Abstract
The lncRNA H19 and its mature product miR-675 have recently been shown to be upregulated and promote the progression of gastric cancer. However, the detailed function and underlying molecular mechanism of H19/miR-675 in the carcinogenesis of gastric cancer remains unclear. In this study, we found that H19 depended on miR-675 to enhance the proliferation and invasion of gastric cancer AGS cells, and the expression of miR-675 was positively correlated with H19 in patients with gastric cancer. Subsequently, the tumor-suppressor runt domain transcription factor 1 (RUNX1) was confirmed to be a downstream molecule of H19/miR-675 axis, since overexpression of H19 or miR-675 significantly decreased RUNX1 expression in AGS cells, and knockdown of H19 or miR-675 enhanced RUNX1 expression. More importantly, a series of assays further demonstrated that introduction of RUNX1 abrogated H19/miR-675-induced Akt/mTOR pathway activation and the following cellular proliferation and invasion of AGS cells. To our knowledge, this is the time to demonstrate that RUNX1 serves as a link between H19/miR-675 axis and Akt/mTOR signaling and is a pivotal mediator in gastric cancer progression induced by H19/miR-675. Thus, our study provides important clues for understanding the key roles of lncRNA-miRNA functional network and identifying new therapeutic targets for gastric cancer.
Key words: H19, miR-675, Runt domain transcription factor 1 (RUNX1), Akt/mTOR pathway, Gastric cancer
INTRODUCTION
Gastric cancer was the fifth most common malignancy (952,000 cases, 6.8% of the total) and the third leading cause of cancer-related death in both sexes worldwide (723,000 deaths, 8.8% of the total) in 2012 (1). Due to multiple risk factors involved in the progression of gastric cancer, especially Helicobacter pylori infection, adequate surgery, as the cornerstone of gastric cancer treatment, has only slightly improved the disease-free survival over the last few decades, and locoregional control for advanced disease remains very difficult (2). Therefore, it is essential to further elucidate the underlying molecular mechanisms of gastric cancer progression and thus identify new prognostic biomarkers and therapeutic targets for this disease.
In recent years, emerging evidence has strongly suggest that long noncoding RNAs (lncRNAs), defined as endogenous cellular RNAs of more than 200 nucleotides in length, play critical roles in a wide repertoire of biological processes that could contribute to disease occurrence (3,4). More importantly, numerous studies have confirmed that multiple lncRNAs are involved in the regulation of cancer progression (5–11). For example, the expression of lncRNA loc285194 is associated with the pathogenesis of colon and breast cancers. As a p53 transcription target, loc285194 inhibits tumor cell growth both in vitro and in vivo, in part through repressing the expression of oncogene miR-211 (12). Prostate cancer gene expression marker 1 (PCGEM1), being a coactivator for both androgen and c-Myc, not only reprograms the androgen network and the central metabolism in a tumor-specific way but also suppresses tumor-suppressor miR-145 expression and thus promotes prostate cancer growth both in vitro and in vivo (13,14). lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) overexpression was correlated with a poor overall survival of patients with hepatocarcinoma (15), and it is also aberrantly upregulated in non-small-cell lung carcinomas, breast cancer, pancreatic cancer, colorectal cancer, and prostate cancer (16–18).
lncRNA H19 is produced from paternally imprinted genes H19 and has been considered as an oncogenic lncRNA in many types of cancers, including hepatocarcinoma, bladder cancer, breast cancer, esophageal cancer, and glioma (19–23). In gastric cancer, studies have found that lncRNA H19 abnormally upregulated (8,24,25) and contributed to the proliferation of gastric cancer cells by inactivating p53 (26) or via miR-675 to inhibit a well-known tumor suppressor, runt domain transcription factor 1 (RUNX1) expression (27,28). However, the precise underlying mechanism of H19 in gastric cancer tumorigenesis still needs to be further explored. The Akt/mTOR signaling axis has been shown to be regulated by RUNX1 (29,30) and is activated in the majority of human cancers, including gastric cancer (31,32). The H19/miR-675 signaling axis also has been shown to promote glioma cell invasion (22). Consequently, we speculated that RUNX1 may play a critical role in the tumorigenesis process by regulating the Akt/mTOR pathway that is activated by H19-derived miR-675.
Thus, in the current study, we explored the clinical feature, pathophysiological roles, and potential mechanism of lncRNA H19 and miR-675 in gastric cancer carcinogenesis. We found that H19 mainly depended on miR-675 to promote gastric cancer progression. Furthermore, we identified that H19-derived miR-675 activates Akt/mTOR pathway via inhibiting RUNX1 expression and thus modulates gastric cell carcinogenesis, which may serve as a potential diagnostic and therapeutic target for gastric cancer.
