Abstract
Understanding the mechanisms underlying the metabolically unhealthy normal weight (MUHNW) and metabolically healthy obese (MHO) phenotypes is important for developing strategies to prevent cardiometabolic diseases. Here, we conducted genome-wide association studies (GWASs) to identify the MUHNW and MHO genetic indices. The study dataset comprised genome-wide single-nucleotide polymorphism genotypes and epidemiological data from 49,915 subjects categorised into four phenotypes—metabolically healthy normal weight (MHNW), MUHNW, MHO, and metabolically unhealthy obese (MUHO). We conducted two GWASs using logistic regression analyses and adjustments for confounding variables (model 1: MHNW versus MUHNW and model 2: MHO versus MUHO). GCKR, ABCB11, CDKAL1, LPL, CDKN2B, NT5C2, APOA5, CETP, and APOC1 were associated with metabolically unhealthy phenotypes among normal weight individuals (model 1). LPL, APOA5, and CETP were associated with metabolically unhealthy phenotypes among obese individuals (model 2). The genes common to both models are related to lipid metabolism (LPL, APOA5, and CETP), and those associated with model 1 are related to insulin or glucose metabolism (GCKR, CDKAL1, and CDKN2B). This study reveals the genetic architecture of the MUHNW and MHO phenotypes in a Korean population-based cohort. These findings could help identify individuals at a high metabolic risk in normal weight and obese populations and provide potential novel targets for the management of metabolically unhealthy phenotypes.
Subject terms: Biomarkers, Endocrinology, Medical research
Introduction
Obesity is associated with numerous metabolic disorders, including type 2 diabetes mellitus1, dyslipidemia2, hypertension3, cardiovascular disease4, and cancers5, which are leading causes of mortality in adults. The ongoing worldwide obesity epidemic constitutes an enormous public health burden6. However, not all individuals with obesity have cardiometabolic complications despite excess adiposity; this phenotype is called metabolically healthy obese (MHO). A commonly used definition for MHO requires individuals to be obese and lack metabolic abnormalities7. Conversely, individuals with normal weight occasionally exhibit metabolic abnormalities that are usually observed in individuals with obesity, and this phenotype is termed metabolically unhealthy normal weight (MUHNW)8. Since there are no universally accepted standard definitions of MHO and MUHNW, the prevalence of these phenotypes heavily depends of the definition that is being used for the characterisation of metabolic health. According to a study conducted using National Health and Nutrition Examination Surveys in the US, 31.7% of obese individuals were metabolically healthy, and 23.5% of normal weight individuals were metabolically unhealthy9. In Korea, an estimated 33%–48% of the population with obesity is reported to be MHO, and 12%–21% of individuals with normal weight are reported to be MUHNW10,11.
Although the mechanisms that determine why some individuals with obesity remain free from metabolic complications while others with normal weight are susceptible to metabolic complications are not fully understood, previous studies have shown the biological mechanisms possibly associated with the MHO and MUHNW phenotypes, apart from demographic factors (e.g., age, sex, and ethnicity) and environmental factors (e.g., physical activity, smoking, and alcohol intake). These studies indicated that reduced abdominal fat mass and increased gluteofemoral fat mass are associated with the metabolically healthy phenotype, whereas elevated abdominal fat mass and lower gluteofemoral fat mass contribute to the metabolically unhealthy phenotype12–14. In addition to fat accumulation and distribution, lipodystrophy, adipogenesis, inflammation, and mitochondrial function are reported to be key contributors to the MHO and MUHNW phenotypes13–17.
Over the past decade, genome-wide association studies (GWASs) have been used to identify genetic variants associated with a wide range of diseases and traits. GWASs have identified various genetic loci associated with adiposity, fat distribution, insulin resistance, and metabolic diseases, including hypertension, dyslipidemia, and type 2 diabetes18–22. Elucidating such genetic variations can provide insights into the proteins and pathways involved in the development of the MHO and MUHNW phenotypes. Indeed, several GWASs on body fat percentage showed that the genetic variants of certain genes, such as IRS1, could be associated with the MUHNW and MHO phenotypes23–25. However, few studies have specifically investigated the genetic variants associated with the MUHNW or MHO phenotypes in Asian populations. Here, we conducted GWASs to identify candidate genes harbouring single-nucleotide polymorphisms (SNPs) associated with the MHO and MUHNW phenotypes in a large Korean population-based cohort.
Results
Clinical characteristics of the study participants
The clinical characteristics of the participants are described in Table 1. The percentages of individuals with metabolically healthy normal weight (MHNW), MUHNW, MHO, and metabolically unhealthy obese (MUHO) phenotypes among the study population were 47.0% (23 466 individuals), 20.8% (10 358 individuals), 14.0% (7 008 individuals), and 18.2% (9 083 individuals), respectively. The cardiometabolic variables differed significantly among the four groups. We observed a clear elevation in cardiometabolic variables in metabolically unhealthy individuals (MUHNW and MUHO). The prevalence of hypertension and diabetes was also significantly higher among participants with the MUHNW or MUHO phenotypes than among those with the MHNW or MHO phenotypes.
Table 1.
