Abstract
Biot2 is a tumor-associated antigen, and it is a novel gene (GenBank EF100607) that was first identified with the SEREX technique and named by our laboratory. It is highly expressed in cancer cells and testis, with low or no expression in normal tissues. In our previous study, RNA interference of human Biot2 can inhibit tumor cell growth, and it is associated with poor prognosis of patients in clinical study; however, the mechanism of Biot2 that effects tumor growth is not yet clear. Here, in this study, we explore further the mechanism of Biot2 by silencing Biot2 in CT26 cells. It provides some theoretical basis for Biot2 as a new target for gene therapy. In CT26 cells, the expression of Biot2 was downregulated by Biot2-shRNA. It also promoted G1 phase arrest, the expression of p16 and p21, and cell apoptosis. In the mouse model, the tumor volume and the expression of PCNA of the Biot2-shRNA group significantly decreased. These results suggest that silencing Biot2 in CT26 cells by RNA interference can inhibit cell growth in vitro and in vivo. It also induces cell cycle arrest in the G1 phase and apoptosis throughout regulation of p16 and p21. Taken together, our data demonstrate that Biot2 can be a potential target of gene therapy.
Key words: Biot2, RNA interference, CT26, Gene therapy
INTRODUCTION
Gene therapy, as an important area of tumor biological treatment, has received more and more attention and application. The purpose of this treatment is to replace or correct the human gene structure and function by its expression. Currently, gene therapy is widely used as an important means of treating cancer and genetic diseases (1). In 2000, Fischer Professor Necker Hospital in Paris, France, treated 17 children with SCID disease with gene therapy; however, after that, three patients had symptoms of leukemia (2–4). Thus, for gene therapy, we need to consider the problem of security. The main body of the target gene in the vector is accomplished by gene transfer vectors; this includes viral vectors and nonviral vectors. For the security of gene therapy, nonviral vectors get more attention and concern, such as targeting of liposomes, polymers, and liposome/polymer/DNA complexes, coupled with combined electric pulses, ultrasound, and other new technologies, greatly improving the efficiency of import, and nonviral vector targeting (5,6) will be an important direction for future development of nonviral vectors.
RNA interference (RNAi) is defined by short double-stranded RNA (dsRNA)-induced specific degradation of homologous mRNA and leads to the phenomenon of posttranscriptional gene silencing (7–9). Owing to the use of RNAi, technology can specifically remove or turn off the expression of specific genes; the technology has been widely used to explore gene function studies and viral infection in malignant tumors (10–12), neurodegenerative diseases (13,14), cardiovascular disease, and other gene therapy (15). RNAi in experimental studies is mainly through small interfering RNA and short hairpin RNA (shRNA) (16,17).
The Biot2 gene was first discovered and identified by the SEREX serological screening assay (18,19) and named Biot2 by our laboratory. RT-PCR results revealed that Biot2 had high expression in the testis tissue but low or no expression in the heart, liver, spleen, lung, and kidney. It gives us a hint that Biot2 may have tumor specificity and maybe a cancer-testis antigen (19). In our previous study, the pcDNA3.1-Biot2-transfected NIH3T3 cell line successfully promoted the proliferation of cells (20,21), and targeting interference mouse Biot2 gene can inhibit the growth of tumor cells in mice (22). Clinical research on human Biot2 genes was by real-time PCR experiments. Biot2 expression levels in cancer are higher than the adjacent tissues in cervical cancer, endometrial cancer, and colorectal cancer (23). By analysis and detection of clinical patient tissue sections, with increasing tumor stages, the levels of Biot2 expression also significantly increased (24,25). Biot2 genes may be related to tumorigenesis, as the higher the Biot2 expression levels, the worse is the prognosis in early patients with colorectal cancer (26). This indicates that human Biot2 may be a novel cancer therapy target and a predictive marker for tumor growth in cancer. The mouse Biot2 gene has high homology with the human Biot2 gene, and we have a preliminary study in Biot2 for tumor growth; however, the molecular mechanism of Biot2 remains poorly understood because of the lack of animal and in vitro models. Because of its specificity to tumors and its important functions, the new gene has attracted increasing attention as a candidate for tumor gene therapy.
MATERIALS AND METHODS
Cell Culture
CT26 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS), 10 mM L-Glu, and 5 mg/ml penicillin/streptomycin.
Animals
Male BALB/c mice aged between 6 and 8 weeks were purchased from HFK Bioscience (Beijing, China). All mice were specifically pathogen-free animals and were maintained in our animal research facility in a dedicated aseptic environment, as approved by the institutional protocol and guidelines. All of the mice that were involved in this study were approved by the Animal Care and Use Committee of our institute.
