Abstract
Metastasis is the primary cause of cancer-related death all over the world. Metastasis is a process by which cancer spreads from the place at which it first arose to distant locations in the body. It is well known that several steps are necessary for this process, including cancer cell epithelial–mesenchymal transition (EMT), cell migration, resistance to anoikis, and angiogenesis. Therefore, investigating the molecular mechanism of regulating cancer metastasis progress may provide helpful insights in the development of efficient diagnosis and therapeutic strategy. Recent studies have indicated that long noncoding RNAs (lncRNAs) play important roles in cancer metastasis. lncRNAs are the nonprotein coding RNAs that have a size longer than 200 nucleotides. More and more studies have indicated that lncRNAs are involved in a broad range of biological processes and are associated with many diseases, such as cancer. The role of lncRNAs in cancer metastasis has been widely studied; however, lncRNAs are mainly involved in the EMT process on the current literature. This review focuses on the mechanisms underlying the role of lncRNAs in cancer metastasis.
Key words: Noncoding RNAs, Long noncoding RNAs (lncRNAs), Cancer metastasis, Gene regulation
INTRODUCTION
Cancer metastasis is a complex process involving the spread of a tumor to distant parts of the body from its original site (1). More evidence indicated that metastasis has occurred in 60% to 70% of cancer patients by the time of diagnosis. Cancer metastasis is the most common cause of death in cancer patients. Many molecular mechanisms enable tumor cells to infiltrate the process of cancer metastasis. Besides protein-coding genes, some noncoding RNAs may participate in cancer metastasis, such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) (2,3). There have been many reviews about miRNAs in cancer metastasis (4,5). This review will focus on lncRNAs in cancer metastasis.
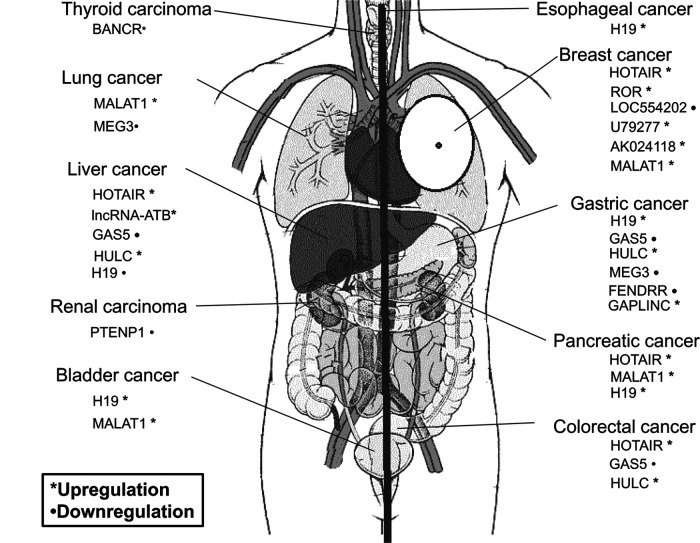
lncRNAs are non-protein-coding RNA molecules longer than 200 nt in length, are poorly conserved, and capable of regulating gene expression at various levels, including histone modification, transcription, and/or posttranscriptional regulation. They act as activators, decoys, guides, or scaffolds for their interacting proteins, DNA and RNA (6,7). Increasing evidence has suggested that lncRNAs could play critical roles in almost all the biological processes, including stem cell maintenance, cell proliferation, cell apoptosis, cell invasion, and metastasis (8–10). More and more studies have revealed that lncRNAs may be involved in almost all the human cancers (Fig. 1). A well-studied lncRNA HOTAIR is significantly increased in breast cancer patients, whose expression is strongly predictive of cancer metastasis and death (9). HOTAIR could change the cell expression profile involving cancer metastasis, leading to breast cancer metastasis by associating with polycomb repressive complex 2 (PRC2) (11). Besides breast cancer, HOTAIR was also observed to be aberrantly expressed in colon, liver, pancreatic cancers, and gastrointestinal stromal tumors and contributes to their metastasis (12–15). The study of the metastasis-related lncRNAs represents a new approach that may enhance our understanding of the molecular mechanisms modulating the metastatic cascade. This review will focus on lncRNAs in the cancer metastatic process.
Figure 1.
lncRNAs may be involved in almost all the human cancers.
REGULATION OF lncRNAs
Transcriptional Regulation of lncRNAs
With the development of high-throughput genomic technologies like lncRNA microarray and RNA sequencing, many lncRNAs have been discovered in recent years. However, because of the high false-positive rates of predictive algorithms for transcription factor binding sites of lncRNAs, little is known about the transcriptional regulation of the lncRNA genes yet. Based on ChIP-Seq peak lists of transcription factors from ENCODE, some databases for decoding the transcriptional regulation of lncRNA have been developed (http://mlg.hit.edu.cn/tf2lncrna; http://deepbase.sysu.edu.cn/chipbase/). The predicated transcription factors of lncRNAs need to be validated.