Clinical characteristics of the study participants categorised into the four obesity phenotypes.
| MHNW (n = 23 466) |
MUHNW (n = 10 358) |
MHO (n = 7 008) |
MUHO (n = 9 083) |
p-value | p-value1 | p-value2 | p-value3 | |
|---|---|---|---|---|---|---|---|---|
| Age (years) | 51.4 ± 7.0 | 56 ± 7.6 | 52.9 ± 7.9 | 55.3 ± 7.8 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| Sex | < 0.001 | < 0.001 | < 0.001 | < 0.001 | ||||
| Male (%) | 27.8 | 39.5 | 38.5 | 47.3 | ||||
| Female (%) | 72.1 | 60.5 | 61.5 | 52.7 | ||||
| BMI (kg/m2) | 22.1 ± 1.8 | 22.9 ± 1.5 | 26.8 ± 1.7 | 27.4 ± 2.1 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| SBP (mmHg) | 116.7 ± 12.9 | 128.1 ± 14.3 | 122.2 ± 13.3 | 131.1 ± 14.2 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| DBP (mmHg) | 72.5 ± 8.9 | 78.7 ± 9.4 | 76.0 ± 9.2 | 80.8 ± 9.5 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| FPG (mg/dL) | 89.0 ± 11.1 | 102.6 ± 25.4 | 90.9 ± 11.7 | 105.1 ± 25.7 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| Total cholesterol (mg/dL) | 195.8 ± 33.3 | 200.3 ± 38.1 | 201.6 ± 34 | 201.3 ± 37.5 | < 0.001 | < 0.001 | 0.020 | < 0.001 |
| TG (mg/dL) | 89.9 ± 43.6 | 167.7 ± 104.7 | 105.1 ± 50.1 | 186.4 ± 113 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| HDL-C (mg/dL) | 59.1 ± 12.7 | 48.2 ± 12.0 | 55 ± 11.1 | 46.1 ± 10.6 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| Hypertension (%) | 1 516 (6.4) | 3 313 (32.0) | 911 (13.0) | 3 765 (41.4) | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| Diabetes (%) | 351 (1.5) | 1 430 (13.8) | 110 (1.6) | 1 320 (14.5) | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| Regular exerciser (%) | 9 071 (39.4) | 4 120 (40.6) | 2 726 (39.8) | 3 453 (38.7) | 0.401 | 0.039 | 0.179 | 0.265 |
| Smoker (%) | < 0.001 | < 0.001 | < 0.001 | < 0.001 | ||||
| Current smoker (%) | 2 215 (9.4) | 1 415 (13.7) | 776 (11.1) | 1 386 (15.3) | ||||
| Former smoker (%) | 2 749 (11.7) | 1 791 (17.3) | 1 244 (17.8) | 2 011 (22.1) | ||||
| Drinker (%) | 3 777 (14.4) | 1 928 (18.6) | 1 341 (19.1) | 2 017 (22.2) | < 0.001 | < 0.001 | < 0.001 | 0.388 |
Data are presented as the mean ± standard deviation or percentage. We obtained p-values by one-way analysis of variance or independent two-sample t-tests for continuous variables or by χ2 tests for categorical variables.
p-values represent the difference in each variable among the MHNW, MUHNW, MHO and MUHO phenotypes.
MHNW, metabolically healthy normal weight; MUHNW, metabolically unhealthy normal weight; MHO, metabolically healthy obese; MUHO, metabolically unhealthy obese; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; FPG, fasting plasma glucose; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol.
p-values1 represent the difference in each variable between the MHNW and MUHNW phenotypes.
p-values2 represent the difference in each variable between the MHO and MUHO phenotypes.
p-values3 represent the difference in each variable between the MUHNW and MHO phenotypes.
Genetic regions associated with metabolically unhealthy phenotypes in model 1 (MHNW versus MUHNW) and model 2 (MHO versus MUHO)
We conducted a GWAS for model 1 to identify the genetic factors associated with the metabolically unhealthy phenotype among the normal weight groups and for model 2 to identify the genetic factors associated with the metabolically unhealthy phenotype among the obese groups.
The lead SNPs and clusters of SNPs in the regions associated with the metabolically unhealthy phenotype in models 1 and 2 are shown in Tables 2-1 and 2-2, respectively. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using logistic regression analysis after adjusting for age, sex, exercise status, smoking status, alcohol intake, body mass index (BMI), and principle component (PC) 1 and PC2. SNPs in LPL, APOA5, and CETP exhibited significant association with the risk of metabolically unhealthy phenotypes in both normal weight and obese individuals (in models 1 and 2). SNPs in GCKR, ABCB11, CDKAL1, CDKN2B, NT5C2, and APOC1 were significantly associated with the risk of metabolically unhealthy phenotypes only in normal weight individuals (in model 1 only).
Table 2.
