Abstract
Acetaminophen (APAP) overdose is the leading cause of drug-induced liver injury, and its prognosis depends on the balance between hepatocyte death and regeneration. Sirtuin 6 (SIRT6) has been reported to protect against oxidative stress-associated DNA damage. But whether SIRT6 regulates APAP-induced hepatotoxicity remains unclear. In this study, the protein expression of nuclear and total SIRT6 was up-regulated in mice liver at 6 and 48 h following APAP treatment, respectively. Sirt6 knockdown in AML12 cells aggravated APAP-induced hepatocyte death and oxidative stress, inhibited cell viability and proliferation, and downregulated CCNA1, CCND1 and CKD4 protein levels. Sirt6 knockdown significantly prevented APAP-induced NRF2 activation, reduced the transcriptional activities of GSTμ and NQO1 and the mRNA levels of Nrf2, Ho-1, Gstα and Gstμ. Furthermore, SIRT6 showed potential protein interaction with NRF2 as evidenced by co-immunoprecipitation (Co-IP) assay. Additionally, the protective effect of P53 against APAP-induced hepatocytes injury was Sirt6-dependent. The Sirt6 mRNA was significantly down-regulated in P53−/− mice. P53 activated the transcriptional activity of SIRT6 and exerted interaction with SIRT6. Our results demonstrate that SIRT6 protects against APAP hepatotoxicity through alleviating oxidative stress and promoting hepatocyte proliferation, and provide new insights in the function of SIRT6 as a crucial docking molecule linking P53 and NRF2.
KEY WORDS: Acetaminophen, Hepatotoxicity, SIRT6, NRF2, P53
Abbreviations: AAV, adeno-associated virus; ALF, acute liver failure; ALT, serum alanine aminotransferase; APAP, acetaminophen; AST, aspartate aminotransferase; ARE, antioxidant response element; BCA, bicinchoninic acid; BrdU, bromodeoxyuridine; CCK-8, cell counting kit-8; CCNA1, cyclin A1; CCND1, cyclin D1; CDK4, cyclin-dependent kinase 4; Co-IP, co-immunoprecipitation; CYP450, cytochromes P450; DCF, dichlorofluorescein; Dox, doxorubicin; ECL, electrochemiluminescence; GSH, glutathione; GSTα, glutathianone S-transferase α; GSTμ, glutathione S-transferase μ; H&E, hematoxylin and eosin; H3K56ac, histone H3 Nε-acetyl-lysines 56; H3K9ac, histone H3 Nε-acetyl-lysines 9; HO-1, heme oxygenase-1; KEAP1, Kelch-like ECH-associated protein 1; LDH, lactate dehydrogenase; NAPQI, N-acetyl p-benzoquinone imine; NQO1, NAD(P)H quinone dehydrogenase 1; NRF2, nuclear factor erythroid 2-related factor 2; ROS, reactive oxygen species; siRNA, small interfering RNA; SIRT6, sirtuin 6
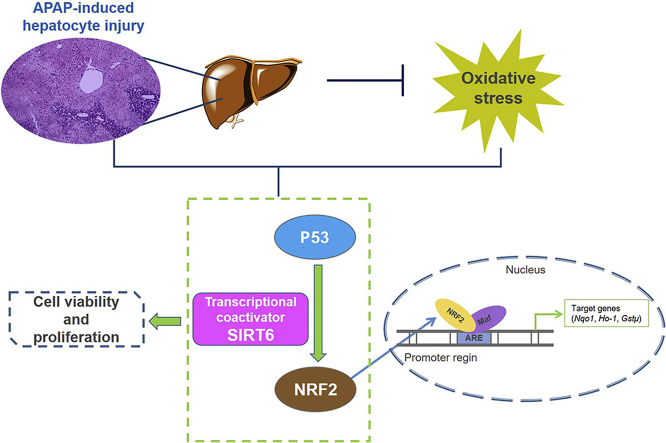
Graphical abstract
Sirtuin 6 as a transcriptional coactivator of P53 and nuclear factor erythroid 2-related factor 2 (NRF2) protects against acetaminophen-induced hepatotoxicity through alleviating oxidative stress and promoting hepatocyte proliferation.
1. Introduction
Acetaminophen (APAP) is a widely used over-the-counter drug with potent analgesic and antipyretic properties across the world1. APAP is safe and effective at recommended doses, but overdose of APAP may cause liver injury progressing to acute liver failure (ALF) and death. APAP-induced hepatotoxicity is the leading cause of ALF and a persistent important public health concern in many countries2. The formation of reactive metabolites N-acetyl-p-benzoquinone imine (NAPQI) by cytochromes P450 (CYP450) is the critical event of APAP hepatotoxicity3. Excess NAPQI after APAP overdoses depletes glutathione (GSH) and covalently binds to intracellular proteins including mitochondrial proteins to form APAP adducts, which leads to mitochondrial dysfunction, generation of reactive oxygen species (ROS), release of mitochondrial cell death factors, ultimately resulting in nuclear DNA fragmentation and hepatocyte death4.
Despite extensive studies, the mechanisms of APAP-induced liver injury and hepatocyte death are not fully elucidated. Mitochondrial oxidative stress is considered to be the predominant cellular event in APAP-induced liver injury5. Cellular stress and liver injury induced by APAP overdose are subsequently followed by compensatory liver repair and regeneration. The balance between hepatocytes death and regeneration determines the outcome of APAP hepatotoxicity6. Thus, it can be realized to treat APAP-induced liver injury through intervening oxidative stress and liver regeneration. And it is essential to find potential targets with simultaneous regulation on the above molecular events.
Sirtuins have emerged as key regulators of many important biological processes with either histone deacetylase or mono-ribosyltransferase activity. SIRT6, one of the 7 mammalian homologs of yeast Sir2, is responsible for deacetylation of histone H3 Nε-acetyl-lysines 9 (H3K9ac) and 56 (H3K56ac). SIRT6 functions on several important transcription factors and regulatory proteins, which regulate DNA repair, cell proliferation, inflammation, tumor and aging7. Recently, sirtuin 6 (SIRT6) has been reported to safeguard cells such as human mesenchymal stem cells and retinal ganglion cells from oxidative stress8, 9, 10, suggesting that SIRT6 plays a role in redox-related cellular homeostasis regulation. SIRT6 is also involved in DNA damage repair by deacetylation of H3K56 at the damaged DNA region11,12. The function of SIRT6 on cell proliferation varies in different cells. Overexpressed SIRT6 is reported to cause massive apoptosis in a variety of cancer cell lines but not in normal, non-transformed cells13. Additionally, SIRT6 deletion promotes the proliferation of hematopoietic stem cell and multiple myeloma14,15, but inhibits the proliferation and enhances the apoptosis of Huh-7 cells8.
