Abstract
The family Apiaceae includes many important vegetables and medicinal plants. Auxin response factors (ARFs) play critical roles in regulating plant growth and development. Here, we performed a comprehensive analysis of the ARF gene family in three Apiaceae species, celery, coriander, and carrot, and compared the results with the ARF gene family of lettuce, Arabidopsis, and grape. We identified 156 ARF genes in all six species and 89 genes in the three Apiaceae species, including 28, 34, and 27 in celery, coriander, and carrot, respectively. The paralogous gene number in coriander was far greater than that in carrot and celery. Our analysis revealed that ARF genes of the three Apiaceae species in 34 branches of the phylogenetic tree underwent significant positive selection. Additionally, our findings indicated that whole-genome duplication played an important role in ARF gene family expansion. Coriander contained a greater number of ARF genes than celery and carrot because of more gene duplications and less gene losses. We also analyzed the expression of ARF genes in three tissues by RNA-seq and verified the results by quantitative real-time PCR. Furthermore, we found that several paralogous genes exhibited divergent expression patterns. Overall, this study provides a valuable resource for exploring how ARF family genes regulate plant growth and development in other plants. Since this is the first report of the ARF gene family in Apiaceae, our results will serve as a guide for comparative and functional analyses of ARF and other gene families in Apiaceae.
Keywords: ARF gene family, orthologous and paralogous genes, gene duplication and loss, expression pattern, Apiaceae species, phylogenetic analysis
Introduction
Celery (Apium graveolens), coriander (Coriandrum sativum), and carrot (Daucus carota) are three typical members of Apiaceae family. They grow all over the world and are especially famous for their fragrance and medicinal value. Celery is a multipurpose plant, which used as a vegetable and medicinal herb for treating diseases (Ballmer-Weber et al., 2002; Maljaei et al., 2019). Coriander contains many bioactive phytochemicals and has also been used as a traditional medicine (Prachayasittikul et al., 2018). Carrot is one of the most important root vegetables around the world and is valued for its high content of beta-carotene, an essential component of vitamin A (Ahmad et al., 2019). Above all, these three Apiaceae crops are of high economical value, given their medicinal and edible properties, and therefore are a major source of income for growers.
The phytohormone auxin plays a very significant role in regulating not only plant developmental processes, such as apical dominance, later root initiation, and vascular differentiation, but also in cellular processes, including cell division, expansion, and differentiation (Wang et al., 2007; Xing et al., 2011). Auxin Response Factors (ARFs) regulate the expression of auxin-responsive genes during plant growth. The auxin response signal is transmitted to related response genes via auxin response elements (AuxREs) (Wan et al., 2014).
ARF1 was the first ARF gene identified in Arabidopsis thaliana using a yeast one-hybrid screen (Ulmasov et al., 1997). A total of 23 ARF genes have been identified in Arabidopsis to date (Okushima et al., 2005). With the sequencing of genomes, more ARF family genes have been detected in plants. For example, 39 ARF genes were identified in poplar (Populus trichocarpa) (Kalluri et al., 2007), 25 in rice (Oryza sativa) (Wang et al., 2007), 19 in grape (Vitis vinifera) (Wan et al., 2014), 19 in sweet orange (Citrus × sinensis) (Li et al., 2015), 31 in Chinese cabbage (Brassica rapa subsp. pekinensis) (Mun et al., 2012), 20 in pineapple (Ananas comosus) (Su et al., 2017), 20 in barley (Hordeum vulgare) (Tombuloglu, 2019), 31 in maize (Zea mays) (Xing et al., 2011), 17 in tomato (Solanum lycopersicum) (Kumar et al., 2011), 31 in apple (Malus domestica) (Luo et al., 2014), 17 in physic nut (Jatropha curcas) (Tang et al., 2018), and 19 in pepper (Capsicum annuum) (Zhang et al., 2017). However, a comprehensive analysis of ARF gene family in Apiaceae species has not yet been reported. Recently, more sequencing data of Apiaceae have been released, thus serving as a valuable resource for further analysis of the ARF gene family (Iorizzo et al., 2016; Song et al., 2020b).
In this study, we performed a comprehensive and systematic analysis of the ARF gene family in three representative species of Apiaceae (celery, coriander, and carrot), with the aim to (i) identify ARF gene family members; (ii) classify these members based on phylogenetic relationship; (iii) map ARF genes to chromosomes; (iv) identify the paralogous and orthologous genes; (v) explore ARF gene loss and duplication; and (vi) explore the gene expression patterns in three tissues of celery and coriander.
Materials and Methods
Genome Sequence Retrieval and ARF Gene Identification
Whole genome sequences of coriander and celery were retrieved from the Coriander Genome Database (CGDB1) (Song et al., 2020a). The genome sequence of Arabidopsis was downloaded from the TAIR database2. Genome sequences of carrot (v2.0), lettuce (v5.0), and grape (Genoscope.12X) were downloaded from Phytozome (Jaillon et al., 2007; Iorizzo et al., 2016; Reyes-Chin-Wo et al., 2017).
The Pfam database was used to identify ARF family genes using the identifier PF06507 with an e-value < 1e-4 (Punta et al., 2012). Furthermore, the conserved domain database (CDD) and simple modular architecture research tool (SMART) were used to verify the identified genes (Marchler-Bauer et al., 2009; Letunic et al., 2012).
