Abstract
Purpose
Accumulated evidence has shown that microRNAs (miRNAs) are closely related with the regulation of autophagy, which plays vital roles in fungal keratitis (FK). Microarray data showed elevated expression of miR-665-3p in mouse corneal tissues after infection with Fusarium solani (F. solani). Here, we investigated the effect of miR-665-3p in regulating autophagy in experimental F. solani keratitis and determined the potential molecular mechanisms involved.
Methods
In this article, we established an in vivo mouse model of FK and an in vitro model of corneal stromal cells by inoculating with F. solani. We divided them into the following six groups: control, chloroquine (CQ), rapamycin (Rapa), miR-665-3p antagomir (ant-665), miR-665-3p agomir (miR-665), and the negative control group (miR-NC). The levels of autophagy were detected by electron microscopy, Western blotting, and immunofluorescence. Then, we used a dual-luciferase reporter assay to determine the binding of miR-665-3p to the autophagy-related gene (ATG)5 3'UTR. Detection of IL-1β protein levels and hematoxylin and eosin (H&E) staining of corneal tissues were used to observe the effect of miR-665-3p on inflammation in FK.
Results
Here, we showed that inhibition of miR-665-3p expression in FK upregulated autophagy and alleviated inflammation. Nevertheless, the opposite results were found by overexpressing miR-665-3p. Additionally, ATG5 was a direct target gene for miR-665-3p.
Conclusions
Together, our data demonstrated that miR-665-3p might be involved in F. solani keratitis of mice by regulating autophagic pathways and inflammation.
Keywords: fungal keratitis, autophagy, miR-665-3p, ATG5, inflammation
Fungal keratitis (FK) is an infectious corneal disease, which is caused by fungi resulting in high incidence of blindness.1 Recently, the incidence of FK has been increasing.2 According to previous research, Fusarium solani (F. solani) is the main pathogenic microorganism that causes FK in China.3 The production of mycotoxins and the limitations of antifungal drugs (e.g. narrow antimicrobial spectrum and poor water solubility) are associated with the prognosis of FK.4,5 In addition, the incidence of corneal perforation is still high and requires accurate diagnosis and proper treatment.6 To date, the pathogenesis of FK has not been clearly elucidated.
MicroRNAs (miRNAs), which are small noncoding NA molecules, could negatively regulate the expression level of genes by binding to the 3′-untranslated region (3′UTR).7 Moreover, miRNAs have been found to regulate autophagy by modulating inflammation, apoptosis, proliferation, aging, and other vital pathophysiological processes.8 Recently, dysregulation of the expression of miRNAs has been reported in many eye-related diseases, such as age-related macular degeneration and diabetic keratopathy.9–12 Our preliminary studies found that miR-665-3p was upregulated in fungal infected corneal tissue of mouse. Li et al.13 observed that the inhibition of miR-665 can reduce inflammation and apoptosis during intestinal ischemia/reperfusion (I/R) by restoring autophagic flux. It has been demonstrated that autophagy participates in the clearance of bacteria and toxins from infected cells and plays an important role in cellular homeostasis via inflammation regulation in various diseases.14–16 The initiation, elongation, and regulation of autophagosome formation rely on autophagy-related genes (ATGs).17,18 ATG-mediated autophagy exerts an important role in the control of inflammatory signaling.19 Notably, recent studies found that autophagy has a significant role in the occurrence of FK.20 Therefore, we speculated that dysregulated miR-665-3p expression in FK may be related to changes in autophagic levels and activation of inflammation.
In this article, a mouse model of FK was established. Here, we detected changes in autophagy and the inflammatory response by regulating the level of miR-665-3p. We demonstrated that the inhibition of miR-665-3p in F. solani keratitis of mice can promote autophagy and reduce inflammation, which will provide a new idea for the pathogenesis and treatment of FK from the perspective of miRNAs.
Materials and Methods
Experimental Animals and Treatment
BALB/C mice (6–8 weeks old) were obtained from the Experimental Animal Center of Jinan Pengyue. The animals and the experimental process were in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. The fungus F. solani (AS 3.1829) was provided by the China General Microbiological Culture Collection Center (CGMCC, Beijing, China). Mice were divided into the following six groups randomly before infection with F. solani (n = 6/group): control, chloroquine (CQ), rapamycin (Rapa), miR-665-3p antagomir (ant-665), miR-665-3p agomir (miR-665), and the negative control (miR-NC) group. The FK model was established according to a previous protocol.21 In brief, three 1 mm incisions were made with a sterile 26 needle to scarify the mouse central corneas under systemic anesthetization with pentobarbital (50 mg/kg). F. solani (5 µL, 1 × 108 CFUs) was applied locally to the ocular surface, and then soft contact lenses were used to cover the eye surface. Slit lamp microscopy was used to observe the condition of corneal lesions every day. The clinical scores were evaluated as per Wu et al.22 Eye samples and corneal tissue were collected at a suitable time for the following experiments.
