Abstract
The synthesis and trafficking of iron-sulfur (Fe-S) clusters in both prokaryotes and eukaryotes requires coordination within an expanding network of proteins that function in the cytosol, nucleus, mitochondria, and chloroplasts in order to assemble and deliver these ancient and essential cofactors to a wide variety of Fe-S-dependent enzymes and proteins. This review focuses on the evolving roles of two ubiquitous classes of proteins that operate in this network: CGFS glutaredoxins and BolA proteins. Monothiol or CGFS glutaredoxins possess a Cys-Gly-Phe-Ser active site that coordinates an Fe-S cluster in a homodimeric complex. CGFS glutaredoxins also form [2Fe-2S]-bridged heterocomplexes with BolA proteins, which possess an invariant His and an additional His or Cys residue that serve as cluster ligands. Here we focus on recent discoveries in bacteria, fungi, humans, and plants that highlight the shared and distinct roles of CGFS glutaredoxins and BolA proteins in Fe-S cluster biogenesis, Fe-S cluster storage and trafficking, and Fe-S cluster signaling to transcriptional factors that control iron metabolism.
Keywords: Glutaredoxin, BolA, iron-sulfur cluster biogenesis, iron homeostasis, iron regulation, glutathione
1. Introduction
Transition metal trafficking, utilization, and regulation are critical processes for all organisms because transition metals serve as cofactors for a wide variety of essential pathways. In particular, iron is one of the most common cofactors for metalloenzymes owing to its abundance in nature and its favorable chemical properties [1]. This versatile redox-active metal can achieve multiple oxidation states, coordination geometries, and cofactor configurations in biological systems, in the form of mono- and di-iron centers, heme, and iron-sulfur (Fe-S) clusters. As such, iron metalloproteins play diverse, indispensable roles in cellular respiration, oxygen transport, nucleotide biosynthesis and repair, drug metabolism, biosynthesis of amino acids, proteins, cofactors, and vitamins, and many others [2]. Organisms must acquire and store iron in order to maintain sufficient levels to supply these iron-dependent pathways while minimizing harmful side reactions and mismetallation events that can occur with iron overaccumulation. The pathways for trafficking and storing iron as well as assembling iron cofactors involves coordination within an ever-growing network of proteins. Here we focus on two widespread protein families, the CGFS-type monothiol glutaredoxin (Grx) proteins and the BolA proteins, that specifically function in Fe-S cluster biogenesis, Fe-S cluster trafficking and storage, and iron sensing and regulation.
2. Class II monothiol CGFS glutaredoxins bind Fe-S clusters
Grxs are members of the ubiquitous thioredoxin (Trx) superfamily found in all domains of life. All Grxs harbor the canonical Trx fold consisting of a four-stranded anti-parallel β-sheet flanked by three α-helices (Fig. 1). Grxs can be classified into two main groups based on their active site motifs that are highly conserved between prokaryotes and eukaryotes. Additional less common Grx classes exist that are specific to terrestrial plants (Class III), eukaryotes (Class IV), or cyanobacteria (Classes V-VI) [3]. Class I Grxs with a classical dithiol CPY/FC motif catalyze reversible thiol-disulfide exchange reactions using the redox-active tripeptide glutathione (GSH) as a substrate (Fig. 1A). As such, they catalyze oxidation and reduction of disulfide bonds as well as mixed disulfides between protein thiols and GSH [4]. Dithiol Grxs thus have important roles in thiol redox control, redox signaling, and oxidative stress responses. Class II Grxs are also known as monothiol or CGFS-type Grxs since they harbor one or more Grx folds with a conserved Cys-Gly-Phe-Ser active site motif. The Grx domain in CGFS Grxs has two additional α-helices at the N- and C-termini (Fig. 1B). In contrast to Class I Grxs, Class II Grxs are enzymatically inactive in standard thiol-disulfide oxidoreductase assays [5]. Instead, these Grxs are implicated in iron metabolism due to their ability to reversibly bind Fe-S clusters. Two Grx monomers typically ligate a [2Fe-2S]2+ cluster at their interface using Cys ligands provided by the CGFS active sites and two GSH molecules. The GSH bound to the Fe-S cluster is nestled in the same GSH binding pocket that serves to hold GSH in the Grx1-GSH mixed disulfide [6]. This structural arrangement is demonstrated by the X-ray crystal structure of Escherichia coli Grx4 in its [2Fe-2S]-bridged homodimeric form (Fig. 1B, right). Interestingly, a recent report using a combination of structural analysis, mutagenesis, enzymatic assays, and in vivo functional complementation studies to evaluate the structure/function differences between Class I and Class II Grxs, revealed that the CGFS Grxs bind GSH in a slightly different orientation that favors [2Fe-2S] cluster binding over oxidoreductase activity [7]. These authors established that the GSH binding mode is dictated by the active site residues and the length of the loop region located between a conserved Lys group at the end of the first ß-sheet and the N-terminal active site Cys (Fig. 1). Class II Grxs have a longer loop than Class I Grxs, which subtly alters the orientation of the Lys residue and its interactions with GSH, which in turn influences the reactivity of the GSH thiol, the position of the bound Fe-S cluster, and the relative orientation of the protein monomers in the [2Fe-2S]-bridged homodimer [7]. Some CGFS Grxs may also bind cubane [4Fe-4S]2+ and linear [3Fe-4S]2+ clusters under certain conditions [8], although three-dimensional structures of these forms are lacking. Of note, formation of the cubane [4Fe-4S]2+ cluster has only been demonstrated for Saccharomyces cerevisiae Grx5 via in vitro reconstitution in the absence of GSH. Presumably, an additional Cys near the C-terminus that is partially conserved in some CGFS Grxs (e.g. Cys84 in E. coli Grx4, Fig. 1B and Fig. 2) serves as a cluster ligand in this conformation [8].
Figure 1.
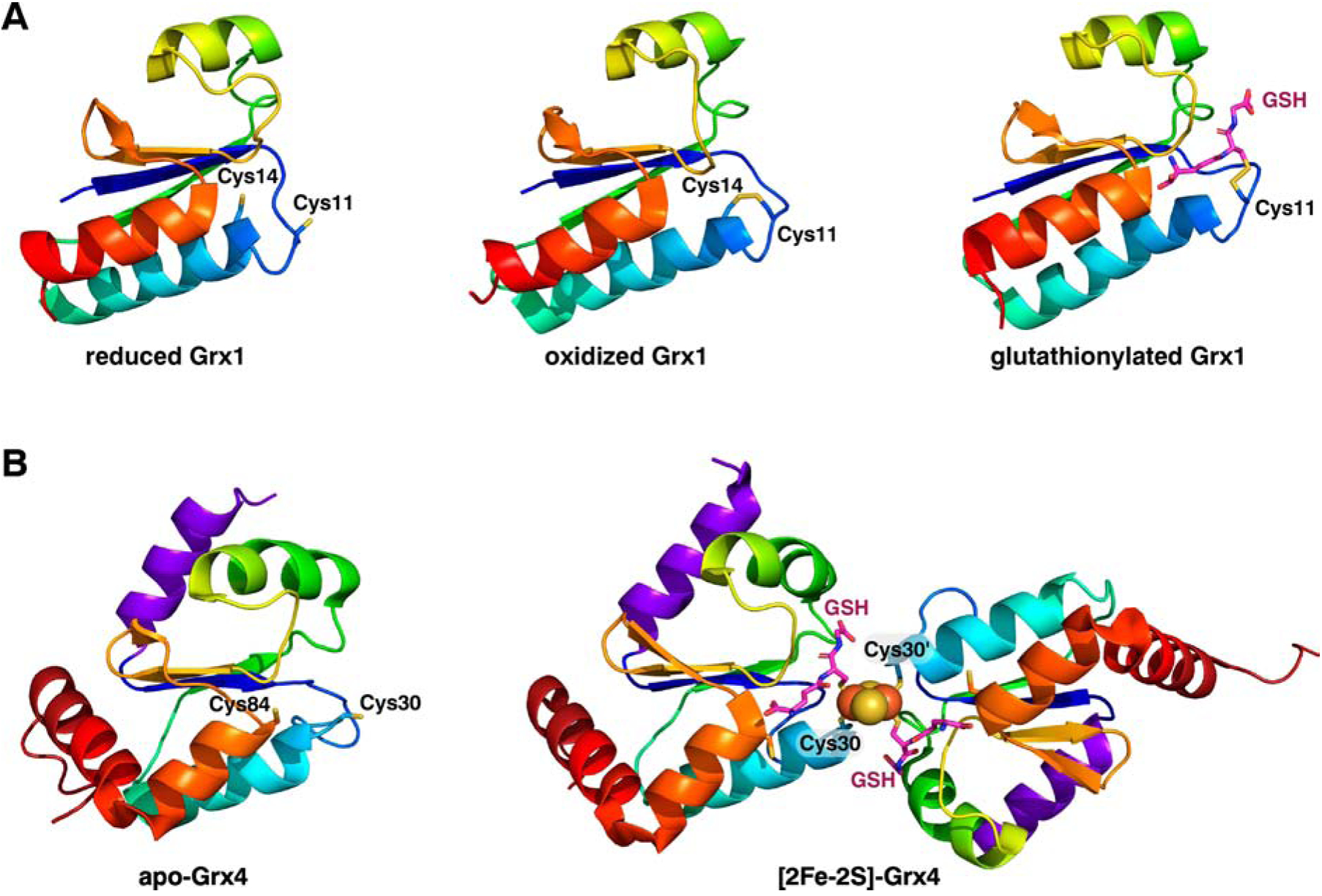
Structures of a typical class I dithiol Grx (A) and a class II monothiol Grx (B). A, NMR structures of E. coli Grx1 in the reduced form (PDB entry 1EGR), the disulfide oxidized form (PDB entry 1EGO), and the glutathionylated form (PDB entry 1GRX). NMR structure of E. coli Grx4 in the apo form (PDB entry 1YKA) and the X-ray crystal structure of the [2Fe-2S]-GSH2-bridged homodimer form (PDB entry 2WCI). The variable-length loop dictating Grx function is located between ß1 (in dark blue) and the N-terminal active site Cys (Cys-11 in Grx1 and Cys 30 in Grx4) [7].
Figure 2.
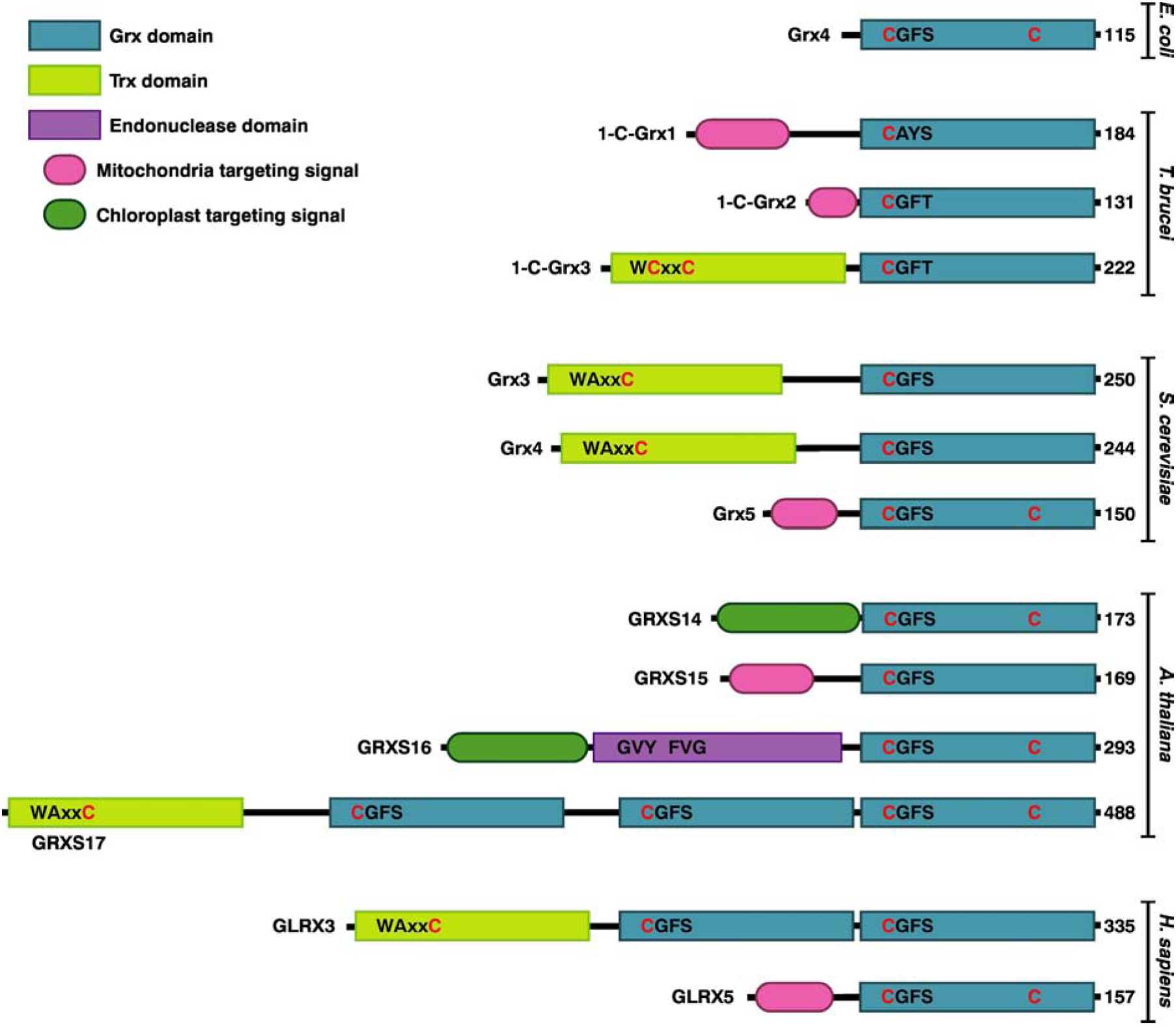
Domain structures of Class II Grxs from bacteria (Escherichia coli), trypanosomes (Trypanosoma brucei), yeast (Saccharomyces cerevisiae), plants (Arabidopsis thaliana), and mammals (Homo sapiens). The conserved cysteines in the Trx and Grx domains are shown in red. Additional conserved residues are shown in black. In eukaryotes, mitochondrial CGFS Grxs are single domain while cytosolic versions are multi-domain. Plants additionally have both single and multi-domain CGFS Grxs located in the chloroplast. Please note that the active site motifs of both cytosolic and mitochondrial Class II Grxs from trypanosomes (CAYS and CGFT) deviate from the canonical CGFS motif [117].
CGFS Grxs can be further classified into single and multidomain Grxs (Fig. 2). Single domain Grxs are found in bacteria, archaea, and the mitochondria and chloroplasts of eukaryotes (Fig. 3). Multidomain Grxs, which are typically localized to the cytosol and/or nucleus of eukaryotic organisms, have one Trx-like domain fused with one or more Grx domains, although other Grx fusion combinations are found in specific organisms [9]. For example, chloroplast-localized GRXS16 proteins in photosynthetic organisms have an N-terminal endonuclease domain fused to the C-terminal Grx-like domain [10]. The Trx-like domain in Trx-Grx fusions usually includes a variation of the classical WCGPC Trx active site motif (Fig. 2).
Figure 3.