Phylogenetic Analysis of ARFs
Amino acid sequences of ARFs of celery, coriander, carrot, Arabidopsis, lettuce, and grape were used for phylogenetic analysis. First, sequences were aligned using ClustalW (Li, 2003). Then, the multiple sequence alignment was restored in the PHYLIP format. Finally, a phylogenetic tree was constructed using the maximum likelihood (ML) method with IQ-TREE (Nguyen et al., 2015), based on JTT + F + R8 model, with 1,000 bootstrap replications.
Chromosomal Location, Gene Structure, and Conserved Motif Analysis of ARF Genes
The chromosomal location of each ARF gene was retrieved from general feature format (gff) file, and the chromosome number, start position and end position of each gene were extracted using a Perl script. The chromosome information file was submitted to the MapChart to display the distribution of each gene (Voorrips, 2002).
The structure of ARF genes was drawn using Gene Structure Displayer Server 2.0 (GSDS) (Hu et al., 2015). The gff file of ARF gene was submitted to GSDS to illustrate the positions of exons, introns, and untranslated regions (UTRs). Conserved motif analysis was performed using the Multiple Expression motifs (Em) for Motif Elicitation (MEME) (Bailey et al., 2009).
Analysis of ARF Gene Duplication and Loss
Orthologous and paralogous genes were identified using the OrthoMCL, with an e-value of 1e-5 (Li et al., 2003). The relationship among ARF gene orthologs and paralogs was illustrated using the Circos (Krzywinski et al., 2009). Gene duplication and gene loss analyses were performed using the Notung2.9 (Stolzer et al., 2012).
Analysis of Collinearity and Duplication Type
The MCScanX was used to conduct collinearity analysis (Wang et al., 2012). Amino acid sequences were analyzed using the Blastp, with an e-value set at 1.0 × 10–5. Then, collinear blocks were detected by submitting the whole genome gff and Blastp result to the MCScanX. The duplicate_gene_classifier sub-program was used to identify the duplication type.
Evolutionary Analysis of ARF Genes
Coding sequences (CDSs) of orthologous ARF gene pairs were aligned using the ClustalW, and the alignment file was transformed into the axt format file. The synonymous (Ks) and non-synonymous (Ka) substitution rates were calculated using the Ka/Ks_calculator 2.0 (Wang et al., 2010). The divergence time (T) was estimated using the equation, T = Ks/2r. The “r” represents neutral substitution rate (5.2 × 10–9 substitutions per site per year) (Song et al., 2020b).
Selective Pressure Analysis of ARF Gene Family
The selective pressure analysis was performed using the PAML4.9 (Yang, 2007). The ML method and codon substitution models were adopted to test the likelihood rate of positive selection. Firstly, CDSs of ARF genes were aligned using the ClustalW. Then, each branch of the phylogenetic tree constructed by the PhyML3.0 using amino acid sequences was analyzed to speculate ω (the ratio of non-synonymous to synonymous distances) (Guindon et al., 2010). The M0, M1, M7, and M8 models were used to calculate variation sites.
ARF Gene Expression Analysis Using RNA-Seq
RNA-seq data of ARF gene expression in three tissues (root, petiole, and leaf) of celery and coriander (each with three replicates) were obtained from our previous study (Song et al., 2020a). The RNA-seq data were deposited in the Genome Sequence Archive (GSA) of the BIG Data Center3 under the accession numbers CRA001996 and CRA001658. The expression data expressed as Fragments Per Kilobase of transcript sequence per Millions base pairs (FPKM) were log2-transformed for cluster analysis, as described previously (Song et al., 2014b). Hierarchical clustering analysis was conducted using the TBtools (Chen et al., 2020).
Verification of RNA-Seq Data
The RNA-seq data of celery and coriander ARF genes were verified by quantitative real-time PCR (qRT-PCR). Total RNA was extracted from each sample using the RNA Kit (Tiangen, Beijing, China), and the mRNA was transcribed into cDNA using the PrimeScript cDNA Synthesis Kit (TaKaRa, Dalian, China). The resulting cDNA was used as a template for qRT-PCR, which was performed on the CFX96TM Real-Time System (Bio-Rad, Beijing, China) using sequence-specific primers (Supplementary Table 1), with three replicates for each gene as described previously (Song et al., 2014a, 2016).
Results
Identification, Phylogenetic Analysis, and Classification of ARF Genes
We identified 28, 34, and 27 ARF genes in celery, coriander, and carrot, respectively, and renamed these genes according to their order on chromosomes (Supplementary Tables 2, 3). Lettuce shows the closest relationship with Apiaceae, and its genome sequence has been released (Reyes-Chin-Wo et al., 2017). Additionally, the ARF gene family of Arabidopsis and grape has been analyzed in detail (Okushima et al., 2005; Wei et al., 2006; Wan et al., 2014). Comparative analysis of the ARF genes of celery, coriander, and carrot with the Arabidopsis, lettuce, and grape genomes revealed 22, 26, and 19 ARF genes in the latter three species, respectively.