Cell Culture
Mouse corneas were immersed in Dispase II (15 mg/mL) for 12 hours at 4°C. On the next day, the separated corneal epithelium was gently peeled off along the limbus. The tear off the descemet and endothelium of the cornea were done with micro tweezers, and the remaining corneal stroma were cut into small pieces. Then, the tissues were washed with sterile PBS and digested in 3 mg/mL type I collagenase for 2 hours with shaking. The corneal stromal cells were grown in Dulbecco's modified eagle medium/nutrient mixture F-12 Ham (DMEM F-12; Sigma, USA) with 10% fetal bovine serum (FBS) at 37°C, 5% CO2. The cell medium was replaced every 48 hours.
miRNA Microarray
Total RNA was extracted from corneal tissues using TRIzol reagent (Invitrogen) and purified via a mirVana miRNA Isolation Kit (AM1560; Ambion, Austin, TX, US). Briefly, 200 ng RNA was dephosphorylated and labeled with pCp-Cy3 using the Agilent miRNA labeling reagent. Labeled RNA was processed for microarray hybridization to the miRNA array (Agilent Technologies, Santa Clara, CA, USA). The Agilent array was designed with eight identical arrays per slide (8 × 60 K format), with each array containing probes interrogating 2549 human mature miRNAs from miRBase R21.0. Microarray slides were scanned with an Agilent microarray scanner (Agilent Technologies), then the scans were analyzed with Agilent feature extraction software version 10.10. The differentially expressed genes were selected using fold-change thresholds of ≥ 2 or ≤ −2 and adjusted P value < 0.05. Hierarchical clustering was calculated using average-linkage clustering and visualized using TreeView software (Stanford University, Stanford, CA, USA).
Real-Time Quantitative PCR
Total miRNA was extracted from mouse corneal stromal cells or corneal tissues via the miRNeasy Mini Kit (Qiagen, Germany). All specific primers were synthesized by Invitrogen (Shanghai, China). The relative expression level of miR-665-3p was analyzed by the 2−ΔΔCt method with normalization to the U6 internal control.
Transmission Electron Microscopy
The samples were postfixed with 1% osmium tetroxide (OsO4) at room temperature for 2 hours and dehydrated through an alcohol gradient (50%–100%). Sections (60 nm) were cut with an ultramicrotome, and then stained with uranyl acetate for 15 minutes. The sections were poststained for 5 minutes with lead citrate and then air-dried overnight. Images were viewed on a transmission electron microscope (HT7700; Hitachi, Japan).
Luciferase Reporter Assay
Bioinformatics tools (miRWalk, Microt4, miRanda, miRDB, miRMap, and TargetScan) were applied to predict the target genes of miR-665-3P. The 3'UTRs of wild-type and mutant ATG5 (ATG5-WT and ATG5-MUT) were amplified and then inserted into the pmiR-GLO vector. Human embryonic kidney 293T (HEK293T) cells were cotransfected with pmirGLO-ATG5-3'UTR-WT or pmirGLO-ATG5-3'UTR-MUT and miR-665-3p mimics or NC for 48 hours. Finally, we used a dual-luciferase reporter assay system to detect the luciferase activity.
Hematoxylin-Eosin Staining
Eyeballs were obtained from the different groups of mice and fixed with 4% formaldehyde. Then, the corneal tissues were dehydrated through a graded ethanol series, embedded in paraffin, afterward, 5 µm thick sections were cut. Finally, sections were stained with hematoxylin and eosin (H&E) to observe the pathological changes under a microscope.
Western Blot Assay
To determine the change in autophagy, we detected the expression of the autophagy-associated markers LC3II/I and P62 by Western blotting. Protein samples from mouse corneal stromal cells or corneal tissues were extracted using RIPA buffer reagent (Thermo Fisher Scientific, Germany) and separated using 12.5% SDS-PAGE gels. After electrophoresis, the separated proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and blocked with 5% defatted milk powder (Applygen, Beijing, China). Primary antibodies, including anti-LC3 I/II (L7543; Sigma, St. Louis, MO, USA), anti-p62 (P0067; Sigma), anti-IL-1β (ab9722; Abcam, Cambridge, UK), anti-ATG5 (A0856; Sigma), and anti-GAPDH (AF7021; Affinity Biosciences, Cincinnati, OH, USA), were incubated overnight at 4°C. After washing with PBS containing 0.05% Tween 20 (Bio-Rad, Hercules, CA, USA) three times, the membranes were incubated with specific secondary antibodies for 2 hours. Finally, the proteins were quantified using the ECL detection reagent (WBKLS0100; Millipore, Billerica, MA, USA).
AD-mRFP-GFP-LC3 for Autophagic Detection
Mouse corneal stroma cells were seeded in a plate and reached 50% to 70% confluence before transfection. The mRFP-GFP-LC3 adenovirus vectors (HanBio, Shanghai, China) were used to monitor autophagic flux. The cells were incubated with adenovirus at an MOI of 500 for 6 hours. The cells were treated with CQ, Rapa, miR-665 agomir, miR-665 antagomir, or miR-NC at the specified time points. Autophagic flux analysis was performed by calculating GFP and mRFP point numbers.
Immunofluorescence
The extracted eyeballs or mouse corneal stromal cells were fixed in 4% paraformaldehyde. Next, samples were treated with 0.1% Triton X-100 for 5 minutes and blocked using 5% BSA for 1 hour. Afterward, incubated with rabbit anti-p62 (P0067; Sigma) and rabbit anti-ATG5 (A0856; Sigma) at 4°C overnight, and the sections were washed and incubated with Alexa Fluor 488 goat anti-rabbit (ab150077; Abcam, Cambridge, UK) or Alexa Fluor 594 goat anti-rabbit (ab150080; Abcam) secondary antibodies in the dark for 1 hour at 37°C. Isotype IgG was used as the negative control and nuclei were visualized with DAPI (Beyotime Biotechnology, Shanghai, China). Images were captured with a fluorescence microscope.
