Abstract
The observation of bile duct paucity is an important diagnostic finding in children, occurring in roughly 11% of pediatric liver biopsies. Alagille syndrome (ALGS) is a well-defined syndromic form of intrahepatic bile duct paucity that is accompanied by a number of other key features, including cardiac, facial, ocular, and vertebral abnormalities. In the absence of these additional clinical characteristics, intrahepatic bile duct paucity results in a broad differential diagnosis that requires supplementary testing and characterization. Nearly 30 years after ALGS was first described, genetic studies identified a causative gene, JAGGED1, which spearheaded over two decades of research aimed to meticulously delineate the molecular underpinnings of ALGS. These advancements have characterized ALGS as a genetic disease and led to testing strategies that offer the ability to detect a pathogenic genetic variant in almost 97% of individuals with ALGS. Having a molecular understanding of ALGS has allowed for the development of numerous in vitro and in vivo disease models, which have provided hope and promise for the future generation of gene-based and protein-based therapies. Generation of these disease models has offered scientists a mechanism to study the dynamics of bile duct development and regeneration, and in doing so, produced tools that are applicable to the understanding of other congenital and acquired liver diseases.
Keywords: Alagille syndrome (ALGS), bile duct paucity, JAG1, NOTCH2, Notch signaling, liver disease, cholestasis
Alagille syndrome (ALGS)
Overview and clinical manifestations
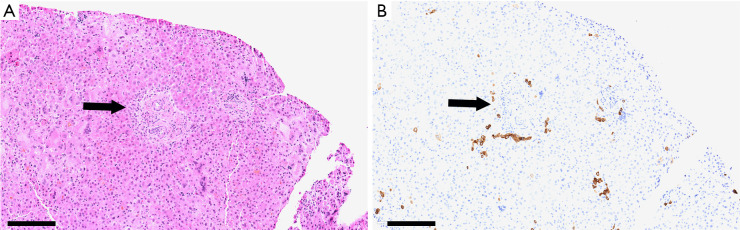
In 1969, Daniel Alagille first described several families manifesting cholestatic jaundice, and went on to publish a larger series characterizing the clinical phenotype of bile duct hypoplasia with cardiac manifestations (1). Around the same time, Watson and Miller observed a syndrome that they called arteriohepatic dysplasia, characterized by familial neonatal liver disease and pulmonary artery stenosis (2). Clinical criteria of arteriohepatic dysplasia, or ALGS as it is more commonly known, were later described by Alagille et al., and include bile duct paucity (Figure 1), cholestasis, cardiac involvement (heart murmur, peripheral pulmonic stenosis, and/or structural heart defects), characteristic facial appearance, eye findings (most commonly posterior embryotoxon), and skeletal anomalies (most commonly butterfly vertebrae) (3). Renal and vascular findings are also relatively common and are considered to be primary manifestations of the syndrome. ALGS has a strong genetic component with dominant inheritance and variable penetrance, and in 1997 the gene JAGGED1 (JAG1), was identified as the first disease gene (4,5). Genetic testing is now a key component of the diagnostic algorithm for ALGS.
Figure 1.
Liver biopsy from a 5-month old cholestatic infant with ALGS. (A) Hematoxylin and eosin stain of liver biopsy shows a portal tract (arrow) with branches of hepatic artery and portal vein but no bile duct present. (B) Cytokeratin 19 staining highlights minimal ductular reaction around portal tracts (arrow) throughout the biopsy. Scale bar =200 µM. ALGS, Alagille syndrome.
Genetics
The genetics of ALGS have been very well-defined and show a clear pattern of autosomal dominant Mendelian inheritance, with pathogenic variants identified in one of two genes in close to 97% of cases (6). These two genes, JAG1 and NOTCH2, encode a ligand-receptor pair, respectively, that participate in the Notch signaling pathway and are specifically critical in the formation of bile ducts, where JAG1 expression in the portal vein mesenchyme has been shown to be necessary in driving NOTCH2-expressing hepatoblasts to adopt a cholangiocyte fate (7,8). Failure to activate this signaling, which could arise from the presence of a pathogenic variant in either JAG1 or NOTCH2, drives hepatoblasts toward the hepatocyte lineage resulting in bile duct paucity, one of the classic features of ALGS (7-10).
JAG1
Pathogenic variants are most commonly identified in JAG1, occurring in 94.3% of individuals with ALGS (6). The overwhelming majority of JAG1 pathogenic variants are protein-truncating or result in whole or partial gene deletions or complex structural variants. A smaller percentage of individuals with ALGS have been found to have JAG1 missense variants (13%) (6). This observation has led to the hypothesis that ALGS is caused by haploinsufficiency of JAG1, and additional support for this pathophysiologic mechanism includes the finding that individuals with whole gene deletions have been shown to have phenotypes indistinguishable from those with intragenic variants, including missense variants (5,11-13).