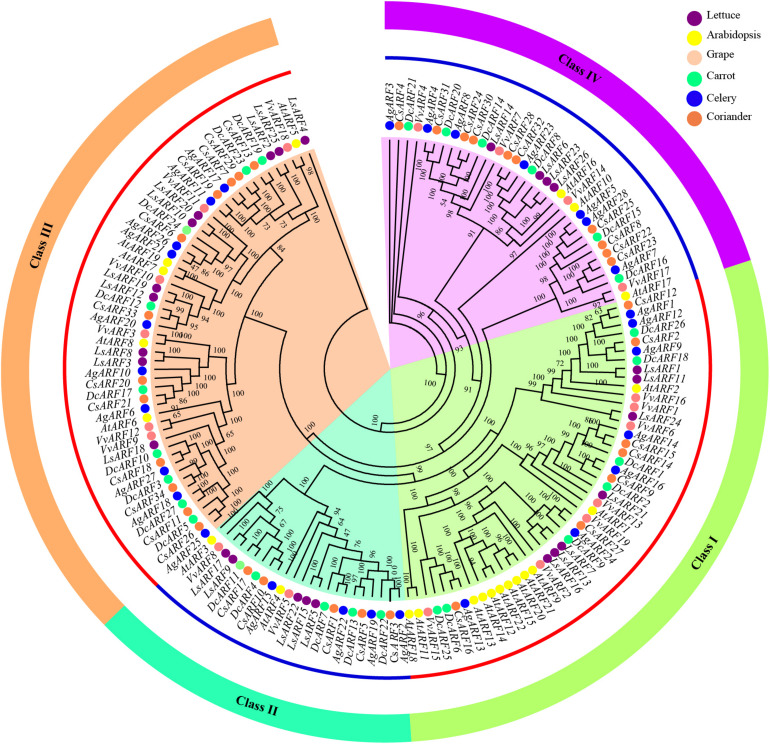
To explore the evolutionary history and relationship of the ARF gene family, we constructed a phylogenetic tree using 156 ARF amino acid sequences from six species, including Arabidopsis, lettuce, grape, celery, coriander, and carrot (Figure 1). According to the phylogenetic analysis, all ARFs were divided into four classes (I − IV), based on the topology and classification in grape. Classes I and III contained a greater number of ARFs than classes II and IV. Interestingly, the number of Arabidopsis ARFs in one branch of class I was notably higher than that of other species, and eight Arabidopsis ARFs (AtARF9, AtARF12–15, AtARF20–22) clustered together.
FIGURE 1.
Phylogenetic analysis of ARF amino acid sequences in three Apiaceae species (carrot, celery, and coriander), lettuce, grape, and Arabidopsis. The phylogenetic tree was generated using the IQ-TREE software with the maximum likelihood (ML) based on the JTT + F + R8 model and 1,000 bootstrap replications.
We further constructed another phylogenetic tree using ARF sequences of celery, coriander, and carrot (Supplementary Figure 1). ARFs were also divided into four classes. The classification of ARFs among three Apiaceae species was highly consistent with that of six plant species (Figure 1). Additionally, classification within the Apiaceae showed that class III contained the highest number of ARFs (31), followed by class IV (23), class I (21), and class II (14).
ARF Gene Structure Analysis and Conserved Motif Identification
To further explore the conservation of ARF family genes, we analyzed the gene structure and motifs. Full-length cDNA sequences of ARF genes were compared with the corresponding genomic sequences using the GSDS program (Supplementary Figure 2A). The number of exons in ARF genes varied from 1 − 15. In class I, most ARF genes contained 10–14 exons, with the exception of AgARF1 and CsARF16. In class III, most ARF genes harbored 10–15 exons, except AgARF26 and CsARF29. In class IV, ARF genes contained only 3–4 exons, which was far less than the number of exons in other classes. We found that the position and number of exons and introns in genes belonging to the same class or subclass were similar. This finding supports the phylogenetic relationship of ARF family genes.
We also analyzed conserved motifs in ARF genes using MEME. Eight motifs were detected (Supplementary Figure 2B), of which four (motifs 1, 2, 3, and 6) were common to almost all ARF genes. In classes I–III, most ARF genes contained motifs 1–4 and motifs 6–8. Interestingly, AgARF1 contained only motif 4. In class IV, all ARF genes carried motifs 1–4 and motifs 6–8, except CsARF30. However, motif 5 was absent from almost all genes in class IV. In conclusion, motif 5 was lost in most ARF genes of celery, carrot, and coriander, while motifs 1–4 and motifs 6–8 were highly conserved in three Apiaceae species.
Chromosomal Distribution of ARF Family Genes
In celery, 25 out of 28 ARF genes were unevenly distributed on 10 chromosomes, and three genes could not be mapped to any chromosome (Supplementary Figure 3A). Chromosomes 3 and 11 harbored the highest number of ARF genes (four genes), but no gene was detected on chromosome 8. Several genes, such as AgARF4, AgARF8, AgARF9, AgARF12, and AgARF25, were located at the end of the chromosomes 2, 3, 4, 5, and 11, respectively.