DNA Methylation Regulation of lncRNAs
Accumulating evidence has uncovered the underlying cross talk between lncRNAs and DNA methylation regulatory network. lncRNA Dum epigenetically silences its neighboring gene, Dppa2, in cis through recruiting methylation enzymes Dnmt1, Dnmt3a, and Dnmt3b. Also, several lncRNAs have been found to be deregulated in cancers due to epigenetic changes. As we all know, the imprinted genes regulated by DNA methylation of either maternal or paternal alleles is very important for embryonic development. Many lncRNAs are imprinted genes such as H19 and MEG3. H19 contains a differentially methylated region (DMR) in its promoter and differentially methylated according to parental inheritance. Normally, the paternal allele of H19 is silent by DNA methylation, while the maternal allele is activated result from DNA unmethylated. H19 is overexpressed in many human cancers because of DNA methylation regulation, such as esophageal cancer, lung cancer, breast cancer, and bladder cancer (16–21). Besides H19, some tumor-suppressive lncRNAs are downregulated in many cancers with high CpG methylation of the promoter, such as lncRNA MEG3 (22–25) and LOC554202 (26,27). A lncRNA NBAT-1 (neuroblastoma-associated transcript-1) was explored as a prognostic biomarker in neuroblastoma, and the expression of NBAT-1 was regulated by CpG methylation (28). Altogether, lncRNA as an epigenetic regulator in gene expression is deregulated in many malignant diseases due to aberrant DNA methylation. Same as coding genes, there are hypermethylation of tumor-suppressor lncRNAs and hypomethylation of oncolncRNAs contributing to cancer development.
lncRNAs AND CANCER METASTASIS
lncRNAs have been reported to directly regulate the metastatic process both in vitro and in vivo in many human cancers (Table 1).
Table 1.
Metastasis-Regulating lncRNAs
| lncRNA/Properties | Functional Mechanism | Type of Cancer | Reference(s) |
|---|---|---|---|
| HOTAIR | |||
| Promote metastasis | Targets PRC2 | Breast cancer | 9 |
| Promote metastasis | Reprogramming of PRC2 | Colon cancer | 12 |
| Promote invasion | Suppress interferon-related genes | Pancreatic cancer | 13 |
| Biomarker of recurrence | NA | Liver cancer | 42 |
| Promote metastasis | NA | Lung cancer | 43 |
| Prognostic marker | NA | Esophageal cancer | 44 |
| Biomarkers of poor survival | EMT | Gastric cancer | 47 |
| MALAT1 | |||
| Promote metastasis | HPV infection | Cervical cancer | 36 |
| Promote metastasis | Activating Wnt signaling | Bladder cancer | 34,37 |
| Promote metastasis | EMT | Lung cancer | 29,38 |
| Biomarker of poor prognosis | NA | Colorectal cancer | 35 |
| Promote migration and invasion | EMT | Pancreatic cancer | 32 |
| Poor prognosis | NA | Glioma | 30 |
| GAS-5 | |||
| Prognostic marker | NA | Cervical cancer | 71 |
| Prognostic biomarker | Related to p53 expression | Colorectal cancer | 74,75 |
| Prognostic biomarker | NA | Hepatocellular carcinoma | 73 |
| Prognostic biomarker | Regulating E2F1 and P21 expression | Gastric cancer | 72 |
| H19 | |||
| Suppress metastasis | Targeting TGFBI | Prostate cancer | 62 |
| Promote invasion and migration | Increasing HMGA2-mediated EMT | Pancreatic adenocarcinoma | 59 |
| Promote invasion and metastasis | Direct upregulation of ISM1 | Gastric cancer | 54 |
| Promote migration | EMT | Bladder cancer | 64–66 |
| Promote invasion | Deriving miR-675 | Glioma | 61 |
| Suppress migration and invasion | AKT/GSK-3β/Cdc25A signaling pathway | Hepatocellular cancer | 63,67 |
| MEG3 | |||
| Relative poor prognosis | NA | Gastric cancer | 81 |
| Relative poor prognosis | NA | Pituitary adenomas | 78 |
| Poor clinical outcome | NA | Tongue carcinoma | 83 |
| Poor clinical outcome | Affecting p53 expression | Lung cancer | 84 |
| HULC | |||
| Prognostic biomarker | Interaction with microRNA-372 | Liver cancer | 88,90 |
| Promote invasion | NA | Gastric cancer | 91 |
| LincRNA-RoR | |||
| Promote metastasis | EMT | Breast cancer | 95 |
| Promote invasion | Targeting ARF6 | Triple-negative breast cancer | 96 |
| lncRNA-ATB | |||
| Promote invasion and metastasis | EMT and autocrine induction of IL-11 | Liver cancer | 97-98 |
| PTENP1 | |||
| Reduce invasion and metastasis | Functions as a competing endogenous RNA | Clear cell renal cell carcinoma | 100–102 |
| FENDRR | |||
| Prognosis biomarker | Affecting fibronectin1 expression | Gastric cancer | 105 |
| GAPLINC | |||
| Prognosis biomarker | Regulates CD44-dependent cell invasiveness | Gastric cancer | 106,107 |
| EBIC | |||
| Promote invasion | Binding to EZH2 and repressing E-cadherin | Cervical cancer | 108 |
PRC2, polycomb repressive complex 2; EMT, epithelial–mesenchymal transition.
Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1)
MALAT1 was first significantly associated with high metastatic potential and poor prognosis in early stage non-small cell lung cancer patients (29). MALAT1 was upregulated in a variety of human cancers including colorectal, pancreatic, prostate, glioma, and bladder cancers (30–35). In cervical cancer, MALAT1 dysregulation was correlated with HPV infection, and MALAT1 could participate in cervical cancer cell proliferation and invasion (36). The epithelial–mesenchymal transition (EMT) is an important process in cancer metastasis in which epithelial cells lose their properties to become migratory and invasive mesenchymal cells. TGF-β, which is an EMT activator, induces MALAT1 expression in bladder cancer cells (37). MALAT1 decreases the expression of E-cadherin and increases the expression of N-cadherin and fibronectin by associating with suppressor of zeste 12 (SUZ12). Another study found that MALAT1 promoted the EMT process through activating the Wnt signaling pathway in bladder cancer and contributing to this cancer metastasis (34). Besides bladder cancer, MALAT1 is increased in lung cancer and promotes lung cancer brain metastasis by inducing EMT (38). MALAT1 is involved in cancer metastasis mainly through EMT. MALAT1 may participate in cancer migration and invasion by other mechanisms. Miyagawa et al. found that MALAT1 could regulate gene expression in transcriptional and/or posttranscriptional levels in HeLa cells (39). Some studies imply that MALAT1 may participate in the regulation of alternate splicing by modulating the level of active serine/arginine (SR) splicing protein (40), although precisely how this may contribute to tumor metastasis is still unknown.
HOX Antisense Intergenic RNA (HOTAIR)
HOTAIR is a noncoding RNA located in the mammalian HOXC locus on chromosome 12, which was found to be overexpressed in metastatic breast cancers (9,41). In hepatocellular cancer, high HOTAIR expression increased the risk of recurrence after liver transplant therapy (42). HOTAIR may represent a novel prognosis marker for non-small cell lung cancer, esophageal squamous cell carcinoma, endometrial carcinoma, and cervical cancer (43–46). HOTAIR could also act as a potential predictor for overall survival in patients with gastric cancer, and knockdown of HOTAIR could reduce invasiveness and reverse the EMT process in gastric cancer cells (47). HOTAIR was demonstrated to be involved in gene expression associated with the PRC2 complex responsible for H3K27 methylation. HOTAIR functions as a guide interacting with PRC2 results in a genome-wide retargeting of the PRC2 complex (9,48). The retargeting of PRC2 silenced the HOXD locus, which is located on chromosome 2 in breast epithelial cells (9). Further study indicated that HOTAIR not only functions as a guide by binding to PRC2, it appears to act as a molecular scaffold by binding at least two distinct histone modification complexes. HOTAIR could modulate H3K27 methylation by binding PRC2 and H3K4 demethylation by binding LSD1 complex (49). In short, HOTAIR has the ability to modulate the cancer epigenome by binding different histone modification complexes and to reprogram chromatin states to promote cancer metastasis.
H19
The H19 gene is a 2.3-kb RNA product that does not code protein (50). It is transcribed by RNA polymerase II, spliced, and polyadenylated. The H19 gene is an imprinting gene that is expressed exclusively in one parental allele (51). H19 is increased in many cancers including esophageal cancer, breast cancer, bladder cancer, ovarian serous epithelial cancer, gastric cancer, and lung cancer (16,18,52–58). H19 is involved in cancer metastasis maybe through EMT progress (59) by antagonizing miRNAs or epigenetic regulation. H19 is a precursor for miR-675, and multiple inducers of EMT upregulate H19 and miR-675 (60–62). TGF-β upregulated Slug, H19, and miR-675 through the PI3K/AKT pathway (63). H19 expression was upregulated remarkably in primary pancreatic ductal adenocarcinoma (PDAC), which subsequently metastasized. H19 increased the expression of HMGA2, which is involved in EMT through antagonizing let-7 and promoted PDAC cell invasion and migration (59). In bladder cancer, H19 levels are remarkably increased in bladder cancer tissues, and H19 may be used as an oncodevelopmental marker for bladder cancer (64). Further study indicated that H19 could activate the Wnt/β-catenin signal pathway and decrease the expression of E-cadherin by associating with enhancer of zeste homolog 2 (EZH2) in bladder cancer cells (65,66). H19 could promote cancer metastasis mainly involving the EMT process.
On the other hand, H19 was downregulated in intratumoral hepatocellular carcinoma (HCC) tissues and predicted HCC prognosis (63). H19 can suppress the expression of EMT markers by activating the miR-200 family and contributing to HCC metastatic inhibition (67). Furthermore, H19 activated miR-200 family expression potentially by increasing histone acetylation associated with the protein complex hnRNP U/PCAF/RNAPol II (68). The unconventional expression patterns of H19 may be due to the tissue specificity, and the mechanism underlying the unconventional expression still needs to be further investigated. Like miRNAs, lncRNA may play different functions in various cancers through different pathways. So a detailed understanding of lncRNA in tumorigenesis is very important.
