Abstract
Acute myeloid leukemia (AML) is the most lethal hematological malignancy, and the occurrence of chemoresistance prevents the achievement of complete remission following the standard therapy. MicroRNAs have been extensively investigated as critical regulators of hematopoiesis and leukemogenesis, and they represent a promising strategy for AML therapy. In this study, we identified miR-34b as a novel regulator in myeloid proliferation and apoptosis of leukemic cells. We found that miR-34b was developmentally upregulated in plasma and myeloid cells of healthy subjects, while it was significantly reduced in blood samples of patients with AML and AML cell lines. Moreover, the miR-34b mimicked transfection-mediated restoration of miR-34b inhibited cell viability and promoted cell apoptosis of HL-60 and OCI-AML3 cell lines. Using a miRNA predicting algorithm miRanda, we selected a potent target heat shock transcription factor 1 (HSF1) since that is a master regulator of the heat shock response and is associated with cancer aggressiveness and dissemination. In contrast to the level of miR-34b, HSF1 was highly expressed in blood samples of patients with AML and AML cell lines. The luciferase reporter assay revealed that miR-34b directly targeted the HSF1 gene. HSF1 silencing exhibited comparable inhibitory effects on AML cell proliferation and survival. The upregulated HSF1 elevated the activation of the Wnt–β-catenin pathway. In conclusion, miR-34b suppressed AML cell proliferation and survival by targeting HSF1, in turn leading to the inactivation of Wnt–β-catenin pathway, which may highlight a new therapeutic approach for AML.
Key words: miR-34b, Acute myeloid leukemia (AML), Heat shock transcription factor 1 (HSF1), Wnt pathway
INTRODUCTION
Acute myeloid leukemia (AML) is the most common acute leukemia in adults, and it ranks among the most lethal hematological malignancy. As a clonal neoplasm derived from myeloid progenitor cells, AML is characterized by the uncontrolled proliferation and accumulation of immature and dysfunctional hematopoietic progenitors accompanied by blockage in normal hematopoiesis (1,2). The initial goal of treatment is the achievement of complete remission, and allogeneic stem cell transplantation offers the best prospect of cure for many cases of AML (3). However, owing to the occurrence of chemoresistance in leukemia cells, the majority of patients do not achieve complete remission or relapse following the standard therapy (4). Despite extensive efforts that have been put forward into the understanding of the pathogenesis of AML, it is still difficult to translate the current knowledge into viable treatments, and AML remains an incurable disease with a high mortality rate (5). Therefore, understanding the underlying mechanisms and developing new therapies for AML are urgently needed.
MicroRNA (miRNA) is a class of small noncoding endogenous RNAs with approximately 19 to 25 nucleotides (nt), which is incorporated into the RNA-induced silencing complex (RISC), which aids in generating their repressive functions. The mature miRNA then binds to 3′-UTR of the target mRNA and causes either degradation or inhibition of the mRNA at the posttranscriptional level (6). An overwhelming number of studies have revealed the multiple functions of miRNAs in cell survival, migration, differentiation, and hematopoiesis (7). Moreover, alterations in the miRNA expression profile are involved in the initiation and progression of human tumors (8). During the last few decades, studies concerning the role of miRNA in AML pathogenesis have surfaced, and their applications to AML diagnosis, as well as treatment, have attracted much more attention. However, few studies have been performed to elaborate on the role of miRNA in AML, and the mechanisms implicated in the regulatory role of miRNA are not yet fully identified due to the high level of heterogeneity in patients with different subtypes of AML (7,9).
miR-34b is a known tumor-suppressor gene in the p53 tumor-suppressor network that belongs to the evolutionary conserved miR-34 family. miR-34 is downregulated in various human tumors and has an antiproliferative potential in cancer cell lines (10). It has been demonstrated that miR-34a is one of the most significantly downregulated miRNAs in AML patients with a complex karyotype, and its clinically low expression predicts chemotherapy resistance and inferior outcome for AML (11). Pigazzi et al. reported that miR-34b directly targeted cyclic AMP-responsive element-binding protein (CREB) to restore cell cycle abnormalities in AML (12). Although efforts have been made to identify miR-34b target genes, less information is currently available on their targets and functions in acute leukemia. In this study, we will elaborate on the role of miR-34b and the function of the predicted target heat shock transcription factor 1 (HSF1) in AML cell lines, which may represent a new underlying mechanism and therapeutic strategy for AML.
