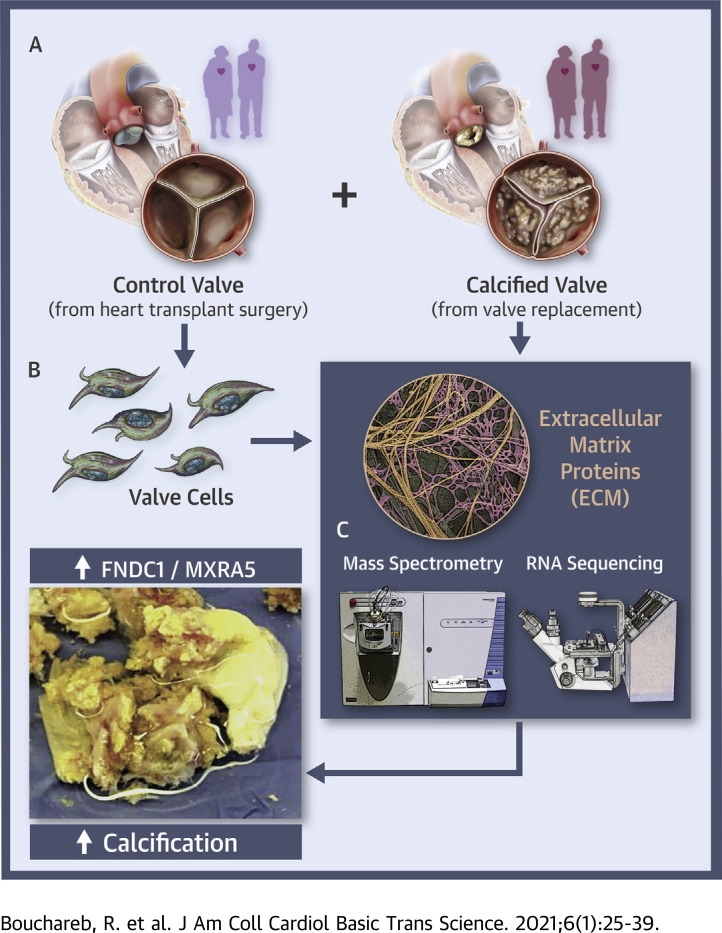
Visual Abstract
Key Words: aortic stenosis, calcified aortic valves, ECM, metabolism, proteomics, RNA-Seq
Abbreviations and Acronyms: AS, aortic stenosis; EC, endothelial cell; ECM, extracellular matrix; FN, fibronectin; FNDC1, fibronectin type III domain containing 1; hVIC, human valve interstitial cell; KEGG, Kyoto Encyclopedia of Genes and Genomes; LDL, low-density lipoprotein; MetS, metabolic syndrome; MXRA5, matrix-remodeling-associated protein 5; PBS, phosphate-buffered saline; RNAseq, RNA sequencing; TAVc, calcified tricuspid aortic valve; TAVn, noncalcified tricuspid aortic valve; VAHC, calcified human aortic valve; VAHN, normal human aortic valve
Highlights
-
•
ECM proteins play an important role in maintaining the structural architecture and the mechanical behavior of the aortic valve.
-
•
Network analysis highlights a strong connection between metabolic markers and ECM proteins.
-
•
MXRA5 and FNDC1 were identified as new biomarkers of aortic stenosis in 2 independent cohorts
Summary
This study analyzed the expression of extracellular matrix (ECM) proteins during aortic valve calcification with mass spectrometry, and further validated in an independent human cohort using RNAseq data. The study reveals that valve calcification is associated with significant disruption in ECM and metabolic pathways, and highlights a strong connection between metabolic markers and ECM remodeling. It also identifies FNDC1 and MXRA5 as novel ECM biomarkers in calcified valves, electing them as potential targets in the development and progression of aortic stenosis.
The prevalence of aortic stenosis (AS) increases with age, ranging from 3% to 5% in the population over 65 years of age (1,2). Age, sex, smoking, hypertension, obesity, type 2 diabetes, and metabolic syndrome (MetS) have been linked to valvular calcification (3). Currently, surgical valve, balloon valvuloplasty, and transcatheter aortic valve replacement and implantation are the standard treatments for AS (4,5). There are no proven pharmacological treatments to reverse or stop the progression of AS.
In this study, we explored the prospective association of extracellular matrix (ECM) protein alterations with changes in metabolic proteins during aortic valve calcification. There are currently no studies that have documented such a crosstalk. Indeed, MetS is a powerful and independent predictor of rapid AS progression (6) and affects the ECM structure in adipose tissue (7).
The ASTRONOMER (Aortic Stenosis Progression Observation Measuring Effects of Rosuvastatin) study has identified MetS as an independent predictor of faster AS progression (8). Furthermore, a recent study has shown that MetS may play a role during the early stages of AS development (9). Accumulation of ECM proteins is 1 hallmark of metabolic dysfunction in adipose tissue (10). In addition, MetS is known to affect the ECM structure of aortic valves (7). MetS may affect aortic valve ECM through the overaccumulation of advanced glycation end-products (11). Indeed, advanced glycation end-product accumulation within the ECM has been shown to induce collagen cross-linking in animals (12). In addition, a pathological expansion of the ECM with macrophage recruitment and increased protein expression has been observed in adipose tissue of obese patients (13). An expansion of the ECM of aortic valves is requisite to provide enough space for hypertrophic adipocytes (13). In parallel, valvular ECM remodeling may disturb the regulation of cell metabolism and lipid synthesis (14).
To date, however, no studies have documented the crosstalk between the ECM of aortic valves and the possible involvement of metabolic proteins in aortic valve calcification. Proteomic approaches have previously been used to investigate proteins that are involved in the pathogenesis of AS (15,16). Indeed, ECM protein expression alteration has been identified in calcified valvular tissue (17). However, the current study explored for the first time to our knowledge the modulation of ECM proteins during aortic valve calcification and their potential to interact with proteins involved in MetS. We present ECM proteins identified using proteomics performed on human aortic valves and isolated human interstitial cells during the cell calcification process. We further have validated our proteomics findings with RNA sequencing (RNAseq) of valve tissue in an independent cohort of patients with AS. The study shows that the disruption of ECM proteins in aortic valves is more pronounced in the calcified stage and is associated with down-regulation of metabolic enzymes. We also have identified matrix-remodeling-associated protein 5 (MXRA5) and fibronectin type III domain containing 1 (FNDC1) as new ECM biomarkers of calcified AS in different human cohorts. Further studies are required to validate the translational use of these 2 markers to assess the severity of AS in the clinic.
Methods
An expanded Methods section is provided in the Supplemental Appendix.
Protein extraction
Calcified human aortic valves (VAHC) were obtained from aortic valve replacement surgeries at Mount Sinai Hospital, New York, New York. Control aortic valves were obtained from heart transplantation surgeries. To ensure consistency between samples, all patient extractions were performed at the same time. The study was approved by the institutional review board of the Icahn School of Medicine at Mount Sinai, and all subjects gave written informed consent before surgery. All cases in this study were seen and assessed in the Mount Sinai surgery department. Inclusion criteria for entry into the study were a minimum age of 18 years, consenting to participate, and fluency in English. All samples were washed with cold phosphate-buffered saline (PBS), collected, and then kept on ice. All tissues were sectioned according to their Warren score (18). ECM proteins were extracted from score 1 (fibrotic) and score 4 (highly calcified) aortic valve tissues (Figure 1). All tissues were frozen at −80°C.
Figure 1.