Growth Arrest-Specific 5 (GAS5)
GAS5 is a lncRNA that was originally isolated from mouse genomic DNA, and this gene is a potential tumor-suppressor gene highly expressed in saturation density-arrested cells (69). GAS5 could fuse to the Bcl6 gene as a result of t(1;3)(q25;q27) in B-cell lymphoma (70). GAS5 is a prognostic biomarker in cervical cancer, colorectal cancer, hepatocellular carcinoma, and gastric cancer (71–74). Although GAS5 has been found as a tumor-suppressor gene in many cancers, the mechanism of this gene involved in tumorigenesis is still not very clear. A recent study found that GAS5-associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer (75). The main function of GAS5 in cancer is cell apoptosis, and there has been no study about GAS5 in cancer metastasis until recently.
Maternally Expressed 3 (MEG3)
MEG3 is an imprinted lncRNA gene expressed in the maternal allele. Imprinting of this gene is mediated through cytosine methylation-controlled binding protein CTCF (76). MEG3 is silent in many cancer cells because of DNA methylation (77–79). miR-29 and miR-148 can modulate DNA methyltransferase (DNMT) 1 and 3, increasing expression of MEG3 in hepatocellular cancer and gastric cancer, respectively (80,81). MEG3 is a relatively poor prognosis in gastric cancer, pituitary adenomas, tongue squamous cell carcinoma, and lung cancer (22,82–84). Yin et al. found that the lower expression of MEG3 was remarkably correlated with low histological grade, deep tumor invasion in colorectal cancer (85). However, the mechanism of MEG3 underlying cancer metastasis is not very clear. Further study indicated that MEG3 might suppress tumor proliferation through p53-dependent and/or p53-independent pathways (84,86).
Highly Upregulated in Liver Cancer (HULC)
HULC was first identified in hepatocellular cancer and is also expressed in colorectal carcinomas that metastasize to the liver (87,88). IGF2 mRNA-binding protein 1 (IGF2BP1) could regulate the expression of HULC at the posttranscriptional level (89). HULC was upregulated by PKA pathway or transcriptional factor CREB in liver cancer. Upregulated HULC may function as an endogenous sponge by binding to many miRNAs, including miR-372 (90). miR-372 represses the translation of the kinase PRKACB, leading to increased levels of PRKACB (90). PRKACB activates CREB, which upregulated HULC by phosphorylation, therefore leading to increased expression of HULC. The HULC-miR-372-PRKACB-CREB-HULC regulation loop plays an important role in cancer metastasis. Zhao et al. determined that silencing of HULC effectively reversed the EMT phenotype in human gastric cancer (91).
lncRNA-RoR
lncRNA-RoR is a lncRNA that suppresses p53 translation by direct interaction with the heterogeneous nuclear ribonucleoprotein I (hnRNP I) (92,93). The main function of lncRNA-ROR was the maintenance of induced pluripotent stem cells (iPSCs) and embryonic stem cells or involvement in tumor genesis (8,94). Hou et al. discovered that lncRNA-ROR was increased in breast tumor tissues, and ROR regulated EMT progress by functioning as a competing endogenous RNA for miR-205 in human mammary epithelial cells (95).
Triple-negative (ER−, HER2−, PR−) breast cancer (TNBC) is an aggressive disease with a poor prognosis because of no available therapeutic strategies. Silencing of miRNA-145 may be a defining marker of TNBC. RoR is dramatically upregulated in TNBC and functions as a competitive endogenous RNA sponge for miR-145. ARF6, a target gene of miR-145, is a regulator of breast tumor cell invasion and metastasis. Mechanistically, ARF6 regulates E-cadherin localization and impacts cell–cell adhesion (96). These studies reveal a lincRNA-ROR/miR-145/ARF6 pathway that regulates metastasis in TNBCs. In short, lncRNA-ROR mainly functions as a key competing endogamous miRNA sponge contributing to cancer metastasis.
Other lncRNAs
lncRNA-ATB was first identified in the metastases of hepatocellular carcinoma (HCC) and is activated by TGF-β. lncRNA-ATB could promote cancer cell invasion by competitively binding the miR-200 family. miR-200 could decrease the expression of ZEB1 and ZEB2, which are EMT inducers. lncRNA-ATB promotes cancer cell invasion by inducing EMT process. In addition, lncRNA-ATB could promote the organ colonization of disseminated tumor cells by binding to IL-11 mRNA and triggering the STAT3 signaling pathway (97,98).
PTENP1 is a pseudogene of the tumor-suppression gene PTEN (99). PTENP1 is downregulated in clear cell renal cell carcinoma (ccRCC) due to DNA methylation (100). PTENP1 was deleted in human melanoma (101). PTENP1 and PTEN are direct targets of miR-21, and their expression is suppressed by miR-21 in ccRCC cell lines (102). The expression of PTENP1 and PTEN in tissues is correlated, and both expressions are inversely correlated with miR-21 expression. Patients with ccRCC with no PTENP1 expression have a lower survival rate. Overexpression of PTENP1 in cells expressing miR-21 reduces cell proliferation, invasion, tumor growth, and metastasis, recapitulating the phenotypes induced by PTEN expression.
lncRNA LOC554202 is a host gene of miR-31. miR-31 and LOC554202 are both down expressed in the TNBC cell lines because of promoter methylation (26). Inhibition of Loc554202 could decrease the migration and invasion of breast cancer cell (27).