MATERIALS AND METHODS
Gastric Cancer Tissue and Cell Culture
Cancer tissue samples were obtained from Renmin Hospital of Wuhan University and Central Hospital of Enshi Autonomous Prefecture. Samples were immediately preserved in RNA fixer (Bioteke, Beijing, China) after removal from the body and stored at −80°C until further use. This study was approved by the Ethics Committee of Renmin Hospital and Central Hospital of Enshi Autonomous Prefecture. The patients’ characteristics are detailed in Table 1. The human gastric cancer cell line AGS and the immortalized human gastric epithelial mucosa cell line GES-1 were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were maintained in RPMI-1640 medium (Hyclone) with 10% fetal bovine serum (FBS; Gibco) and cultured in a humidified incubator with 5% CO2 at 37°C.
Table 1.
The Clinical Characteristics of the Gastric Cancer Patients
| Variables | No. (n = 114) | Percent |
|---|---|---|
| Age (years) | ||
| <60 | 38 | 33.3% |
| ≥60 | 76 | 66.7% |
| Gender | ||
| Male | 83 | 72.8% |
| Female | 31 | 27.2% |
| Diameter (cm) | ||
| <5 | 67 | 58.8% |
| ≥5 | 47 | 41.2% |
| CEA | ||
| Positive | 71 | 62.3% |
| Negative | 43 | 37.7% |
| CA19-9 | ||
| Positive | 58 | 50.9% |
| Negative | 56 | 49.1% |
| Differentiation | ||
| Well | 11 | 9.7% |
| Moderate | 47 | 41.2% |
| Poor | 56 | 49.1% |
| Lymphatic metastasis | ||
| Absent | 31 | 27.2% |
| Present | 83 | 72.8% |
| Invasion | ||
| Tis&T1–T3 | 37 | 32.5% |
| T4 | 77 | 67.5% |
| Distal metastasis | ||
| Absent | 71 | 62.3% |
| Present | 43 | 37.7% |
| TNM stage | ||
| 0–I | 25 | 21.9% |
| II | 22 | 19.3% |
| III | 24 | 21.1% |
| IV | 43 | 37.7% |
All samples consist of 23 endoscopic biopsy samples and 91 surgery samples from patients with gastric cancer.
RNA Extraction and Quantitative Real-Time PCR
RNA was extracted from cells using TRIzol (Invitrogen) following the manufacturer’s protocol. To detect the levels of miR-675 in cells, reverse transcription (RT) was conducted with the Applied Biosystems TaqManH MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The primers for the miR-675 were purchased from Life Technologies (USA). The human U6 RNA was amplified in parallel as an internal control. For mRNA detection, the primers used in this study were as follows: H19 (forward: 5′-TACAACCACTGCACTACCTG-3′, reverse: 5′-TGGAATGCTTGAAGGCTGCT-3′) (33); RUNX1 (forward: 5′-CCGAGAACCTCGAAGACATC-3′, reverse: 5′-GATGGTTGGATCTGCCTTGT-3′); and GAPDH as an internal control (forward: 5′-ACCTGACCTGCCGTCTAGAA-3′, reverse: 5′-TCCACCACCCTGTTGCTGTA-3′) (27). The ABI StepOne Plus (Applied Biosystems, Foster City, CA, USA) was used to perform the amplification reaction. Each experiment was performed in triplicate, and the data were analyzed by the 2-ΔΔCt method.
Transfection
H19 cDNA (GenBank accession No. NR_002196.1) and RUNX1 cDNA (GenBank accession No. BC136381) were constructed into the multiple cloning sites of pcDNA3.1 vector (Invitrogen) according to Zhuang et al. (27). Knockdown expressions of H19/RUNX1 were performed by transfection with specific siRNA or its negative control (H19: 4390771; RUNX1: AM16708; Life Technologies), respectively. For the enhanced or knockdown expressions of miR-675, miR-675 mimic, inhibitor, and negative control were purchased from Life Technologies. A total of 5 × 105 cells were plated in six-well plate for 24 h and then transfected with 30 nM of miR-675 mimic/inhibitor/negative control or 2 µg of each respective plasmid with Lipofectamine 2000 (Invitrogen) for 48 h. The cells were then subjected to RNA/protein extraction or further functional assays.
Cell Proliferation Assay
For the cell proliferation assay, 5 × 104 cells were plated in a 24-well plate and transfected with related plasmids or oligonucleotides, and cell proliferation was determined using a CyQUANT cell proliferation assay (Life Technologies) according to the manufacturer’s protocol. The fluorescence intensity was measured using a fluorescence microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Colony Formation Assay
Soft agar plates were prepared in six-well plates with a bottom layer of 0.5% agar in serum-free RPMI-1640 medium. The cells were first seeded in 35-mm tissue culture dishes for 24 h and then transfected with different agents. After trypsinization, 1,000 cells mixed with 0.35% agar in 10% FBS-supplemented RPMI-1640 medium were seeded as the top agar layer onto the agar plates. The cells were then incubated in a 37°C incubator for 3 weeks. The number of colonies was counted after the colonies were stained with 0.1% crystal violet and washed extensively with PBS.