Illustration of ECM Pipeline Extraction From Human Aortic Valve Tissue
(A) Macroscopic analysis of calcified valve tissue. (B) Dissection of fibrotic stage tissue. (C) Dissection of calcified stage tissue. (D) Diagram to compare proteome between noncalcified, low-calcified, and highly calcified tissue from the same aortic valve. (E) ECM extraction process. (F) Alizarin Red staining of noncalcified valve versus (G) staining of calcified valve with calcium nodes highlighted with yellow rectangles the corresponding dispersive x-ray energy graphs showing the calcium peak (circled) are below each section. Proteins were isolated from the calcified (yellow rectangle) and noncalcified (green) stages of the same valve tissue. Masson trichrome staining of (H) a calcified valve and (I) a control noncalcified valve. The high-calcium samples were taken from the area indicated by the black highlighted box (I) and the low-calcium samples from the area indicated by the red highlighted box. ECM = extracellular matrix; LC-MS/MS = liquid chromatography tandem mass spectrometry.
Protein extraction was performed from 35 mg of tissue and incubated with 175 μl of guanidine buffer (4 mol/l GuHCl, 50 mmol/l Na-acetate, 25 mmol/l EDTA, protease inhibitors, and phosphatase inhibitors, pH = 5.8). Samples were then vortexed at 1,500 rpm for 48 h at room temperature, and protein concentration was measured using bicinchoninic acid protein reagents.
Exclusion criteria
The control valves were obtained from individuals undergoing heart transplantation without evidence of aortic valve disease and with normal aortic valve function determined by echocardiography. Calcified aortic valves were collected from aortic valve replacement surgeries. Tissues were collected consecutively; only tricuspid aortic valves were collected. Patients with endocarditis, congenital heart disease, or rheumatic AS were excluded.
Cell isolation
Noncalcified human aortic valves (VAHN) were collected from 2 different patients undergoing heart transplantation surgery. Valve leaflets were macroscopically and histologically normal, without any signs of calcification. Valves were collected in cold sterile PBS and kept on ice until the start of the digestion. Samples were first washed with sterile 1× PBS, scraped to remove the endothelial cells, then washed 3 times in cold 1× PBS. The tissues were incubated in 1 mg/ml collagenase I for 30 min at 37°C, washed in PBS, segmented into small pieces, and then reincubated in 4.5 mg/ml collagenase I for an additional 30 min at 37°C. Dulbecco's Modified Eagle Medium with 10% fetal bovine serum was added to stop the enzymatic activity of the collagenase. Cells were then centrifuged at 150 ×g for 5 min, washed twice in Dulbecco's Modified Eagle Medium–10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, Massachusetts), 100 U/ml penicillin (Thermo Fisher Scientific), and 100 μg/ml streptomycin (Thermo Fisher Scientific), and plated onto 6-well plates (19). Cells were serum-starved for 24 h before protein collection for mass spectrometry processing. The clinical characteristics of patients used to isolate aortic valve cells are described in Supplemental Table 1.
Statistical methods
Proteome and secretome analysis
The proteome and secretome analyses were performed using Perseus v.1.6.6.0 software (20). Before uploading the data to Perseus, the proteins were filtered based on a 1% false discovery rate. Only proteins assigned to a given protein group (Master protein) with a minimum of 2 unique proteins were included for the Perseus analysis. This filter was performed using R v.3.6.2 through R Studio v.1.2.5019 software (R Foundation for Statistical Computing, Vienna, Austria). Contaminated proteins were filtered out. The data were log2 transformed and normalized using z-score normalization. Proteins present in at least 1 of the groups analyzed were kept for statistical testing. Student's t-test was applied to compare each pair of groups. Proteins with a false discovery rate <0.05 between 2 groups were considered significant. The Venn diagrams were plotted using the web-based tool InteractiVenn (21). The plots were created using R v.3.6.2 software, and publication quality images were generated with Inkscape software (Free Software Foundation, Boston, Massachusetts) when possible.
Statistical analyses for in vitro studies
For the aforementioned in vitro studies, statistical analyses were performed with the Prism software package (GraphPad Version 7, GraphPad Software, San Diego, California). Differences between 2 means were assessed by a 2-tailed paired or unpaired Student's t-test. Values of p < 0.05 were considered significant. For evaluating morphological, histological, and survival data, analysis of variance followed by Bonferroni correction was used to compare 3 groups. When sample number is not suffice to test for normality, nonparametric tests (Mann-Whitney U test or 1-way analysis of variance followed by Holm-Sidak’s test analysis) were used.
Results
Clinical characteristic of human samples
A total of 5 cohorts were used. Proteomic analysis was performed on 3 different cohorts of both males and females: 1) calcified explanted aortic valve (n = 7); versus 2) age-matched control noncalcified valves (n = 8); and 3) cellular and secreted proteins from isolated human aortic valve cells (n = 2). The results were validated with RNAseq from an independent human cohort (n = 18). The ECM proteome and secretome were also explored during human valve cell calcification in vitro. Human valve cells were isolated from control valves of the patients described in Table 1. The clinical characteristics of patients with AS (VAHC; n=7) and control aortic valves (human noncalcified aortic valve [VAHN]; n = 8) are shown in Table 1. The mean ages were 66 ± 4.3 years and 57 ± 6.0 years for VAHC and VAHN, respectively. In addition, 57% of patients in the VAHC group were male versus 75% in the VAHN group. The average peak gradient was 44.5 ± 13 mm Hg for the VAHC group. Both groups had similar lipid profiles (Table 1). The demographic characteristics of the RNAseq external validation cohort have previously been published (22).
Table 1.
Clinical Characteristics of the Proteomics Human Cohorts
| VAHN (n = 8) | VAHC (n = 7) | p Value | |
|---|---|---|---|
| Male | 66 | 57 | 0.24 |
| Age, yrs | 57 ± 6 | 66 ± 4.3 | 0.29 |
| Weight, kg | 168.5 ± 9.4 | 165.11 ± 19.3 | 0.85 |
| Smoking | 46 | 54 | 0.65 |
| Metabolic markers | |||
| Cholesterol, mg/dl | 158 ± 32 | 161.5 ± 28 | 0.60 |
| HDL, mg/dl | 94.5 ± 14 | 43.7 ± 4.4 | 0.07 |
| LDL, mg/dl | 74.5 ± 49 | 84.8 ± 27 | 0.56 |
| Echocardiography | |||
| Peak gradient, mm Hg | ND | 44.5 ± 13 | ND |
| Mean gradient, mm Hg | ND | 29.5 ± 18 | ND |
| Peak aortic jet velocity, m/s | 1.3 ± 0.4 | 3.7 ± 0.5 | <0.001 |
| Ejection fraction, % | 51 ± 6 | 51 ± 13 | 0.50 |
Values are n, mean ± SD, or %.
HDL = high-density lipoprotein; LDL = low-density lipoprotein; ND = not determined; VAHC = calcified aortic valve; VAHN = normal aortic valve.