Statistical Analysis
Student's t-test was applied to evaluate statistical significance, and a P value < 0.05 was considered of statistical significance. Data were expressed as the mean ± standard deviation (mean ± SD). All data were analyzed in GraphPad Prism version 8.0 (La Jolla, CA, USA).
Results
miR-665-3p Was Upregulated in Fungal Keratitis
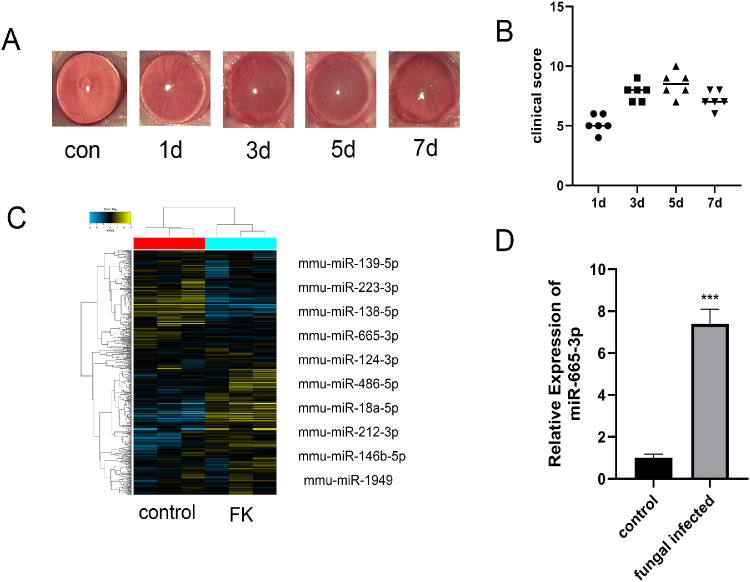
All eyes that were inoculated with F. solani showed typical signs of FK, such as corneal edema and opacity and stromal infiltration. Corneal inflammation was gradually aggravated after inoculation, reached its peak at day 5 postinfection, and then gradually decreased; thus, we used the mouse cornea infected by F. solani at 5 days for a follow-up experiment (Figs. 1A, 1B). To determine the expression profile of miRNA in FK, we assessed the normal and F. solani-infected mouse corneal tissues by Affymetrix Gene Chip miRNA4.0. The results indicated that 149 miRNAs (P ≤ 0.05, FC ≥ 2) were differentially expressed between the two groups, and 88 miRNAs were upregulated (Fig. 1C). PCR was applied to detect the expression of the top 10 differentially expressed miRNAs. In particular, the expression of miR-665-3p was increased significantly in the mice with FK compared with the normal mice (Fig. 1D).
Figure 1.
miR-665-3p was upregulated in fungal keratitis. (A, B) Corneal inflammation was gradually aggravated after fungal infection in mice, reaching its peak on day 5. (C) The heatmap shows the top 10 differentially expressed miRNAs between the two groups. (D) The expression of miR-665-3p was increased in the infected group. n = 6, ***P < 0.001.
miR-665-3p Negatively Regulated ATG5
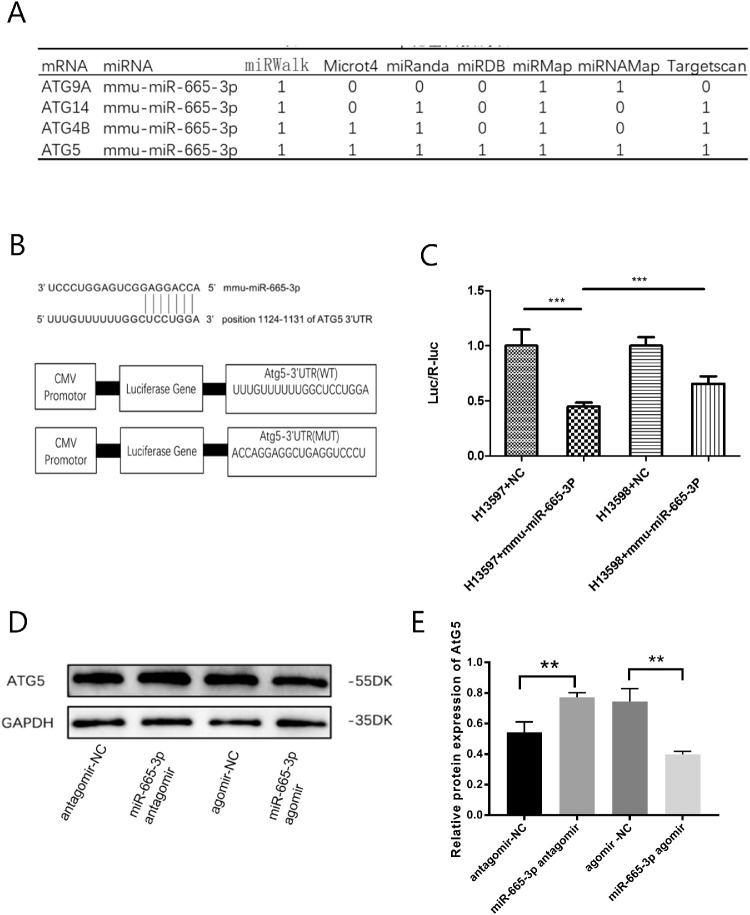
Bioinformatics tools (miRWalk, Microt4, miRanda, miRDB, miRMap, and TargetScan) were applied to predict target genes of miR-665-3P. The data showed that ATG9A, ATG14, ATG4B, and ATG5 might be potential target genes of miR-665-3p, and ATG5 had the strongest correlation (Fig. 2A). As displayed in Figure 2B, the 3′UTR of ATG5 contains the complementary site for the seed region of miR-665-3p. Luciferase reporter assays were performed in 293T cells. The results showed that cotransfection with miR-665-3p and the ATG5-3′UTR-WT resulted in significantly decreased luciferase activity. When the binding site was mutated, the luciferase activity showed some recovery (Fig. 2C). To further explore the potential targeting relationship between the two, we further detected the regulatory effect of miR-665-3p on ATG5 by using Western blotting in mouse corneal stromal cells. Treatment with a miR-665-3p antagomir could increase the expression of the ATG5 protein compared with miR-NC treatment, while opposite trends were observed after treatment with a miR-665-3p agomir (Figs. 2D, 2E).