Reagents
Mouse anti-cyclinD1 (1:1,000), and rabbit anti-p21 (1:1,000) were purchased from Protein Tech (Suzhou, China). Rabbit anti-CDK2 (1:1,000), rabbit anti-p16 (1:1,000), rabbit anti-p27 (1:1,000), and mouse anti-PCNA (1:1,000) were purchased from BOSTER (China). Rabbit anti-caspase-3 (1:1,000), rabbit anti-AKT (1:1,000), and rabbit anti-p-AKT (1:1,000) were bought from Abcam (Cambridge, UK). Rabbit anti-p53 and mouse anti-β-actin were bought from Santa Cruz Biotechnology (Heidelberg, Germany). TransIT®-2020 transfection reagent was purchased from Mirus (USA). A Cell-Light™ EdU Apollo®567 In Vitro Imaging Kit was purchased from Riobo (Guangdong, China). Cationic liposomes were produced by Sichuan University, State Key Laboratory of Biotherapy. The recombinant plasmid Biot2-shRNA was bought from Jingsai (Wuhan, China). The plasmid was prepared using the Endofree Plasmid Giga kit (Qiagen, Chatsworth, CA, USA).
Transient Transfections
CT26 cells were seeded in six-well plates at a density of 1 × 105 cells/well. Twenty-four hours later, 2.5 ml complete growth medium was added per well. Transfections were carried out using 250 µl of reduced-serum medium with 2.5 µg shBiot2 according to the protocol for the Trans IT-2020 reagent. Cells were incubated for 24–48 h before use.
Quantification of shBiot2 by RT-PCR
Total RNA was extracted from the transfected cells with the RNA simple total RNA kit according to the manufacturer’s instructions, and 1 µg of RNA was used to synthesize cDNA using the first-strand cDNA synthesis kit. The RT-PCR was performed using an RT-PCR kit (Life Technologies) according to the manufacturer’s protocol. Data were normalized according to the level of GAPDH expression in each sample. Briefly, 1 µg of total RNA was used as the template and then reverse-transcribed by using an Oligo primer. The cDNA was further amplified in the RT-PCR with a universal reverse primer and a specific forward primer. The sequences of the primers were as follows: Biot2_F1: 5′-AAAATGAAATGTGCAAAGCATCC-3′ and Biot2_R1: 5′-TAGGCAGGTCACCAATGAAG-3′; M-GAPDH F1: 5′-TCCACCACCCTGTTGCTGTA-3′, M-GAPDH R1: 5′-ACCACAGTCCATGCCATCAC-3′. The reaction solution consisted of 2.0 µl diluted RT-PCR product, 0.5 µM of each paired primer. The PCR procedure included predenaturation at 94°C for 1 min and then 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final extension of 10 min at 72°C. All amplifications were carried out within the linear range of the assay. The reaction products were separated on 2% agarose gel and stained with 1 mg/ml Goldview and quantified by IS-1000 digital imaging system; the results were confirmed in at least three replicate experiments.
EDU Cell Proliferation Assay
CT26 cells (1 × 104 cells/well) were seeded in 24-well plates (n = 3 wells per treatment) and then transfected 24 h later with shBiot2. EdU assays were then carried out according to the manufacturer’s protocol. Cells were imaged under an inverted fluorescence microscope.
Cell Cycle Analysis by Flow Cytometry
CT26 cells (1 × 106) were harvested and washed twice with cold phosphate-buffered saline (PBS). Cells then were stained with 500 µl of propidium iodide (PI) staining solution (50 mg/ml PI, 0.1% Triton X-100, 200 mg/ml DNase-free RNase in PBS) for 30 min at room temperature in the dark. Ten thousand events per sample were acquired using a FACS-scan flow cytometer (Becton-Dickinson, San Jose, CA, USA), and the percentage of cells in G0/G1, S, G2/M, and sub-G2/M phases of the cell cycle were determined using CELL Quest software (Becton-Dickinson).
Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick-End Labeling Assay (TUNEL)
In order to evaluate the number of cells undergoing apoptosis, TUNEL staining was performed with the use of an in situ cell death detection kit (DeadEnd™ Fluorometric TUNEL System, Promega, Madison, WI, USA) following the instructions of the manufacturer. Five fields were randomly selected and analyzed under a light microscope (200×). The apoptosis index was obtained by counting the number of apoptotic cells in each field.
Western Blotting
All protein samples for Western blot analysis were resolved by SDS-PAGE on 12% gels and then transferred to nitrocellulose membranes, which were then blocked for 1 h at room temperature in Tris-buffered saline (TBS) containing 0.1% Tween 20 and 5% fat-free milk. All primary antibody incubations were performed overnight at 4°C. Secondary antibody incubations were carried out at room temperature for 1 h. Secondary antibodies conjugated with horseradish peroxidase were used for detection by enhanced chemiluminescence (Super Signal; Pierce, Rockford, IL, USA) or ECL Plus (Amersham Pharmacia Biotech, Buckinghamshire, UK) according to the manufacturer’s instructions.
