Abstract
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy, accounting for 90% of all ovarian cancer. Dysregulation of miRNAs is associated with several types of EOC. In the current research, we aimed to study the role of abnormal expression of miR-146a in the development of EOC and to elucidate the possible molecular mechanisms. Compared with control samples, mRNA expression of miR-146a was significantly decreased in EOC tissues and cell lines. Overexpression of miR-146a prohibited cell proliferation, enhanced apoptosis, and increased sensitivity to chemotherapy drugs in EOC cells. In contrast, downregulation of miR-146a promoted cell proliferation, suppressed apoptosis, and decreased sensitivity to chemotherapy drugs in EOC cells. Overexpression of miR-146a increased the reactive oxygen species (ROS) level and decreased SOD2 mRNA and protein expression. Downregulation of miR-146a increased SOD2 mRNA and protein expression. Overexpression of SOD2 significantly inhibited miR-146a mimics-induced suppression of cell proliferation and the increase of apoptosis and chemosensitivity. In conclusion, we identify miR-146a as a potential tumor suppressor in patients with EOC. miR-146a downregulates the expression of SOD2 and enhances ROS generation, leading to increased apoptosis, inhibition of proliferation, and enhanced sensitivity to chemotherapy. The data demonstrate that the miR-146a/SOD2/ROS pathway may serve as a novel therapeutic target and prognostic marker in patients with EOC.
Key words: miR-146a, SOD2, Epithelial ovarian cancer (EOC), Proliferation, Apoptosis, Chemosensitivity
INTRODUCTION
Ovarian cancer is the third most common gynecological malignancy worldwide (1,2). Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy, accounting for 90% of all ovarian cancer (1,2). The genomic and epigenomic atlas of EOC varies significantly with tumor histotype, grade, stage, sensitivity to chemotherapy, and prognosis (3). It is estimated that 70% of patients have an advanced stage of the disease at diagnosis, when the tumor has disseminated beyond the ovaries to the pelvic and abdominal organs (4). Although surgical and radio- or chemicotoxic therapies have been applied in most types of tumors, the 5-year survival rate of ovarian cancer remains only 30% (5). The molecular mechanism underlying EOC is not clear. It is urgent to investigate molecular pathogenesis and therapeutic targets and search for novel biomarkers of EOC.
MicroRNAs (miRNAs) are defined as a large class of small (20–24 nucleotides) single-stranded and noncoding RNAs that are involved in gene regulation through binding to the 3′-untranslated region (3′-UTR) of their target mRNAs, resulting in mRNA degradation or translation inhibition (6,7). These kinds of RNAs play a significant role in various physiological and pathophysiological processes (8,9). It is estimated that one third of all mammalian genes were targeted and regulated by miRNAs (10,11). Moreover, miRNAs can function either as oncomiRNAs, by targeting tumor suppressors, or as tumor suppressors by targeting oncogenes (6,12). Thus, it is believed that inactivation of oncogenic miRNAs (13) or restoration of tumor-suppressor miRNAs (14) may be an effective avenue for cancer treatment. It is well known that dysregulation of miRNAs is involved in many fundamental processes of cancer (15). We have found that miR-164a was abnormally expressed in ovarian cancer. However, the exact role of miR-164a in the development of ovarian cancer is still unknown.
In the present study, we aimed to explore whether abnormal expression of miR-164a played a role in the pathogenesis of EOC and to study the possible mechanisms. We found that miR-164a level was reduced in EOC tissues and cell lines; overexpression of miR-164a inhibited EOC cell proliferation and enhanced apoptosis and sensitivity to chemotherapy drugs through a decrease in superoxide dismutase 2 (SOD2).
MATERIALS AND METHODS
Chemicals and Materials
β-Actin and SOD2 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Paclitaxel and DHE were procured from Sigma-Aldrich. All of the other chemicals used were of the highest grade available commercially.
Cell Lines and Culture
Human EOC cell lines, OVCAR3, CAOV3, and HEY cells were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in DMEM (Gibco-BRL, USA) supplemented with 10% FBS (Gibco-BRL), 100 U/ml penicillin, and 100 µg of streptomycin at 37°C in a humidified incubator with 5% CO2. Human ovarian surface epithelial (HOSE) normal cells were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and maintained in ovarian epithelial cell medium supplemented with 1× ovarian epithelial cell growth supplement (ScienCell Research Laboratories) at 37°C in a humidified incubator with 5% CO2.
Transfection of Plasmids
The miR-146a mimics, inhibitors, and scrambles and SOD2 plasmids were synthesized by Gene Pharma Company (Shanghai, China). Transient transfection of plasmids was conducted using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) system according to the manufacturer’s instructions.