Invasion Assay
Cell invasion was determined by the Transwell assay. The related oligonucleotides were transfected into the cells according to the protocol. After incubated for 48 h, 3 × 104 cells were transferred on the top of the Matrigel-coated invasion chambers (BD Biosciences, USA) in a serum-free RPMI-1640 and RPMI-1640 containing 10% FBS was added to the lower chamber. After 24 h, noninvasion cells were removed, and the invading cells were fixed with 95% ethanol, stained with 0.1% crystal violet, and photographed (×100). Tests were repeated via three independent experiments.
Western Blotting
The oligonucleotides were transfected into the cells. Proteins were extracted from cells with RIPA lysis buffer (Beyotime, China) and were quantified using a BCA Protein Assay Kit (Beyotime). Thirty micrograms of protein lysates was loaded to SDS-PAGE. The electrophoresed proteins were transferred to PVDF membranes (Millipore, USA). The membrane was blocked in 5% nonfat milk and incubated with diluted antibodies (CST, USA), followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:2,000; Santa Cruz, USA). GAPDH was used as a control (1:1,000; CST, USA).
Statistical Analysis
All experiments were performed three times. Data are presented as the mean ± SD and analyzed by GraphPad Prism V.5.00 software (GraphPad Software, San Diego, CA, USA). Spearman rank test was used to assess the association between H19 and miR-675 expression level. Differences among different groups were tested by one-way ANOVA followed by Neuman–Keuls post hoc test. Two-sided values of p < 0.05 were considered statistically significant.
RESULTS
H19 Positively Correlates With miR-675 Expression in Gastric Cancer Tissues
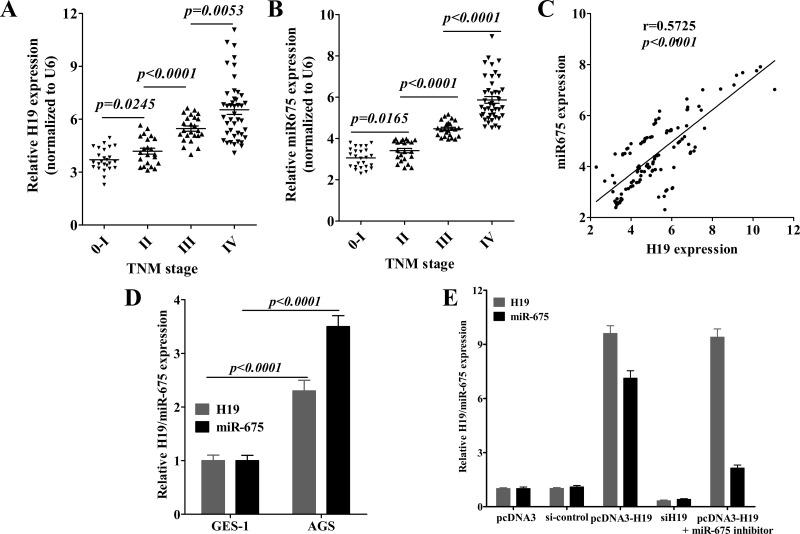
miR-675 upregulation has been reported to be positively correlated with H19 expression in glioma, colorectal cancer, and gastric cancer (22,27,33,34). In order to further confirm this result in a different population of patients with gastric cancer, we initially analyzed the expression of H19 and miR-675 in gastric cancer tissues from patients in Hubei province of China. The expression levels of H19 and miR-675 were significantly upregulated in gastric cancer patients as reported by Zhuang et al. (27), and we further identified that H19 and miR-675 expression were associated with tumor grade (Fig. 1A, B). Correlation analysis further revealed that H19 positively correlated with miR-675 expression in gastric cancer tissues (r = 0.5725, p < 0.0001). Thus, these results further confirmed that, as the derivate of H19, miR-675 may be involved in H19-induced gastric cancer progression.
Figure 1.
H19 and miR-675 are systematically positively correlated with clinical stage in gastric cancer. Relative expression of H19 (A) and miR-675 (B) in different stages of 114 gastric cancer tissues, which were normalized to GAPDH/U6 with respect to normal gastric mucosa. (C) Positive correlation of H19 and miR-675 expression in gastric cancer tissues, r = 0.5725, p < 0.0001, n = 114. (D) Relative H19/miR-675 expression in AGS cells was compared with those of GES-1 cells. (E) AGS cells were transfected with plasmids or oligonucleotides for 48 h. Then the relative expression of H19/miR-675 was validated using quantitative reverse transcription-PCR. Data presented as mean ± SD, n = 3 independent experiments.
miR-675 Is a Pivotal Mediator in H19-Induced Gastric Cancer Progression
To further elucidate the role of miR-675 played in H19-induced gastric cancer progression, we first compared H19 and miR-675 expression between gastric epithelial mucosa cell line GES-1 and gastric cancer cell line AGS. As shown in Figure 1D, H19 and miR-675 were significantly increased in AGS cells. Then we transfected AGS cells with H19 siRNA or pcDNA-H19 and found that deprivation of H19 expression remarkably reduced miR-675 expression (Fig. 1E), while overexpression of H19 induced a high level of miR-675 expression.