MATERIALS AND METHODS
AML Patients
Peripheral blood samples were collected from 16 newly diagnosed AML patients (9 females, 7 males) who did not undergo any therapy or treatment, and 12 age- and gender-matched healthy individuals (6 females, 6 males) were set as controls. The 16 patients with AML included 4 with M2, 3 with M3, 6 with M4, and 5 with M5 disease according to the FAB classification. The mean age of the patients and control subjects was 36.8 ± 7.6 years and 34.3 ± 6.5 years, respectively. The diagnosis of leukemia was made by morphologic and cytochemical studies of bone marrow smears. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki and with approval of the Medical Ethics Committee of Xi’an Central Hospital. The study was conducted according to an institutional review board-approved protocol.
Cell Culture
The human leukemia cell line HL-60 was purchased from American Type Cell Culture Collection (Manassas, VA, USA) and OCI-AML3 from DSMZ (Braunschweig, Germany) and deemed free of mycoplasma and bacterial contaminants. Primary AML blasts or mononuclear cells (MNCs) from peripheral blood samples of healthy donors were isolated by density-gradient centrifugation using the Ficoll-Paque Plus kits (Ficoll, Piscataway, NJ, USA). Blood cells with specific surface markers were further purified from MNCs of healthy donors by cell sorting with antibodies (APC-CD11b, PE-CD14). All antibodies used for flow cytometry were purchased from Abcam (Cambridge, UK). Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml of penicillin G, 100 µg/ml of streptomycin, and 2 mM of l-glutamine, at 37°C in humidified air containing 5% CO2 incubator.
miR-34b Mimic and HSF1 siRNA Transfection
The synthetic miR-34b mimic and their matched negative controls were purchased from GenePharma (Shanghai, China). The siRNA targeting human HSF1 was synthesized by Sangon Biotech Co. Ltd. (Shanghai, China). Transfection of miRNAs and siRNAs was performed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The HL60 and OCI-AML3 cells were grown in plates at a density of 2 × 105 cells/ml and cultured for 24 h, then transfected with miR-34b mimic or nonspecific (NS) miRNA using Lipofectamine 2000 at a final concentration of 40 nM. The effect of the mimic of miR-34b was examined in triplicate at 24 h posttransfection. The siRNA sequence was designed as follows: 5′-TCC TAA GGT TGG CGT TGT A-3′. The siRNA-transfected cells were incubated for 48 h and then harvested for experiments.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
The expression levels of miR-34b in blood samples and cancer cells were measured using RT-qPCR. Total RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was reverse transcribed from 1.0 µg total RNA using special stem-loop primers and the mirVana reverse transcription kit (Ambion Inc., Austin, TX, USA), followed by qPCR using the SYBR® Premix Ex TaqTM II (Perfect Real Time, Takara) at Applied Biosystems 7500HT (CA, USA). U6 small nuclear RNA was used as an internal control for miR-34b. Data analyses were performed using the 2−ΔΔCt method. Each PCR was performed in triplicate. To determine HSF1 and HSP27 expression, cDNA was synthesized from ∼1 µg total RNA using the M-MLV RTase cDNA Synthesis kit (Takara Biotechnology, Dalian, China) according to the manufacturer’s instructions. The resultant cDNA was used as a template in the RT-PCR amplifications using a SYBR Green Real-Time PCR Master Mix kit (Takara Biotechnology). GAPDH was used to normalize the target gene expression levels. HSF1 primers (sense: 5′-CAT GAA GCA TGA GAA TGA GGC T-3′, antisense: 5′-ACTG CAC CAG TGA GAT CAG GA-3′) were used with GAPDH primers (sense 5′-GCC AAA AGG GTC ATC ATC TC-3′, antisense 5′-GTA GAG GCA GGG ATG ATG TTC-3′) as the internal control. Fold changes in mRNA expression levels were calculated using the comparative threshold cycle (Ct) method. Aliquots of the PCR products were separated on 1.5% agarose gels, and PCR fragments were visualized by ethidium bromide staining.
Western Blot
Total protein was extracted with RIPA lyse buffer containing 1 mmol/L of PMSF, 1 mmol/L of NaF, and 1 mmol/L of Na3VO4. The cell lysates were separated using polyacrylamide gel electrophoresis (PAGE) and subsequently transferred to a PVDF membrane by electrotransfer. Afterward, the membrane was blocked for 1 h and probed with primary antibodies against HSF1 (1:1,000), β-catenin (1:500), and β-actin (1:300) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) followed by a horseradish peroxidase (HRP)-conjugated secondary antibody (Abcam). Blots were visualized using Amersham Western blot detection reagent (GE Healthcare, Piscataway, NJ, USA). Blot density of each band was quantified by Gel imaging system and Quantity One 4.62 software (Bio-Rad, Hercules, CA, USA).
