Abstract
Background/objectives
COVID-19 followed a mortal course in some young patients without any underlying factors, however, it followed a very benign course in some very older individuals with multiple comorbidities. These observations question if some genetic factors may be related to the vulnerability and poor prognosis of the disease. In this study, we aimed to investigate whether MBL2 gene B variant at codon 54 (rs1800450) were related to the variabilities in clinical course of this infection.
Methods
284 PCR-confirmed COVID-19 patients and 100 healthy controls were included in the study. COVID-19 patients were subdivided according to the clinical features and clinical characteristics were analyzed. DNAs of all patients and controls were examined for the codon 54 A/B (gly54asp: rs1800450) variation in exon 1 of the MBL2 gene.
Results
In univariate analysis, BB genotype of MBL2 gene was more common among COVID-19 cases compared with controls (10.9% vs 1.0%, respectively; OR = 12.1, 95%CI = 1.6–90.1, p = 0.001). Multivariate analyses, adjusted for age, sex and MBL genetic variants, revealed that when compared with the COVID-19 patients that had AA genotype (reference), the patients that had BB or AB genotypes suffered from a higher risk for severe disease (for BB genotype, odds ratio (OR) = 5.3, p < 0.001; for AB genotype, OR = 2.9, p = 0.001) and for ICU need (for BB genotype, OR = 19.6, p < 0.001; for AB genotype, OR = 6.9, p = 0.001). On the other hand, there was not any significant difference between the genotype variants in terms of mortality at 28 days or development of secondary bacterial infection.
Conclusion
The B variants of MBL2 gene at codon 54, which were associated with lower MBL2 levels, were related to a higher risk for a more severe clinical course of COVID-19 infection in some respects. Our findings may have potential future implications, e.g. for use of MBL protein as potential therapeutics or prioritize the individuals with B variants during vaccination strategies.
Keywords: MBL2, Gene mutation, COVID-19, Hyperinflammation, Cytokine, SARS-CoV-2
1. Introduction
Coronavirus epidemic (COVID-19) emerged in late 2019 (Huang et al., 2020) and is identified as caused by a coronavirus species named Serious Acute Respiratory Syndrome Type 2 Coronavirus 2 (SARSCOV2) (Zheng, 2020). Epidemiologically, it has rapidly progressed to a pandemic as declared by World Health Organization (WHO) on March 11, 2020 (https://covid19.who.int/, 2021). To date, the number of affected individuals has increased to nearly 30 million, and deaths over 900,000 as the global toll (https://www.who.int/emergencies/diseases/novel-coronavirus-2019, 2021). Although the clinical and imaging findings of the disease have been relatively well defined with rapid publications from pandemic centers, there are still unknowns for prognostic factors and underlying causes of the variable clinical course of the disease; i.e. asymptomatic in most but very severe in about 5–15% of the affected population (Sharma et al., 2020; Hu et al., 2020). The researchers, overall, tried to identify the poor prognostic factors by observational studies. Poor prognostic factors have been reported as high viral load, male gender, underlying chronic diseases, especially hypertension, and multiple co-morbidities, obesity, and advanced age (Sharma et al., 2020; Pujadas et al., 2020; Tamara and Tahapary, 2020). However, surprisingly, evidence to date has indicated that fatality has also been observed in young cases without any underlying factors identified so far (https://www.who.int/emergencies/diseases/novel-coronavirus-2019, 2021). On the other hand, some very older individuals with multiple co-morbidities also follow a very benign course (https://www.aa.com.tr/en/europe/113-year-old-spanish-woman-beatscoronavirus/1837442, 2021). Furthermore, some cases clustered within a family have also been reported (Ikitimur et al., 2020). These observations question whether some genetic factors are related to the vulnerability and poor prognosis of the disease. Such factors can be responsible for the susceptibility or resistance to SARS-CoV-2 infection. Genes that have been linked with response to infections involve the genes that take role in coding of virus entry receptors, co-receptors, or receptor-modifying enzymes. Specific cytokines are also known to affect and thereby regulate viral disease severity. Genetic defects related to virus sensing, signaling in response to viruses, the activity of antiviral restriction factors, or proper initiation of T cell responses, have also been reported to be associated with enhanced severity of numerous viral infections and their outcomes (Kenney et al., 2017). MBL2 gene is one of the innate immune system genes. It encodes the soluble mannosebinding lectin (MBL) or mannose-binding protein (MBP) which is secreted by the liver as part of the acute-phase response. The protein encoded belongs to the collectin family and is an important element in the innate immune system (Fig. 1 ) (https://www.genome.jp/kegg/pathway.html, 2021). The specific protein recognizes and binds to mannose and N-acetylglucosamine on many microorganisms, including viruses, such as influenza virus, HIV, and SARS-CoV; and also other microorganisms, such as bacteria and yeasts. This specific binding activates the classical complement pathway. Mannose-binding lectin is a large macromolecule that has a bouquet-like structure. The basic structural subunit of MBL is a homotrimer of MBL polypeptides, entwined in a triple helix. Each single polypeptide chain has four domains: (1) a 21-amino acid N-terminal cysteine-rich region involved in oligomerization, (2) a 59-amino acid collagen-like domain, (3) a 30-amino acid α-helical, hydrophobic coiled-coil neck domain, which is crucial for initiating the oligomerization, and (4) a 188-amino acid C-terminal carbohydrate recognition domain (Shen et al., 2020). The homozygous mutation, i.e., BB, results in the overall absence of the MBL2 protein, whereas heterozygous mutation; i.e., AB, results in a 5 to 10-fold reduction in serum MBL2 protein level when compared to the wild type, i.e., AA. The deficiency of MBL2 gene has been reported as associated with susceptibility to infectious and autoimmune diseases (O'Leary et al., 2016).