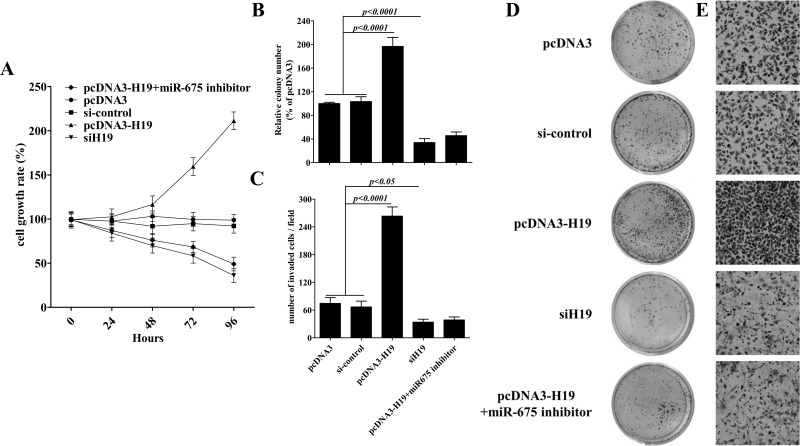
As miR-675 inhibitor downregulated pcDNA-H19-induced miR-675 expression (Fig. 1E), next we examined these kinds of treatment in gastric cancer progression. As shown in Figure 2A–E, miR-675 inhibitor rescued the promotion in the cellular proliferation, colony formation, and invasiveness of AGS cells induced by pcDNA-H19 transfection, and the declining level was even similar with H19 siRNA. Thus, these results suggested that H19 regulates the gastric cancer progression via miR-675.
Figure 2.
H19/miR-675 promotes proliferation, colony formation, and invasion of AGS cells. (A) AGS cells were plated in a 24-well plate and transfected with plasmids or oligonucleotides, and then the cell number was determined at the indicated time points using a CyQUANT cell proliferation assay. AGS cells, 1 × 103, treated the same as (A) and were seeded in soft agar for evaluating the ability of colony formation. Representative images show staining of colonies in each treatment (D). The colony formation efficiencies of indicated treatments were summarized (B). Cells were examined for cell invasion in 24-well plates with Transwell chambers. Migrated cells were stained with crystal violet and counted. Six random fields were photographed (shown in E) and summarized (C) per upper chamber in each group. Data presented as mean ± SD, n = 3 independent experiments.
The Expression of RUNX1 Is Regulated by H19-Derived miR-675 in Gastric Cancer Cells
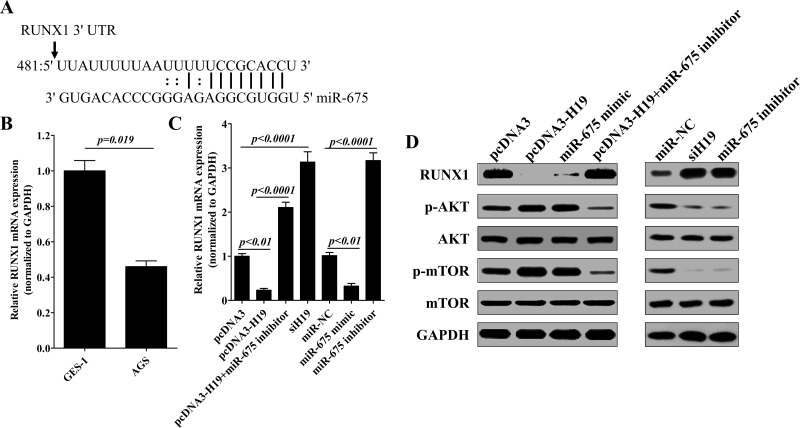
RUNX1 is an important tumor suppressor gene; as shown in Figure 3A, it was predicted to be a direct target of miR-675 (http://www.microrna.org/microrna) and has been identified by Zhuang et al. (27). In our study, we found that both H19 and miR-675 significantly decreased RUNX1 expression in AGS cells, and miR-675 inhibitor significantly rescued RUNX1 expression that decreased by ectopic H19 expression (Fig. 3C and D). Thus, these results demonstrated that H19 depend on miR-675 to inhibit RUNX1 expression.
Figure 3.
