Abstract
Most clinically approved cancer therapies are potent and toxic small molecules that are limited by severe off-target toxicities and poor tumor-specific localization. Over the past few decades, attempts have been made to load chemotherapies into liposomes, which act to deliver the therapeutic agent directly to the tumor. Although liposomal encapsulation has been shown to decrease toxicity in human patients, reliance on passive targeting via the enhanced permeability and retention (EPR) effect has left some of these issues unresolved. Recently, investigations into modifying the surface of liposomes via covalent and/or electrostatic functionalization have offered mechanisms for tumor homing and subsequently controlled chemotherapeutic delivery. A wide variety of biomolecules can be utilized to functionalize liposomes such as proteins, carbohydrates, and nucleic acids, which enable multiple directions for cancer cell localization. Importantly, when nanoparticles are modified with such molecules, care must be taken as not to inactivate or denature the ligand. Peptides, which are small proteins with <30 amino acids, have demonstrated the exceptional ability to act as ligands for transmembrane protein receptors overexpressed in many tumor phenotypes. Exploring this strategy offers a method in tumor targeting for cancers such as glioblastoma multiforme, pancreatic, lung, and breast based on the manifold of receptors overexpressed on various tumor cell populations. In this review, we offer a comprehensive summary of peptide-functionalized liposomes for receptor-targeted cancer therapy.
I. INTRODUCTION
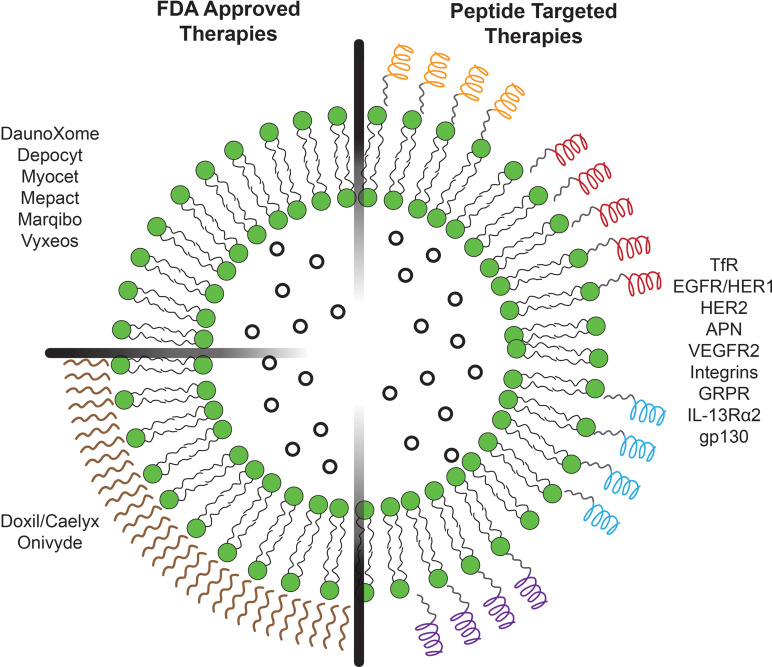
Liposomes have been at the forefront of drug delivery research for the past few decades, following their discovery in 1965 by Alec Bangham.1,2 Since then, they have been delineated into categories including small unilamellar vesicles (SUVs < 100 nm), medium unilamellar vesicles (MUVs 100–250 nm), large unilamellar vesicles (LUVs > 250 nm), and giant unilamellar vesicles (GUVs)—with the majority of drug delivery studies focusing on SUVs.3 Composed of a concentric hydrophobic phospholipid bilayer compartmentalizing an aqueous core from its aqueous environment, liposomes offer a plethora of controlled delivery applications for different classes of drugs.4 In brief, hydrophobic drugs such as small molecule chemotherapeutics can be loaded into the lipid lamella, while hydrophilic therapies such as nucleic acids can be loaded into the aqueous core for applications such as gene editing.5,6 The majority of current cancer therapies rely on the systemic administration of chemotherapeutic agents that exhibit off-target effects due to their inability to differentiate between healthy and tumor tissue—this often results in side effects like nausea and fatigue or more severely cardiotoxicity.7–10 To improve the therapeutic index of these agents, drugs can be encapsulated in liposomal membranes, which act as a barrier to decrease toxic effects in healthy tissues. Several liposomal drug formulations have been approved by the FDA, loading chemotherapeutics such as doxorubicin (Doxil), duanorubicin (DaunoXome), cytarabine (Depocyte), vincristine (Marqibo), mifamurtide (Mepact), irinotecan (Onivyde), and daunorubicin/cytarabine (Vyxeos) (Table I).11–15 Some of these nanoparticle drug formulations also take advantage of PEGylation, which is thought to act as a nanoparticle cloak to greatly enhance their circulation time and reduce immune responses.16,17
TABLE I.
Clinically approved liposomal cancer therapies.
| Name | Encapsulated drug | Indications | Year approved | Ref. |
|---|---|---|---|---|
| Doxil/Caelyx | Doxorubicin | HIV-related Kaposi's sarcoma | 1995 | 11, 12, 14, 25 |
| Ovarian cancer | 2005 | |||
| Multiple myeloma | 2008 | |||
| Breast cancer | 2012 | |||
| DaunoXome | Duanorubicin | HIV-related Kaposi's sarcoma | 1996 | |
| Depocyt | Cytarabine/Ara-C | Neoplastic meningitis | 1999 | |
| Myocet | Doxorubicin | Metastatic breast cancer | 2000 | |
| Mepact | Mifamurtide | High-grade, resectable, and non-metastatic osteosarcoma | 2004 | |
| Marqibo | Vincristine | Acute lymphoblastic leukemia | 2012 | |
| Onivyde | Irinotecan | Metastatic pancreatic cancer | 2016 | |
| Vyxeos | Daunorubicin and Cytarabine | High-risk acute myeloid leukemia | 2017 | 13 |
Relying on passive localization of nanoparticles to target tissue has many limitations. When in the bloodstream, liposomes are immediately coated with a surface of plasma proteins that form a protein corona.18 Circulation times can often be modulated by the protein species present in the corona, leading to either extended circulation times due to dysopsonins, or more often limiting it through rapid clearance by the reticuloendothelial system (RES) or mononuclear phagocyte system (MPS) with opsonins.19,20 PEGylation of liposomes was thought to circumvent these limitations by shielding protein coronas from forming and subsequently undergoing clearance, but recent studies have shown contradicting evidence with a phenomenon called accelerated blood clearance (ABC) and complement activation-related pseudoallergy (CARPA).21–24 Furthermore, tumor localization heavily relies on the enhanced permeability and retention (EPR) effect, which is the combined phenomenon of leaky vasculature proximal to the tumor and blocked lymphatic drainage within the tumor microenvironment. Unfortunately, although this phenomenon is repeatedly observed in mice, it is highly variable in human patients in part due to cancer heterogeneity.26–30 Therefore, active targeting of tumor tissue through functionalization of liposomes offers an avenue to greatly enhance localization of therapies to minimize collateral damage.31
There are several options for modifying liposomes to enable tumor targeting,30 as many groups have explored the use of peptides, proteins and antibodies,31–38 nucleic acids and aptamers,33–36 and carbohydrates.33,39–42 Each of these biomolecules offers advantages and disadvantages in applicability, such as high biocompatibility and bioactivity, as either liposome-membrane fusogens43,44 or ligands for receptor targeting.25,30 Here, we discuss peptides because they are ideal for liposomal surface modification due to their ease of synthesis, ease of manufacturing at industrial and clinical levels, and chemical versatility.47–49 Overall, these advantageous characteristics promote an immense field for novel innovation and discovery for a plethora of diseases. In this review, we focus on the recent advancements in receptor targeting peptide-functionalized liposomes over the past five years. By peptides acting as ligands with high affinities toward overexpressed cell membrane receptors, liposomes encapsulating chemotherapeutic agents can improve targeting in vitro and in vivo and act as clinically applicable therapies for improved cancer treatment (Fig. 1).
FIG. 1.
Schematic illustration of liposome-encapsulated therapies. (Left) FDA-approved therapies for chemotherapeutic-encapsulated liposomes (top left) without PEGylation and (bottom left) with PEGylation. (Right) Peptide-functionalized liposomes for receptor-targeted chemotherapeutic delivery.