The nuclear factor erythroid 2-related factor 2 (NRF2), one of the master regulators of the antioxidant defense system, mediates cell survival response. It regulates the expression of a battery of genes encoding intracellular detoxifying enzymes and antioxidant proteins through the antioxidant response element (ARE). In response to oxidative/electrophilic stress, NRF2 is released from Kelch-like ECH-associated protein 1 (KEAP1 ) and translocates into the nucleus, and subsequently activates ARE-responsive gene expression16. Several previous studies have demonstrated that activation of the NRF2 signaling pathway serves to protect animals against liver injury produced by various hepatotoxicants, including APAP. NRF2 is reported to alleviate oxidative stress induced by APAP, and many bioactive components activating NRF2–ARE signaling pathway exert protective effects against APAP-induced hepatotoxicity17, 18, 19. Recently, it is reported that SIRT6 activates NRF2–ARE signaling pathway against oxidative stress10,20. Nevertheless, whether SIRT6–NRF2 axis participates in APAP-induced hepatocyte toxicity has not been determined.
The P53 tumor suppressor protein plays a crucial role in modulation of cell cycle, apoptosis, senescence and metabolism in response to diverse stimulus such as cellular stress and DNA damage21. In response to moderate cellular stress and damage, P53 was activated to promote cell survival and repair of genetic damage through target genes, which mediate cell cycle arrest and facilitate DNA repair22. Our previous study found that P53 signaling pathway was associated with compensatory liver regeneration following APAP-induced hepatotoxicity23. Recently, we revealed a novel role of P53 in regulating APAP metabolism and disposition, which serves as a potential new therapeutic target for liver injury induced by APAP. P53 is reported to be closely associated with SIRT6. P53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis and gluconeogenesis by promoting FOXO1 nuclear exclusion24,25. Moreover, SIRT6 directly deacetylates P53 to regulate apoptosis and stress resistance26. However, the complicated relationship among SIRT6, P53 and NRF2 remains largely unclear and how they participate in APAP-induced hepatocyte toxicity has not been determined.
Therefore, the aim of the present study is to explore whether SIRT6 protects against APAP-induced hepatocyte injury and to determine the underlying relationships among SIRT6, P53 and NRF2 involved in this hepato-protection.
2. Materials and methods
2.1. Animal experiments
Male C57BL/6 mice (6–8 weeks old) were obtained from Laboratory Animal Center of Sun Yat-sen University (Guangzhou, China). P53 knockout and paired wild-type mice (19–20 g) were established by National Institutes for Food and Drug Control and Beijing Biocytogen Co., Ltd. (Beijing, China) and purchased from the National Center of Laboratory Rodents (Beijing, China). The mice were maintained under controlled conditions (22–24 °C, 55%–60% humidity, and 12-h light/dark cycle) and fed with standard food and water ad libitum. All experimental procedures were in accordance with the Regulations of Experimental Animal Administration issued by the Ministry of Science and Technology of the People's Republic of China (http://www.most.gov.cn). All animal protocols were approved by the Ethics Committee on the Care and Use of Laboratory Animals of Sun Yat-sen University (Guangzhou, China). The mice were acclimatized one week prior to the experiment. APAP solution was made fresh in 0.9% saline at 40 mg/mL, and C57BL/6 mice were administered a single dose of 400 mg/kg APAP by intraperitoneal injection. All C57BL/6 mice were killed at 0, 6, 12, 24, 48 and 72 h after APAP treatment. Serum samples and liver tissues were harvested. Additionally, P53+/+ and P53−/− mice (n = 5) without APAP treatment were killed, and liver tissues were obtained to investigate Sirt6 mRNA expression. A portion of the liver was immediately fixed in 10% buffered formalin for histologic analysis and the remaining tissues were flash frozen in liquid nitrogen and stored at −80 °C for further use.
2.2. Histological and biochemical assessment
Formalin-fixed liver tissues were subjected to dehydration in serial concentrations of ethanol and xylene and then embedded in paraffin. Three-micrometer thick sections were cut and stained with hematoxylin and eosin (H&E) according to a standard protocol. H&E-stained liver sections were observed using a LEICA DM5000B microscope (Leica, Heidelberg, Germany) and used for necrosis scoring.
Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activities were determined using commercially-available kits (Kefang biotech, Guangzhou, China) on a Beckman Synchron CX5 Clinical System according to the manufacturer's instructions.
2.3. Cell culture and treatment
AML12 cells, a non-tumorigenic mouse hepatocyte cell line, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DMEM/F12 containing 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 5 ng/mL selenium, 40 ng/mL dexamethasone and 10% FBS at standard cell culture conditions (5% CO2, 95% air). Negative small interfering RNA (siRNA) or siRNA targeting at Nrf2 and Sirt6 (100 nmol/L; RiboBio, Guangzhou, China) were transiently transfected using lipofectamine 2000 siRNA Transfection Reagent (Roche Diagnostics, Mannheim, Germany). Cells were treated with doxorubicin (Dox, 125–500 nmol/L) or APAP solution (5 mmol/L) at 24 h after siRNA transfection and incubated for 48 h.
Small interfering RNA (siRNA) was transfected to decrease Sirt6 level in AML12 cells. The knockdown efficiency of Sirt6 siRNA was validated by investigating protein expression of SIRT6 using Western blot analysis (Supporting Information Fig. S1). Three different siRNA sequences of Sirt6 were compared and the highest one with knockdown efficiency >80% (sequence: GTGCATGTTTCGTATAAGT) was chosen in the current study.
2.4. Cell viability assay
Cell viability assay was performed according to our previous reported procedures27. Briefly, after Sirt6 siRNA transfection and APAP treatment, AML12 cells were mixed with 10 μL of Cell Counting Kit-8 (CCK-8) solution per well and incubated for 1 h at 37 °C. The amount of formazan dye generated by cellular dehydrogenase activity was measured for absorbance at 450 nm by a microplate reader (Molecular Devices, Sunnyvale, CA, USA). The optical density value of each well represented the survival/proliferation of AML12 cells.
2.5. ROS detection
ROS detection was performed according to our previous reported procedures27. Briefly, intracellular ROS accumulation was measured with an oxidation-sensitive fluorescent probe, 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (Beyotime Biotechnology, Shanghai, China), which is oxidized to a fluorescent compound 2,7-dichlorofluorescein (DCF) in the presence of ROS. AML12 cells transfected with Sirt6 siRNA were treated with APAP solution for 24 h. The medium was removed and the cells were washed three times with PBS followed by incubation with 10 mmol/L DCFH-DA for 30 min at 37 °C. DCF intensity was detected using the Flex Station 3 (Molecular Devices) at an excitation wavelength of 488 nm and an emission wavelength of 525 nm. The protein concentration for each sample was used for normalization. Fluorescence images were photographed with ArrayScan VTI high content screening system (Thermo Fisher Scientific, Rockford, IL, USA).
2.6. Bromodeoxyuridine assay
Proliferation capacity was investigated by bromodeoxyuridine (BrdU) assay using BrdU (colorimetric) kit (Roche Applied Science, CA, USA) according to our previous reported procedures27. The cells were transfected for 48 h, labeled with BrdU for 24 h, and then fixed in situ and incubated with an anti-BrdU antibody for 1.5 h. The absorbance was detected at a wavelength of 370 nm with a reference wavelength of 492 nm.