Animal Studies
BALB/c mice received subcutaneous implantations of a total of 100 µl of 1 × 105 of CT26 cells in their rear left footpad. The interval between the extraction of ascites and the final inoculation into the mice was less than 1 h.
When the mean diameter of the tumors reached 3 mm 5 days after inoculation, the mice were separated at random into two groups: the Lip-HK group (n = 10 mice) received 10 µg of the HK plasmid and 50 µg liposome complexes (final volume = 100 µl), and the Lip-Biot2-shRNA group (n = 10 mice) received 10 µg of the shRNA plasmid and 50 µg liposome complexes (final volume = 100 µl). Mice were treated once daily for 10 days via injections into the tail vein.
Tumor size was monitored by measuring the largest and perpendicular diameters using a caliper once every 3 days. Tumor volume was calculated according to the following formula: V = length × width2 × 0.52. All of the data are represented as means ± standard error (SE). After 25 days of treatment, mice were sacrificed and were isolated and fixed in 4% of neutral formalin for histological analysis.
Immunohistochemical Staining
Polycolonal goat anti-mouse PCNA (1:200; Santa Cruz Biotechnology) was used for immunohistochemistry (IHC) and was applied to paraffin-embedded material. The IHC staining was performed using the labeled streptavidin–biotin method. Sections of primary tumor were deparaffinized in xylol and rehydrated in graded alcohol series. Antigen retrieval was carried out by autoclaving sections in retrieval buffer (10 mM pH 6.0 EDTA citrate buffer) for 3 min. Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide at room temperature for 15 min. Nonspecific binding of reagents was quenched by incubation for 20 min in 5% normal serum, as determined by secondary antibodies. Sections were then probed with primary antibodies, after which the sections were incubated overnight at 4°C, followed by incubation with biotinylated secondary antibodies at 37°C for 45 min, washed with a slight agitation in PBS for 15 min, and incubated with avidin–biotin–horseradish peroxidase complex at 37°C for 45 min. Cell nuclei were gently counterstained with reformative Gill’s hematoxylin, and slides were dehydrated and mounted.
The results regarding microvessel density (MVD) and lymphatic microvessel density (LMVD) were expressed as the average value of five fields with the highest number of microvessels in each tumor section. Microvessel counting was performed with the use of a light microscope (400× magnification).
Statistical Analysis
All assays were conducted three times and found to be reproducible. Statistical significance was defined as p < 0.05. Results were expressed as mean ± standard deviation (SD). Statistical correlation of data between groups is checked for significance by Student’s t test. These analyses were performed using SPSS 13.0 software.
RESULTS
RNA Interference Reduced Biot2 Expression and Inhibited the Proliferation of CT26 Cells
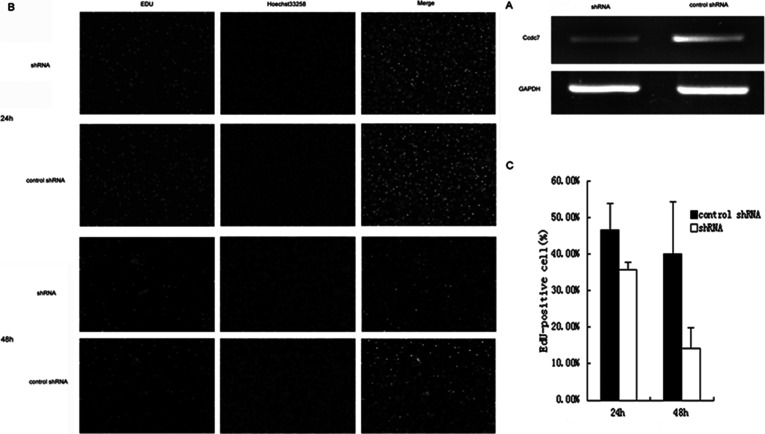
To investigate the interference efficiency of Biot2-shRNA, we transfected CT26 cells with sh-Biot2. As shown using real-time reverse transcription-polymerase chain reaction (RT-PCR), the expression of Biot2 decreased compared with that in the control cells after 24 h of growth (Fig. 1A). Early studies found that Biot2 gene downregulation can inhibit tumor cell growth. In this experiment, to further confirm the effects of sh-Biot2 on proliferation in CT26 cells, we used EdU assays to measure cell numbers and found a 10.78% ± 1.95% and 25.92% ± 5.59% decrease in growth compared with the control after 24 h and 48 h growth (p < 0.05) (Fig. 1B, C).
Figure 1.
