Abstract
We previously discovered (−)-DHMEQ as a selective inhibitor of NF-κB, and it was shown to suppress many cancer and inflammation models in animals. (−)-DHMEQ directly binds to NF-κB components to inhibit DNA binding, and moreover, it often inhibits nuclear translocation of NF-κB. The mechanism of inhibiting nuclear translocation has been elucidated for RelB, a main noncanonical NF-κB component. However, it was not elucidated for p65, a main canonical NF-κB component. In the present research, we studied how (−)-DHMEQ inhibits nuclear localization of p65. First, (−)-DHMEQ inhibited p65 nuclear accumulation in adult T-cell leukemia MT-2 cells in which canonical p65 is constitutively activated. But there was no change in the stability and importin-α3 affinity of p65. Then, we prepared a p65 mutant protein with Arg35Ala and Tyr36Ala (AA) mutations having no DNA-binding ability in HeLa cells. The p65 AA mutant showed reduced nuclear localization without changing the stability and importin affinity. Taken together, the mechanism of inhibition is different between RelB and p65, and inhibition of p65 nuclear localization is likely to be due to the inhibition of DNA binding changing the equilibrium between the nuclear and cytoplasmic amounts of p65.
Key words: (−)-DHMEQ, Canonical NF-κB, Nuclear localization, Adult T-cell leukemia, DNA binding, IκBα
INTRODUCTION
NF-κB is a transcription factor and has a role in immune activation and tissue stability in the physiological state. However, its overactivation often causes inflammation and enhances cancer progression (1). NF-κB promotes expressions of many inflammatory cytokines, antiapoptosis proteins, and adhesion proteins. Extracellular stimulation of macrophages or dendritic cells activates cellular NF-κB to enhance inflammation. NF-κB is often constitutively activated in leukemia and solid cancer cells, including adult T-cell leukemia MT-1 and MT-2 cells (2) and human breast carcinoma MDA-MB-231 cells (3). When NF-κB is activated in cancer cells, it increases drug resistance and secretion of prometastatic proteins, increasing malignancy.
NF-κB/Rel-family proteins include p65, RelB, c-Rel, p50, and p52. Each NF-κB/Rel-family member participates in the formation of different homo- or heterodimers. NF-κB is classified as a canonical one important for innate immunity, general inflammation, and cancer and a noncanonical one important for B-cell maturation and autoimmune diseases. The canonical NF-κB complex is a heterodimer that is typically composed of p65 and p50, the noncanonical one of RelB and p52. In the case of canonical NF-κB, IκBα prevents nuclear translocation by masking the nuclear localization signal (NLS). A cellular stimulant such as TNF-α induces proteasome-mediated degradation of IκB, which results in the liberation of the p65/p50 heterodimer followed by its rapid translocation into the nucleus (4). Dissociation of the NF-κB component from IκB enables it to interact with importin family proteins (5,6). Importins are the major cargo carriers from the cytoplasm into the nucleus. Six importin-α family members (α1, α3, α4, α5, α6, α7, and α8) have been identified in humans. NF-κB, consisting of p65 and p50, is known to be transported into the nucleus preferentially by importin-α3 or importin-α4 (6).
(−)-DHMEQ (Fig. 1A) was designed and synthesized as a specific inhibitor of NF-κB (7). It was widely used in many disease models and shown to suppress various inflammation and cancer models in animal experiments without any toxicity (8). DHMEQ is synthesized as its racemic form (9), which can be separated into (−)- and (+)-DHMEQ (10). After the chiral separation, (−)-DHMEQ is about 10 times more effective than the (+)-isomer (9). As the mechanism of inhibition, first, it was reported to inhibit nuclear translocation of NF-κB (11), but later, it was found to inhibit DNA binding more directly (12). It covalently binds to a specific Cys residue of each NF-κB/Rel family protein, except p52, to inhibit the DNA binding. In the case of p65, (−)-DHMEQ covalently binds to Cys38 selectively (12). On one hand, it binds to Cys144 of RelB to prevent DNA binding (12). Recently, we found that (−)-DHMEQ induces instability of RelB and also decreases the affinity to importin-α5 in addition to the inhibition of DNA binding (13). Moreover, mutation to inhibit DNA binding in RelB resulted in instability and loss of importin-α5 affinity. However, these effects of (−)-DHMEQ have not been elucidated for canonical NF-κB, which is more popular.