H19-derived miR-675 regulates RUNX1 and promotes Akt/mTOR phosphorylation. (A) RUNX1 is potentially targeted by miR-675 using bioinformatic analyses. (B) Relative expression of RUNX1 in AGS cells was compared with GES-1 cells. AGS cells were transfected with plasmids or oligonucleotides for 48 h and then the relative expression of RUNX1 was validated using quantitative reverse transcription-PCR (C). Western blotting analyses were performed to detect the protein levels of RUNX1, Akt, and mTOR (D). Data presented as mean ± SD, n = 3 independent experiments.
miR-675 Promotes AKT/mTOR Pathway Activation via Downregulating RUNX1
The Akt/mTOR signaling axis has been shown to be regulated by RUNX1 (29,30) and is activated in gastric cancer (32). Furthermore, Akt was found to be involved in H19/miR675-mediated carcinogenesis of human hepatocellular carcinoma (35). Here, in AGS cells, we found that both H19 and miR-675 promoted Akt and mTOR phosphorylation, and miR-675 inhibitor significantly suppressed the phosphorylation of Akt and mTOR that was induced by ectopic H19 expression (Fig. 3D). Thus, these results confirmed that H19 depend on miR-675 to enhance the activation of AKT/mTOR signaling.
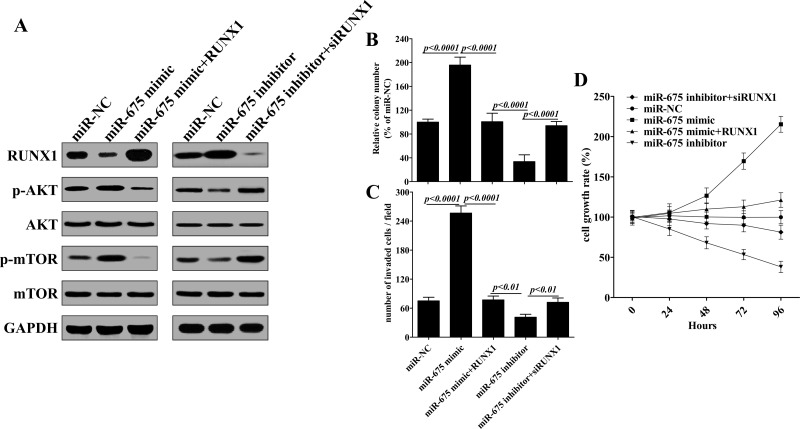
Next, AGS cells were cotransfected with RUNX1 overexpression plasmid and miR-675 mimic, ectopic expression of RUNX1 suppressed the phosphorylation of Akt and mTOR that was induced by miR-675 mimic (Fig. 4A). More importantly, in AGS cells, which cotransfected with RUNX1 siRNA and miR-675 inhibitor, we further noticed that RUNX1 interfered and reversed the suppressive function of miR-675 inhibitor on AKT and mTOR phosphorylation (Fig. 4A). Thus, these results demonstrated that H19-derived miR-675 activates Akt/mTOR signaling pathway by depressing RUNX1 expression.
Figure 4.
H19-derived miR-675 regulates gastric cancer progression via regulating the RUNX1/Akt/mTOR pathway. (A) miR-675 promotes Akt/mTOR phosphorylation via inhibiting RUNX1 expression. AGS cells were plated in a 24-well plate and transfected with plasmids or oligonucleotides for 48 h, and then Western blotting analyses were performed to detect the protein levels of RUNX1, Akt, and mTOR. AGS cells were treated the same as (A); the ability of colony formation (B), invasion (C), and cellular proliferation was examined. Data presented as mean ± SD, n = 3 independent experiments.
Ectopic Expression of RUNX1 Reverses miR-675-Induced Gastric Cancer Progression
As an important tumor suppressor, RUNX1 has been found to downregulate in gastric cancer (27,36). To further confirm that the tumor-promoting effects of miR-675 in gastric cancer cells is the result of the repression of RUNX1, AGS cells were simultaneously transfected with miR-675 mimic and RUNX1. As shown in Figure 4B–D, ectopic expression of RUNX1 suppressed the promotion in cell growth, colony formation, and invasiveness of AGS cells caused by miR-675 mimic. In addition, as for AGS cells, which were transfected with miR-675 inhibitor plus RUNX1-specific siRNA, knockdown of RUNX1 reverses the tumor retardation caused by miR-675 inhibitor. These results strongly indicated that RUNX1 is an important target that was downregulated by miR-675 to promote gastric cancer progression.
DISCUSSION
Recently, accumulating studies have demonstrated that serious lncRNAs are dysregulated in gastric cancer and are closely related with tumorigenesis, metastasis, or prognosis (37), such as the GAS5, SUMO1P3, MEG3, and H19 (34,38–40). Furthermore, the mechanisms underlying the role of lncRNAs in transcriptional regulation are complex (41,42). As for H19, studies have suggested that the oncogenic role of H19 is associated with its function as the precursor of miR-675 (43,44). However, the underlying mechanism remains unclear. In this study, we first established the possible link between H19-derived miR-675 and RUNX1/Akt/mTOR signaling axis in gastric development, which associates lncRNAs with microRNAs and signaling proteins, and this approach may provide valuable clues for the identification of new potential targets for cancer therapy.