Interference of Biot2 gene inhibits CT26 tumor cell growth in vitro. (A) Cells were transfected with control shRNA or shRNA (Biot2-shRNA). After transfection, the expression of control shRNA and shRNA was evaluated using RT-PCR. GAPDH was used as loading controls. (B) At 24 h after transfection with Biot2-shRNA, cell viability was estimated by EdU assay. Newly grown CT26 cells were stained by combined EdU and Hoechst 33258 staining. EdU-stained cells (left) were observed under a fluorescence microscope (200×), and cells stained with Hoechst 33258 (middle) were counted as a proliferation control (%). (C) EdU-positive cells. Each bar represents the mean ± SD of three independent experiments. *p < 0.05, compared with control group cells.
Biot2 Gene Downregulation Induced CT26 Cells G1 Arrest
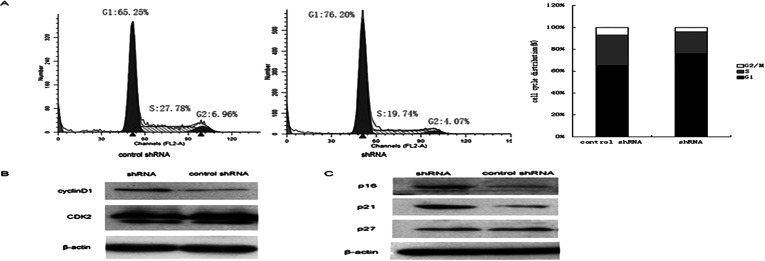
Biot2 gene downregulation inhibited the cell proliferation of CT26; furthermore, we want to know whether targeting interference Biot2 gene can inhibit the CT26 cell proliferation or not. We detected that when Biot2 gene is downregulated, CT26 cell DNA replication and cell cycle change. Transfected Biot2-shRNA and control-shRNA in CT26 cells, 48 h later, detected the DNA replication and cell cycle by flow cytometry. The Biot2-shRNA group had tumor cell cycle arrest in G1 phase. G1 phase cells increased by 11.0% compared to control-shRNA group (Fig. 2A). Then, we examined the expression of associated cell cycle regulatory proteins, kinase, and kinase-dependent kinase inhibitors (CKIs) in the cell cycle. Transfected Biot2-shRNA and the control-shRNA, after 48 h, detected the expression of cell cycle regulatory proteins by Western blot; compared to the control group, p21, p16, and cyclinD1 had significantly increased, and p27 and CDK2 had no significant change (Fig. 2B, C).
Figure 2.
Biot2 genes downregulated arrest of CT26 cell cycle. The effect of Biot2-shRNA on inducing G1 cell cycle arrest in CT26 cells. After treatment with Biot2-shRNA for 48 h, (A) the cell cycle distributions were determined by flow cytometry. (B) The protein expressions of cycle regulatory proteins and cyclin-dependent kinase cyclin D1, CDK2 and (C) the expression of several CKIs, p16, p21, and p27, were measured by Western blotting.
Biot2 Gene Downregulation Promotes CT26 Cell Apoptosis
Programmed cell death usually occurs after cell cycle arrest. Biot2 gene downregulation led to CT26 cell arrest in G1 phase. In order to verify whether the Biot2 gene downregulation also caused CT26 cell apoptosis, we detected the apoptotic cells through a series of apoptosis detection methods. After transfecting Biot2-shRNA and control-shRNA for 48 h, Hoechst/PI double staining was used to detect the apoptotis of CT26 cells. Compared to the control group, the apoptosis rate increased 15.96% ± 0.85% in the Biot2-shRNA group (p < 0.05) (Fig. 3A). TUNEL assay detected apoptosis of CT26 cells; TUNEL-positive rate increased 6.36% ± 0.89% of the Biot2-shRNA group (p < 0.05) (Fig. 3B). Western blot detected the expression of caspase-3, an apoptosis marker protein. Cleaved caspase-3 expression of the Biot2-shRNA group was significantly increased compared to the control group (Fig. 3C).
Figure 3.
Biot2 downregulation leads to CT26 cell apoptosis. shRNA induces apoptosis via downregulating the expression of Biot2 genes in CT26 cells. (A) A representative apoptosis analysis for programmed cell death. Harvesting the cells that transfected for 48 h, then were stained with Hoechst 33258 and PI. Apoptosis cells (100×) (*p < 0.05). (B) TUNEL assay tested CT26 cells apoptosis (100×), nuclear stained with Hoechst 33258 as a control. (C) Western blot to detect the expression of caspase-3 and cleaved caspase-3, β-actin was used as loading control. Results are shown as the mean of three independent experiments (±SD indicated by error bar).