Fig. 1.
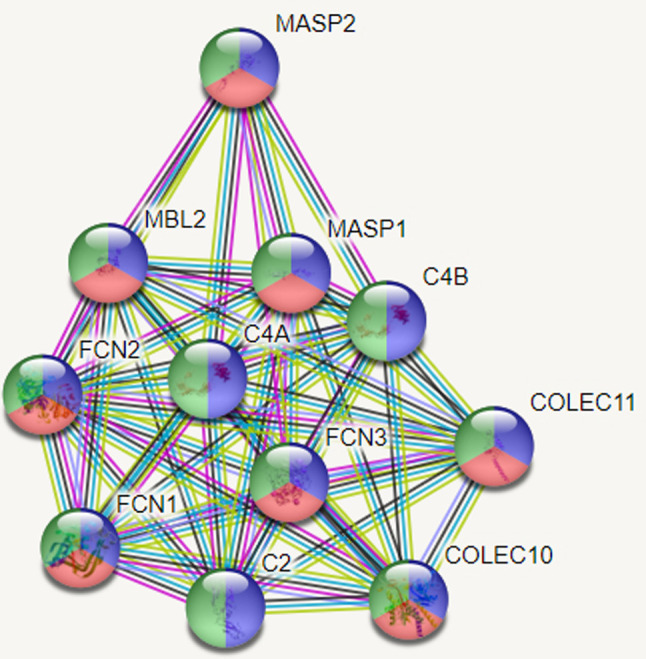
Protein-protein interaction analysis. The proteins with a role in complement activation, complement activation, and lectin pathway of proteins and innate immune response are shown as blue, red and, green, respectively.
In this report, therefore, we aimed to investigate whether MBL2 gene variant (rs1800450) may be related to the variable clinical courses observed interestingly in COVID-19 with a case-control study protocol.
2. Material and methods
2.1. Classification of the clinical course, severe versus mild
We performed laboratory confirmation (RT-PCR examination) of SARS-CoV-2 in certified laboratories where RT-PCR assays were carried out in accordance with the protocol defined by the WHO (World Health Organization, 2020). The patients were considered to have severe disease when any of the following clinical and laboratory parameters are met: a respiratory rate of ≥30/min, suffering from dyspnea, peripheral oxygen saturation of <90%, getting more than 5 L/min nasal oxygen supply, PaO2/FiO2 of ≤300, lactate of >2 mmol/L, hypotension (systolic blood pressure 40 mmHg lower than the usual systolic blood pressure) pulse of >100 beats/min, renal, hepatic, hematological (thrombocytopenia) or cerebral (confusion) dysfunction findings, presence of sepsis or septic shock, skin findings of capillary return disorder such as cutis marmorata and coldness, moderate/severe pneumonia (bilateral infiltration and/or multiple mottling and groundglass opacity), need for anticytokine therapy and/or broad-spectrum antibacterial therapy.
2.2. DNA isolation and analyses
We analyzed rs1800450 (c.161G > A p.Gly54Asp) representing one of the well-studied single nucleotide polymorphisms in the MBL2 gene. The patients were included between April–June 2020 that admitted to the COVID-19 center of a single university hospital. We included 100 healthy volunteers to serve as the control group. The patients and control groups were matched in terms of age and ethnic background. The institutional clinical research ethics committee approved the protocol (21/05/2020–84,539). We isolated DNA from the collected blood by using the Genemark isolation kit (Genemark, USA). All of the patients and controls were examined for the codon 54 A/B (gly54asp: rs rs1800450) variation in exon 1 of the MBL2 gene.