2.7. Colony formation assay
The colony formation ability of the Sirt6 knockdown AML12 cells was assessed using a colony formation assay as described in our previous reported method28. In brief, the transfected cells growing in the logarithmic phase were trypsinized with 0.25% Trypsin-EDTA (Gibco, Carlsbad, CA, USA) and seeded into six-well plates at a density of 5000 cells/well. The cells were maintained in an incubator at 37 °C with 5% CO2 or 14 days. Then the colonies were fixed with 4% formaldehyde and stained with Diff-Quick staining kit (Solarbio Science and Technology Ltd., Beijing, China).
2.8. Co-immunoprecipitation (Co-IP) assay
AML12 cells were cultured according to the method described above and treated with or without APAP solution for 24 h. The protein was extracted following the protocol. Co-immunoprecipitation (Co-IP) was performed using Thermo Scientific Pierce Co-IP kit (Thermo Fisher Scientific, Rockford, IL, USA) according to our previous reported procedures29. Samples were analyzed by Western blotting using anti-IgG (Merk, Kenilworth, NJ, USA, AP124), anti-SIRT6 (Cell Signaling Technology, Shanghai, China, 12486), anti-P53 (Abcam, Cambridge, MA, China, ab26) and anti-NRF2 (Cell Signaling Technology, 12721).
2.9. Luciferase activity assay
The pGL3-basic vectors containing 3.0 kb upstream of the core coding region of the specified genes (120 ng/well), Luc-TK (5 ng/well), and overexpression plasmids (60 ng/well) were transfected to HEK-293T cells in 96-well plates using MegaTrans 1.0 (Origene, Rockville, MD, USA). Luciferase enzymatic activity was measured by a commercial Dual-Luciferase Reporter Assay System Kit (Promega, San Luis Obispo, CA, USA) at 24 h after transfection.
2.10. Quantitative real-time PCR analysis
Total RNA in AML12 cells was extracted using Trizol reagent (Takara, Kusatsu, Shiga, Japan) and reverse transcribed using a PrimeScript RT-PCR kit (Takara) according to the manufacturer's instructions. Real-time PCR analysis was performed using SYBR Premix Ex Taq (Takara) with a 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The ΔΔCT method was utilized to calculate the relative expression of specific genes, which was further normalized to the Gapdh mRNA levels. The sequences of gene-specific primers are listed in Table 1.
Table 1.
Sequences of primers used for real-time PCR.
| Primer | Sequence (5′–3′) |
|---|---|
| Nrf2 Forward | CTTTAGTCAGCGACAGAAGGAC |
| Nrf2 Reverse | AGGCATCTTGTTTGGGAATGTG |
| Gstα Forward | AAGCCCGTGCTTCACTACTTC |
| Gstα Reverse | GGGCACTTGGTCAAACATCAAA |
| Gstμ Forward | ATACTGGGATACTGGAACGTCC |
| Gstμ Reverse | AGTCAGGGTTGTAACAGAGCAT |
| Ho-1 Forward | CACAGCACTATGTAAAGCGTCT |
| Ho-1 Reverse | GTAGCGGGTATATGCGTGGG |
| Gapdh Forward | AGGTCGGTGTGAACGGATTTG |
| Gapdh Reverse | GGGGTCGTTGATGGCAACA |
2.11. Western blot analysis
As described in our previous study23,30, total and nuclear protein extracts from AML12 cells and mouse liver tissues were prepared and quantified by the bicinchoninic acid (BCA) protein assay kits (Thermo Fisher Scientific). Protein extract was separated in 8%–12% SDS-PAGE and electrophoretically transferred onto polyvinylidene fluoride membranes. Phosphorylated proteins were blocked with 5% bovine serum albumin in Tris-buffered saline, and other proteins with 5% nonfat milk in Tris-buffered saline. Membranes were incubated overnight with primary antibodies including anti-SIRT6 (Cell Signaling Technology, 12486), anti-NRF2 (Abcam, ab62352), anti-phosphated-NRF2 (Abcam, ab76026), anti-SIRT1 (Cell Signaling Technology, 9475), anti-SIRT2 (Sangon Biotech, Shanghai, China, D221221), anti-SIRT3 (Sangon Biotech, D221219), anti-SIRT4 (Sangon Biotech, D162208), anti-SIRT5 (Sangon Biotech, D260735), anti-CDK4 (Sangon Biotech, D120396), anti-CCNA1 (Sangon Biotech, D220575), anti-CCND1 (Sangon Biotech, D220509), anti-Lamin B (Sangon Biotech, D190800-0100), anti-GAPDH (Cell Signaling Technology, 5174) and anti-β-actin (Cell Signaling Technology, 4970), followed by secondary horseradish peroxidase conjugated anti-rabbit IgG antibody for 1 h at room temperature. Protein–antibody complexes were detected using an electrochemiluminescence (ECL) kit (Engreen Biosystem, Beijing, China) and exposed to an X-ray film (GE Healthcare, Piscataway, NJ, USA). The intensity of protein bands was analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.12. Statistical analysis
Data are expressed as means ± standard deviation (SD). To determine statistically significant difference between two groups, two-tailed unpaired Student's t-test was carried out using GraphPad Prism 7 (GraphPad Software Inc., San Diego, CA, USA). For multiple comparisons, one-way analysis of variance followed by with Bonferroni post hoc test was used. The difference was considered statistically significant at P < 0.05.