In coriander, 28 out of 34 ARF genes were unevenly distributed on eight chromosomes, while six ARF genes could not be mapped to any chromosome (Supplementary Figure 3B). Chromosome 10 contained the highest number of ARF genes (nine genes), but no ARF gene was found on chromosomes 6 and 8. Two ARF gene clusters were detected at the ends of chromosome 10, which might be caused by gene duplication. CsARF3, CsARF14, and CsARF20 were located very close to CsARF4, CsARF15, and CsARF21, respectively.
In carrot, all 27 ARF genes were mapped to one of the eight chromosomes (Supplementary Figure 3C). Chromosome 2 carried the highest number of ARF genes (seven genes), followed by chromosomes 4 and 5, whereas chromosomes 3 and 8 harbored only one ARF gene each.
Identification of Orthologous and Paralogous Gene Pairs
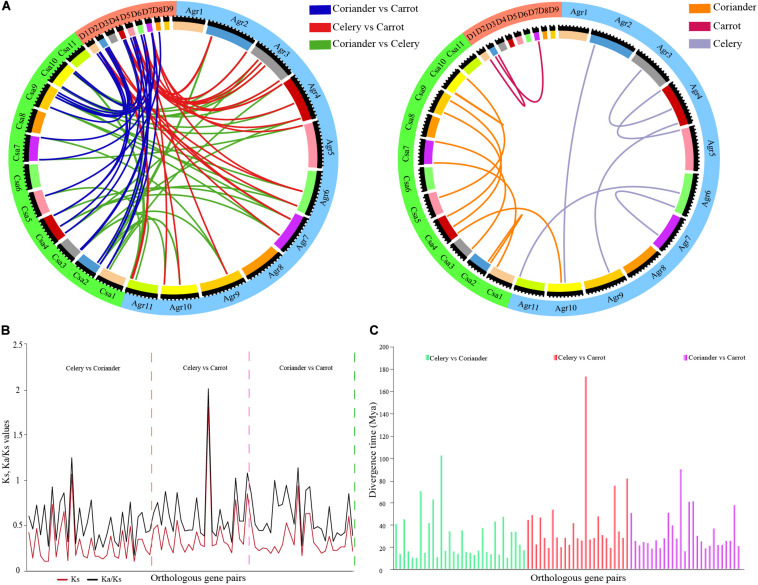
Forty orthologous gene pairs were detected between celery and coriander, while eight gene pairs could not be mapped to any chromosome. Twenty-six orthologous genes were found between celery and carrot, of which one pair could not be mapped. Totally, 29 orthologous gene pairs were identified between coriander and carrot (Figure 2A and Supplementary Table 4). Next, we identified paralogous gene pairs to explore the relationship of ARF genes within species (Supplementary Table 5). A total of eight and 12 paralogous gene pairs were detected in celery and coriander, respectively; however, only two paralogous gene pairs were identified in carrot.
FIGURE 2.
Analysis of paralogous and orthologous ARF gene pairs. (A) Circos plot of ARF gene paralogs and orthologs in three Apiaceae species. (B) Ks and Ka/Ks values of orthologous ARF gene pairs between any two of three Apiaceae species. (C) Divergence time of orthologous ARF gene pairs between any two of three Apiaceae species.
We calculated the Ka/Ks ratios (Figure 2B and Supplementary Table 6) and divergence time for orthologous genes using Ks values (Figure 2C and Supplementary Table 6). The results showed the divergence time of orthologous gene pairs varied from 10.52–102.39 million years between celery and coriander, 19.38–173.48 million years between celery and carrot and 16.40–90.09 million years between coriander and carrot.
Whole-Genome Duplication (WGD) Played a Leading Role in the Expansion of ARF Gene Family in Apiaceae
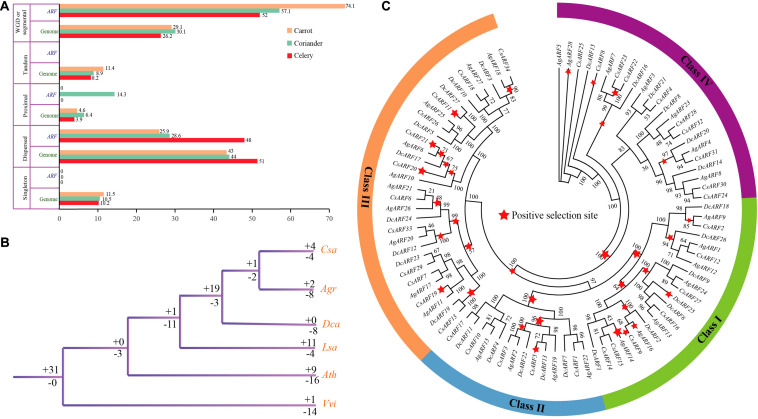
Five gene duplication types were detected, including singleton, dispersed, proximal, tandem, and WGD or segmental duplication (Figure 3A and Supplementary Tables 7, 8). The results indicated that WGD or segmental duplication played a significant role in ARF gene family expansion in Apiaceae species. In celery, coriander, and carrot, 52.0, 57.1, and 74.1% of ARF genes, respectively, arose by WGD or segmental duplication. No singleton or tandem duplication was detected in ARF gene family of these species. Moreover, most ARF genes formed collinear blocks within each genome (Supplementary Table 9).