II. RECEPTOR-TARGETING PEPTIDE LIGANDS
One avenue to actively target cancer cells is through receptor–ligand binding. Many novel peptides have been discovered to act as ligands with high affinity for a plethora of overexpressed receptors on the surface of cancer cells. Therefore, functionalizing liposomes with these sequences offers a method of tumor targeting, with a simultaneous reduction in off-target toxicity. The receptors focused on in this section include the transferrin (TfR), epidermal growth factor (EGFR/HER1 and HER2), gastrin-releasing peptide (GRPR), aminopeptidase N (APN), vascular endothelial growth factor 2 (VEGFR2), and integrin receptors (Table II).
TABLE II.
Peptide ligands for targeting tumor and tumor microenvironment receptors. L denotes L-amino acids, and D denotes D-amino acids.
| Targeted receptor | Peptide name | Peptide sequence | Reported KD (M) | Ref |
|---|---|---|---|---|
| TfR | T7 | HAIYPRH | 2.1 × 10−8 | 50, 51 |
| EGFR/HER1 | GE11 | YHWYGYTPQNVI | 2.2 × 10−8 | 52–54 |
| HER2 | P6.1 | KCCYSL | 3.0–4.5 × 10−8 | 55 |
| HER-2 Peptide | YCDGFYACYMDV | — | 56 | |
| AHNP | FCDGFYACYADV | 3.59 × 10−7 | 57 | |
| APN | NGR | NGR | — | 58 |
| LN | YEVGHRC | 1.0 × 10−7 | 59 | |
| VEGFR2 | STP | SKDEEWHKNNFPLSP | 8.50 × 10−8 | 60, 61 |
| TP | TIDHEWKKTSFPLSF | 5.93 × 10−7 | 60 | |
| S1 | LIDHEWKENYFPLSF | 1.31 × 10−7 | 62 | |
| LA7R | ATWLPPR | 9.29–18.09 × 10−9 | 63–66 | |
| DA7R | ATWLPPR | 8.41 × 10−9 | 63 | |
| Cyclic A7R | CATWLPPR | 6.79 × 10−9 | 66 | |
| Integrins | Linear RGD | RGD | — | 67–72 |
| GRGDS | — | 73 | ||
| Cyclic RGD | c(RGDyC) | — | 74 | |
| c(RGDfK) | — | 75 | ||
| c(RGDf[N-methyl]C) | — | 76 | ||
| c(RGDyK) | — | 77, 78 | ||
| RWrNK | RWrNK | 1.6 × 10−9 | 79 | |
| P1c | CIRTPKISKPIKFELSG | — | 80, 81 | |
| GRPR | Cystabn | FQWAVGH-Sta-L-NH2 | — | 82 |
| IL-13Rα2 | Pep-1 | CGEMGWVRC | — | 83 |
| gp130 | VTW | VTWTPQAWFQWV | — | 84 |
When functionalizing the surface of liposomes with peptides, there are two general strategies for conducting this modification. First, ligands can be covalently conjugated to lipid headgroups or polymer extensions (such as PEG) that project the ligand orthogonally from the liposomal surface.19 More specifically, it is generally understood that there exist four main chemical conjugation methods to achieve these modifications: activated carboxyl groups can react with amino groups to form amide bonds, pyridyldithiols can react with thiols to form disulfide bonds, maleimide derivatives can react with thiols to yield thioether bonds, and p-nitrophenylcarbonyl groups can react with amino groups to form carbamate bonds.30 Alternatively, peptides can adsorb to and/or interpolate into the liposomal surface via electrostatic and/or hydrophobic interactions.19,45 Both these methods present the ligands to their respective receptors and aid in the targeting of specific tumor cells.
A. Transferrin receptor (TfR)
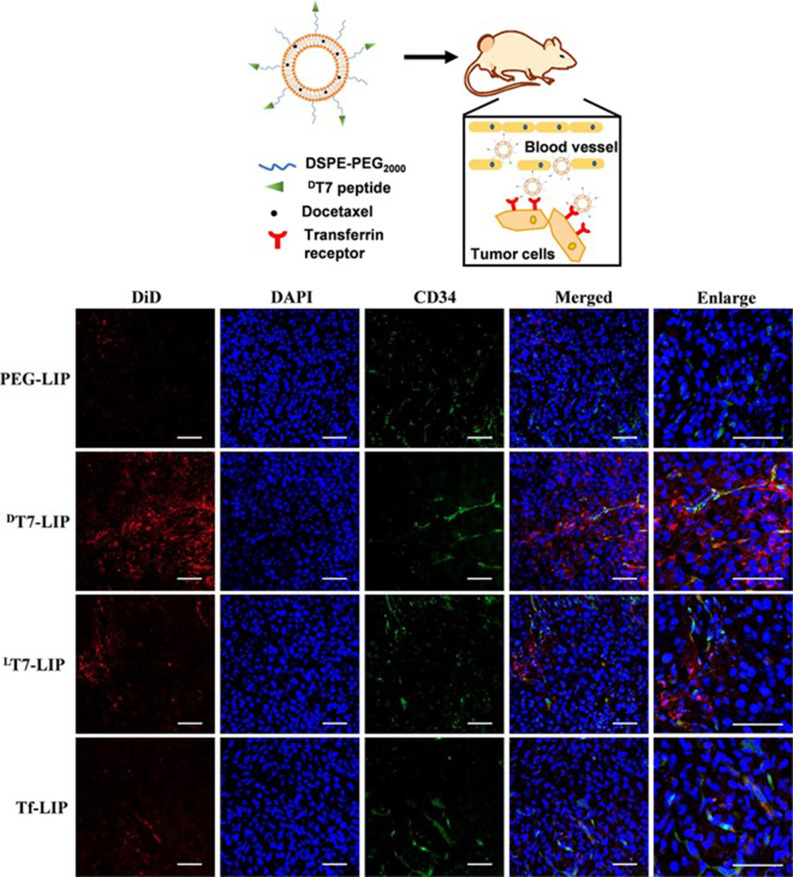
Transferrin receptors (TfRs) are often overexpressed in a variety of cancer types due to the increased metabolic demand for iron, indicating an attractive targeting receptor for cancer therapeutics.85,86 Peptide T7, sequence HAIYPRH, has been widely characterized and shown to exhibit a high binding affinity to TfR. One group compared peptide-modified liposomes to target hepatocellular carcinoma cells with L- and D-enantiomers, DT7 and LT7, as well as a transferrin (Tf)-modified liposome control.50 It would be expected for the D-enantiomer to exhibit a decreased selectivity due to chiral specific receptors; however, they demonstrated increased binding affinity via surface plasmon resonance (SPR) and in vitro studies where DT7-modified liposomes greatly enhanced the targeting ability of liposomes to the cells over LT7 and Tf. Furthermore, the results were replicated in vivo with a significant reduction in tumor growth in mice treated with DT7-modified liposomes loaded with the chemotherapeutic docetaxel. Upon tumor sectioning of 24 h post-injection mice with DiD dye-loaded liposomes, immunofluorescence shows a significant increase in tumor accumulation when nanoparticles are decorated with DT7 (Fig. 2). Another group focused on treating lung cancer and used T7-functionalized liposomes loaded with quercetin to improve localization.43 Their results show that the T7 surface-modified liposomes increased the induction of apoptosis significantly compared to all controls and improved localization of drug-loaded nanoparticles toward A549 cells compared to MRC-5 normal lung fibroblasts. Remarkably, they demonstrated a threefold increase in cytotoxicity of quercetin when loaded into T7-functionalized liposomes compared to the free drug. Importantly, tumor penetration was visualized with fluorescently labled liposomes and quantified to find a significant increase in the penetration depth with T7 surface modifications. Altogether, DT7 offers not only advantages over LT7 due to proteolytic stability but also enhanced tumor targeting, advancing the field of TfR-specific cancer therapeutics.
FIG. 2.
Intratumoral distribution of liposomes encapsulated with DiD in HepG2 xenograft tumors. (Red) DiD-encapsulated liposome. (Blue) DAPI-cell nuclei. (Green) anti-CD34-blood vessels. Scale bar = 40 μm. Reprinted with permission from Tang et al., “A stabilized retro-inverso peptide ligand of transferrin receptor for enhanced liposome-based hepatocellular carcinoma-targeted drug delivery,” Acta Biomater. 83, 379–389 (2019). Copyright 2019 Elsevier.