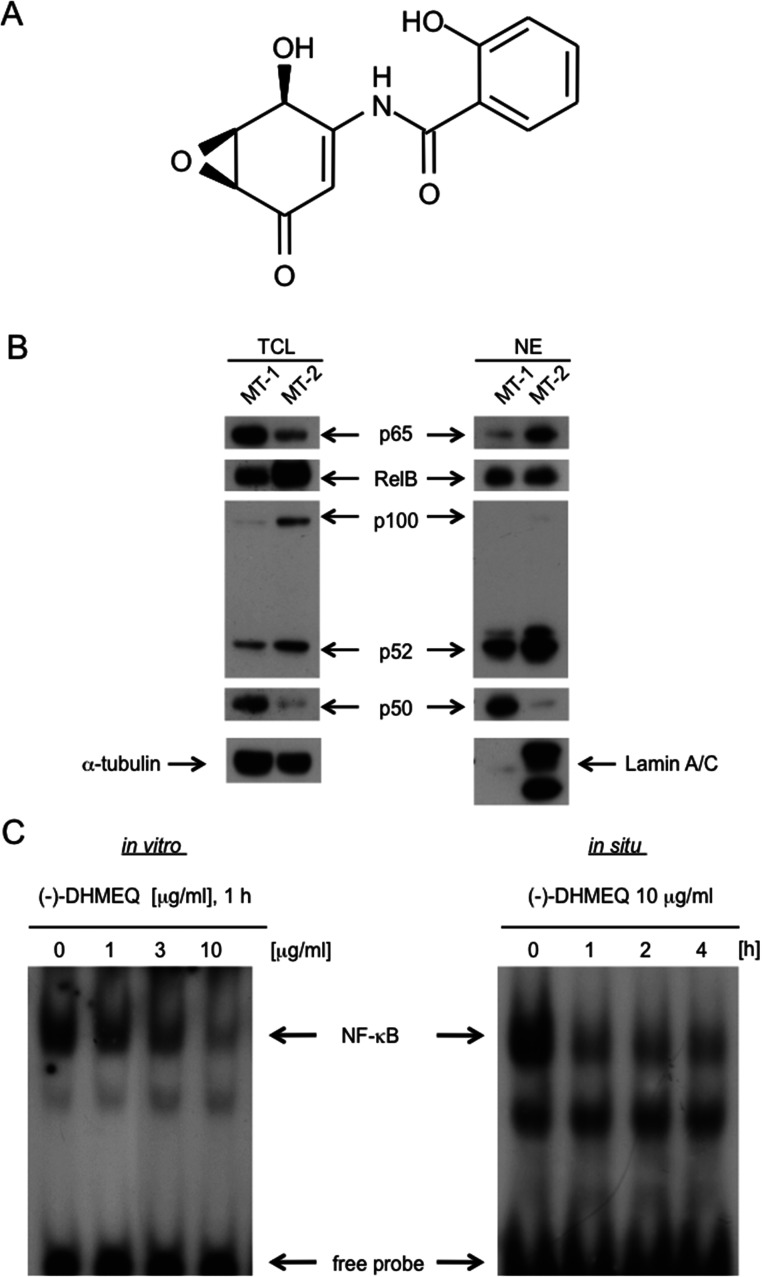
Figure 1.
Cellular localization of p65 in adult T-cell leukemia MT-1 and MT-2 cells. (A) Structure of (−)-DHMEQ. (B) Total and nuclear amount of p65 in MT-1 and MT-2 cells. Total cell lysates (TCL) or nuclear extracts (NE) were analyzed by Western blotting using each antibody. Laminin A/C and α-tubulin were used for nuclear and total cell markers, respectively. (C) Inhibition of NF-κB by (−)-DHMEQ in vitro and in situ. Left: Inhibition of NF-κB-DNA binding by (−)-DHMEQ. Nuclear extracts prepared from MT-2 cells were treated with 10 μg/ml (−)-DHMEQ or DMSO for 1 h, and they were subjected to electrophoretic mobility shift assay (EMSA) with a radiolabeled consensus κB oligo-DNA. Right: Inhibition in cultured cells. Cultured MT-2 cells were treated with 10 μg/ml (−)-DHMEQ for the indicated periods. Nuclear extracts were subjected to EMSA with a radiolabeled consensus κB DNA.
In the present research, we have found that the effect of (−)-DHMEQ on p65 is quite different from that on RelB. (−)-DHMEQ inhibited nuclear translocation of p65, but did not affect the stability and importin-α affinity in p65. The p65 mutant having no DNA-binding ability showed no change in stability and no change in importin affinity.
MATERIALS AND METHODS
Materials
(−)-DHMEQ was synthesized in our laboratory as described previously (9). Mouse monoclonal anti-p65 NF-κB (sc-8008), anti-p50 NF-κB (sc-8414), anti-p52 NF-κB (sc-7386), anti-c-Rel NF-κB (sc-6955), anti-GST (sc-138), and rabbit polyclonal anti-IκBα (sc-203) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit monoclonal anti-RelB (#4922), anti-NF-κB2 p100/p52 (#3017), and polyclonal anti-Lamin A/C (#2032) antibodies were purchased from Cell Signaling (Beverly, MA, USA). Mouse monoclonal anti-FLAG and anti-α-tubulin antibodies were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell Culture
MT-2 cells were grown in RPMI-1640 medium (Nissui, Tokyo, Japan) containing 10% heat-inactivated FBS, 100 μg/ml kanamycin, 100 units/ml penicillin G, 300 μg/ml L-glutamine, and 2.25 g/L NaHCO3. HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Nissui) supplemented with 10% calf serum, 200 μg/ml kanamycin, 100 units/ml penicillin G, 600 μg/ml L-glutamine, and 2.25 g/L NaHCO3.
Preparation of Total Cell Extracts, Cytoplasmic Extracts, and Nuclear Protein Extracts
Whole cell extracts were suspended in lysis buffer [25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1 mM EDTA, 0.1% SDS (w/v), 1% NP-40 (v/v), 1% sodium deoxycholate (w/v), 1 mM PMSF, and 0.3 µM aprotinin], sonicated, and centrifuged for 10 min at 14,000 rpm, and the supernatant was used as the total cell extracts.
Nuclear extracts were prepared according to the method of Andrews and Faller (14). Cells (2 × 105) were grown in 60-mm dishes and incubated with the desired chemicals. They were then harvested and washed with phosphate-buffered saline (PBS), suspended in 400 μl of buffer A [10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, and 0.2 mM PMSF] and incubated on ice for 15 min. Nuclei were pelleted by centrifugation for 5 min at 500 × g, and the supernatant was used as the cytoplasmic fraction. Then, the pellets were resuspended in 400 μl of buffer A and incubated on ice for 15 min. After the incubation, the nucleus fractions were pelleted by centrifugation for 5 min at 500 × g, resuspended in 40 μl of buffer C [20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, and 25% glycerol (v/v)], sonicated, and centrifuged for 5 min at 14,000 rpm. The resulting supernatant was used as the nuclear extract.