2.3. Genotyping of MBL2 gene (gly54asp - rs1800450)
Polymerase chain reaction (PCR) was performed using forward (5′TAGGACAGAGGGCATGCTC-3′) and reverse (5′-CAGGCAGTTTCCTCTGGAAGG 3′) primers in a 25 mL volume, containing 50 ng of DNA, 2 mM of dNTPs, 2 nmoL of each primer, 1.5 mM of MgCl2 and 3 U of Taq polymerase (Qiagen). The product's 349 base pair (bp) were digested with the restriction enzyme BanI (Fermentase) to identify the codon 54 polymorphism. BanI digestion was performed at 37 °C for overnight incubation with 5 U of the enzyme. After enzyme digestion, products were visualized by electrophoresis on 3% agarose gel. The BanI restriction site is present on wild type allele A (269 and 89 bp) and absent on variant allele B (349 bp) (Baysal et al., 2013). We selected 10% of the sample randomly and controlled the method on this sample (Fig. 2 ).
Fig. 2.
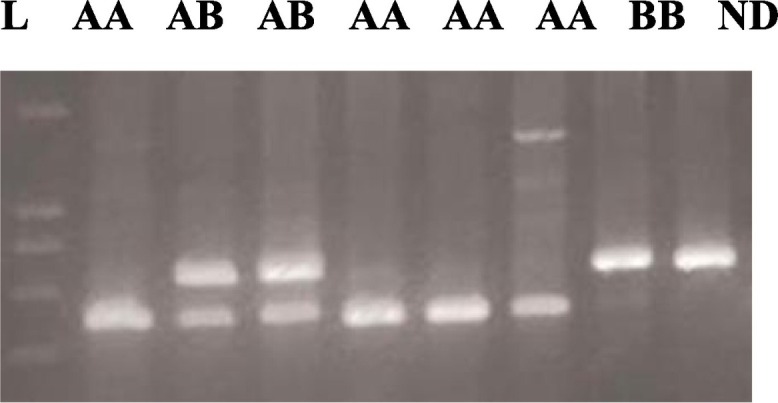
DNA fragments on agarose gel electrophoresis after restriction enzyme digestion of exon 1 of MBL gene. 349 bp PCR product was digested with BanI for codon 54 polymorphism ND). The normal allele (Allele A) is cut into two fragments with BanI, 89 bp, and 260 bp. The variant/mutant allele (Allele B) remains uncut. L: 100 bp DNA ladder ND: non-digested PCR product.
2.4. Statistical analysis
We used IBM SPSS version 21.0 (IBM Corp. released 2012; Armonk, NY, USA) for the statistical analyses. Descriptive statistics included mean, standard deviation, median, minimum, maximum for the continuous variables after assessing the normality, frequency, and percentage for the nominal variables. Pearson Chi-square test with Yates correction or Fisher's exact test were used for 2 × 2 contingency tables when appropriate for non-numerical data. We performed multivariate binary logistic regression analyses to find out the association between different genetic variants of the MBL2 gene with study parameters. The results were adjusted for age and sex. Consequently, we have given the odds ratio (OR) and 95% confidence interval (CI) for the association of MBL gene variants with the study parameters. We used the Hosmer&Lemeshow goodness-of-fit statistic to assess model fit for all regression models. Hardy Weinberg Equilibrium (HWE) was calculated using the De-Finetti program (online HWE and Association Testing- Institut für Humangenetik, Munich, Germany). Statistical significance was accepted as p < 0.05 for the results of all analyses.