Many functional studies have been published to investigate the basis of missense variant pathogenicity, and they have identified cellular trafficking defects, caused by errors in post-translational glycosylation of the mature JAG1 protein, and an inability of mutant JAG1 proteins to bind to and activate NOTCH2 (6,14-17). However, individual analysis of these missense variants has revealed differing degrees of impairment in their functional capacity. For instance, the L37S variant has been well-studied and shows both an inability of the mutant JAG1 to interact with NOTCH2 as well as impaired cellular trafficking, while the G274D variant has been shown to produce both an abnormally-glycosylated protein that is not expressed on the cell surface as well as a protein that can effectively induce Notch signaling (15,18). Other proposed pathogenic missense variants have been studied and found to retain complete wild type function, resulting in their reclassification as benign polymorphisms (15).
These studies have highlighted the importance of functionally validating each missense variant independently, and they have also led to the hypothesis that an alternate disease mechanism could exist for a minority of identified JAG1 variants, namely dominant negative pathogenesis. Studies to consider a dominant negative disease mechanism have shown that in some instances the mutant transcripts (mainly arising from missense alleles rather than protein-truncating alleles) are stably-expressed, and that when the resultant protein products are co-expressed with wild type JAG1 in a NOTCH2 luciferase reporter assay, there is a reduction in fluorescence readout compared to transfection of wild type alone, suggesting that the mutant protein may be able to compete with wild type protein for NOTCH2 binding (19,20). However, the physiological relevance of this in vitro model is debatable since it does not take into account cellular trafficking defects that would prevent JAG1 from interacting with NOTCH2. A separate study has shown that induced pluripotent stem cells (iPSCs) containing a heterozygous deletion of JAG1 resulted in the establishment of liver organoids that appeared normal whereas iPSCs containing a heterozygous nonsense variant were impaired in their ability to generate liver organoids, providing evidence that haploinsufficiency in this model does not alone result in a deficiency in the generation of liver organoids (21). The authors propose that a second genetic effector must be required for cases of ALGS caused by full gene deletions, however this hypothesis is not supported by the existing literature or the finding that a substantial number of individuals with ALGS have been found to carry a gene deletion (13%) while no unaffected individuals have been found to carry a gene deletion (6,21).
NOTCH2
Pathogenic variants are less commonly seen in NOTCH2, occurring in 2.5% of individuals with ALGS and accounting for a total of 19 identified pathogenic variants (6). Unlike with JAG1, the majority of NOTCH2 variants are missense (68%) (6), and the few that have been functionally-studied have shown a reduction in the ability of the resultant protein to undergo activation by JAG1 through luciferase reporter assays (22).
Phenotypic variability
A cardinal feature of ALGS has been the widely-reported variability in clinical phenotype among affected individuals. Classically, a diagnosis of ALGS is made when an individual presents with a minimum of three out of the five major disease features (including liver, heart, eye, vertebrae, and facies), however patients with fewer than three features have been identified on the basis of JAG1/NOTCH2 gene testing, and in all cases of ALGS, the severity of each clinical characteristic can vary greatly from mild to severe (23,24). Indeed, this variability extends to familial studies where related individuals with the same pathogenic variants have been found to have highly disparate disease presentations (13,22,23,25-30). Furthermore, there is no evidence to support any genotype-phenotype correlations, and even the earliest genetic reports have commented on a general lack of distinction between individuals who have whole gene deletions compared to intragenic variants (4,12,31).
This variable expressivity of ALGS has prompted a search for genetic factors that are capable of modifying the effects of pathogenic variants in JAG1 and NOTCH2 to either alleviate or worsen disease features. A number of genetic modifiers have now been proposed, and this field represents an ongoing and active area of research. Work has shown that genes involved in the post-translational addition of sugar moieties (glycosylation and fucosylation) to JAG1 protein can influence JAG1-mediated Notch signaling. The first group of genes identified to function in this capacity was a family of genes called the Fringe genes, which include Lunatic Fringe (LFNG), Radical Fringe (RFNG), and Manic Fringe (MFNG) (32). All three Fringe genes were found to affect the postnatal growth of bile ducts in a mouse model of Jag1 haploinsufficiency that was sensitive to Fringe gene dosage (32). Two subsequent murine studies focused on the liver-specific glycosyltransferase, Rumi (also known as Poglut1), which was found to lead to a dosage-sensitive effect on liver function in mouse models, however these two studies were in disagreement with the overall result of Rumi-mediated genetic modification, with one study providing evidence that Rumi expression was protective of liver function and a second study supporting the opposite effect (33,34). The two studies utilized different mouse models of ALGS, which was likely contributory to their discrepant results, however the overall conclusion that altered glycosylation of JAG1 could function to modify the effects of liver disease was universally supported. Interestingly, in their work, Thakurdas et al. identified a transcription factor, SOX9, that was differentially expressed in livers from animals that were heterozygous for Jag1 compared to livers from animals that were heterozygous for both Jag1 and Poglut1, leading to a new hypothesis, and a new report, with data supporting a role for SOX9 in affecting both the severity and progression of liver disease (35). One final proposed modifier, THBS2, was identified through a GWAS comparing individuals with mild or severe liver disease in whom a JAG1 variant had been identified (36). This study identified a genetic locus upstream of the THBS2 gene and provided evidence suggesting that individuals with a JAG1 variant and increased levels of THBS2 may have an increased risk of developing more severe liver disease (36).