3. Results
3.1. Dynamic change of SIRT6 expression is related to APAP injury-responsive liver repair in mice
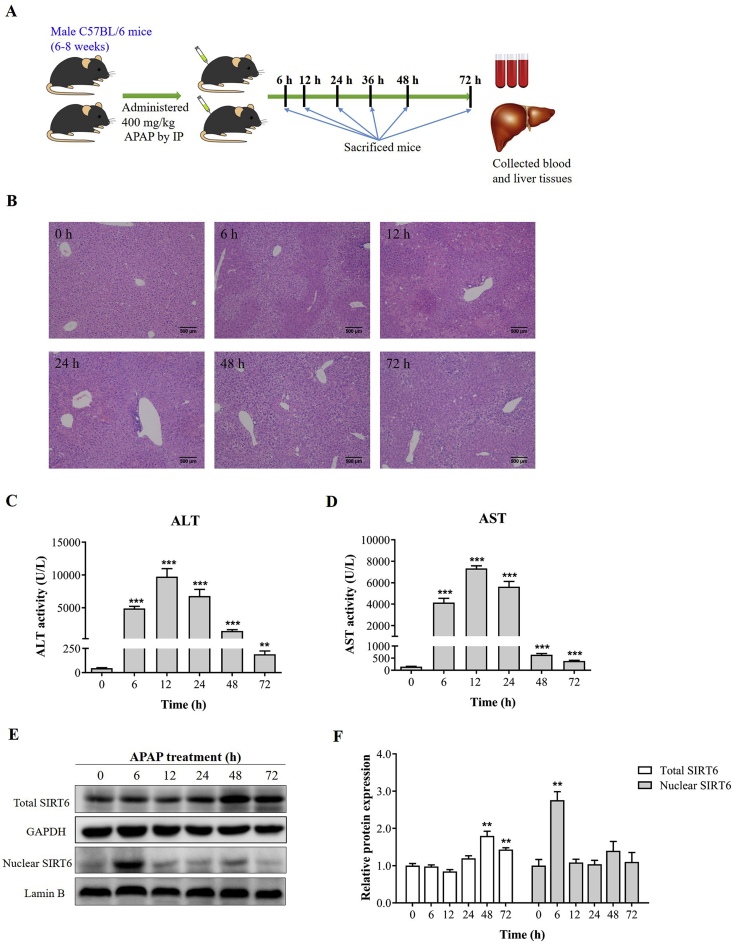
In this study, a single dose of 400 mg/kg APAP solution caused massive hepatic toxicity in mice as evidenced by H&E staining of liver sections (Fig. 1A) and elevated serum ALT and AST activities (Fig. 1B and C). Centrilobular necrosis was evident at 6 h with necrosis of 40%–50% after APAP administration and peaked between 12 and 24 h with necrosis of 60%–70%. Compared with the first 24 h, much less hepatocellular injury and necrosis was observed at 48 h, but was not yet completely resolved at 72 h with 10%–20% necrosis. The ALT and AST levels were increased in parallel to the area of hepatic necrosis. The ALT and AST activities were significantly elevated with a peak at 12 h after APAP dosing, subsequently reduced at later time points, and then tended toward baseline levels by 72 h as previously described23. These results indicated that APAP-induced liver injury was followed by compensatory liver repair in mice.
Figure 1.
Dynamic change of SIRT6 expression is related to APAP injury-responsive liver repair. (A) Male C57BL/6 mice were administered a single dose of 400 mg/kg APAP by intraperitoneal injection and killed at 0, 6, 12, 24, 48 and 72 h after APAP treatment (n = 6). (B) H&E-stained liver sections, (C) serum ALT and (D) AST activities from APAP-treated mice over a time course of 0–72 h after 400 mg/kg APAP challenge (n = 6). (E) Western blot analysis of total and nuclear SIRT6 protein expression in APAP-treated mice at various time points after APAP challenge. (F) Densitometric analysis of Western blots (n = 3−4). Data are expressed as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 versus 0 h.
The protein expression of 7 isoforms involved in sirtuin family was measured to explore whether sirtuins participated in APAP-induced acute liver injury (Supporting Information Fig. S2). The results of Western blot analysis showed that the protein expression of SIRT1, SIRT2, SIRT3, SIRT5, and SIRT6 in liver tissues was slightly downregulated after APAP administration, and SIRT4 and SIRT7 levels were significantly decreased. SIRT1 and SIRT7 levels returned to normal at 72 h, but SIRT6 protein was significantly upregulated at 48 and 72 h. The dynamic change of SIRT6 expression was most prominent than other 6 isoforms. It was further found that the SIRT6 protein levels in nucleus at 6 h following APAP administration were markedly increased compared with basal levels observed in uninjured livers, which at other time points were kept at normal levels (Fig. 1D and E). It suggested that dynamic regulation of SIRT6 was related to APAP injury initiation, progression and liver repair.
3.2. Sirt6 knockdown potentiates APAP-induced hepatocytes damage and oxidative stress injury in AML12 cells
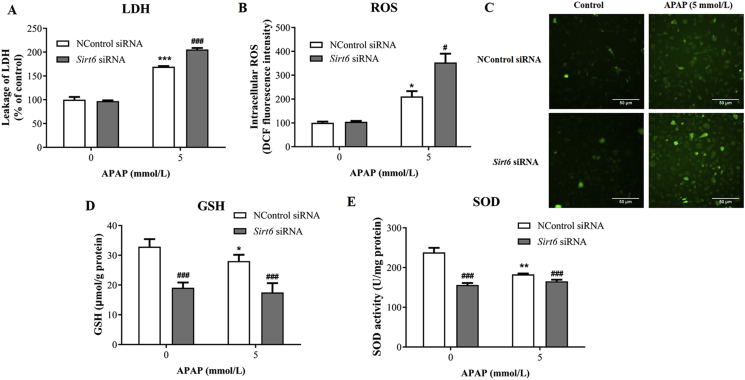
To investigate the role of SIRT6 on APAP-induced hepatocytes damage, short-interfering RNAs (siRNA) was used to knock down Sirt6 in vitro in cultured AML12 cells. As an indicator of cell death, lactate dehydrogenase (LDH) activity was measured in cell lysate and culture medium. Sirt6 knockdown significantly increased LDH activity following APAP exposure in AML12 cells compared with the control siRNA group (Fig. 2A), suggesting that Sirt6 was essential for protection against APAP-induced cytotoxicity.
Figure 2.
Effect of Sirt6 knockdown on APAP-induced hepatocytes damage and oxidative stress injury in vitro. AML12 cells were transfected with Sirt6 siRNA for 24 h and further treated with APAP for 24 h: (A) LDH levels, (B) and (C) ROS levels, (D) GSH levels, and (E) SOD levels were measured respectively. Data are expressed as the mean ± SD, n = 5. *P < 0.05, **P < 0.01, ***P < 0.001 versus the untreated group; #P < 0.05, ###P < 0.001 versus the control siRNA group.
Oxidative stress is crucial to APAP-induced hepatotoxicity. Whether Sirt6 contributed to hepatocellular antioxidant defense in response to APAP was explored using Sirt6-depleted cells. As described previously, APAP treatment caused large amount of ROS production in hepatocytes. Sirt6 knockdown further exacerbated oxidative stress triggered by APAP as evidenced by higher ROS levels (Fig. 2B and C). Additionally, APAP significantly reduced hepatocellular GSH and SOD activity in AML12 cells compared with the control group, which was further aggravated after Sirt6 knockdown (Fig. 2D and E). These results demonstrate that SIRT6 plays an important role in antioxidant stress defense following APAP exposure.