3. Results
A total of 284 cases and 100 controls were enrolled in the study. We outlined the clinical features and treatment regimens of COVID-19 patients in Table 1 . The most common comorbidities were hypertension (24.5%) and diabetes mellitus (12.7%). With the pre-specified classification, the severe disease was observed in 33.1%. In the follow-up, at day 28, mortality had occurred in 9 patients (3.2%), and 6.3% had needed intensive unit care. The prevalence of BB genotype was significantly more common in COVID-19 cases when compared to controls (10.9% vs 1.0%, respectively; OR = 12.1, 95%CI = 1.6–90.1, p = 0.001). The prevalence of AB genotype and the AA genotypes were not statistically different in between the COVID-19 cases and controls (for AB genotype, 25.4% in cases vs 35.0% in controls, p = 0.06; for AA genotype, 63.7% in cases vs 64.0% in controls, p = 0.9). We performed multivariate analyses, adjusted for age, sex and MBL genetic variants, to examine whether different MBL genotypes had an independent relationship with the study parameters (i.e. severe disease, ICU need, mortality at day 28 and development of secondary bacterial infection). When compared with the COVID-19 patients that had AA genotype (reference), the patients that had BB or AB genotypes suffered from a higher risk for severe disease (for BB genotype, OR = 5.3, 95% CI = 2.1–13.4, p < 0.001; for AB genotype, OR = 2.9, 95%CI = 1.5–5.7, p = 0.001) (Table 2 ) and for ICU need (for BB genotype, OR = 19.6, 95% CI = 4.6–83.3, p < 0.001; for AB genotype, OR = 6.9, 95% CI = 1.6–30.4, p = 0.001) (Table 3 ). On the other hand, there was not any significant difference between the genotype variants in terms of mortality at 28 days or development of secondary bacterial infection (Table 4, Table 5 ).
Table 1.
Clinical features and treatment regimens of Covid-19 patients (n = 284).
| Age (years) | 49 (19–92) |
| Sex (female vs males) | 128/ 156 (45.1% vs 54.9%) |
| Co-morbid diseases | |
| Hypertension | 72 (24.5%) |
| Diabetes mellitus | 36 (12.7%) |
| Chronic obstructive pulmonary disease | 30 (10.6%) |
| Coronary artery disease | 17 (6.0%) |
| Congestive heart failure | 5 (1.8%) |
| Solid malignancy | 26 (9.2%) |
| Hematological malignancy | 7 (2.5%) |
| Clinical course | |
| Severe vs mild | 94/190 (33.1% vs 66.9%) |
| Symptoms | |
| Dry cough | 170 (59.9%) |
| Fever | 154 (54.2%) |
| Myalgia | 145 (51.1%) |
| Dyspnea | 94 (33.1%) |
| Nausea/vomitus | 30 (10.6%) |
| Diarrhea | 23 (8.1%) |
| Anosmia/dysgeusia | 13 (4.6%) |
| Sputum | 1 (0.4%) |
| Physical examination findings on admission | |
| Fever | 94/190 (33.1%/66.9%) |
| Body temperature | 36.6 (35–40) |
| Saturation pO2 (peripheral) | 97 (87–100) |
| Systolic blood pressure | 130 (90–240) |
| Diastolic blood pressure | 75 (50–100) |
| Pulse (/min) | 92 (60–160) |
| Respiratory rate (/min) | 16 (12–40) |
| Laboratory evaluation | |
| pH | 7.41 (7.1–8.1) |
| pO2 | 62 (35–86) |
| pCO2 | 40 (23–63) |
| HCO3 | 24 (14–35) |
| Lactate | 1.5 (1–7) |
| Hemoglobin (g/dL) | 13.2 (6.3–18) |
| Leukocyte (/mm3) | 7130 (930–28,300) |
| Thrombocyte (/mm3) | 241,000 (66000–576,000) |
| Lymphocyte (/mm3) | 1390 (270–4520) |
| Lymphopenia (<800/mm3) | 57 (20.1%) |
| BUN (mg/dL) | 13 (5–107) |
| Creatinine (mg/dL) | 0.8 (0.4–6) |
| Glucose (mg/dL) | 106 (68–496) |
| AST (U/L) | 21 (10–409) |
| ALT (U/L) | 21 (2–493) |
| GGT (U/L) | 21 (4–744) |
| ALP (U/L) | 73 (33–400) |
| LDH (U/L) | 194 (78–731) |
| Total protein (g/dL) | 7.4 (5–9) |
| Albumin (g/dL) | 4.0 (2–5) |
| CRP (mg/dL) | 19.5 (1–363) |
| Procalcitonin (ng/mL) | 0.06 (0.2–50.0) |
| Ferritin | 166 (6–6656) |
| D-dimer | 58 (5–35,000) |
| Troponin-T | 4 (3–848) |
| Fibrinogen | 423 (151–1053) |
| INR | 0.9 (0.8–3.8) |
| aPTT | 28 (21–53) |
| Follow-up | |
| Mortality (at 28 days) | 9 (3.2%) |
| Intensive care unit need | 16 (6.3%) |
Data are given as numbers (%) or median (interquartile range).
Table 2.