Representative SNPs identified by GWAS among the significant loci for model 1 (MHNW versus MUHNW) and model 2 (MHO versus MUHO).
| Gene | Region | Chr | Position (bp) | SNP | SNP cluster | Minor allele | MAF | OR (95% CIs) | p-value |
|---|---|---|---|---|---|---|---|---|---|
| 2–1. MHNW (control) versus MUHNW (case) | |||||||||
| GCKR | Intron variant | 2 | 27 734 972 | rs6547692 | rs780096, rs1260326 | A | 0.46 | 0.90 (0.87–0.94) | 1.27E-08 |
| ABCB11 | Intron variant | 2 | 169 803 568 | rs16856261 | rs3755157, rs58512362 | T | 0.37 | 1.12 (1.08–1.16) | 2.07E-09 |
| CDKAL1 | Intron variant | 6 | 20 693 697 | rs138420022 | rs34499031, rs35261542 | A | 0.47 | 1.15 (1.11–1.19) | 1.09E-14 |
| LPL | Downstream gene variant | 8 | 19 865 455 | rs77237194 | rs10096633, rs17482753 | T | 0.12 | 0.83 (0.78–0.87) | 2.56E-12 |
| CDKN2B | Downstream gene variant | 9 | 22 132 729 | rs10965247 | rs10811660, rs10965246 | G | 0.44 | 0.89 (0.86–0.92) | 1.27E-10 |
| NT5C2 | Downstream gene variant | 10 | 104 960 464 | rs113278154 | rs79237883, rs34747231 | T | 0.27 | 0.90 (0.86–0.92) | 4.37E-08 |
| APOA5 | Upstream gene variant | 11 | 116 662 579 | rs651821 | rs662799, rs2075291 | C | 0.30 | 1.47 (1.42–1.53) | 1.68E-90 |
| CETP | Intron variant | 16 | 57 002 663 | rs9926440 | rs17231506, rs821840 | C | 0.31 | 1.16 (1.11–1.20) | 2.95E-14 |
| APOC1 | Intron variant | 19 | 45 420 082 | rs73052335 | rs111789331, rs66626994 | C | 0.10 | 1.24 (1.17–1.32) | 1.24E-13 |
| 2–2. MHO (control) versus MUHO (case) | |||||||||
| LPL | Downstream gene variant | 8 | 19 827 848 | rs10105606 | rs1441766, rs4464984 | A | 0.12 | 0.83 (0.78–0.87) | 1.68E-11 |
| APOA5 | Upstream gene variant | 11 | 116 662 579 | rs651821 | rs662799 | C | 0.30 | 1.43 (1.36–1.51) | 8.55E-44 |
| CETP | Upstream gene variant | 16 | 56 993 886 | rs821840 | rs36229491, rs17231506 | G | 0.17 | 0.83 (0.78–0.88) | 2.02E-10 |
Logistic regression models were adjusted for age, sex, exercise status, smoking status, alcohol intake, body mass index, and PC1 and PC2.
GWAS, genome-wide association study; MHNW, metabolically healthy normal weight; MUHNW, metabolically unhealthy normal weight; MHO, metabolically healthy obese; MUHO, metabolically unhealthy obese; Chr, chromosome; SNP, single-nucleotide polymorphism; MAF, minor allele frequency; OR, odds radio; CI, confidence interval.
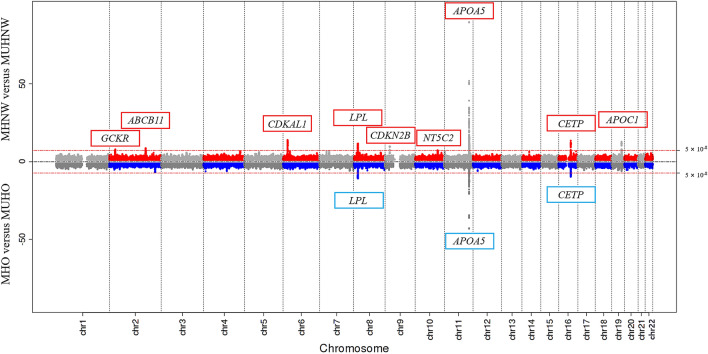
The results of the model 1 and model 2 GWASs are illustrated in a Miami plot (Fig. 1), a format recently developed at Michigan University to describe two comparable GWAS results. We observed genome-wide significant association clusters of GCKR, ABCB11, CDKAL1, LPL, CDKN2B, NT5C2, APOA5, CETP, and APOC1 in model 1 (upper plot) and LPL, APOA5, and CETP in model 2 (lower plot).
Figure 1.
Miami plot of the GWASs for model 1 (MHNW versus MUHNW) and model 2 (MHO versus MUHO). SNP locations are plotted on the x-axis according to their chromosomal position. The −log10(p-values) derived from the logistic regression analysis are plotted on the y-axis. The p-values were adjusted for age, sex, exercise status, smoking status, alcohol intake, body mass index, and PC1 and PC2. The horizontal red line indicates the formal threshold for genome-wide significance at p = 5.00 × 10−8. GWASs, genome-wide association studies; MHNW, metabolically healthy normal weight; MUHNW, metabolically unhealthy normal weight; MHO, metabolically healthy obese; MUHO, metabolically unhealthy obese; PC, principle component; SNP, single-nucleotide polymorphism. The figure was generated using EasyStrata version 8.6 (http://www.genepi-regensburg.de/easystrata).