Cell Viability Determination
The HL60 cells and OCI-AML3 cell viability were analyzed using cell counting kit (CCK)-8 kits (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions after cells were cultured at 24, 48, and 72 h.
Cell Apoptosis
The apoptotic cells were counted using a commercially available FITC Annexin-V Apoptosis Detection Kit (BD Biosciences), performed according to a recommended protocol of the manufacturer. Cells were collected, washed, and dispersed in 100 ml of 1× binding buffer and stained with FITC-conjugated annexin V and propidium iodide (PI). The tagged apoptotic cells were sorted by the FACSCalibur flow cytometer and analyzed using CellQuest software (Accuri C6; BD Biosciences).
Luciferase Assay
A mixture of human HSF1 3′-UTR luciferase reporter plasmid pMIR-LUC-3′-UTR-HSF1-wt or mutant pMIR-LUC-3′-UTR-HSF1-mut, Renilla plasmid (REN), and miR-34b mimics was transfected into HL60 cells using Lipofectamine 2000 (Invitrogen). The negative control miRNA replaced miR-34b in the mixture and was used as the control. After 24 h of culture, luciferase activity was measured using the luciferase assay system (Promega, Madison, WI, USA) with Top Count Microplate Scintillation Counter (Canberra, Meriden, CT, USA). Experiments were performed in triplicate.
Statistical Analysis
Data are presented as mean ± SD. Differences were compared by analysis of variance (ANOVA) followed by the LSD post hoc test. A value of p < 0.05 was considered statistically significant. All statistical tests were performed using SPSS, v. 13.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
miR-34b Is Downregulated in AML Patients and Cell Lines
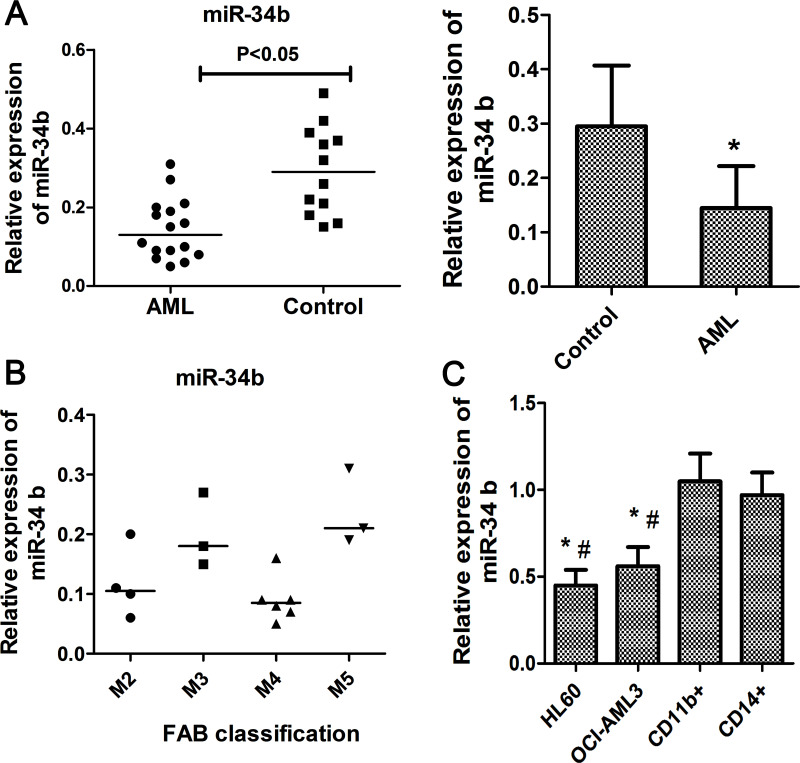
To investigate whether miR-34b was involved in ontogenesis of AML, we analyzed its expression in bone marrow samples from AML patients and healthy volunteers. Sixteen AML patients with a mean age of 34.3 years old and 12 healthy subjects with comparable ages were enrolled in this study. The results showed that miR-34b was downregulated in AML samples compared to the control (Fig. 1A). In addition, the miR-34b was lower in M2 and M4 subtype AML patients compared with M3 and M5 subtype (Fig. 1B). Furthermore, we detected the amount of miR-34b in human leukemia cells HL-60 and OCI-AML3 cells and CD11b+ and CD14+ myeloid cells isolated from peripheral blood samples of healthy donors using RT-qPCR. As shown in Figure 1D, the expression of miR-181b was significantly decreased in human leukemia cell lines OCI-AML3 and HL-60, compared to the normal myeloid cells such as CD11b and CD14 cells, respectively (p < 0.05 for each comparison). These results indicated that miR-34b might be involved in the oncogenesis and progression AML.