FIGURE 3.
Duplication, loss, and positive selection analysis of ARF genes in three Apiaceae species. (A) Percentage of different duplication types for ARF genes and all other genes. (B) Analysis of ARF gene duplication and loss. The “ + ” and “–” symbols indicate ARF gene duplication and loss, respectively, and the values after these symbols represent gene number. (C) Positive selection of ARF genes in celery, coriander, and carrot. Red stars represent positive selection branches.
More Genes Were Lost in Celery and Carrot Than in Coriander After Gene Duplication
In celery, the number of ARF genes lost was greater than that duplicated (8 vs. 2), whereas in coriander, the number of ARF genes lost and duplicated was equal (Figure 3B and Supplementary Figure 2). Interestingly, in carrot, eight genes were lost, but no gene duplication was detected compared with other species. This phenomenon indicates that more genes were lost after the genome duplication in Apiaceae species. In the common ancestor of coriander and celery, one ARF gene duplication and two gene losses were detected. In the common ancestor of coriander, celery, and carrot, 19 genes were duplicated while three genes were lost. This phenomenon indicates that there were more gene duplications in the common ancestor of Apiaceae species compared with the ancestor of lettuce and Apiaceae.
Most Apiaceae ARF Genes Underwent Positive Selection
Strong positive selection was observed at major nodes of the phylogenetic tree, which may have contributed to the divergence of Apiaceae species. A total of 10, 4, 13, and 5 positive selection branches were detected in class I, II, III, and IV, respectively (Figure 3C). The number of positive selection sites was the highest in class III, indicating that ARF genes in class III were under stronger natural selection than those in other classes. Overall, we found that most branches underwent positive selection, which indicates that ARF genes played an important role in the evolution of Apiaceae.
Expression Analysis of ARF Genes
We analyzed the expression patterns of ARF genes in three different tissues of celery and coriander. In celery, 16, 9, and 2 ARF genes showed higher expression levels in the root, petiole, and leaf, respectively (Supplementary Table 10 and Supplementary Figure 5A). The expression level (FPKM values) of three celery genes (AgARF9, AgARF12, AgARF24) was over 100 in the root. Among all AgARF genes, AgARF9 showed the highest expression level in the petiole. However, AgARF26 showed no expression in any of the three tissues, while AgARF1 and AgARF7 showed no expression in the leaf. Overall, genes in the same phylogenetic group or subgroup showed a similar expression patterns.
In coriander, 9, 22, and 1 ARF genes exhibited higher expression in the root, petiole, and leaf, respectively (Supplementary Table 10). Most coriander ARF genes were expressed to higher levels in the petiole than in the other two tissues (Supplementary Figure 5B). CsARF27 showed the highest expression level in the root, while CsARF12 showed the highest expression level in petiole and leaf. However, CsARF22 and CsARF23, which clustered within the same phylogenetic group, were not expressed in any tissue.
To validate the RNA-seq data, the expression of six ARF genes was analyzed by qRT-PCR. The results were consistent with the transcriptome results (Supplementary Figure 6), indicating that our RNA-seq data were reliable.
ARF Gene Paralogs Exhibit Notably Different Expression Patterns
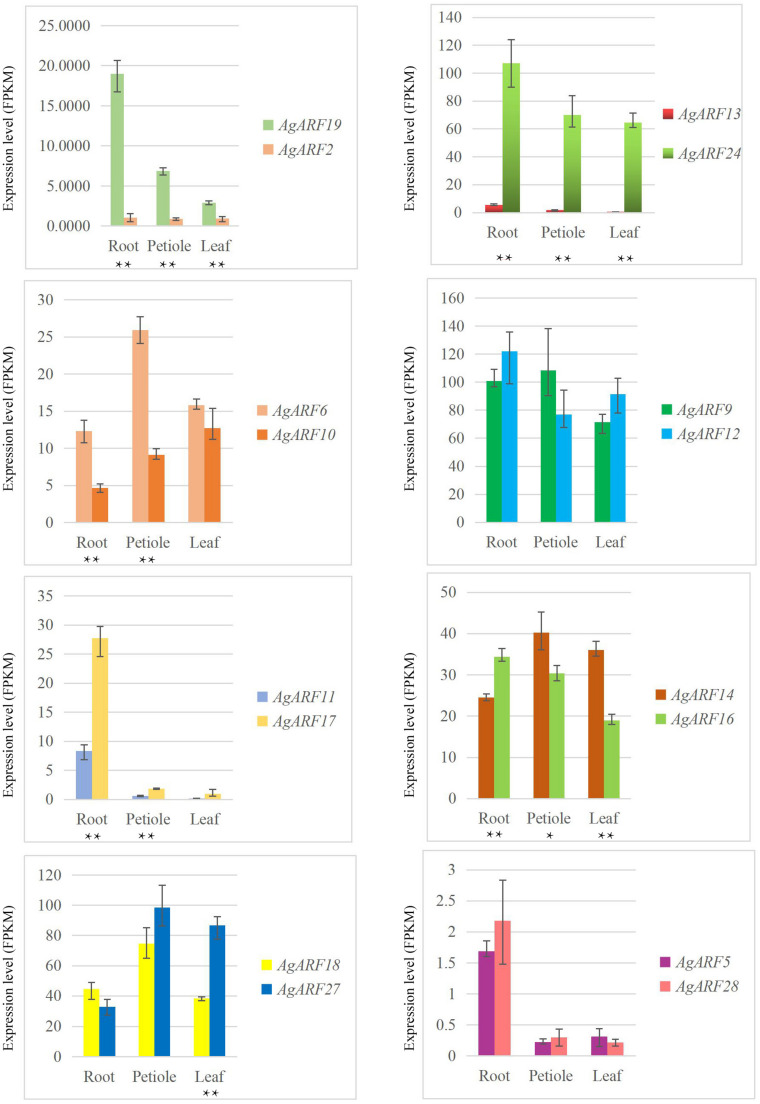
Next, we explored the expression patterns of paralogous genes in celery and coriander (Figure 4 and Supplementary Figure 7). Although most paralogous genes showed similar expression patterns, there were several exceptions. For example, the expression level of AgARF19 was notably higher than that of AgARF2 in all three tissues (Figure 4). Similarly, the expression of AgARF24 was notably higher than that of AgARF13 in three tissues. In coriander, CsARF5, CsARF7, CsARF10, and CsARF32 were expressed to higher levels than CsARF3, CsARF13, CsARF17, and CsARF28, respectively (Supplementary Figure 7). These results suggest that some paralogous ARF genes diverged during the evolution of Apiaceae.