Furthermore, our data showed that H19 expression in tumor tissues was positively correlated with clinical stage. This was consistent with the study conducted by Li et al. (34). They further identified that high expression of H19 in GC was correlated with poor prognosis while showing a little difference with the study of Song et al. (8), as there is no significant difference in their study. Owing to the difficulty in getting individual information of patients in these two studies, the correlation of H19 with the clinical stage may need a further meta-analysis in the future.
As miR-675 is derived from the first exon of H19 (45), miR-675 expression showed a positive correlation with H19 in gastric cancer. Currently, a study has found that miR-675 expression could discriminate adrenocortical carcinomas from adrenocortical adenomas (46) and dramatically promote the progression of hepatocellular carcinoma (47). In addition, H19-derived miR-675 promotes cancer cell proliferation and invasion in different cancers, including colorectal cancer, glioma, and gastric cancer (22,27,33). However, the underlying mechanism of miR-675 and its regulatory network in gastric cancer is still largely unknown. Many targets of miR-675 have been identified in different models of disease, such as Twist1, NOMO1, RB, CALN1, and RUNX1 (27,33,34,44,47).
RUNX1 is an important tumor suppressor and has been found to be a direct target of miR-675 (27); however, it was denied in a study conducted by Li et al. (34). So, in this study, we predicted the target of miR-675 in the website of miRBase and reexamined in AGS cells. Our data showed that H19 and miR-675 regulated RUNX1 expression in mRNA and protein level. In addition, whether RUNX1 is involved in regulating invasion and colony formation of gastric cancer cells is still unclear and which are important abilities that mediated tumor metastasis. In this study, we found that ectopic expression of RUNX1 suppressed these activities caused by miR-675 mimic in gastric cancer cells. Thus, the expression level of RUNX1 played a pivotal role in miR-675-mediated tumor progression.
Akt/mTOR pathway has been reported to be regulated by RUNX1 in leukemia carcinogenesis (29,30). More importantly, it also linked with H19/miR-675 to mediate tumor metastasis (35,48). In this study, by manipulating the expression of H19/miR-675 or RUNX1, we found that H19/miR-675 regulated the phosphorylation of Akt and mTOR, and this effect was dependent on the expression level of RUNX1. In addition, we also observed that introduction of RUNX1 rescued p53 inactivation in response to the overexpression of H19 or miR-675 (data not shown). This may be due to RUNX1, which is required for the stimulation of p53 in response to DNA damage caused by H19/miR-675 (26,49). Further exploration of the role of RUNX1-participated pathways in H19/miR-675-mediated tumor progression would be of interest in future studies.
In conclusion, the major findings of this study can be summarized as follows. (i) H19 regulates the gastric cancer progression via miR-675. (ii) H19-derived miR-675 modulates gastric cancer progression by downregulating the tumor-suppressor RUNX1. (iii) RUNX1 regulates the activation of Akt/mTOR signaling pathway and is an important factor for the tumorigenesis and metastasis of gastric cancer. Thus, our study provides value clues for understanding the regulatory network of H19 in tumor carcinogenesis and identifying new therapeutic targets for the treatment of gastric cancer.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Grant No. 81370562).
REFERENCES
- 1. Ferlay J.; Soerjomataram I.; Dikshit R.; Eser S.; Mathers C.; Rebelo M.; Parkin D. M.; Forman D.; Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136(5):E359–386; 2015. [DOI] [PubMed] [Google Scholar]
- 2. Hartgrink H. H.; Jansen E. P.; van Grieken N. C.; van de Velde C. J. Gastric cancer. Lancet 374(9688):477–490; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen G.; Wang Z.; Wang D.; Qiu C.; Liu M.; Chen X.; Zhang Q.; Yan G.; Cui Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 41(Database issue):D983–986; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lalevee S.; Feil R. Long noncoding RNAs in human disease: Emerging mechanisms and therapeutic strategies. Epigenomics 7(6):877–879; 2015. [DOI] [PubMed] [Google Scholar]
- 5. Ye N.; Wang B.; Quan Z. F.; Cao S. J.; Wen X. T.; Huang Y.; Huang X. B.; Wu R.; Ma X. P.; Yan Q. G. Functional roles of long non-coding RNA in human breast cancer. Asian Pac. J. Cancer Prev. 15(15):5993–5997; 2014. [DOI] [PubMed] [Google Scholar]