3.3. Sirt6 knockdown inhibited hepatocyte proliferation in AML12 cells exposed to APAP
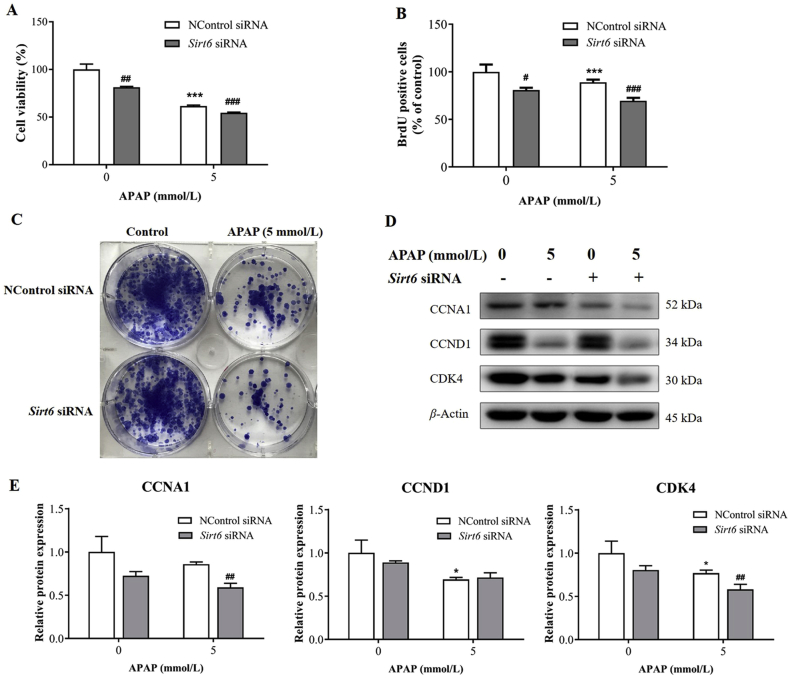
SIRT6 was reported as a novel regulator of cell proliferation. Therefore, experiments were conducted to evaluate the role of SIRT6 on cell proliferation in AML12 cells exposed to APAP. The CCK-8 assay was used to detect cell viability (Fig. 3A), and BrdU assay was carried out to directly determine proliferation capacity of cells with Sirt6 siRNA and APAP treatment (Fig. 3B). Both the viability and proliferation of AML12 cells decreased exposed to 5 mmol/L APAP, which were further exacerbated following Sirt6 siRNA treatment in the Sirt6 siRNA-treated group compared with the control siRNA group. The colony formation assay was further carried out in this study. APAP treatment significantly suppressed the colony-formation ability of AML12 cells as observed by decreased colonies formed by AML12 cells compared with the control group. Sirt6 knockdown further exacerbated the inhibition of colony-formation ability in hepatocytes after APAP exposure (Fig. 3C).
Figure 3.
Knockdown of Sirt6 inhibited hepatocellular proliferation in AML12 cells exposed to APAP. (A) Cell viability assay was determined using Cell Counting Kit-8 in AML12 cells after Sirt6 siRNA transfection and APAP treatment. (B) Proliferation capacity of AML12 cells was investigated by BrdU assay. (C) Colony formation assay of AML12 cells was stained by Diff-Quick after cultured for 7 days. (D) Western blot and (E) densitometric analysis of CCNA1, CCND1 and CDK4 proteins were measured. Data are expressed as the mean ± SD, n = 5. *P < 0.05, **P < 0.01, ***P < 0.001 versus the untreated group; #P < 0.05, ##P < 0.01, ###P < 0.001 versus the control siRNA group.
The expression of cell cycle associated proteins including CCNA1, CCND1 and CDK4 in AML12 cells after APAP treatment was investigated by Western blot analysis. APAP significantly decreased the protein levels of CCND1 and CDK4 compared with those in the control group (Fig. 3D and E). AML12 cells transfected by Sirt6 siRNA exhibited reduced CCNA1, CCND1 and CDK4 protein levels. However, Sirt6 knockdown promoted the downregulation of CCNA1 and CDK4 protein levels caused by APAP. These results demonstrate that Sirt6 knockdown inhibits hepatocyte proliferation in AML12 cells exposed to APAP.
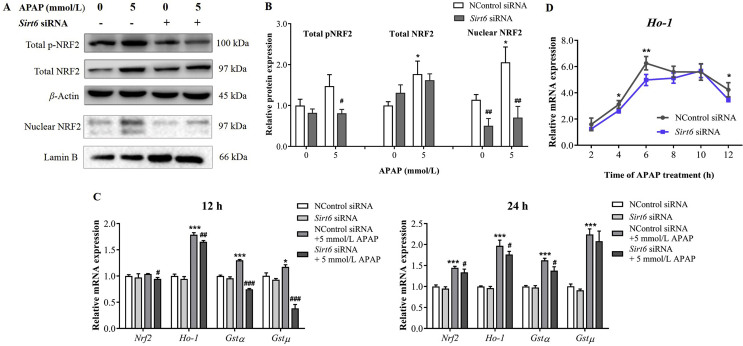
3.4. Knockdown of Sirt6 inhibited NRF2 activation and its downstream target gene expression
The expression levels of total NRF2, total phosphorylated NRF2, and nuclear NRF2 were measured in AML12 cells exposed to APAP after Sirt6 siRNA transfection. APAP exposure upregulated the levels of total and nuclear NRF2 and p-NRF2 (Fig. 4A and B). However, Sirt6 knockdown significantly inhibited the phosphorylation and nuclear translocation of NRF2 induced by APAP, but had no effect on total NRF2 protein expression compared with the control group. These results suggest that SIRT6 may regulate the activation of NRF2 and promote nuclear translocation of NRF2 to initiate transcription of Nrf2 downstream genes.
Figure 4.
Effects of knockdown of Sirt6 on the expressions of NRF2 and its downstream genes. Western blot (A) and densitometric analysis (B) of total NRF2, nuclear NRF2, and pNRF2 in AML12 cells were performed after transfected with Sirt6 siRNA for 24 h and further treated with APAP for 24 h. (C) qRT-PCR analysis was performed to measure the Nrf2, Ho-1, Gstα, Gstμ mRNA level after transfection with Sirt6 siRNA and further treated with APAP for 12 and 24 h, respectively. (D) Dynamic changes of Ho-1 mRNA expression were measured after transfection with Sirt6 siRNA and further treated with APAP. Data are expressed as the mean ± SD, n = 5. *P < 0.05, **P < 0.01, ***P < 0.001 versus the untreated group; #P < 0.05, ##P < 0.01, ###P < 0.001 versus the control siRNA group.
Further, the mRNA expression of Nrf2 and ARE-driven detoxification and antioxidant genes was investigated by qPCR analysis after Sirt6 siRNA and APAP treatment in AML12 cells. APAP treatment upregulated Nrf2, Ho-1, Gstα and Gstμ mRNA levels in a time-dependent manner (Fig. 4C). Nrf2 mRNA expression was upregulated at 24 h after APAP treatment. And Ho-1, Gstα and Gstμ mRNA levels in AML12 cells were significantly increased following APAP exposure for 12 and 24 h. The mRNA upregulation of these genes by APAP were prevented by Sirt6 knockdown. Dynamic changes of Ho-1 mRNA after APAP exposure were observed at a time course of 12 h. The Ho-1 mRNA expression was significantly upregulated with a peak at 6 h after APAP treatment, and subsequently reduced at later time points. AML12 cells transfected by Sirt6 siRNA exhibited the peak value of Ho-1 mRNA level at 10 h followed by APAP exposure, indicating that Sirt6 knockdown inhibited the NRF2 activation and thus delayed transcription of Nrf2 downstream genes.