Cell Proliferation and Viability
Cell proliferation was detected using a CCK8 Assay Kit according to the manufacturer’s instructions. Absorbance was detected at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA). Cell viability was determined by MTT assay. Absorbance was measured at 570 nm using a microplate reader (Bio-Rad).
TUNEL Assay
TdT-mediated dUTP nick-end labeling (TUNEL) staining was used to detect apoptotic cell death. TUNEL assay was performed using an in situ cell death detection kit (Roche, Germany) according to the manufacturer’s protocols. Apoptosis was analyzed using flow cytometry, and percentage of apoptotic cells was shown.
RNA Isolation and Real-Time PCR
Total RNA was isolated from tissues or cell lines using TIANGEN Total RNA Isolation Kit (TIANGEN, Beijing, China) according to the manufacturer’s protocols. RNA (500 ng) was reverse transcribed to synthesize cDNA using the Takara FirstStrand cDNA Synthesis Kit (Takara, Japan). Quantitative real-time polymerase chain reaction (PCR) was performed to precisely quantify miR-146a and SOD2. Real-time PCR was conducted using SYBR Green reagents (Roche, Germany) in Bio-Rad CFX96 real-time PCR system. The 2−ΔΔCT method was used to measure relative mRNA expression compared to β-actin. Amplification conditions were initial step at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 63°C for 30 s, and then extension at 72°C for 10 s.
Protein Extraction and Western Blot
Total proteins were collected from cell lines using RIPA lysis buffer according to the manufacturer’s protocols. Cell lysates were centrifuged at 25,000 × g for 30 min at 4°C, and the protein concentrations in supernatants were measured using a BCA kit (Pierce, Rockford, IL, USA). Samples equal to 20 µg of protein were separated using SDS-PAGE. Then proteins were transferred to NC membranes (Millipore, Billerica, MA, USA) and then blocked with 8% nonfat milk. After that, membranes were incubated with primary SOD2 antibodies overnight at 4°C. After washing for four times, the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody (Pierce) at 37°C for 1 h. Protein bands were visualized with the ECL and captured using Bio-Rad Imaging Systems (Bio-Rad).
Statistical Analysis
The data are shown as the means ± SEM, and a value of p < 0.05 was considered significant. All experiments were conducted at least in triplicate. Data analysis was performed using Graphpad Prism 5.0 software. One-way analysis of variance (ANOVA) was used to measure the significance between more than two groups. Student’s t-test was conducted to measure the significance between two groups.
RESULTS
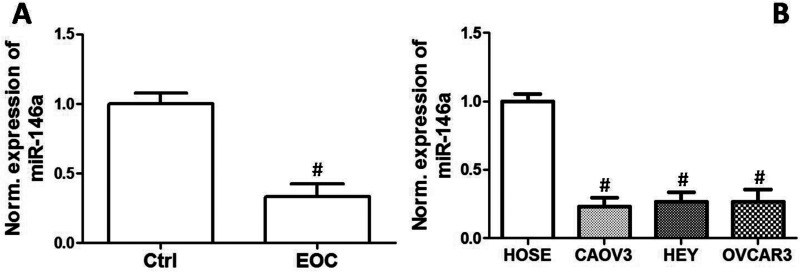
miR-146a Level Was Decreased in EOC Cell Lines
We compared the expression of miR-146a between HOSE cells and three human EOC cell lines, CAOV3, OVCAR3, and HEY cells. We found that mRNA expression of miR-146a was markedly decreased in EOC cell lines, compared with that in HOSE cells (Fig. 1).
Figure 1.
Expression levels of miR-146a in EOC cell lines. Relative expression of miR-146a in HOSE, OVCAR3, CAOV3, and HEY cell lines. #p < 0.05 indicates statistical significance, compared with control.