U79277, AK024118, BC040204, and AK000974 have been identified using lncRNA expression profiling, and the expression of these four lncRNAs are associated with the survival times for breast cancer patients (103).
lncRNA FENDRR is an essential regulator of heart and body development in mouse (104). This lncRNA was downregulated in gastric cancer tissues resulting from histone deacetylation, and this downregulation was correlated with tumor invasion and lymphatic metastasis. Further study indicated that FENDRR could suppress gastric cancer cell invasion and migration by decreasing the expression of FN1 and MMP2/MMP9, which are all metastasis-related genes (105).
GAPLINC (gastric adenocarcinoma predictive long intergenic noncoding RNA) is overexpressed in gastric cancer tissues, and GAPLINC overexpression defines a subgroup of patients with very poor survival. Mechanistic investigations revealed that GAPLINC regulates CD44 as a molecular decoy for miR-211, a microRNA that targets both CD44 and GAPLINC (106,107).
lncRNA-EBIC (EZH2-binding lncRNA in cervical cancer) was upregulated in cervical cancer. lncRNA-EBIC could promote the migration and invasion of cervical cancer cells by binding to EZH2. EBIC/EZH2 decreases the expression of E-cadherin, which is a key molecular in cervical cancer metastasis (108).
lncRNA AND miRNA INTERACTIONS IN CANCER METASTASIS
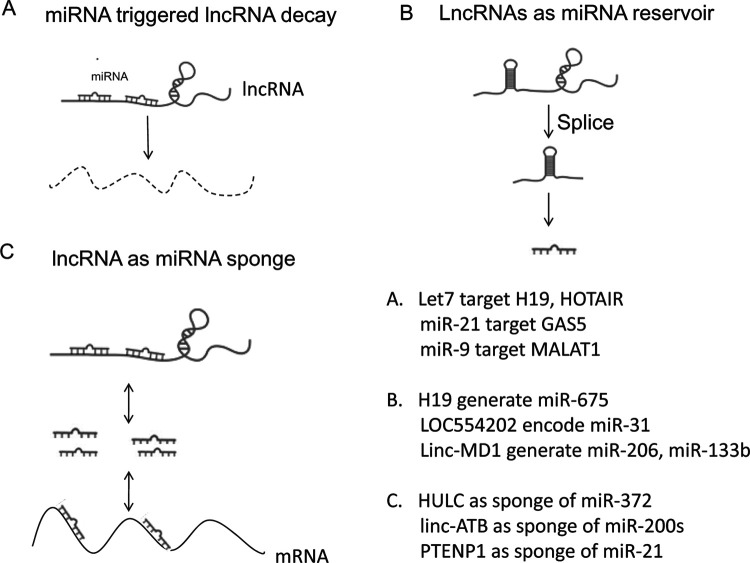
miRNA-Triggered lncRNA Decay
miRNAs and lncRNAs are all noncoding RNAs, and the abundance of numerous lncRNAs is controlled by miRNAs (Fig. 2A). More evidence indicated that miRNAs can regulate over one third of the protein-coding genes by binding to their 3′ untranslated region (UTR); more studies indicated that miRNAs can also target lncRNAs and trigger lncRNA decay. The lethal-7 (let-7) gene family was first discovered as a key developmental regulator and is a direct regulator of oncogene RAS in human cancers (109). Except for Ras gene, let-7 could also regulate lncRNAs. H19 is regulated by four let-7 genes (let-7a, let-7b, let-7g, let-7i) (110,111). Another lncRNA HOTAIR was decreased by let-7, and this regulation is recruited to the RNA-binding protein HuR; this suggested that the mechanism of lncRNA decay through HuR-enhanced microRNA interactions may be widespread. Another most studied miRNA, miR-21, has been found as an oncogene in various types of cancers (112). Zhang et al. found that lncRNA GAS5 is regulated by miR-21 through the pathway involving the RNA-induced silencing complex (RISC) in breast cancer cells (113). lncRNA MALAT1 is targeted by miR-9 also involving the RISC (114). lncRNAs and miRNAs are noncoding RNAs involved in gene regulation. A better understanding of the interactions between microRNAs and lncRNAs will provide new insights into mechanisms underlying various aspects of tumor process including metastasis.
Figure 2.
lncRNA and miRNA interactions in cancer metastasis. (A) Numerous lncRNAs are controlled by miRNAs and trigger lncRNA decay. (B) lncRNAs could generate miRNAs. (C) lncRNAs can compete with miRNAs for binding to target mRNAs.
lncRNAs: Reservoir of miRNAs
Some lncRNAs are involved in cancer metastasis by generating miRNAs (Fig. 2B). H19 can generate miR-675, a process that is repressed by HuR in mice (115,116). H19 could promote cancer metastasis by deriving miR-675, including prostate cancer, gastric cancer, and glioma (61,62,117,118). In contrast, down expression of H19 and miR-675 could promote the migration and invasion of human hepatocellular carcinoma cells (63). Similarly, lncRNA LOC554202 could encode miR-31, and promoter methylation of lncRNA LOC554202 leads to decreased miR-31 and thus contributes to breast cancer invasion and metastasis (26,119). Linc-MD1 generates miR-206 and miR-133b from an intron and an exon, respectively. miR-206 could suppress the migration of breast cancer cells by direct targeting of coronin 1C, which is an actin-binding protein (120). miR-133b is a potential new prognostic marker for human colorectal cancer (121). All of these studies confirm that some lncRNAs serve as a reservoir of miRNAs and could have a dual regulatory output. Besides generating miRNAs, lncRNA can also regulate the miRNA biogenesis. Liz et al. found that lncRNA Uc.283+A regulated pri-miRNA-195 maturation at the level of Drosha processing (122).