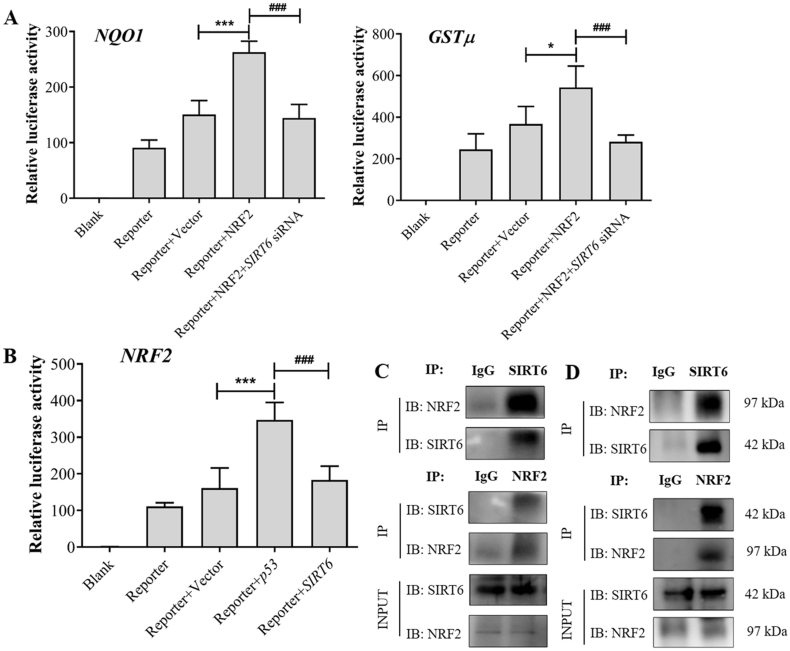
NRF2 plasmid significantly increased the luciferase reporter activity of NRF2 target genes including GSTμ and NQO1, which were prevented by SIRT6 knockdown, suggesting that SIRT6 knockdown reduced GSTμ and NQO1 transcriptional activities (Fig. 5A). A luciferase reporter assay further confirmed the regulation of NRF2 transcription by SIRT6. SIRT6 siRNA prevented the upregulation of NRF2 luciferase reporter activity by P53, which had no significant difference with the control group (Fig. 5B). Additionally, the functional interaction between SIRT6 and NRF2 was further confirmed by Co-IP assay using the endogenous proteins from AML12 cells treated with or without APAP for 24 h. Co-IP analysis showed that SIRT6 had the potential to interact with NRF2 in AML12 cells, which was not due to APAP treatment (Fig. 5C and D). These results demonstrate that SIRT6 could interact with NRF2 and regulate its downstream transcriptional activity.
Figure 5.
SIRT6 interacts with NRF2 and regulates its downstream transcriptional activity. (A) Effects of SIRT6 knockdown on NQO1 and GSTμ transcriptional activity were investigated. Data are expressed as the mean ± SD, n = 5. *P < 0.05, ***P < 0.001 versus the Reporter + Vector group; ###P < 0.001 versus the Reporter + NRF2 group. (B) Luciferase analysis was performed to detect the NRF2 transcriptional activity after transfected with pcDNA-SIRT6. Data are expressed as the mean ± SD, n = 5. ***P < 0.001 versus the Reporter + Vector group; ###P < 0.001 versus the Reporter + P53 group. (C) and (D) Co-IP analysis was performed to detect the potential interaction between SIRT6 and NRF2 in AML12 cells treated without APAP (C) or with APAP for 24 h (D).
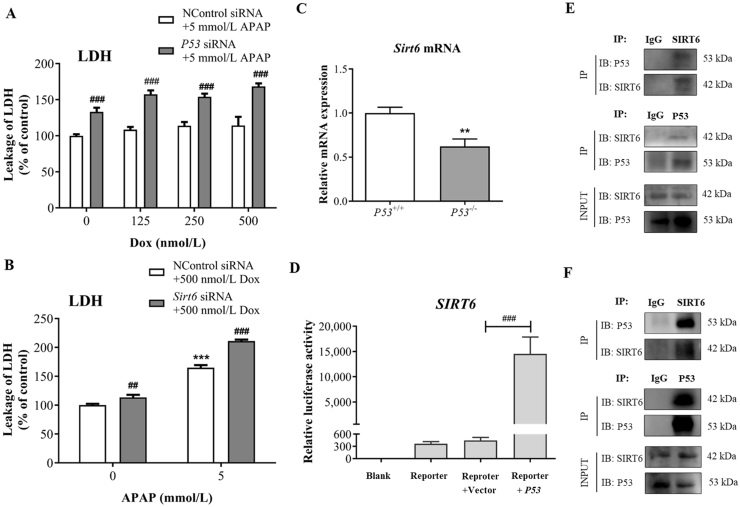
3.5. The protective effect of P53 against APAP-induced hepatocytes injury is Sirt6-dependent
As described in our previous study31, P53 activator Dox could ameliorate the intracellular ROS induced by APAP treatment. We confirmed that P53 siRNA significantly increased LDH levels in AML12 cells treated by APAP compared with the control siRNA group, which could not be reversed by Dox (Fig. 6A). Sirt6 knockdown also elevated the LDH levels before or after APAP exposure at the presence of Dox, suggesting the protective effect of P53 against APAP-induced hepatocytes injury is Sirt6-dependent (Fig. 6B). To further confirm the relationship between P53 and SIRT6, we compared the gene expression of Sirt6 between P53+/+ and P53−/− mice. Sirt6 mRNA levels of liver tissues were lower in P53−/− mice compared with that in P53+/+ mice (Fig. 6C).
Figure 6.
The protective effect of P53 against APAP-induced hepatocytes injury is Sirt6-dependent. (A) and (B) LDH levels in AML12 cells transfected with P53 siRNA or Sirt6 siRNA following APAP and Dox treatment for 24 h were detected by the lactate dehydrogenase assay. Data are expressed as the mean ± SD, n = 5. ***P < 0.001 versus the untreated group; ###P < 0.001 versus the control siRNA group. (C) qRT-PCR analysis was performed to detect Sirt6 mRNA expression in P53−/− and P53+/+ mice. Data are expressed as the mean ± SD, n = 5. **P < 0.01 versus the P53+/+ group. (D) Luciferase analysis was performed to detect the SIRT6 transcriptional activity after transfection with pcDNA-P53. Data are expressed as the mean ± SD, n = 5. ***P < 0.001 versus the Reporter + Vector group. (E) and (F) Co-IP analysis was performed to detect the potential interaction between SIRT6 and P53 in AML12 cells treated without APAP (E) or with APAP for 24 h (F).