Figure 1.
miR-34b was downregulated in AML patients and cell lines. (A) Relative expression of miR-34b in blood samples of AML patients (n = 16) and healthy controls (n = 12). *p < 0.05 versus control. (B) miR-34b level in AML patients with different subtypes according to the FAB classification. (D) miR-34b expression in AML cell lines (HL-60 and OCI-AML3) and normal blood cells from healthy donors (n = 12). Each experiment was repeated in triplicate. *p < 0.05 versus CD11b cell, #p < 0.05 versus CD14 cell.
miR-34b Upregulation Inhibits AML Cell Survival
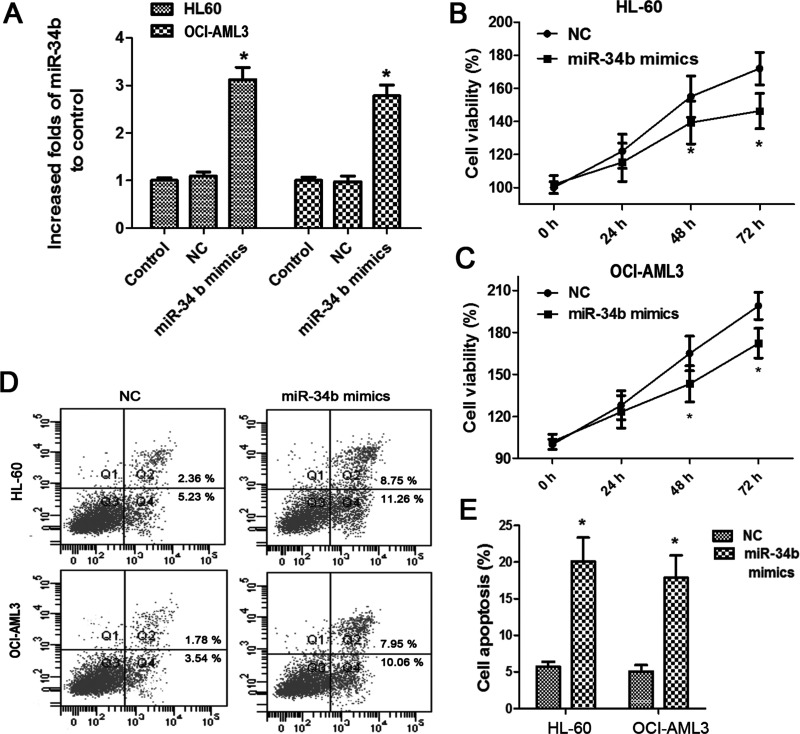
To further evaluate the role of miR-34b on AML progression, we restored the miR-34b expression by transfecting its mimics into HL60 and OCI-AML3 cells, and it was shown that miR-34b was markedly elevated in either cells after transfections (Fig. 2A). Moreover, we found that miR-34b upregulation significantly suppressed cell viability at 48 h posttransfection, and the inhibitory effects of miR-34b seemed to be more obvious in HL-60 cells (Fig. 2B, C). Moreover, we detected cell apoptosis in the two cell lines using flow cytometry. The mean apoptotic cells reached 20.06% in HL-60 cells and 17.6% in OCI-AML3 cells after miR-34b upregulation, indicating that miR-34b was implicated in AML cell death (Fig. 2D, E).
Figure 2.
miR-34b upregulation inhibited AML cell viability and cell survival. (A) Evaluation of miR-34b expression after HL-60 and OCI-AML3 cells were transfected with miR-34b mimics. (B, C) Effects of miR-34b upregulation on the cell viability of HL-60 and OCI-AML3. (D, E) Effects of miR-34b upregulation on cell apoptosis of HL-60 and OCI-AML. Each experiment was repeated in triplicate. *p < 0.05 versus NC.