Western Blotting
Whole cell extracts, cytoplasmic extracts, and nuclear extracts were boiled in Laemmli loading buffer [58.3 mM Tris-HCl (pH 6.8), 1.7% SDS (w/v), 6% glycerol (v/v), and 0.83% 2-mercaptoethanol] and separated on 10–12.5% SDS-polyacrylamide gel electrophoresis. Then the proteins were transferred to 200 mA for 1 h onto Hybond-P membranes (GE Healthcare). Nonspecific binding was blocked for 30 min at room temperature with TBST buffer [20 mM Tris-HCl (pH 7.6), 135 mM NaCl, and 0.1% Tween 20 (v/v)] containing 3% (w/v) nonfat milk. After a short washing in TBST, the membrane was incubated in a 1:1,000 dilution of either primary antibody overnight at 4°C. Primary antibodies were diluted in 3% nonfat milk. In the case when anti-p65 NF-κB antibody was used, it was diluted in solution 1 (Toyobo, Osaka, Japan). After washing for 10–30 min in TBST, the bound antibodies were visualized with horseradish peroxidase-conjugated antibodies against rabbit or mouse IgG (diluted at 1:5,000 in TBST containing 3% nonfat milk) using the ECL Western blotting system (GE Healthcare).
Semiquantitative RT-PCR
Total cellular RNA from cells was prepared with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. cDNAs were synthesized from total RNA by using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) in a total volume of 20 μl. The resultant cDNA was amplified by PCR with rTaq DNA polymerase (Takara Bio, Shiga, Japan) using the following primers: total p65, 5′-CAGGCTCCTGTGCGTGTCTC-3′ (forward) and 5′-CTGGCTGATCTGCCCAGAAG-3′ (reverse); exo-p65, 5′-TACAAGGATGACGACGATAAGGCC-3′ (forward) and 5′-TAGAAGCCATCCCGGCAGTC-3′ (reverse); IκBα, 5′-CAAGGAGCTGCAGGAGATCC-3′ (forward) and 5′-CCAAGGACACCAAAAGCTCC-3′ (reverse); β-actin, 5′-CTTCGAGCAAGAGATGGCCA-3′ (forward) and 5′-CCAGACAGCACTGTGTTGGC-3′ (reverse). The expression of β-actin was measured as an internal control.
Electrophoretic Mobility Shift Assay (EMSA)
DNA-binding reactions and electrophoretic mobility shift assays were carried out as described previously (13). Samples of 20 μ1 containing 5 μg of nuclear cell extracts were incubated with 1 μg poly(dI–dC), and 10,000 cpm 32P-labeled probe (oligonucleotide containing NF-κB binding site), binding buffer [15 mM Tris-HCl (pH 7.0), 75 mM NaCl, 1.5 mM EDTA, 1 mM DTT, 7.5% glycerol (v/v), and 1.5% NP-40 (v/v)] for 20 min at room temperature in this mixture. DNA–protein complexes were separated from free DNA on 4% native polyacrylamide gels in TBE buffer [22.5 mM Tris-HCl (pH 8.3) and 0.5 mM EDTA]. The DNA probes used for NF-κB binding were purchased from Promega (Madison, WI, USA). The following sequences were used as NF-κB probes: 5′-AGTTGAGGGGACTTTCCCAGG C-3′ and 5′-GCCTGGGAAAGTCCCCTCAACT-3′. These oligonucleotides were labeled with [γ-32P]ATP (3000 Ci/mmol; GE Healthcare, Little Chalfont, UK) by use of T4 polynucleotide kinase (Takara, Ohtsu, Japan) and purified by passage through a NICK column (GE Healthcare).
Preparation of Designed Plasmids
The cDNA encoding full-length importin-α3 was digested with BamHI and XhoI. It was inserted into BamHI and XhoI sites of pGEX-6P-1 vector (GE Healthcare). The following primers were used: 5′-TTTTTTCTCGAGATGGCGGACAACGAGAAA-3′ (forward) and 5′-TTTTTTGCGGCCGCCTAAAACTGGAACCCT-3′ (reverse). The cDNA encoding full-length IκBα was digested with BamHI and NotI. It was inserted into BamHI and NotI sites of pGEX-6P-1 vector (GE Healthcare). The following primers were used: 5′-TTTTTTGGATCCATGTTCCAGGCGGCCGAGCGCCCC-3′ (forward) and 5′-TTTTTTGCGGCCGCTCATAACGTCAGACGCTGGCCT-3′ (reverse). The cDNA encoding full-length p65 was digested with BamHI and XhoI. It was inserted into BamHI and XhoI sites of Flag-tagged pCMV-tag2B vector (Stratagene, La Jolla, CA, USA). The following primers were used: 5′-TTTTGGATCCATGGACGAACTGTTCCCCCTCATC-3′ (forward) and 5′-TTTTCTCGAGTTAGGAGCTGATCTGACTCAGCAG-3′ (reverse). The cDNA encoding c-Myc-tagged full-length IκBα was EcoRI and NotI. It was inserted into EcoRI and NotI sites of pCI-neo vector (Promega). The following primers were used: 5′-TCAGAAGAGGATCTGATGTTCCAGGCGGCC-3′ (forward 1), 5′-GAACAGAAACTCATCTCAGAAGAGGATCTG-3′ (forward 2), 5′-TTTTTTGAATTCATGGAACAGAAACTCATC-3′ (forward 3), and 5′-TTTTTTGCGGCCGCTCATAACGTCAGACGCTGGCCT-3′ (reverse). The cDNA encoding c-Myc-tagged full-length IκBα was amplified in three installments.