Multivariate regression analysis for association of MBL gene variants with severe and mild disease.
| MBL Genotype | Severe disease (n = 94) | Mild disease (n = 190) | OR | 95% CI | p |
|---|---|---|---|---|---|
| AA | 44 (46.8%) | 137 (72.1%) | REFERENCE | ||
| AB | 29 (30.3%) | 43 (22.6%) | 2.9 | 1.5–5.7 | 0.001⁎ |
| BB | 21 (22.4%) | 10 (5.3%) | 5.3 | 2.1–13.4 | <0.001⁎ |
Dependent variable was: severe vs mild disease, independent variables were age, sex and MBL genetic variants.
Hosmer&Lemeshow Model ꭓ2 (8) = 4.40; p = 0.819.
significant association.
Table 3.
Multivariate regression analysis for association of MBL gene variants for intensive care unit.
| MBL Genotype | ICU need (n = 18) | No ICU need (n = 266) | OR | 95% CI | p |
|---|---|---|---|---|---|
| AA | 3 (33.3%) | 178 (66.9%) | REFERENCE | ||
| AB | 6 (66.7%) | 66 (24.8%) | 6.9 | 1.61–30.4 | 0.009⁎ |
| BB | 9 (50%) | 22 (8.3%) | 19.6 | 4.6–83.3 | <0.001⁎ |
Dependent variable was: intensive care need, independent variables were age, sex and MBL genetic variants.
Hosmer&Lemeshow Model ꭓ2 (8) = 5.489; p = 0.704.
ICU: intensive care unit.
significant association.
Table 4.
Multivariate regression analysis for association of MBL gene variants for early mortality and survival (n = 284).
| MBL Genotype | Mortality at 28 days (n = 9) | Survived at 28 days (n = 275) | p |
|---|---|---|---|
| AA | 0 (0%) | 181 (65.8%) | REFERENCE |
| AB | 3 (33.3%) | 69 (25.1%) | 0.99 |
| BB | 6 (67.7%) | 25 (9.1%) | 0.99 |
Dependent variable was: mortality at 28 days, independent variables were age, sex and MBL genetic variants.
Hosmer&Lemeshow Model ꭓ2 (8) = 0.76; p = 0.999.
Table 5.
Multivariate regression analysis for association of MBL gene variants with secondary bacterial infection in the follow-up.
| MBL Genotype | SBI (n = 27) | No SBI (n = 257) | OR | 95% CI | p |
|---|---|---|---|---|---|
| AA | 11 (40.7%) | 170 (66.1%) | REFERENCE | ||
| AB | 9 (33.3%) | 63 (24.5%) | 2.72 | 0.99–7.50 | 0.05 |
| BB | 7 (25.9%) | 24 (9.4%) | 2.81 | 0.90–8.75 | 0.07 |
Dependent variable was: development of secondary bacterial infection, independent variables were age, sex and MBL genetic variants.
Hosmer&Lemeshow Model ꭓ2 (8) = 10.05; p = 0.262.
SBI: secondary bacterial infection.
4. Discussion
The main findings of this case-control study were as follows: in univariate analysis, the MBL BB genotype was significantly more common among COVID-19 cases when compared with controls. Multivariate analyses revealed that when compared with the COVID-19 patients that had AA genotype, the patients that had BB or AB genotypes suffered from a higher risk for severe disease and for ICU need (Table 2, Table 3). On the other hand, there was not any significant difference between the genotype variants in terms of mortality at 28 days or development of secondary bacterial infection (Table 4, Table 5) MBL2 gene exon1 polymorphisms have three variants: B, C, and D variants and they are single-nucleotide polymorphism. They are known as structural variants because they modify the structure of the protein and the assembly of MBL oligomers, lead to the formation of smaller non-functional oligomers ( [17,18]. This affects binding avidity and have a possible functional implication because the prolonged interaction can facilitate self-activation of MBL-associated serine proteases 1 to activate the complement cascade more efficiently [19]. Another consequence of MBL variants is increased susceptibility to degradation by metalloproteases [20]. On the other hand, high MBL levels may facilitate the infection of intracellular pathogens into host cells through C3b receptors and accordingly, some studies have shown that MBL deficiency indicated protection in some infectious diseases [17]. Hence, in the present, there are contradictory findings for associations of MBL2 variants with susceptibility to infections. The MBL2 BB variant results in the overall absence of MBL protein whereas MBL2 AB variant results in a 5 to 10-fold decrease in MBL protein compared to the AA genotype. Our results suggest that, deficient or decreased MBL2 protein due to these variable genotypes is related to acquiring COVID-19 infection and more severe clinical course as shown by higher risk to develop severe disease and ICU need. Moreover, the risks for developing severe disease and ICU need were higher with the BB genotype when compared to the AB genotype (OR = 5.3 vs 2.9 for severe disease and OR = 19.6 vs 6.9 for ICU need, respectively). Hence, this study suggests that as the MBL protein level decreases, the course of COVID-19 becomes more severe in some respects. This finding may contribute to our understanding why some robust younger individuals follow a serious or fatal course and some multimorbid older individuals follow a benign course. MBL2 gene B variant polymorphism may have a role in these cases.