miR-34b Targets HSF1 in AML Cells
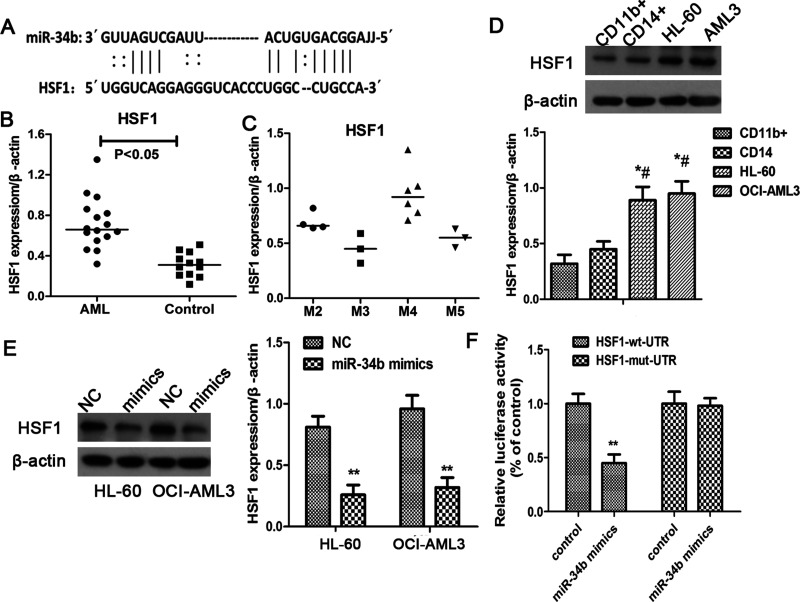
Analysis of miR-34b-predicted target genes was performed using the algorithm miRanda. The algorithm produced a list of hundreds of target genes for miR-34b. In this study, we focused on the HSF1 gene for its significantly increased expression in blood sample of AML patients as reported by Newman et al. (13) (Fig. 3A). We evaluated the expression of HSF1 in AML patients and healthy subjects, and it also suggested that HSF1 was significantly elevated in AML patients compared to the controls, especially in the M2 and M4 subclass patients, while the M3 and M5 AML patients showed relative lower HSF1 level (Fig. 3B, C). Similarly, leukemia cells had more HSF1 than normal blood cells (Fig. 3D). Moreover, we found that the miR-34b overexpressed cells showed significantly reduced HSF1 expression either in HL-60 or OCI-AML3 cells (p < 0.05) (Fig. 3E). Therefore, in order to confirm whether miR-34b directly binds to the HSF 3′-UTR region, we performed a luciferase assay and found that miR-34b upregulation significantly suppressed the luciferase activity in HSF1 wild-type vector in HL-60 cells, while it did not display inhibitory effects on cells transfected with the corresponding mutant HSF1 plasmid (Fig. 3F). These data demonstrated that HSF1 was a direct target of miR-34b in AML cells.
Figure 3.
miR-34b targeted to HSF1 in AML cells. (A) Predicted binding site of miR-34b on target gene HSF1 using the algorithm miRanda. (B) Relative expression of HSF1 in blood sample of AML patients (n = 16) and healthy controls (n = 12). (C) HSF1 protein level in AML patients with different subtypes according to the FAB classification. (D) miR-34b expression in AML cell lines (HL-60 and OCI-AML3) and normal blood normal blood cells from healthy donors (n = 12). Each experiment was repeated in triplicate. *p < 0.05 versus CD11b cell, #p < 0.05 versus CD14 cell. (E) HSF1 expression in AML cells in response to the administration of miR-34b mimics. (F) Luciferase assay analysis of the binding of miR-34b on HSF1 3′-UTR. HSF-wt-UTR and HSF1-mut-UTR reporter plasmids together with miR-34b mimics were transfected into HL-60 cells, and the relative luciferase activities were determined.