- 6. Silva J. M.; Perez D. S.; Pritchett J. R.; Halling M. L.; Tang H.; Smith D. I. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics 95(6):355–362; 2010. [DOI] [PubMed] [Google Scholar]
- 7. Perez D. S.; Hoage T. R.; Pritchett J. R.; Ducharme-Smith A. L.; Halling M. L.; Ganapathiraju S. C.; Streng P. S.; Smith D. I. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum. Mol. Genet. 17(5):642–655; 2008. [DOI] [PubMed] [Google Scholar]
- 8. Song H.; Sun W.; Ye G.; Ding X.; Liu Z.; Zhang S.; Xia T.; Xiao B.; Xi Y.; Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J. Transl. Med. 11:225; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bhan A.; Mandal S. S. Long noncoding RNAs: Emerging stars in gene regulation, epigenetics and human disease. Chem. Med. Chem. 9(9):1932–1956; 2014. [DOI] [PubMed] [Google Scholar]
- 10. Wapinski O.; Chang H. Y. Long noncoding RNAs and human disease. Trends Cell Biol. 21(6):354–361; 2011.21550244 [Google Scholar]
- 11. Zhang X.; Gejman R.; Mahta A.; Zhong Y.; Rice K. A.; Zhou Y.; Cheunsuchon P.; Louis D. N.; Klibanski A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 70(6):2350–2358; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Q.; Huang J.; Zhou N.; Zhang Z.; Zhang A.; Lu Z.; Wu F.; Mo Y. Y. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 41(9):4976–4987; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hung C. L.; Wang L. Y.; Yu Y. L.; Chen H. W.; Srivastava S.; Petrovics G.; Kung H. J. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA 111(52):18697–18702; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He J. H.; Zhang J. Z.; Han Z. P.; Wang L.; Lv Y. B.; Li Y. G. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J. Exp. Clin. Cancer Res. 33:72; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai M. C.; Yang Z.; Zhou L.; Zhu Q. Q.; Xie H. Y.; Zhang F.; Wu L. M.; Chen L. M.; Zheng S. S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 29(3):1810–1816; 2012. [DOI] [PubMed] [Google Scholar]
- 16. Liu J. H.; Chen G.; Dang Y. W.; Li C. J.; Luo D. Z. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac. J. Cancer Prev. 15(7):2971–2977; 2014. [DOI] [PubMed] [Google Scholar]
- 17. Ji P.; Diederichs S.; Wang W.; Boing S.; Metzger R.; Schneider P. M.; Tidow N.; Brandt B.; Buerger H.; Bulk E.; Thomas M.; Berdel W. E.; Serve H.; Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22(39):8031–8041; 2003. [DOI] [PubMed] [Google Scholar]
- 18. Lin R.; Maeda S.; Liu C.; Karin M.; Edgington T. S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26(6):851–858; 2007. [DOI] [PubMed] [Google Scholar]
- 19. Adriaenssens E.; Dumont L.; Lottin S.; Bolle D.; Lepretre A.; Delobelle A.; Bouali F.; Dugimont T.; Coll J.; Curgy J. J. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 153(5):1597–1607; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ariel I.; Miao H. Q.; Ji X. R.; Schneider T.; Roll D.; de Groot N.; Hochberg A.; Ayesh S. Imprinted H19 oncofetal RNA is a candidate tumour marker for hepatocellular carcinoma. Mol. Pathol. 51(1):21–25; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo M.; Li Z.; Wang W.; Zeng Y.; Liu Z.; Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333(2):213–221; 2013. [DOI] [PubMed] [Google Scholar]
- 22. Shi Y.; Wang Y.; Luan W.; Wang P.; Tao T.; Zhang J.; Qian J.; Liu N.; You Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One 9(1):e86295; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vennin C.; Spruyt N.; Dahmani F.; Julien S.; Bertucci F.; Finetti P.; Chassat T.; Bourette R. P.; Le Bourhis X.; Adriaenssens E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 6(30):29209–239223; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J.; Song Y. X.; Wang Z. N. Non-coding RNAs in gastric cancer. Gene 560(1):1-8; 2015. [DOI] [PubMed] [Google Scholar]
- 25. Li P. F.; Chen S. C.; Xia T.; Jiang X. M.; Shao Y. F.; Xiao B. X.; Guo J. M. Non-coding RNAs and gastric cancer. World J. Gastroenterol. 20(18):5411–5419; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang F.; Bi J.; Xue X.; Zheng L.; Zhi K.; Hua J.; Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 279(17):3159–3165; 2012. [DOI] [PubMed] [Google Scholar]
- 27. Zhuang M.; Gao W.; Xu J.; Wang P.; Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 448(3):315–322; 2014. [DOI] [PubMed] [Google Scholar]
- 28. Fijneman R. J.; Anderson R. A.; Richards E.; Liu J.; Tijssen M.; Meijer G. A.; Anderson J.; Rod A.; O’Sullivan M. G.; Scott P. M.; Cormier R. T. Runx1 is a tumor suppressor gene in the mouse gastrointestinal tract. Cancer Sci. 103(3):593–599; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuka G.; Kantner H. P.; Grausenburger R.; Inthal A.; Bauer E.; Krapf G.; Kaindl U.; Kauer M.; Dworzak M. N.; Stoiber D.; Haas O. A.; Panzer-Grumayer R. Silencing of ETV6/RUNX1 abrogates PI3K/AKT/mTOR signaling and impairs reconstitution of leukemia in xenografts. Leukemia. 26(5):927–933; 2012. [DOI] [PubMed] [Google Scholar]