Identification of ECM proteins in calcified valve tissue
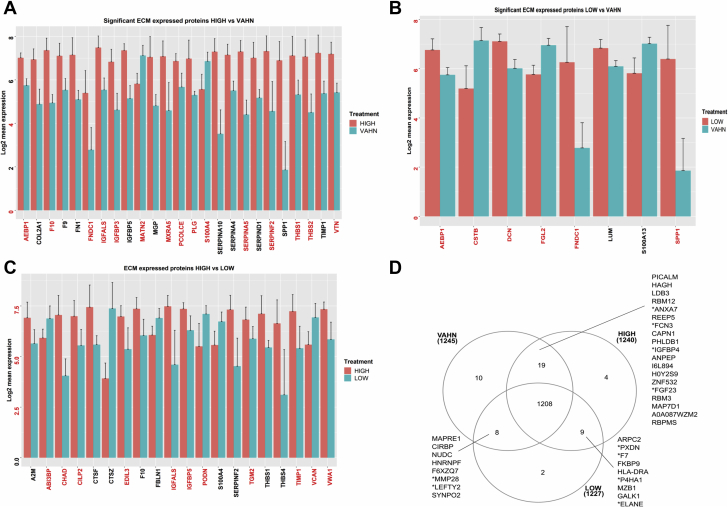
AS is a disease characterized by fibrosis and calcification of the aortic valve. ECM proteins are the major components of the aortic valve. In order to better understand AS transition from the normal to symptomatic phase, we compared the expression of ECM proteins of human valve tissues from calcified stages (high calcium) (Figures 1A, 1C, and 1D) and fibrotic stages (low calcium) (Figures 1A, 1B, and 1D) of the same calcified aortic valve and nondiseased valve tissue (control). Tissue was sectioned according to Warren score (18). The high calcified sections (scores 3 to 4) were identified by the presence of calcium nodes and dystrophic fibrosa as shown in Figures 1H and 1I. However, those with low calcium were more fibrotic with no apparent calcium nodes as shown with Alizarin Red staining (Figures 1F and 1G). Calcification was confirmed with scanning electron microscopy coupled energy dispersive x-ray analysis (Figures 1F and 1G). The energy dispersive x-ray analysis showed a peak of calcium in calcified valve tissue. Proteins were extracted in guanidine buffer (Figure 1E), which was very efficient in extracting and preserving ECM proteins. Next, 1,027 proteins with valid values in at least 70% of the samples were used to perform the comparison between the groups. ECM proteins were identified from the total number of proteins using the Matrisome Project software. Interestingly, ECM proteins represented 75% of the top 20 significantly expressed proteins. The ECM proteins matrilin2 (MATN2) and S100 calcium binding protein A4 (S100A4) were decreased in calcified VAHC tissue compared with control VAHN valves (Figure 2A), whereas AEBP1, F1, FNDC1, IGFALS, IGFB3, MXRA5, PCOLCE, PLG, SERPINA5, SRPINF2, THSB1, THSB2, and VTN were overexpressed in VAHC valves compared with VAHN control valves (Figure 2A).
Figure 2.
Representation of the Top 20 Significant Proteins in the 3 Disease Stages
Core matrisome proteins are highlighted in red. (A) Significant ECM proteins between highly calcified (HIGH) and control valve (VAHN) tissues. (B) Significant ECM proteins between low-calcified (LOW) and control valve tissues. (C) Significant ECM proteins between high- and low-calcified tissues from the same valve are labeled with asterisks. (D) Venn diagram showing the comparison of the proteomes in the 3 different stages. ECM = extracellular matrix; VAHN = normal human aortic valve.
ECM protein expression in fibrotic valve tissue
When we compared fibrotic valve tissues to control valve tissues, we observed that there were fewer ECM proteins present in the top 20 of the most significantly expressed proteins. Adipocyte enhancer-binding protein 1 (AEBP1) was highly expressed in low calcified/fibrotic tissue compared with control valves (Figure 2B). FNDC1 and osteopontin (SPP1), 2 ECM products that play a role in bone matrix calcification, were highly expressed in low calcified tissue compared with control valves (Figure 2B). Decorin, a proteoglycan that preferentially retains low-density lipoprotein (LDL) (23), was highly increased in the fibrotic tissue of calcified valves (Figures 2B and 3B).
Figure 3.
Heat Map Representation of ECM Proteins in the 3 Different Stages
(A) Comparing highly calcified tissue (VAHC-HIGH) to control valve (VAHN) tissue. (B) Low calcified tissue (VAHC-LOW) compared with VAHN. (C) Low calcified stage compared with high calcified stage tissue in the same valve. Abbreviations as in Figure 2.
We then mapped ECM proteins that were overexpressed in calcified versus fibrotic tissues from the same aortic valve. Our results showed heterogeneity of protein expression within the same valve tissue. The proteomics analysis identified cartilage matrix chondroadherin proteoglycan (CHAD), CLIP2, EDIL3, IGFALS, IGFBP5, TGM2, TMP1, and VWFA1 as overexpressed in the calcified tissue compared with the fibrotic tissue (Figures 2C, 2D, and 3). Figure 2D lists examples of differentially expressed proteins that are commonly intersected in VAHN and low calcium, VAHC and low calcium, and VAHC and VAHN.
We further determined commonly expressed ECM proteins in isolated human valve interstitial cells (hVICs) and calcified tissue (VAHC). Of the top 30 proteins expressed, we identified 13 ECM proteins, from which 5 proteins (F10, F2, VTN, SERPIND1, and FN1) were dually expressed in both hVICs and VAHC (Table 2). Additionally, SERPINS proteins and coagulation factors were the dominant proteins.
Table 2.
Top 30 Proteins Expressed in Valve Tissue and in Cells
| Accession | Gene Symbol | Log2 Fold Change High VAHN |
q Value High VAHN | Mean |
ECM | Proteome Mean |
Secretome Mean |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VAHN | Low | High | 0 Days | 3 Days | 7 Days | 0 Days | 3 Days | 7 Days | |||||
| P00742 | F10 | 2.422 | 0 | 4.941 | 6.046 | 7.362 | Matrisome-associated | ND | ND | ND | 7.882 | 4.327 | 4.382 |
| P24593 | IGFBP5 | 2.210 | 0 | 5.148 | 6.299 | 7.358 | Core matrisome | ND | ND | ND | ND | ND | ND |
| P00734 | F2 | 1.972 | 0 | 5.133 | 5.955 | 7.106 | Matrisome-associated | 6.892 | 6.717 | 5.974 | 7.042 | 5.534 | 6.670 |
| P05154 | SERPINA5 | 2.893 | 0 | 4.407 | 6.359 | 7.301 | Matrisome-associated | ND | ND | ND | ND | ND | ND |
| Q8IUX7 | AEBP1 | 1.264 | 0 | 5.750 | 6.763 | 7.015 | Core matrisome | ND | ND | ND | ND | ND | ND |
| Q13228 | SELENBP1 | −2.250 | 0 | 7.461 | 5.617 | 5.211 | 6.920 | 6.331 | 6.488 | 4.620 | 5.668 | 7.573 | |