Liver disease in ALGS
Liver disease in ALGS, like the other associated clinical manifestations, is highly variable, ranging from mild biochemical abnormalities to profound cholestasis associated with malnutrition, fat-soluble vitamin deficiencies, refractory pruritus, and xanthomas. In some large series, the rate of progression to liver transplantation was found to be 20% to 30% (27), although more recent reports suggest the numbers are higher, especially in patients presenting with cholestatic liver disease early in life (37). Patients with ALGS are also at risk for development of hepatocellular carcinoma (38).
ALGS patients with chronic cholestasis in early childhood present a management dilemma regarding the timing of interventions such as liver transplantation, since cholestasis is known to resolve or improve in a subset of individuals. Several studies have addressed this question through the identification of laboratory and clinical biomarkers that are predictive of later outcome. Kamath et al. analyzed laboratory data from 33 ALGS patients, stratified into mild and severe hepatic outcome groups, at greater than 10 years of age. They found that total bilirubin >6.5 mg/dL, conjugated bilirubin >4.5 mg/dL, and cholesterol >520 mg/dL in these children before 5 years of age were predictive of severe liver disease later in life (39). As has been found in other studies, JAG1 mutation type did not have any relationship to outcome. In a larger study, analysis of clinical data on 144 patients with a confirmed clinical or genetic diagnosis of ALGS (67 mild and 77 severe) determined that severe long-term hepatic outcome could be predicted by a model including presence of fibrosis on liver biopsy obtained before 5 years of age, presence of xanthomas on physical examination and total bilirubin >3.8 mg/dL between 1 and 2 years of age (40). Taken together, these data indicated that prolonged hyperbilirubinemia beyond the first few years of life in ALGS portends a more severe hepatic outcome.
Kamath et al. recently published a study of the outcomes of childhood cholestasis in 293 ALGS patients enrolled in a longitudinal study through the NIH-funded Childhood Liver Disease Research Network (ChiLDReN) (37). This cohort of patients was followed at pediatric referral centers, with evidence of liver involvement as a criterion for enrollment in the study. The study found that markers of cholestasis, such as total bilirubin and cholesterol, tended to peak in infancy and normalize over time, and xanthomas and pruritus improved. However, the authors found a high burden of complications related to portal hypertension by 20 years of age, with 36% cumulative incidence of ascites, GI bleeding in 16%, and splenomegaly in 50%. By age 20 years, 40% of the cohort met criteria for clinically evident portal hypertension [as defined in (41)]. Overall transplant-free survival in this cohort at the age of 18.5 years was 24%. These data suggest that in ALGS, cholestasis peaks in the first years of life but portal hypertension commonly develops later, with associated complications, and often requires liver transplantation during childhood. In contrast, signs of portal hypertension often manifest earlier in other cholestatic liver diseases, biliary atresia in particular. In one study of long-term outcomes in 219 children with biliary atresia surviving with native liver, splenomegaly was reported in 56% and thrombocytopenia in 64% at a mean age of 10 years (42). In another study of 163 biliary atresia patients surviving with native liver at a mean age of 9.2 years, 43 (26%) had already experienced at least one complication of portal hypertension, and 37 (23%) met criteria for definite portal hypertension based on platelet count and spleen size (43). Further studies will be necessary to understand the differences in pathophysiology that lead to disease progression in ALGS and other cholestatic disorders.
Growth failure is a common clinical manifestation in ALGS that was observed from the earliest descriptions of the syndrome (1,3) and is often severe enough to be an indication for liver transplantation. Growth deficits are likely multi-factorial in ALGS, related to the underlying genetic etiology, as well as cholestatic liver disease, malnutrition, vitamin deficiencies, and in some cases renal or cardiac disease. Several studies have shown more significant growth deficits in ALGS as compared with other cholestatic diseases such as biliary atresia. In one study, ALGS children had stunting of linear growth, low body weight and fat and muscle stores (44). Despite these growth deficits, resting energy expenditure has been found to be normal in ALGS (45). One large study examined outcomes of liver transplantation in 91 patients with ALGS compared with 236 with biliary atresia as reported in the Studies of Pediatric Liver Transplantation database (46). Growth failure was listed as a primary or secondary indication for liver transplantation in the majority of the ALGS patients. Growth failure was more profound in the ALGS patients, with a height z-score of -3 at the time of transplant in ALGS as compared with -1 in the biliary atresia group. The ALGS patients did demonstrate good catch-up growth post-transplant, but their parameters did not normalize, indicating a genetic component to their growth deficits.
ALGS therapy: current recommendations and future directions
Clinical treatments
In general, therapy for ALGS-related liver disease is focused on supportive care for nutrition, fat-soluble vitamin deficiencies, and cholestasis. Pruritus of cholestasis is one of the most difficult problems to treat, and also one of the most disruptive to patients’ quality of life. Medical treatment of pruritus typically involves a combination of approaches, including choleretics, antihistamines, and rifampin and opiate antagonists such as naltrexone. Recently, sertraline has also been studied as an alternative treatment for pruritus in ALGS and other cholestatic disorders (47). In the most refractory cases, surgical biliary diversion may be considered.