- 30. Edwards H.; Xie C.; LaFiura K. M.; Dombkowski A. A.; Buck S. A.; Boerner J L.; Taub J. W.; Matherly L. H.; Ge Y. RUNX1 regulates phosphoinositide 3-kinase/AKT pathway: Role in chemotherapy sensitivity in acute megakaryocytic leukemia. Blood 114(13):2744–2752; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karar J.; Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 4:51; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tapia O.; Riquelme I.; Leal P.; Sandoval A.; Aedo S.; Weber H.; Letelier P.; Bellolio E.; Villaseca M.; Garcia P.; Roa J. C. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 465(1):25–33; 2014. [DOI] [PubMed] [Google Scholar]
- 33. Tsang W. P.; Ng E. K.; Ng S. S.; Jin H.; Yu J.; Sung J. J.; Kwok T. T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 31(3):350–358; 2010. [DOI] [PubMed] [Google Scholar]
- 34. Li H.; Yu B.; Li J.; Su L.; Yan M.; Zhu Z.; Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5(8):2318–2329; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lv J.; Ma L.; Chen X. L.; Huang X. H.; Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3beta/Cdc25A signaling pathway. J. Huazhong Univ. Sci. Technolog. Med. Sci. 34(3):363–369; 2014. [DOI] [PubMed] [Google Scholar]
- 36. Sakakura C.; Hagiwara A.; Miyagawa K.; Nakashima S.; Yoshikawa T.; Kin S.; Nakase Y.; Ito K.; Yamagishi H.; Yazumi S.; Chiba T.; Ito Y. Frequent downregulation of the runt domain transcription factors RUNX1, RUNX3 and their cofactor CBFB in gastric cancer. Int. J. Cancer 113(2):221–228; 2005. [DOI] [PubMed] [Google Scholar]
- 37. Gan L.; Xu M.; Zhang Y.; Zhang X.; Guo W. Focusing on long noncoding RNA dysregulation in gastric cancer. Tumour Biol. 36(1):129–141; 2015. [DOI] [PubMed] [Google Scholar]
- 38. Sun M.; Jin F. Y.; Xia R.; Kong R.; Li J. H.; Xu T. P.; Liu Y. W.; Zhang E. B.; Liu X. H.; De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 14:319; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun M.; Xia R.; Jin F.; Xu T.; Liu Z.; De W.; Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 35(2):1065–1073; 2014. [DOI] [PubMed] [Google Scholar]
- 40. Mei D.; Song H.; Wang K.; Lou Y.; Sun W.; Liu Z.; Ding X.; Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med. Oncol. 30(4):709; 2013. [DOI] [PubMed] [Google Scholar]
- 41. Ponting C. P.; Oliver P. L.; Reik W. Evolution and functions of long noncoding RNAs. Cell 136(4):629–641; 2009. [DOI] [PubMed] [Google Scholar]
- 42. Wang K. C.; Chang H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43(6):904–914; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim S. J.; Park S. E.; Lee C.; Lee S. Y.; Jo J. H.; Kim J. M.; Oh Y. K. Alterations in promoter usage and expression levels of insulin-like growth factor-II and H19 genes in cervical carcinoma exhibiting biallelic expression of IGF-II. Biochim. Biophys. Acta 1586(3):307–315; 2002. [DOI] [PubMed] [Google Scholar]
- 44. Gao W. L.; Liu M.; Yang Y.; Yang H.; Liao Q.; Bai Y.; Li Y. X.; Li D.; Peng C.; Wang Y. L. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1). RNA Biol. 9(7):1002–1010; 2012. [DOI] [PubMed] [Google Scholar]
- 45. Keniry A.; Oxley D.; Monnier P.; Kyba M.; Dandolo L.; Smits G.; Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 14(7):659–665; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmitz K. J.; Helwig J.; Bertram S.; Sheu S. Y.; Suttorp A. C.; Seggewiss J.; Willscher E.; Walz M. K.; Worm K.; Schmid K. W. Differential expression of microRNA-675, microRNA-139-3p and microRNA-335 in benign and malignant adrenocortical tumours. J. Clin. Pathol. 64(6):529–535; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hernandez J. M.; Elahi A.; Clark C. W.; Wang J.; Humphries L. A.; Centeno B.; Bloom G.; Fuchs B. C.; Yeatman T.; Shibata D. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann. Surg. Oncol. 20(Suppl. 3):S625–635; 2013. [DOI] [PubMed] [Google Scholar]
- 48. Matouk I. J.; Raveh E.; Abu-lail R.; Mezan S.; Gilon M.; Gershtain E.; Birman T.; Gallula J.; Schneider T.; Barkali M.; Richler C.; Fellig Y.; Sorin V.; Hubert A.; Hochberg A.; Czerniak A. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta 1843(7):1414–1426; 2014. [DOI] [PubMed] [Google Scholar]
- 49. Wu D.; Ozaki T.; Yoshihara Y.; Kubo N.; Nakagawara A. Runt-related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 protein in response to DNA damage through complex formation and acetylation. J. Biol. Chem. 288(2):1353–1364; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]