Discussion
Our study aimed to identify the genetic variations that differentiated the MHO phenotype from the MUHNW phenotype. Determining the genetic characteristics associated with the MHO and MUHNW phenotypes will enable us to pinpoint the biological mechanisms driving these two paradoxical conditions and develop approaches to prevent cardiometabolic diseases. Over the past decade, several genomic studies have identified numerous genetic variants associated with adiposity in the context of a favourable cardiometabolic profile; many of these loci are located in or near genes involved in adipogenesis, fat distribution, and insulin signalling18,19. However, few studies have fully utilised genome-wide genetic variants to characterise the MHO and MUHNW phenotypes. Furthermore, the majority of genetic studies have analysed populations of European ancestry, and limited data are available from Asian populations.
We found that LPL, APOA5, and CETP were associated with metabolically unhealthy phenotypes among both normal weight and obese individuals. These three genes are related to lipid metabolism. LPL, located on 8p21.3, encodes a key lipolysis regulator. In addition, LPL may link insulin resistance to atherosclerosis because it controls the delivery of free fatty acids to muscles, adipose tissues, and vascular wall macrophages26. APOA5, a member of the APOA4/APOC3/APOA1 gene cluster, is located on chromosome 11q23.3 and plays important roles in lipid metabolism, particularly for triglycerides (TGs) and TG-rich lipoproteins. There is considerable evidence supporting the association between APOA5 SNPs, such as rs662799 and rs651821, and an increased risk of obesity and metabolic syndrome27. Consistent with previous results, we found that rs662799 and rs651821 in APOA5 were associated with metabolically unhealthy phenotypes. CETP, located on chromosome 16q13, encodes the CETP protein that shuttles TGs and cholesteryl ester between lipoproteins28. The expression of CETP genetic variants has been associated with variations in high-density lipoprotein cholesterol (HDL-C) levels in various ethnic groups29. Our results support the link between certain genetic variants and metabolically unhealthy phenotypes.
We also found distinct genetic variants that were associated with metabolically unhealthy phenotypes among normal weight and obese individuals. GCKR, ABCB11, CDKAL1, CDKN2B, NT5C2, and APOC1 were associated with metabolically unhealthy phenotypes in individuals with normal weight but not in those with obesity. Of these, GCKR, CDKAL1, and CDKN2B are related to insulin or glucose metabolism. GCKR, which maps to chromosome 2p23.3, encodes a protein involved in the regulation of glucokinase activity; the protein modulates glucose balance and glucose-stimulated insulin secretion30. Some variants of GCKR are associated with insulin, fasting glucose, and TG levels and with susceptibility to type 2 diabetes mellitus31,32. Genetic variants of CDKAL1, which maps to chromosome 6p22.3, are strongly linked to an increased risk of developing type 2 diabetes and obesity33. CDKAL1 is associated with proinsulin conversion and insulin response upon glucose stimulation34 and is necessary for normal mitochondrial morphology and adipose tissue function33. In recent studies, the CDKAL1 rs7754840 variant was shown to be associated with increased waist circumference and waist-to-hip ratio in Chinese Han patients35, and the CDKAL1 rs2206734 polymorphism was shown to be an independent predictor of the MUHNW phenotype in Chinese children36. Similarly, our study suggested that CDKAL1 SNPs are important predictors of the MUHNW phenotype. CDKN2B, which is located in chromosome 9p21, plays a role in the deterioration of insulin secretion by participating in the regulation of pancreatic β-cell proliferation and function37,38. Studies across varied ethnicities and geographical locations showed that polymorphisms at the CDKN2A locus are related to type 2 diabetes mellitus development39–41. ACBC11 located in chromosome 2q24 encodes an ATB-binding cassette transporter. Previous studies have shown that variations in ABCB11 are associated with increased fasting glucose levels42–44. NT5C2 encodes a hydrolase that plays a key role in cellular purine metabolism and uric acid regulation45,46. SNPs in NT5C2 have been associated with hypertension47–49. APOC1 encodes a member of the apolipoprotein C1 family that plays an important role in HDL-C and very low density lipoprotein metabolism. Previous studies have indicated that some variants of APOC1 are associated with metabolic abnormalities50,51. Our findings suggested that strategies for protecting against complications related to metabolically unhealthy phenotypes might differ for individuals with normal weight and those with obesity. Further studies to identify the potential interactions of these candidate genes using biological and mechanical analyses are required.
Previous GWASs have identified various genetic variants associated with diverse metabolic diseases such as metabolic syndrome52,53, dyslipidemia54,55, and obesity56,57. However, only a limited number of studies have specifically revealed the genetic loci associated with the MUHNW and MHO phenotypes. We performed GWASs for the metabolically unhealthy phenotypes divided into the normal weight and obesity groups to identify the genes and SNPs associated with the MUHNW and MHO phenotypes. The present findings contribute to our understanding of the genetic architecture of the MUHNW and MHO phenotypes.
There are several limitations to consider in the interpretation of our results. The findings were not replicated. Furthermore, since the current study was performed in a Korean population, the findings may not apply to non-Asian populations. Replication studies and studies in other populations are necessary to confirm our findings and determine their applicability to ethnically diverse groups. In addition, there are no universally accepted standard definitions of MHO and MUHNW; we employed definitions that have been widely used in previous studies58,59. Despite these potential limitations, we believe our GWASs provide valuable data on the genetic characteristics associated with the MUHNW and MHO phenotypes in a large population-based cohort.