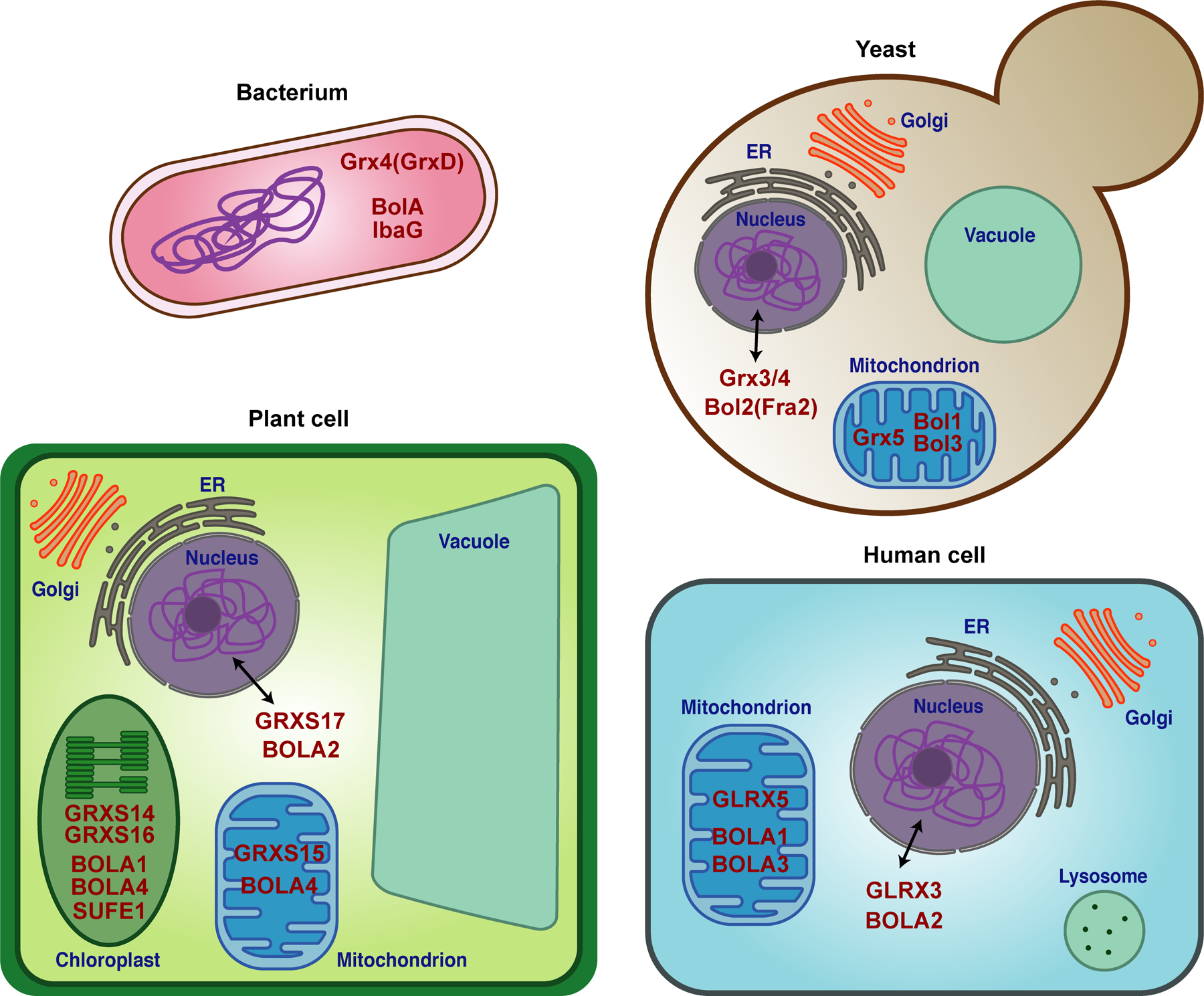
Diagrams depicting the subcellular localization of Grx and BolA homologs (dark red text) in bacteria, yeast, plant, and human cells.
3. BolA proteins are functional partners for CGFS Grxs
A strong connection between CGFS Grxs and the BolA protein family was first discovered using bioinformatics to mine genomes and functional genomics databases. Comparative genome analyses revealed that CGFS Grxs and BolA proteins co-occur in almost all sequenced genomes and are often found in close proximity in bacterial genomes, suggesting a cooperative function [11, 12]. Furthermore, proteomics approaches such as comprehensive yeast-two hybrid analyses and mass spectrometry of native protein complexes have identified physical interactions between these proteins in a wide variety of organisms [13–27]. Similar to the CGFS Grxs, BolA proteins are common in both prokaryotes and eukaryotes [25, 28]. All BolA proteins share a similar α1β1β2α2α3β3α4 fold with four α-helices and three β-sheets (Fig. 4A). Two of these α-helices (α2 and α3) are involved in a helix-turn-helix motif that is commonly found in nucleic acid binding proteins [29]. Phylogenetic analyses led to classification of BolA proteins into four main groups: BolA1, BolA2, BolA3, and BolA4 classes [25]. The BolA1 class is the most widespread and found in all organisms analyzed, with the exception of archaea and cyanobacteria. This class includes the first characterized member of this family, the E. coli BolA protein, which was named due the round, ball-like cell shape induced by its overexpression [30]. The BolA2 and BolA3 classes are exclusively limited to eukaryotes and can be distinguished by differences in conserved residues implicated in Fe-S cluster binding. The BolA4 class is found only in photosynthetic eukaryotes and prokaryotes including archaea and cyanobacteria [25]. In eukaryotes, BolA proteins are co-localized to the same subcellular compartments as CGFS Grxs. BolA2 proteins are typically found in the nucleo-cytoplasmic compartment, while BolA1, BolA3, and BolA4 proteins are localized in the mitochondria and/or chloroplast (Fig. 3).
Figure 4.
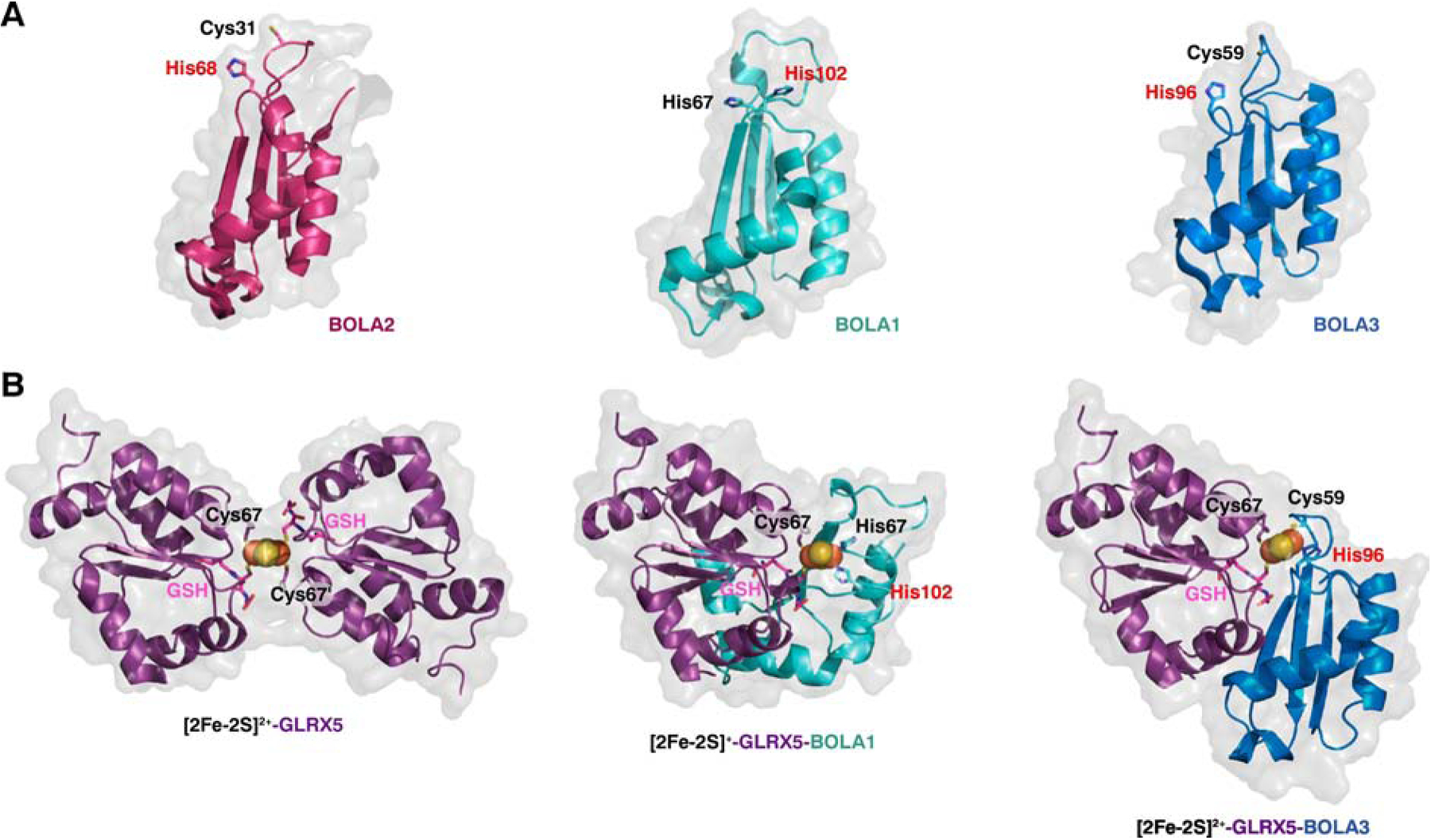
Structures of human apo-BOLA proteins, [2Fe-2S]-GLRX5 homodimer, and [2Fe-2S]-GLRX5-BOLA1/3 heterodimers. The invariant His in BolA proteins is shown as red text. (A) Human apo-BOLA2 modeled on NMR structure of mouse BOLA2 (PDB entry 1V9J) (left) and NMR structures of human apo-BOLA1 (PDB entry 5LCI) (center) and human apo-BOLA3 (PDB entry 2NCL) (right). (B) X-ray crystal structure of human [2Fe-2S]-GLRX5 homodimer (PDB entry 2WUL) and structural models of [2Fe-2S]-GLRX5-BOLA1 (center) and [2Fe-2S]-GLRX5-BOLA3 (right) heterodimers [35]. The GLRX5 monomers in the heterodimer structures in the middle and right were aligned with the left protomer of the [2Fe-2S]-GLRX5 homodimer using PyMol [118].
4. Structural Studies of Grx-BolA Interactions
Although there are currently no X-ray crystal structures available for any Grx-BolA complexes, a number of other structural, spectroscopic, biophysical, and biochemical techniques have been used to probe the dynamic physical interactions between these functional partners. Earlier studies relied on spectroscopic tools such as UV-visible absorption, circular dichroism (CD), resonance Raman, electron paramagnetic resonance (EPR), and Mossbauer spectroscopy to interrogate the coordination geometry, oxidative state, ligand identity, and stability of the Fe-S clusters bound by Grx-BolA heterocomplexes (reviewed in [11, 31, 32]). Moreover, titrations monitored via optical spectroscopy and gel filtration chromatography revealed that almost all BolA proteins can rapidly form 1:1 [2Fe-2S]-bridged complexes with individual Grx domains by displacing a Grx monomer and GSH from the Fe-S coordination sphere in Grx homodimers [11, 33, 34]. These spectroscopic approaches coupled with site-directed mutagenesis helped pinpoint the specific residues utilized by both Grx and BolA proteins to ligate the cluster. In all cases, the Grx partner binds the cluster using the strictly conserved Cys in the CGFS active site and a Cys from GSH. Likewise, all BolA proteins possess an invariant His located at the N-terminal end of the 3rd β-sheet that serves as a cluster ligand (Table 1, Fig. 4A). The other ligand provided by BolA is located in a loop between the 1st and 2nd β-sheet (known as the H/C loop) and is either a Cys or His depending on the specific BolA class. For homologs in the BolA1 and BolA4 classes, this loop contains a His, while proteins in the BolA2 and BolA3 classes all have a Cys [25, 28, 29, 35]. In several cases, substitution of this variable BolA ligand does not strongly impact the spectroscopic signature of the Grx-BolA complex [33, 35, 36] nor disrupt the function of the BolA protein in vivo [19, 36], suggesting that this ligand may be weakly coordinated and/or more easily exchangeable. Nevertheless, in most cases, the [2Fe-2S] bound in Grx-BolA heterocomplexes is less labile and more stable to reduction than the corresponding Grx [2Fe-2S]-bridged homodimers (Table 1). Although a few exceptions exist, including the E. coli [2Fe-2S]-Grx4-IbaG complex (discussed in section 5) [33] and the human [2Fe-2S]-GLRX5-BOLA3 complex (discussed below) [35], in which the [2Fe-2S] cluster is less stable compared to Grx4 or GLRX5 [2Fe-2S]-bridged homodimers, respectively.
Table 1.
Grx-BolA [2Fe-2S] complexes analyzed by structural and/or spectroscopic methods. The invariant His in BolA proteins is shown in bold.
| Organism | Grx partner | BolA partner | [2Fe-2S] stabilized or destabilizeda | BolA ligands | Reference |
|---|---|---|---|---|---|
| E. coli | Grx4 (GrxD) |
BolA | Stabilized | NDb | [42] |
| IbaG | Destabilized | His27 His63 |
[33] | ||
| S. cerevisiae | Grx3 | Bol2 (Fra2) |
Stabilized | Cys66 His103 |
[36, 37, 116] |
| Grx4 | Stabilized | Cys66 His103 |
[36, 116] | ||
| S. pombe | Grx4 | Fra2 | Stabilized | NDb | [23] |
| Human | GLRX3 (Grx3) |
BOLA2 | Stabilized | Cys31 His68 |
[107, 108] |
| GLRX5 (Grx5) |
BOLA1 | Stabilized | His67 His102 |
[35, 38] | |
| BOLA3 | Destabilized | Cys59 His96 |
[35, 38] | ||
| A. thaliana | GRXS14 | BOLA1 | Stabilized | His109 His144 |
[29] |
Refers to cluster stability in [2Fe-2S] Grx-BolA heterocomplexes relative to corresponding [2Fe-2S] Grx homodimers.
Not determined
More recently, several NMR studies have mapped the specific interaction faces between BolA proteins and CGFS Grxs in the presence or absence of the [2Fe-2S] cluster. The first study examined the atomic-level interactions between the A. thaliana chloroplastic proteins GRXS14 and BOLA1 and revealed for the first time the Rieske-type Cys2His2 Fe-S coordination found in Grx-BolA heterodimers involving BolA1 and BolA4 class proteins [29]. Interestingly, this study concluded that the interaction surfaces for the apo heterodimers are different from the holo heterodimers. The apo proteins exhibit a low μM binding affinity and chemical shift mapping revealed that the helix-turn-helix motif of apo-BolA interacts with the C-terminal region of apo-GRXS14. These regions do not include the Fe-S cluster ligands from either protein suggesting that the binding interfaces for the apo heterodimers are distinct from the interfaces for the holo heterodimers [29]. A similar conclusion was drawn from NMR studies examining the interaction between the cytosolic S. cerevisiae proteins Grx3 and Fra2 (recently renamed Bol2). Two independent surface plasmon resonance (SPR) analyses measured a 20–30 μM binding affinity between the two apo proteins [33, 37]. The interaction face for Fra2/Bol2 was identified by chemical shift mapping as the helix-turn-helix motif, similar to the plant BolA homolog. The Grx3 binding face for the apo complex was not reciprocally mapped by NMR; however, binding affinity measurements of Grx3 mutants suggested that the C-terminal α-helix was required for formation of the apo complex, again similar to the plant homolog [37].
In contrast, NMR analysis of the human mitochondrial binding partners GLRX5-BOLA1 and GLRX5-BOLA3 demonstrated that the apo proteins in each heterodimer pair interacted via similar regions involved in Fe-S cluster binding [38]. Apo-BOLA1 and apo-BOLA3 both bound to apo-GLRX5 with low μM affinity via an interaction surface that is centered around the invariant His [19, 38]. Likewise, the GLRX5 region involved in binding apo-BOLA1 and apo-BOLA3 was mapped to the region surrounding the GSH binding site, rather than the C-terminus as demonstrated for the plant and yeast homologs. These authors also mapped chemical shift changes upon formation of the holo-heterocomplexes. Similar changes were noted for the region around the BOLA1/BOLA3 invariant His upon holo-complex formation compared to the apo-complexes, with additional chemical shift changes detected for His67 of BOLA1 and Cys59 of BOLA3 in the holo-complexes. From the 15N-labeled GLRX5 spectra, they concluded that the same regions shifted in the apo interactions were altered in the holo-complexes with the addition of the conserved Cys67. Furthermore, these regions were identical to the interacting residues in GLRX5 homodimers, demonstrating that the same GLRX5 cluster ligands and binding surfaces are utilized for both homo and heterodimers [19, 38].