A luciferase reporter assay was performed to further confirm the regulation of P53 in SIRT6 transcription. The results indicated that P53 plasmid significantly increased the luciferase reporter activity of SIRT6 (Fig. 6D). Additionally, the molecular link between SIRT6 and P53 was further investigated by Co-IP assay using the endogenous proteins from AML12 cells treated with or without APAP for 24 h. Co-IP analysis indicated that the potential interaction between P53 and SIRT6 existed in AML12 cells was independent of APAP treatment (Fig. 6E and F). These results demonstrate that P53 regulates SIRT6 transcriptional activity and potentially interacts with SIRT6.
4. Discussion
Sirtuins, as a family of NAD+-dependent class III histone deacetylases that catalyze post-translational modifications of proteins, are considered to be master regulators of several cellular processes, including DNA repair, cell proliferation, inflammation, tumor and aging7. We had investigated the dynamic changes of mRNA and protein expression of seven mammalian sirtuins (SIRT1–7) in APAP injured liver of mice. The change of SIRT6 expression was closely associated with the injury initiation, progression, regression and repair induced by APAP compared with other 6 sirtuins. SIRT6 functions on several important transcription factors and regulatory proteins9,10 and thus regulates a series of biological processes including DNA repair and oxidative stress, which are related with APAP hepatotoxicity. However, the effect of SIRT6 on APAP-induced liver injury remains unknown.
Several cell lines including mouse primary hepatocytes, AML12 and LO2 cells were used to screen the concentrations of APAP for further in vitro experiment according to previous literatures31, 32, 33. AML12 cells were selected considering of easy to stably establishing APAP-injured hepatocyte models and possessing good proliferation capacity. The viability of AML12 cells between 70% and 80% at 24 h after APAP treatment was used to determine the concentration of APAP, and thus 5 mmol/L APAP was selected. In this study, APAP treatment resulted in hepatocellular oxidative stress and death as observed by large amount of ROS production, reduced GSH level and SOD activity, and increased LDH level in AML12 cells. The dynamic changes of GSH, H2O2 and MDA levels in mice after APAP treatment had also been investigated in our previous study23. However, Sirt6 knockdown further aggravated oxidative damage caused by APAP with higher ROS and LDH levels, lower GSH level and SOD activity, suggesting that SIRT6 may alleviate APAP-induced oxidative injury. NRF2, as a master regulator of the antioxidant defense system, plays a protective role in APAP-induced liver injury. Whether SIRT6 regulated NRF2 to counteract oxidative stress caused by APAP was further explored in this study. We found that Sirt6 knockdown significantly prevented APAP-induced NRF2 phosphorylation and reduced NRF2 transferring to the nucleus to initiate transcription of Nrf2 downstream genes in AML12 cells. The phosphorylation of NRF2 leads to the escape or release of NRF2 from KEAP1 in response to oxidative stress34,35. NRF2 is thus stabilized and translocates to the nucleus, causing a coordinated activation of ARE-responsive gene expression34,35. Nrf2, Ho-1, Gstα and Gstμ mRNA levels in AML12 cells were significantly upregulated following APAP exposure, which were prevented by Sirt6 knockdown. Moreover, Sirt6 knockdown delayed transcription of Nrf2 downstream genes with the peak value of Ho-1 mRNA level at 10 h. We further verified that SIRT6 knockdown prevented the upregulation of NRF2 luciferase reporter activity by P53, and reduced GSTμ and NQO1 transcriptional activities. Additionally, SIRT6 potentially interacted with NRF2 in AML12 cells. These results indicated that knockdown of SIRT6 inhibited NRF2 activation and its downstream target gene expression.
Hepatocyte proliferation is crucial to liver repair and determines the final outcome of APAP-induced liver injury. SIRT6 has been confirmed as a key factor to regulate cell proliferation in AML12 cells exposed to APAP. Sirt6 deficiency inhibited cell viability and proliferation, and exacerbated the inhibition of colony-formation ability in AML12 cells after APAP exposure, indicating that Sirt6 knockdown inhibited hepatocyte proliferation. Additionally, the expression of cell cycle associated proteins including CCNA1, CCND1 and CDK4 was further decreased by Sirt6 knockdown following APAP treatment.
The protective effect of P53 on APAP-induced liver toxicity was verified in our previous study31. Some studies show that P53 is closely associated with SIRT6 as evidenced by that P53 not only cooperated with SIRT6 by promoting FOXO1 nuclear exclusion24,25, but also could be directly deacetylated by SIRT6. Whether the protective effect of P53 was related to SIRT6 remained unknown and was further investigated in this study. The results indicated that Sirt6 knockdown prevented P53 to alleviate hepatocyte damage and oxidative stress induced by APAP, which could not be reversed by P53 activator Dox. The Sirt6 mRNA was significantly downregulated in P53−/− mice compared with that in P53+/+ mice. Furthermore, P53 could activate the transcriptional activity of SIRT6 and interact with SIRT6 in AML12 cells. These results suggest the protective effect of P53 against APAP-induced hepatocytes injury is Sirt6-dependent. However, the roles of SIRT6 in P53–SIRT6–NRF2 axis during APAP-induced liver injury should be further verified in vivo using Sirt6−/− mice or Adeno-associated virus (AAV) Sirt6 shRNA-treated mice in the future.
In our previous study, dynamic and coordinated regulation of NRF2–ARE and P53/P21 signaling pathways was confirmed to be associated with compensatory liver regeneration following APAP-induced liver injury in mice.17 Additionally, P53 was found to promote NRF2 activation and the transcription of its downstream target genes related to APAP detoxification31. In summary, this study demonstrates that SIRT6 protects against APAP hepatotoxicity through alleviating oxidative stress and promoting hepatocyte proliferation. SIRT6 may be a crucial molecular event linking P53 and NRF2 in APAP hepatotoxicity.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grants 81603185, 81973392, and 82025034), the Natural Science Foundation of Guangdong Province (Grant 2020A1515011452, China), the National Key Research and Development Project (Grant 2017YFE0109900, China), and the Sanming Project of Medicine in Shenzhen (SZSM201406007 and SZSM201606088, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.06.016.
Author contributions
Research design: Xiaomei Fan, Min Huang, Huichang Bi. Experiments conducting: Yanying Zhou, Tingying Jiao, Wenzhou Li, Panpan Chen, Yixin Chen, Yiming Jiang. Data analysis: Yanying Zhou, Pan Chen, Jiahong Sun, Lihuan Guan, Yajie Wen. Manuscript writing and revising: Xiaomei Fan, Huichang Bi.