lncRNAs: Sponge of miRNAs
lncRNAs could generate miRNAs, and lncRNAs could also compete with miRNAs for binding to target mRNAs (Fig. 2C). miRNAs binding to lncRNAs could result in lncRNA decay or just as a miRNA sponge. Lnc-MD1 enhanced the expression of MAML1 and MEF2C mRNAs by functioning as an endogenous sponge for miR-133, and miR-135 triggered muscle differentiation in mouse and human myoblasts (123). HULC was found to function as a miRNA sponge, contributing to hepatic cancer metastasis (90). lncRNA-ATB could stimulate EMT through sequestering miR-200s in liver cancer (97). PTENP1 is a pseudogene of tumor-suppressor gene PTEN, and they share similar 3′ UTR for the same miRNAs. PTENP1 could decrease the effect of posttranscriptional inhibition of PTEN by functioning as competing endogenous RNA (99,100). In human melanoma, specific mutations in 3′ UTR of PTENP1 could affect the expression of PTEN, contributing to cancer metastasis (101).
In sum, expanding evidence had revealed that lncRNAs and miRNAs work together to regulate gene expression by complex posttranscriptional mechanisms. All of these studies highlighted the increasing complexity of ncRNA-mediated regulatory networks. More examples of lncRNAs regulating gene expression by competing or cooperating with miRNAs are expected to emerge. Together, microRNAs and lncRNAs contribute to a robust and dynamic control of cancer metastasis.
lncRNAs AS DIAGNOSTICS AND THERAPEUTIC BIOMARKERS IN CANCER
Cancer is one of the diseases with a high mortality rate because of metastasis. It is very hard to look for early diagnosis and therapeutic targets for this disease. With the development of molecular mechanism studies, personalized medicine is entering into the era for cancer. More and more accurate and meaningful diagnosis and prognosis markers for cancer are coming because of greater understanding of molecular alterations. In addition to genetic changes, additional epigenetic alterations including DNA methylation, histone modification, and microRNA and lncRNA expression can provide other clinical information. lncRNA as an epigenetic regulator may participate in gene regulation at the transcriptional or posttranscriptional level, and lncRNA may be a potential marker for cancer diagnosis and therapy.
Many studies have found that lncRNAs were aberrantly regulated in many cancers and associated with cancer metastasis. lncRNAs can be potential novel biomarkers for cancer diagnosis and therapy. Using lncRNA array or RNA sequencing, many lncRNAs have been found as cancer prognosis markers. lncRNA-ATB was activated by TGF-β in HCC, and the expression of lncRNA-ATB was associated with cancer prognosis. Higher HOTAIR expression increased the risk of recurrence after liver transplant therapy. GAPLINC overexpression defines a subgroup of patients with gastric cancer with very poor survival. These lncRNA expressions are associated with cancer patient prognosis and are all detected in tumor tissues. Like circulating miRNAs, lncRNAs are also detectable in the sputum, blood, and urine of cancer patients. For example, lncRNA DD3, which is specifically expressed in the prostate, has been developed as a marker for prostate cancer, and this lncRNA has higher specificity than serum prostate-specific antigen (PSA) (124–126). Similarly, the lncRNA HULC, which is highly expressed in liver cancer, can be detected in the blood of cancer patients (87). These lncRNAs may be potential noninvasive diagnosis targets for cancer.
Use of lncRNAs as therapeutic targets for cancer is just beginning (127). Although the exact function of lncRNA in cancer is not very clear, some lncRNAs are candidates for therapeutic intervention. Many lncRNAs may form a secondary structure and play important roles by binding to a protein complex; this may provide a means of intervention (128). HOTAIR regulates gene expression by binding to PRC2 or LSD1 complexes; preventing this binding will decrease the metastatic potential of breast cancer cells (129). lncRNAs function as a tumor suppressor or oncogene by interaction with DNA, miRNA, and protein. It has been suggested that RNA-induced transcriptional gene silencing (TGS) or activation could be a potential therapeutic strategy (130).
lncRNAs are dysregulated in many human cancers, and this may provide a therapeutic target for transgene-mediated treatment. Among these lncRNAs, H19 is overexpressed in many cancers (60,117,131). So a series of studies want to regulate the expression of H19 for cancer treatment primarily in bladder cancer. BC-819 is a double-stranded DNA plasmid that carries the gene for diphtheria toxin-A under regulation of the H19 promoter sequence. Most clinical trials have investigated the efficacy and toxicity of BC-819 for bladder cancer treatment (132). A phase IIb clinical trial in patients with intermediate risk nonmuscle invasive bladder cancer treated with BC-819 reported that BC-819 could prevent new tumor growth and ablate marker lesions (133). In order to improve the therapeutic efficacy, a double promoter plasmid with H19 and IGF2-P4 regulatory sequences was constructed. The double promoter plasmid exhibited enhanced anticancer activity compared to the single promoter expression vector in bladder cancer (134,135). Besides bladder cancer, BC-819 was used to treat other cancers, such as pancreatic cancer, ovarian cancer, and heterotopic cancer (136–138). In short, combining conventional chemotherapy and lncRNA transgene treatment might be a new therapeutic option for cancer patients.