Although BOLA1 and BOLA3 use similar binding surfaces to interact with GLRX5, structural models of the holo-heterocomplexes based on the NMR data demonstrate that the two BOLA homologs have somewhat different orientations relative to the GLRX5 binding partner (Fig. 4B). Further analysis of the redox properties and Fe-S cluster stabilities of GLRX5-BOLA1 and GLRX5-BOLA3 holo-heterocomplexes revealed significant differences between the two mitochondrial BOLA homologs. Relative to the [2Fe-2S]-GLRX5 homodimer, BOLA1 binding stabilizes the cluster while BOLA3 binding destabilizes the cluster [38]. These observations are reminiscent of the opposing effects of bacterial BolA and IbaG proteins on E. coli [2Fe-2S]-Grx4 stability (see section 5 below). The high stability of [2Fe-2S]-GLRX5-BOLA1 is attributed to the relative inaccessibility (~ 3% accessible) of the bound cluster (Fig. 4B) [35]. Surprisingly, the reconstituted cluster in GLRX5-BOLA1 is found in the S = 1/2 reduced state ([2Fe-2S]1+), in contrast with all other purified and reconstituted Grx-BolA heterocomplexes that are isolated in the S = 0 oxidized state ([2Fe-2S]2+). The significance of this unique redox state is not clear, but based on these findings and in vivo analysis of BOLA1 function in human cells [28], the [2Fe-2S]-GLRX5-BOLA1 complex is proposed to perform a redox or electron transfer function in mammalian cells (discussed further in section 6.2). In contrast, the [2Fe-2S]2+ cluster in the GLRX5-BOLA3 heterodimer is more solvent exposed (~ 20% accessible) (Fig. 4B) and significantly more labile than the cluster in the GLRX5-BOLA1 complex [35].
5. Roles for CGFS Grxs and BolA proteins in bacteria
Genetic studies in E. coli provided the first functional connection between CGFS Grxs, BolA-like proteins, and bacterial Fe-S cluster biogenesis pathways (reviewed in [11, 12]). The E. coli genome includes a single CGFS Grx, namely Grx4 (encoded by the grxD gene), and two BolA proteins BolA and IbaG (formerly known as YrbA), which are classified as BolA1 and BolA4 proteins, respectively. ΔgrxD and ΔibaG strains each display synthetic lethality with mutations in the isc (iron sulfur cluster) operon, which encodes the housekeeping Fe-S cluster assembly pathway in E. coli (recently reviewed in [39]). The Isc pathway is composed of proteins that function together to assemble and deliver [2Fe-2S] and [4Fe-4S] cluster cofactors to target proteins. The isc operon encodes the core components of this assembly system, including the cysteine desulfurase IscS that mobilizes sulfide, the ferredoxin Fdx that donates electrons for sulfur reduction on IscS, the scaffold protein IscU on which the cluster is built, the HscAB chaperone system that facilitates ATP-dependent cluster release from IscU, and the carrier protein IscA that delivers clusters to target proteins. The E. coli genome also encodes a parallel pathway for Fe-S cluster assembly known as the Suf (sulfur utilization factor) system, which operates during oxidative stress and iron starvation conditions in which the Isc system is more vulnerable (reviewed in [40, 41]). Either the Isc or the Suf pathway must be functional for E. coli survival. The fact that ΔgrxD and ΔibaG strains exhibit aggravating genetic interactions with isc components suggests that the products of these genes function in the parallel Suf pathway for Fe-S cluster assembly. Furthermore, ΔgrxD strains are hypersensitive to iron depletion similar to suf mutants, and the grxD gene is upregulated during iron starvation [6, 11, 12]. Synthetically lethal genetic interactions for ΔbolA strains have not been reported; however, BolA was shown to interact with Grx4 in vivo [42].
To better understand the potential roles of Grx4, BolA, and IbaG in bacterial Fe-S cluster biogenesis, several biochemical studies have examined their protein-protein and metal-protein interactions. In addition to a [2Fe-2S]-bridged homodimer (see Fig. 1), Grx4 also forms [2Fe-2S]-bridged heterodimers with both BolA and IbaG [33, 42]. The heterodimers can be formed by titration of [2Fe-2S]-Grx4 with either apo-BolA or apo-IbaG. In addition, both BolA and IbaG bind apo-Grx4 with 3–5 μM affinity in the absence of Fe-S cluster coordination, suggesting a possible undefined function for the apo complexes. While the specific ligands utilized by BolA for cluster binding were not identified, IbaG is proposed to employ two conserved His ligands to bind the cluster [33]. Interestingly, interaction of BolA or IbaG with Grx4 has opposite effects on the Fe-S cluster stability. BolA binding to Grx4 stabilizes the [2Fe-2S] cluster [42], while IbaG binding destabilizes it, especially in the presence of GSH [33]. The significance of these differing biophysical properties is unclear. In vitro Fe-S cluster transfer assays have demonstrated that both E. coli Grx4 homodimer and Grx4-BolA heterodimer are able to donate Fe-S clusters to apo-Fdx [42]. However, the Grx4 homodimer, which binds the Fe-S cluster less stably than the Grx4-BolA heterodimer, is significantly more efficient at this process. More recently, fluorescently labelled E. coli Grx4 was used to investigate its function as an intermediate carrier in Fe-S cluster transfer reactions [43]. In this study, the multi-step process of Fe-S cluster biosynthesis and trafficking was examined using the IscS-IscU complex to synthesize [2Fe-2S] clusters with Grx4 serving as a fluorescent reporter to provide real-time kinetic information on cluster trafficking. These results demonstrated that Grx4 mediates rapid and efficient cluster transfer from the IscS-IscU complex to multiple target proteins via direct ligand exchange that does not involve a GSH-bound cluster intermediate. These findings support earlier biochemical studies revealing that a CGFS Grx from Azotobacter vinlandii serves as an intermediate carrier accepting a [2Fe-2S] cluster from the IscU scaffold (facilitated by the HscAB chaperone system in this case) and rapidly delivering this cluster to IscA [44, 45]. IscA, in turn, is proposed to deliver Fe-S clusters to apo target proteins. Interestingly, these authors also noted that cluster transfer between A-type Fe-S assembly proteins (e.g. IscA) and CGFS Grxs is reversed in the presence of a BolA protein. More specifically, they showed that S. cerevisiae apo Bol2-Grx3 can receive a [2Fe-2S] cluster from A. vinlandii NifIscA [44]. Although these donor:acceptor proteins are not physiological partners, these findings echo the E. coli Grx4/BolA/IbaG biochemical studies mentioned above demonstrating that BolA proteins modulate the cluster binding and transfer properties of CGFS Grxs [33, 42].
Apart from the synthetic lethality studies and low iron hypersensitivity of ΔgrxD strains mentioned above, additional strong genetic evidence for the roles of Grx4, BolA, and/or IbaG in E. coli Fe-S cluster biogenesis has been somewhat lacking. One recent study suggests that BolA plays a role in insertion of the N1b [2Fe-2S] cluster into E. coli respiratory complex I because this particular cluster is undetectable in electron paramagnetic resonance (EPR) analysis of membrane preparations from ΔbolA strains [46]. However, loss of this cluster only slightly impacts the measured NADH:ubiquinone oxidoreductase activity of this complex in ΔbolA strains. Similarly, ibaG deletion does not significantly alter complex I activity, but does lead to a measurable decrease in the stability of respiratory complex I in membrane preparations. In contrast, grxD deletion has no effect on complex I activity but does lead to a 25% reduction of the succinate oxidase activity of respiratory complex II, which contains three Fe-S clusters. This activity is further reduced by deletion of bolA. Furthermore, ΔbolA and ΔibaG single mutants possess normal succinate oxidase activity, while the ΔbolA ΔibaG double mutant exhibits a 50% reduction in activity. Taken together, these results suggest that BolA, IbaG, and Grx4 may have overlapping and interdependent roles in the insertion of Fe-S clusters into E. coli respiratory complex II [46].
Additional genetic and biochemical studies have been performed with a variety of bacteria to unlock the specific functions of BolA proteins independent of Grx4. Earlier studies in E. coli indicated that BolA served as a transcriptional regulator with pleiotropic effects that impacted cell morphology, stress responses, biofilm formation, membrane permeability, and cell motility [30, 47, 48]. Similar roles have also been proposed in a variety of other bacteria. For example, in the filamentous cyanobacterium Fremyella diplosiphon, bolA depletion impacts cell morphology and leads to increased ROS levels, while bolA overexpression decreases ROS levels and increases filament length [49, 50]. In Shewanella oneidensis, bolA overexpression increases biofilm formation but does not impact cell morphology [51]. In addition, bolA deletion leads to decreased virulence of the enteric pathogen Salmonella enterica serovar Typhimurium, [52], while bolA overexpression triggered an increase in metabolites involved in stress response and virulence [53].
In contrast, neither bolA deletion nor overexpression was found to impact cellular morphology in the gram-negative pathogen Vibrio cholerae. Instead, a strong connection between IbaG function, cellular morphology, and Fe-S cluster metabolism was recently discovered, shedding new light on the specific function of this BolA homolog [54]. Similar to the E. coli ibaG gene, the V. cholerae ibaG gene is located near genes involved in peptidoglycan synthesis and cellular envelope maintenance. Accordingly, V. cholerae ΔibaG strains exhibit distorted cell shapes, intestinal colonization defects, and marked sensitivity to cell envelope stressors. These phenotypes are all attributed to alterations in the peptidoglycan and lipopolysaccharide levels in the cell envelope. Affinity-tagged IbaG was also found to copurify with at least 19 different Fe-S-cluster binding proteins, including the Fe-S cluster assembly factors IscU, IscS, NfuA, and ErpA. Most intriguingly, a physical interaction between IbaG and IspG was revealed by both copurification and bacterial two-hybrid analysis [54]. IspG is a [4Fe-4S]-containing enzyme that catalyzes an essential step in the synthesis of isoprenoids required for peptidoglycan precursor assembly. This discovery suggests that IbaG functions in assembly or trafficking of clusters to IspG. Although E. coli ΔibaG strains have not been characterized with similar cell envelope deficiencies [55], the synthetic lethality observed between ibaG and Fe-S assembly pathways and the confirmed Fe-S cluster binding ability of E. coli IbaG [33] supports this hypothesis. Whether or not Grx4 partners with IbaG in this function is unknown.
6. Roles for CGFS Grxs and BolA proteins in mitochondria
6.1. Grx5, Bol1, and Bol3 function in mitochondrial Fe-S assembly and trafficking in yeast.
In eukaryotes, single-domain Grxs are co-localized to the mitochondrial matrix with BolA proteins from the BolA1, BolA3, and/or BolA4 classes (Fig. 3). Since mitochondria are descendants of endosymbiotic bacteria, they inherited many components of the iron-sulfur cluster assembly (ISC) machinery described in section 5. Genetic studies in the model yeast S. cerevisiae first identified Grx5 as a critical component of the mitochondrial ISC system (recently reviewed in [56, 57]). Deletion of grx5 led to reduced activity of Fe-S-dependent enzymes, oxidative damage, and dysregulation of iron homeostasis causing mitochondrial iron overaccumulation. Complementation of these phenotypes could be achieved with Grx5 homologs from both prokaryotes and eukaryotes, revealing that the function of CGFS Grxs is evolutionarily conserved from bacteria to humans. Similar phenotypes were noted in grx5Δ mutants from the fission yeast Schizosaccharomyces pombe, while in vivo interactions analysis identified a physical interaction between S. pombe Grx5 and the A-type scaffold proteins Isa1 and Isa2. Putting together information gleaned from both in vivo and in vitro studies, the current model for the mitochondrial Fe-S cluster assembly pathway places Grx5 as a central player linking the early and late-acting ISC machinery [56, 57]. The early ISC machinery initially assembles a [2Fe-2S] cluster on the scaffold protein Isu1 using iron, sulfide released from cysteine, and electrons donated from a ferredoxin (Fig. 5). The [2Fe-2S] cluster is then released from Isu1 via the Hsp40-Hsp70 chaperone system (Jac1 and Ssq1), which interacts with both Isu1 and Grx5 to facilitate cluster transfer [58].
Figure 5.
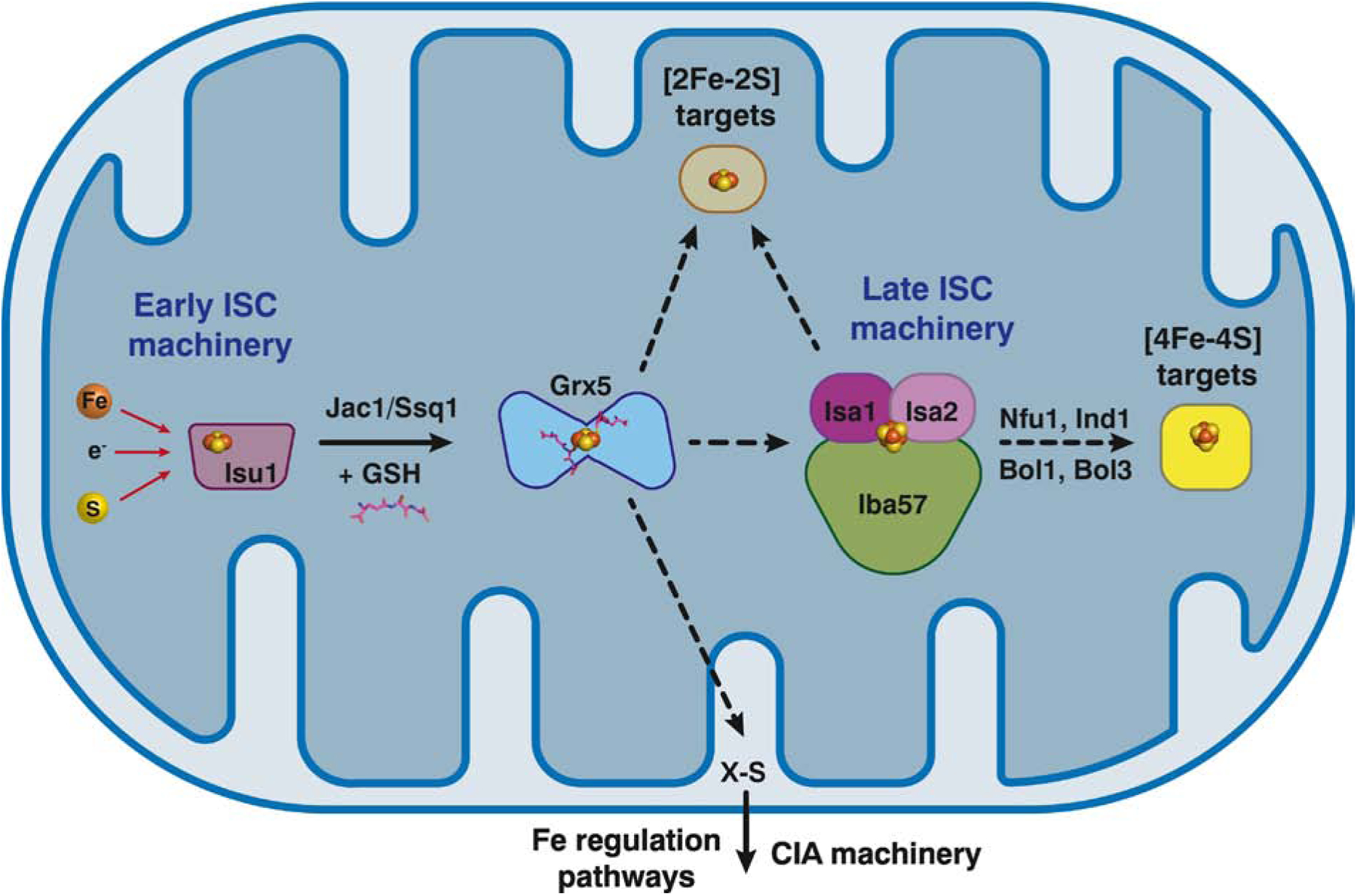
Diagram depicting the ISC pathway in yeast mitochondria highlighting the central role of Grx5 and the proposed roles of Bol1/Bol3 proteins. A [2Fe-2S] cluster is assembled on the scaffold protein Isu1 by the early ISC machinery, which is then delivered to the intermediate carrier protein Grx5. The chaperone/co-chaperone complex Jac1/Ssq1 is proposed to assist in release of the cluster from Isu1. Grx5 may deliver [2Fe-2S] clusters to apo targets in the mitochondria and is also required for formation of an unidentified sulfur-containing species (X-S) that exported to the cytosol. X-S is used by the CIA machinery for nuclear/cytosolic Fe-S cluster assembly and is required to relay the cellular iron status to iron-responsive transcription factors in the nucleus. In addition, Grx5 interacts with and transfers [2Fe-2S] clusters to the late ISC machinery, a complex composed of Isa1, Isa2, and Iba57, which then synthesizes [4Fe-4S] clusters via coupling of two [2Fe-2S] clusters. Nfu1, Ind1, Bol1, and Bol3 serve as targeting factors that interact with Grx5 and/or the late ISC machinery to deliver [4Fe-4S] clusters to specific apo acceptor proteins.