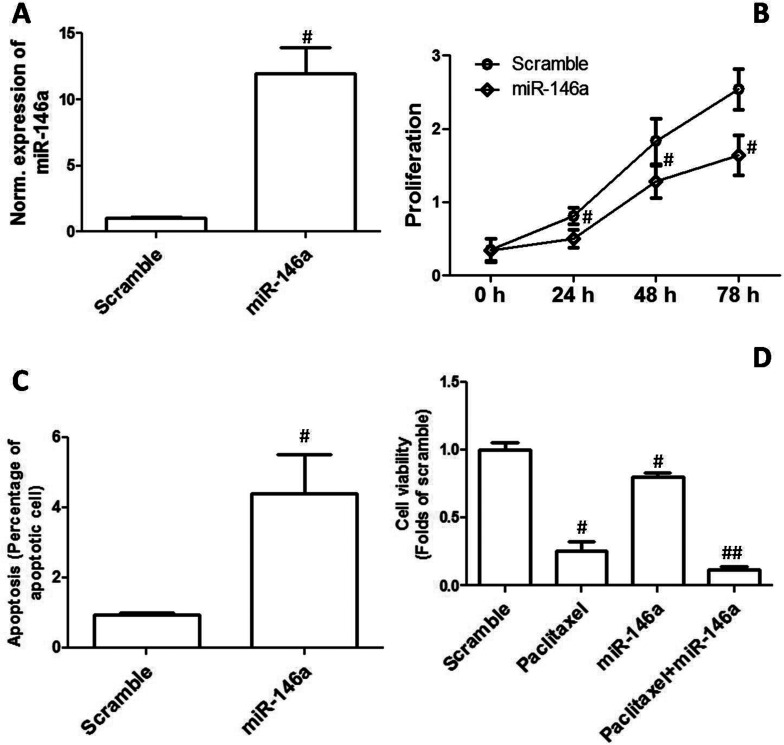
Overexpression of miR-146a Inhibited Proliferation and Enhanced Apoptosis and Chemosensitivity
To explore the possible role of downregulation of miR-146a in the development of EOC, CAOV3 cells were transfected with miR-146a mimics. As shown in Figure 2A, the results confirmed that miR-146a expression was significantly increased after the transfection of miR-146a mimics. As shown in Figure 2B, overexpression of miR-146a remarkably inhibited cell proliferation in CAOV3 cells. In addition, the effect of miR-146a mimics transfection on apoptosis was examined. We showed that overexpression of miR-146a markedly increased the percentage of apoptotic cells in the CAOV3 cell line (Fig. 2C). Moreover, the effect of miR-146a mimics on the sensitivity to chemotherapy drugs in OVCAR3 cells was determined by MTT assay. Figure 2D shows that incubation with 400 ng/ml paclitaxel for 48 h and overexpression of miR-146a significantly decreased cell viability. Overexpression of miR-146a significantly enhanced paclitaxel-induced decrease in cell viability (Fig. 2D).
Figure 2.
Overexpression of miR-146a inhibited proliferation and enhanced apoptosis and chemosensitivity. CAOV3 cells were transfected with miR-146a mimics. (A) Relative mRNA expression of miR-146a is shown. (B) Cell proliferation was determined, and the growth curve is shown. (C) Apoptosis was determined, and percentage of apoptotic cells is shown. (D) OVCAR3 cells were transfected with miR-146a mimics and then treated by 400 ng/ml paclitaxel for 48 h. After that, cell viability was assessed by MTT. #p < 0.05 indicates statistical significance, compared with scramble. ##p < 0.05 indicates statistical significance, compared with paclitaxel and miR-146a.
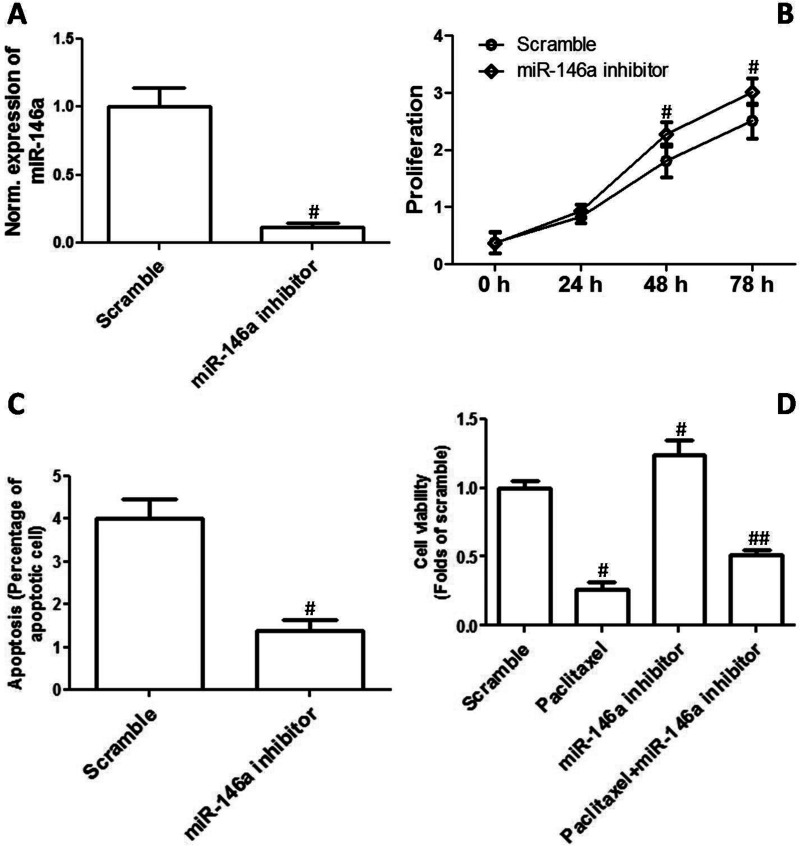
Decrease in miR-146a Increased Proliferation and Inhibited Apoptosis and Chemosensitivity