HSF1 Is Associated With AML Cell Survival by Activating Wnt Signaling
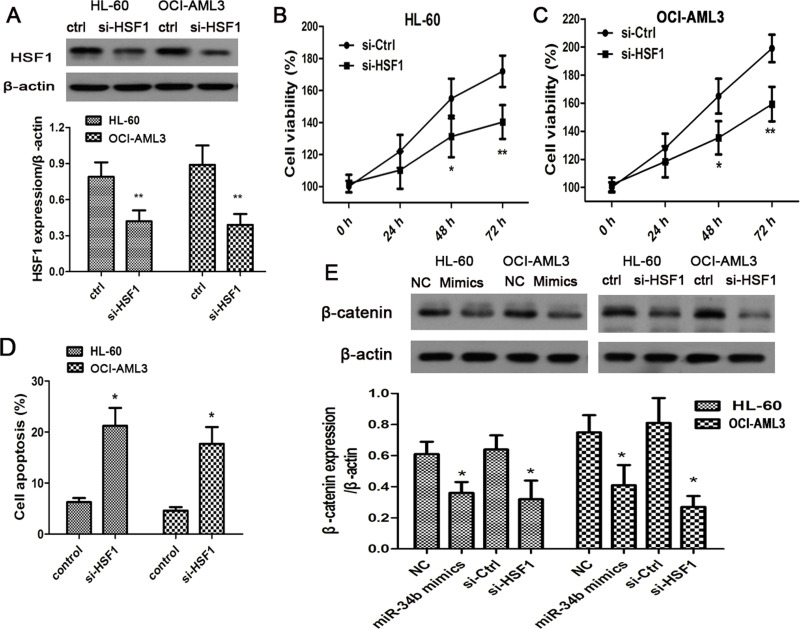
As reflected by the above results, miR-34b targeted HSF1 and played an important role in the progression of AML. We then intended to clarify the roles of HSF1 in cancer cell survival and the potential mechanisms. HSF siRNA was transfected in the two cell lines, and the protein levels of HSF1 were reduced (Fig. 4A). Meanwhile, their cell viability was efficiently prohibited at 24 h after the transfection (Fig. 4B, C). In addition, cell apoptosis was accelerated in both cells and in HSF1-silenced cells (Fig. 4D). It was proven that HSF1 regulates the Wnt signaling cascade in breast cancer cells. We then sought to investigate the role of HSF1 in modulating Wnt signaling to promote the survival of AML cells. In both the miR-34b mimics and the HSF1 siRNA injected cells, β-catenin, classical molecule in Wnt signaling pathway, were all inhibited in AML cells (Fig. 4E).
Figure 4.
miR-34b inactivated Wnt signaling pathway by suppressing HSF1. (A) Western blotting analysis of HSF1 expression in AML cell lines following the HSF1 siRNA introduction. (B, C) Effects of HSF1 silencing on the cell viability of HL-60 and OCI-AML3. (D) AML cell apoptosis before and after treatment with HSF1 siRNA. (E) Western blotting analysis of β-catenin expression in AML cell lines following the transfection of miR-34b mimics or HSF1 siRNA. Each experiment was repeated in triplicate. *p < 0.05 versus control or NC.
DISCUSSION
During the past few decades, the role of microRNA in AML pathogenesis and its availability in the early diagnosis and prognosis of AML has attracted much more attention. However, the understanding of the role of this bulk of miRNAs and their target gene in the initiation and progression is far from satisfactory due to the high level of heterogeneity in patients with different subtypes of AML. In our study, we demonstrated that miR-34b is routinely suppressed in blood samples of AML patients and leukemia cell lines. The deficiency of miR-34b in cancer cells reversely lead to the increase in its target gene HSF1, which eventually activated Wnt signaling to promote cancer cell survival.
miR-34b expression is documented to be abnormal in many types of leukemia, such as AML (12), chronic lymphocytic leukemia (CLL) (14), and acute promyelocytic leukemia (15). In addition, in accordance with other reports, we also confirmed that miR-34b was less expressed in blood samples of AML patients and myeloid leukemia cell lines when compared with healthy bone marrow. It was demonstrated that miR-34b is a tumor suppressor that was frequently methylated, which contributed to its downregulation in CLL (16). However, whether miR-134b is epigenetically inactivated and contributes to its downregulation in AML still requires further study. Accumulating evidence pointed to forced expression of miR-34b in suppressing cell proliferation and its clonogenic potential in various cancer cells (17,18). Our study revealed that miR-34b restoration inhibited cell growth and survival in AML cell lines.
Heat shock transcription factor 1 (HSF1) is the master regulator of the heat shock response, which is overexpressed in multiple types of human cancers and is associated with cancer aggressiveness and dissemination (19). It is clarified that HSF1 could enhance the survival of cancer cells exposed to various stressors and could serve as an independent diagnostic or prognostic biomarker (20). In addition, HSF1 is surfacing as a potential target in leukemia therapy. The recent findings demonstrated that targeting HSF1 could disrupt HSP90 chaperone function in chronic lymphocytic leukemia and inhibit the HSF pathway, contributing to the inhibition of AML progression (21,22). In the current study, HSF silencing also exhibited anticancer capability in AML cells. Furthermore, we confirmed that miR-34b directly targeted HSF1 to control its cellular abundance, and miR-34b expression deficiency failed to manage the levels of HSF1.