Wang et al. recently conducted an analysis of surgical interruption of the enterohepatic circulation as a treatment for pediatric cholestasis (48). They identified 58 children with inherited cholestasis [20 with ALGS and 38 with forms of progressive familial intrahepatic cholestasis (PFIC)] who had undergone biliary diversion or ileal exclusion surgeries at centers participating in ChiLDReN. The majority of the ALGS patients were reported to have improvement in pruritus scores and reduction in serum cholesterol but not bilirubin levels. There was also a trend towards improvement in xanthomas in ALGS.
A new class of drugs under study for treatment of pruritus in ALGS are the apical, sodium-dependent bile acid transporter (ASBT) inhibitors. These drugs work to interrupt the enterohepatic circulation through potent chemical inhibition of the ileal bile acid transporters, thereby acting as a form of medical biliary diversion. ChiLDReN conducted a placebo-controlled clinical trial of the ASBT inhibitor maralixibat (Mirum Pharmaceuticals, Foster City, CA, USA) in 37 children with ALGS and found that some doses resulted in significantly reduced pruritus (49). There was no statistically significant reduction in serum bile acids with the active drug in this study, and adverse events were similar between those receiving active drug and placebo. Another study of maralixibat in ALGS patients included a randomized drug withdrawal and long-term open label extension. In this study, the ASBT inhibitor maralixibat was found to be well-tolerated and associated with significant and durable improvement in pruritus scores and serum bile acids (50). A separate Phase 2 study of the ASBT inhibitor odevixibat (A4250; Albireo Pharma, Inc., Boston, MA, USA) enrolled 24 children with cholestatic liver disease, including 6 with ALGS and found that the majority of patients experienced improvements in pruritus and sleep disturbance and reduction in serum bile acids (51). These drugs have promise for treatment of pruritus in children with cholestatic liver disease and are currently undergoing further study.
Experimental research
ALGS provides a unique opportunity for the development of novel therapeutic approaches that would target Notch pathway signaling to promote cholangiocyte differentiation. Unlike structural heart defects that occur during early embryogenesis, intrahepatic bile duct development continues postnatally as the liver grows. This hypothesis is supported by the work of Libbrecht et al, who performed a detailed study of a liver explant in a 17-year-old individual with ALGS (52). They found normally formed bile ducts in the hilar area, with complete absence of bile ducts in the periphery, suggesting a lack of branching and elongation of bile ducts during postnatal liver growth. Theoretically, it would be possible to target this process and augment Notch signaling sufficiently to allow new bile ducts to develop and thereby improve manifestations of cholestatic liver disease. These therapeutic approaches might be specific to JAG1 mutation types or include global methods to increase expression of the wildtype JAG1 allele. Exploitation of genetic modifiers could present another opportunity to fine-tune Notch signaling at the cellular level. Research efforts are underway to develop such personalized therapeutic approaches that could ameliorate the clinical manifestations in ALGS. In support of these types of therapeutic investigations, a number of both in vivo and in vitro models of ALGS have been developed to better understand the biology of this disease.
Animal models
Numerous mouse models have been described that attempt to recapitulate the human phenotypic spectrum of ALGS, and these have been very thoroughly reviewed in a recent publication (53). These models have focused heavily on creating perturbations in the Notch signaling pathway, leading predominantly to the generation of various knockout strains of both Jag1 and Notch1. Not surprisingly, given the universal importance of Notch signaling across many developmental processes, homozygous deletion of Jag1 was quickly shown to be embryonic lethal (54). Although original heterozygous Jag1 mouse models did not mimic any of the clinical features of ALGS (54), subsequent reports found that breeding these animals on a different genetic background (C57BL/6) produced heterozygous animals with human disease features (33). In an effort to identify alternate models that give rise to an ALGS phenotype, mouse models incorporating compound heterozygous loss of both Jag1 and Notch2 were developed that displayed features of human disease, including hepatic, cardiac, eye, and kidney defects (55).
To circumvent the issues surrounding lethality in Jag1 knockout animals, various animals engineered by conditional knockout have been described (10,56), and serendipitously these models have also helped to define the biological role and cellular context for Jag1 by confirming its cell-specific expression in the portal vein mesenchyme (9). Selective knockout of Jag1 in SM22α-positive cells in the portal vein mesenchyme give rise to many of the hepatic features of ALGS, including bile duct paucity and liver failure (9).
Although a majority of studies aimed to identify mouse models of ALGS have utilized gene knockout as a tool to decrease and/or remove Jag1 expression, models of Jag1 missense variants have also been characterized (57,58). Interestingly these mice were the products of N-ethyl-N-nitrosurea (ENU) screens that were selected for study due to having unique head-bobbing phenotypes, and subsequently were found to have missense variants in Jag1 that affected development of the inner ear and other organs. Of the described models, homozygous expression of the missense variant H268Q, which gives rise to the aptly named Nodder mice (Jag1Ndr), has been carefully analyzed and shown to recapitulate many of the features of ALGS, including delayed biliary differentiation and morphogenesis and jaundice (58). Of particular, value is that these animals are able to survive into adulthood, which has been advantageous in showing that biliary maintenance is also defective in this model.