In summary, our GWASs provide several insights into the genetic architecture of metabolically unhealthy individuals. We found that LPL, APOA5, and CETP were associated with the metabolically unhealthy phenotypes in individuals with normal weight or obesity. GCKR, ABCB11, CDKAL1, CDKN2B, NT5C2, and APOC1 were associated with metabolically unhealthy in individuals with normal weight but not in those with obesity. Our study provides an understanding of the genetic architecture of the MUHNW and MHO phenotypes in a Korean population-based cohort. Although this study remains to be validated in a larger cohort and followed up with investigation on the pathophysiological pathways involved, our findings could help identify metabolically high-risk individuals in normal weight and obese populations and provide potential novel targets for the management of metabolically unhealthy phenotypes.
Methods
Study overview and study participants
The current study used Korean Genome and Epidemiology Study (KoGES) Health Examination data. The cohort consisted of male and female community dwellers recruited from the national health examinee registry (aged 40–79 years at baseline). Eligible participants were asked to volunteer via on-site invitation, letters, telephone calls, media campaign, or community conferences. The responders were invited to visit the survey sites, including medical schools, hospitals, and health institutions, for an interview in which they answered a questionnaire administered by trained staff and underwent physical examination. We collected information on their past medical history, smoking history, alcohol consumption, and physical activity during a health interview. We defined a regular exerciser as an individual who participated in vigorous physical activity more than three times per week. Current smokers were individuals who smoked ≥ 100 cigarettes in their lifetime and were smoker at the time of the study, and former smokers were individuals who smoked ≥ 100 cigarettes in their lifetime but were non-smokers at the time of the study. A drinker was defined as an individual consuming alcoholic beverages at least twice a week. The KoGES participants were all of Korean ethnicity. The detailed history and profile of KoGES were previously published60.
In total, 58 701 participants, for whom genome-wide SNP genotype data were available, were included in the KoGES Health Examination dataset. Of these, we excluded the participants who had a history of cancer, thyroid disease, stroke, and/or myocardial infarction (n = 6 965). Participants aged 75 years or older (n = 23) and those with missing data for BMI, blood pressure, fasting plasma glucose, TG, and/or HDL-C levels, and/or exercise/smoking/alcohol were excluded (n = 1 798). After these exclusions, 49 915 participants were included in the final analysis. Written informed consent was obtained from all participants. This research project was approved by the Institutional Review Board of Theragen Etex (approval number: 700062-20190819-GP-006-02). In addition, the study complied with the ethical principles of the Declaration of Helsinki.
Measurement of anthropometric and laboratory data
Weight and height were measured to the nearest 0.1 kg and 0.1 cm, respectively, with participants wearing light indoor clothing and no shoes. We calculated the BMI as body weight (kg) divided by the square of height (m2). Systolic and diastolic blood pressure (SBP and DBP, respectively) were measured twice with a standardised mercury sphygmomanometer (Baumanometer Standby; W.A. Baum, New York, NY, USA). Blood samples were obtained in the morning after overnight fasting. We measured fasting plasma glucose, total cholesterol, TG, and HDL-C levels with an automatic analyser (ADIVA 1650; Siemens, Tarrytown, NY, USA).
Definition of study phenotypes
We defined a participant with obesity as an individual with BMI ≥ 25 kg/m2 based on the Asia–Pacific regional guidelines of the World Health Organization and International Obesity Task Force61. Normal weight was defined as BMI < 25 kg/m2. A metabolically healthy individual was a participant with less than two of the following four metabolic traits: elevated blood pressure (SBP/DBP ≥ 130/85 mm Hg or taking antihypertensive medication); impaired fasting plasma glucose (≥ 100 mg/dL), diagnosis of diabetes mellitus, and/or prescription for antidiabetic medication; high plasma TG (≥ 150 mg/dL); and low HDL-C (< 40 mg/dL in men or < 50 mg/dL in women). According to these criteria, study participants were categorised into one of four groups: (1) MHNW: BMI < 25 kg/m2 and less than two metabolic risk factors; (2) MUHNW: BMI < 25 kg/m2 and at least two metabolic risk factors; (3) MHO: BMI ≥ 25 kg/m2 and less than two metabolic risk factors; (4) MUHO: BMI ≥ 25 kg/m2 and at least two metabolic risk factors.
Genotyping and quality-control
The genotype data were graciously provided by the Centre for Genome Science, Korea National Institute of Health and were produced using a Korea Biobank Array (Affymetrix, Santa Clara, CA, USA)62. The experimental results of the Korea Biobank Array were filtered using the following quality-control criteria: call rate, > 97%; minor allele frequency, > 1%; Hardy–Weinberg equilibrium, p < 1 × 10−5. After quality-control filtering, the experimental phenotypes were used to analyse the genotype datasets from the 1 000 Genome Phase 1 and 2 Asian panel. The GWASs identified 7 975 321 SNPs on chromosomes 1 to 22.