B. Epidermal growth factor receptor (EGFR)
Erythroblastic leukemia viral oncogene homolog (ERBB) receptors are a family of four transmembrane receptor tyrosine kinases (RTKs) that are often overexpressed during the malignant transformation of healthy cells.87,88 The four receptors within this family include epidermal growth factor receptor (EGFR) or HER1, as well as HER2, HER3, and HER4. In 2005, the peptide sequence YHWYGYTPQNVI, known as GE11, was designed using phage display by the labs of Yuhong Xu and Jianren Gu and exhibited targeting affinities for EGFR.52,53 Recently, this sequence became used for liposomal conjugation to exploit its EGFR-targeting capabilities. In 2008, Song et al. showed the preliminary targeting capabilities of GE11-functionalized, unloaded liposomes.89 In 2014, Cheng and colleagues functionalized liposomes encapsulating doxorubicin (Dox) and demonstrated increased drug delivery with a 2.6-fold reduction in IC50 of the modified particles compared to the nontargeted control.54 Further, tumor accumulation was exhibited by a 2.2-fold greater fluorescence intensity of the GE11 liposomes compared to the untargeted control. More recently, in 2017, Xu et al. focused on the localized delivery of docetaxel (DTX) and an siRNA responsible for silencing the ABCG2 gene regulating multidrug resistance (MDR).52 In vitro, they showed a reduction in the IC50 of DTX in Hep-2 laryngeal cancer cells when treated with GE11 liposomes compared to treatment with free drug. Furthermore, in vivo Hep-2 xenograft-bearing mice demonstrated that the GE11 liposomes greatly enhanced tumor growth inhibition compared to the non-targeted liposomes.
Ringhieri and coworkers designed liposomes coated with the HER2-targeting peptide P6.1, sequence KCCYSL, for the treatment of breast cancer.55 Liposomes were modified with the monomers, dimers, and tetramers and treated against the under- and over-expressing HER2 breast cancer cells MDA-MB-231 and BT-474, respectively. The results showed that tetrameric P6.1-conjugated liposomes had a 10 times greater degree of cellular binding and uptake in BT-474 compared to MDA-MB-231 cells, highlighting their targeting ability to cells with overexpressed HER2. Furthermore, the tetrameric peptide yielded greater binding and uptake than other peptide forms. Gratifyingly, the levels present were comparable to that of the clinically approved anti-HER2 antibody Herceptin. Often, peptides are compared against Herceptin as the current standard of localization to HER2.55,56 Using the antibody as a template, Shi et al. synthesized an HER-2 peptide analog, sequence YCDGFYACYMDV, which showed increased tumoral delivery of doxorubicin-loaded and pH-sensitive liposomes toward multidrug-resistant MCF-7 breast cancer cells.56 In this study, they showed a strong increase in mitochondrial localization compared to other organelles, which is important for increasing mitochondrial-driven apoptosis. Zahmatkeshan and coworkers also studied HER2 targeting with a different peptide and Anti-HER2/neu peptide (AHNP) with sequence FCDGFYACYADV, this time to improve localization of doxorubicin-loaded PEGylated liposomes.57 Their results demonstrated that increasing the density of functionalized peptides significantly increases tumor homing and uptake in two HER2 overexpressing lines, SKBR3 and TUBO. When tested in a TUBO breast cancer mouse model, liposomes functionalized with 100 and 200 ligands showed a similar tumor growth delay and a significantly longer life expectancy.
C. Aminopeptidase N (APN)
Aminopeptidase N (APN) has been studied extensively due to its overexpression on the surface of cancer cells, most commonly seen in aggressively growing phenotypes. APN is a zinc metalloenzyme found in the plasma membrane, responsible for cleaving N-terminal neutral amino acids; this can lead to functions relating to migration and invasion or metastasis in tumor cells.90 Therefore, exploitation of this overexpression can lead to tumor targeting of nanoparticles. A widely studied peptide known to selectively bind to APN is the tripeptide sequence NGR, which has been appended onto the surfaces of liposomes to target multiple tumor types.91–93 Huang and collaborators presented an NGR-modified liposome for improved glioma targeting and the delivery of combretastatin A4.58 Their formulation showed success in targeting in vitro when treating U87-MG human glioma tumor cells for the inhibition of cellular migration and reduction in vasculogenic mimicry (VM). Subsequently, in vivo results with U87-MG orthotopic tumor-bearing mice demonstrated enhanced targeting of cancer cells with both anti-tumor and anti-VM activities.
Jia et al. utilized a novel peptide with a high affinity for APN called LN, sequence YEVGHRC, to functionalize liposomes loaded with doxorubicin to treat HepG2 cells.59 In vitro, LN conjugation greatly enhanced the cell internalization of doxorubicin-loaded liposomes; these results were reproduced in vivo in a subcutaneous HepG2 xenograft BALB/c nude mouse model with significantly greater tumor accumulation and decreased tumor growth. The implications of peptide-modified liposomes to target APN, particularly with novel sequences such as LN, are increasing in importance paralleling the rise in aggressive and drug-resistant tumors.
D. Vascular endothelial growth factor receptor 2 (VEGFR2)
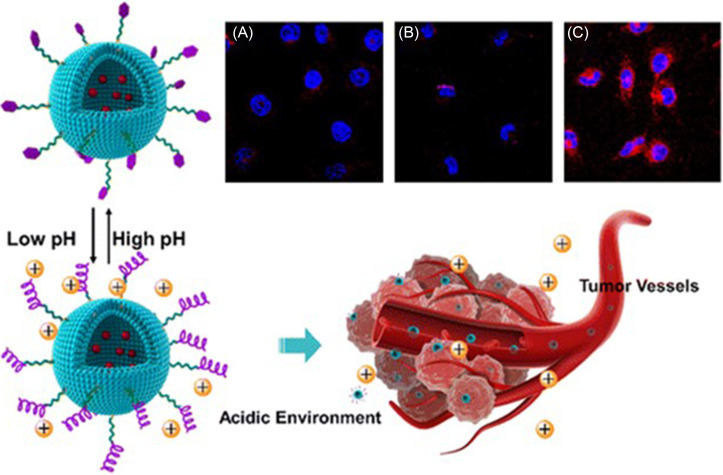
Vascular endothelial growth factor receptor 2 (VEGFR2) is a well-characterized and overexpressed angiogenesis marker commonly found in newly formed tumor vessels.94–96 Activation of this tyrosine kinase receptor has been shown to increase proliferation and migration and subsequently lead to the metastasis of tumors. Zhigyuan Hu's group has investigated peptides that can target VEGFR2 for functionalizing liposomal nanoparticles and directing them to the tumor microenvironment.60–62 Qian et al. first used an imprinting microarray to optimize a peptide sequence to accomplish this, arriving at the peptide STP with sequence SKDEEWHKNNFPLSP.60 STP became activated in low pH tumor microenvironments by conforming to an alpha-helix with the tri-amino acid section Asp–Glu–Glu enabling strong binding to VEGFR2. In vivo, STP successfully recognized and penetrated human umbilical vein endothelial cells (HUVECs) in the tumor microenvironment. Another peptide characterized by Qian was TP, with the sequence TIDHEWKKTSFPLSF, which exhibited similar VEGFR2 targeting at a neutral pH compared to non-VEGFR2 overexpressing cells. Han et al. utilized the more successful STP peptide and used it to functionalize liposomes loaded with doxorubicin (STP-LS-DOX) for pH-responsive targeted drug delivery (Fig. 3).61 STP-LS-DOX exhibited limited HUVEC internalization at a neutral pH [Fig. 3(a)] and without the targeting peptide (LS-DOX) [Fig. 3(b)], but demonstrated significant accumulation in acidic conditions [Fig. 3(c)]. Furthermore, STP-LS-DOX was able to target HT-29 colon adenocarcinoma xenografts in mice through VEGFR2 targeting in tumor endothelial cells, and achieved high targeting efficiency indicated by significant induction of apoptosis of the tumor compared to controls. Further refinement of STP via microarray screening led to a second-generation peptide sequence with high affinity for VEGFR2 called S1, sequence LIDHEWKENYFPLSF.62 Using SPR imaging, S1 was determined to exhibit a KD value of 131 nM, which is comparable to the VEGFR2 poly-antibody. Furthermore, S1 was able to preferentially localize to VEGFR2-overexpressing cells (HUVECs) over non-VEGFR2-expressing cells (293 T). When conjugated to doxorubicin-loaded liposomes, the inhibitory effects of doxorubicin were significantly higher in VEGFR2-overexpressing cells, demonstrating preferential localization. Moreover, the targeted distribution was reproduced in HT-29 tumor-bearing mice.
FIG. 3.