Biot2 Gene Downregulation Inhibition of CT26 Cell Proliferation and Promotion of Cell Apoptosis May Be Relevant With p53 Gene
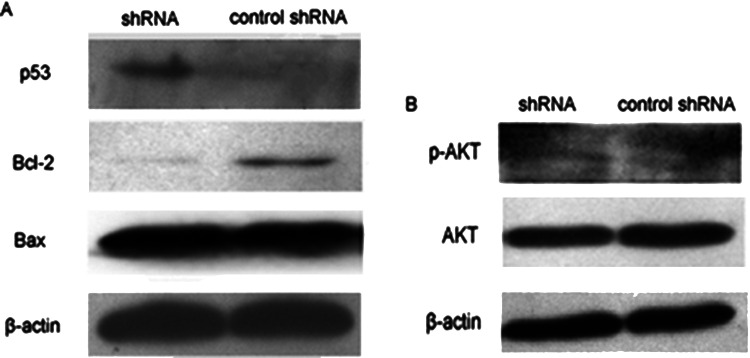
We conclude that Biot2 gene downregulation inhibited CT26 cell proliferation and apoptosis, and may be related to two factors, p53 and AKT: p21 mainly through the upstream p53 and AKT-mediated pathway expression protein. In order to verify the speculation, transfected Biot2-shRNA and control-shRNA, 48 h later, used Western blot to detect expression of the proteins. Compared with the control group, the p53 protein of the Biot2-shRNA group significantly increased, and the expression of apoptosis family members Bcl-2 decreased significantly, too, but proapoptotic family member Bax expression did not change significantly (Fig. 4A); p-AKT also did not change significantly (Fig. 4B). These data show that inhibiting of CT26 cell proliferation and promoting cell apoptosis are likely due to the increase in p53 activity.
Figure 4.
Biot2-shRNA inhibits cell proliferation and promotes apoptosis of CT26, resulting in p53 upregulation. CT26 cells were transfected with Biot2-shRNA for 48 h, then analyzed by Western blotting to determine the protein level of P53, Bcl-2, Bax (A) and the protein expression of AKT and p-AKT (B). These proteins were assayed by Western blotting. Equal amounts of total cellular protein (20 µg) were resolved by 12% SDS-PAGE. β-Actin was used as loading control.
Biot2 Gene Downregulation Inhibits CT26 Cell Growth In Vivo
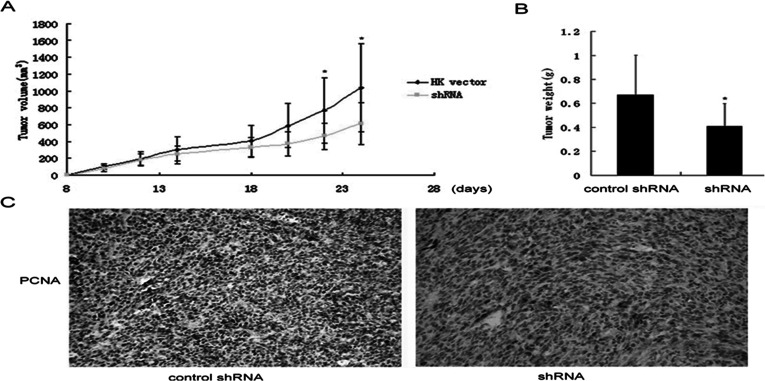
Because of these dramatic effects of Biot2 knockdown in cells, we next used a mouse model to investigate the effects of Biot2 inhibition in tumors. Mice bearing CT26-derived tumors were treated daily with Lip-HK or Lip-CCDC7-shRNA, and tumor size was analyzed. Importantly, tumors in the Biot2-shRNA group were significantly smaller than those in the control group. Moreover, animals treated with Lip-Biot2-shRNA showed a significant decrease in the average tumor volume when compared with the controls (p < 0.05) (Fig. 5A). After 25 days, the mice were sacrificed and the tumors were weighed. Wet weight of the tumors in the control group was 0.67 ± 0.32 g; the other group was 0.41 ± 0.16 g (p < 0.05) (Fig. 5B). The Biot2-shRNA group tumors weighed significantly less than the controls. IHC staining of proliferating cell nuclear antigen (PCNA) expression in the tumor was found. PCNA-positive cells in the Biot2-shRNA group were significantly reduced compared to the controls (Fig. 5C). Thus, these data supported that knockdown of Biot2 by RNA interference suppressed the proliferation and oncogenic potential of colorectal cancer cells and decreased tumor formation in mouse.
Figure 5.
Biot2-shRNA inhibits the tumor growth in vivo. (A) Curve of tumor growth. Lip-Biot2-shRNA inhibits the growth of tumors. Mice were treated with HK or Biot2-shRNA, and tumors were measured as described in Materials and Methods. Tumor volumes are shown as the means ± standard errors (SEs); *p < 0.05 relative to controls. (B) Tumor weight. Mice were sacrificed, and tumors were collected and weighed immediately. Data show the means ± SEs; *p < 0.05. (C) IHC staining of PCNA in tumor samples. Typical images were taken under a light microscope (magnification 400×).