Moreover, we found that HSF1 linked to the activated β-catenin pathway. It is well established that canonical Wnt–β-catenin pathway is essential for self-renewal, growth, and survival of AML cells, and new therapeutic approaches targeting β-catenin showed promising preclinical activity against ALL (23). Fanelli et al. reported a specific interaction between β-catenin and Hsp27 (19). HSF1 is known to regulate the expression of heat shock proteins (HSPs) including HSP90, HSP70, and HSP27. Therefore, HSP27 may mediate the regulation of HSF1 on the canonical Wnt–β-catenin pathway. However, the exact mechanism needs more investigation. Interestingly, HSF1 was suggested to participate in regulating microRNAs (miRNAs) and long noncoding RNA (lncRNA) expression (24). Whether there is a feedback loop between miR-34b and HSF1 still awaits further confirmation.
In conclusion, our study demonstrated that miR-34b was downregulated in blood samples of AML patients and AML cell lines, accompanied with the increased expression of HSF1. miR-34b restoration inhibited cell viability and survival by directly targeting HSF1, which inactivated the Wnt–β-catenin pathway and HSP27 may mediate this inactivation. Therefore, miR-34b restoration may represent an alternative strategy for the treatment of AML.
REFERENCES
- 1. Lu F.; Zhang J.; Ji M.; Li P.; Du Y.; Wang H., Zang S.; Ma D.; Sun X.; Ji C. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int. J. Oncol. 45:383–392; 2014. [DOI] [PubMed] [Google Scholar]
- 2. Sengsayadeth S.; Savani B. N.; Blaise D.; Malard F.; Nagler A.; Mohty M. Reduced intensity conditioning allogeneic hematopoietic cell transplantation for adult acute myeloid leukemia in complete remission—A review from the Acute Leukemia Working Party of the EBMT. Haematology 100:859–869; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ossenkoppele G. J.; Janssen J. J.; van de Loosdrecht A. A. Risk factors for relapse after allogeneic transplantation in acute myeloid leukemia. Haematology 101:20–25; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu Y.; Cao C.; Gong X.; Rong L. Inhibition of ERK5 enhances cytarabine-induced apoptosis in acute myeloid leukemia cells. Int. J. Clin. Exp. Med. 8:6446–6455; 2015. [PMC free article] [PubMed] [Google Scholar]
- 5. Kadia T. M.; Ravandi F.; Cortes J.; Kantarjian H. New drugs in acute myeloid leukemia. Ann. Oncol. 27(5):770–778; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heuston E. F.; Lemon K. T.; Arceci R. J. The beginning of the road for non-coding RNAs in normal hematopoiesis and hematologic malignancies. Front. Genet. 2:94; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Volinia S.; Galasso M.; Costinean S.; Tagliavini L.; Gamberoni G.; Drusco A.; Marchesini J.; Mascellani N.; Sana M. E.; Abu Jarour R.; Desponts C.; Teitell M.; Baffa R.; Aqeilan R.; Iorio M. V.; Taccioli C.; Garzon R.; Di Leva G.; Fabbri M.; Catozzi M.; Previati M.; Ambs S.; Palumbo T.; Garofalo M.; Veronese A.; Bottoni A.; Gasparini P.; Harris C. C.; Visone R.; Pekarsky Y.; de la Chapelle A.; Bloomston M.; Dillhoff M.; Rassenti L. Z.; Kipps T. J.; Huebner K.; Pichiorri F.; Lenze D.; Cairo S.; Buendia M. A.; Pineau P.; Dejean A.; Zanesi N.; Rossi S.; Calin G. A.; Liu C. G.; Palatini J.; Negrini M.; Vecchione A.; Rosenberg A.; Croce C. M. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 20:589–599; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spizzo R.; Nicoloso M. S.; Croce C. M.; Calin G. A. SnapShot: MicroRNAs in cancer. Cell 137(3):586; 2009. [DOI] [PubMed] [Google Scholar]