CAOV3 cells were transfected with miR-146a inhibitors to evaluate the effect of inhibition of miR-146a on EOC cell proliferation and chemosensitivity. The results shown in Figure 3A confirmed that miR-146a expression was substantially decreased by its inhibitors. As shown in Figure 3B, downregulation of miR-146a remarkably promoted cell proliferation in CAOV3 cells. In addition, the effect of miR-146a inhibitor transfection on apoptosis was examined. We showed that downregulation of miR-146a markedly decreased the percentage of apoptotic cells in the CAOV3 cell line (Fig. 3C). Moreover, the effect of miR-146a inhibitors on the sensitivity to chemotherapy drugs in OVCAR3 cells was determined by MTT assay. Figure 3D shows that a decrease in miR-146a significantly inhibited paclitaxel-induced decrease in cell viability.
Figure 3.
Decrease in miR-146a increased proliferation and inhibited apoptosis and chemosensitivity. CAOV3 cells were transfected with miR-146a inhibitors. (A) Relative mRNA expression of miR-146a is shown. (B) Cell proliferation was determined, and the growth curve is shown. (C) Apoptosis was determined, and percentage of apoptotic cells is shown. (D) OVCAR3 cells were transfected with miR-146a inhibitors and then treated by 400 ng/ml paclitaxel for 48 h. After that, cell viability was assessed by MTT. #p < 0.05 indicates statistical significance, compared with scramble. ##p < 0.05 indicates statistical significance, compared with paclitaxel and miR-146a.
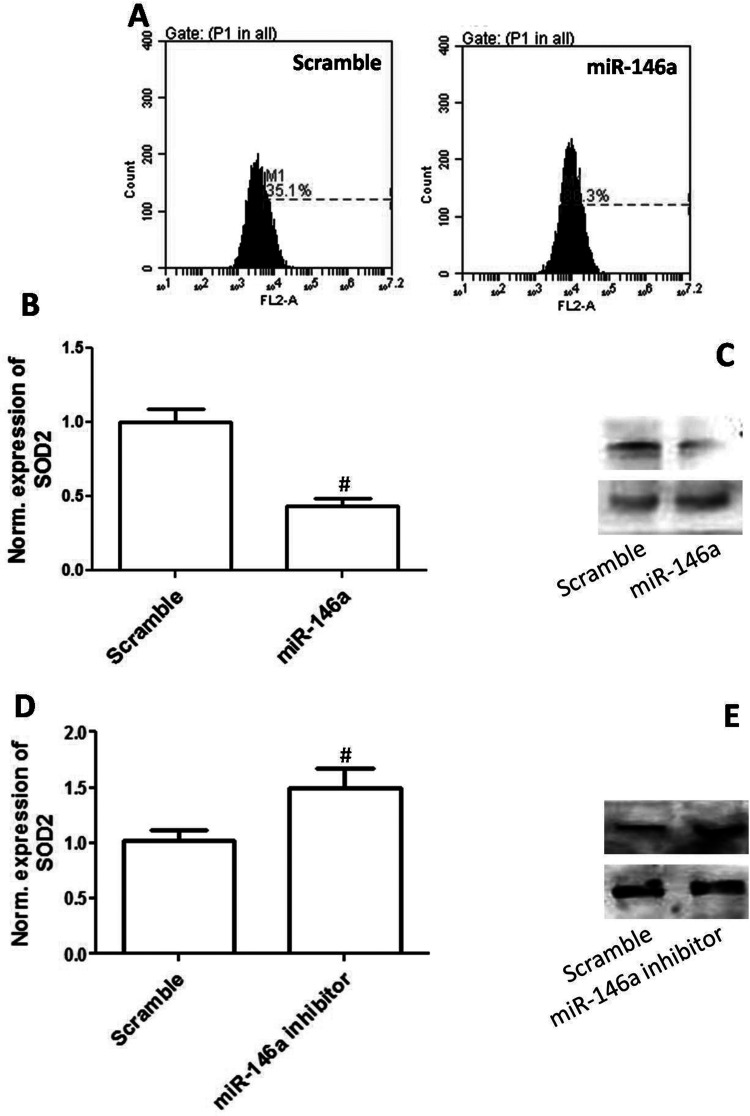
Overexpression of miR-146a Promoted ROS Generation and Decreased SOD Expression
To test whether oxidative stress was involved in miR-146a-mediated cytotoxicity to EOC cells, we determined the effect of overexpression of miR-146a on reactive oxygen species (ROS) generation. As shown in Figure 4A, overexpression of miR-146a significantly increased DHE-positive cells, indicating that miR-146a promoted ROS generation. We also evaluated the effect of upregulation or downregulation of miR-146a on SOD2 expression. In Figure 4B and C, the results show that overexpression of miR-146a significantly decreased mRNA and protein expression of SOD2 in CAOV3 cells. As shown in Figure 4D and E, downregulation of miR-146a significantly increased mRNA and protein expression of SOD2 in CAOV3 cells. The results indicated that SOD2 may be a target of miR-146a and overexpression of miR-146a promoted ROS generation through inhibition of SOD2.