DISCUSSION
Gene therapy is an important research direction, as the current biological treatment of cancer has been widely used; however, its existence also has its own problems. Security and targeted gene therapy are two issues that need to be considered. Researchers are constantly looking for new carrier transport systems and new gene therapy, hoping to solve these problems. In this study, we targeted interference of the Biot2 gene, Biot2 expression downregulated in CT26 cells. We also explored the possibility of Biot2 gene as a therapy target in vitro and in vivo. From our results, we found that interference of the Biot2 gene can inhibit the proliferation of CT26 cells in vitro and in vivo. Therefore, these data provide important insights into the role of Biot2, which plays a key role in the growth of CT26 tumors, and it is a potential target for gene therapy.
Cell cycle regulation plays an important role in the growth of tumor cells (27,28). During the cell’s carcinogenesis, the cycle regulation of the expression of proteins and CKI cycle disorders may cause abnormality and the abnormal expression of tumor-suppressor genes, which cause cells to become cancerous (29,30). G1 cell cycle arrest is a key part of the regulation. G1 is regulated by a variety of regulatory proteins and CKIs, such as cyclinD1, cyclinE, p16, p21, p27, and so on. Among the above, CKIs are especially critical and important in the regulation of G1 (30,31). In our experiments, after Biot2 gene downregulation, CT26 cells were arrested in G1 phase; the two major proteins p16 and p21 in G1 phase expression increased, but there was no significant change in p27. The result indicates that the cycle arrest is caused mainly by p16 and p21. p16 and p21 are the key regulated proteins during the G1 phase, and they play a key regulatory role in the development and progression of tumors. In 50% of human tumors, the deletion, point mutation, and methylation of the p16 gene caused genetic inactivation, and overexpression of p16 had a poor prognosis of tumors (32). p21 has also been reported to have a role in many tumors (33,34).
Cells will slowly die after the tumor cell cycle arrest. Therefore, we studied the Biot2 gene downregulation and CT26 cell apoptosis occurrence. We found that the Biot2 gene downregulation can promote CT26 cell apoptosis. Therefore, it may provide a reliable basis for the Biot2 gene as a therapy target.
Cyclin-dependent kinase inhibitor p21 is mainly regulated by p53 and AKT pathways; the two pathways are critical for inhibiting tumor growth and promoting apoptosis. In our study, we examined the expression of p53 and p-AKT protein; when the Biot2 gene was downregulated, p53 gene expression increased, but the p-AKT did not change significantly. In addition, we also tested the expression of the inhibitor of apoptosis protein family member Bcl-2 and proapoptotic family member Bax; Bcl-2 decreased significantly and Bax did not. p53 as an important tumor-suppressor gene has been the focus of tumor biology treatment. p53 is the cell cycle regulation station from G1 to S phase transition, and it regulated downstream protein p21 and cyclin-dependent kinase to arrest the cell cycle in G1 phase. Before DNA replication, failed accurately repaired DNA will lead to p53-dependent apoptosis. p53 plays an important role in tumor growth; when abnormally expressed or mutated, cells begin malignant transformation. In the experimental studies, after Biot2 gene downregulation, p53 expression increased. We hypothesize that CT26 cell growth inhibition and apoptosis are likely due to the increase in p53 activity, but it needs further research. Moreover, we found that the expression of Bcl-2 increased, but not in Bax. Thus, the specific mechanism of Biot2 downregulation leading to CT26 cells undergoing apoptosis remains to be further studied.
By this study, we demonstrated the possibility of the Biot2 gene as cancer therapy and its role in tumor growth. Biot2 downregulation can inhibit the growth of cells by experiments in vitro and in vivo. However, it should be noted that this study has some disadvantages. First of all, the result is only for one of our cell lines. In order to prove the role of Biot2 in tumors, we also need to repeat our studies in other tumor cell lines. Second, the specific mechanism of Biot2 downregulation leading to CT26 cell apoptosis is not clear. Is it mitochondria mediated or receptor mediated? It is also our future research direction. Finally, the specific mechanism of Biot2 genes and p53 is not very deep; it needs more experiments to explore. The evidence we have to prove the possibility of Biot2 as a target therapy gene may not be enough, so further studies in this direction will be summarized in our next study. But through the studies, it is confirmed that some of the features and functions of Biot2 genes are slowly being understood and also provides some basis for gene therapy of Biot2 gene.