In addition to mice, zebrafish models of ALGS have also been described. The extrahepatic and intrahepatic biliary system has been well-characterized in these animals (59-61), and both morpholino and transgenic perturbation of jagged genes has been successful in replicating the spectrum of clinical features associated with the disease (60,62). Zebrafish express three jagged genes (jag1a, jag1b, and jag2b) and targeted morpholino knockdown of both jag1b and jag2b has been shown to phenocopy ALGS, with fish displaying liver, kidney, pancreatic, craniofacial, and cardiac defects (60). This result was corroborated using transgenic and knockout zebrafish lines that further showed a necessary and sufficient role for Jag1b and Jag2b as the sole ligands responsible for inducing all duct cell specification within the liver (62).
Cell-based models
Multiple approaches have been undertaken to identify physiologically-relevant in vitro models in which to study the hepatic features of ALGS. These have involved the generation of bi-potent liver progenitor cells from liver tissues (63), the derivation of liver progenitors from hepatocytes via chemical conversion (64), and the utilization of both embryonic stem cells and iPSCs to produce 3D organoid cultures (21,65-68). For a thorough overview of these individual systems, we recommend a recently published review article (69).
Liver tissue is difficult to access, and there is an allure to being able to derive 3D systems from iPSCs, which can be generated from numerous tissue types that are more accessible, including peripheral blood and fibroblasts. Indeed, 3D cultures produced from stem cells have been shown to form ducts and exhibit properties of fluid secretion, showing promise in their ability to mimic physiologically-relevant hepatic properties (65-68). A more recent report details the generation of human hepatic organoids derived from iPSCs from both healthy individuals as well as individuals with ALGS (21). Importantly, this system was shown to recapitulate both liver development and the capacity for regeneration, two aspects that are particularly important for the study of ALGS. The authors were able to show ALGS patient-derived organoids were defective in their ability to form duct structures and were generally less efficient at forming organoids than those derived from controls (21).
Non-syndromic bile duct paucity
Bile duct paucity is defined as a bile duct to hepatic artery ratio of less than 0.5 in an adequate liver biopsy specimen (at least 10 portal tracts). The differential diagnosis of bile duct paucity in the absence of other clinical manifestations of ALGS is broad and includes other metabolic and genetic disorders, infection, immune/inflammatory disorders, drug-associated vanishing bile duct syndrome, endocrinopathies and idiopathic (Table 1) (70). The increasing availability of genetic testing for inherited cholestatic disorders will result in the identification of a definitive diagnosis in a higher percentage of cases than in the past. A child found to have bile duct paucity on liver biopsy should undergo a directed evaluation depending on other presenting signs. Genetic testing might include either a next generation sequencing cholestasis panel (available at several different laboratories), or exome sequencing. It should also be noted that patients with mutations in NOTCH2, but not JAG1, have been reported to present with isolated bile duct paucity (22).
Table 1. Disorders associated with bile duct paucity in children (70).
| Syndromic bile duct paucity—Alagille syndrome |
| Non-syndromic bile duct paucity |
| Metabolic and genetic disorders |
| Alpha-1 antitrypsin deficiency |
| BSEP deficiency (ABCB11) |
| MDR3 deficiency (ABCB4) |
| HNF3β deficiency |
| Cystic fibrosis |
| Peroxisomal disorders |
| Niemann pick type C |
| ARC syndrome (arthrogryposis-renal-cholestasis) |
| Kabuki syndrome (rare) |
| Chromosomal abnormalities (trisomy 17, 18 or 21) (rare) |
| Infection |
| Congenital cytomegalovirus infection |
| Congenital syphilis (rare) |
| Congenital rubella (rare) |
| Inflammatory and immune disorders |
| Graft versus host disease |
| Chronic hepatic allograft rejection |
| Sclerosing cholangitis (primary or secondary) |
| Biliary atresia (late) |
| Sarcoidosis (rare) |
| Other |
| Drug- or antibiotic-associated vanishing bile duct syndrome |
| Panhypopituitarism (rare) |
| Idiopathic |
In one recent report of a single center experience, bile duct paucity was present in about 11% of pediatric liver biopsies. Over a median follow up period of 11 months, about 50% improved and the other 50% progressed. The best predictor of poor outcome in this cohort was the presence of advanced fibrosis on initial liver biopsy (70). Overall, clinical outcome in children with bile duct paucity is largely dependent on the underlying etiology.
Acknowledgments
The authors would like to thank Dr. Benjamin Wilkins for his assistance in preparing the figure of liver histology in ALGS.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Luca Fabris and Mario Strazzabosco) for the series “Recent Advances in Rare Liver Diseases” published in Translational Gastroenterology and Hepatology. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tgh-2020-03). The series “Recent Advances in Rare Liver Diseases” was commissioned by the editorial office without any funding or sponsorship. MAG reports personal fees from Retrophin, outside the submitted work. KML reports grants and personal fees from Mirum Pharma, grants and personal fees from Albireo Pharma, other from Retrophin, outside the submitted work.