Figure 4.
Overexpression of miR-146a increased ROS generation and inhibited SOD2 expression. CAOV3 cells were transfected with miR-146a mimics. (A) ROS level was detected by DHE (a superoxide probe, 10 µM) and analyzed by flow cytometry. (B) Relative mRNA expression of miR-146a is shown. (C) Protein expression of miR-146a is shown. CAOV3 cells were transfected with miR-146a inhibitors. (D) Relative mRNA expression of miR-146a is shown. (E) Protein expression of miR-146a is shown. #p < 0.05 indicates statistical significance, compared with scramble.
Reduction of SOD2 Was Involved in miR-146a-Mediated Cytotoxicity to EOC Cells
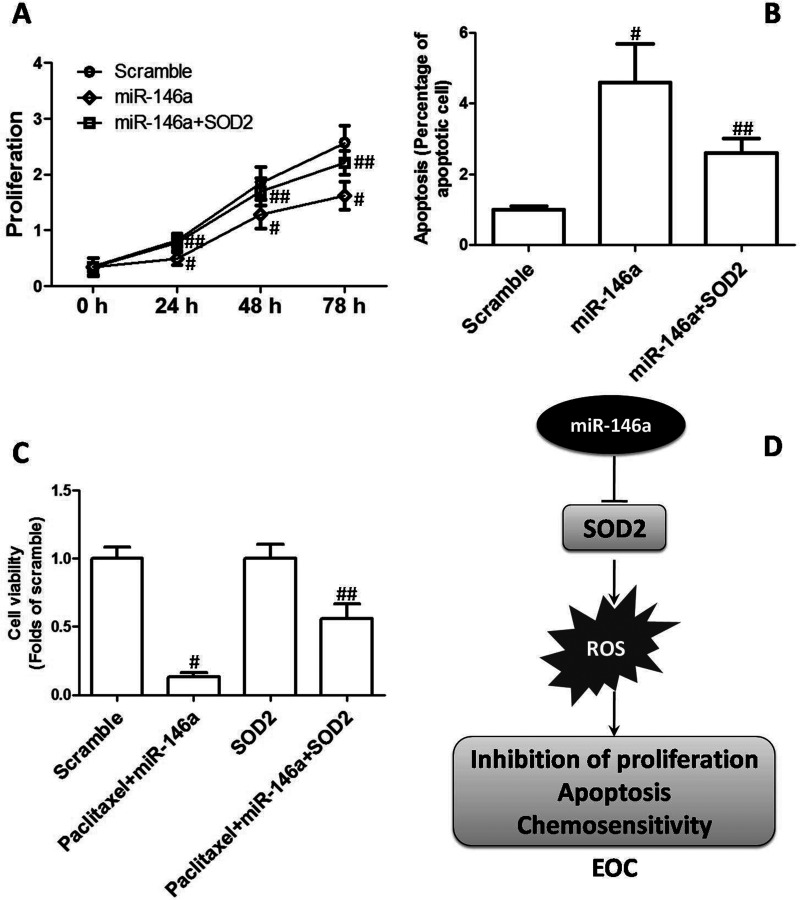
To explore whether reduction of SOD2 and ROS generation was involved in miR-146a-mediated inhibition of EOC cell proliferation and enhancement of chemosensitivity, cells were transfected with SOD plasmids. We found that overexpression of SOD2 significantly suppressed miR-146a-induced inhibition of cell proliferation in CAOV3 cells (Fig. 5A). Moreover, miR-146a-induced increase in apoptosis was notably blocked by SOD2 plasmids (Fig. 5B). Furthermore, overexpression of SOD2 markedly reversed miR-146a-enhanced sensitivity to paclitaxel in OVCAR3 cells (Fig. 5C). The results demonstrated that reduction of SOD2 was involved in miR-146a-mediated cytotoxicity to EOC cells (Fig. 5D).