Collectively, these studies indicated that lncRNA may be a potential target for cancer diagnostics and therapies.
CONCLUSIONS
Recently, with the development of high-throughput array technologies, such as microarray and RNA sequencing, something important hidden in the variety of noncoding RNAs makes researchers deeply investigate the pathogenesis of cancer. lncRNA is dysregulated in many human cancers, and this may be a new hallmark in cancer. Although there are more and more studies about lncRNAs in cancer, the exact function of lncRNA in cancer genesis is still unsolved. Cancer metastasis is the leading cause of cancer patient death, and lncRNAs participate in cancer metastasis. In this review, we highlight the character of lncRNA in cancer metastasis. lncRNAs participate in cancer metastasis mainly by interaction with miRNAs, epigenetic gene regulation, and involvement in EMT progress. Finally, we note that lncRNA may be a potential diagnostic and therapeutic marker in cancer. Thus, more effects are needed to better elucidate the function and critical mechanisms of cancer-specific lncRNAs in the progression of cancer metastasis. In the future, integration of lncRNA biology into cancer biology may further deepen our understanding of the mechanisms of cancer metastasis and provide novel applications for efficient, rapid, and specific diagnosis and treatments.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (No. 31100936).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Gupta G. P.; Massague J. Cancer metastasis: Building a framework. Cell 127:679–695; 2006. [DOI] [PubMed] [Google Scholar]
- 2. Shi Z.; Wei Q.; She J. MicroRNAs in gastric cancer metastasis. Crit. Rev. Eukaryot. Gene Expr. 24:39–53; 2014. [DOI] [PubMed] [Google Scholar]
- 3. Shen X. H.; Qi P.; Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod. Pathol. 28(1):4–13; 2015. [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y.; Yang P.; Wang X. F. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol. 24:153–160; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bouyssou J. M.; Manier S.; Huynh D.; Issa S.; Roccaro I. A. M.; Ghobrial M. Regulation of microRNAs in cancer metastasis. Biochim. Biophys. Acta 1845:255–265; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mercer T. R.; Mattick J. S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20:300–307; 2013. [DOI] [PubMed] [Google Scholar]
- 7. Wang K. C.; Chang H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43:904–914; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loewer S.; Cabili M. N.; Guttman M.; Loh Y. H.; Thomas T.; Park I. H.; Garber M.; Curran M.; Onder T.; Agarwal S.; Manos P. D.; Datta S.; Lander E. S.; Schlaeger T. M.; Daley G. Q.; Rinn J. L. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42:1113–1117; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta R. A.; Shah N.; Wang K. C.; Kim J.; Horlings H. M.; Wong D. J.; Tsai M. C.; Hung T.; Argani P.; Rinn J. L.; Wang Y.; Brzoska P.; Kong B.; Li R.; West R. B.; van de Vijver M. J.; Sukumar S.; Chang H. Y. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Venkatraman A.; He X. C.; Thorvaldsen J. L.; Sugimura R.; Perry J. M.; Tao F.; Zhao M.; Christenson M. K.; Sanchez R.; Yu J. Y.; Peng L.; Haug J. S.; Paulson A.; Li H.; Zhong X. B.; Clemens T. L.; Bartolomei M. S.; Li L. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 500:345–349; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoon J. H.; Abdelmohsen K.; Kim J.; Yang X.; Martindale J. L.; Tominaga-Yamanaka K.; White E. J.; Orjalo A. V.; Rinn J. L.; Kreft S. G.; Wilson G. M.; Gorospe M. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 4:2939; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xue Y.; Gu D.; Ma G.; Zhu L.; Hua Q.; Chu H.; Tong N.; Chen J.; Zhang Z.; Wang M. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis 30(2):303–310; 2015. [DOI] [PubMed] [Google Scholar]
- 13. Kim K.; Jutooru I.; Chadalapaka G.; Johnson G.; Frank J.; Burghardt R.; Kim S.; Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32:1616–1625; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geng Y. J.; Xie S. L.; Li Q.; Ma J.; Wang G. Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 39:2119–2128; 2011. [DOI] [PubMed] [Google Scholar]
- 15. Niinuma T.; Suzuki H.; Nojima M.; Nosho K.; Yamamoto H.; Takamaru H.; Yamamoto E.; Maruyama R.; Nobuoka T.; Miyazaki Y.; Nishida T.; Bamba T.; Kanda T.; Ajioka Y.; Taguchi T.; Okahara S.; Takahashi H.; Nishida Y.; Hosokawa M.; Hasegawa T.; Tokino T.; Hirata K.; Imai K.; Toyota M.; Shinomura Y. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 72:1126–1136; 2012. [DOI] [PubMed] [Google Scholar]