The [2Fe-2S]-bridged Grx5 homodimer is thought to direct the cluster to a variety of different pathways (Fig. 5). [2Fe-2S]-Grx5 may deliver the cluster to specific [2Fe-2S] target proteins, although this particular pathway is not well defined [56]. The early ISC machinery and Grx5 are also required for formation of an undefined sulfur-containing compound, known as XS, that is exported from the mitochondria and required for nuclear/cytosolic Fe-S cluster assembly via the CIA pathway, as well as for signaling iron bioavailability to the transcription factors that control iron metabolism (Fig. 5 and see section 8.1) [56, 59]. Finally, experimental evidence suggests that Grx5 delivers [2Fe-2S] clusters to the late acting ISC machinery (Isa1, Isa2, and Iba57), which is required for the maturation of mitochondrial [4Fe-4S] cluster-containing proteins [56] as well as some [2Fe-2S] clusters proteins [60]. To generate a [4Fe-4S] cluster, the Isa1/Isa2/Iba57 complex is proposed to combine two [2Fe-2S] clusters via reductive coupling, as demonstrated for the human homologs [61]. [4Fe-4S] clusters generated by the late ISC machinery are then delivered to specific apo targets by a variety of targeting factors that include Nfu1, Ind1, Bol1, and Bol3.
Additional details about the specific function of Bol1 and Bol3 in mitochondrial Fe-S cluster trafficking were unveiled in two recent reports [19, 38]. Unlike the severe Fe-S deficiency phenotypes observed for grx5Δ strains, single deletion of the bol1 or bol3 gene has little or no effect on Fe-S cluster enzyme activity nor growth on respiratory media [19]. However, the Bol1/3 proteins apparently have partially overlapping functions because the bol1Δbol3Δ double mutant exhibits a respiratory growth defect caused by reduced activity and deficient Fe-S cluster incorporation into succinate dehydrogenase [38]. In addition, this mutant exhibits markedly decreased lipoylation of mitochondrial enzymes, reflecting a defect in the [4Fe-4S]-binding protein lipoic acid synthase (LIAS) [19, 38]. Proteomic analysis of affinity purified proteins demonstrated that Bol3 forms a complex with the Fe-S cluster maturation factor Nfu1, and both Bol3 and Nfu1 interact with the same specific [4Fe-4S] target proteins, including aconitase, succinate dehydrogenase, and lipoic acid synthase [19]. Taken together, these results suggest that Bol3 functions with Nfu1 in the later steps of the ISC pathway by moving [4Fe-4S] clusters from the Isa1/Isa2 complex to specific mitochondrial target proteins. In contrast, Bol1 interacts with Grx5 but not Nfu1 nor Fe-S client proteins in vivo, reflecting somewhat divergent structural and/or functional features. Nevertheless, the functional interactions of both Bol proteins are dependent on an invariant His residue identified as a critical Fe-S binding ligand in all BolA homologs (His 93 in Bol1 and His101 in Bol3) [11, 29, 33, 35], supporting the proposal that Fe-S binding is essential to their function [19].
6.2. Defects in human mitochondrial BOLA3 and GLRX5 cause rare neurological and hematological disorders.
The pathway for assembling Fe-S clusters in the mitochondria of human cells shares many parallels with the yeast system described in section 6.1. All of the critical ISC components identified in yeast have human orthologs that are shown to perform similar functions (recently reviewed in [57, 62]). As such, human GLRX5 is proposed to have a central role accepting [2Fe-2S] clusters from the early ISC machinery and delivering these cofactors to [2Fe-2S] target proteins or the late acting Fe-S cluster carrier proteins ISCA1 and ISCA2 (human orthologs of yeast Isa1 and Isa2) [61]. Likewise, BOLA3 is proposed to function as a secondary carrier that mediates delivery of [4Fe-4S] clusters to LIAS. Another model proposes that the [2Fe-2S] GLRX5-BOLA3 complex shown in Figure 4B transfers two clusters to apo-NFU1 forming a [4Fe-4S]-bridged NFU1 homodimer that bypasses the ISCA1/ISCA2 complex to deliver [4Fe-4S] clusters to specific targets such as aconitase and LIAS [63]. The specific role of BOLA1 is less clear than BOLA3 since knockdown of BOLA1 in mammalian cells did not yield a clear Fe-S cluster biogenesis defect, but did have a measurable impact on the redox state of mitochondrial GSH pools [28]. Based on this result and the unusual redox state and stability of the purified, reconstituted [2Fe-2S]1+ GLRX5-BOLA1 complex, BOLA1 is proposed to function with GLRX5 in redox homeostasis or electron transfer [35]. More clues are available to delineate the specific functions of GLRX5 and BOLA3 in mitochondria due to their causative role in the development of some rare human diseases.
6.2.1. Consequences of human GLRX5 deficiency.
Due to its critical function in Fe-S cluster biogenesis, mutations in the GLRX5 gene have been shown to cause two rare autosomal recessive diseases with divergent phenotypes: sideroblastic anemia-3 (SIDBA-3) and childhood onset spasticity with hyperglycinemia (SPAHGC) (Table 2). Three cases of SIDBA-3 have been reported in the literature with clinical symptoms centered around the inability of erythrocytes to produce heme, resulting in adult-onset sideroblastic anemia [64–67]. Sideroblasts are red blood cell precursors with iron-overloaded mitochondria that appear as a ring around the nucleus. Additional symptoms include enlargement of the liver and spleen, type II diabetes, and systemic iron overload that manifests as increased transferrin saturation and serum ferritin levels, and liver iron accumulation.
Table 2.
Diseases linked to mutations in human GLRX and BOLA genes.
| Gene | Disease | Clinical symptoms | Tissues impacted | Reference |
|---|---|---|---|---|
| GLRX5 | Sideroblastic anemia-3 (SIDBA-3) | Adult-onset anemia; systemic iron overload; hepatosplenomegaly | erythrocytes, liver, spleen | [64–67] |
| Childhood onset spasticity with hyperglycinemia (SPAHGC) | Non-ketotic hyperglycinemia; slowly progressive spasticity; leukodystrophy and/or spinal lesions; visual defects and mild learning disabilities in some patients | Central nervous system | [71–73] | |
| BOLA3 | Multiple mitochondrial dysfunctions syndrome-2 with hyperglycinemia (MMDS2) | Severe developmental regression in infancy characterized by seizures, spasticity, and poor head control; hypotonia; lactic acidosis and elevated glycine levels in serum and cerebrospinal fluid; optic atrophy, cardiomyopathy, and leukodystrophy in some patients; death in childhood. | Central nervous system | [73, 75–81] |
The biochemical basis of this disease can be explained by GLRX5’s role in mitochondrial Fe-S cluster biogenesis. Patients with this disease have no detectible GLRX5 protein or express mutant forms that are apparently unable to fully support Fe-S cluster biogenesis [65–67]. As a result, mitochondrial aconitase and complex I activities in the respiratory chain are decreased, and the enzyme ferrochelatase, which catalyzes the last iron insertion step in heme biosynthesis, is destabilized in the absence of its bound [2Fe-2S] cluster [68]. Furthermore, low production of Fe-S clusters triggers an iron starvation response because the [4Fe-4S]-binding cytosolic aconitase is predominantly shifted to the apo form that possesses mRNA binding activity, known as iron regulatory protein 1 (IRP1). IRP1 together with its homolog IRP2 function to regulate intracellular iron homeostasis via post-transcriptional regulation of genes involved iron trafficking and metabolism [69]. IRP binding blocks translation of mRNA encoding the iron storage protein ferritin, while stabilizing transcripts encoding the transferrin receptor TfR1, thereby promoting iron uptake and mobilization leading to systemic iron overload. In addition, IRP binding destabilizes mRNA encoding the erythroid-specific heme biosynthetic enzyme ∂-aminolevulinic acid synthase 2 (ALAS2), further hindering heme production and causing mitochondrial iron accumulation. The GLRX5 defect is thought to specifically impact erythroid cells because these cells have high heme requirements, express relatively high levels of GLRX5 [65, 70], and are the only cell type that expresses the IRP-regulated ALAS2.
The other autosomal recessive human disorder linked to GLRX5 mutations is known as childhood onset spasticity with hyperglycinemia (SPAHGC), which is a form of variant non-ketotic hyperglycinemia (Table 2) [71–73]. Unlike the hematological symptoms associated with SIDBA-3, this disease primarily impacts the central nervous system. Three patients with this disease have been identified, each exhibiting slowly progressive spasticity in childhood that gives rise to walking difficulties. Brain and spinal cord imaging revealed leukodystrophy (myelin sheath defects) and lesions in the upper spinal cord. One patient also exhibited vision problems and another had mild learning and concentration difficulties [72, 73]. Laboratory tests for all patients revealed increased glycine levels in the serum with mildly increased or normal levels in the cerebrospinal fluid, and normal serum lactate levels. The biochemical basis of the neurological symptoms and test results for SPAHGC has been identified as a deficiency in mitochondrial lipoic acid biosynthesis [72]. Lipoic acid is an essential cofactor for the multiprotein glycine cleavage system (GCS), which allows the degradation of glycine in the liver, kidney, and brain. In the absence of this activity, glycine accumulates to toxic levels causing neurological damage. The activities of GCS and the lipoic acid-dependent enzyme pyruvate dehydrogenase complex (PDHc) were both markedly reduced in patients with SPAHCG due to defective lipoylation of these enzymes [73]. As mentioned earlier, the link between lipoic acid deficiency and GLRX5 function lies in the mitochondrial enzyme LIAS. LIAS is a member of the radical SAM (s-adenosyl methionine) enzyme family and requires two [4Fe-4S] clusters for its activity. The fact that GLRX5 mutations apparently impact LIAS maturation suggests that GLRX5 has a specific role in delivery of Fe-S clusters to this mitochondrial protein.
Interestingly, the contrasting phenotypes observed for SIDBA-3 vs. SPAHCG patients may be due to the specific GLRX5 mutations that cause each disease [74]. For one patient with SIDBA-3, the disease was caused by a splicing defect that led to undetectable GLRX5 protein in western blots [64, 65]. The other two patients had compound heterozygous missense mutations in conserved residues (K101Q/L148S in one patient [66] and C69Y/M128K in another [67]) which interfere with either Fe-S cluster binding by GLRX5 or possibly interactions with protein binding partners. The SPAHCG patients all harbored a deletion of the highly conserved Lys51 residue located on the surface of the protein. This deletion does not impact GLRX5 proteins levels nor lead to heme and respiratory chain defects and iron dysregulation as noted in the SIDBA-3 patients. Rather, the observed phenotypes suggest that the absence of Lys51 in GLRX5 may specifically impair Fe-S cluster insertion into LIAS [66, 73].
6.2.2. Consequences of human BOLA3 deficiency.
BOLA3 is the only human BolA homolog that has been directly linked to human disease. BOLA3 deficiencies are rare but produce fatal consequences. Homozygous mutation in the BOLA3 gene causes the severe autosomal recessive disorder known as multiple mitochondrial dysfunctions syndrome-2 with hyperglycinemia (MMDS2) (Table 2) [73, 75–81]. Fifteen patients have been diagnosed with MMDS2 to date, exhibiting symptoms that resemble SPAHCG patients with GLRX5 mutations, including spasticity, leukodystrophy, and hyperglycinemia caused by defects in mitochondrial lipoic acid biosynthesis. However, the pathology of this disease is usually compounded by more severe symptoms, including seizures, lactic acidosis, hypotonia, and cardiomyopathy, eventually leading to death in childhood. Biochemical analysis of patient cells with this disease revealed severe deficiencies of GCS, PDHc and the α-ketoglutarate dehydrogenase complex (αKGDHc) [73, 75–80]. As mentioned above, GCS, PDHc and αKGDHc are not Fe-S cluster binding proteins, but require lipoic acid as a cofactor, which in turn is dependent on the [4Fe-4S] cluster binding enzyme LIAS. Most patients also exhibited reduced complex I and II activities in the respiratory chain [75–77, 79, 81], which require Fe-S cluster cofactors. However, mitochondrial and cytosolic aconitase activities are typically normal in this disease, thus dysregulation of iron metabolism and iron overload due to increased IRP mRNA binding activity are not observed. Taken together, these results suggest that BOLA3 is required for specifically trafficking [4Fe-4S] clusters to complex I, succinate dehydrogenase (complex II), and LIAS, which synthesizes lipoic acid as a cofactor for GCS and the α-keto acid dehydrogenase enzymes PDHc and αKGDHc.
6.3. Fe-S cluster assembly in plant mitochondria
Recent studies in plants have also shed light on the roles of CGFS Grxs and BolA proteins in mitochondria. The plant mitochondrial CGFS Grx, namely GRXS15, plays a critical role in plant growth and development since its deficiency causes embryonic lethality in Arabidopsis thaliana [82]. Furthermore, Arabidopsis GRXS15 partially complements the growth defect in grx5Δ yeast cells suggesting a conservation of function across species. Down-regulation of GRXS15 in plant lines causes a growth defect, decreased respiration, lowered aconitase activity, a defect in lipoic acid-dependent enzymes, and reduced abundance of mitochondrial LIAS [83], which mirrors some of the biochemical effects observed for human SPAHCG patients with GLRX5 deficiencies (section 6.2.1). Recombinant GRXS15 has the ability to bind a [2Fe-2S] as a homodimer, and transfer the cluster to mitochondrial ferredoxin 1 [25]. Thus, GRXS15 is proposed to be required for trafficking Fe-S clusters to specific targets, including LIAS. The consequences of deletion or deficiency of mitochondrial BOLA4 have not yet been established in plants, so the potential function of this BolA homolog is unclear. However, GRXS15 was also shown to interact with BOLA4 in plant cells via bimolecular fluorescence complementation, and expression of plant BOLA proteins in yeast can rescue the respiratory growth defect of bol1Δbol3Δ cells and partially recover the activity of complex II and lipoic-acid dependent enzymes (see section 6.1) [84]. Taken together, these results suggest that GRXS15 and BOLA4 function together in Fe-S cluster biogenesis as demonstrated for their yeast and human homologs [25].