Schematic illustration of STP-decorated, doxorubicin-loaded liposomes in response to pH changes for targeting the tumor microenvironment. Confocal micrographs of HUVEC cells with stained nuclei (blue) and doxorubicin (red) treated with (a) STP-LS-DOX at pH 7.4, (b) LS-DOX at pH 5.8, and (c) STP-LS-DOX at pH 5.8. Adapted with permission from Han et al., ACS Appl. Mater. Interfaces 8(29), 18658–18663 (2016). Copyright 2016 American Chemical Society.
The peptide A7R, which was discovered by Binétruy-Tournaire and coworkers in 2000 via phage display, has been well characterized to bind with high affinity to VEGFR2 and neuropilin-1 (NRP-1).97,98 To improve stability and efficacy of the peptide, sequence ATWLPPR, in targeting tumors, modifications were made to A7R with great success. Cao et al. conjugated cysteine to the N-terminus of A7R to form A7RC and used it to modify paclitaxel-loaded liposomes; MDA-MB-231 breast cancer xenografts in vivo showed improved accumulation of the chemotherapeutic and greater inhibitory effects compared to the unmodified liposome control.64 Ying and colleagues further improved the efficacy and stability of the peptide through cyclization of the sequence (cA7R)66 and incorporation of D-amino acids,63 respectively. Cysteine was conjugated to A7R at the N-terminus to create an amide bond, and it was shown to bind with high affinity to VEGFR2 with a KD value of 6.79 nM.66 When tested in vitro in VEGFR2-overexpressing endothelial (HUVEC) and glioma (U87) cells, fluorescein-labeled cA7R showed over 80% and 90% positive targeting and internalization, respectively. The localization results were recapitulated in vivo with U87 xenograft tumors, demonstrating improved localization of cA7R compared to linear A7R. Furthermore, cA7R-conjugated liposomes encapsulating doxorubicin showed significantly improved tumor volume reduction compared to nanoparticles with the linear peptide, doxorubicin, or doxorubicin alone. In parallel studies, DA7R-modified liposomes were loaded with doxorubicin and used to treat subcutaneous tumor models. Using the D-enantiomer demonstrated increased proteolytic stability, significantly inhibited tumor growth, and significantly greater intracellular accumulation of the chemotherapeutic compared to the L-enantiomer or liposomal control.63 Most recently, the group modified myristic acid to DA7R to improve blood–brain barrier (BBB) penetration in treating glioma; the results showed that when conjugated to liposomes, there was increased cell internalization, tumor and angiogenesis homing, and improved therapeutic outcome of the doxorubicin treatment.65
Important findings from these studies targeting VEGFR2 are the profound binding affinities seen with the sequences STP, S1, and A7R. When peptides have a KD value within the same order of magnitude as antibodies, it is reasonable to expect selective binding activity that would significantly help localize modified liposomes to specific tumor cells for anticancer drug delivery. As seen with many of these peptides, there has been a surprising increase in targeting capabilities when switching the chirality to D-enantiomers; this area of research should be further explored to elucidate possible mechanisms for this activity.
E. Integrin αvβ3 and α5β1 receptors
Integrin αvβ3 and α5β1 receptors are common targets on the cell membrane of cancer cells, due to their increased expression which contributes to cell adhesion, motility, invasion, and metastasis.99,100 The tripeptide RGD motif, arginine–glycine–aspartic acid, is perhaps the most common sequence to achieve high binding affinity and high selectivity. Many studies have been conducted, and reviews have been written on utilizing the linear RGD peptide and cyclic analogs.25,101–103 Below, we briefly describe the most recent advances made in both these categories, as well as sequences that target integrins not related to the RGD motif.
1. Linear RGD
Linear RGD has been one of the most widely studied ligand appended to liposomes for improved tumor delivery. Zuo et al. functionalized docetaxel-loaded liposomes,67 Sonali et al. docetaxel and quantum dot-loaded liposomes,68 Wen et al. shikonin-loaded liposomes,69 and Tang et al. gemcitabine-loaded liposomes.70 Other groups focus on including pH-responsive materials to increase specificity in the acidic tumor microenvironment. Zhang and colleagues co-functionalized liposomes with RGD and a pH-responsive antimicrobial peptide DH6L9 to selectively target tumor spheroids.71 Veneti et al. incorporated a pH-triggered elastin-like peptide linker (VPGVG)n between the liposome and the RGD ligand to enhance peptide-cell interactions in acidic solutions.72 Parallel research effort has inserted spacers between the peptide ligand and the liposomal surface and studied its effect on peptide-receptor affinity. While most use a conventional PEG spacer, the potential immunogenicity of PEG polymers has urged the exploration of alternative linkers. For example, Veneti et al. used (VPGVG)n that acted as a pH-responsive linker,72 while Suga et al. used the linker (SG)n to greatly improve not only targeting but also intratumoral distribution.73
2. Cyclic RGD
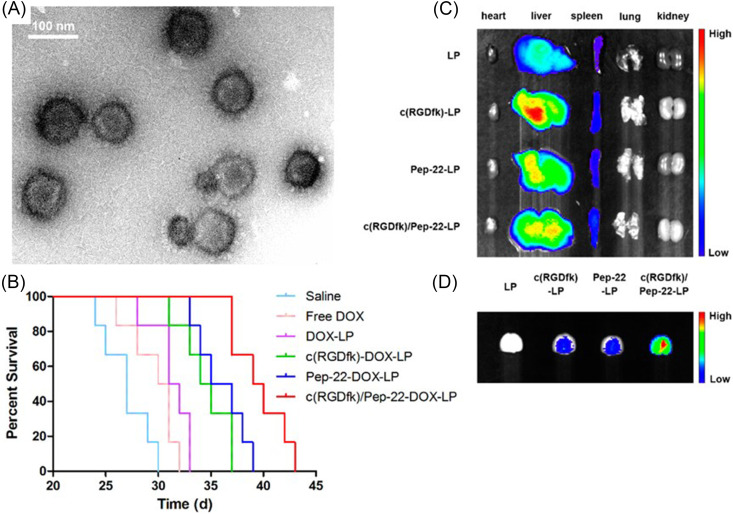
Similar to the widespread use of linear RGD peptide ligands, cyclic RGD is often used to functionalize liposomes to improve targeting efficacy and stability. Kang et al. functionalized sodium borocaptate-loaded liposomes with c(RGDyC) to improve boron neutron capture therapy.74 Notably, when compared to control liposomes, the modified nanoparticles were significantly more toxic, with significantly greater delivery efficiency toward αvβ3 expressing cells. Chen et al. demonstrated that co-functionalized doxorubicin-loaded liposomes with peptide-22 and c(RGDfK) improved tumor localization in vivo with a reduction in the IC50 in vitro when treating glioma.75 They showed that the surface co-modification was not only stable via transmission electron microscopy (TEM) [Fig. 4(a)], but also improved survival time [Fig. 4(b)], decreased liver accumulation [Fig. 4(c)], and increased tumor localization [Fig. 4(d)] in glioma-bearing mice compared to single modified liposomal controls. In a similar approach, Belhadj et al. co-modified liposomes with p-hydroxybenzoic acid (pHA) and c(RGDyK) to target both dopamine receptors and αvβ3 integrins, respectively, on the vasculature of the blood–brain barrier (BBB) and blood–brain tumor barrier (BBTB).77 In parallel studies, pHA was linked to c(RGDyK) and conjugated to doxorubicin-loaded liposomes with a PEG linker, c(RGDyK)-pHA-PEG-liposome.78 Impressively, the results showed a significant increase in the survival time of mice with glioma when treated with functionalized particles (36.5 days) compared to the surface unmodified control (26.5 days). Amin et al. modified the peptide ligand to explore a greater hydrophobicity by adding an N-methyl, c(RGDf(N-methyl)C) and found it to increase the circulation time and decrease binding to normal cells.76
FIG. 4.
(a) Transmission electron microscopy (TEM) was conducted on c(RGDfK)/Pep-22-DOX-LP, indicating the uniform size and morphology of dually modified liposomes. (b) Kaplan–Meier survival curve of glioma-bearing mice when treated with unmodified liposomes LP, liposomes functionalized with the peptide c(RGDfk)-LP or Pep-22-LP, and co-functionalized liposomes with both c(RGDfk)/Pep-22-LP showing that the combination imparts a lower intensity of localization to the liver as well as a greater intensity of localization to the tumor. Ex vivo excised (c) organs and (d) brains in mice with glioma after 24 h post-injection. Adapted with permission from Chen et al., ACS Appl. Mater. Interfaces 9(7), 5864–5873 (2017). Copyright 2017 American Chemical Society.