Statistical analysis
We compared the clinical characteristics of the study participants with different phenotypes using one-way analysis of variance for continuous variables and χ2 tests for categorical variables.
This study included two GWASs. The first GWAS was conducted on metabolically unhealthy individuals from the normal weight groups (model 1: MHNW [control] and MUHNW [case]). The second GWAS was conducted on metabolically unhealthy individuals from the obesity groups (model 2: MHO [control] and MUHO [case]).
We performed PC analysis to reduce the bias of the genomic data according to regions where the samples were collected. Based on this analysis, we obtained PC1 and PC2, which were used as covariates for statistical analysis. All GWASs were conducted using logistic regression analysis after adjusting for age, sex, exercise status, smoking status, alcohol intake, BMI, and PC1 and PC2 as covariates, using PLINK version 1.90. We calculated the ORs and 95% CIs for the GWASs. The SNPs listed in the tables are representative SNPs of approximately 50 kbp with significant p-values (p < 5.00 × 10–8). Significant associations were defined by genome-wide significance level p-values < 5.00 × 10–8.
Acknowledgements
This study was conducted with bioresources from the National Biobank of Korea of the Centers of Disease Control and Prevention of the Republic of Korea (2019-059). This work was supported by the Bio and Medical Technology Development Programme through the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning (NRF2018R1D1A1B07049223) and the Technology Innovation Programme (20002781, A Platform for Prediction and Management of Health Risk Based on Personal Big Data and Lifelogging) funded by the Ministry of Trade, Industry & Energy (Republic of Korea).
Author contributions
Conceptualization, J.-M.P. and J.-W.L.; methodology, K.-W.H.; software, Y.S.; validation, J.-E.C., Y.-J.K. and S.-J.K.; formal analysis, D.-H.P.; investigation, J.-M.P.; resources, J.K.; data curation, Y.S.; writing—original draft preparation, J.-M.P. and D.-H.P.; writing—review and editing, J.-W.L. and K.-W.H.; visualization, D.-H.P.; supervision, J.K.; project administration, J.-W.L.; funding acquisition, K.-W.H. All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jae-Min Park and Da-Hyun Park.
Contributor Information
Ji-Won Lee, Email: indi5645@yuhs.ac.
Kyung-Won Hong, Email: kyungwon.hong@therabio.kr.
References
- 1.Lazar MA. How obesity causes diabetes: not a tall tale. Science. 2005;307:373–375. doi: 10.1126/science.1104342. [DOI] [PubMed] [Google Scholar]
- 2.Klop B, Elte JW, Cabezas MC. Dyslipidemia in obesity: mechanisms and potential targets. Nutrients. 2013;5:1218–1240. doi: 10.3390/nu5041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seravalle G, Grassi G. Obesity and hypertension. Pharmacol. Res. 2017;122:1–7. doi: 10.1016/j.phrs.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Kachur S, Lavie CJ, de Schutter A, Milani RV, Ventura HO. Obesity and cardiovascular diseases. Minerva Med. 2017;108:212–228. doi: 10.23736/S0026-4806.17.05022-4. [DOI] [PubMed] [Google Scholar]
- 5.De Pergola G, Silvestris F. Obesity as a major risk factor for cancer. J. Obes. 2013;2013:291546. doi: 10.1155/2013/291546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tremmel M, Gerdtham UG, Nilsson PM, Saha S. Economic burden of obesity: a systematic literature review. Int. J. Environ. Res. Public Health. 2017;14:435. doi: 10.3390/ijerph14040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith GI, Mittendorfer B, Klein S. Metabolically healthy obesity: facts and fantasies. J. Clin. Invest. 2019;129:3978–3989. doi: 10.1172/JCI129186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefan N, Schick F, Häring HU. Causes, characteristics, and consequences of metabolically unhealthy normal weight in humans. Cell Metab. 2017;26:292–300. doi: 10.1016/j.cmet.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Wildman RP, et al. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: prevalence and correlates of 2 phenotypes among the US population (NHANES 1999–2004) Arch. Intern. Med. 2008;168:1617–1624. doi: 10.1001/archinte.168.15.1617. [DOI] [PubMed] [Google Scholar]
- 10.Lee HY, et al. Metabolic health is more closely associated with decrease in lung function than obesity. PLoS ONE. 2019;14:e0209575. doi: 10.1371/journal.pone.0209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K. Metabolically obese but normal weight (MONW) and metabolically healthy but obese (MHO) phenotypes in Koreans: characteristics and health behaviors. Asia Pac. J. Clin. Nutr. 2009;18:280–284. [PubMed] [Google Scholar]
- 12.Chen GC, et al. Association between regional body fat and cardiovascular disease risk among postmenopausal women with normal body mass index. Eur. Heart J. 2019;40:2849–2855. doi: 10.1093/eurheartj/ehz391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefan N. Causes, consequences, and treatment of metabolically unhealthy fat distribution. Lancet Diabetes Endocrinol. 2020;8:616–627. doi: 10.1016/S2213-8587(20)30110-8. [DOI] [PubMed] [Google Scholar]