Site-Directed Mutagenesis
Site-directed mutagenesis for the p65 Cys38Ser (p65 C38S) mutant and p65 Arg35Ala, Tyr36Ala (p65 AA) mutant was carried out by the overlap extension method (15). The following primers were used (altered nucleotides are underlined in bold): p65 C38S mutant, 5′-CCGCTACAAGTCCGAGGGGCGCTCCGCGG-3′ (forward) and 5′-CCGCGGAGCGCCCCTCGGACTTGTAGCGG-3′ (reverse); p65 AA mutant, 5′-CAGCGGGGCATGCGCTTCGCCGCCAAGTGCGAG-3′ (forward) and 5′-GGAGCGCCCCTCGCACTTGGCGGCGAAGCGCAT-3′ (reverse). The cDNA was inserted into BamHI and XhoI sites of Flag-tagged pCMV-tag2B vector (Stratagene).
Recombinant Protein Purification
Recombinant proteins were expressed in E. coli BL21 (Promega) as GST fusion proteins by induction at 25°C for 2.5 h with 100 μM isopropyl-1-thio-β-D-galactopyranoside (GE Healthcare). The bacteria were lysed in a sonication buffer [PBS containing 0.1% NP-40 (v/v), 1 μM DTT, and 1% protease inhibitors (v/v) (Nacalai Tesque, Kyoto, Japan)], sonicated for 10 min on ice, and centrifuged for 10 min at 14,000 rpm at 4°C. The supernatant was mixed with 500 μl of equilibrated glutathione-Sepharose 4B (GE Healthcare) at 4°C for 1 h followed by washing five times with the sonication buffer. For preparation of cell lysates, MT-1 cells and HeLa cells transfected with plasmids were incubated with or without (−)-DHMEQ. The cells were then lysed in L buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 0.1% protease inhibitors (v/v) (Nakalai Tesque)] on ice for 30 min and centrifuged for 10 min at 14,000 rpm at 4°C.
Transfection With Plasmids
HeLa cells (1.5 × 105 cells/dish) were grown in 60-mm dishes. The cells were transfected with the desired DNA by using Lipofectamine LTX (Invitrogen, Grand Island, NY, USA) as described by the manufacturer. After 24 h, the transfected cells were treated with the desired chemical prior to Western blotting.
GST Pull-Down Assay
GST fusion proteins were expressed in E. coli BL21 and purified using glutathione-Sepharose beads. Briefly, the GST fusion proteins were coupled to glutathione beads at 4°C for 1 h and then washed 10 times with the L buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1% (w/v), Triton X-100]. The immobilized GST fusion proteins were incubated with the cell lysates for 2 h at 4°C. The beads were washed with the L buffer five times, and then the protein complexes were separated in 10% SDS-PAGE, followed by Western blotting.
RESULTS
Constitutive Activation of p65 in Adult T-Cell Leukemia MT-2 Cells
Both adult T-cell leukemia (ATL)-derived cell lines MT-1 and MT-2 show constitutively activated NF-κB activity (16), possibly due to the viral Tax activity (17). We compared p65 nuclear localization between MT-1 and MT-2 cells. As a result, p65 was found to be more accumulated in the nucleus of MT-2 cells than in MT-1 cells (Fig. 1B). So we employed MT-2, and (−)-DHMEQ inhibited the DNA binding of NF-κB prepared from the nuclear extract of MT-2 cells as shown in Figure 1B. It also inhibited NF-κB when added to the cultured MT-2 cells (Fig. 1C). Thus, we employed MT-2 cell line to study the effect of (−)-DHMEQ on canonical NF-κB nuclear translocation.
Inhibition of p65 Nuclear Translocation by (−)-DHMEQ in MT-2 Cells
(−)-DHMEQ lowered the nuclear localization of p65 in a dose- and time-dependent manner (Fig. 2A, B). The total p65 amount did not change by addition of (−)-DHMEQ (Fig. 2C). Therefore, (−)-DHMEQ will likely not affect the stability of p65. Next, we studied how (−)-DHMEQ inhibits nuclear translocation. Importin-α3 is necessary for the TNF-α-induced nuclear translocation of p65 (6). Employing the GST pull-down assay, we found that treatment of p65 with (−)-DHMEQ did not reduce the binding affinity of p65 to importin-α3, as shown in Figure 2D. This observation is quite different from the effect of (−)-DHMEQ on RelB, in which binding of (−)-DHMEQ to RelB markedly reduced the affinity to importin-α5 (13).
Figure 2.
Inhibition of p65 nuclear accumulation by (−)-DHMEQ in MT-2 cells. (A) Inhibition of p65 nuclear accumulation by (−)-DHMEQ (time course). The cells were treated with (−)-DHMEQ for the indicated periods. The cytoplasmic (CE) or nuclear (NE) extracts were analyzed by Western blotting using each antibody. (B) Inhibition of p65 nuclear accumulation by (−)-DHMEQ in MT-2 cells (dose dependency). (C) Effect of (−)-DHMEQ on total amount of p65. Total cell lysates were analyzed by Western blotting using antibodies against p65. (D) Effect of (−)-DHMEQ on the affinity of p65 to importin-α3. Total cell lysates from MT-2 cells treated with 10 μg/ml (−)-DHMEQ were subjected to GST pull-down assays using the purified recombinant GST fusion proteins. Coprecipitated p65 was detected by Western blotting. Application of equal amounts of GST-fusion input proteins was confirmed by Western blotting with antibodies against GST, p65, and α-tubulin.