MBL2 gene is one of the innate immune system genes. Its product, mannose-binding lectin is a well-characterized C-type lectin. Innate recognition of virus proteins is one of the vital components of the immune response to viral agents. The family of lectins A composes a component of this immune recognition; pattern recognition receptors (PRRs) which serve to recognize viral pathogen-associated molecular patterns (PAMPs) that include viral glycoproteins (Mason and Tarr, 2015). MBL is a soluble, Ca2 + −dependent protein of the collectin family-characterized as C-type lectins with collagenous domains–encoded by the MBL2 gene on human chromosome 10q11.2–10q21 (Mason and Tarr, 2015; Sastry et al., 1989). While MBL is primarily expressed in the liver and secreted into the blood, lower expression has been detected in other tissues such as mammalian muscle tissue and brain after an immune challenge (Singh et al., 2011). Regarding the role of MBL with viruses, one should note that MBL interacts with several of them, but not all. It happens through a Ca2 + −dependent manner. As an example, MBL interacts with several strains of human immunodeficiency virus-1 (HIV-1) and it is able to bind and neutralize diverse strains of HIV-1 (Saifuddin et al., 2000). MBL also neutralizes influenza A virus (Kase et al., 1999), hepatitis C virus (Brown et al., 2010), and notably the severe acute respiratory syndrome coronavirus (SARS-CoV) (Zhou et al., 2010; Ip et al., 2005), Dengue virus (Avirutnan et al., 2011), and West Nile virus (Fuchs et al., 2010), in vitro. Accordingly, recombinant MBL has been suggested to have therapeutic potential against the Ebola virus infection (Michelow et al., 2011). Considering our results, one can suggest that there may be a similar therapeutic potential of recombinant MBL against the SARSCoV-2 infection, which is the major infection that affects the globe currently with a very significant impact. This area warrants future research.
MBL deficiency-associated alleles have been reported to correlate with increased susceptibility to infectious diseases. Nevertheless, this relationship is complex. As an example, similar to our findings suggesting a worse prognostic role of BB/AB variant in COVID-19, HIV-1 patients carrying the MBL-B allele had higher viral loads in their sera. This finding suggests a decreased MBL-mediated viral elimination (Vallinoto et al., 2006). Higher MBL levels conferred protection. However, on the contrary, some studies found no association between variant alleles and susceptibility to HIV-1 infection or its progression (Malik et al., 2003; Lian et al., 2004; Nielsen et al., 1995). Conflicting evidences are also valid for other viruses such as HCV (Brown et al., 2008; Vallinoto et al., 2009; Esmat et al., 2012), and HBV infections (Höhler et al., 1998). Hence, we cannot put forward a definitive role of MBL genetic variants in the SARS-CoV-2 infection. The association of the MBL gene variant with SARS infection (Ip et al., 2005; Tu et al., 2015; Zhang et al., 2005) deserves special attention as SARS-CoV-2 has a very close similarity with the former SARS infection in 2010. In a study that involved 352 cases with SARS and 392 control subjects, MBL gene polymorphisms were found associated with susceptibility to SARS-CoV infection. The authors suggested that this might be explained by the reduced expression of functional MBL secondary to having the codon 54 variant as in line with the previous pathophysiological studies (Zhang et al., 2005).
Our findings have some future implications. The observation that MBL mutations are associated with increased susceptibility to SARS-CoV-2 and SARS-CoV infections could alter transmission precautions and clinical practices as well. The BB variant individuals seem to be at higher risk for SARS infections. Therefore, general screenings to detect MBL mutations and MBL deficiency may be used to defend against the outbreaks provided that this approach would be cost-effective. Moreover, vaccination efforts are humbly put forward all around the World and these results may imply the selection of candidates for the vaccination with studies for confirmation of the effectiveness of vaccines in such high-risk groups. Another point is that, as mentioned above, MBL replacement therapy, as has been used in patients with repeated infections, could have a potential for decreasing severity of or susceptibility to the disease (Valdimarsson et al., 1998). Of course, the usefulness of this intervention in SARS infections need to be investigated but seems to deserve it.