- 14.Karpe F, Pinnick KE. Biology of upper-body and lower-body adipose tissue–link to whole-body phenotypes. Nat. Rev. Endocrinol. 2015;11:90–100. doi: 10.1038/nrendo.2014.185. [DOI] [PubMed] [Google Scholar]
- 15.Eigentler T, Lomberg D, Machann J, Stefan N. Lipodystrophic nonalcoholic fatty liver disease induced by immune checkpoint blockade. Ann. Intern. Med. 2020;172:836–837. doi: 10.7326/L19-0635. [DOI] [PubMed] [Google Scholar]
- 16.Stefan N, Häring HU, Schulze MB. Metabolically healthy obesity: the low-hanging fruit in obesity treatment? Lancet Diabetes Endocrinol. 2018;6:249–258. doi: 10.1016/S2213-8587(17)30292-9. [DOI] [PubMed] [Google Scholar]
- 17.Iacobini C, Pugliese G, Blasetti Fantauzzi C, Federici M, Menini S. Metabolically healthy versus metabolically unhealthy obesity. Metabolism. 2019;92:51–60. doi: 10.1016/j.metabol.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Loos RJF, Kilpeläinen TO. Genes that make you fat, but keep you healthy. J. Intern. Med. 2018;284:450–463. doi: 10.1111/joim.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang LO, Loos RJF, Kilpeläinen TO. Evidence of genetic predisposition for metabolically healthy obesity and metabolically obese normal weight. Physiol. Genomics. 2018;50:169–178. doi: 10.1152/physiolgenomics.00044.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heid IM, Winkler TW. A multitrait GWAS sheds light on insulin resistance. Nat. Genet. 2016;49:7–8. doi: 10.1038/ng.3758. [DOI] [PubMed] [Google Scholar]
- 21.Miyaki K, et al. The combined impact of 12 common variants on hypertension in Japanese men, considering GWAS results. J. Hum. Hypertens. 2012;26:430–436. doi: 10.1038/jhh.2011.50. [DOI] [PubMed] [Google Scholar]
- 22.Abe S, et al. Association of genetic variants with dyslipidemia. Mol. Med. Rep. 2015;12:5429–5436. doi: 10.3892/mmr.2015.4081. [DOI] [PubMed] [Google Scholar]
- 23.Yaghootkar H, et al. Genetic evidence for a link between favorable adiposity and lower risk of type 2 diabetes, hypertension, and heart disease. Diabetes. 2016;65:2448–2460. doi: 10.2337/db15-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat. Commun. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilpeläinen TO, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat. Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mead JR, Ramji DP. The pivotal role of lipoprotein lipase in atherosclerosis. Cardiovasc. Res. 2002;55:261–269. doi: 10.1016/S0008-6363(02)00405-4. [DOI] [PubMed] [Google Scholar]
- 27.Su X, Kong Y, Peng DQ. New insights into apolipoprotein A5 in controlling lipoprotein metabolism in obesity and the metabolic syndrome patients. Lipids Health Dis. 2018;17:174. doi: 10.1186/s12944-018-0833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansen M, Puetz G, Hoffmann MM, Winkler K. A mathematical model to estimate cholesterylester transfer protein (CETP) triglycerides flux in human plasma. BMC Syst. Biol. 2019;13:12. doi: 10.1186/s12918-019-0679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Rios A, et al. Beneficial effect of CETP gene polymorphism in combination with a Mediterranean diet influencing lipid metabolism in metabolic syndrome patients: CORDIOPREV study. Clin. Nutr. 2018;37:229–234. doi: 10.1016/j.clnu.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes Silva L, Vangipurapu J, Kuulasmaa T, Laakso M. An intronic variant in the GCKR gene is associated with multiple lipids. Sci. Rep. 2019;9:10240. doi: 10.1038/s41598-019-46750-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onuma H, et al. The GCKR rs780094 polymorphism is associated with susceptibility of type 2 diabetes, reduced fasting plasma glucose levels, increased triglycerides levels and lower HOMA-IR in Japanese population. J. Hum. Genet. 2010;55:600–604. doi: 10.1038/jhg.2010.75. [DOI] [PubMed] [Google Scholar]
- 32.Sparsø T, et al. The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia. 2008;51:70–75. doi: 10.1007/s00125-007-0865-z. [DOI] [PubMed] [Google Scholar]
- 33.Palmer CJ, et al. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol. Metab. 2017;6:1212–1225. doi: 10.1016/j.molmet.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stancáková A, et al. Association of 18 confirmed susceptibility loci for type 2 diabetes with indices of insulin release, proinsulin conversion, and insulin sensitivity in 5,327 nondiabetic Finnish men. Diabetes. 2009;58:2129–2136. doi: 10.2337/db09-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang J, Guan RC, Zhao Y, Chen Y. Obesity-related loci in TMEM18, CDKAL1 and FAIM2 are associated with obesity and type 2 diabetes in Chinese Han patients. BMC Med. Genet. 2020;21:65. doi: 10.1186/s12881-020-00999-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, et al. Interaction between early environment and genetic predisposition instigates the metabolically obese, normal weight phenotype in children: findings from the BCAMS study. Eur. J. Endocrinol. 2020;182:393–403. doi: 10.1530/EJE-19-0755. [DOI] [PubMed] [Google Scholar]