ACKNOWLEDGMENTS
This study was funded by the National Key Basic Research Program of China (2010 CB 529900) and the National Natural Science Foundation of China (81102062, 30801048). All authors have contributed directly to the planning, execution, or analysis of the work reported or to the writing of the paper.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Matsushime H.; Ewen M. E.; Strom D. K.; Kato J. Y.; Hanks S. K.; Roussel M. F.; Sherr C. J. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71(2):323–334; 1992. [DOI] [PubMed] [Google Scholar]
- 2. Kelly B. L.; Wolfe K. G.; Roberts J. M. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc. Natl. Acad. Sci. USA 95(5):2535–2540; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Z.; Wang C.; Prendergast G. C.; Pestell R. G. Cyclin D1 functions in cell migration. Cell Cycle 5(21):2440–2442; 2006. [DOI] [PubMed] [Google Scholar]
- 4. Fu M.; Wang C.; Li Z.; Sakamaki T.; Pestell R. G. Cyclin D1: Normal and abnormal functions. Endocrinology 145(12):5439–5447; 2004. [DOI] [PubMed] [Google Scholar]
- 5. Li Z.; Wang C.; Jiao X.; Lu Y.; Fu M.; Quong A. A.; Dye C.; Yang J.; Dai M.; Ju X.; Zhang X.; Li A.; Burbelo P.; Stanley E. R.; Pestell R. G. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol. Cell. Biol. 26(11):4240–4256; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Z.; Jiao X.; Wang C.; Jum X.; Lum Y.; Yuan L.; Lisanti M. P.; Katiyar S.; Pestell R. G. Cyclin D1 induction of cellular migration requires p27(KIP1). Cancer Res. 66(20):9986–9994; 2006. [DOI] [PubMed] [Google Scholar]
- 7. Zhou X.; Zhang Z.; Yang X.; Chen W.; Zhang P. Inhibition of cyclin D1 expression by cyclin D1 shRNAs in human oral squamous cell carcinoma cells is associated with increased cisplatin chemosensitivity. Int. J. Cancer 124(2):483–489; 2009. [DOI] [PubMed] [Google Scholar]
- 8. Biliran H. Jr.; Wang Y.; Banerjee S.; Xu H.; Heng H.; Thakur A.; Bollig A.; Sarkar F. H.; Liao J. D. Overexpression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgene-expressing pancreatic tumor cell line. Clin. Cancer Res. 11(16):6075–6086; 2005. [DOI] [PubMed] [Google Scholar]
- 9. Kuroda Y.; Sakai A.; Tsuyama N.; Katayama Y.; Munemasa S.; Asaoku H.; Okikawa Y.; Nakaju N.; Mizuno M.; Ogawa K.; Nishisaka T.; Matsui H.; Tanaka H.; Kimura A. Ectopic cyclin D1 overexpression increases chemosensitivity but not cell proliferation in multiple myeloma. Int. J. Oncol. 33(6):1201–1206; 2008. [PubMed] [Google Scholar]
- 10. Roy P. G.; Pratt N.; Purdie C. A.; Baker L.; Ashfield A.; Quinlan P.; Thompson A. M. High CCND1 amplification identifies a group of poor prognosis women with estrogen receptor positive breast cancer. Int. J. Cancer 127(2):355–360; 2010. [DOI] [PubMed] [Google Scholar]
- 11. Wang M. T.; Chen G.; An S. J.; Chen Z. H.; Huang Z. M.; Xiao P.; Ben X. S.; Xie Z.; Chen S. L.; Luo D. L.; Tang J. M.; Zhang X. C.; Wu Y. L. Prognostic significance of cyclinD1 amplification and the co-alteration of cyclinD1/pRb/ppRb in patients with esophageal squamous cell carcinoma. Dis. Esophagus 127(2):664–670; 2012. [DOI] [PubMed] [Google Scholar]
- 12. Zhao X.; Song T.; He Z.; Tang L.; Zhu Y. A novel role of cyclinD1 and p16 in clinical pathology and prognosis of childhood medulloblastoma. Med. Oncol. 27(3):985–991; 2010. [DOI] [PubMed] [Google Scholar]
- 13. Sui L.; Dong Y.; Ohno M.; Sugimoto K.; Tai Y.; Hando T.; Tokuda M. Implication of malignancy and prognosis of p27(kip1), Cyclin E, and Cdk2 expression in epithelial ovarian tumors. Gynecol. Oncol. 27(3):56–63; 2001. [DOI] [PubMed] [Google Scholar]
- 14. Yue H.; Jiang H. Y. Expression of cell cycle regulator p57kip2, cyclinE protein and proliferating cell nuclear antigen in human pancreatic cancer. World J. Gastroenterol. 11(32):5057–5060; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zolota V.; Sirinian C.; Melachrinou M.; Symeonidis A.; Bonikos D. S. Expression of the regulatory cell cycle proteins p21, p27, p14, p16, p53, mdm2, and cyclin E in bone marrow biopsies with acute myeloid leukemia. Correlation with patients’ survival. Pathol. Res. Pract. 203(4):199–207; 2007. [DOI] [PubMed] [Google Scholar]