Preparation of p65 Mutant Defective in DNA Binding and Expression in HeLa Cells
We have prepared two mutants of p65, one without DNA binding and another with stronger DNA-binding abilities. It was reported that p65 with its Arg35 mutated to Ala cannot bind to DNA (18). Arg35 and Tyr36 in p65 are essential amino acids for DNA binding, forming hydrogen bonds (18). Cys38 exists just between Arg35/Tyr36 and DNA, which would clearly explain how (−)-DHMEQ inhibits p65-DNA binding. In the case of p52, its Arg54 mutated to Ala, and its Tyr55 to Ala cannot bind to DNA (19), and these two residues are conserved in all the Rel family proteins. Thus, we newly prepared Arg35Ala and Tyr36Ala double mutant (AA mutant) that should not bind to DNA. We then transfected wild, AA mutant, or Cys38Ser mutant DNA into HeLa cells. As shown in Figure 3A and B (normalized for the protein input), the AA mutant largely lost the DNA-binding ability, while the Cys38Ala mutant showed higher DNA-binding ability, as reported previously (20).
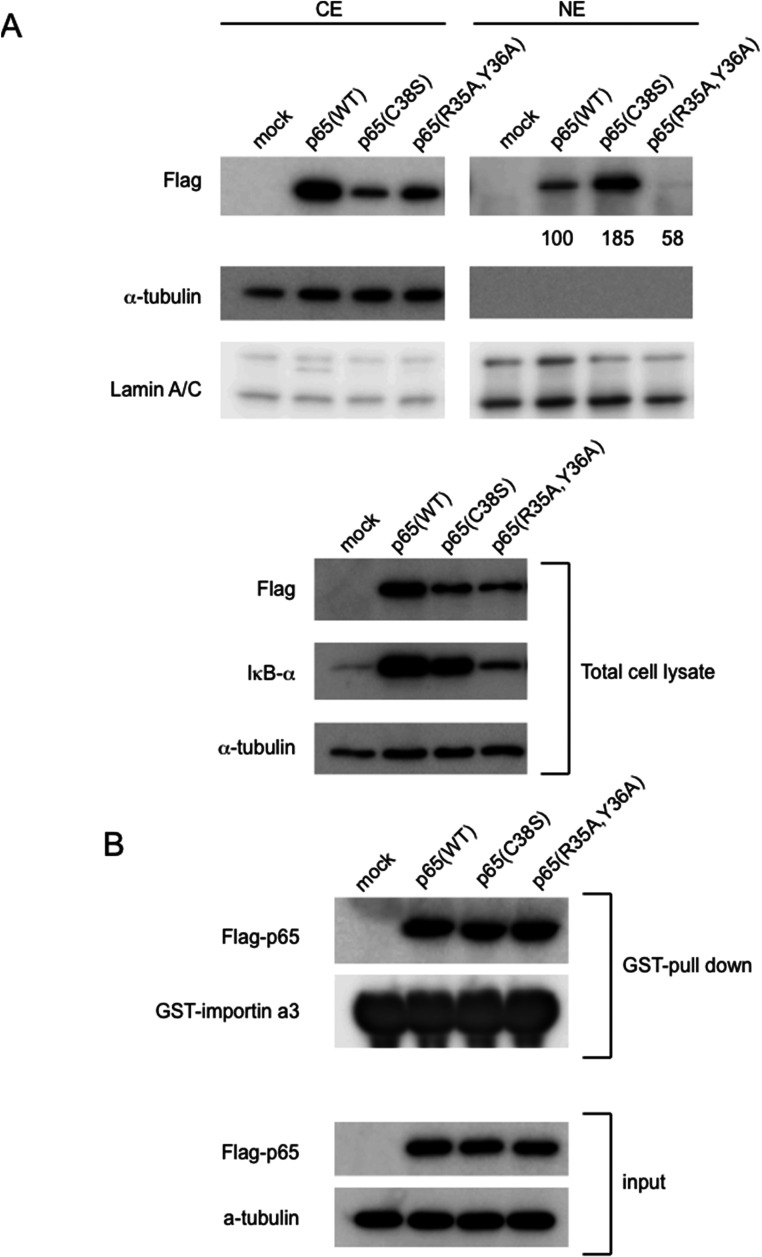
Figure 3.
Preparation of Arg35Ala and Tyr36Ala double mutant (AA mutant) of p65 and transfection to HeLa cells. (A) p65 AA mutant loses DNA-binding ability. HeLa cells were transiently transfected with plasmids encoding Flag epitope-tagged p65 (WT) and two mutants (Cys38Ser and AA mutant). As a control, a vector plasmid (mock) was transfected (EMSA, upper). The total extracts were analyzed by Western blotting using antibody against Flag (middle). Each content of tubulin is also shown (lower) (B) Quantification of DNA-binding activity. For quantification, the band intensities were normalized to α-tubulin. Equal amounts (5 μg) of nuclear extracts were subjected to EMSA with a radiolabeled consensus κB site oligo DNAs. The band intensities were normalized with the intensities of Flag-p65 protein levels.