- 9. Shahjahani M.; Khodadi E.; Seghatoleslami M.; Asl J. M.; Golchin N.; Zaieri Z. D.; Saki N. Rare cytogenetic abnormalities and alteration of microRNAs in acute myeloid leukemia and response to therapy. Oncol. Rev. 9:261; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bagchi A.; Mills A. A. The quest for the 1p36 tumor suppressor. Cancer Res. 68:2551–2556; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rucker F. G.; Russ A. C.; Cocciardi S.; Kett H.; Schlenk R. F.; Botzenhardt U.; Langer C.; Krauter J.; Fröhling S.; Schlegelberger B.; Ganser A.; Lichter P.; Zenz T.; Döhner H.; Döhner K.; Bullinger L. Altered miRNA and gene expression in acute myeloid leukemia with complex karyotype identify networks of prognostic relevance. Leukemia 27:353–361; 2013. [DOI] [PubMed] [Google Scholar]
- 12. Pigazzi M.; Manara E.; Baron E.; Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 69:2471–2478; 2009. [DOI] [PubMed] [Google Scholar]
- 13. Newman B.; Liu Y.; Lee H. F.; Sun D.; Wang Y. HSP90 inhibitor 17-AAG selectively eradicates lymphoma stem cells. Cancer Res. 72:4551–4561; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deneberg S.; Kanduri M.; Ali D.; Bengtzen S.; Karimi M.; Qu Y.; Kimby E.; Mansouri L.; Rosenquist R.; Lennartsson A.; Lehmann S. microRNA-34b/c on chromosome 11q23 is aberrantly methylated in chronic lymphocytic leukemia. Epigenetics 9:910–917; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng H. Y.; Wan T. S.; So C. C.; Chim C. S. Epigenetic inactivation of DAPK1, p14ARF, mir-34a and -34b/c in acute promyelocytic leukaemia. J. Clin. Pathol. 67:626–631; 2014. [DOI] [PubMed] [Google Scholar]
- 16. Wang L. Q.; Kwong Y. L.; Wong K. F.; Kho C. S.; Jin D. Y.; Tse E.; Rosèn A.; Chim C. S. Epigenetic inactivation of mir-34b/c in addition to mir-34a and DAPK1 in chronic lymphocytic leukemia. J. Translat. Med. 12:52; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Majid S.; Dar A. A.; Saini S.; Shahryari V.; Arora S.; Zaman M. S.; Chang I.; Yamamura S.; Tanaka Y.; Chiyomaru T.; Deng G.; Dahiya R. miRNA-34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin. Cancer Res. 19:73–84; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiroki E.; Suzuki F.; Akahira J.; Nagase S.; Ito K.; Sugawara J.; Miki Y.; Suzuki T.; Sasano H.; Yaegashi N. MicroRNA-34b functions as a potential tumor suppressor in endometrial serous adenocarcinoma. Int. J. Cancer 131:E395–404; 2012. [DOI] [PubMed] [Google Scholar]
- 19. Fanelli M. A.; Montt-Guevara M.; Diblasi A. M.; Gago F. E.; Tello O.; Cuello-Carrion F. D.; Callegari E.; Bausero M. A.; Ciocca D. R. P-cadherin and beta-catenin are useful prognostic markers in breast cancer patients; beta-catenin interacts with heat shock protein Hsp27. Cell Stress Chaperones 13:207–220; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao Y.; Xue Y.; Zhang L.; Feng X.; Liu W.; Zhang G. Higher heat shock factor 1 expression in tumor stroma predicts poor prognosis in esophageal squamous cell carcinoma patients. J. Transl. Med. 13:338; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harada M.; Benito J.; Yamamoto S.; Kaur S.; Arslan D.; Ramirez S.; Jacamo R.; Platanias L.; Matsushita H.; Fujimura T.; Kazuno S.; Kojima K.; Tabe Y.; Konopleva M. The novel combination of dual mTOR inhibitor AZD2014 and pan-PIM inhibitor AZD1208 inhibits growth in acute myeloid leukemia via HSF pathway suppression. Oncotarget 6:37930–37947; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ganguly S.; Home T.; Yacoub A.; Kambhampati S.; Shi H.; Dandawate P.; Padhye S.; Saluja A. K.; McGuirk J.; Rao R. Targeting HSF1 disrupts HSP90 chaperone function in chronic lymphocytic leukemia. Oncotarget 6:31767–31779; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fiskus W.; Sharma S.; Saha S.; Shah B.; Devaraj S. G.; Sun B.; Horrigan S.; Leveque C.; Zu Y.; Iyer S.; Bhalla K. N. Pre-clinical efficacy of combined therapy with novel beta-catenin antagonist BC2059 and histone deacetylase inhibitor against AML cells. Leukemia 29:1267–1278; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chou S. D.; Murshid A.; Eguchi T.; Gong J.; Calderwood S. K. HSF1 regulation of beta-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene 34:2178–2188; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]