3. Non-RGD ligands
A few groups have also experienced success modifying liposomes with non-RGD-based ligands. Zhang and colleagues discovered a novel linear peptide RWrNK, which exhibited twofold higher binding affinity to αvβ3 than RGD and its mimetic peptides, Cilengitide, and demonstrated a higher sensitivity, specificity, and permeability of the BBB and BBTB.79 Wu et al. of the Liu group first discovered a novel peptide sequence named P1c, sequence CIRTPKISKPIKFELSG, and its ability to target αvβ3-rich tumor cells.80 The group continued to examine the sequence, and Xu et al. found that P1c and PEG co-decorated liposomes loaded with doxorubicin were able to reduce tumor angiogenesis and significantly inhibit tumor growth, while simultaneously reducing hepatotoxicity in vivo.81
F. Other receptors
Many other receptors have been studied as potential targets for peptide-ligand localization. Although less frequently studied, these receptors have demonstrated promising tumor targeting results and are briefly described below.
1. Gastrin-releasing peptide receptor (GRPR)
Gastrin-releasing peptide receptor (GRPR) is another transmembrane protein, a G protein-coupled receptor, which is highly overexpressed in many types of cancer ranging from pancreatic cancer, glioma, and lung cancer.104–106 Although there have been many discoveries of peptides to target GRPR, such as BBN7–14,105 GB-6,105 and AN-215,106 functionalizing liposomes for localized drug delivery using this receptor has been relatively underexplored. Akbar and colleagues were able to use a D-enantiomeric GRPR antagonist peptide known as cystabn with the γ-amino acid statine (Sta), sequence FQWAVGH-Sta-L-NH2, to treat small-cell lung cancer.82 When conjugated to DSPE-PEG2000 lipids and formed into liposomes, the nanoparticles demonstrated increased localization to GRPR over-expressing A549 cells in vitro.
2. Interleukin-13 receptor α2 (IL-13Rα2)
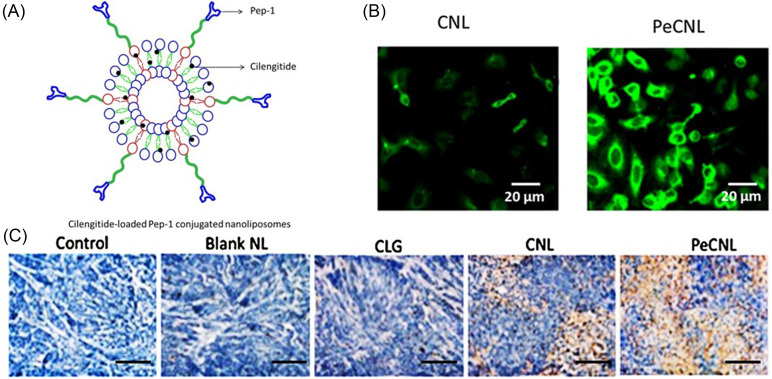
Interleukin-13 receptor α2 (IL-13Rα2) is an overexpressed plasma membrane protein commonly found in glioblastoma multiforme. Using phage display, the peptide sequence CGEMGWVRC, named Pep-1, was discovered to tightly bind to the receptor and penetrate the BBB and BBTB.107 Recognizing the applicability of Pep-1 for targeted liposomal delivery, Jiao et al. functionalized liposomes loaded with Cilengitide to improve spatial distribution.83 Remarkably, cellular uptake was improved from 47.5% to 89.8% when Pep-1 was conjugated to the liposome surface [Fig. 5(a)], and in vivo studies exhibited a significant reduction in the tumor volume by targeted formulations in U87-bearing xenograft mice. Notably, Ki67 immunohistochemical analysis of tumor sections from mice treated with Cilengitide loaded Pep-1 liposomes indicated that cell proliferation was greatly reduced compared to controls [Fig. 5(b)]. These advancements in novel peptide design and discovery as seen with Pep-1 are important in targeting IL-13Rα2 for increasing penetrability of the BBB and BBTB to lessen the difficulty of traversing these difficult barriers.
FIG. 5.
(a) Structural design of Cilengitide-loaded Pep-1-conjugated liposomes. (b) Confocal micrographs of human glioma cells treated with liposomes before (left) and after (right) Pep-1 conjugation. Coumarin-6 (green) was used as a tracking agent. Scale bar = 20 μm. (c) Ki67 immunohistochemical analysis of sectioned tumors from mice. Control-untreated; blank NL-blank liposome; CLG-free cilengitide; CNL-cilengitide loaded liposome; and PeCNL-Pep-1-conjugated cilengitide-loaded liposome. Reprinted with permission from Jiao et al., “Pep-1 peptide-functionalized liposome to enhance the anticancer efficacy of cilengitide in glioma treatment,” Colloids Surf., B 158, 68–75 (2017). Copyright 2017 Elsevier.
3. Glycoprotein 130 (gp130)
The plasma membrane signal transducing receptor glycoprotein 130 (gp130) is utilized in many cellular functions and has been shown to form a hexameric high-affinity receptor complex with interleukin-11 receptor α with higher expression in glioma cells.108,109 Using phage display, Wu et al. discovered a 12 amino acid sequence named VTW, sequence VTWTPQAWFQWV, with high binding affinity to the gp130/IL-11Rα complex.108 Suga et al. used this sequence to functionalize PEGylated liposomes with the previously described (SG)n linker to enhance targeting of glioma cells.84 Their results demonstrated a selective association of VTW-K3-(SG)5/PEGylated liposomes with U251MG glioma cells for enhanced uptake compared to other control ligand sequences.
III. FUTURE DIRECTIONS
Over the past five years, many advancements have been made regarding chemotherapy-loaded liposomes conjugated with various peptide ligands for tumor-localized therapy. Whether targeting the tumor cell membrane receptors or tumor microenvironment vasculature receptors, success has been demonstrated with increased cellular uptake, tumoral localization, and efficient drug delivery. Although the only clinically approved liposomal therapeutics are either unmodified or PEGylated therapies, numerous clinical trials in all phases are rapidly progressing toward implementing peptide-functionalized liposomes.25 Some of the highlights include multi-functional and multi-loaded liposomes for enhanced delivery and combinatorial therapeutic activity, a topic not discussed in detail in this review. These formulations assist in augmenting poor distribution and greatly reduce off-target effects; however, the ubiquitous limitations of systemic clearance by immune cells and multidrug resistance (MDR) remain. Current technologies exhibit difficulties in circumventing limitations of protease degradation, as serum proteins that bind to foreign peptides and proteins bind to and subsequently degrade the sequences conjugated to liposomal surfaces, which decrease drug delivery efficiency.18 Furthermore, the discovery of highly specific sequences is limited by large empirical screenings, which are costly and laborious, impeding the profound targeting potential and complete reduction in off-target delivery.
Further investigations in engineering highly efficient, novel, and synergistic therapies are required for the continuous improvement of targeted cancer therapies. For example, structure-vs-function studies should be conducted to elucidate the peptide secondary structure, enabling the design and optimization of sequences to increase binding affinities. Also, expanding the exploration into novel sequences using methods such as microarray chips, phage display, and machine learning will greatly advance the field of peptide targeting. As discussed throughout the review, chirality has shown importance in D-enantiomers increasing serum-stabilizing properties and should be further explored as results contraindicate the expectation for chirality to decrease binding affinity. Synergistic therapies also offer numerous advantages in decreasing drug dose requirements and improving efficacy. In the past, peptides have been shown to synergize with chemotherapies,46 which presents a largely underexplored field. Within the next few years, this effort in the field of peptide-modified liposomes will have great success in implementing these technologies in clinical settings to improve cancer patient outcome.
AUTHORS' CONTRIBUTIONS
M. R. A. conceptualized and wrote the paper; M. R. A., M. J. M., and S. H. M reviewed and edited the paper; and M. R. A. and M. J. M acquired the funding. All the authors have read and agreed to the published version of this manuscript.
ACKNOWLEDGMENTS
Funding for this work was generously provided by a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a U. S. National Institutes of Health (NIH) Director's New Innovator Award (No. DP2 TR002776), a grant from the American Cancer Society (No. 129784-IRG-16–188-38-IRG), the National Institutes of Health (Nos. NCI R01 CA241661, NCI R37 CA244911, and NIDDK R01 DK123049), and a 2018 AACR-Bayer Innovation and Discovery Grant, Grant No. 18–80-44-MITC (to M. J. M.). M. R. A. was funded by the National Science Foundation Graduate Research Fellowship (No. DGE 1845298).