References
- 1.Alagille D, Odievre M, Gautier M, et al. Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental, and sexual development, and cardiac murmur. J Pediatr 1975;86:63-71. 10.1016/S0022-3476(75)80706-2 [DOI] [PubMed] [Google Scholar]
- 2.Watson GH, Miller V. Arteriohepatic dysplasia: familial pulmonary arterial stenosis with neonatal liver disease. Arch Dis Child 1973;48:459-66. 10.1136/adc.48.6.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alagille D, Estrada A, Hadchouel M, et al. Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): review of 80 cases. J Pediatr 1987;110:195-200. 10.1016/S0022-3476(87)80153-1 [DOI] [PubMed] [Google Scholar]
- 4.Li L, Krantz ID, Deng Y, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet 1997;16:243-51. 10.1038/ng0797-243 [DOI] [PubMed] [Google Scholar]
- 5.Oda T, Elkahloun AG, Pike BL, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet 1997;16:235-42. 10.1038/ng0797-235 [DOI] [PubMed] [Google Scholar]
- 6.Gilbert MA, Bauer RC, Rajagopalan R, et al. Alagille syndrome mutation update: Comprehensive overview of JAG1 and NOTCH2 mutation frequencies and insight into missense variant classification. Hum Mutat 2019;40:2197-220. 10.1002/humu.23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanimizu N, Miyajima A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. J Cell Sci 2004;117:3165-74. 10.1242/jcs.01169 [DOI] [PubMed] [Google Scholar]
- 8.Kodama Y, Hijikata M, Kageyama R, et al. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology 2004;127:1775-86. 10.1053/j.gastro.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 9.Hofmann JJ, Zovein AC, Koh H, et al. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development 2010;137:4061-72. 10.1242/dev.052118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loomes KM, Russo P, Ryan M, et al. Bile duct proliferation in liver-specific Jag1 conditional knockout mice: effects of gene dosage. Hepatology 2007;45:323-30. 10.1002/hep.21460 [DOI] [PubMed] [Google Scholar]
- 11.Saleh M, Kamath BM, Chitayat D. Alagille syndrome: clinical perspectives. Appl Clin Genet 2016;9:75-82. 10.2147/TACG.S86420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spinner NB, Colliton RP, Crosnier C, et al. Jagged1 mutations in alagille syndrome. Hum Mutat 2001;17:18-33. [DOI] [PubMed] [Google Scholar]
- 13.Krantz ID, Colliton RP, Genin A, et al. Spectrum and frequency of jagged1 (JAG1) mutations in Alagille syndrome patients and their families. Am J Hum Genet 1998;62:1361-9. 10.1086/301875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer RC, Laney AO, Smith R, et al. Jagged1 (JAG1) mutations in patients with tetralogy of Fallot or pulmonic stenosis. Hum Mutat 2010;31:594-601. 10.1002/humu.21231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrissette JD, Colliton RP, Spinner NB. Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum Mol Genet 2001;10:405-13. 10.1093/hmg/10.4.405 [DOI] [PubMed] [Google Scholar]
- 16.Tada M, Itoh S, Ishii-Watabe A, et al. Functional analysis of the Notch ligand Jagged1 missense mutant proteins underlying Alagille syndrome. FEBS J 2012;279:2096-107. 10.1111/j.1742-4658.2012.08595.x [DOI] [PubMed] [Google Scholar]
- 17.Guarnaccia C, Dhir S, Pintar A, et al. The tetralogy of Fallot-associated G274D mutation impairs folding of the second epidermal growth factor repeat in Jagged-1. FEBS J 2009;276:6247-57. 10.1111/j.1742-4658.2009.07333.x [DOI] [PubMed] [Google Scholar]
- 18.Lu F, Morrissette JJ, Spinner NB. Conditional JAG1 mutation shows the developing heart is more sensitive than developing liver to JAG1 dosage. Am J Hum Genet 2003;72:1065-70. 10.1086/374386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyer-Di Ponio J, Wright-Crosnier C, Groyer-Picard MT, et al. Biological function of mutant forms of JAGGED1 proteins in Alagille syndrome: inhibitory effect on Notch signaling. Hum Mol Genet 2007;16:2683-92. 10.1093/hmg/ddm222 [DOI] [PubMed] [Google Scholar]
- 20.Boyer J, Crosnier C, Driancourt C, et al. Expression of mutant JAGGED1 alleles in patients with Alagille syndrome. Hum Genet 2005;116:445-53. 10.1007/s00439-005-1262-7 [DOI] [PubMed] [Google Scholar]
- 21.Guan Y, Xu D, Garfin PM, et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017;2. 10.1172/jci.insight.94954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamath BM, Bauer RC, Loomes KM, et al. NOTCH2 mutations in Alagille syndrome. J Med Genet 2012;49:138-44. 10.1136/jmedgenet-2011-100544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guegan K, Stals K, Day M, et al. JAG1 mutations are found in approximately one third of patients presenting with only one or two clinical features of Alagille syndrome. Clin Genet 2012;82:33-40. 10.1111/j.1399-0004.2011.01749.x [DOI] [PubMed] [Google Scholar]