Figure 2.
miR-665-3p negatively regulated ATG5. (A) The potential targets of miR-665-3p were predicted by integrating the results of different algorithms. (B) The 3'UTR of ATG5 contains the complementary site for the seed region of miR-665-3p. (C) A luciferase reporter assay was used to determine whether miR-665-3p directly targeted the ATG5 3′UTR in HEK293T cells. (D, E) Treatment with a miR-665-3p antagomir increased the expression of ATG5 protein compared with miR-NC, whereas opposite trends were observed after treatment with a miR-665-3p agomir in mouse corneal stromal cells. n = 6; **P < 0.01, ***P < 0.001.
Autophagy Flux was Impaired in Fusarium solani Keratitis
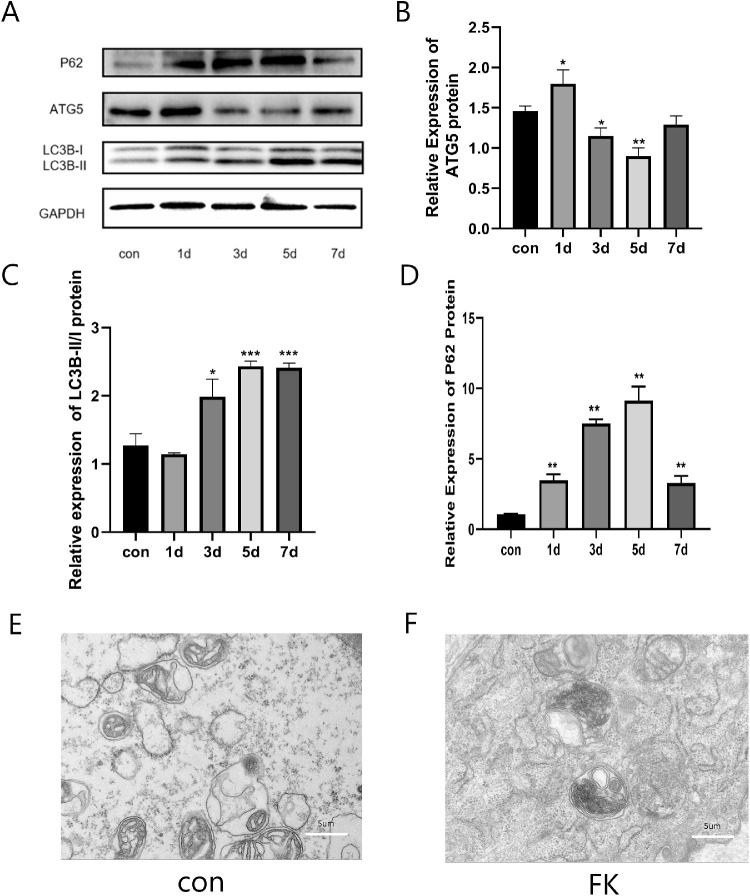
The ratio of LC3II/I and P62 protein expression levels were increased significantly in the F. solani-infected mouse corneal tissues and reached a peak on days 5, whereas ATG5 protein expression levels were decreased at the fifth day (Figs. 3A–D). On day 5 postinfection, we detected the accumulation of autophagosomes in the mouse cornea by transmission electron microscopy. We only observed the higher number of autophagosomes in the control group, but the number of autophagosomes decreased after F. solani infection, and there was no significant difference in morphology and nature (Figs. 3E, 3F). These results suggested that autophagy flux was impaired in the F. solani-infected mouse corneas.
Figure 3.
Autophagy flux was impaired in Fusarium solani keratitis. (A–D) The ratio of LC3II/I and P62 protein expression levels were increased significantly in the F. solani-infected mouse corneal tissues and reached a peak on day 5, whereas the ATG5 protein expression levels were decreased at the fifth day. (E, F) Transmission electron microscopy (TEM) showed autophagosomes in both groups at day 5. n = 6; *P < 0.05, **P < 0.01, ***P < 0.001.