The authors declare no conflict of interest.
Note: This paper is part of the special issue on Functional Biomaterials.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Allen T. M. and Cullis P. R., Adv. Drug Delivery Rev. 65, 36 (2013). 10.1016/j.addr.2012.09.037 [DOI] [PubMed] [Google Scholar]
- 2. Beltrán-Gracia E., López-Camacho A., Higuera-Ciapara I., Velázquez-Fernández J. B., and Vallejo-Cardona A. A., Cancer Nanotechnol. 10, 11 (2019). 10.1186/s12645-019-0055-y [DOI] [Google Scholar]
- 3. Accardo A., Mannucci S., Nicolato E., Vurro F., Diaferia C., Bontempi P., Marzola P., and Morelli G., Drug Delivery Transl. Res. 9, 215 (2019). 10.1007/s13346-018-00606-x [DOI] [PubMed] [Google Scholar]
- 4. Mitchell M. J., Billingsley M. M., Haley R. M., et al. “ Engineering precision nanoparticles for drug delivery,” Nat. Rev. Drug Discov. (published online, 2020). 10.1038/s41573-020-0090-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olusanya T. O. B., Ahmad R. R. H., Ibegbu D. M., Smith J. R., and Elkordy A. A., Molecules 23, 907 (2018). 10.3390/molecules23040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Minotti G., Menna P., Salvatorelli E., Cairo G., and Gianni L., Pharmacol. Rev. 56, 185 (2004). 10.1124/pr.56.2.6 [DOI] [PubMed] [Google Scholar]
- 7. Han X., Mitchell M. J., and Nie G., “ Nanomaterials for therapeutic RNA delivery,” Matter 3(6), 1948–1975 (2020). 10.1016/j.matt.2020.09.020 [DOI] [Google Scholar]
- 8. Steichen S. D., Caldorera-Moore M., and Peppas N. A., Eur. J. Pharm. Sci. 48, 416 (2013). 10.1016/j.ejps.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albini A., Pennesi G., Donatelli F., Cammarota R., Flora S. D., and Noonan D. M., J. Natl. Cancer Inst. 102, 14 (2010). 10.1093/jnci/djp440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lefrak E. A., Piťha J., Rosenheim S., and Gottlieb J. A., Cancer 32, 302 (1973). [DOI] [PubMed] [Google Scholar]
- 11. Bulbake U., Doppalapudi S., Kommineni N., and Khan W., Pharmaceutics 9, 12 (2017). 10.3390/pharmaceutics9020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raj S., Khurana S., Choudhari R., Kesari K. K., Kamal M. A., Garg N., Ruokolainen J., Das B. C., and Kumar D., “ Specific targeting cancer cells with nanoparticles and drug delivery in cancer therapy,” Semin. Cancer Biol. (published online, 2019). 10.1016/j.semcancer.2019.11.002 [DOI] [PubMed] [Google Scholar]
- 13. Krauss A. C., Gao X., Li L., Manning M. L., Patel P., Fu W., Janoria K. G., Gieser G., Bateman D. A., Przepiorka D., Shen Y. L., Shord S. S., Sheth C. M., Banerjee A., Liu J., Goldberg K. B., Farrell A. T., Blumenthal G. M., and Pazdur R., Clin. Cancer Res. 25, 2685 (2019). 10.1158/1078-0432.CCR-18-2990 [DOI] [PubMed] [Google Scholar]
- 14. Anselmo A. C. and Mitragotri S., Bioeng. Transl. Med. 1, 10 (2016). 10.1002/btm2.10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harrington K. J., Mohammadtaghi S., Uster P. S., Glass D., Peters A. M., Vile R. G., and Stewart J. S. W., Clin. Cancer Res. 7, 243 (2001). [PubMed] [Google Scholar]
- 16. Gabizon A. and Martin F., Drugs 54, 15 (1997). 10.2165/00003495-199700544-00005 [DOI] [PubMed] [Google Scholar]
- 17. Fenton O. S., Olafson K. N., Pillai P. S., Mitchell M. J., and Langer R., Adv. Mater. 30(29), 1705328 (2018). 10.1002/adma.201705328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caracciolo G., Nanomedicine 11, 543 (2015). 10.1016/j.nano.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 19. Riaz M. K., Riaz M. A., Zhang X., Lin C., Wong K. H., Chen X., Zhang G., Lu A., and Yang Z., Int. J. Mol. Sci. 19, 195 (2018). 10.3390/ijms19010195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guan J., Shen Q., Zhang Z., Jiang Z., Yang Y., Lou M., Qian J., Lu W., and Zhan C., Nat. Commun. 9, 2982 (2018). 10.1038/s41467-018-05384-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohamed M., Abu Lila A. S., Shimizu T., Alaaeldin E., Hussein A., Sarhan H. A., Szebeni J., and Ishida T., Sci. Technol. Adv. Mater. 20, 710 (2019). 10.1080/14686996.2019.1627174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsieh Y. C., Wang H. E., Lin W. W., Roffler S. R., Cheng T. C., Su Y. C., Li J. J., Chen C. C., Huang C. H., Chen B. M., Wang J. Y., Cheng T. L., and Chen F. M., Theranostics 8, 3164 (2018). 10.7150/thno.22164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Børresen B., Henriksen J. R., Clergeaud G., Jørgensen J. S., Melander F., Elema D. R., Szebeni J., Engelholm S. A., Kristensen A. T., Kjær A., Andresen T. L., and Hansen A. E., ACS Nano 12, 11386 (2018). 10.1021/acsnano.8b06266 [DOI] [PubMed] [Google Scholar]
- 24. El Sayed M. M., Takata H., Shimizu T., Kawaguchi Y., Abu Lila A. S., Elsadek N. E., Alaaeldin E., Ishima Y., Ando H., Kamal A., Sarhan H. A., and Ishida T., J. Controlled Release 323, 102 (2020). 10.1016/j.jconrel.2020.04.011 [DOI] [PubMed] [Google Scholar]
- 25. Belfiore L., Saunders D. N., Ranson M., Thurecht K. J., Storm G., and Vine K. L., J. Controlled Release 277, 1 (2018). 10.1016/j.jconrel.2018.02.040 [DOI] [PubMed] [Google Scholar]
- 26. Danhier F., J. Controlled Release 244, 108 (2016). 10.1016/j.jconrel.2016.11.015 [DOI] [PubMed] [Google Scholar]
- 27. Nel A., Ruoslahti E., and Meng H., ACS Nano 11, 9567 (2017). 10.1021/acsnano.7b07214 [DOI] [PubMed] [Google Scholar]
- 28. Fang J., Islam W., and Maeda H., Adv. Drug Delivery Rev. 244, 108–121 (2020). [DOI] [PubMed] [Google Scholar]
- 29. Park J., Choi Y., Chang H., Um W., Ryu J. H., and Kwon I. C., Theranostics 9, 8073 (2019). 10.7150/thno.37198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torchilin V. P., Nat. Rev. Drug Discovery 4, 145 (2005). 10.1038/nrd1632 [DOI] [PubMed] [Google Scholar]
- 31. Allen T. M., Brandeis E., Hansen C. B., Kao G. Y., and Zalipsky S., Biochim. Biophys. Acta-Biomembr. 1237, 99 (1995). 10.1016/0005-2736(95)00085-H [DOI] [PubMed] [Google Scholar]
- 32. Torchilin V., Expert Opin. Drug Delivery 5, 1003 (2008). 10.1517/17425247.5.9.1003 [DOI] [PubMed] [Google Scholar]
- 33. Mitchell M., Jain R., and Langer R., “ Engineering and physical sciences in oncology: Challenges and opportunities,” Nat. Rev. Cancer 17, 659–675 (2017). 10.1038/nrc.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yingchoncharoen P., Kalinowski D. S., and Richardson D. R., Pharmacol. Rev. 68, 701 (2016). 10.1124/pr.115.012070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riley R. S., June C. H., Langer R., et al. “ Delivery technologies for cancer immunotherapy,” Nat. Rev. Drug Discov. 18, 175–196 (2019). 10.1038/s41573-018-0006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moosavian S. A. and Sahebkar A., Cancer Lett. 448, 144 (2019). 10.1016/j.canlet.2019.01.045 [DOI] [PubMed] [Google Scholar]