- 24.Turnpenny PD, Ellard S. Alagille syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet 2012;20:251-7. 10.1038/ejhg.2011.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhorne-Pollet S, Deleuze JF, Hadchouel M, et al. Segregation analysis of Alagille syndrome. J Med Genet 1994;31:453-7. 10.1136/jmg.31.6.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elmslie FV, Vivian AJ, Gardiner H, et al. Alagille syndrome: family studies. J Med Genet 1995;32:264-8. 10.1136/jmg.32.4.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emerick KM, Rand EB, Goldmuntz E, et al. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology 1999;29:822-9. 10.1002/hep.510290331 [DOI] [PubMed] [Google Scholar]
- 28.Shulman SA, Hyams JS, Gunta R, et al. Arteriohepatic dysplasia (Alagille syndrome): extreme variability among affected family members. Am J Med Genet 1984;19:325-32. 10.1002/ajmg.1320190215 [DOI] [PubMed] [Google Scholar]
- 29.Izumi K, Hayashi D, Grochowski CM, et al. Discordant clinical phenotype in monozygotic twins with Alagille syndrome: Possible influence of non-genetic factors. Am J Med Genet A 2016;170A:471-5. 10.1002/ajmg.a.37429 [DOI] [PubMed] [Google Scholar]
- 30.Kamath BM, Bason L, Piccoli DA, et al. Consequences of JAG1 mutations. J Med Genet 2003;40:891-5. 10.1136/jmg.40.12.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crosnier C, Driancourt C, Raynaud N, et al. Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology 1999;116:1141-8. 10.1016/S0016-5085(99)70017-X [DOI] [PubMed] [Google Scholar]
- 32.Ryan MJ, Bales C, Nelson A, et al. Bile duct proliferation in Jag1/fringe heterozygous mice identifies candidate modifiers of the Alagille syndrome hepatic phenotype. Hepatology 2008;48:1989-97. 10.1002/hep.22538 [DOI] [PubMed] [Google Scholar]
- 33.Thakurdas SM, Lopez MF, Kakuda S, et al. Jagged1 heterozygosity in mice results in a congenital cholangiopathy which is reversed by concomitant deletion of one copy of Poglut1 (Rumi). Hepatology 2016;63:550-65. 10.1002/hep.28024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Valdivia R, Takeuchi H, Samarghandi A, et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 2011;138:1925-34. 10.1242/dev.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams JM, Huppert KA, Castro EC, et al. Sox9 is a modifier of the liver disease severity in a mouse model of Alagille syndrome. Hepatology 2020;71:1331-49. 10.1002/hep.30912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai EA, Gilbert MA, Grochowski CM, et al. THBS2 Is a Candidate Modifier of Liver Disease Severity in Alagille Syndrome. Cell Mol Gastroenterol Hepatol 2016;2:663-75.e2. 10.1016/j.jcmgh.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamath BM, Ye W, Goodrich NP, et al. Outcomes of Childhood Cholestasis in Alagille Syndrome: Results of a Multicenter Observational Study. Hepatology Communications 2020. doi: . 10.1002/hep4.1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhadri VA, Stormon MO, Arbuckle S, et al. Hepatocellular carcinoma in children with Alagille syndrome. J Pediatr Gastroenterol Nutr 2005;41:676-8. 10.1097/01.mpg.0000179759.60048.c4 [DOI] [PubMed] [Google Scholar]
- 39.Kamath BM, Munoz PS, Bab N, et al. A longitudinal study to identify laboratory predictors of liver disease outcome in Alagille syndrome. J Pediatr Gastroenterol Nutr 2010;50:526-30. 10.1097/MPG.0b013e3181cea48d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouzaki M, Bass LM, Sokol RJ, et al. Early life predictive markers of liver disease outcome in an International, Multicentre Cohort of children with Alagille syndrome. Liver Int 2016;36:755-60. 10.1111/liv.12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bass LM, Shneider BL, Henn L, et al. Clinically Evident Portal Hypertension: An Operational Research Definition for Future Investigations in the Pediatric Population. J Pediatr Gastroenterol Nutr 2019;68:763-7. 10.1097/MPG.0000000000002333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng VL, Haber BH, Magee JC, et al. Medical status of 219 children with biliary atresia surviving long-term with their native livers: results from a North American multicenter consortium. J Pediatr 2014;165:539-46 e2. [DOI] [PMC free article] [PubMed]
- 43.Shneider BL, Abel B, Haber B, et al. Portal hypertension in children and young adults with biliary atresia. J Pediatr Gastroenterol Nutr 2012;55:567-73. 10.1097/MPG.0b013e31826eb0cf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rovner AJ, Schall JI, Jawad AF, et al. Rethinking growth failure in Alagille syndrome: the role of dietary intake and steatorrhea. J Pediatr Gastroenterol Nutr 2002;35:495-502. 10.1097/00005176-200210000-00007 [DOI] [PubMed] [Google Scholar]
- 45.Rovner AJ, Stallings VA, Piccoli DA, et al. Resting energy expenditure is not increased in prepubertal children with Alagille syndrome. J Pediatr 2006;148:680-2. 10.1016/j.jpeds.2005.12.019 [DOI] [PubMed] [Google Scholar]
- 46.Kamath BM, Yin W, Miller H, et al. Outcomes of liver transplantation for patients with Alagille syndrome: the studies of pediatric liver transplantation experience. Liver Transpl 2012;18:940-8. 10.1002/lt.23437 [DOI] [PubMed] [Google Scholar]