Our study has some limitations. The most important point is the narrow patient population included in the study. The contribution to the literature can be increased with a study design to be conducted in a larger population. The sample size is too small to draw convincing conclusions from statistical inference, particularly for early mortality (n = 9), intensive care need (n = 18), and secondary bacterial infection (n = 27). Apart from this, treatment preferences and MBL2 association can be studied in more detail in a larger patient group with subgroups, even thrombosis secondary to inflammation can be investigated. In this case-control study, the BB variant was significantly more common among COVID-19 cases when compared with controls. However, one should consider that this was an unadjusted model because the sampling process of the study was observational and, therefore, we did not have detailed data to eliminate all possible confounding factors due to the study design. Nevertheless, this analysis suggests a strong relationship between the presence of BB genotype and acquiring COVID-19 infection.
5. Conclusion
In summary, it is the first time that an association between an MBL polymorphism and increased susceptibility and a more severe course of SARS-CoV-2 infection has been shown by this study. Future studies are needed in different populations, preferably with a higher case number. Nevertheless, the information on genetic factors that contribute to the pathogenesis and severity of COVID-19 disease is currently been highly warranted. These insights have the potential to contribute to the control of the outgoing pandemic and improve treatment outcomes.
Author statements
All authors have read and approved the version to be submitted. All authors had access to all the study data, take responsibility for the accuracy of the analysis, and had authority over manuscript preparation and the decision to submit the manuscript for publication.
The authors affirm that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as originally planned (and, if relevant, registered) have been explained.
The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
The authors didn't receive any writing assistance.
This study was financially partially supported by the Istanbul University Scientific Research Project (TSG-2020-36785).
The study protocol was approved by the Istanbul University, Istanbul Medical School ethics committee (21/05/2020-84539).
This study conducted according to the guidelines laid down in the Declaration of Helsinki.
This manuscript, including related data and tables has not been previously published.
Data sharing statement
All data relevant to the study are included in the uploaded article.
Declaration of Compeitng Interest
None.
Acknowledgements
Authors would like to thank Merve Oren and Mustafa Pehlivan for their contribution to statistical analyses, and Akın Tekcan for his contribution to bioinformatics analysis.
Written permission has been obtained from Merve Oren, Mustafa Pehlivan and Akin Tekcan
References
- Avirutnan P., Hauhart R.E., Marovich M.A., Garred P., Atkinson J.P., Diamond M.S. Complement-mediated neutralization of dengue virus requires mannose-binding lectin. mBio. 2011;2(6) doi: 10.1128/mBio.00276-11. Published 2011 Dec 13. doi:10.1128/mBio.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal E., et al. The polymorphisms of the MBL2 and MIF genes associated with Pediatric Cochlear Implant Patients. Int. J. Pediatr. Otorhinolaryngol. 2013;77(3):338–340. doi: 10.1016/j.ijporl.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Brown E.E., et al. MBL2 and hepatitis C virus infection among injection drug users. BMC Infect. Dis. 2008;8:57. doi: 10.1186/1471-2334-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.S., et al. Specific interaction of hepatitis C virus glycoproteins with mannan binding lectin inhibits virus entry. Protein Cell. 2010;1(7):664–674. doi: 10.1007/s13238-010-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmat S., et al. Serum mannan-binding lectin in egyptian patients with chronic hepatitis C: its relation to disease progression and response to treatment. Hepat. Mon. 2012;12(4):259–264. doi: 10.5812/hepatmon.704. doi:10.5812/hepatmon.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A., et al. Direct complement restriction of flavivirus infection requires glycan recognition by mannose-binding lectin. Cell Host Microbe. 2010;8(2):186–195. doi: 10.1016/j.chom.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhler T., et al. No association between mannose-binding lectin alleles and susceptibility to chronic hepatitis B virus infection in german patients. Exp. Clin. Immunogenet. 1998;15:130–133. doi: 10.1159/000019064. [DOI] [PubMed] [Google Scholar]
- https://covid19.who.int/
- https://www.aa.com.tr/en/europe/113-year-old-spanish-woman-beatscoronavirus/1837442
- https://www.genome.jp/kegg/pathway.html