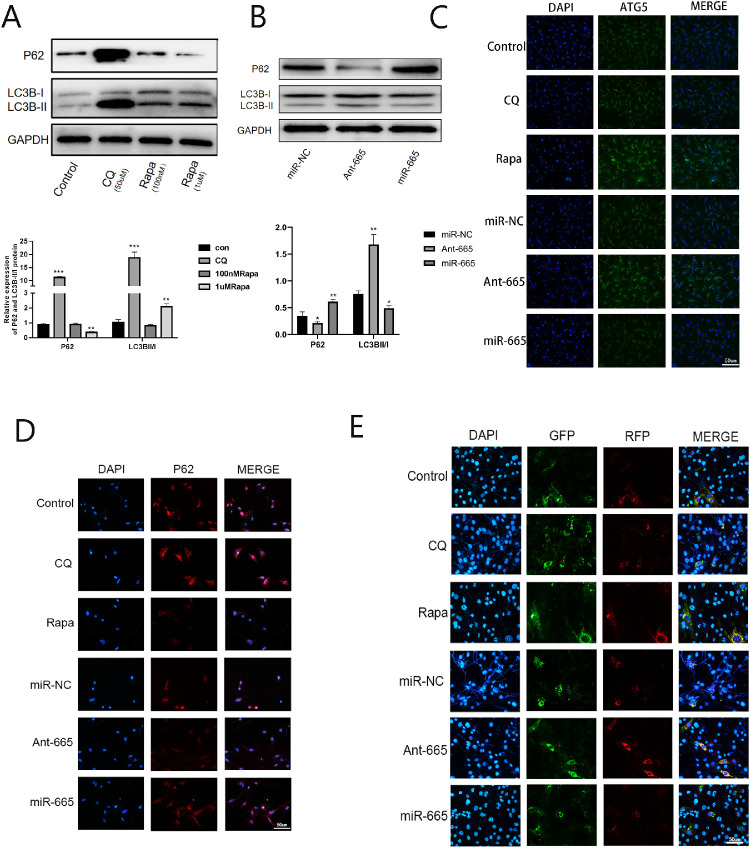
Inhibition of miR-665-3p Enhanced Autophagy In Vivo and In Vitro
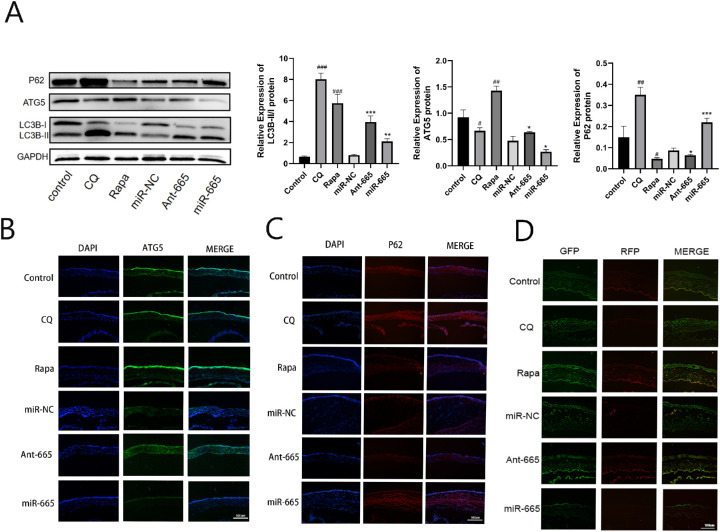
To better understand the effects of miR-665-3p on autophagy, we performed in vivo experiments using a mouse model of F. solani-induced keratitis. Both the Western blotting and immunofluorescence results showed that treatment with a miR-665-3p antagomir and Rapa could promote the ATG5 protein expression, whereas the protein level of p62 was decreased. At the same time, the LC3B-II/I ratio increased compared with the control group. In contrast, opposite results were obtained in the corneas of mice treated with an miR-665-3p agomir. We used CQ as an autophagic inhibitor, which led to increase in the LC3B-II/I ratio and p62 protein as well as to decrease in ATG5 protein levels. Then, the mRFP-GFP-LC3 adenovirus construct was used to investigate autophagic flux in vivo. The fluorescence intensity of the yellow band (spot aggregation) in the Rapa and miR-665-3p antagomir group was stronger than that of the control group, and that was weakened after being treated with miR-665-3p agomir or CQ (Fig. 4). To further validate the data shown above, we conducted in vitro experiments using the corneal stroma cells infected with F. solani and observed similar results (Fig. 5).
Figure 4.
Inhibition of miR-665-3p stimulated autophagic flux in F. solani-infected corneas. (A) The protein levels of p62, ATG5, and LC3B were detected by Western blotting. (B, C) The images of ATG5 and p62 expression showed by immunofluorescence. (D) Autophagic flow was observed after mRFP-GFP-LC3 adenovirus transfection. n = 6; # P < 0.05, ## P < 0.01, ### P < 0.001 vs. the control group; *P < 0.05, **P < 0.01, ***P < 0.001 vs. the miR-NC group.
Figure 5.