| P00352 | ALDH1A1 | −2.667 | 0.002 | 7.374 | 5.008 | 4.706 | 7.221 | 6.153 | 5.971 | ND | ND | ND | |
| P00325 | ADH1B | −1.976 | 0.002 | 7.364 | 5.661 | 5.387 | ND | ND | ND | ND | ND | ND | |
| P12277 | CKB | −2.699 | 0.002 | 7.494 | 5.230 | 4.794 | 6.214 | 6.593 | 6.678 | ND | ND | ND | |
| G3V2W1 | SERPINA10 | 3.783 | 0.002 | 3.515 | 5.913 | 7.299 | Matrisome-associated | ND | ND | ND | ND | ND | ND |
| A0A087WWT3 | A0A087WWT3 | 4.592 | 0.002 | 3.655 | 5.971 | 8.247 | ND | ND | ND | ND | ND | ND | |
| P18669 | PGAM1;LOC643576 | −1.975 | 0.002 | 7.347 | 6.026 | 5.371 | 6.806 | 6.506 | 6.541 | 6.420 | 6.063 | 7.168 | |
| P10451 | SPP1 | 5.037 | 0.002 | 1.859 | 6.393 | 6.897 | Core matrisome | ND | ND | ND | ND | ND | ND |
| A0A0G2JPR0 | LOC100294156 | 2.541 | 0.002 | 4.885 | 5.272 | 7.426 | ND | ND | ND | ND | ND | ND | |
| P07358 | C8B | 1.148 | 0.002 | 6.013 | 6.156 | 7.162 | ND | ND | ND | ND | ND | ND | |
| Q00688 | FKBP3 | 3.991 | 0.002 | 3.628 | 6.070 | 7.620 | 6.710 | 6.352 | 6.693 | ND | ND | ND | |
| P02765 | P02765 | 2.492 | 0.003 | 4.641 | 6.252 | 7.133 | 6.357 | 6.559 | 6.907 | 3.991 | 3.495 | 6.530 | |
| P04004 | VTN | 1.769 | 0.004 | 5.423 | 6.335 | 7.192 | Core matrisome | ND | ND | ND | 7.439 | 6.038 | 5.649 |
| P40925 | MDH1 | −2.051 | 0.004 | 7.425 | 5.264 | 5.373 | 6.646 | 6.629 | 6.654 | 6.650 | 5.971 | 7.070 | |
| P35858 | IGFALS | 1.945 | 0.004 | 5.542 | 4.602 | 7.488 | Core matrisome | ND | ND | ND | ND | ND | ND |
| P30086 | PEBP1 | −1.795 | 0.004 | 7.239 | 6.197 | 5.444 | 6.851 | 6.363 | 6.665 | 6.074 | 6.030 | 7.255 | |
| Q15847 | ADIRF;C10orf116 | −1.578 | 0.004 | 7.209 | 6.229 | 5.630 | ND | ND | ND | ND | ND | ND | |
| P29622 | SERPINA4 | 1.642 | 0.005 | 5.507 | 5.955 | 7.149 | Matrisome-associated | ND | ND | ND | ND | ND | ND |
| P68371 | TUBB4B | −0.882 | 0.006 | 7.025 | 6.460 | 6.143 | 6.694 | 6.589 | 6.645 | 6.941 | 5.155 | 6.965 | |
| Q9ULC3 | RAB23 | 3.020 | 0.006 | 4.742 | 5.442 | 7.763 | 6.873 | 6.461 | 6.493 | ND | ND | ND | |
| P02458 | COL2A1 | 2.052 | 0.007 | 4.887 | 6.535 | 6.940 | Core matrisome | ND | ND | ND | ND | ND | ND |
| P06727 | APOA4 | 1.945 | 0.007 | 5.131 | 5.940 | 7.076 | ND | ND | ND | ND | ND | ND | |
| P05546 | SERPIND1 | 1.836 | 0.009 | 5.174 | 5.776 | 7.011 | Matrisome-associated | ND | ND | ND | 7.505 | 5.551 | 5.334 |
| P02751 | FN1 | 2.048 | 0.008 | 5.099 | 6.213 | 7.148 | Core matrisome | 6.904 | 6.566 | 6.409 | 6.279 | 6.412 | 6.986 |
| P07437 | TUBB | −1.662 | 0.009 | 7.224 | 6.238 | 5.561 | 6.807 | 6.484 | 6.620 | 7.006 | 6.033 | 6.657 | |
Abbreviations as in Table 1.
Protein networks in calcified and fibrotic tissues
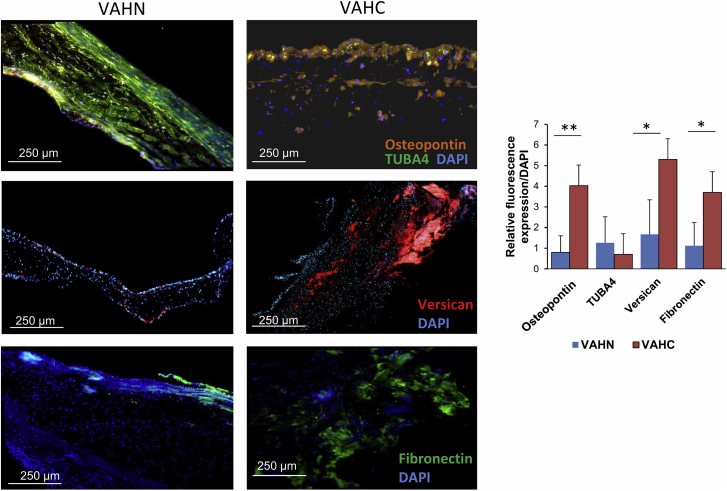
In order to better understand the involvement of ECM proteins in the aortic valve calcification process, we established a network analysis of the significant proteins. Figure 4 highlights the core matrisome proteins and the matrisome-associated proteins. We observed that the network is more complex in the calcific stage compared with the fibrotic phase. We also confirmed the overexpression of pro-calcifying ECM proteins, such as collagen (COL2A1), fibronectin 1 (FN1), and SPP1 in calcified tissue (Figure 4A). The overexpression of FN1 and SPP1 was confirmed using immunofluorescence in calcified valve tissue compared with control tissue (Figure 5). Both ECM proteins primarily interact with the complement cascade proteins (C4B, C5, and C8B) and thrombospondin 1 (THSB1), an adhesive glycoprotein that plays a role in platelet activation. Moreover, we observed a down-regulation of proteins that are involved in metabolic pathways such as aldehyde dehydrogenase 1 family member A1 (ALDH1), α-enolase 1 (ENO1), and oxidative stress protein superoxide dismutase 1 (SOD1) in the VAHC group (Figure 4B).
Figure 4.
Network Analysis Surrounding ECM Proteins
(A) Network of significant proteins when comparing high calcified to control valve tissues (VAHN). (B) The network between low-calcified tissue and control valves. Orange circle outline = core matrisome protein, yellow circle outline = matrisome-associated protein, green dot = up-regulated, red dot = down-regulated. Abbreviations as in Figure 2.
Figure 5.
IF Staining of Selected Proteins
Immunofluorescence (IF) staining (left) and quantification (right) show the up-regulation of osteopontin, versican, and fibronectin, and down-regulation of TUBA4 in calcified (VAHC) versus noncalcified (VAHN) valves (∗p < 0.05, ∗∗p < 0.01).
Pathway identification in different disease stages
In an effort to identify the pathways that are involved in the aortic valve calcification process, we found an overabundance of differentially regulated proteins for pathways within KEGG (Kyoto Encyclopedia of Genes and Genomes) in control valves compared with calcific and fibrotic stages (Table 3). The KEGG analysis showed that the metabolic pathways are the most overrepresented pathways, in particular the PI3K-Akt signaling pathway (Table 3), consistent with our earlier observations showing an overall down-regulation in metabolic proteins (Figure 4). Other pathways that are also significantly represented are the ECM–receptor interaction and complement/coagulation cascades, and the platelet activation pathway (Table 3). These findings suggest that metabolic changes are potentially linked to ECM alterations leading to platelet activation. This is conceivably true because platelets play a role in the valve calcification process (24).
Table 3.