- 37. Cao Z., Tong R., Mishra A., Xu W., Wong G. C. L., Cheng J., and Lu Y., Angew. Chem.-Int. Ed. 48, 6494 (2009). 10.1002/anie.200901452 [DOI] [PubMed] [Google Scholar]
- 38. Lopez A. and Liu J., Langmuir 34, 15000 (2018). 10.1021/acs.langmuir.8b01368 [DOI] [PubMed] [Google Scholar]
- 39. Jones M. N., Adv. Drug Delivery Rev. 13, 215 (1994). 10.1016/0169-409X(94)90013-2 [DOI] [Google Scholar]
- 40. Zhang H., Ma Y., and Sun X.-L., Med. Res. Rev. 30, 270 (2010). 10.1002/med.20171 [DOI] [PubMed] [Google Scholar]
- 41. Mitchell M. J., Castellanos C. A., and King M. R., “ Nanostructured surfaces to target and kill circulating tumor cells while repelling leukocytes,” J. Nanomater. 2012, 831263. 10.1155/2012/831263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wayne E. C., Chandrasekaran S., Mitchell M. J., Chan M. F., Lee R. E., Schaffer C. B., and King M. R., “ TRAIL-coated leukocytes that prevent the bloodborne metastasis of prostate cancer,” J. Control. Release 223, 215–223 (2016). 10.1016/j.jconrel.2015.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitchell M. J., Wayne E., Rana K., Schaffer C. B., and King M. R., “ TRAIL-coated leukocytes that kill cancer cells in the circulation,” Proc. Natl. Acad. Sci. USA 111(3), 930–935 (2014). 10.1073/pnas.1316312111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han N. K., Shin D. H., Kim J. S., Weon K. Y., Jang C. Y., and Kim J. S., Int. J. Nanomed. 11, 1413 (2016). 10.2147/IJN.S95850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aronson M. R., Simonson A. W., Orchard L. M., Llinás M., and Medina S. H., Acta Biomater. 80, 269 (2018). 10.1016/j.actbio.2018.09.025 [DOI] [PubMed] [Google Scholar]
- 46. Aronson M. R., Dahl E. S., Halle J. A., Simonson A. W., Gogal R. A., Glick A. B., Aird K. M., and Medina S. H., Cell. Mol. Bioeng. 13, 447–461 (2020). 10.1007/s12195-020-00626-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simonson A. W., Aronson M. R., and Medina S. H., Molecules 25, 2751 (2020). 10.3390/molecules25122751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Craik D. J., Fairlie D. P., Liras S., and Price D., Chem. Biol. Drug Des. 81, 136 (2013). 10.1111/cbdd.12055 [DOI] [PubMed] [Google Scholar]
- 49. Henninot A., Collins J. C., and Nuss J. M., J. Med. Chem. 61, 1382 (2018). 10.1021/acs.jmedchem.7b00318 [DOI] [PubMed] [Google Scholar]
- 50. Tang J., Wang Q., Yu Q., Qiu Y., Mei L., Wan D., Wang X., Li M., and He Q., Acta Biomater. 83, 379 (2019). 10.1016/j.actbio.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 51. Riaz M. K., Zhang X., Wong K. H., Chen H., Liu Q., Chen X., Zhang G., Lu A., and Yang Z., Int. J. Nanomed. 14, 2879 (2019). 10.2147/IJN.S192219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu W.-W., Liu D., Cao Y., and Wang X., Int. J. Nanomed. 12, 6461 (2017). 10.2147/IJN.S129946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Z., Zhao R., Wu X., Sun Y., Yao M., Li J., Xu Y., and Gu J., FASEB J. 19, 1978 (2005). 10.1096/fj.05-4058com [DOI] [PubMed] [Google Scholar]
- 54. Cheng L., Huang F. Z., Cheng L. F., Zhu Y. Q., Hu Q., Li L., Wei L., and Chen D. W., Int. J. Nanomed. 9, 921 (2014). 10.2147/IJN.S53310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ringhieri P., Mannucci S., Conti G., Nicolato E., Fracasso G., Marzola P., Morelli G., and Accardo A., Int. J. Nanomed. 12, 501 (2017). 10.2147/IJN.S113607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi M., Zhang J., Li X., Pan S., Li J., Yang C., Hu H., Qiao M., Chen D., and Zhao X., Int. J. Nanomed. 13, 4209 (2018). 10.2147/IJN.S163858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zahmatkeshan M., Gheybi F., Rezayat S. M., and Jaafari M. R., Eur. J. Pharm. Sci. 86, 125 (2016). 10.1016/j.ejps.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 58. Huang D., Zhang S., Zhong T., Ren W., Yao X., Guo Y., Duan X. C., Yin Y. F., Zhang S. S., and Zhang X., Oncotarget 7, 43616 (2016). 10.18632/oncotarget.9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jia X., Han Q., Wang Z., Qian Y., Jia Y., Wang W., and Hu Z., Biomater. Sci. 5, 417 (2017). 10.1039/C6BM00898D [DOI] [PubMed] [Google Scholar]
- 60. Qian Y., Wang W., Wang Z., Han Q., Jia X., Yang S., and Hu Z., Chem. Commun. 52, 5690 (2016). 10.1039/C6CC01302C [DOI] [PubMed] [Google Scholar]
- 61. Han Q., Wang W., Jia X., Qian Y., Li Q., Wang Z., Zhang W., Yang S., Jia Y., and Hu Z., ACS Appl. Mater. Interfaces 8, 18658 (2016). 10.1021/acsami.6b05678 [DOI] [PubMed] [Google Scholar]
- 62. Han Q., Jia X., Qian Y., Wang Z., Yang S., Jia Y., Wang W., and Hu Z., J. Mater. Chem. B 4, 7087 (2016). 10.1039/C6TB01823H [DOI] [PubMed] [Google Scholar]
- 63. Ying M., Shen Q., Liu Y., Yan Z., Wei X., Zhan C., Gao J., Xie C., Yao B., and Lu W., ACS Appl. Mater. Interfaces 8, 13232 (2016). 10.1021/acsami.6b01300 [DOI] [PubMed] [Google Scholar]
- 64. Cao J., Wang R., Gao N., Li M., Tian X., Yang W., Ruan Y., Zhou C., Wang G., Liu X., Tang S., Yu Y., Liu Y., Sun G., Peng H., and Wang Q., Biomater. Sci. 3, 1545 (2015). 10.1039/C5BM00161G [DOI] [PubMed] [Google Scholar]
- 65. Ying M., Wang S., Zhang M., Wang R., Zhu H., Ruan H., Ran D., Chai Z., Wang X., and Lu W., ACS Appl. Mater. Interfaces 10, 19473 (2018). 10.1021/acsami.8b05235 [DOI] [PubMed] [Google Scholar]
- 66. Ying M., Shen Q., Zhan C., Wei X., Gao J., Xie C., Yao B., and Lu W., J. Controlled Release 243, 86 (2016). 10.1016/j.jconrel.2016.09.035 [DOI] [PubMed] [Google Scholar]
- 67. Zuo T., Guan Y., Chang M., Zhang F., Lu S., Wei T., Shao W., and Lin G., Colloids Surf., B 147, 90 (2016). 10.1016/j.colsurfb.2016.07.056 [DOI] [PubMed] [Google Scholar]
- 68. Sonali, Singh R. P., Sharma G., Kumari L., Koch B., Singh S., Bharti S., Rajinikanth P. S., Pandey B. L., and Muthu M. S., Colloids Surf., B 147, 129 (2016). 10.1016/j.colsurfb.2016.07.058 [DOI] [PubMed] [Google Scholar]
- 69. Wen X., Li J., Cai D., Yue L., Wang Q., Zhou L., Fan L., Sun J., and Wu Y., Molecules 23, 268 (2018). 10.3390/molecules23020268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tang Z., Feng W., Yang Y., and Wang Q., Drug Des. Dev. Ther. 13, 3281 (2019). 10.2147/DDDT.S211168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang Q., Lu L., Zhang L., Shi K., Cun X., Yang Y., Liu Y., Gao H., and He Q., Sci. Rep. 6, 19800 (2016). 10.1038/srep19800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Veneti E., Tu R. S., and Auguste D. T., Bioconjug. Chem. 27, 1813 (2016). 10.1021/acs.bioconjchem.6b00205 [DOI] [PubMed] [Google Scholar]