Inhibition of miR-665-3p stimulated autophagic flux in the corneal stroma cells infected with F. solani.(A, B) The protein levels of p62 and LC3B were detected by Western blotting. (C, D) The images of ATG5 and p62 expression showed by immunofluorescence. (E) Autophagic flow was observed after mRFP-GFP-LC3 adenovirus transfection. n = 6; *P < 0.05, **P < 0.01, ***P < 0.001.
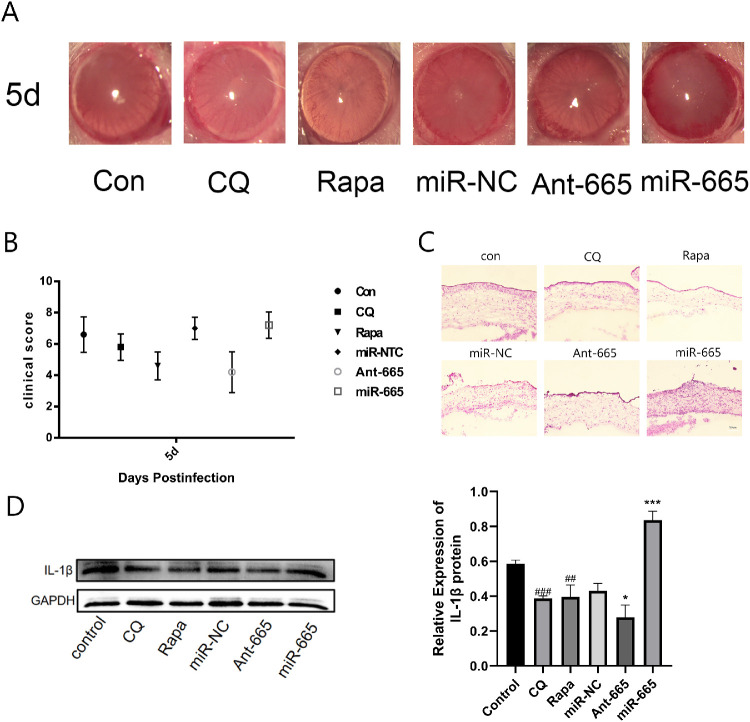
Inhibition of miR-665-3p Alleviated Corneal Inflammation After F. solani Infection
The corneal changes were observed under a slit lamp. We found that after miR-665-3p inhibitor and Rapa treatment, corneal opacification and edema were evidently alleviated (Fig. 6A). Subsequently, the clinical scores were determined for each group. The results showed that the scores for the miR-665-3p inhibitor and Rapa treatment group were lower than that of control group significantly (Fig. 6B). In addition, H&E staining showed that the corneas of the miR-665-3p inhibitor and Rapa-treated group showed decreased inflammatory cell infiltration, whereas overexpression of miR-665-3p aggravated the inflammatory state (Fig. 6C). After the miR-665-3p inhibitor and Rapa treatment, the expression level of IL-1β was significantly decreased, whereas expression of the IL-1β protein was increased by overexpressing miR-665-3p (Fig. 6D).
Figure 6.
Inhibition of miR-665-3p alleviated corneal inflammation after F. solani infection. (A, B) Evaluation and clinical scoring of corneal fungal keratitis after different treatments. (C) H&E staining images for inflammation in corneal tissues. (D) IL-1β protein levels were determined in F. solani-infected mice corneas after different treatment. n = 6; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. the control group; *P < 0.05, **P < 0.01, ***P < 0.001 vs. the miR-NC group.
Discussion
FK is a corneal disease that is caused by pathogenic fungi, among which F. solani is the most reported strain in many countries, with a range from 25% to 73.3%.23 However, due to its rapid progression, there is no effective treatment to control this disease. Exploring the pathogenesis of FK is crucial for the prevention of adverse complications and for the disease treatment. Recent evidence has demonstrated that the altered expression of miRNAs is correlated with human corneal diseases, suggesting that they might play an important regulatory role in pathogenesis.24–26 Notably, Hemadevi et al.27 identified miRNA expression profiles in human corneal tissue infected with pathogenic fungi for the first time, suggesting that some miRNAs highly expressed in FK may play an important regulatory role in corneal inflammation as well as wound healing. We also found upregulation of miR-665-3p expression in mouse keratitis after F. solani infection. Previously, miR-665-3p was identified to function in inflammatory diseases.13,28 However, the specific pathogenic role of miR-665-3p in FK remains unclear.
To explore the functional mechanisms of miR-665-3p in FK, we screened its target genes by a bioinformatics tool. We found that the seed sequences of miR-665-3p were complementary to the 3′UTR of ATG5. Moreover, fluorescence reporter analysis showed that miR-665-3p caused a significant decrease in the luciferase activity of a wild-type ATG5 3′UTR reporter, but some recovery of luciferase activity was observed when the binding site was mutated. Furthermore, it was verified in corneal stromal cells that miR-665-3p can negatively regulate the level of ATG5 protein. All of these findings implied that ATG5 is a direct target gene of miR-665-3p. ATG5, as an essential autophagy-related regulatory protein, can participate in autophagosome formation.29 Recent findings showed that miR-665-3p inhibition can reduce inflammation and apoptosis during intestinal I/R by restoring autophagic flux.13 Thus, we postulated that miR-665-3p may be an important autophagy-related miRNA in F. solani keratitis of mice, with ATG5 as its target gene, and miR-665-3p may participate in autophagy as an autophagic inhibitor.