7. GRXS14/16 and BOLA1/4 may function with the SUF pathway in chloroplast Fe-S cluster biogenesis
In lieu of the ISC pathway found in mitochondria, chloroplasts utilize the alternate SUF Fe-S cluster assembly pathway that bears resemblance to the bacterial Suf system. However, the basic principles of de novo Fe-S cluster assembly are the same: 1) Fe-S clusters are built on a scaffold system by combining sulfur mobilized from cysteine with iron and electrons provided by donors; and 2) clusters are delivered to specific client proteins via a network of trafficking and maturation factors (recently reviewed in [85]). Plant chloroplasts house two CGFS Grxs, GRXS14 and GRXS16, and two BolA proteins, BOLA1 and BOLA4 (Fig. 2, 3). In addition, chloroplasts harbor the SUFE1 protein which possesses a sulfurtransferase domain typical of SufE proteins as well as a C-terminal BolA domain. As a sulfurtransferase, SufE1 activates the cysteine desulfurase activity of Nfs2, which mobilizes sulfur from cysteine in the first phase of chloroplast Fe-S cluster assembly. The specific function of the BolA domain in SUFE1 is unclear. Both chloroplast Grxs were shown to interact with both BolA proteins and SUFE1 via bimolecular fluorescence complementation experiments in plant cells and yeast two-hybrid assays [25]. Similar to their bacterial, yeast, and human homologs, chloroplast GRXS14 and GRXS16 can form [2Fe-2S] bridged homodimers [86] or [2Fe-2S]-bridged heterodimers in concert with BolA proteins [29]. Furthermore, anaerobic CD spectroscopy studies confirmed that the chloroplast-specific intermediate carrier protein Nfu2 can deliver [2Fe-2S] clusters to GRXS14 but not GRXS16 [87], while GRXS14 can rapidly transfer [2Fe-2S] clusters to the Fe-S cluster carrier protein SufA1 [44] as well as ferredoxin [86]. Nevertheless, the physiological relevance of these in vitro findings is still unclear. Due to the likely redundancy between the chloroplast Grxs, single grxs14 or grxs16 mutants do not yield strong phenotypes under the growth conditions tested [88]. Parallel studies with bolA1 and bolA4 mutant plants have not yet been published so the impact of bolA1/4 deficiency on plant growth and function is unknown. However, similar to the mitochondrial BolAs and Grxs from plants, the chloroplast orthologs can complement the growth phenotypes of yeast grx5Δ or bol1Δbol3Δ strains, respectively [84, 86]. These results hint to a conserved function for these proteins in Fe-S cluster targeting and insertion.
8. Roles for CGFS Grxs and BolA proteins in the eukaryotic cytosol
The eukaryotic cytosol typically houses one BolA homolog and one or two CGFS Grxs. Cytosolic BolA proteins are classified in the BolA2 subfamily that contain a conserved Cys residue located in the H/C loop and the invariant His residue near the C-terminus (Fig. 4A). Unlike their mitochondrial homologs, cytosolic CGFS Grxs are multidomain proteins with a Trx-like domain at the N-terminus, usually with a modified monothiol active site (WAxxC), and one to three CGFS Grx domains in tandem at the C-terminus (Fig. 2). Roles for these nuclear/cytosolic proteins in iron metabolism have been well studied in both yeast and human cells with recent highlights outlined below.
8.1. Grx3/4 and Bol2 function in regulation of iron metabolism in yeasts and other fungi.
The first connection between cytosolic CGFS, BolA proteins, and iron regulation arose from genetic studies in S. cerevisiae (reviewed in [11, 89, 90]). The cytosolic CGFS paralogs Grx3 and Grx4 were identified as intracellular iron trafficking factors that function with the BolA2 protein Fra2 (recently renamed Bol2) to control the activity of the low iron sensing transcription factor Aft1 and its paralog Aft2. Aft1 and Aft2 undergo nucleocytoplasmic shuttling in response to iron, binding DNA and activating expression of iron uptake and storage gene when iron is low (Fig. 6A, top), then dissociating from DNA and moving to the cytosol when iron levels are sufficient (Fig. 6A, bottom). Grx3 and Grx4 form [2Fe-2S]-bridged heterodimers with Bol2 that specifically transfer a [2Fe-2S] cluster to Aft2 that alters its oligomeric state from monomer to dimer [91, 92]. Bol2 plays a clear role in facilitating this cluster transfer since [2Fe-2S]-bridged Grx3 homodimers are unable to quantitatively deliver the cluster to Aft2 under the same conditions. DNA binding assays revealed that [2Fe-2S]-bridged Aft2 homodimers formed after cluster transfer have lower DNA binding affinity than apo Aft2 monomers, which presumably leads to deactivation of Aft2 target genes [92]. Due to the high sequence homology and overlapping functions of Aft1 and Aft2, a similar mechanism is proposed for control of Aft1 activity. In vivo Fe-S cluster binding to Grx3/4-Bol2 heterodimers, in turn, is dependent on the mitochondrial ISC pathway, GSH, and the mitochondrial ABC transporter Atm1 in order to export the undefined sulfur-containing X-S species required for synthesis of nuclear/cytosolic Fe-S clusters. Interestingly, the CIA pathway is not required for signaling iron bioavailability to Aft1 or Aft2, suggesting that Fe-S clusters are assembled or delivered to Grx3/4-BolA complexes via an alternate pathway [93, 94].
Figure 6.
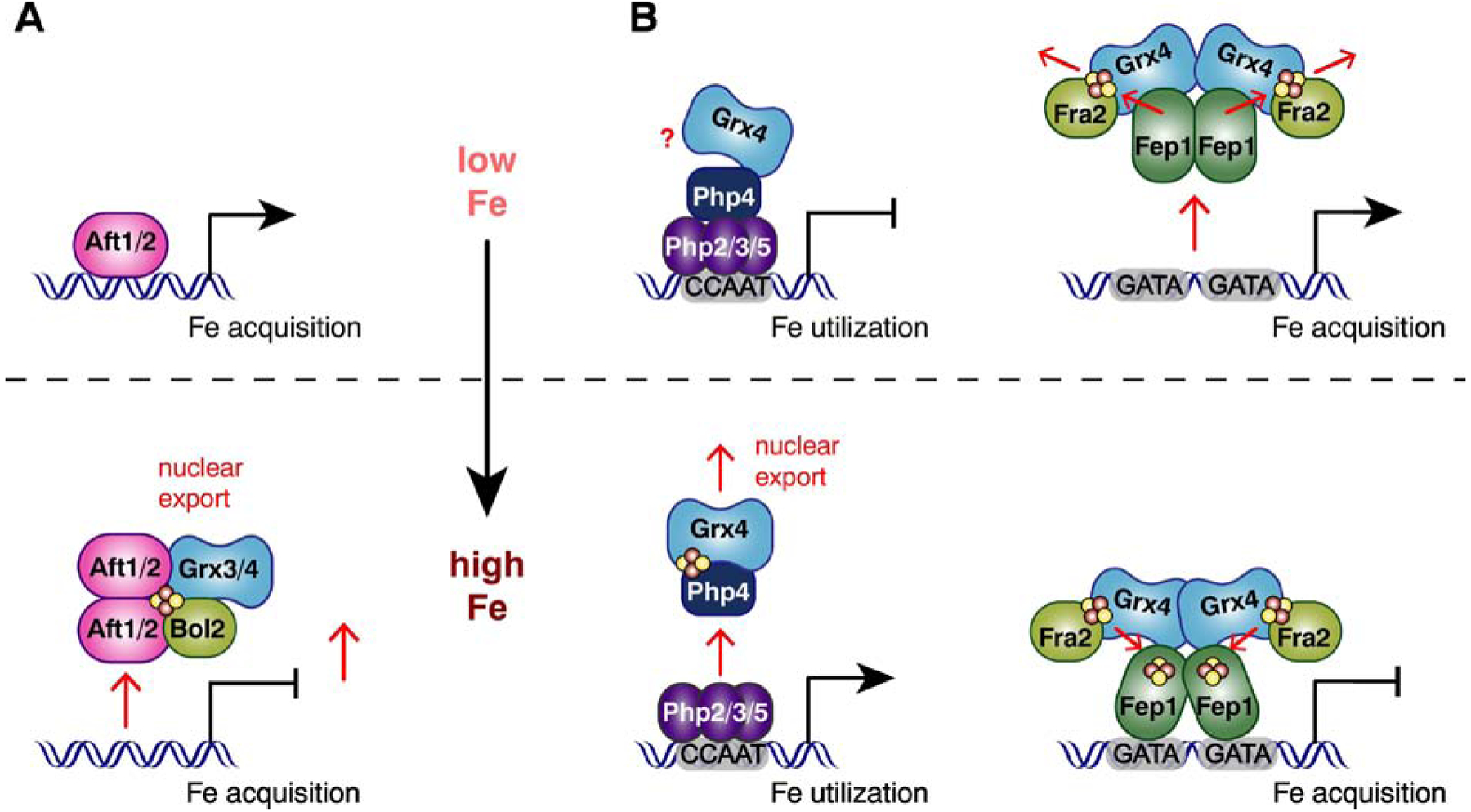
Models for iron-responsive transcriptional activation by Aft1/2 in S. cerevisiae (A) and iron-responsive transcriptional repression by Fep1 and Php4 in S. pombe (B). When iron levels are low in S. cerevisiae (A, top), Aft1 and Aft2 bind DNA and activate expression of iron acquisition genes. During conditions of iron sufficiency (A, bottom), Grx3 and Grx4 form Fe-S-bridged heterodimers with Bol2 and deliver [2Fe-2S] clusters to Aft1/2, which promotes their DNA dissociation, dimerization, and export to the cytosol. This change, in turn, leads to deactivation of Aft1/2-regulated genes. During iron deficiency in S. pombe (B, top), Php4 binds to Php2/3/5 which represses transcription of genes encoding proteins in non-essential, iron-utilizing pathways. There are conflicting reports whether or not Grx4 maintains an interaction with Php4 during iron deficiency. In either case, Grx4 does not interfere in Php4’s association with Php2/3/5. Fep1 dissociates from its target genes when iron is low, leading to the expression of iron acquisition genes. The dissociation of Fep1 from DNA is presumably triggered by [2Fe-2S] cluster transfer from Fep1 to the Grx4-Fra2(Bol2) heterodimer. When iron levels increase (B, bottom), Grx4 and Php4 bind a [2Fe-2S] cluster that promotes dissociation of Php4 from Php2/3/5 and its nuclear export, leading to derepression of the Php4 regulon. In parallel, Fep1 maintains an interaction with Grx4-Fra2 and binds [2Fe-2S] clusters that facilitate its DNA binding activity and repression of its target genes.
The fission yeast S. pombe is evolutionary divergent from S. cerevisiae, utilizing a different set of iron-responsive transcription factors that do not share homology with Aft1/Aft2 (Fig. 6B). Nevertheless, a cytosolic CGFS Grx and/or BolA protein control the transcriptional activity of these DNA binding proteins. Iron homeostasis in S. pombe is primarily regulated by two repressors, Fep1 and Php4, that are responsible for controlling iron acquisition and iron utilization, respectively (recently reviewed in [89, 95]). Fep1 is a GATA-type transcription factor that binds to GATA-containing sequences in promoters of iron uptake genes in iron replete conditions, thereby repressing their expression to avoid iron overload. Php4 binds to a heterotrimeric CCAAT-binding complex composed of Php2, Php3, and Php5. Under iron replete conditions, the Php2/3/5 complex activates expression of iron utilization genes by binding to CCAAT sequences in their promoters. When iron levels drop, Php4 binds to the Php2/3/5 complex, triggering the switch from activator to repressor. Consequently, non-essential iron utilization pathways are downregulated to conserve intracellular iron pools. Php4 was shown to form a [2Fe-2S] cluster binding complex with S. pombe Grx4 via two conserved Cys residues [96], which promotes its dissociation from Php2/3/5 and subsequent nuclear export (Fig. 6B, bottom) [89]. Whether or not Grx4 maintains an interaction with Php4 under iron deficiency is somewhat unclear. One study using yeast two-hybrid assays demonstrated that the Trx domain of Grx4 maintains a strong interaction with Php4 in cells cultured in iron deficiency media [97], while another study using co-immunoprecipitation found that the Grx4-Php4 interaction is weakened in cells grown in iron-limited media [23]. In any case, regulation of Php4 activity in vivo is dependent on mitochondrial Fe-S cluster biogenesis and GSH, similar to Aft1/2 regulation in S. cerevisiae, which likely reflects the requirement for the GSH-ligated [2Fe-2S]-Grx4 complex to regulate Php4 activity [23, 89, 95]. However, there are conflicting reports regarding the role of the S. pombe Bol2 ortholog (known as Fra2) in controlling Php4 function. One study found that fra2 deletion did not impact nucleocytoplasmic shuttling or transcriptional regulation by Php4, concluding that Fra2 does not function with Grx4 to inhibit Php4 in response to iron [98]. Meanwhile, another study found that Php4 represses expression of target genes somewhat more rapidly in fra2Δ strains compared to WT when cells are switched to iron deficient conditions, suggesting that Fra2 may play a minor role in retention of Php4 in the cytosol [23]. However, a direct physical interaction between Fra2 and Php4 has not been demonstrated. Nevertheless, both studies concluded that the activity of the Fep1 repressor clearly involves post-translational regulation by both Grx4 and Fra2 (Fig. 6B). Grx4 modulates Fep1 activity via specific protein-protein interactions and requires Cys-172 in the CGFS motif to inhibit Fep1 activity when iron is scarce [95]. Fra2 also coimmunoprecipitates in a complex with Fep1 and Grx4 [98] and co-regulates the iron-dependent inhibition of Fep1 activity in vivo [23, 98]. Furthermore, purified recombinant Fep1 and the Grx4-Fra2 complex possess UV-visible spectra that are suggestive of [2Fe-2S] binding proteins [23]. Additional EPR analysis confirmed that recombinant S. pombe Fep1 binds Fe-S clusters, although the specific cluster type ([4Fe-4S], [3Fe-4S], or [2Fe-2S]) was unclear [99]. Based on the genetic studies and DNA binding and Fe-S transfer assays, the current regulation model suggests that during iron sufficiency, Fep1 binds [2Fe-2S] clusters that favor DNA binding and repression of its target genes (Fig. 6B, bottom) [89, 95]. Grx4 and Fra2 maintain an interaction with Fep1 under both low and high iron conditions [98], but it has not been established whether or not Grx4 or Fra2-Grx4 delivers the Fe-S clusters to Fep1. However, Fep1 binds DNA constitutively in grx4Δ strains suggesting that Grx4 is not essential for Fe-S cluster delivery to Fep1 [23, 100]. Conversely, iron starvation is proposed to cause loss of the bridging [2Fe-2S] cluster between Grx4 and Fra2, triggering [2Fe-2S] cluster transfer from Fep1 to Grx4-Fra2 [23], which in turn inactivates the repressor activity of Fep1 (Fig. 6B, top). The role of S. pombe Grx4-Fra2 serving as a cluster acceptor is reminiscent of previous studies mentioned earlier demonstrating that S. cerevisiae Grx3-Bol2 accepts a [2Fe-2S] cluster from the A-type carrier protein NifIscA [44].