Unlike in the case of RelB (13), the p65 AA mutant cells showed similar protein and mRNA amounts as wild-type and Cys38Ser cells, and the stability of p65 did not change by AA mutation.
Decrease in p65 Nuclear Accumulation by AA Mutation in HeLa Cells
Next we compared the nuclear accumulation of each mutant in HeLa cells. As in the case of RelB, the p65 AA mutant cells showed much lowered protein amounts in the nucleus (Fig. 4A). The Cys38Ser mutant of p65 showed higher accumulation than the wild type. Then, we examined the affinity to importin-α3 that is essential for p65 nuclear translocation. Unlike in the case of RelB, the affinity to importin-α3 did not change in p65, as shown in Figure 4B.
Figure 4.
Effect of AA mutation on p65 nuclear accumulation, stability, and affinity to importin-α3 in HeLa cells. (A) Effect on nuclear accumulation. The cytoplasmic (CE) and nuclear (NE) extracts were analyzed by Western blotting using antibodies against Flag and IκBα. Total cell lysates were analyzed by Western blotting using antibodies against Flag and IκBα. (B) Effect on affinity to importin-α3. Total cell lysates were subjected to GST pull-down assay using the purified recombinant GST fusion proteins. Coprecipitated p65 was detected by Western blotting using anti-p65 antibody. Equal amounts of GST-fusion proteins and input proteins were confirmed by Western blotting with antibodies against GST, p65, and α-tubulin.
Involvement of IκBα in the Regulation of p65 Subcellular Localization in Breast Carcinoma MDA-MB-231 Cells
These observations suggest that nuclear accumulation of p65 would be regulated by its DNA-binding ability. If so, there should be an effective export system in the cells. IκBα contributes to the cytoplasmic localization and nuclear export of canonical NF-κB (21,22). Then, we studied the effect of IκBα on the inhibition of p65 nuclear translocation by (−)-DHMEQ. The breast carcinoma MDA-MB-231 cell line has constitutively activated p65, and (−)-DHMEQ inhibited the NF-κB activity in EMSA (23). However, (−)-DHMEQ could not inhibit p65 nuclear translocation, as shown in Figure 5A, and p65 nuclear translocation rather increased by treatment with (−)-DHMEQ in 0.5–2.0 h. Both p65 and IκBα amounts did not change in the total lysate. We then prepared c-Myc-tagged IκBα and overexpressed this protein in MDA-MB-231 cells. As a result, (−)-DHMEQ inhibited p65 nuclear accumulation, as shown in Figure 5B. Thus, IκBα would be involved in the inhibition of p65 nuclear translocation by (−)-DHMEQ. Loss of DNA binding by (−)-DHMEQ should change this equilibrium in the presence of IκBα (Fig. 5C).
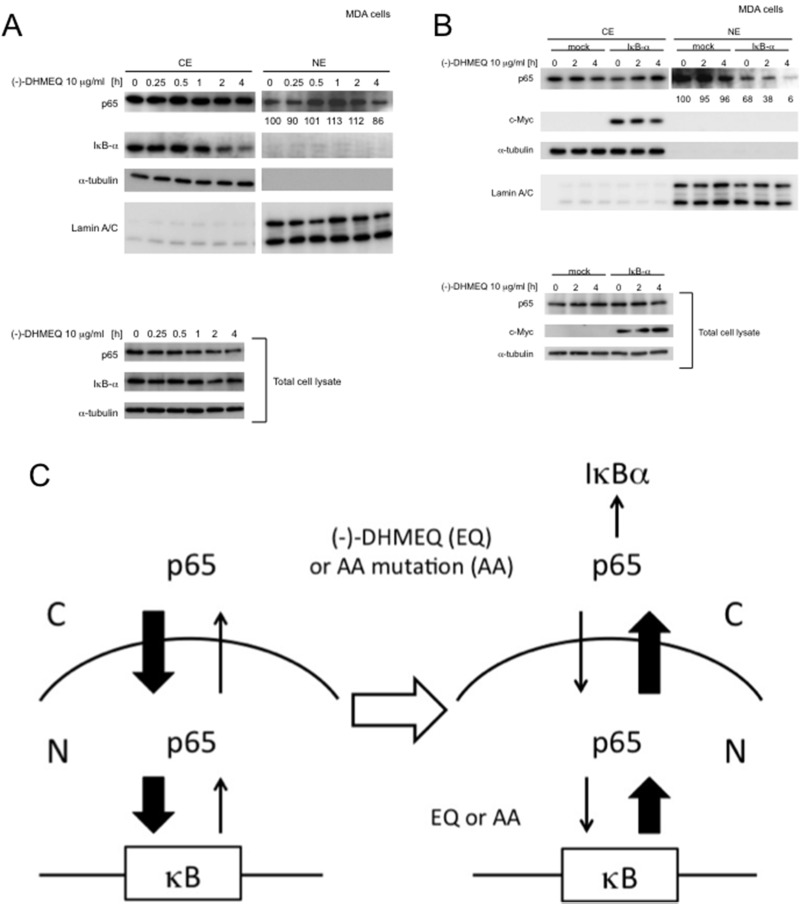
Figure 5.