Autophagy, which is finely regulated by protein complexes formed by a series of ATGs, is an automatic defense system of cells.30 As one hotspot of biomedical research, its important role in diabetic keratopathy, glaucoma, and other eye diseases has been discussed previously.11,12,31 In addition, autophagy can actively respond to the invasion of pathogens, such as bacteria and viruses, and these pathogens are subsequently eliminated directly or by regulating innate and adaptive immunity.32–35 Recent findings indicated that autophagy level changes in corneal tissue infected by Aspergillus fumigatus and exerts anti-inflammatory effects on the innate immune response of A. fumigatus-induced keratitis.20 The results of our study also showed that autophagy is impaired in the corneal tissue of mice infected with F. solani. To determine the effect of miR-665-3p on autophagy, we inhibited the expression of miR-665-3p in FK and found that it has the same effect as the autophagy inducer rapamycin, which can increase the ATG5 protein and LC3B-II/I ratio, whereas the protein level of p62 was decreased. However, the opposite results were obtained following miR-665-3p overexpression. CQ suppressed autophagic flux by inhibiting the fusion of autophagosomes and lysosomes, which led to an increased LC3B-II/I ratio and enhanced p62 protein levels, as well as decreased ATG5 protein levels.
The above results suggested that miR-665-3p may regulate autophagy by targeting ATG5 in F. solani keratitis of mice and then affect the inflammatory response. IL-1β is one of the most promising proinflammatory cytokines and can improve host defense against pathogenic microorganisms by activating various reactions.36 It has previously been demonstrated that IL-1β is a pivotal proinflammatory cytokine in FK.37,38 Our research demonstrated that the miR-665-3p inhibitor and autophagic inducer rapamycin can reduce the expression of IL-1β in the corneal tissues of mice infected with F. solani and significantly reduce corneal inflammation, whereas overexpression of miR-665-3p will aggravate the inflammatory state. We found that after the treatment of miR-665-3p antagomir, the colonies were significantly reduced, but not completely cleared (unpublished data). The data in our article, including H&E staining and the detection of IL-1β, fully show that inhibition of miR-665-3p can effectively reduce the inflammatory reaction of mouse cornea after fungal infection. Taken together, a miR-665-3p inhibitor may be an anti-inflammasome agent to control the inflammatory responses of F. solani keratitis of mice.
Our major findings include the following: (1) the expression of miR-665-3p was upregulated and autophagy flux was impaired in the corneal tissues of mice infected with F. solani, (2) ATG5 is a direct target gene of miR-665-3p, and (3) inhibition of miR-665-3p enhanced autophagy and alleviated corneal inflammation. Thus, we propose that miR-665-3p inhibitors may be involved in F. solani keratitis of mice by regulating autophagic pathways and inflammation.
Acknowledgments
Supported by the National Natural Science Foundation of China, Beijing, China (grant 81870636). The author(s) have no proprietary or commercial interest in any materials discussed in this article.
Disclosure: Q. Guo, None; Y. Lin, None; J. Hu, None
References
- 1. Li X, Yuan M, Yin R, et al.. Histone deacetylase inhibitor attenuates experimental fungal keratitis in mice. Sci Rep. 2019; 9(1): 9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao C, Liu G, Li X, et al.. Inflammatory mechanism of Rumenitis in dairy cows with subacute ruminal acidosis. BMC Vet Res. 2018; 14(1): 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xie L, Zhong W, Shi W, Sun S.. Spectrum of fungal keratitis in north China. Ophthalmology. 2006; 113(11): 1943–1948. [DOI] [PubMed] [Google Scholar]
- 4. Kuo M-T, Chen J-L, Hsu S-L, Chen A, You H-L. An omics approach to diagnosing or investigating fungal keratitis. Int J Mol Sci. 2019; 20(15): 3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ortega-Rosales A, Quizhpe-Ocampo Y, Montalvo-Flores M, Burneo-Rosales C, Romero-Ulloa G.. A case of fungal keratitis due to Fusarium solani after an indigenous healing practice. IDCases. 2019; 18: e00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mande SS, Ranjith K, Sharma S, et al.. Correction: Alterations in the gut bacterial microbiome in fungal keratitis patients. PLoS One. 2019; 14(1): e0211757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Small EM, Olson EN.. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011; 469(7330): 336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ross SA, Davis CD.. The emerging role of microRNAs and nutrition in modulating health and disease. Annu Rev Nutr. 2014; 34: 305–336. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Q, He C, Li R, Ke Y, Sun K, Wang J.. miR-708 and miR-335-3p inhibit the apoptosis of retinal ganglion cells through suppressing autophagy [published online ahead of print July 19, 2020]. J Mol Neurosci, 10.1007/s12031-020-01648-y. [DOI] [PubMed] [Google Scholar]
- 10. Lian C, Lou H, Zhang J, et al.. MicroRNA-24 protects retina from degeneration in rats by down-regulating chitinase-3-like protein 1. Exp Eye Res. 2019; 188: 107791. [DOI] [PubMed] [Google Scholar]