More recently, a number of studies have elucidated the role of cytosolic CGFS Grxs in controlling iron regulation in pathogenic fungi (recently reviewed in [89, 95, 101]). The filamentous fungi Aspergillus nidulans and Aspergillus fumigatus possess orthologs of Php4 and Fep1 known as HapX and SreA, respectively. Both regulators from A. fumigatus were shown to physically and functionally interact with the cytosolic CGFS glutaredoxin GrxD to coordinate the transcriptional response to iron starvation. Furthermore, recombinant HapX binds a [2Fe-2S] cluster in the presence or absence of GrxD, which is proposed to regulate its DNA binding activity [102]. The same research group also demonstrated that the mitochondrial ISC pathway and GSH are both essential for regulation of iron metabolism while the CIA pathway is dispensible [102], which parallels similar findings in S. cerevisiae. Taken together, these studies underscore the importance of CGFS Grxs and Fe-S cluster biogenesis in regulating iron metabolism in A. fumigatus. Clear connections between Fe-S cluster biogenesis, CGFS Grxs, and iron regulation have also been made in the opportunistic pathogen Candida albicans. The phenotypes of a C. albicans grx3Δ/Δ mutant demonstrate that Grx3 is essential for regulation of iron homeostasis and resistance to oxidative stress [103]. Furthermore, deficiency of the C. albicans mitochondrial ISC pathway causes cellular iron accumulation, oxidative stress, and decreased virulence, thus linking mitochondrial Fe-S cluster biogenesis with iron regulation [104]. A more recent comprehensive study using virulence assays, proteomic profiling, chromatin immunoprecipitation and other methods further clarified the critical role of C. albicans Grx3 in iron regulation [27]. These authors showed that Grx3 is required for growth in low iron media and for C. albicans virulence in mouse models. Grx3-interacting proteins identified by proteomic analysis included Bol2/Fra2, Fe-S cluster assembly and redox homeostasis factors, and iron-responsive transcription factors, as well as proteins involved in amino acid biosynthesis and metabolism. Furthermore, co-immunoprecipitation analysis confirmed that Grx3 interacts with the GATA-type repressor Sfu1 (an ortholog of S. pombe Fep1) in vivo in an iron-independent manner and impacted the DNA binding and transcriptional repressor activity of Sfu1 in an iron-dependent manner. Grx3 was also shown to interact with Hap43 (an ortholog of S. pombe Php4) in an iron-independent manner while impacting nucleocytoplasmic shuttling of this transcription factor in response to iron [27]. These results mirror the functional interactions reported for the S. pombe orthologs of Grx3, Sfu1, and Hap43 (S. pombe Grx4, Fep1, and Php4, respectively), suggesting conserved mechanisms of post-translational control by CGFS Grxs for these types of iron-responsive transcriptional repressors. Finally, the fungal pathogen Cryptococcus neoformans also relies on a cytosolic CGFS Grx to control the activity of its iron-responsive transcriptional repressor Cir1. Cir1 is an ortholog of S. pombe Fep1, Aspergillus SreA, and C. albicans Sfu1, and thus represses expression of iron uptake systems during iron sufficiency. C. neoformans Grx4 is essential for the virulence of this pathogen and was recently shown to physically and functionally interact with Cir1, serving as a master regulator of iron-responsive genes [105]. The Fe-S binding domain of Grx4 is essential for this regulatory function, suggesting that Fe-S binding modulates the activity of Cir1, as proposed for its S. pombe ortholog Fep1 (Fig. 6B). Notably, for each of these pathogenic fungi, the specific roles of cytosolic Bol2 proteins in iron regulation have not yet been addressed.
8.2. GLRX3 and BOLA2 serve as [2Fe-2S] chaperones in mammalian cells
A clear role for trafficking [2Fe-2S] clusters in the cytosol has also been establishing for human GLRX3 and BOLA2. Depletion of GLRX3 in human cell lines was shown to disrupt cellular iron metabolism, causing defects in cytosolic iron-sulfur cluster assembly that induce an iron starvation response. Furthermore, Grx3 knockdown in zebrafish impairs heme biosynthesis, likely reflecting a defect in iron trafficking [6, 106]. Similar to their yeast counterparts, human GLRX3 and BOLA2 can form heterocomplexes, although human GLRX3 has two tandem GRX domains rather than one (Fig. 2). Consequently, one BolA2 protein binds to each of the GRX domains generating a heterotrimer bridged by two [2Fe-2S] clusters [107, 108]. GLRX3 and BOLA2 have been shown to deliver [2Fe-2S] clusters to CIA components in in vitro studies. Fe-S cluster transfer assays indicate that the [2Fe-2S]2-GLRX32 homodimer and the [2Fe-2S]2-GLRX3-BOLA22 heterotrimer are capable of transferring two [2Fe-2S] clusters to two different Fe-S cluster binding sites in CIAPIN1 (also known as anamorsin), an essential component of the CIA machinery (Fig. 7). In both cases, the N-terminal TRX domain of GLRX3 specifically recognizes the N-terminal region of CIAPIN1, facilitating the transfer [107, 109]. More recently, [2Fe-2S]2-GLRX32 was demonstrated to function in vitro in the maturation of the Fe-S cluster scaffold protein NUBP1 in the CIA pathway, delivering [2Fe-2S]2+ clusters that are converted to [4Fe-4S]2+ clusters in the presence of GSH [110]. The details of the CIAPIN1-GLRX3-BOLA2 binding interactions were further elucidated in human cell lines [111]. GLRX3-BOLA2 were shown to form an Fe-S cluster dependent binding interaction in cells that increased 6–8-fold with increasing cellular iron levels. CIAPIN1 and its functional binding partner NDOR1 coimmunoprecipitate with GLRX3 and with BOLA2, although the CIAPIN1-BOLA2 interaction is weaker and dependent on the presence of GLRX3. However, Fe incorporation into CIAPIN1 in cells is dependent on the presence of both GLRX3 and BOLA2, while the opposite is not true, i.e. formation of the Fe-S-bridged GLRX3-BOLA2 complex is not dependent on CIAPIN1. Furthermore, the Fe-S binding residues in GLRX3 are essential for GLRX3 binding to both CIAPIN1 and BOLA2. Taken together, these studies provide strong cell-based evidence that the GLRX3-BOLA2 complex functions as a [2Fe-2S] chaperone that delivers Fe-S clusters to specific targets that include CIAPIN1 in the CIA pathway [111] (Fig. 7).
Figure 7.
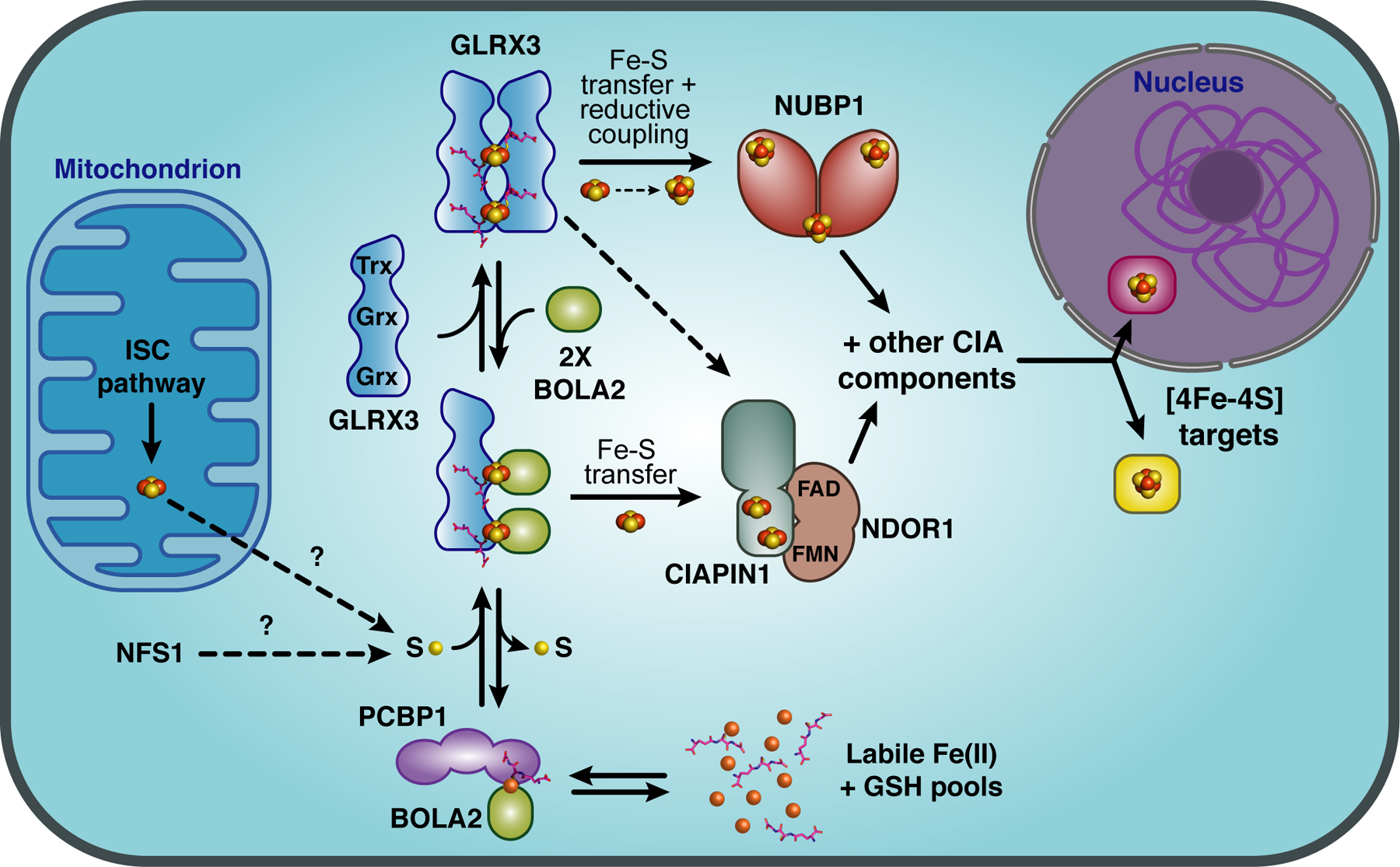
Model for the roles of GLRX3 and BOLA2 in mammalian cytosolic iron trafficking. The cytosolic iron chaperone PCBP1 acquires Fe(II) from the labile iron pool and forms an Fe(II)-bridged complex with BOLA2 that requires GSH binding for stability. This complex is proposed to interact with apo-GLRX3 to form a GLRX3-BOLA2 heterocomplex. Each Grx domain in GLRX3 forms a [2Fe-2S]-binding complex with a BOLA2 monomer and GSH leading to formation of a [2Fe-2S]2-bridged GLRX3-BOLA22-GSH2 heterotrimer. The sulfur source required to form the [2Fe-2S] clusters is unclear, possibly provided by the cytosolic isoform of the cysteine desulfurase NFS1 or exported from the mitochondrial ISC pathway. Both the [2Fe-2S]2-GLRX3-BOLA22 heterotrimer and the [2Fe-2S]2-GLRX32 homodimer can deliver [2Fe-2S] clusters to the CIA Fe-S cluster assembly protein CIAPIN1 in vitro, although both GLRX3 and BOLA2 are required for Fe incorporation into CIAPIN1 in cell-based assays. [2Fe-2S]2-GLRX32 may also deliver clusters to the CIA scaffold protein NUBP1, which requires reductive coupling of [2Fe-2S]2+ clusters to form [4Fe-4S]2+ clusters. CIAPIN1, its functional electron transfer partner NDOR1, and NUBP1 are all components of the CIA machinery that assembles and delivers [4Fe-4S] clusters to target proteins in the cytosol and nucleus.
The details of cytosolic iron trafficking to Fe-S cluster proteins in human cells were further elucidated in a recent study that revealed a functional and physical interaction between BOLA2 and the multifunctional poly r(C)-binding protein PCBP1 [112]. PCBP1 was previously characterized as a metallochaperone that binds and delivers Fe2+ to the intracellular iron storage protein ferritin as well as other cytosolic iron enzymes (recently reviewed in [113]). Proteomic analysis of PCBP1-interacting proteins and co-immunoprecipitation studies confirmed an interaction between PCBP1 and BOLA2 that is independent of GLRX3. Moreover, the interaction between PCBP1 and BOLA2 was enhanced in the absence of GLRX3 suggesting that PCBP1 and GLRX3 compete for binding to BOLA2. Interestingly, both iron and GSH are required to stabilize the PCBP1-BOLA2 complex, while inorganic sulfide and Fe-S cluster binding are not required. Formation of this complex as well as the [2Fe-2S]-bridged GLRX3-BOLA2 complex is dependent on the conserved Cys31 and His68 in human BOLA2. Based on this evidence, the authors propose that the PCBP1-Fe-GSH-BOLA2 complex provides iron for the formation of the [2Fe-2S]2-GLRX3-BOLA22 complex in human cells, linking the cytosolic iron trafficking and Fe-S cluster biogenesis pathways [112] (Fig. 7). Interestingly, the human BOLA2 gene maps to a chromosomal region that is prone to duplications, resulting in variable copy numbers of this gene in the human population, ranging from three to eight diploid copies [114]. By correlating BOLA2 copy number with blood-related phenotypes, a recent study discovered that individuals with low BOLA2 copy numbers are more prone to iron-deficiency anemia, further supporting the model that BOLA2 functions in maintaining human iron homeostasis [115].
Conclusions and Future Outlook
The past few years have seen a plethora of new studies that have revealed key insights and crucial details about the molecular structures and functions of CGFS Grxs and BolA proteins found in prokaryotic organisms and in different subcellular compartments of eukaryotic organisms. CGFS Grxs and BolA proteins are implicated in Fe-S cluster trafficking and biogenesis in prokaryotes, although more biochemical and genetic studies are required to fully reveal the specific functions of these proteins in these organisms. In contrast, strong physiological evidence exists for the role of CGFS Grxs in mitochondria as central carrier proteins that link the early and late machinery for Fe-S biogenesis in yeast, plants, and humans. Similar phenotypes are observed across species when mitochondrial CGFS Grxs deficient or absent, suggesting a strong conservation of function. In general, mitochondrial BOLA deletions have somewhat weaker and more diffuse phenotypes (with the exception of the human BOLA3 deficiency symptoms), partially owing to the overlapping functions of multiple mitochondrial BolA paralogs, making the assignment of their specific functions more difficult. Additional cellular and genetic studies are required to more carefully tease out the individual roles of mitochondrial BolA proteins. Even less is known about the in vivo function of chloroplast Grx and BolA proteins due to the redundancy of function between Grx or BolA paralogs in the same compartment and the lack of specific single and double mutants for study. Although the considerable structural and biophysical information provided by analysis of these plant proteins has certainly driven this field forward. Finally, recent studies in yeast and human cells have clearly shown essential roles for cytosolic Grx and BolA proteins in trafficking [2Fe-2S] clusters to Fe-S cluster dependent enzymes and transcription factors that control iron metabolism. Many of the mechanistic details of these trafficking pathways are still missing, representing a ripe area of future study. In addition, a focus on cytosolic CGFS Grxs and BolA proteins controlling iron regulation and virulence in pathogenic fungi is just recently emerging, with many molecular details about their functional roles unexamined.
Highlights.
Glutaredoxins with a CGFS active site and BolA proteins are ubiquitous in nature
CGFS glutaredoxins form iron-sulfur bridged complexes with BolA proteins
BolA proteins function with CGFS glutaredoxins in iron-sulfur cluster assembly and trafficking
CGFS glutaredoxin-BolA complexes regulate iron homeostasis in fungi
Mutations in GLRX5 and BOLA3 genes cause human mitochondrial diseases
Acknowledgements
Research in the C. Outten laboratory is funded by grant number R35 GM118164 from the National Institute of General Medical Sciences of the National Institutes of Health.