Figure 5.
Role of reduction of SOD2 in miR-146a-mediated inhibition of EOC. CAOV3 cells were transfected with miR-146a mimics with or without plasmids expressing SOD2. (A) Cell proliferation was determined, and the growth curve is shown. (B) Apoptosis was determined, and percentage of apoptotic cells is shown. (C) OVCAR3 cells were transfected with miR-146a mimics with or without plasmids expressing SOD2 and then treated by 400 ng/ml paclitaxel for 48 h. After that, cell viability was assessed by MTT. (D) Role of reduction of SOD2 in miR-146a-mediated inhibition of EOC. #p < 0.05 indicated statistical significance, compared with scramble. ##p < 0.05 indicated statistical significance, compared with paclitaxel + miR-146a.
DISCUSSION
Dysregulation of microRNA is associated with the pathogenesis and development of ovarian cancer (16–20). Previous studies have shown that miR-146a acted as a tumor suppressor. For example, in squamous cell carcinoma patients, Lerner et al. found that downregulation of miR-146a in the blood of patients correlated with the occurrence of distant metastasis, and inhibition of miR-146a dramatically promoted its proliferation and migration potential (21). Li et al. discovered that downregulation of miR-146a expression in breast tissue was related to the development and deterioration of breast cancer, and miR-146a might act as a potential biomarker for young women with breast cancer (22). However, little is known about the role of miR-146a in the pathogenesis of EOC.
We have previously found that miR-146a was downregulated in EOC tissues (data not shown). In the present study, we showed that miR-146a was downregulated in EOC cell lines, indicating a potential tumor-suppressive role. To test whether downregulation of miR-146a was involved in EOC cell line proliferation, CAOV3 cells were transfected with miR-146a mimics or inhibitors. The effect of miR-146a mimics or inhibitors on cell proliferation and apoptosis was determined. Overexpression of miR-146a inhibited proliferation and enhanced apoptosis. In contrast, decrease in miR-146a increased proliferation and inhibited apoptosis. Moreover, the effect of upregulation or downregulation of miR-146a on paclitaxel-induced cytotoxicity was evaluated. The results showed that overexpression of miR-146a enhanced paclitaxel-induced decrease in cell viability. In contrast, paclitaxel-induced cytotoxicity was significantly inhibited by the downregulation of miR-146a. Apoptotic cell death is an important programmed death. Abnormal apoptosis plays a key role in the development of multiple types of cancer, and targeting apoptosis is considered to be a substantial avenue for cancer therapy (23–26). A large number of studies have shown that microRNA could alter apoptosis through either directly interacting with apoptotic pathway members or indirectly influencing apoptosis-inducing stress signals (27,28). The results in the present study indicated that downregulation of miR-146a contributed to CAOV3 cell proliferation and ameliorated sensitivity to chemotherapy. Reduction of apoptosis participated in the downregulation of miR-146a-mediated enhancement of CAOV3 cell proliferation and reduction of sensitivity to chemotherapy.
Oxidative stress is an important stimulus of apoptotic cell death, and a large amount of ROS generation is considered to be involved in the antitumor effect of various chemotherapy drugs (29,30). In the present study, we also examined whether ROS generation was involved in miR-146a-mediated inhibition of CAOV3 cell proliferation and the increase in sensitivity to chemotherapy. We showed that overexpression of miR-146a significantly increased ROS level. miR-146a-mediated increase of ROS may be attributed to reduction of SOD2. Our results showed that overexpression of miR-146a significantly decreased SOD2 expression while inhibition of miR-146a significantly increased SOD2 expression, indicating SOD2 was a major target of miR-146a. A previous study has shown that miR-146a could regulate SOD2 expression in PC12 cells (31). Thus, it is suggested that SOD2 may be a primary target under different stressful or pathological conditions. SOD2 is a pivotal antioxidant enzyme that mainly functions in the mitochondria (32) and converts highly reactive superoxide (O2 •−) to hydrogen peroxide (H2O2) and oxygen (O2). Abnormal expression of SOD2 is believed to be associated with several types of cancers (33,34). In ovarian clear cell carcinoma, SOD2 functions as a protumorigenic and prometastatic agent (35). Combined with these literatures, the data demonstrate that SOD2 may be an important regulator of EOC development via regulation of ROS level, and miR-146a plays a suppressive role in EOC via targeting SOD2.