- 11. Hu J, Huang Y, Lin Y, Lin J.. Protective effect inhibiting the expression of miR-181a on the diabetic corneal nerve in a mouse model. Exp Eye Res. 2020; 192: 107925. [DOI] [PubMed] [Google Scholar]
- 12. Hu J, Hu X, Kan T.. MiR-34c participates in diabetic corneal neuropathy via regulation of autophagy. Invest Ophthalmol Vis Sci. 2019; 60(1): 16–25. [DOI] [PubMed] [Google Scholar]
- 13. Li Z, Wang G, Feng D, et al.. Targeting the miR-665-3p-ATG4B-autophagy axis relieves inflammation and apoptosis in intestinal ischemia/reperfusion. Cell Death Dis. 2018; 9(5): 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nazio F, Cecconi F.. Autophagy up and down by outsmarting the incredible ULK. Autophagy. 2017; 13(5): 967–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi AMK, Ryter SW, Levine B.. Autophagy in human health and disease. N Engl J Med. 2013; 368(7): 651–662. [DOI] [PubMed] [Google Scholar]
- 16. Rubinsztein DC, Codogno P, Levine B.. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012; 11(9): 709–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakoh-Nakatogawa M, Matoba K, Asai E, et al.. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol. 2013; 20(4): 433–439. [DOI] [PubMed] [Google Scholar]
- 18. Wible DJ, Chao H-P, Tang DG, Bratton SB.. ATG5 cancer mutations and alternative mRNA splicing reveal a conjugation switch that regulates ATG12-ATG5-ATG16L1 complex assembly and autophagy. Cell Discov. 2019; 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G.. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014; 15(2): 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li C, Li C, Lin J, et al.. The role of autophagy in the innate immune response to fungal keratitis caused by aspergillus fumigatus infection. Invest Ophthalmol Vis Sci. 2020; 61(2): 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu J, Hu Y, Chen S, et al.. Role of activated macrophages in experimental Fusarium solani keratitis. Exp Eye Res. 2014; 129: 57–65. [DOI] [PubMed] [Google Scholar]
- 22. Wu TG, Wilhelmus KR, Mitchell BM.. Experimental keratomycosis in a mouse model. Invest Ophthalmol Vis Sci. 2003; 44(1): 210–216. [DOI] [PubMed] [Google Scholar]
- 23. Todokoro D, Suzuki T, Tamura T, et al.. Efficacy of luliconazole against broad-range filamentous fungi including Fusarium solani species complex causing fungal keratitis. Cornea. 2019; 38(2): 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matthaei M, Hu J, Kallay L, et al.. Endothelial cell microRNA expression in human late-onset Fuchs’ dystrophy. Invest Ophthalmol Vis Sci. 2014; 55(1): 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mulik S, Bhela S, Rouse BT.. Potential function of miRNAs in herpetic stromal keratitis. Invest Ophthalmol Vis Sci. 2013; 54(1): 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009; 28(2): 87–116. [DOI] [PubMed] [Google Scholar]
- 27. Boomiraj H, Mohankumar V, Lalitha P, Devarajan B.. Human corneal microRNA expression profile in fungal keratitis. Invest Ophthalmol Vis Sci. 2015; 56(13): 7939–7946. [DOI] [PubMed] [Google Scholar]
- 28. Zhang X, Feng Y, Li J, et al.. MicroRNA-665-3p attenuates oxygen-glucose deprivation-evoked microglial cell apoptosis and inflammatory response by inhibiting NF-κB signaling via targeting TRIM8. Int Immunopharmacol. 2020; 85: 106650. [DOI] [PubMed] [Google Scholar]
- 29. Zheng W, Xie W, Yin D, Luo R, Liu M, Guo F.. ATG5 and ATG7 induced autophagy interplays with UPR via PERK signaling. Cell Commun Signal. 2019; 17(1): 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cabrera S, Maciel M, Herrera I, et al.. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy. 2015; 11(4): 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li R, Jin Y, Li Q, Sun X, Zhu H, Cui H.. MiR-93-5p targeting PTEN regulates the NMDA-induced autophagy of retinal ganglion cells via AKT/mTOR pathway in glaucoma. Biomed Pharmacother. 2018; 100: 1–7. [DOI] [PubMed] [Google Scholar]
- 32. Kwon DH, Song HK.. A structural view of xenophagy, a battle between host and microbes. Mol Cells. 2018; 41(1): 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jang YJ, Kim JH, Byun S.. Modulation of autophagy for controlling immunity. Cells. 2019; 8(2): 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larabi A, Barnich N, Nguyen HTT.. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020; 16(1): 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rovetta AI, Peña D, Hernández Del Pino RE, et al.. IFNG-mediated immune responses enhance autophagy against Mycobacterium tuberculosis antigens in patients with active tuberculosis. Autophagy. 2014; 10(12): 2109–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang L, Mosoian A, Schwartz ME, et al.. HIV infection modulates IL-1β response to LPS stimulation through a TLR4-NLRP3 pathway in human liver macrophages. J Leukoc Biol. 2019; 105(4): 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Underhill DM, Pearlman E.. Immune interactions with pathogenic and commensal fungi: a two-way street. Immunity. 2015; 43(5): 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao X, Zhao G, Li C, et al.. LOX-1 and TLR4 affect each other and regulate the generation of ROS in A. fumigatus keratitis. Int Immunopharmacol. 2016; 40: 392–399. [DOI] [PubMed] [Google Scholar]