Representation of KEGG Pathways in 3 Different Stages of the Disease
| KEGG Pathway | VAHN.FDR | HIGH.FDR | LOW.FDR |
|---|---|---|---|
| Glycolysis/gluconeogenesis | 2.96 × 10−9 | 2.66 × 10−9 | 2.19 × 10−9 |
| Citrate cycle (TCA cycle) | 8.49 × 10−5 | 7.98 × 10−5 | 7.40 × 10−5 |
| Fatty acid degradation | 0.0003 | 0.001 | 0.0009 |
| Oxidative phosphorylation | 0.0002 | 0.0002 | 0.0002 |
| Valine, leucine, and isoleucine degradation | 0.0002 | 0.0005 | 5.00E-04 |
| Arginine and proline metabolism | 0.0182 | 0.0062 | 0.0059 |
| Tryptophan metabolism | 0.0071 | 0.0068 | 0.0065 |
| beta-Alanine metabolism | 0.0064 | 0.02 | 0.0194 |
| Glutathione metabolism | 0.0002 | 0.0007 | 0.0007 |
| Pyruvate metabolism | 2.18 × 10−6 | 2.03 × 10−6 | 7.86 × 10−6 |
| Glyoxylate and dicarboxylate metabolism | 0.0137 | 0.0133 | 0.0126 |
| Propanoate metabolism | 0.0019 | 0.007 | 0.0067 |
| Sulfur metabolism | 0.0291 | 0.0285 | 0.0275 |
| Metabolic pathways | 0.0001 | 7.61 × 10−5 | 7.40 × 10−5 |
| Carbon metabolism | 3.14 × 10−14 | 9.58 × 10−14 | 7.74 × 10−14 |
| Biosynthesis of amino acids | 5.76 × 10−6 | 5.31 × 10−6 | 4.90 × 10−6 |
| HIF-1 signaling pathway | 0.0028 | 0.0027 | 0.0025 |
| Endocytosis | 1.58 × 10−5 | 6.24 × 10−6 | 5.44 × 10−6 |
| Phagosome | 1.60 × 10−8 | 1.24 × 10−8 | 9.78 × 10−9 |
| PI3K-Akt signaling pathway | 2.27 × 10−5 | 1.01 × 10−5 | 3.27 × 10−5 |
| Necroptosis | 0.031 | 0.0294 | NA |
| Cardiac muscle contraction | 0.0152 | 0.0142 | 0.0134 |
| Focal adhesion | 1.03 × 10−13 | 2.86 × 10−14 | 7.74 × 10−14 |
| ECM-receptor interaction | 1.65 × 10−10 | 2.90 × 10−11 | 1.16 × 10−10 |
| Complement and coagulation cascades | 3.00 × 10−21 | 5.18 × 10−23 | 2.48 × 10−22 |
| Platelet activation | 0.0093 | 0.0038 | 0.0079 |
| Antigen processing and presentation | 0.0002 | 0.0002 | 0.0002 |
| Fc gamma R-mediated phagocytosis | 0.0011 | 0.0004 | 0.0003 |
| Leukocyte transendothelial migration | 0.0405 | 0.0387 | 0.0364 |
| Regulation of actin cytoskeleton | 7.25 × 10−7 | 2.36 × 10−7 | 4.62 × 10−7 |
| Vasopressin-regulated water reabsorption | 0.012 | 0.0116 | 0.0108 |
| Cholesterol metabolism | 1.51 × 10−5 | 1.21 × 10−5 | 1.19 × 10−5 |
ECM = extracellular matrix; FDR = false discovery rate; HIGH = high calcification; KEGG = Kyoto Encyclopedia of Genes and Genomes; LOW = low calcification; TCA = tricarboxylic acid; VAHN = normal human aortic valve.
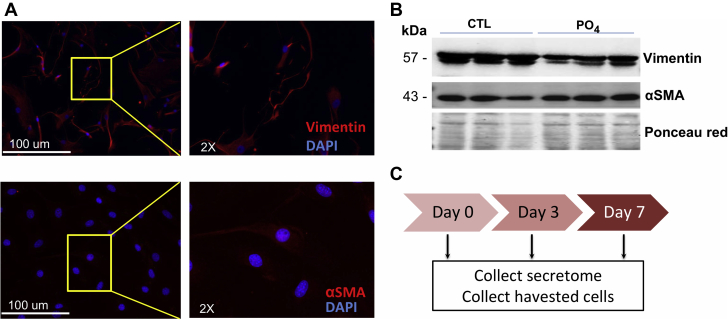
Expression of ECM proteins during valve cells calcification in vitro
To identify which ECM proteins define the osteoblastic transition, we explored the expression of ECM proteins during the calcification process in vitro. hVICs were isolated from noncalcified valves and maintained until passage 7. Cells were positive for vimentin and α-SMA (Figures 6A and 6B). hVICs were treated with the calcifying medium for 7 days, and the secretome was collected at 3 different time points: day 0, day 3, and day 7 (Figure 6C). We then validated the cell calcification efficacy with Alizarin Red staining, which shows the formation of calcium nodes (Figures 7A and 7B), and with scanning electron microscopy with x-ray analysis, which confirms the hydroxyapatite nature of the observed calcium nodes in phosphate-treated cells (Figures 7C and 7D). Importantly, the calcium concentration was 4-fold more pronounced in the phosphate-treated cells compared with the nontreated cells (Figures 7C and 7D). Our in vitro results showed the efficiency of the calcifying medium treatment in inducing calcium nodes that are similar to their pathological formation in AS patients.
Figure 6.
Characterization of Human Aortic Valve Cells
(A) Isolated human valve cells are positive for vimentin and αSMA as shown with both immunofluorescence and Western blotting. Boxed areas (left) are shown at higher magnification on the right. (B) Treatment of cells with phosphate-based calcifying medium enhanced the expression of α-SMA. (C) Diagram showing the process followed to screen extracellular matrix expression during valve calcification. CTL = control.
Figure 7.
Human Valve Cell Calcification In Vitro
(A) Human valve interstitial cells (hVICs) were treated with phosphate-based calcifying medium (CM) for 7 days. Calcium nodes were stained with Alizarin Red and observed with a polarized light microscope. The dashed-line boxes indicate the regions shown at higher magnification in the solid-line boxes. (B) The quantification of calcium was performed with Arsenazo III comparing control (CTL) with cells treated with CM. Scanning electron microscopy analyses shows (C) noncalcified cells and (D) calcified nodes in phosphate-treated cells. Boxed areas (left) are shown at higher magnification on the right. Calcium nodes are highlighted with an artificial green color. Calcium nodes were revealed with energy dispersive x-ray to show their hydroxyapatite nature. Calcium nodes were revealed with energy dispersive x-ray, in black eclipse, to show their hydroxyapatite nature (C, D). ∗∗∗p < 0.001.
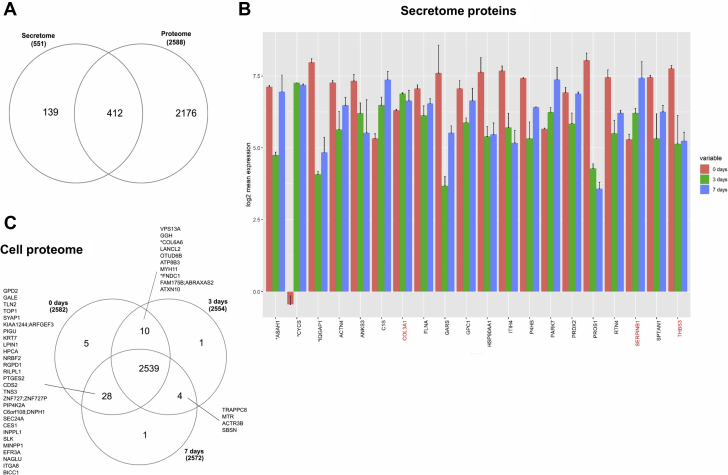
We then analyzed the expression changes of secreted and cellular proteins during cell calcification. The Venn diagram shows the expression of 139 secretome-specific and 2,176 proteome-specific proteins (Figure 8A) with 412 intersected proteins between secreted and cellular proteins after 7 days of calcification. The analyses of the top 20 secreted proteins showed the presence of 3 ECM proteins, COL3A1, THBS3, and neutrophil serine protease inhibitor, a protein that plays an essential role in the regulation of the activity of inflammatory caspases (25) (Figure 8B). However, no ECM proteins were expressed in the top 20 significant cellular proteins. The Venn diagram of cellular proteins showed more proteins (i.e., 28 proteins) in the intersection between the baseline (day 0) and calcified cells on the seventh day of calcification (Figure 8C). COL6A6 and FNDC1 were the 2 ECM proteins expressed at the early calcification stage between the baseline and the third day (Figure 8C).