In conclusion, we identified miR-146a as a potential tumor suppressor in patients with EOC. miR-146a downregulates expression of SOD2 and enhances ROS generation, leading to increased apoptosis, inhibition of proliferation, and enhanced sensitivity to chemotherapy (Fig. 5D). Overall, the data demonstrate that miR-146a/SOD2/ROS pathways may serve as novel therapeutic targets and prognostic markers in patients with EOC.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jemal A.; Bray F.; Center M. M.; Ferlay J.; Ward E.; Forman D. Global cancer statistics. CA Cancer J. Clin. 61:69–90; 2011. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R.; Ma J.; Zou Z.; Jemal A. Cancer statistics, 2014. CA Cancer J. Clin. 64:9–29; 2014. [DOI] [PubMed] [Google Scholar]
- 3. Bai H.; Cao D.; Yang J.; Li M.; Zhang Z.; Shen K. Genetic and epigenetic heterogeneity of epithelial ovarian cancer and the clinical implications for molecular targeted therapy. J. Cell. Mol. Med.; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang W.; Liu R.; Tang C.; Xi Q.; Lu S.; Chen W.; Zhu L.; Cheng J.; Chen Y.; Wang W.; Zhong J.; Deng Y. PFTK1 regulates cell proliferation, migration and invasion in epithelial ovarian cancer. Int. J. Biol. Macromol. 85:405–416; 2016. [DOI] [PubMed] [Google Scholar]
- 5. Naora H.; Montell D. J. Ovarian cancer metastasis: Integrating insights from disparate model organisms. Nat. Rev. Cancer 5:355–366; 2005. [DOI] [PubMed] [Google Scholar]
- 6. Sayed D.; Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 91:827–887; 2011. [DOI] [PubMed] [Google Scholar]
- 7. Bartel D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116:281–297; 2004. [DOI] [PubMed] [Google Scholar]
- 8. Bartel D. P. MicroRNAs: Target recognition and regulatory functions. Cell 136:215–233; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen C. Z.; Li L.; Lodish H. F.; Bartel D. P. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86; 2004. [DOI] [PubMed] [Google Scholar]
- 10. Orellana E. A.; Kasinski A. L. MicroRNAs in cancer: A historical perspective on the path from discovery to therapy. Cancers (Basel) 7:1388–1405; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garzon R.; Marcucci G.; Croce C. M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 9:775–789; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang L.; Yue Y.; Wang X.; Jin H. Function and clinical potential of microRNAs in hepatocellular carcinoma. Oncol. Lett. 10:3345–3353; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Medina P. P.; Nolde M.; Slack F. J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467:86–90; 2010. [DOI] [PubMed] [Google Scholar]
- 14. Lujambio A.; Calin G. A.; Villanueva A.; Ropero S.; Sanchez-Cespedes M.; Blanco D.; Montuenga L. M.; Rossi S.; Nicoloso M. S.; Faller W. J.; Gallagher W. M.; Eccles S. A.; Croce C. M.; Esteller M. A. microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 105:13556–13561; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Leva G.; Garofalo M.; Croce C. M. MicroRNAs in cancer. Annu. Rev. Pathol. 9:287–314; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katz B.; Trope C. G.; Reich R.; Davidson B. MicroRNAs in ovarian cancer. Hum. Pathol. 46:1245–1256; 2015. [DOI] [PubMed] [Google Scholar]
- 17. Zou J.; Yin F.; Wang Q.; Zhang W.; Li L. Analysis of microarray-identified genes and microRNAs associated with drug resistance in ovarian cancer. Int. J. Clin. Exp. Pathol. 8:6847–6858; 2015. [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu T.; Yuan J.; Wang Y.; Gong C.; Xie Y.; Li H. MiR-661 contributed to cell proliferation of human ovarian cancer cells by repressing INPP5J expression. Biomed. Pharmacother. 75:123–128; 2015. [DOI] [PubMed] [Google Scholar]
