Google Scholar]
- Wang Z, Liu N, Shi S, Liu S, and Lin H (2016). The Role of PIWIL4, an Argonaute Family Protein, in Breast Cancer. J. Biol. Chem 291, 10646–10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DM, Hong XW, Wen X, Han XR, Wang S, Wang YJ, Shen M, Fan SH, Zhuang J, Zhang ZF, et al. (2019). MCL1 gene silencing promotes senescence and apoptosis of glioma cells via inhibition of the PI3K/Akt signaling pathway. IUBMB Life 71, 81–92. [DOI] [PubMed] [Google Scholar]
- Ye Y, Yin D-T, Chen L, Zhou Q, Shen R, He G, Yan Q, Tong Z, Issekutz AC, Shapiro CL, et al. (2010). Identification of Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS ONE 5, e13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Bellon M, and Nicot C (2018). FBXW7: a critical tumor suppressor of human cancers. Mol. Cancer 17, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuniati L, Scheijen B, van der Meer LT, and van Leeuwen FN (2019). Tumor suppressors BTG1 and BTG2: Beyond growth control. J. Cell. Physiol 234, 5379–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P-P, Yao M-J, Chang S-Y, Gou L-T, Liu M-F, Qiu Z-L, and Yuan X-B (2015a). Novel function of PIWIL1 in neuronal polarization and migration via regulation of microtubule-associated proteins. Mol. Brain 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data generated in this study are available in GEO database under the accession number GEO: GSE160437