Figure 8.
ECM Expression During Valve Cell Calcification In Vitro
(A) Venn diagram showing cellular and secreted proteins during calcification. (B) Top 20 secreted proteins expressed, with comparison of their expression at 0, 3, and 7 days of calcifying medium treatment. ECM proteins are labeled in red. (C) Venn diagram showing cellular protein expression at 0, 3, and 7 days. ECM = extracellular matrix.
Validation of proteomics data with transcriptomics profiling
In order to validate our proteomics data, we used an independent Canadian cohort of an RNA sequencing dataset (Table 4). RNAseq was performed on 9 calcified valves (TAVc) versus 8 control valves (TAVn). In the RNAseq experiment, 499 genes were differentially expressed, with 329 up-regulated and 170 down-regulated genes in TAVc compared with TAVn (26). Interestingly, 8 proteins from our proteomics analysis were significantly altered in the RNAseq gene dataset of TAVc versus TAVn (Table 4), 6 of which were ECM proteins (SPP1, VTN, THBS2, MATN2, FNDC1, and MXRA5). These findings provide strong support to our hypothesis that ECM alterations are of paramount importance in the process of valve cell calcification. The enrichment in the fibrotic proteins (SPP1, THBS2, and FNDC1) supports the concept that ECM proteins are driving the fibrocalcific nature of AS. The up-regulation of MXRA5 is of particular interest because it may be involved in anti-fibrotic remodeling during the progression of AS, which will be explored in future investigations.
Table 4.
The Significant Proteins (N = 8) in the Comparison HIGH Versus VAHN That Were Validated at the Mrna Level in the Rnaseq Analysis
| Accession | Gene Name | RNAseq Fold Change TAVc – TAVn |
Log2 Fold Change HIGH_VAHN |
q Value HIGH_VAHN | Proteome |
Secretome |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 Days | 3 Days | 7 Days | 0 Days | 3 Days | 7 Days | |||||
| P00352 | ALDH1A1 | −2.90 | −2.66 | 0.000 | 7.22 | 6.15 | 5.97 | ND | ND | ND |
| P10451 | SPP1∗ | 15.39 | 5.037 | 0.000 | ND | ND | ND | ND | ND | ND |
| P04004 | VTN∗ | −2.63 | 1.76 | 0.000 | ND | ND | ND | 7.44 | 6.04 | 5.65 |
| P35442 | THBS2∗ | 2.91 | 2.56 | 0.010 | 6.67 | 5.74 | 7.12 | 8.01 | 3.99 | 4.20 |
| P08571 | CD14 | 1.87 | 1.58 | 0.020 | ND | ND | ND | ND | ND | ND |
| O00339 | MATN2∗ | −1.95 | −1.30 | 0.020 | ND | ND | ND | ND | ND | ND |
| Q4ZHG4 | FNDC1∗ | 3.69 | 2.61 | 0.030 | ND | ND | ND | ND | ND | ND |
| Q9NR99 | MXRA5∗ | 2.73 | 2.49 | 0.040 | ND | ND | ND | ND | ND | ND |
Discussion
In this study, we analyzed the expression of ECM proteins in human calcified aortic valves and during aortic valve calcification with mass spectrometry. The proteomics results were further validated in an independent human cohort with calcified valves using RNAseq datasets. Our study focused on both valve cell–secreted ECM proteins and valve tissue–expressed/isolated proteins. Our current findings are particularly distinguished from prior studies (15, 16, 17) because of the method of ECM isolation, the number and diversity of the ECM proteins identified, and the complex networking between proteomics and RNA sequencing we used. Importantly, the proteomics data in this work were validated in a completely independent human cohort from Canada and reveal FNDC1 and MXRA5 as new ECM biomarkers for calcified aortic valves. Thus, the study documents that valve calcification is associated with significant disruption in ECM proteins. This is of particular interest because ECM organization and VIC compartmentalization are hallmarks of normal valve development (27).
The aortic valve has a dynamic structure to regulate the unidirectional blood flux from the left ventricle to the rest of the body. The structural flexibility of the valve is achieved by 3 organized ECM layers arranged according to the blood flow direction. As such, the ECM proteins are exposed to an incredible amount of pressure and mechanical stress (19,28). A modification in the hemodynamics surrounding the valve may affect ECM organization and disrupt its normal function (28). It has previously been shown that in pathological aortic valve, gene expression level varies according to the state of mineralization (29). So, in order to understand the transition from a normal to a symptomatic stage, we compared the expression of ECM proteins from both the calcified and fibrotic stages of the same aortic valve, and nondiseased valve tissues. However, we recognize that the interpatient variability is more important than intrapatient (same valve), especially when the cohort size is small. We have confirmed the presence of the known aortic valve ECM proteins such as collagen, elastin, fibronectin, and proteoglycans (15). In addition, we have observed an increase in the expression of FNDC1 in both low and high calcified valves. FNDC1 contains the main functional components of a fibronectin (FN) domain called the type III conserved FN domain, which led to the hypothesis that FNDC1 may possess a similar function to that of FN (30). FN itself is required by osteoblasts to form mineralized nodules in vitro (31). The overexpression of FNDC1 in aortic valve tissue has been reported by others (32). However, the implication of FNDC1 in the calcification process of the aortic valve is still unknown. In parallel, ABI3BP, the paralog of FNDC1, was significantly increased in both disease stages. The gene expression level of FNDC1 is higher in osteoporosis (30), suggesting that FNDC1 may be implicated in the paradoxical mineralization in females where bone resorption and valve calcification are predominant (33). Chondrocyte and ECM calcification proteins were expressed in both calcified and fibrotic stages. CHAD, a cartilage matrix marker, was highly expressed in the calcified stage relative to the fibrotic stage in the same tissue. SPP1 is involved in the attachment of osteoclasts to the mineralized bone matrix. The validation of these markers by our proteomics data clearly demonstrates the accuracy of our data analysis.
Furthermore, we studied the modulation of the ECM during valve calcification in vitro using an efficient phosphate-based calcifying medium previously validated by our group (24). Secreted proteins were collected at 3 different time points: baseline (day 0), 3 and 7 days of calcification in vitro. Similar to the tissue analysis, FNDC1 expression was particularly altered and up-regulated after 3 days of calcifying medium treatment. The implication of FNDC1 in cell calcification has never been studied before, to our knowledge.
We have validated our proteomics data with RNA sequencing of valve tissue samples from an independent human cohort. Our analyses confirmed the expression of FNDC1, SPP1, VTN, THBS2, MATN2, and MXRA5 in both the proteomics and the RNAseq datasets. Only MATN2 expression was down-regulated. Matrilins are multidomain extracellular adaptor proteins with 2 von Willebrand factor A-like domains (34). MATN2 forms filaments by interacting with itself and with other ECM proteins to perform an adaptor function in the supramolecular organization of the ECM (34). In long bones, MATN-1 and MATN-3 are present in all cartilage regions, in contrast to MATN2, which is expressed in the proliferative and upper hypertrophic zones. Further studies are needed to better understand the function of MATN2 in AS. MXRA5 is another interesting protein that we found to be overexpressed in calcified disease. The same pattern was observed in both RNAseq and proteomic analyses. MXRA5 has anti-fibrotic and anti-inflammatory properties in response to TGFβ1 stimulation in kidney disease (35). It is therefore conceivable to speculate that MXRA5 is up-regulated during the mineralization process as a compensatory mechanism to offset ECM overproduction. However, additional future studies are required to tingle out the implication of MXRA5 in regulating the calcification process of the aortic valve.