FIGURE 4.
Relative expression level (FPKM) of celery ARF paralogous gene pairs in the root, petiole, and leaf. Asterisks indicate significant differences (*P < 0.05, **P < 0.01).
Discussion
Functions of ARF Genes in Apiaceae
ARF genes exist in most plant species and play key roles in plant growth and development (Li et al., 2016). Most ARF proteins contain three domains: a B3-type DNA-binding domain (DBD) at N-terminus, a variable middle region comprising an activation domain (AD) and a repression domain (RD), and a C-terminal dimerization domain (Tombuloglu, 2019). These domains facilitate interactions with Aux/IAA proteins (Mun et al., 2012; Li et al., 2015). Depending on these domains, ARFs act as transcriptional activators or repressors (Finet et al., 2013).
In Arabidopsis, the function of most ARFs has been studied. ARF1 regulates flower development (Ellis et al., 2005); ARF2 connects the ethylene and auxin signaling pathways to regulate hypocotyl bending (Okushima et al., 2005); ARF7 and ARF19 regulate lateral root formation (Okushima et al., 2007). The characterization of ARF gene function in Arabidopsis facilitates the functional analysis of ARF genes in Apiaceae. For example, the Apiaceae ARF genes in class II clustered together with AtARF3 and AtARF4. Therefore, we speculate that most genes in class II might be related to the floral meristem and reproductive organs. Here, we used bioinformatics to predict the function on a large scale. However, further experimentation is needed to determine the ARF gene function in Apiaceae.
Here, we found that some ARF gene paralogs exhibit notably different expression patterns, indicating the functions these paralogs diverged during the evolution of Apiaceae. This result is consistent with previous reports on the functional divergence of paralogous genes (Wang et al., 2013a; Soria et al., 2014). In fact, gene duplicates undergo one of four evolutionary fates, including conservation, subfunctionalization, neofunctionalization, and specialization, according to previous reports (Blanc and Wolfe, 2004; Assis and Bachtrog, 2013; Wang et al., 2016). Therefore, this study lays a foundation for further studies on the function of paralogous ARF genes in Apiaceae.
ARFs Interact With Aux/IAA Proteins to Regulate Auxin-Responsive Gene Expression
Both ARF and Aux/IAA proteins act as transcription factors that regulate the expression of auxin-responsive genes (Kumar et al., 2015). Aux/IAA genes contain several highly conserved domains, and the structure of ARF and Aux/IAA proteins is similar at the C-terminal. Both ARFs and Aux/IAAs harbor the CTD, domain III, and domain IV. Auxin responses rely on ARF–Aux/IAA interactions, which are mediated by CTD (Wang et al., 2013b).
Aux/IAA proteins function as transcriptional repressors, while ARFs function as either activators or repressors for regulating auxin-responsive genes (Liscum and Reed, 2002). The function of ARF proteins is determined by variable middle region (Liscum and Reed, 2002). ARFs are released from Aux/IAAs to repress/activate the expression of auxin-responsive genes (Wu et al., 2017). Here, we identified all ARF genes present in three Apiaceae species. This will enable the analysis of interactions between ARF and Aux/IAA proteins in Apiaceae.
Systematic and Comprehensive Analysis of ARF Gene Family in Apiaceae Species
Although the ARF gene family was previously investigated in many plants, there was no report of it in Apiaceae. Whole-genome sequences of celery, coriander, and carrot were released recently (Iorizzo et al., 2016; Song et al., 2020b), which greatly facilitated this study.
To understand the evolution of ARF genes, we constructed a phylogenetic tree using ARF amino acid sequences of celery, coriander, carrot, Arabidopsis, lettuce, and grape. Gene structure and conserved motif analyses revealed that genes in the same group or subgroup showed similar features. The number of paralogous ARF gene pairs in celery (8) and coriander (12) was notably more than that in carrot (2). Moreover, based on collinearity analysis, we found that WGD had a significant impact on ARF gene family expansion in Apiaceae.