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Hu Y., et al. Prevalence and severity of corona virus disease 2019 (COVID-19): A systematic review and meta-analysis. J. Clin. Virol. 2020;127:104371. doi: 10.1016/j.jcv.2020.104371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikitimur H., et al. Determining host factors contributing to disease severity in a family cluster of 29 hospitalized SARS-CoV-2 patients: could genetic factors be relevant in the clinical course of COVID-19? J. Med. Virol. 2020;93(1):357–365. doi: 10.1002/jmv.26106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip W.K., et al. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;191(10):1697–1704. doi: 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kase T., et al. Human mannan-binding lectin inhibits the infection of influenza A virus without complement. Immunology. 1999;97(3):385–392. doi: 10.1046/j.1365-2567.1999.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.D., et al. Human genetic determinants of viral diseases. Annu. Rev. Genet. 2017;51:241–263. doi: 10.1146/annurev-genet-120116-023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Y.C., et al. Immunological analysis in paediatric HIV patients at different stages of the disease. Scand. J. Immunol. 2004;60(6):615–624. doi: 10.1111/j.0300-9475.2004.01492.x. [DOI] [PubMed] [Google Scholar]
- Malik S., et al. Absence of association between mannose-binding lectin gene polymorphisms and HIV-1 infection in a Colombian population. Immunogenetics. 2003;55(1):49–52. doi: 10.1007/s00251-003-0550-4. [DOI] [PubMed] [Google Scholar]
- Mason C.P., Tarr A.W. Human lectins and their roles in viral infections. Molecules. 2015;20(2):2229–2271. doi: 10.3390/molecules20022229. Published 2015 Jan 29. doi:10.3390/molecules20022229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelow I.C., et al. High-dose mannose-binding lectin therapy for Ebola virüs infection. J. Infect. Dis. 2011;203(2):175–179. doi: 10.1093/infdis/jiq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.L., et al. The level of the serum opsonin, mannan-binding protein in HIV 1 antibody-positive patients. Clin. Exp. Immunol. 1995;100(2):219–222. doi: 10.1111/j.1365-2249.1995.tb03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary N.A., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E., et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020;8(9) doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifuddin M., et al. Interaction of mannose-binding lectin with primary isolates of human immunodeficiency virus type 1. J. Gen. Virol. 2000;81(Pt 4):949–955. doi: 10.1099/0022-1317-81-4-949. [DOI] [PubMed] [Google Scholar]
- Sastry K., et al. The human mannose-binding protein gene. Exon structure reveals its evolutionary relationship to a human pulmonary surfactant gene and localization to chromosome 10. J. Exp. Med. 1989;170(4):1175–1189. doi: 10.1084/jem.170.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., et al. Clinical characteristics and differential clinical diagnosis of novel coronavirus disease 2019 (COVID-19) Coronavirus Disease 2019 (COVID-19). 2020;55-70 Published 2020 Apr 30. doi:10.1007/978-981-15-4814-7_6. [Google Scholar]
- Shen W., et al. Association between polymorphisms in mannose-binding lectin 2 gene with pulmonary tuberculosis susceptibility. Hereditas. 2020;157(1):33. doi: 10.1186/s41065-020-00146-w. Published 2020 Aug 3. doi:10.1186/s41065-020-00146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K.K., et al. Expression of mannose binding lectin in HIV-1-infected brain: implications for HIV-related neuronal damage and neuroAIDS. Neurobehav. HIV Med. 2011;3:41–52. doi: 10.2147/NBHIV.S19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamara A., Tahapary D.L. Obesity as a predictor for a poor prognosis of COVID-19: A systematic review. Diabetes Metab Syndr. 2020;14(4):655–659. doi: 10.1016/j.dsx.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X., et al. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J. Inf. Secur. 2015;71(1):101–109. doi: 10.1016/j.jinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdimarsson H., et al. Reconstitution of opsonizing activity by infusion of mannan-binding lectin (MBL) to MBL-deficient humans. Scand. J. Immunol. 1998;48(2):116–123. doi: 10.1046/j.1365-3083.1998.00396.x. [DOI] [PubMed] [Google Scholar]
- Vallinoto A.C., et al. Mannose-binding lectin gene polymorphism and its impact on human immunodeficiency virus 1 infection. Mol. Immunol. 2006;43(9):1358–1362. doi: 10.1016/j.molimm.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Vallinoto A.C., et al. Mannose-binding lectin gene polymorphisms are not associated with susceptibility to hepatitis C virus infection in the Brazilian Amazon region. Hum. Immunol. 2009;70(9):754–757. doi: 10.1016/j.humimm.2009.06.014. [DOI] [PubMed] [Google Scholar]
- World Health Organization . Interim guidance; 2020. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases.https://www.who.int/publications/i/item/10665-331501 [Google Scholar]
- Zhang H., et al. Association between mannose-binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;192(8):1355–1361. doi: 10.1086/491479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., et al. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannosebinding lectin through multiple mechanisms. J. Virol. 2010;84(17):8753–8764. doi: 10.1128/JVI.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