We further explored the prospective association of ECM protein alterations with changes in metabolic proteins during aortic valve calcification. There are currently no studies that have documented such crosstalk between the ECM and metabolism in aortic valve calcification. Indeed, MetS is a powerful and independent predictor of rapid AS progression (6) and affects the ECM structure in adipose tissue (7). In parallel, ECM remodeling may disrupt the regulation of cell metabolism and lipid synthesis (14). Our proteomics analyses revealed that the metabolic pathways were altered with a significant reduction in the expression of key metabolic proteins such as SOD1, ALDH1, and ENO1. Although our data do not directly implicate ECM proteins in the modulation of metabolic proteins and vice versa, they nevertheless highlight strong potential interactions between these 2 classes because the KEGG network analysis showed that ECM proteins and the metabolic pathways are the most overrepresented. This observation further supports the hypothesis implicating metabolic disorders in the progression of AS (8).
Furthermore, our analysis documented the presence of apolipoproteins in the same node as ECM proteins, raising the possibility of lipid-ECM interaction, potentially amplifying the inflammatory process in the calcified stage. Indeed, proteoglycan accumulations promote lipoprotein retention (23), such as oxidized LDL, which is a key stimulator of cell mineralization through the activation of an inflammatory response (36,37). We have previously shown that phospholipase A2 (LP-PLA2) uses oxidized phospholipids, which are transported by Lp(a), as a substrate to produce lysophosphatidylcholine (LPC) (38,39). Also, through nonenzymatic pathways, the oxidation of LDLs generates LPC, a highly reactive metabolite with pro-osteogenic properties that is present in mineralized aortic valves (36,40). It is also worth mentioning here that alterations in fatty acid metabolism, and other processes such as oxidative stress, in valve ECM are also implicated in the maladaptive function of endothelial cells (ECs) (41), which play an important role in regulating permeability and maintaining valve homeostasis (42). EC dysfunction in aortic valves is associated with lipid and inflammatory cells infiltration (42). Once inside the valve, macrophages release proteases that lead to ECM degradation and disrupt the normal structure of the valve matrix (42). Further investigations are needed to unveil pathway crosstalk between ECs and VICs.
Study limitations
The main limitation of our study was the absence of a complete clinical data information and blood samples. However, the validation of our results in an independent human cohort strengthens our findings. The exploration of ECM proteins in vitro was limited to 2 biological replicates and 3 experimental replications. Isolating cells from control noncalcified valves was difficult. Indeed, control valves were collected from heart transplantation surgery during unusual hours, very often overnight, limiting access to fresh tissue to isolate cells in the lab. We collected control valves only from heart transplantation surgery because hearts from sudden death patients are mainly reserved for transplantation and could not be used for research purposes. Because we did not have the entire aortic valve, we were not able to perform RNAseq on our patients. In our local cohort, we used the tissue mainly for ECM protein extraction, histological section, and electron microscopy screening. However, the validation of our data in a different cohort has strengthened our finding. Finally, further studies are necessary to study the association of FNCD1 and MXRA5 with the clinical outcomes in patients with AS.
Conclusions
This study represents a comprehensive analysis of the modulation of ECM proteins during aortic valve calcification. We used a proteomic approach to explore ECM proteins in human calcified valve tissue and human valve cells. We validated key ECM proteins in 2 different human cohorts of AS. This work highlights a strong connection between metabolic markers and ECM remodeling and identifies FNDC1 and MXRA5 as novel ECM biomarkers in calcified valves, suggesting they are potential targets in the development and progression of AS.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: There is an urgent need to identify new therapeutic targets to stop or reverse aortic valve calcification. The ECM plays an important role in maintaining the structural architecture and mechanical behavior of the valve. Its disruption contributes to valve mineralization.
TRANSLATIONAL OUTLOOK 1: The current study identifies FNDC1 and MXRA5 as novel ECM markers in human calcified valves, electing them as potential translational targets in the development and progression of aortic stenosis.
TRANSLATIONAL OUTLOOK 2: Further studies with a larger number of patients are required to confirm this finding.
Author Disclosures
Dr. Bouchareb was supported in part by a National Institutes of Health (NIH) grant T32 HL7824-19; Dr. Lebeche was supported by National Institutes of Health grant R01HL137220. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For an expanded Methods section and a supplemental table, please see the online version of this paper.
Appendix
References
- 1.Stewart B.F., Siscovick D., Lind B.K. Clinical factors associated with calcific aortic valve disease. Cardiovascular Health Study. J Am Coll Cardiol. 1997;29:630–634. doi: 10.1016/s0735-1097(96)00563-3. [DOI] [PubMed] [Google Scholar]
- 2.Rajamannan N.M., Evans F.J., Aikawa E. Calcific aortic valve disease: not simply a degenerative process: a review and agenda for research from the National Heart and Lung and Blood Institute Aortic Stenosis Working Group. Executive summary: calcific aortic valve disease-2011 update. Circulation. 2011;124:1783–1791. doi: 10.1161/CIRCULATIONAHA.110.006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briand M., Lemieux I., Dumesnil J.G. Metabolic syndrome negatively influences disease progression and prognosis in aortic stenosis. J Am Coll Cardiol. 2006;47:2229–2236. doi: 10.1016/j.jacc.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 4.Owens D.S., Katz R., Takasu J., Kronmal R., Budoff M.J., O'Brien K.D. Incidence and progression of aortic valve calcium in the Multi-Ethnic Study of Atherosclerosis (MESA) Am J Cardiol. 2010;105:701–708. doi: 10.1016/j.amjcard.2009.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenaweser P., Pilgrim T., Kadner A. Clinical outcomes of patients with severe aortic stenosis at increased surgical risk according to treatment modality. J Am Coll Cardiol. 2011;58:2151–2162. doi: 10.1016/j.jacc.2011.05.063. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann S., Walther T., Kempfert J. Mechanical strain and the aortic valve: influence on fibroblasts, extracellular matrix, and potential stenosis. Ann Thorac Surg. 2009;88:1476–1483. doi: 10.1016/j.athoracsur.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 7.Sonbol H.S. Extracellular matrix remodeling in human disease. J Microsc Ultrastruct. 2018;6:123–128. doi: 10.4103/JMAU.JMAU_4_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capoulade R., Clavel M.A., Dumesnil J.G. ASTRONOMER Investigators. Impact of metabolic syndrome on progression of aortic stenosis: influence of age and statin therapy. J Am Coll Cardiol. 2012;60:216–223. doi: 10.1016/j.jacc.2012.03.052. [DOI] [PubMed] [Google Scholar]
- 9.Testuz A., Nguyen V., Mathieu T. Influence of metabolic syndrome and diabetes on progression of calcific aortic valve stenosis. Int J Cardiol. 2017;244:248–253. doi: 10.1016/j.ijcard.2017.06.104. [DOI] [PubMed] [Google Scholar]