- 19. Braza-Boils A.; Mari-Alexandre J.; Gilabert J.; Sanchez-Izquierdo D.; Espana F.; Estelles A.; Gilabert-Estelles J. MicroRNA expression profile in endometriosis: Its relation to angiogenesis and fibrinolytic factors. Hum. Reprod. 29:978–988; 2014. [DOI] [PubMed] [Google Scholar]
- 20. Zhang X.; Liu P.; Zhang B.; Mao H.; Shen L.; Ma Y. Inhibitory effects of STAT3 decoy oligodeoxynucleotides on human epithelial ovarian cancer cell growth in vivo. Int. J. Mol. Med. 32:623–628; 2013. [DOI] [PubMed] [Google Scholar]
- 21. Lerner C.; Wemmert S.; Bochen F.; Kulas P.; Linxweiler M.; Hasenfus A.; Heinzelmann J.; Leidinger P.; Backes C.; Meese E.; Urbschat S.; Schick B. Characterization of miR-146a and miR-155 in blood, tissue and cell lines of head and neck squamous cell carcinoma patients and their impact on cell proliferation and migration. J. Cancer Res. Clin. Oncol.; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y.; Xu Y.; Yu C.; Zuo W. Associations of miR-146a and miR-146b expression and breast cancer in very young women. Cancer Biomark. 15:881–887; 2015. [DOI] [PubMed] [Google Scholar]
- 23. Chen L.; Cui H. Targeting glutamine induces apoptosis: A cancer therapy approach. Int. J. Mol. Sci. 16:22830–22855; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su Z.; Yang Z.; Xu Y.; Chen Y.; Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer 14:48; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koff J. L.; Ramachandiran S.; Bernal-Mizrachi L. A time to kill: Targeting apoptosis in cancer. Int. J. Mol. Sci. 16:2942–2955; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang C.; Freter C. Lipid metabolism, apoptosis and cancer therapy. Int. J. Mol. Sci. 16:924–949; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao C.; Li T.; Han B.; Yue W.; Shi L.; Wang H.; Guo Y.; Lu Z. DDAH1 deficiency promotes intracellular oxidative stress and cell apoptosis via a miR-21-dependent pathway in mouse embryonic fibroblasts. Free Radic. Biol. Med. 92:50–60; 2016. [DOI] [PubMed] [Google Scholar]
- 28. Wang X. Z.; Hang Y. K.; Liu J. B.; Hou Y. Q.; Wang N.; Wang M. J. Over-expression of microRNA-375 inhibits papillary thyroid carcinoma cell proliferation and induces cell apoptosis by targeting ERBB2. J. Pharmacol. Sci.; in press. [DOI] [PubMed] [Google Scholar]
- 29. Ray T.; Chakrabarti M. K.; Pal A. Hemagglutinin protease secreted by V. Cholerae induced apoptosis in breast cancer cells by ROS mediated intrinsic pathway and regresses tumor growth in mice model. Apoptosis 21:143–154; 2016. [DOI] [PubMed] [Google Scholar]
- 30. Banerjee K.; Basu S.; Das S.; Sinha A.; Biswas M. K.; Choudhuri S. K. Induction of intrinsic and extrinsic apoptosis through oxidative stress in drug resistant cancer by a newly synthesized Schiff base copper chelate. Free Radic. Res. 2016 Jan 5:1–55. [DOI] [PubMed] [Google Scholar]
- 31. Ji G.; Lv K.; Chen H.; Wang T.; Wang Y.; Zhao D.; Qu L.; Li Y. MiR-146a regulates SOD2 expression in H2O2 stimulated PC12 cells. PLoS One. 8:e69351; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pani G.; Koch O. R.; Galeotti T. The p53-p66shc-Manganese Superoxide Dismutase (MnSOD) network: A mitochondrial intrigue to generate reactive oxygen species. Int. J. Biochem. Cell Biol. 41:1002–1005; 2009. [DOI] [PubMed] [Google Scholar]
- 33. Konzack A.; Kietzmann T. Manganese superoxide dismutase in carcinogenesis: Friend or foe? Biochem. Soc. Trans. 42:1012–1016; 2014. [DOI] [PubMed] [Google Scholar]
- 34. Papa L.; Hahn M.; Marsh E. L.; Evans B. S.; Germain D. SOD2 to SOD1 switch in breast cancer. J. Biol. Chem. 289:5412–5416; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hemachandra L. P.; Shin D. H.; Dier U.; Iuliano J. N.; Engelberth S. A.; Uusitalo L. M.; Murphy S. K.; Hempel N. Mitochondrial superoxide dismutase has a protumorigenic role in ovarian clear cell carcinoma. Cancer Res. 75:4973–4984; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]