IκBα would be essential for inhibition of p65 nuclear accumulation by (−)-DHMEQ in MDA-MB-231 cells. (A) (−)-DHMEQ does not inhibit nuclear accumulation in MDA-MB-231 cells. MDA-MB-231 (MDA) cells were treated with 10 μg/ml (−)-DHMEQ for the indicated periods. The CE and NE were analyzed by Western blotting using antibodies against p65 and IκBα. Total cell lysates were also analyzed by Western blotting using antibodies against p65 and IκBα. (B) Recovery of sensitivity to (−)-DHMEQ by overexpression of IκBα. MDA-MB-231 cells were transiently transfected with plasmids encoding c-Myc epitope-tagged IκBα. As a control, a vector plasmid was transfected. After 24 h of transfection, the cells were treated with 10 μg/ml (−)-DHMEQ for the indicated periods. The cytoplasmic (CE) or nuclear (NE) extracts were analyzed as in (A). Total cell lysates were also analyzed by Western blotting using antibodies against c-Myc and p65. (C) Possible mechanism for inhibition of p65 nuclear accumulation by (−)-DHMEQ or AA mutation. C, cytoplasm; N, nucleus.
DISCUSSION
There are a lot of similarities in p65 and RelB as members of Rel family proteins, for example, possessing DNA-binding sites as Arg35Tyr36 and Arg141Tyr142, respectively, in the similar conformation. The DNA-binding abilities of both proteins are inhibited by (−)-DHMEQ by its binding to Cys38 in p65 and to Cys144 in RelB (12). It is an unexpected finding that the behavior of each protein is very different, except for the loss of DNA binding. We previously reported that (−)-DHMEQ treatment or AA mutation in RelB results in the decreases of cellular stability and importin affinity (13). On the other hand, we have found in the present research that (−)-DHMEQ treatment or AA mutation in p65 never changes the cellular stability and importin affinity.
Although (−)-DHMEQ binding or AA mutation does not lower the cellular stability or affinity to importin-α3, these treatments decrease nuclear accumulation of p65. To explain this observation, we assume that there would be equilibrium between cytoplasmic and nuclear p65, and its DNA binding should shift it to the nuclear localization. Then, loss of DNA binding should shift it to the cytoplasmic localization, as shown in Figure 5C. For this mechanism, a p65 export system is essential. One of the known factors for p65 cytoplasmic localization is IκBα, which exists in the cytoplasm (24). We found that loss of DNA-binding ability in p65 by (−)-DHMEQ did not lower the nuclear localization in highly malignant breast carcinoma MDA-MB-231 cells (Fig. 5A). NF-κB is constitutively and strongly activated in this cell line. We have assumed that the IκBα/p65 ratio is too low to export full p65 in this cell line, and we carried out overexpression of IκBα by DNA transfection. Overexpression of this protein resulted in the increase in sensitivity to (−)-DHMEQ on inhibition of nuclear accumulation (Fig. 5B). This observation would support the hypothesis of nuclear/cytoplasmic p65 equilibrium.
There are several exportins, including exportin-1 (CRM1), in the general protein exporting system. CRM1 would also contribute to the nuclear export of NF-κB (25). IκBα would be necessary for CRM1-mediated nuclear export of the IκBα/Rel protein complex (26, 27).
In conclusion, we have demonstrated that (−)-DHMEQ treatment or AA mutation resulted in inhibition of nuclear accumulation. The inhibitory mechanism would be mainly due to the change of IκBα-mediated nuclear/cytoplasmic equilibrium (Fig. 5C). (−)-DHMEQ is now being developed as an anti-inflammatory and anticancer agent. These findings may lead to a better understanding of the therapeutic activity of (−)-DHMEQ.
ACKNOWLEDGMENTS
The authors wish to thank Dr. Masatoshi Takeiri, Kyoto University, for the valuable suggestions. This work was supported in part by grants from the programs Grants-in-Aid for Scientific Research (B 23310163 and C 26350975). This work was also supported in part by MEXT-Supported Program for the Strategic Research Foundation at Private Universities, which is for Aichi Medical University 2011–2015 (S1101027).
REFERENCES
- 1. Karin M.; Greten F. R. NF-κB: Linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5:749–759; 2005. [DOI] [PubMed] [Google Scholar]
- 2. Sun S. C.; Ballard D. W. Persistent activation of NF-κB by the tax transforming protein of HTLV-1: Hijacking cellular IkappaB kinases. Oncogene 18:6948–6458; 1999. [DOI] [PubMed] [Google Scholar]
- 3. Nakshatri H.; Bhat-Nakshatri P.; Martin D. A.; Goulet R. J. Jr.; Sledge G. W. Jr. Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Mol. Cell. Biol. 17:3629–3639; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen L. F.; Greene W. C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell. Biol. 5:392–401; 2004. [DOI] [PubMed] [Google Scholar]
- 5. Fagerlund R.; Melen K.; Cao X.; Julkunen I. NF-κB p52, RelB and c-Rel are transported into the nucleus via a subset of importin alpha molecules. Cell Signal. 20:1442–1451; 2008. [DOI] [PubMed] [Google Scholar]