- 73. Suga T., Kato N., Hagimori M., Fuchigami Y., Kuroda N., Kodama Y., Sasaki H., and Kawakami S., Mol. Pharm. 15, 4481 (2018). 10.1021/acs.molpharmaceut.8b00476 [DOI] [PubMed] [Google Scholar]
- 74. Kang W., Svirskis D., Sarojini V., McGregor A. L., Bevitt J., and Wu Z., Oncotarget 8, 36614 (2017). 10.18632/oncotarget.16625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen C., Duan Z., Yuan Y., Li R., Pang L., Liang J., Xu X., and Wang J., ACS Appl. Mater. Interfaces 9, 5864 (2017). 10.1021/acsami.6b15831 [DOI] [PubMed] [Google Scholar]
- 76. Amin M., Mansourian M., Koning G. A., Badiee A., Jaafari M. R., and Ten Hagen T. L. M., J. Controlled Release 220, 308 (2015). 10.1016/j.jconrel.2015.10.039 [DOI] [PubMed] [Google Scholar]
- 77. Belhadj Z., Zhan C., Ying M., Wei X., Xie C., Yan Z., and Lu W., Oncotarget 8, 66889 (2017). 10.18632/oncotarget.17976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Belhadj Z., Ying M., Cao X., Hu X., Zhan C., Wei X., Gao J., Wang X., Yan Z., and Lu W., J. Controlled Release 255, 132 (2017). 10.1016/j.jconrel.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 79. Zhang L., Shan X., Meng X., Gu T., Guo L., An X., Jiang Q., Ge H., and Ning X., Mol. Pharm. 16, 3977 (2019). 10.1021/acs.molpharmaceut.9b00602 [DOI] [PubMed] [Google Scholar]
- 80. Wu G., Wang X., Deng G., Wu L., Ju S., Teng G., Yao Y., Wang X., and Liu N., J. Magn. Reson. Imaging 34, 395 (2011). 10.1002/jmri.22620 [DOI] [PubMed] [Google Scholar]
- 81. Xu W., Yan X., Liu N., and Wu G., RSC Adv. 8, 25575 (2018). 10.1039/C8RA05014G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Akbar M. J., Ferreira P. C. L., Giorgetti M., Stokes L., and Morris C. J., Beilstein J. Nanotechnol. 10, 2553 (2019). 10.3762/bjnano.10.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jiao Z., Li Y., Pang H., Zheng Y., and Zhao Y., Colloids Surf., B 158, 68 (2017). 10.1016/j.colsurfb.2017.03.058 [DOI] [PubMed] [Google Scholar]
- 84. Suga T., Watanabe M., Sugimoto Y., Masuda T., Kuroda N., Hagimori M., and Kawakami S., J. Drug Delivery Sci. Technol. 49, 668 (2019). 10.1016/j.jddst.2018.12.037 [DOI] [Google Scholar]
- 85. Shen Y., Li X., Dong D., Zhang B., Xue Y., and Shang P., Am. J. Cancer Res. 8, 916 (2018). [PMC free article] [PubMed] [Google Scholar]
- 86. Choudhury H., Pandey M., Chin P. X., Phang Y. L., Cheah J. Y., Ooi S. C., Mak K. K., Pichika M. R., Kesharwani P., Hussain Z., and Gorain B., Drug Delivery Transl. Res. 8, 1545 (2018). 10.1007/s13346-018-0552-2 [DOI] [PubMed] [Google Scholar]
- 87. Hsu J. L. and Hung M. C., Cancer Metastasis Rev. 35, 575 (2016). 10.1007/s10555-016-9649-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang H., Berezov A., Wang Q., Zhang G., Drebin J., Murali R., and Greene M. I., J. Clin. Invest. 117, 2051 (2007). 10.1172/JCI32278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Song S., Liu D., Peng J., Sun Y., Li Z., Gu J. R., and Xu Y., Int. J. Pharm. 363, 155 (2008). 10.1016/j.ijpharm.2008.07.012 [DOI] [PubMed] [Google Scholar]
- 90. Amin S. A., Adhikari N., and Jha T., J. Med. Chem. 61, 6468 (2018). 10.1021/acs.jmedchem.7b00782 [DOI] [PubMed] [Google Scholar]
- 91. Pastorino F., Brignole C., Paolo D. D., Perri P., Curnis F., Corti A., and Ponzoni M., Small 15, e1804591 (2019). 10.1002/smll.201804591 [DOI] [PubMed] [Google Scholar]
- 92. Pasqualini R., Koivunen E., Kain R., Lahdenranta J., Sakamoto M., Stryhn A., Ashmun R. A., Shapiro L. H., Arap W., and Ruoslahti E., Cancer Res. 60, 722–727 (2000). [PMC free article] [PubMed] [Google Scholar]
- 93. Pastorino F., Brignole C., Marimpietri D., Cilli M., Gambini C., Ribatti D., Longhi R., Allen T. M., Corti A., and Ponzoni M., Cancer Res. 63, 7400 (2003). [PubMed] [Google Scholar]
- 94. Simons M., Gordon E., and Claesson-Welsh L., Nat. Rev. Mol. Cell Biol. 17, 611 (2016). 10.1038/nrm.2016.87 [DOI] [PubMed] [Google Scholar]
- 95. Lian L., Li X. L., Xu M. D., Li X. M., Wu M. Y., Zhang Y., Tao M., Li W., Shen X. M., Zhou C., and Jiang M., BMC Cancer 19, 183 (2019). 10.1186/s12885-019-5322-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. De Palma M., Biziato D., and Petrova T. V., Nat. Rev. Cancer 17, 457 (2017). 10.1038/nrc.2017.51 [DOI] [PubMed] [Google Scholar]
- 97. Binétruy-Tournaire R., Demangel C., Malavaud B., Vassy R., Rouyre S., Kraemer M., Plouët J., Derbin C., Perret G., and Mazié J. C., EMBO J. 19, 1525 (2000). 10.1093/emboj/19.7.1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Starzec A., Ladam P., Vassy R., Badache S., Bouchemal N., Navaza A., du Penhoat C. H., and Perret G. Y., Peptides 28, 2397 (2007). 10.1016/j.peptides.2007.09.013 [DOI] [PubMed] [Google Scholar]
- 99. Ruoslahti E., Annu. Rev. Cell Dev. Biol. 12, 697 (1996). 10.1146/annurev.cellbio.12.1.697 [DOI] [PubMed] [Google Scholar]
- 100. Ahmad K., Lee E. J., Shaikh S., Kumar A., Rao K. M., Park S. Y., Jin J. O., Han S. S., and Choi I., “ Targeting integrins for cancer management using nanotherapeutic approaches: Recent advances and challenges,” Semin. Cancer Biol. (published online, 2019). 10.1016/j.semcancer.2019.08.030 [DOI] [PubMed] [Google Scholar]
- 101. Kuang H., Ku S. H., and Kokkoli E., Adv. Drug Delivery Rev. 110–111, 80 (2017). 10.1016/j.addr.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 102. Alipour M., Baneshi M., Hosseinkhani S., Mahmoudi R., Jabari Arabzadeh A., Akrami M., Mehrzad J., and Bardania H., J. Biomed. Mater. Res., Part A 108, 839 (2020). 10.1002/jbm.a.36862 [DOI] [PubMed] [Google Scholar]
- 103. Asati S., Pandey V., and Soni V., Int. J. Pept. Res. Ther. 25, 49 (2019). 10.1007/s10989-018-9728-3 [DOI] [Google Scholar]
- 104. Moody T. W. and Korman L. Y., Ann. N. Y. Acad. Sci. 547, 351 (1988). 10.1111/j.1749-6632.1988.tb23902.x [DOI] [PubMed] [Google Scholar]
- 105. Tu Y., Tao J., Wang F., Liu P., Han Z., Li Z., Ma Y., and Gu Y., Biomater. Sci. 8, 2682 (2020). 10.1039/D0BM00162G [DOI] [PubMed] [Google Scholar]
- 106. Szepeshazi K., Schally A. V., Nagy A., and Halmos G., Pancreas 31, 275 (2005). 10.1097/01.mpa.0000175892.97036.a7 [DOI] [PubMed] [Google Scholar]
- 107. Pandya H., Gibo D. M., Garg S., Kridel S., and Debinski W., Neuro-Oncol. 14, 6 (2012). 10.1093/neuonc/nor141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wu C., Lo S. L., Boulaire J., Hong M. L. W., Beh H. M., Leung D. S. Y., and Wang S., J. Controlled Release 130, 140 (2008). 10.1016/j.jconrel.2008.05.015 [DOI] [PubMed] [Google Scholar]
- 109. Bravo J., EMBO J. 19, 2399 (2000). 10.1093/emboj/19.11.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.