Abbreviations
- ALAS2
∂-aminolevulinic acid synthase 2
- CD
Circular dichroism
- CGFS
Cys-Gly-Phe-Ser
- CIA
Cytosolic iron-sulfur assembly
- Fe-S
Iron-sulfur
- GCS
Glycine cleavage system
- GRX
Glutaredoxin
- GSH
Glutathione
- EPR
Electron paramagnetic resonance
- αKGDHc
α-Ketoglutarate dehydrogenase complex
- IRP1
Iron regulatory protein 1
- IRP2
Iron regulatory protein 2
- ISC
Iron-sulfur cluster
- LIAS
Lipoic acid synthase
- MMDS2
multiple mitochondrial dysfunctions syndrome-2 with hyperglycinemia
- PDHc
Pyruvate dehydrogenase complex
- SAM
S-adenosyl methionine
- SIDBA-3
Sideroblastic anemia-3
- SPAHGC
Childhood onset spasticity with hyperglycinemia
- SUF
Sulfur utilization factor
- TRX
Thioredoxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Waldron KJ, Rutherford JC, Ford D, Robinson NJ, Metalloproteins and metal sensing, Nature, 460 (2009) 823–830. [DOI] [PubMed] [Google Scholar]
- 2.Dlouhy AC, Outten CE, The iron metallome in eukaryotic organisms, Met Ions Life Sci, 12 (2013) 241–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couturier J, Jacquot JP, Rouhier N, Evolution and diversity of glutaredoxins in photosynthetic organisms, Cell Mol Life Sci, 66 (2009) 2539–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao Z, La Fontaine S, Bush AI, Wedd AG, Molecular mechanisms of glutaredoxin enzymes: Versatile hubs for thiol-disulfide exchange between protein thiols and glutathione, J Mol Biol, 431 (2019) 158–177. [DOI] [PubMed] [Google Scholar]
- 5.Liedgens L, Zimmermann J, Waschenbach L, Geissel F, Laporte H, Gohlke H, Morgan B, Deponte M, Quantitative assessment of the determinant structural differences between redox-active and inactive glutaredoxins, Nat Commun, 11 (2020) 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berndt C, Lillig CH, Glutathione, glutaredoxins, and iron, Antioxid Redox Signal, 27 (2017) 1235–1251. [DOI] [PubMed] [Google Scholar]
- 7.Trnka D, Engelke AD, Gellert M, Moseler A, Hossain MF, Lindenberg TT, Pedroletti L, Odermatt B, de Souza JV, Bronowska AK, Dick TP, Muhlenhoff U, Meyer AJ, Berndt C, Lillig CH, Molecular basis for the distinct functions of redox-active and FeS-transfering glutaredoxins, Nat Commun, 11 (2020) 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Bandyopadhyay S, Shakamuri P, Naik SG, Huynh BH, Couturier J, Rouhier N, Johnson MK, Monothiol glutaredoxins can bind linear [Fe3S4]+ and [Fe4S4]2+ clusters in addition to [Fe2S2]2+ clusters: Spectroscopic characterization and functional implications, J Am Chem Soc, 135 (2013) 15153–15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouhier N, Couturier J, Johnson MK, Jacquot JP, Glutaredoxins: roles in iron homeostasis, Trends Biochem Sci, 35 (2010) 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Liu S, Feng Y, Liu JZ, Chen Y, Pham K, Deng H, Hirschi KD, Wang X, Cheng N, Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts, Proc Natl Acad Sci U S A, 110 (2013) 9565–9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Outten CE, Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis, Biochemistry, 51 (2012) 4377–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couturier J, Przybyla-Toscano J, Roret T, Didierjean C, Rouhier N, The roles of glutaredoxins ligating Fe-S clusters: Sensing, transfer or repair functions?, Biochim Biophys Acta, 1853 (2015) 1513–1527. [DOI] [PubMed] [Google Scholar]
- 13.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL Jr., White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM, A protein interaction map of Drosophila melanogaster, Science, 302 (2003) 1727–1736. [DOI] [PubMed] [Google Scholar]
- 14.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M, Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry, Nature, 415 (2002) 180–183. [DOI] [PubMed] [Google Scholar]
- 15.Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, Chessman K, Pal S, Cromar G, Papoulas O, Ni Z, Boutz DR, Stoilova S, Havugimana PC, Guo X, Malty RH, Sarov M, Greenblatt J, Babu M, Derry WB, Tillier ER, Wallingford JB, Parkinson J, Marcotte EM, Emili A, Panorama of ancient metazoan macromolecular complexes, Nature, 525 (2015) 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, Wrobel RL, Cho H, Kremer LS, Alston CL, Gromek KA, Dolan BK, Ulbrich A, Stefely JA, Bohl SL, Werner KM, Jochem A, Westphall MS, Rensvold JW, Taylor RW, Prokisch H, Kim JP, Coon JJ, Pagliarini DJ, Mitochondrial protein interaction mapping identifies regulators of respiratory chain function, Mol Cell, 63 (2016) 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pourhaghighi R, Ash PEA, Phanse S, Goebels F, Hu LZM, Chen S, Zhang Y, Wierbowski SD, Boudeau S, Moutaoufik MT, Malty RH, Malolepsza E, Tsafou K, Nathan A, Cromar G, Guo H, Abdullatif AA, Apicco DJ, Becker LA, Gitler AD, Pulst SM, Youssef A, Hekman R, Havugimana PC, White CA, Blum BC, Ratti A, Bryant CD, Parkinson J, Lage K, Babu M, Yu H, Bader GD, Wolozin B, Emili A, BraInMap elucidates the macromolecular connectivity landscape of mammalian brain, Cell Syst, 10 (2020) 333–350 e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luck K, Kim DK, Lambourne L, Spirohn K, Begg BE, Bian W, Brignall R, Cafarelli T, Campos-Laborie FJ, Charloteaux B, Choi D, Cote AG, Daley M, Deimling S, Desbuleux A, Dricot A, Gebbia M, Hardy MF, Kishore N, Knapp JJ, Kovacs IA, Lemmens I, Mee MW, Mellor JC, Pollis C, Pons C, Richardson AD, Schlabach S, Teeking B, Yadav A, Babor M, Balcha D, Basha O, Bowman-Colin C, Chin SF, Choi SG, Colabella C, Coppin G, D’Amata C, De Ridder D, De Rouck S, Duran-Frigola M, Ennajdaoui H, Goebels F, Goehring L, Gopal A, Haddad G, Hatchi E, Helmy M, Jacob Y, Kassa Y, Landini S, Li R, van Lieshout N, MacWilliams A, Markey D, Paulson JN, Rangarajan S, Rasla J, Rayhan A, Rolland T, San-Miguel A, Shen Y, Sheykhkarimli D, Sheynkman GM, Simonovsky E, Tasan M, Tejeda A, Tropepe V, Twizere JC, Wang Y, Weatheritt RJ, Weile J, Xia Y, Yang X, Yeger-Lotem E, Zhong Q, Aloy P, Bader GD, De Las Rivas J, Gaudet S, Hao T, Rak J, Tavernier J, Hill DE, Vidal M, Roth FP, Calderwood MA, A reference map of the human binary protein interactome, Nature, 580 (2020) 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melber A, Na U, Vashisht A, Weiler BD, Lill R, Wohlschlegel JA, Winge DR, Role of Nfu1 and Bol3 in iron-sulfur cluster transfer to mitochondrial clients, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM, Kaplan J, Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis, J Biol Chem, 283 (2008) 10276–10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF, Global landscape of protein complexes in the yeast Saccharomyces cerevisiae, Nature, 440 (2006) 637–643. [DOI] [PubMed] [Google Scholar]
- 22.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y, A comprehensive two-hybrid analysis to explore the yeast protein interactome, Proc Natl Acad Sci U S A, 98 (2001) 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Encinar del Dedo J, Gabrielli N, Carmona M, Ayté J, Hidalgo E, A cascade of iron-containing proteins governs the genetic iron starvation response to promote iron uptake and inhibit iron storage in fission yeast, PLoS Genet, 11 (2015) e1005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arabidopsis Interactome Mapping Consortium, Evidence for network evolution in an Arabidopsis interactome map, Science, 333 (2011) 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couturier J, Wu HC, Dhalleine T, Pégeot H, Sudre D, Gualberto JM, Jacquot JP, Gaymard F, Vignols F, Rouhier N, Monothiol glutaredoxin-BolA interactions: redox control of Arabidopsis thaliana BolA2 and SufE1, Mol Plant, 7 (2014) 187–205. [DOI] [PubMed] [Google Scholar]
- 26.Iñigo S, Durand AN, Ritter A, Le Gall S, Termathe M, Klassen R, Tohge T, De Coninck B, Van Leene J, De Clercq R, Cammue BP, Fernie AR, Gevaert K, De Jaeger G, Leidel SA, Schaffrath R, Van Lijsebettens M, Pauwels L, Goossens A, Glutaredoxin GRXS17 associates with the cytosolic iron-sulfur cluster assembly pathway, Plant Physiol, 172 (2016) 858–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alkafeef SS, Lane S, Yu C, Zhou T, Solis NV, Filler SG, Huang L, Liu H, Proteomic profiling of the monothiol glutaredoxin Grx3 reveals its global role in the regulation of iron dependent processes, PLoS Genet, 16 (2020) e1008881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willems P, Wanschers BF, Esseling J, Szklarczyk R, Kudla U, Duarte I, Forkink M, Nooteboom M, Swarts H, Gloerich J, Nijtmans L, Koopman W, Huynen MA, BOLA1 is an aerobic protein that prevents mitochondrial morphology changes induced by glutathione depletion, Antioxid Redox Signal, 18 (2013) 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roret T, Tsan P, Couturier J, Zhang B, Johnson MK, Rouhier N, Didierjean C, Structural and spectroscopic insights into BolA-glutaredoxin complexes, J Biol Chem, 289 (2014) 24588–24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guinote IB, Moreira RN, Barahona S, Freire P, Vicente M, Arraiano CM, Breaking through the stress barrier: the role of BolA in Gram-negative survival, World J Microbiol Biotechnol, 30 (2014) 2559–2566. [DOI] [PubMed] [Google Scholar]
- 31.Rey P, Taupin-Broggini M, Couturier J, Vignols F, Rouhier N, Is there a role for glutaredoxins and BOLAs in the perception of the cellular iron status in plants?, Front Plant Sci, 10 (2019) 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albetel AN, Outten CE, Characterization of glutaredoxin Fe-S cluster-binding interactions using circular dichroism spectroscopy, Methods Enzymol, 599 (2018) 327–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dlouhy AC, Li H, Albetel AN, Zhang B, Mapolelo DT, Randeniya S, Holland AA, Johnson MK, Outten CE, The Escherichia coli BolA protein IbaG forms a histidine-ligated [2Fe-2S]-bridged complex with Grx4, Biochemistry, 55 (2016) 6869–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen S, Rao B, Wachnowsky C, Cowan JA, Cluster exchange reactivity of [2Fe-2S] cluster-bridged complexes of BOLA3 with monothiol glutaredoxins, Metallomics, 10 (2018) 1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasta V, Giachetti A, Ciofi-Baffoni S, Banci L, Structural insights into the molecular function of human [2Fe-2S] BOLA1-GRX5 and [2Fe-2S] BOLA3-GRX5 complexes, Biochim Biophys Acta Gen Subj, 1861 (2017) 2119–2131. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Mapolelo DT, Dingra NN, Keller G, Riggs-Gelasco PJ, Winge DR, Johnson MK, Outten CE, Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast, J Biol Chem, 286 (2011) 867–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chi CB, Tang Y, Zhang J, Dai YN, Abdalla M, Chen Y, Zhou CZ, Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3, J Mol Biol, 430 (2018) 1235–1248. [DOI] [PubMed] [Google Scholar]
- 38.Uzarska MA, Nasta V, Weiler BD, Spantgar F, Ciofi-Baffoni S, Saviello MR, Gonnelli L, Mühlenhoff U, Banci L, Lill R, Mitochondrial Bol1 and Bol3 function as assembly factors for specific iron-sulfur proteins, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baussier C, Fakroun S, Aubert C, Dubrac S, Mandin P, Py B, Barras F, Making iron-sulfur cluster: structure, regulation and evolution of the bacterial ISC system, Adv Microb Physiol, 76 (2020) 1–39. [DOI] [PubMed] [Google Scholar]
- 40.Outten FW, Recent advances in the Suf Fe-S cluster biogenesis pathway: Beyond the Proteobacteria, Biochim Biophys Acta, 1853 (2015) 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérard J, Ollagnier de Choudens S, Iron-sulfur clusters biogenesis by the SUF machinery: close to the molecular mechanism understanding, J Biol Inorg Chem, 23 (2018) 581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeung N, Gold B, Liu NL, Prathapam R, Sterling HJ, Willams ER, Butland G, The E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes, Biochemistry, 50 (2011) 8957–8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vranish JN, Das D, Barondeau DP, Real-time kinetic probes support monothiol glutaredoxins as intermediate carriers in Fe-S cluster biosynthetic pathways, ACS Chem Biol, 11 (2016) 3114–3121. [DOI] [PubMed] [Google Scholar]
- 44.Mapolelo DT, Zhang B, Randeniya S, Albetel AN, Li H, Couturier J, Outten CE, Rouhier N, Johnson MK, Monothiol glutaredoxins and A-type proteins: partners in Fe-S cluster trafficking, Dalton Trans, 42 (2013) 3107–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shakamuri P, Zhang B, Johnson MK, Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins, J Am Chem Soc, 134 (2012) 15213–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burschel S, Kreuzer Decovic D, Nuber F, Stiller M, Hofmann M, Zupok A, Siemiatkowska B, Gorka M, Leimkühler S, Friedrich T, Iron-sulfur cluster carrier proteins involved in the assembly of Escherichia coli NADH:ubiquinone oxidoreductase (complex I), Mol Microbiol, 111 (2019) 31–45. [DOI] [PubMed] [Google Scholar]
- 47.Dressaire C, Moreira RN, Barahona S, Alves de Matos AP, Arraiano CM, BolA is a transcriptional switch that turns off motility and turns on biofilm development, mBio, 6 (2015) e02352–02314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moreira RN, Dressaire C, Barahona S, Galego L, Kaever V, Jenal U, Arraiano CM, BolA is required for the accurate regulation of c-di-GMP, a central player in biofilm formation, mBio, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh SP, Montgomery BL, Regulation of BolA abundance mediates morphogenesis in Fremyella diplosiphon, Front Microbiol, 6 (2015) 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh SP, Montgomery BL, Morphogenes bolA and mreB mediate the photoregulation of cellular morphology during complementary chromatic acclimation in Fremyella diplosiphon, Mol Microbiol, 93 (2014) 167–182. [DOI] [PubMed] [Google Scholar]
- 51.Silva AV, Edel M, Gescher J, Paquete CM, Exploring the effects of bolA in biofilm formation and current generation by Shewanella oneidensis MR-1, Front Microbiol, 11 (2020) 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mil-Homens D, Barahona S, Moreira RN, Silva IJ, Pinto SN, Fialho AM, Arraiano CM, Stress response protein BolA influences fitness and promotes Salmonella enterica serovar Typhimurium virulence, Appl Environ Microbiol, 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graça-Lopes G, Graça G, Barahona S, Moreira RN, Arraiano CM, Gonçalves LG, NMR-metabolomics shows that BolA is an important modulator of Salmonella typhimurium metabolic processes under virulence conditions, Metabolites, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleurie A, Zoued A, Alvarez L, Hines KM, Cava F, Xu L, Davis BM, Waldor MK, A Vibrio cholerae BolA-like protein is required for proper cell shape and cell envelope integrity, mBio, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guinote IB, Moreira RN, Freire P, Arraiano CM, Characterization of the BolA homolog IbaG: a new gene involved in acid resistance, J Microbiol Biotechnol, 22 (2012) 484–493. [DOI] [PubMed] [Google Scholar]
- 56.Braymer JJ, Lill R, Iron-sulfur cluster biogenesis and trafficking in mitochondria, J Biol Chem, 292 (2017) 12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lill R, Freibert SA, Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis, Annu Rev Biochem, 89 (2020) 471–499. [DOI] [PubMed] [Google Scholar]