- 10.Williams A.S., Kang L., Wasserman D.H. The extracellular matrix and insulin resistance. Trends Endocrinol Metab. 2015;26:357–366. doi: 10.1016/j.tem.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri J., Bains Y., Guha S. The role of advanced glycation end products in aging and metabolic diseases: bridging association and causality. Cell Metab. 2018;28:337–352. doi: 10.1016/j.cmet.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spadaccio C., Mozetic P., Nappi F. Cells and extracellular matrix interplay in cardiac valve disease: because age matters. Basic Res Cardiol. 2016;111:16. doi: 10.1007/s00395-016-0534-9. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Ojeda F.J., Mendez-Gutierrez A., Aguilera C.M., Plaza-Diaz J. Extracellular matrix remodeling of adipose tissue in obesity and metabolic diseases. Int J Mol Sci. 2019;20:4888. doi: 10.3390/ijms20194888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romani P., Brian I., Santinon G. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat Cell Biol. 2019;21:338–347. doi: 10.1038/s41556-018-0270-5. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez-Llamas G., Martin-Rojas T., de la Cuesta F. Modification of the secretion pattern of proteases, inflammatory mediators, and extracellular matrix proteins by human aortic valve is key in severe aortic stenosis. Mol Cell Proteomics. 2013;12:2426–2439. doi: 10.1074/mcp.M113.027425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaden J.J., Dempfle C.E., Grobholz R. Inflammatory regulation of extracellular matrix remodeling in calcific aortic valve stenosis. Cardiovasc Pathol. 2005;14:80–87. doi: 10.1016/j.carpath.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Rojas T., Mourino-Alvarez L., Alonso-Orgaz S. iTRAQ proteomic analysis of extracellular matrix remodeling in aortic valve disease. Sci Rep. 2015;5:17290. doi: 10.1038/srep17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren B.A., Yong J.L. Calcification of the aortic valve: its progression and grading. Pathology. 1997;29:360–368. doi: 10.1080/00313029700169315. [DOI] [PubMed] [Google Scholar]
- 19.Bouchareb R., Boulanger M.C., Fournier D., Pibarot P., Messaddeq Y., Mathieu P. Mechanical strain induces the production of spheroid mineralized microparticles in the aortic valve through a RhoA/ROCK-dependent mechanism. J Mol Cell Cardiol. 2014;67:49–59. doi: 10.1016/j.yjmcc.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Tyanova S., Temu T., Sinitcyn P. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 21.Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guauque-Olarte S., Messika-Zeitoun D., Droit A. Calcium signaling pathway genes RUNX2 and CACNA1C are associated with calcific aortic valve disease. Circ Cardiovasc Genet. 2015;8:812–822. doi: 10.1161/CIRCGENETICS.115.001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neufeld E.B., Zadrozny L.M., Phillips D., Aponte A., Yu Z.X., Balaban R.S. Decorin and biglycan retain LDL in disease-prone valvular and aortic subendothelial intimal matrix. Atherosclerosis. 2014;233:113–121. doi: 10.1016/j.atherosclerosis.2013.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouchareb R., Boulanger M.C., Tastet L. Activated platelets promote an osteogenic programme and the progression of calcific aortic valve stenosis. Eur Heart J. 2019;40:1362–1373. doi: 10.1093/eurheartj/ehy696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y.J., Kim S., Choi Y. SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat Immunol. 2019;20:276–287. doi: 10.1038/s41590-018-0303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guauque-Olarte S., Droit A., Tremblay-Marchand J. RNA expression profile of calcified bicuspid, tricuspid, and normal human aortic valves by RNA sequencing. Physiol Genomics. 2016;48:749–761. doi: 10.1152/physiolgenomics.00041.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinton R.B., Jr., Lincoln J., Deutsch G.H. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res. 2006;98:1431–1438. doi: 10.1161/01.RES.0000224114.65109.4e. [DOI] [PubMed] [Google Scholar]
- 28.Arjunon S., Rathan S., Jo H., Yoganathan A.P. Aortic valve: mechanical environment and mechanobiology. Ann Biomed Eng. 2013;41:1331–1346. doi: 10.1007/s10439-013-0785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mkannez G., Gagne-Ouellet V., Jalloul Nsaibia M. DNA methylation of a PLPP3 MIR transposon-based enhancer promotes an osteogenic programme in calcific aortic valve disease. Cardiovasc Res. 2018;114:1525–1535. doi: 10.1093/cvr/cvy111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Y., Wei R., Yuan Z. Rutin suppresses FNDC1 expression in bone marrow mesenchymal stem cells to inhibit postmenopausal osteoporosis. Am J Transl Res. 2019;11:6680–6690. [PMC free article] [PubMed] [Google Scholar]
- 31.Bentmann A., Kawelke N., Moss D. Circulating fibronectin affects bone matrix, whereas osteoblast fibronectin modulates osteoblast function. J Bone Miner Res. 2010;25:706–715. doi: 10.1359/jbmr.091011. [DOI] [PubMed] [Google Scholar]
- 32.Padang R., Bagnall R.D., Tsoutsman T., Bannon P.G., Semsarian C. Comparative transcriptome profiling in human bicuspid aortic valve disease using RNA sequencing. Physiol Genomics. 2015;47:75–87. doi: 10.1152/physiolgenomics.00115.2014. [DOI] [PubMed] [Google Scholar]
- 33.Babanin V.S., Minushkina L.O. [Calcification of valvular heart structures and calcified aortic stenosis in postmenopausal women] Kardiologiia. 2011;51:43. [PubMed] [Google Scholar]
- 34.Korpos E., Deak F., Kiss I. Matrilin-2, an extracellular adaptor protein, is needed for the regeneration of muscle, nerve and other tissues. Neural Regen Res. 2015;10:866–869. doi: 10.4103/1673-5374.158332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poveda J., Sanz A.B., Fernandez-Fernandez B. MXRA5 is a TGF-beta1-regulated human protein with anti-inflammatory and anti-fibrotic properties. J Cell Mol Med. 2017;21:154–164. doi: 10.1111/jcmm.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouchareb R., Mahmut A., Nsaibia M.J. Autotaxin derived from lipoprotein(a) and valve interstitial cells promotes inflammation and mineralization of the aortic valve. Circulation. 2015;132:677–690. doi: 10.1161/CIRCULATIONAHA.115.016757. [DOI] [PubMed] [Google Scholar]
- 37.Cheng H., Yao Q., Song R. Lysophosphatidylcholine activates the Akt pathway to upregulate extracellular matrix protein production in human aortic valve cells. J Surg Res. 2017;213:243–250. doi: 10.1016/j.jss.2017.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nsaibia M.J., Boulanger M.C., Bouchareb R. OxLDL-derived lysophosphatidic acid promotes the progression of aortic valve stenosis through a LPAR1-RhoA-NF-kappaB pathway. Cardiovasc Res. 2017;113:1351–1363. doi: 10.1093/cvr/cvx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmut A., Mahjoub H., Boulanger M.C. Circulating Lp-PLA2 is associated with high valvuloarterial impedance and low arterial compliance in patients with aortic valve bioprostheses. Clin Chim Acta. 2016;455:20–25. doi: 10.1016/j.cca.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Mahmut A., Boulanger M.C., El Husseini D. Elevated expression of lipoprotein-associated phospholipase A2 in calcific aortic valve disease: implications for valve mineralization. J Am Coll Cardiol. 2014;63:460–469. doi: 10.1016/j.jacc.2013.05.105. [DOI] [PubMed] [Google Scholar]
- 41.Theodorou K., Boon R.A. Endothelial cell metabolism in atherosclerosis. Front Cell Dev Biol. 2018;6:82. doi: 10.3389/fcell.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leopold J.A. Cellular mechanisms of aortic valve calcification. Circ Cardiovasc Interv. 2012;5:605–614. doi: 10.1161/CIRCINTERVENTIONS.112.971028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.