In conclusion, we conducted a comprehensive analysis of the ARF gene family in three Apiaceae species. Our results provide a strong foundation for comparative and functional analyses of the ARF gene family in plants.
Data Availability Statement
The RNA-seq data were deposited in the Genome Sequence Archive (GSA) of the BIG Data Center (http://bigd.big.ac.cn/gsa) under the accession numbers CRA001996 and CRA001658.
Author Contributions
XS conceived the project and was responsible for the project initiation. XS and QP supervised and managed the project and research. XS, QP, NL, QY, TW, SF, KG, and TY led the data collection and bioinformatics analyses. XS, QP, NL, XF, ZJ, RZ, and ZW organized, wrote, and revised the manuscript. All authors read and revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (31801856), the Hebei Province Higher Education Youth Talents Program (BJ2018016), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 18KJB210010), and the Agricultural Science and Technology Innovation Project of Science and Technology Plan of Suzhou (No. SNG2017057).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.590535/full#supplementary-material
References
- Ahmad T., Cawood M., Iqbal Q., Arino A., Batool A., Tariq R. M. S., et al. (2019). Phytochemicals in Daucus carota and Their Health Benefits-Review Article. Foods 8:424. 10.3390/foods8090424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R., Bachtrog D. (2013). Neofunctionalization of young duplicate genes in Drosophila. Proc. Natl. Acad. Sci. U S A 110 17409–17414. 10.1073/pnas.1313759110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmer-Weber B. K., Hoffmann A., Wuthrich B., Luttkopf D., Pompei C., Wangorsch A., et al. (2002). Influence of food processing on the allergenicity of celery: DBPCFC with celery spice and cooked celery in patients with celery allergy. Allergy 57 228–235. 10.1034/j.1398-9995.2002.1o3319.x [DOI] [PubMed] [Google Scholar]
- Blanc G., Wolfe K. H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691. 10.1105/tpc.021410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools - an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020:009. [DOI] [PubMed] [Google Scholar]
- Ellis C. M., Nagpal P., Young J. C., Hagen G., Guilfoyle T. J., Reed J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132 4563–4574. 10.1242/dev.02012 [DOI] [PubMed] [Google Scholar]
- Finet C., Berne-Dedieu A., Scutt C. P., Marletaz F. (2013). Evolution of the ARF gene family in land plants: old domains, new tricks. Mol. Biol. Evol. 30 45–56. 10.1093/molbev/mss220 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorizzo M., Ellison S., Senalik D., Zeng P., Satapoomin P., Huang J., et al. (2016). A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48 657–666. 10.1038/ng.3565 [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Noel B., Policriti A., Clepet C., Casagrande A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467. 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Kalluri U. C., Difazio S. P., Brunner A. M., Tuskan G. A. (2007). Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 7:59. 10.1186/1471-2229-7-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Agarwal P., Pareek A., Tyagi A. K., Sharma A. K. (2015). Genomic Survey. Gene Expression, and Interaction Analysis Suggest Diverse Roles of ARF and Aux/IAA Proteins in Solanaceae. Plant Mole. Biol. Rep. 33 1552–1572. 10.1007/s11105-015-0856-z [DOI] [Google Scholar]
- Kumar R., Tyagi A. K., Sharma A. K. (2011). Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genom. 285 245–260. 10.1007/s00438-011-0602-7 [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40 D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. B. (2003). ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 19 1585–1586. 10.1093/bioinformatics/btg192 [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Jr., Roos D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. B., OuYang W. Z., Hou X. J., Xie L. L., Hu C. G., Zhang J. Z. (2015). Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 6:119. 10.3389/fpls.2015.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. B., Xie Z. Z., Hu C. G., Zhang J. Z. (2016). A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 7:47. 10.3389/fpls.2016.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E., Reed J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49 387–400. 10.1007/978-94-010-0377-3_10 [DOI] [PubMed] [Google Scholar]
- Luo X. C., Sun M. H., Xu R. R., Shu H. R., Wang J. W., Zhang S. Z. (2014). Genomewide identification and expression analysis of the ARF gene family in apple. J. Genet. 93 785–797. 10.1007/s12041-014-0462-0 [DOI] [PubMed] [Google Scholar]
- Maljaei M. B., Moosavian S. P., Mirmosayyeb O., Rouhani M. H., Namjoo I., Bahreini A. (2019). Effect of Celery Extract on Thyroid Function; Is Herbal Therapy Safe in Obesity? Int. J. Prev. Med. 10:55 10.4103/ijpvm.ijpvm_209_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., et al. (2009). CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37 D205–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J. H., Yu H. J., Shin J. Y., Oh M., Hwang H. J., Chung H. (2012). Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol. Genet. Genomics 287 765–784. 10.1007/s00438-012-0718-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19 118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Overvoorde P. J., Arima K., Alonso J. M., Chan A., Chang C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. 10.1105/tpc.104.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachayasittikul V., Prachayasittikul S., Ruchirawat S., Prachayasittikul V. (2018). Coriander (Coriandrum sativum): A promising functional food toward the well-being. Food Res. Int. 105 305–323. 10.1016/j.foodres.2017.11.019 [DOI] [PubMed] [Google Scholar]
- Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., et al. (2012). The Pfam protein families database. Nucleic. Acids. Res. 40 290–301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S., Wang Z., Yang X., Kozik A., Arikit S., Song C., et al. (2017). Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 8:14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. M., Liu T. K., Duan W. K., Ma Q. H., Ren J., Wang Z., et al. (2014b). Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis). Genomics 103 135–146. 10.1016/j.ygeno.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Song X., Liu G., Duan W., Liu T., Huang Z., Ren J., et al. (2014a). Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genomics 289 541–551. 10.1007/s00438-014-0833-5 [DOI] [PubMed] [Google Scholar]
- Song X., Liu G., Huang Z., Duan W., Tan H., Li Y., et al. (2016). Temperature expression patterns of genes and their coexpression with LncRNAs revealed by RNA-Seq in non-heading Chinese cabbage. BMC Genomics 17:297. 10.1186/s12864-016-2625-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Nie F., Chen W., Ma X., Gong K., Yang Q., et al. (2020a). Coriander Genomics Database: a genomic, transcriptomic, and metabolic database for coriander. Horticul. Res. 7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Wang J., Li N., Yu J., Meng F., Wei C., et al. (2020b). Deciphering the high-quality genome sequence of coriander that causes controversial feelings. Plant Biotechnol. J. 18 1444–1456. 10.1111/pbi.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria P. S., McGary K. L., Rokas A. (2014). Functional divergence for every paralog. Mol. Biol. Evol. 31 984–992. 10.1093/molbev/msu050 [DOI] [PubMed] [Google Scholar]
- Stolzer M., Lai H., Xu M., Sathaye D., Vernot B., Durand D. (2012). Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Wang L., Li W., Zhao L., Huang X., Azam S. M., et al. (2017). Genome-Wide Identification of Auxin Response Factor (ARF) Genes Family and its Tissue-Specific Prominent Expression in Pineapple (Ananas comosus). Tropical Plant Biol. 10 86–96. 10.1007/s12042-017-9187-6 [DOI] [Google Scholar]
- Tang Y., Bao X., Liu K., Wang J., Zhang J., Feng Y., et al. (2018). Genome-wide identification and expression profiling of the auxin response factor (ARF) gene family in physic nut. PLoS One 13:e0201024 10.1371/journal.pone.0201024.t001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombuloglu H. (2019). Genome-wide analysis of the auxin response factors (ARF) gene family in barley (Hordeum vulgare L.). J. Plant Biochem. Biotechnol. 28 14–24. 10.1007/s13562-018-0458-6 [DOI] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276 1865–1868. 10.1126/science.276.5320.1865 [DOI] [PubMed] [Google Scholar]
- Voorrips R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Wan S., Li W., Zhu Y., Liu Z., Huang W., Zhan J. (2014). Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Rep. 33 1365–1375. 10.1007/s00299-014-1622-7 [DOI] [PubMed] [Google Scholar]
- Wang D., Pei K., Fu Y., Sun Z., Li S., Liu H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394 13–24. 10.1016/j.gene.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteomics Bioinform. 8 77–80. 10.1016/s1672-0229(10)60008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Marowsky N. C., Fan C. (2013a). Divergent Evolutionary and Expression Patterns between Lineage Specific New Duplicate Genes and Their Parental Paralogs in Arabidopsis thaliana. PLoS One 8:e72362. 10.1371/journal.pone.0072362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tao F., Marowsky N. C., Fan C. (2016). Evolutionary Fates and Dynamic Functionalization of Young Duplicate Genes in Arabidopsis Genomes. Plant Physiol. 172 427–440. 10.1104/pp.16.01177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Hagen G., Guilfoyle T. J. (2013b). ARF-Aux/IAA interactions through domain III/IV are not strictly required for auxin-responsive gene expression. Plant signal. Behav. 8 e24526–e24526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 e49. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H. B., Cui B. M., Ren Y. L., Li J. H., Liao W. B., Xu N. F., et al. (2006). Research Progresses on Auxin Response Factors. J. Int. Plant Biol. 48 622–627. 10.1111/j.1744-7909.2006.00280.x [DOI] [Google Scholar]
- Wu W., Liu Y., Wang Y., Li H., Liu J., Tan J., et al. (2017). Evolution Analysis of the Aux/IAA Gene Family in Plants Shows Dual Origins and Variable Nuclear Localization Signals. Int. J. Mol. Sci. 18:2107. 10.3390/ijms18102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Pudake R. N., Guo G., Xing G., Hu Z., Zhang Y., et al. (2011). Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12:178. 10.1186/1471-2164-12-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang H., Cao N., Dong C., Shang Q. (2017). Genome-wide Identification and Expression of ARF Gene Family during Adventitious Root Development in Hot Pepper (Capsicum annuum). Horticultural Plant J. 3 151–164. 10.1016/j.hpj.2017.07.001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data were deposited in the Genome Sequence Archive (GSA) of the BIG Data Center (http://bigd.big.ac.cn/gsa) under the accession numbers CRA001996 and CRA001658.