- 16. Gil J.; Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Mol. Cell. Biol. 7(9):667–677; 2006. [DOI] [PubMed] [Google Scholar]
- 17. Kim W. Y.; Sharpless N. E. The regulation of INK4/ARF in cancer and aging. Cell 127(2):265–275; 2006. [DOI] [PubMed] [Google Scholar]
- 18. Naora H.; Yang Y. Q.; Montz F. J.; Seidman J. D.; Kurman R. J.; Roden R. B. A serologically identified tumor antigen encoded by a homeobox gene promotes growth of ovarian epithelial ceils. Proc. Natl. Acad. Sci. USA 98(7):14060–14065; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J. L.; Dunbar P. R.; Gileadi U.; Jäger E.; Gnjatic S.; Nagata Y.; Stockert E.; Panicali D. L.; Chen Y. T.; Knuth A.; Old L. J.; Cerundolo V. Identification of NY-ESO-l pepfide analogues capable of improved stimulation of tumor-reactive CTL. J. lmmunol. l165(2):1948–955; 2001. [DOI] [PubMed] [Google Scholar]
- 20. Wang C. T.; Zhang P.; Peng F.; Ruan X. Z.; Yang H. S. Effects of the novel tumor/testis antigen, Biot2, on proliferation of NIH3T3 cell. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 23(8):704–706; 2007. [PubMed] [Google Scholar]
- 21. Wang C. T.; Zhang P.; Peng F.; Yang H. S. Construction of the prokaryotic expression vector and expression of murine testis-specific gene Biot2. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 23(5):406–408; 2007. [PubMed] [Google Scholar]
- 22. Wang C. T.; Zhang P.; Wang Y. S.; Ruan X. Z.; Li Z. Y.; Peng F.; Yang H. S.; Wei Y. Q. RNA interference against Biot2, a novel mouse testis-specific gene, inhibits the growth of tumor cells. Cell Mol. Biol. Lett. 14(3):363–376; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yangmei S.; Xiang H.; Hongxin D.; Hanshuo Y.; Feng P.; Yuping X.; Yuquan W.; Xia Z. Expression of human Biot2 and its potential function on carcinogenesis in endometrial cancer. Acta Obstet. Gynecol. Scand. 86(12):1503–1509; 2007. [DOI] [PubMed] [Google Scholar]
- 24. Wang H.; Zhang P.; Wang C. T. Analysis of expression pattern of a novel testis-highly expressed gene Biot2-L and the primary study on its role in testis development. Sichuan Da Xue Xue Bao Yi Xue Ban. 40(5):853–856; 2009. [PubMed] [Google Scholar]
- 25. Shen Y. M.; He X.; Deng H. X.; Xie Y. P.; Wang C. T.; Wei Y. Q.; Zhao X. Overexpression of the hBiot2 gene is associated with development of human cervical cancer. Oncol. Rep. 25(1):75–80; 2011. [PubMed] [Google Scholar]
- 26. Shen Y. M.; Arbman G.; Sandström P.; Gullstrand P.; Wei Y. Q.; Zhang H.; Rosell J.; Olsson B.; Peng F.; Yang H. S.; Wang C. T.; Sun X. F. Novel gene hBiot2 is an independent prognostic factor in colorectal cancer patients. Oncol. Rep. 27(2):376–382; 2012. [DOI] [PubMed] [Google Scholar]
- 27. Sherr C. J.; Roberts J. M. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18(22):2699–2711; 2004. [DOI] [PubMed] [Google Scholar]
- 28. Esteller M.; Corn P. G.; Baylin S. B.; Herman J. G. A gene hypermethylation profile of human cancer. Cancer Res. 61(8):3225–3229; 2001. [PubMed] [Google Scholar]
- 29. Ortega S.; Malumbres M.; Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim. Biophys. Acta 1602(1):73–87; 2002. [DOI] [PubMed] [Google Scholar]
- 30. Gonzalez S.; Serrano M. A new mechanism of inactivation of the INK4/ARF locus. Cell Cycle 5(13):1382–1384; 2006. [DOI] [PubMed] [Google Scholar]
- 31. Sharpless N. E. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat. Res. 576(1–2):22–38; 2005. [DOI] [PubMed] [Google Scholar]
- 32. Lang J. C.; Borchers J.; Danahey D.; Smith S.; Stover D. G.; Agrawal A.; Malone J. P.; Schuller D. E; Weghorst C. M.; Holinga A. J.; Lingam K.; Patel C. R.; Esham B. Mutational status of overexpressed p16 in head and neck cancer: Evidence for germline mutation of p16/p14ARF. Int. J. Oncol. 21(2):401–408; 2002. [DOI] [PubMed] [Google Scholar]
- 33. Sherr C. J.; Roberts J. M. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13(12):1501–1512; 1999. [DOI] [PubMed] [Google Scholar]
- 34. Iaquinta P. J.; Lees J. A. Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 19(6):649–657; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]