- 58.Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Mühlenhoff U, The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation, Mol Biol Cell, 24 (2013) 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Miao R, Bertram S, Jia X, Ward DM, Kaplan J, A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast, J Biol Chem, 287 (2012) 35709–35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez LA, Gomez-Gallardo M, Diaz-Perez AL, Cortes-Rojo C, Campos-Garcia J, Iba57p participates in maturation of a [2Fe-2S]-cluster Rieske protein and in formation of supercomplexes III/IV of Saccharomyces cerevisiae electron transport chain, Mitochondrion, 44 (2019) 75–84. [DOI] [PubMed] [Google Scholar]
- 61.Brancaccio D, Gallo A, Mikolajczyk M, Zovo K, Palumaa P, Novellino E, Piccioli M, Ciofi-Baffoni S, Banci L, Formation of [4Fe-4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery, J Am Chem Soc, 136 (2014) 16240–16250. [DOI] [PubMed] [Google Scholar]
- 62.Maio N, Rouault TA, Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis, Trends Biochem Sci, 45 (2020) 411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nasta V, Suraci D, Gourdoupis S, Ciofi-Baffoni S, Banci L, A pathway for assembling [4Fe-4S](2+) clusters in mitochondrial iron-sulfur protein biogenesis, FEBS J, (2019). [DOI] [PubMed] [Google Scholar]
- 64.Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L, Levi S, Iolascon A, The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload, Blood, 110 (2007) 1353–1358. [DOI] [PubMed] [Google Scholar]
- 65.Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA, Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts, J Clin Invest, 120 (2010) 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu G, Guo S, Anderson GJ, Camaschella C, Han B, Nie G, Heterozygous missense mutations in the GLRX5 gene cause sideroblastic anemia in a Chinese patient, Blood, 124 (2014) 2750–2751. [DOI] [PubMed] [Google Scholar]
- 67.Daher R, Mansouri A, Martelli A, Bayart S, Manceau H, Callebaut I, Moulouel B, Gouya L, Puy H, Kannengiesser C, Karim Z, GLRX5 mutations impair heme biosynthetic enzymes ALA synthase 2 and ferrochelatase in human congenital sideroblastic anemia, Mol Genet Metab, 128 (2019) 342–351. [DOI] [PubMed] [Google Scholar]
- 68.Crooks DR, Ghosh MC, Haller RG, Tong WH, Rouault TA, Posttranslational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery, Blood, 115 (2010) 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rouault TA, The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways, Biometals, 32 (2019) 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI, Tübingen 2000 Screen Consortium, Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis, Nature, 436 (2005) 1035–1039. [DOI] [PubMed] [Google Scholar]
- 71.Chiong MA, Procopis P, Carpenter K, Wilcken B, Late-onset nonketotic hyperglycinemia with leukodystrophy and an unusual clinical course, Pediatr Neurol, 37 (2007) 283–286. [DOI] [PubMed] [Google Scholar]
- 72.Wei SH, Weng WC, Lee NC, Hwu WL, Lee WT, Unusual spinal cord lesions in late-onset non-ketotic hyperglycinemia, J Child Neurol, 26 (2011) 900–903. [DOI] [PubMed] [Google Scholar]
- 73.Baker PR 2nd, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, Aicher J, Creadon-Swindell G, Geiger E, MacLean KN, Lee WT, Deshpande C, Freckmann ML, Shih LY, Wasserstein M, Rasmussen MB, Lund AM, Procopis P, Cameron JM, Robinson BH, Brown GK, Brown RM, Compton AG, Dieckmann CL, Collard R, Coughlin CR 2nd, Spector E, Wempe MF, Van Hove JL, Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5, Brain, 137 (2014) 366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu G, Wang Y, Anderson GJ, Camaschella C, Chang Y, Nie G, Functional analysis of GLRX5 mutants reveals distinct functionalities of GLRX5 protein, J Cell Biochem, 117 (2016) 207–217. [DOI] [PubMed] [Google Scholar]
- 75.Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH, Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes, Am J Hum Genet, 89 (2011) 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haack TB, Rolinski B, Haberberger B, Zimmermann F, Schum J, Strecker V, Graf E, Athing U, Hoppen T, Wittig I, Sperl W, Freisinger P, Mayr JA, Strom TM, Meitinger T, Prokisch H, Homozygous missense mutation in BOLA3 causes multiple mitochondrial dysfunctions syndrome in two siblings, J Inherit Metab Dis, 36 (2013) 55–62. [DOI] [PubMed] [Google Scholar]
- 77.Lebigot E, Gaignard P, Dorboz I, Slama A, Rio M, de Lonlay P, Héron B, Sabourdy F, Boespflug-Tanguy O, Cardoso A, Habarou F, Ottolenghi C, Thérond P, Bouton C, Golinelli-Cohen MP, Boutron A, Impact of mutations within the [Fe-S] cluster or the lipoic acid biosynthesis pathways on mitochondrial protein expression profiles in fibroblasts from patients, Mol Genet Metab, 122 (2017) 85–94. [DOI] [PubMed] [Google Scholar]
- 78.Nikam RM, Gripp KW, Choudhary AK, Kandula V, Imaging phenotype of multiple mitochondrial dysfunction syndrome 2, a rare BOLA3-associated leukodystrophy, Am J Med Genet A, 176 (2018) 2787–2790. [DOI] [PubMed] [Google Scholar]
- 79.Nishioka M, Inaba Y, Motobayashi M, Hara Y, Numata R, Amano Y, Shingu K, Yamamoto Y, Murayama K, Ohtake A, Nakazawa Y, An infant case of diffuse cerebrospinal lesions and cardiomyopathy caused by a BOLA3 mutation, Brain Dev, 40 (2018) 484–488. [DOI] [PubMed] [Google Scholar]
- 80.Stutterd CA, Lake NJ, Peters H, Lockhart PJ, Taft RJ, van der Knaap MS, Vanderver A, Thorburn DR, Simons C, Leventer RJ, Severe leukoencephalopathy with clinical recovery caused by recessive BOLA3 mutations, JIMD Rep, 43 (2019) 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohda M, Tokuzawa Y, Kishita Y, Nyuzuki H, Moriyama Y, Mizuno Y, Hirata T, Yatsuka Y, Yamashita-Sugahara Y, Nakachi Y, Kato H, Okuda A, Tamaru S, Borna NN, Banshoya K, Aigaki T, Sato-Miyata Y, Ohnuma K, Suzuki T, Nagao A, Maehata H, Matsuda F, Higasa K, Nagasaki M, Yasuda J, Yamamoto M, Fushimi T, Shimura M, Kaiho-Ichimoto K, Harashima H, Yamazaki T, Mori M, Murayama K, Ohtake A, Okazaki Y, A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies, PLoS Genet, 12 (2016) e1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moseler A, Aller I, Wagner S, Nietzel T, Przybyla-Toscano J, Mühlenhoff U, Lill R, Berndt C, Rouhier N, Schwarzlander M, Meyer AJ, The mitochondrial monothiol glutaredoxin S15 is essential for iron-sulfur protein maturation in Arabidopsis thaliana, Proc Natl Acad Sci U S A, 112 (2015) 13735–13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ströher E, Grassl J, Carrie C, Fenske R, Whelan J, Millar AH, Glutaredoxin S15 is involved in Fe-S cluster transfer in mitochondria influencing lipoic acid-dependent enzymes, plant growth, and arsenic tolerance in Arabidopsis, Plant Physiol, 170 (2016) 1284–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uzarska MA, Przybyla-Toscano J, Spantgar F, Zannini F, Lill R, Muhlenhoff U, Rouhier N, Conserved functions of Arabidopsis mitochondrial late-acting maturation factors in the trafficking of ironsulfur clusters, Biochim Biophys Acta Mol Cell Res, 1865 (2018) 1250–1259. [DOI] [PubMed] [Google Scholar]
- 85.Przybyla-Toscano J, Roland M, Gaymard F, Couturier J, Rouhier N, Roles and maturation of iron-sulfur proteins in plastids, J Biol Inorg Chem, 23 (2018) 545–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N, Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters, EMBO J, 27 (2008) 1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao H, Subramanian S, Couturier J, Naik SG, Kim SK, Leustek T, Knaff DB, Wu HC, Vignols F, Huynh BH, Rouhier N, Johnson MK, Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins, Biochemistry, 52 (2013) 6633–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rey P, Becuwe N, Tourrette S, Rouhier N, Involvement of Arabidopsis glutaredoxin S14 in the maintenance of chlorophyll content, Plant Cell Environ, 40 (2017) 2319–2332. [DOI] [PubMed] [Google Scholar]
- 89.Gupta M, Outten CE, Iron-sulfur cluster signaling: The common thread in fungal iron regulation, Curr Opin Chem Biol, 55 (2020) 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martínez-Pastor MT, Perea-Garcia A, Puig S, Mechanisms of iron sensing and regulation in the yeast Saccharomyces cerevisiae, World J Microbiol Biotechnol, 33 (2017) 75. [DOI] [PubMed] [Google Scholar]
- 91.Li H, Outten CE, The conserved CDC motif in the yeast iron regulator Aft2 mediates iron-sulfur cluster exchange and protein-protein interactions with Grx3 and Bol2, J Biol Inorg Chem, 24 (2019) 809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poor CB, Wegner SV, Li H, Dlouhy AC, Schuermann JP, Sanishvili R, Hinshaw JR, Riggs-Gelasco PJ, Outten CE, He C, Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2, Proc Natl Acad Sci U S A, 111 (2014) 4043–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Outten CE, The role of Fe-S clusters in yeast iron regulation, in: Rouault T (Ed.) Iron-Sulfur Clusters in Chemistry and Biology, Walter de Gruter GmbH, Berlin, 2017, pp. 161–185. [Google Scholar]
- 94.Lill R, Srinivasan V, Mühlenhoff U, The role of mitochondria in cytosolic-nuclear iron-sulfur protein biogenesis and in cellular iron regulation, Curr Opin Microbiol, 22 (2014) 111–119. [DOI] [PubMed] [Google Scholar]
- 95.Brault A, Mourer T, Labbé S, Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts, IUBMB Life, 67 (2015) 801–815. [DOI] [PubMed] [Google Scholar]
- 96.Dlouhy AC, Beaudoin J, Labbé S, Outten CE, Schizosaccharomyces pombe Grx4 regulates the transcriptional repressor Php4 via [2Fe-2S] cluster binding, Metallomics, 9 (2017) 1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vachon P, Mercier A, Jbel M, Labbé S, The monothiol glutaredoxin Grx4 exerts an iron-dependent inhibitory effect on Php4 function, Eukaryot Cell, 11 (2012) 806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jacques JF, Mercier A, Brault A, Mourer T, Labbé S, Fra2 is a co-regulator of Fep1 inhibition in response to iron starvation, PLoS One, 9 (2014) e98959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim HJ, Lee KL, Kim KD, Roe JH, The iron uptake repressor Fep1 in the fission yeast binds Fe-S cluster through conserved cysteines, Biochem Biophys Res Commun, 478 (2016) 187–192. [DOI] [PubMed] [Google Scholar]
- 100.Jbel M, Mercier A, Labbé S, Grx4 monothiol glutaredoxin is required for iron limitation-dependent inhibition of Fep1, Eukaryot Cell, 10 (2011) 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martínez-Pastor MT, Puig S, Adaptation to iron deficiency in human pathogenic fungi, Biochim Biophys Acta Mol Cell Res, 1867 (2020) 118797. [DOI] [PubMed] [Google Scholar]
- 102.Misslinger M, Scheven MT, Hortschansky P, López-Berges MS, Heiss K, Beckmann N, Heigl T, Hermann M, Kruger T, Kniemeyer O, Brakhage AA, Haas H, The monothiol glutaredoxin GrxD is essential for sensing iron starvation in Aspergillus fumigatus, PLoS Genet, 15 (2019) e1008379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang D, Dong Y, Yu Q, Kai Z, Zhang M, Jia C, Xiao C, Zhang B, Zhang B, Li M, Function of glutaredoxin 3 (Grx3) in oxidative stress response caused by iron homeostasis disorder in Candida albicans, Future Microbiol, 12 (2017) 1397–1412. [DOI] [PubMed] [Google Scholar]
- 104.Dong Y, Zhang D, Yu Q, Zhao Q, Xiao C, Zhang K, Jia C, Chen S, Zhang B, Zhang B, Li M, Loss of Ssq1 leads to mitochondrial dysfunction, activation of autophagy and cell cycle arrest due to iron overload triggered by mitochondrial iron-sulfur cluster assembly defects in Candida albicans, Int J Biochem Cell Biol, 85 (2017) 44–55. [DOI] [PubMed] [Google Scholar]
- 105.Attarian R, Hu G, Sanchez-León E, Caza M, Croll D, Do E, Bach H, Missall T, Lodge J, Jung WH, Kronstad JW, The monothiol glutaredoxin Grx4 regulates iron homeostasis and virulence in Cryptococcus neoformans, MBio, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haunhorst P, Hanschmann EM, Brautigam L, Stehling O, Hoffmann B, Muhlenhoff U, Lill R, Berndt C, Lillig CH, Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation, Mol Biol Cell, 24 (2013) 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Banci L, Camponeschi F, Ciofi-Baffoni S, Muzzioli R, Elucidating the molecular function of human BOLA2 in GRX3-dependent anamorsin maturation pathway, J Am Chem Soc, 137 (2015) 16133–16143. [DOI] [PubMed] [Google Scholar]
- 108.Li H, Mapolelo DT, Randeniya S, Johnson MK, Outten CE, Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2, Biochemistry, 51 (2012) 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Banci L, Ciofi-Baffoni S, Gajda K, Muzzioli R, Peruzzini R, Winkelmann J, N-terminal domains mediate [2Fe-2S] cluster transfer from glutaredoxin-3 to anamorsin, Nat Chem Biol, 11 (2015) 772–778. [DOI] [PubMed] [Google Scholar]
- 110.Camponeschi F, Prusty NR, Heider SAE, Ciofi-Baffoni S, Banci L, GLRX3 Acts as a [2Fe-2S] Cluster Chaperone in the Cytosolic Iron-Sulfur Assembly Machinery Transferring [2Fe-2S] Clusters to NUBP1, J Am Chem Soc, 142 (2020) 10794–10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frey AG, Palenchar DJ, Wildemann JD, Philpott CC, A glutaredoxin•BolA complex serves as an iron-sulfur cluster chaperone for the cytosolic cluster assembly machinery, J Biol Chem, 291 (2016) 22344–22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patel SJ, Frey AG, Palenchar DJ, Achar S, Bullough KZ, Vashisht A, Wohlschlegel JA, Philpott CC, A PCBP1-BolA2 chaperone complex delivers iron for cytosolic [2Fe-2S] cluster assembly, Nat Chem Biol, 15 (2019) 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Philpott CC, Patel SJ, Protchenko O, Management versus miscues in the cytosolic labile iron pool: The varied functions of iron chaperones, Biochim Biophys Act Mol Cell Res, 1867 (2020) 118830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nuttle X, Giannuzzi G, Duyzend MH, Schraiber JG, Narvaiza I, Sudmant PH, Penn O, Chiatante G, Malig M, Huddleston J, Benner C, Camponeschi F, Ciofi-Baffoni S, Stessman HA, Marchetto MC, Denman L, Harshman L, Baker C, Raja A, Penewit K, Janke N, Tang WJ, Ventura M, Banci L, Antonacci F, Akey JM, Amemiya CT, Gage FH, Reymond A, Eichler EE, Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility, Nature, 536 (2016) 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Giannuzzi G, Schmidt PJ, Porcu E, Willemin G, Munson KM, Nuttle X, Earl R, Chrast J, Hoekzema K, Risso D, Mannik K, De Nittis P, Baratz ED, C. p, Herault Y, Gao X, Philpott CC, Bernier RA, Kutalik Z, Fleming MD, Eichler EE, Reymond A, The human-specific BOLA2 duplication modifies iron homeostasis and anemia predisposition in chromosome 16p11.2 autism individuals, Am J Hum Genet, 105 (2019) 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE, The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation, Biochemistry, 48 (2009) 9569–9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Comini MA, Krauth-Siegel RL, Bellanda M, Mono- and dithiol glutaredoxins in the trypanothione-based redox metabolism of pathogenic trypanosomes, Antioxid Redox Signal, 19 (2013) 708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.The PyMOL Molecular Graphics System, Version 2.4.0 Schrödinger, LLC.