- 47.Thebaut A, Habes D, Gottrand F, et al. Sertraline as an Additional Treatment for Cholestatic Pruritus in Children. J Pediatr Gastroenterol Nutr 2017;64:431-5. 10.1097/MPG.0000000000001385 [DOI] [PubMed] [Google Scholar]
- 48.Wang KS, Tiao G, Bass LM, et al. Analysis of surgical interruption of the enterohepatic circulation as a treatment for pediatric cholestasis. Hepatology 2017;65:1645-54. 10.1002/hep.29019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shneider BL, Spino C, Kamath BM, et al. Placebo-Controlled Randomized Trial of an Intestinal Bile Salt Transport Inhibitor for Pruritus in Alagille Syndrome. Hepatol Commun 2018;2:1184-98. 10.1002/hep4.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzales E, Sokal EM, Hardikar W, et al. Durability of Treatment Effect with Long-Term Maralixibat in Children with Alagille Syndrome: 4-year safety and efficacy. Hepatology 2019;70:1479A. [Google Scholar]
- 51.Sturm E, Baumann U, Lacaille F, et al. The Ileal Bile Acid Transport Inhibitor A4250 Reduced Pruritus and Serum Bile Acid Levels in Children with Cholestatic Liver Disease and Pruritus: Final Results from a Multiple-Dose, Open-label, Multinational Study. Hepatology 2017;66:647A. [Google Scholar]
- 52.Libbrecht L, Spinner NB, Moore EC, et al. Peripheral bile duct paucity and cholestasis in the liver of a patient with Alagille syndrome: further evidence supporting a lack of postnatal bile duct branching and elongation. Am J Surg Pathol 2005;29:820-6. 10.1097/01.pas.0000161325.36348.25 [DOI] [PubMed] [Google Scholar]
- 53.Van Hul N, Lendahl U, Andersson ER. Mouse Models for Diseases in the Cholangiocyte Lineage. Methods Mol Biol 2019;1981:203-36. 10.1007/978-1-4939-9420-5_14 [DOI] [PubMed] [Google Scholar]
- 54.Xue Y, Gao X, Lindsell CE, et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet 1999;8:723-30. 10.1093/hmg/8.5.723 [DOI] [PubMed] [Google Scholar]
- 55.McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 2002;129:1075-82. [DOI] [PubMed] [Google Scholar]
- 56.Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet 2006;2:e4. 10.1371/journal.pgen.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vrijens K, Thys S, De Jeu MT, et al. Ozzy, a Jag1 vestibular mouse mutant, displays characteristics of Alagille syndrome. Neurobiol Dis 2006;24:28-40. 10.1016/j.nbd.2006.04.016 [DOI] [PubMed] [Google Scholar]
- 58.Andersson ER, Chivukula IV, Hankeova S, et al. Mouse Model of Alagille Syndrome and Mechanisms of Jagged1 Missense Mutations. Gastroenterology 2018;154:1080-95. 10.1053/j.gastro.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorent K, Moore JC, Siekmann AF, et al. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn 2010;239:855-64. 10.1002/dvdy.22220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorent K, Yeo SY, Oda T, et al. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development 2004;131:5753-66. 10.1242/dev.01411 [DOI] [PubMed] [Google Scholar]
- 61.Hinton DE, Couch JA. Architectural pattern, tissue and cellular morphology in livers of fishes: relationship to experimentally-induced neoplastic responses. EXS 1998;86:141-64. 10.1007/978-3-0348-8853-0_4 [DOI] [PubMed] [Google Scholar]
- 62.Zhang D, Gates KP, Barske L, et al. Endoderm Jagged induces liver and pancreas duct lineage in zebrafish. Nat Commun 2017;8:769. 10.1038/s41467-017-00666-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299-312. 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katsuda T, Kawamata M, Hagiwara K, et al. Conversion of Terminally Committed Hepatocytes to Culturable Bipotent Progenitor Cells with Regenerative Capacity. Cell Stem Cell 2017;20:41-55. 10.1016/j.stem.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 65.Dianat N, Dubois-Pot-Schneider H, Steichen C, et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology 2014;60:700-14. 10.1002/hep.27165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Assuncao TM, Sun Y, Jalan-Sakrikar N, et al. Development and characterization of human-induced pluripotent stem cell-derived cholangiocytes. Lab Invest 2015;95:1218. 10.1038/labinvest.2015.99 [DOI] [PubMed] [Google Scholar]
- 67.Ogawa M, Ogawa S, Bear CE, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol 2015;33:853-61. 10.1038/nbt.3294 [DOI] [PubMed] [Google Scholar]
- 68.Sampaziotis F, Cardoso de Brito M, Madrigal P, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol 2015;33:845-52. 10.1038/nbt.3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghanekar A, Kamath BM. Cholangiocytes derived from induced pluripotent stem cells for disease modeling. Curr Opin Gastroenterol 2016;32:210-5. 10.1097/MOG.0000000000000260 [DOI] [PubMed] [Google Scholar]
- 70.Meena BL, Khanna R, Bihari C, et al. Bile duct paucity in childhood-spectrum, profile, and outcome. Eur J Pediatr 2018;177:1261-9. 10.1007/s00431-018-3181-3 [DOI] [PubMed] [Google Scholar]