- 6. Fagerlund R.; Kinnunen L.; Kohler M.; Julkunen I.; Melen K. NF-κB is transported into the nucleus by importin α3 and importin α4. J. Biol. Chem. 280:15942–15951; 2005. [DOI] [PubMed] [Google Scholar]
- 7. Matsumoto N.; Ariga A.; To-e S.; Nakamura H.; Agata N.; Hirano S.; Inoue J.; Umezawa K. Synthesis of NF-κB activation inhibitors derived from epoxyquinomicin C. Bioorg. Med. Chem. Lett. 10:865–869; 2000. [DOI] [PubMed] [Google Scholar]
- 8. Umezawa K. Peritoneal NF-κB as a possible molecular target for suppression of various cancers and inflammation. Forum Immunopathol. Dis. Ther. 4:63–77; 2013. [Google Scholar]
- 9. Suzuki Y.; Sugiyama C.; Ohno O.; Umezawa K. Preparation and biological activities of optically active dehydroxymethylepoxyquinomicin, a novel NF-κB inhibitor. Tetrahedron 60:7061–7066; 2004. [Google Scholar]
- 10. Hamada M.; Niitsu Y.; Hiraoka C.; Kozawa I.; Higashi T.; Shoji M.; Umezawa K.; Sugai T. Chemoenzymatic synthesis of (2S,3S,4S)-form, the physiologically active stereoisomer of dehydroxymethylepoxyquinomicin (DHMEQ), a potent inhibitor on NF-κB. Tetrahedron 66:7083–7087; 2010. [Google Scholar]
- 11. Ariga A.; Namekawa J.; Matsumoto N.; Inoue J.; Umezawa K. Inhibition of TNF-α-induced nuclear translocation and activation of NF-κB by dehydroxymethyl-epoxyquinomicin. J. Biol. Chem. 277:27625–27630; 2002. [DOI] [PubMed] [Google Scholar]
- 12. Yamamoto M.; Horie R.; Takeiri M.; Kozawa I.; Umezawa K. Inactivation of NF-κB components by covalent binding of (-)-dehydroxymethylepoxyquinomicin to specific cysteine residues. J. Med. Chem. 51:5780–5788; 2008. [DOI] [PubMed] [Google Scholar]
- 13. Takeiri M.; Horie K.; Takahashi D.; Watanabe M.; Horie R.; Simizu S.; Umezawa K. Involvement of DNA binding domain in the cellular stability and importin affinity of NF-κB component RelB. Org. Biomol. Chem. 10:3053–3059; 2012. [DOI] [PubMed] [Google Scholar]
- 14. Andrews N. C.; Faller D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ho S. N.; Hunt H. D.; Horton R. M.; Pullen J. K.; Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59; 1989. [DOI] [PubMed] [Google Scholar]
- 16. Watanabe M.; Ohsugi T.; Shoda M.; Ishida T.; Aizawa S.; Maruyama-Nagai M.; Utsunomiya A.; Koga S.; Yamada Y.; Kamihira S.; Okayama A.; Kikuchi H.; Uozumi K.; Yamaguchi K.; Higashihara M.; Umezawa K.; Watanabe T.; Horie T. Dual targeting of transformed and untransformed HTLV-1-infected T cells by DHMEQ, a potent and selective inhibitor of NF-kappaB, as a strategy for chemoprevention and therapy of adult T-cell leukemia. Blood 106:2462–2471; 2005. [DOI] [PubMed] [Google Scholar]
- 17. Sun S. C.; Elwood J.; Beraud C.; Greene W. C. Human T-cell leukemia virus type I Tax activation of NF-kappa B/Rel involves phosphorylation and degradation of I kappa B alpha and RelA (p65)-mediated induction of the c-rel gene. Mol. Cell Biol. 14:7377–7384; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toledano M. B.; Ghosh D.; Trinh F.; Leonard W. J. N-terminal DNA-binding domains contribute to differential DNA-binding specificities of NF-kappa B p50 and p65. Mol. Cell Biol. 13:852–860; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fusco A. J.; Savinova O. V.; Talwar R.; Kearns J. D.; Hoffmann A.; Ghosh G. Stabilization of RelB requires multidomain interactions with p100/p52. J. Biol. Chem. 283:12324–12332; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia-Pineres A. J.; Castro V.; Mora G.; Schmidt T. J.; Strunck E.; Pahl H. L.; Merfort I. Cysteine 38 in p65/NF-κB plays a crucial role in DNA binding inhibition by sesquiterpene lactones. J. Biol. Chem. 276:39713–39720; 2001. [DOI] [PubMed] [Google Scholar]
- 21. Thanos D.; Maniatis T. NF-kappa B: A lesson in family values. Cell 80:529–532; 1995. [DOI] [PubMed] [Google Scholar]
- 22. Lee S. H.; Hannink M. The N-terminal nuclear export sequence of IκBα is required for Ran GTP-dependent binding to CRM1. J. Biol. Chem. 276:23599–23606; 2001. [DOI] [PubMed] [Google Scholar]
- 23. Matsumoto G.; Namekawa J.; Muta M.; Nakamura T.; Bando H.; Tohyama K.; Toi M.; Umezawa K. Targeting of NF-κB pathways by DHMEQ, a novel inhibitor of breast carcinomas. Anti-tumor and anti-angiogenic activity in vivo. Clin. Cancer Res. 11:1287–1293; 2005. [PubMed] [Google Scholar]
- 24. Tam W. F.; Lee L. H.; Davis L.; Sen R. Cytoplasmic sequestration of rel proteins by IkappaB alpha requires CRM1-dependent nuclear export. Mol. Cell. Biol. 20:2269–2284; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee S. H.; Hannink M. The N-terminal nuclear export sequence of IkappaBalpha is required for RanGTP-dependent binding to CRM1. J. Biol. Chem. 276:23599–23606; 2001. [DOI] [PubMed] [Google Scholar]
- 26. Huang T. T.; Kudo N.; Yoshida M.; Miyamoto S. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc. Natl. Acad. Sci. USA 97:1014–1019; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shih V. F.; Davis-Turak J.; Macal M.; Huang J. Q.; Ponomarenko J.; Kearns J. D.; Yu T.; Fagerlund R.; Asagiri M.; Zuniga E. I.; Hoffmann A. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-kappaB pathways. Nat. Immunol. 13:1162–1170; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]