- 37.Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in β-cell proliferation restricts the capacity of β-cell regeneration in mice. Diabetes. 2009;58:1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong Y, Sharma RB, Nwosu BU, Alonso LC. Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia. 2016;59:1579–1593. doi: 10.1007/s00125-016-3967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikitin AG, et al. Association of polymorphic markers of genes FTO, KCNJ11, CDKAL1, SLC30A8, and CDKN2B with type 2 diabetes mellitus in the Russian population. PeerJ. 2017;5:e3414. doi: 10.7717/peerj.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong X, Xing X, Hong J, Zhang X, Yang W. Genetic variants associated with lean and obese type 2 diabetes in a Han Chinese population: a case-control study. Medicine (Baltimore) 2016;95:e3841. doi: 10.1097/MD.0000000000003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis JP, et al. Association analysis in african americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes. 2008;57:2220–2225. doi: 10.2337/db07-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen WM, et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J. Clin. Invest. 2008;118:2620–2628. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose CS, et al. A variant in the G6PC2/ABCB11 locus is associated with increased fasting plasma glucose, increased basal hepatic glucose production and increased insulin release after oral and intravenous glucose loads. Diabetologia. 2009;52:2122–2129. doi: 10.1007/s00125-009-1463-z. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi F, et al. Common variants at the GCK, GCKR, G6PC2-ABCB11 and MTNR1B loci are associated with fasting glucose in two Asian populations. Diabetologia. 2010;53:299–308. doi: 10.1007/s00125-009-1595-1. [DOI] [PubMed] [Google Scholar]
- 45.Ipata PL, Tozzi MG. Recent advances in structure and function of cytosolic IMP-GMP specific 5'nucleotidase II (cN-II) Purinergic Signal. 2006;2:669–675. doi: 10.1007/s11302-006-9009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh HR, Choi YJ, Yoo NJ, Lee SH. Leukemia relapse-associated mutation of NT5C2 gene is rare in de novo acute leukemias and solid tumors. Pathol. Oncol. Res. 2016;22:223–224. doi: 10.1007/s12253-015-9965-0. [DOI] [PubMed] [Google Scholar]
- 47.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, et al. Genome-wide association study meta-analysis of long-term average blood pressure in East Asians. Circ. Cardiovasc. Genet. 2017;10:e001527. doi: 10.1161/CIRCGENETICS.116.001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R, et al. Effects of apoC1 genotypes on the hormonal levels, metabolic profile and PAF-AH activity in Chinese women with polycystic ovary syndrome. Lipids Health Dis. 2018;17:77. doi: 10.1186/s12944-018-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avery CL, et al. A phenomics-based strategy identifies loci on APOC1, BRAP, and PLCG1 associated with metabolic syndrome phenotype domains. PLoS Genet. 2011;7:e1002322. doi: 10.1371/journal.pgen.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong SW, Chung M, Park SJ, Cho SB, Hong KW. Genome-wide association study of metabolic syndrome in koreans. Genomics Inform. 2014;12:187–194. doi: 10.5808/GI.2014.12.4.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tekola-Ayele F, et al. Genome-wide association study identifies African-ancestry specific variants for metabolic syndrome. Mol. Genet. Metab. 2015;116:305–313. doi: 10.1016/j.ymgme.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hubacek JA, Adamkova V, Lanska V, Dlouha D. Polygenic hypercholesterolemia: examples of GWAS results and their replication in the Czech-Slavonic population. Physiol. Res. 2017;66:S101–S111. doi: 10.33549/physiolres.933580. [DOI] [PubMed] [Google Scholar]
- 55.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, et al. A genome-wide association study on obesity and obesity-related traits. PLoS ONE. 2011;6:e18939. doi: 10.1371/journal.pone.0018939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malhotra A, et al. A genome-wide association study of BMI in American Indians. Obesity (Silver Spring) 2011;19:2102–2106. doi: 10.1038/oby.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung CH, et al. The risk of incident type 2 diabetes in a Korean metabolically healthy obese population: the role of systemic inflammation. J. Clin. Endocrinol. Metab. 2015;100:934–941. doi: 10.1210/jc.2014-3885. [DOI] [PubMed] [Google Scholar]
- 59.Cordola Hsu AR, et al. Sociodemographic and metabolic risk characteristics associated with metabolic weight categories in the Women's Health Initiative. Cardiovasc. Endocrinol. Metab. 2020;9:42–48. doi: 10.1097/XCE.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim Y, Han BG. Cohort profile: The Korean genome and epidemiology study (KoGES) consortium. Int. J. Epidemiol. 2017;46:e20. doi: 10.1093/ije/dyv316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.World Health Organization . The Asia-Pacific Perspective: Redefining Obesity and Its Treatment. Sydney: Health Communications Australia; 2000. [Google Scholar]
- 62.Moon S, et al. The Korea Biobank array: design and identification of coding variants associated with blood biochemical traits. Sci. Rep. 2019;9:1382. doi: 10.1038/s41598-018-37832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]