Conflicts of interest
The authors declare that they have no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Budnitz D.S., Lovegrove M.C., Crosby A.E. Emergency department visits for overdoses of acetaminophen-containing products. Am J Prev Med. 2011;40:585–592. doi: 10.1016/j.amepre.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 2.Larson A.M., Polson J., Fontana R.J., Davern T.J., Lalani E., Hynan L.S. Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology. 2005;42:1364–1372. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 3.Kwon D., Kim S.M., Correia M.A. Cytochrome P450 endoplasmic reticulum-associated degradation (ERAD): therapeutic and pathophysiological implications. Acta Pharm Sin B. 2020;10:42–60. doi: 10.1016/j.apsb.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramachandran A., Jaeschke H. Mechanisms of acetaminophen hepatotoxicity and their translation to the human pathophysiology. J Clin Transl Res. 2017;3:157–169. doi: 10.18053/jctres.03.2017S1.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du K., Ramachandran A., Jaeschke H. Oxidative stress during acetaminophen hepatotoxicity: sources, pathophysiological role and therapeutic potential. Redox Biol. 2016;10:148–156. doi: 10.1016/j.redox.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhushan B., Walesky C., Manley M., Gallagher T., Borude P., Edwards G. Pro-regenerative signaling after acetaminophen-induced acute liver injury in mice identified using a novel incremental dose model. Am J Pathol. 2014;184:3013–3025. doi: 10.1016/j.ajpath.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beauharnois J.M., Bolivar B.E., Welch J.T. Sirtuin 6: a review of biological effects and potential therapeutic properties. Mol Biosyst. 2013;9:1789–1806. doi: 10.1039/c3mb00001j. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C., Yu Y., Huang Q., Tang K. SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Mol Med Rep. 2019;20:1575–1582. doi: 10.3892/mmr.2019.10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan H., Guan D., Liu X., Li J., Wang L., Wu J. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 2016;26:190–205. doi: 10.1038/cr.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michishita E., Mccord R.A., Boxer L.D., Barber M.F., Hong T., Gozani O. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toiber D., Erdel F., Bouazoune K., Silberman D.M., Zhong L., Mulligan P. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Meter M., Mao Z., Gorbunova V., Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011;10:3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Diao D., Shi Z., Zhu X., Gao Y., Gao S. SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell. 2016;18:495–507. doi: 10.1016/j.stem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Cea M., Cagnetta A., Adamia S., Acharya C., Tai Y.T., Fulciniti M. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood. 2016;127:1138–1150. doi: 10.1182/blood-2015-06-649970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi M., Yamamoto M. Molecular mechanisms activating the Nrf2–Keap1 pathway of antioxidant gene regulation. Antioxid Redox Sign. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 17.Fan X., Jiang Y., Wang Y., Tan H., Zeng H., Wang Y. Wuzhi tablet (Schisandra Sphenanthera extract) protects against acetaminophen-induced hepatotoxicity by inhibition of CYP-mediated bioactivation and regulation of NRF2–ARE and p53/p21 pathways. Drug Metab Dispos. 2014;42:1982–1990. doi: 10.1124/dmd.114.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Zhang S., Cheng H., Lv H., Cheng G., Ci X. Nrf2-mediated liver protection by esculentoside A against acetaminophen toxicity through the AMPK/Akt/GSK3β pathway. Free Radic Biol Med. 2016;101:401–412. doi: 10.1016/j.freeradbiomed.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y.M., Wang Y., Tan H.S., Yu T., Fan X.M., Chen P. Schisandrol B protects against acetaminophen-induced acute hepatotoxicity in mice via activation of the NRF2/ARE signaling pathway. Acta Pharmacol Sin. 2016;37:382–389. doi: 10.1038/aps.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J., Sun W., Song Y., Liu J., Xue F., Gong K. SIRT6 protects retinal ganglion cells against hydrogen peroxide-induced apoptosis and oxidative stress by promoting Nrf2/ARE signaling via inhibition of Bach1. Chem Biol Interact. 2019;300:151–158. doi: 10.1016/j.cbi.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Braeuning A., Schwarz M. Regulation of expression of drug-metabolizing enzymes by oncogenic signaling pathways in liver tumors: a review. Acta Pharm Sin B. 2020;10:113–122. doi: 10.1016/j.apsb.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan X., Chen P., Tan H., Zeng H., Jiang Y., Wang Y. Dynamic and coordinated regulation of KEAP1-NRF2-ARE and p53/p21 signaling pathways is associated with acetaminophen injury responsive liver regeneration. Drug Metab Dispos. 2014;42:1532–1539. doi: 10.1124/dmd.114.059394. [DOI] [PubMed] [Google Scholar]
- 24.Li M., Hou T., Gao T., Lu X., Yang Q., Zhu Q. p53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis. Cell Death Dis. 2018;9:941. doi: 10.1038/s41419-018-0984-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P., Tu B., Wang H., Cao Z., Tang M., Zhang C. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc Natl Acad Sci U S A. 2014;111:10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood M., Rymarchyk S., Zheng S., Cen Y. Trichostatin A inhibits deacetylation of histone H3 and p53 by SIRT6. Arch Biochem Biophys. 2018;638:8–17. doi: 10.1016/j.abb.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan L., Chen Y., Wang Y., Zhang H., Fan S., Gao Y. Effects of carnitine palmitoyltransferases on cancer cellular senescence. J Cell Physiol. 2019;234:1707–1719. doi: 10.1002/jcp.27042. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Wang Y., Huang Y., Zeng H., Hu B., Guan L. PPARα regulates tumor cell proliferation and senescence via a novel target gene carnitine palmitoyltransferase 1C. Carcinogenesis. 2017;38:474–483. doi: 10.1093/carcin/bgx023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y., Feng D., Ma X., Fan S., Gao Y., Fu K. Pregnane X receptor regulates liver size and liver cell fate by Yes-associated protein activation in mice. Hepatology. 2019;69:343–358. doi: 10.1002/hep.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng X., Li X., Xu C., Jiang F., Mo Y., Fan X. Schisandra sphenanthera extract (Wuzhi Tablet) protects against chronic-binge and acute alcohol-induced liver injury by regulating the NRF2–ARE pathway in mice. Acta Pharm Sin B. 2017;7:583–592. doi: 10.1016/j.apsb.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J., Wen Y., Zhou Y., Jiang Y., Chen Y., Zhang H. p53 attenuates acetaminophen-induced hepatotoxicity by regulating drug-metabolizing enzymes and transporter expression. Cell Death Dis. 2018;9:536. doi: 10.1038/s41419-018-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuta K., Yoshida Y., Ogura S., Kurahashi T., Kizu T., Maeda S. Gab1 adaptor protein acts as a gatekeeper to balance hepatocyte death and proliferation during acetaminophen-induced liver injury in mice. Hepatology. 2016;63:1340–1355. doi: 10.1002/hep.28410. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed M.M., Wang T., Luo Y., Ye S., Wu Q., Guo Z. Aldo-keto reductase-7A protects liver cells and tissues from acetaminophen-induced oxidative stress and hepatotoxicity. Hepatology. 2011;54:1322–1332. doi: 10.1002/hep.24493. [DOI] [PubMed] [Google Scholar]
- 34.Niture S.K., Khatri R., Jaiswal A.K. Regulation of Nrf2—an update. Free Radic Biol Med. 2014;66:36–44. doi: 10.1016/j.freeradbiomed.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H.C., Nguyen T., Pickett C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.