- 16. Hibi K.; Nakamura H.; Hirai A.; Fujikake Y.; Kasai Y.; Akiyama S.; Ito K.; Takagi H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 56:480–482; 1996. [PubMed] [Google Scholar]
- 17. Kondo M.; Takahashi T. [Altered genomic imprinting in the IGF2 and H19 genes in human lung cancer]. Nihon Rinsho. 54:492–496; 1996. [PubMed] [Google Scholar]
- 18. Lottin S.; Adriaenssens E.; Dupressoir T.; Berteaux N.; Montpellier C.; Coll J.; Dugimont T.; Curgy J. J. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23:1885–1895; 2002. [DOI] [PubMed] [Google Scholar]
- 19. Kanduri C.; Kanduri M.; Liu L.; Thakur N.; Pfeifer S.; Ohlsson R. The kinetics of deregulation of expression by de novo methylation of the h19 imprinting control region in cancer cells. Cancer Res. 62:4545–4548; 2002. [PubMed] [Google Scholar]
- 20. Byun H. M.; Wong H. L.; Birnstein E. A.; Wolff E. M.; Liang G.; Yang A. S. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 67:10753–10758; 2007. [DOI] [PubMed] [Google Scholar]
- 21. Matouk I. J.; Raveh E.; Abu-lail R.; Mezan S.; Gilon M.; Gershtain E.; Birman T.; Gallula J.; Schneider T.; Barkali M.; Richler C.; Fellig Y.; Sorin V.; Hubert A.; Hochberg A.; Czerniak A. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta 1843:1414–1426; 2014. [DOI] [PubMed] [Google Scholar]
- 22. Sun M.; Xia R.; Jin F.; Xu T.; Liu Z.; De W.; Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 35:1065–1073; 2014. [DOI] [PubMed] [Google Scholar]
- 23. McMurray E. N.; Schmidt J. V. Identification of imprinting regulators at the Meg3 differentially methylated region. Genomics 100:184–194; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anwar S. L.; Krech T.; Hasemeier B.; Schipper E.; Schweitzer N.; Vogel A.; Kreipe H.; Lehmann U. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One 7:e49462; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benetatos L.; Hatzimichael E.; Dasoula A.; Dranitsaris G.; Tsiara S.; Syrrou M.; Georgiou I.; Bourantas K. L. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk. Res. 34:148–153; 2010. [DOI] [PubMed] [Google Scholar]
- 26. Augoff K.; McCue B.; Plow E. F.; Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer 11:5; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi Y.; Lu J.; Zhou J.; Tan X.; He Y.; Ding J.; Tian Y.; Wang L.; Wang K. Long non-coding RNA Loc554202 regulates proliferation and migration in breast cancer cells. Biochem. Biophys. Res. Commun. 446:448–453; 2014. [DOI] [PubMed] [Google Scholar]
- 28. Pandey G. K.; Mitra S.; Subhash S.; Hertwig F.; Kanduri M.; Mishra K.; Fransson S.; Ganeshram A.; Mondal T.; Bandaru S.; Ostensson M.; Akyurek L. M.; Abrahamsson J.; Pfeifer S.; Larsson E.; Shi L.; Peng Z.; Fischer M.; Martinsson T.; Hedborg F.; Kogner P.; Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26:722–737; 2014. [DOI] [PubMed] [Google Scholar]
- 29. Ji P.; Diederichs S.; Wang W.; Boing S.; Metzger R.; Schneider P. M.; Tidow N.; Brandt B.; Buerger H.; Bulk E.; Thomas M.; Berdel W. E.; Serve H.; Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22:8031–8041; 2003. [DOI] [PubMed] [Google Scholar]
- 30. Ma K. X.; Wang H. J.; Li X. R.; Li T.; Su G.; Yang P.; Wu J. W. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 36(5):3355–3359; 2015. [DOI] [PubMed] [Google Scholar]
- 31. Zheng H. T.; Shi D. B.; Wang Y. W.; Li X. X.; Xu Y.; Tripathi P.; Gu W. L.; Cai G. W.; Cai S. J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 7:3174–3181; 2014. [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J. H.; Chen G.; Dang Y. W.; Li C. J.; Luo D. Z. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac. J. Cancer Prev. 15:2971–2977; 2014. [DOI] [PubMed] [Google Scholar]
- 33. Ren S.; Liu Y.; Xu W.; Sun Y.; Lu J.; Wang F.; Wei M.; Shen J.; Hou J.; Gao X.; Xu C.; Huang J.; Zhao Y. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 190:2278–2287; 2013. [DOI] [PubMed] [Google Scholar]
- 34. Ying L.; Chen Q.; Wang Y.; Zhou Z.; Huang Y.; Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 8:2289–2294; 2012. [DOI] [PubMed] [Google Scholar]
- 35. Xu C.; Yang M.; Tian J.; Wang X.; Li Z. MALAT-1: A long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 39:169–175; 2011. [DOI] [PubMed] [Google Scholar]
- 36. Jiang Y.; Li Y.; Fang S.; Jiang B.; Qin C.; Xie P.; Zhou G.; Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol. Lett. 7:2135–2141; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan Y.; Shen B.; Tan M.; Mu X.; Qin Y.; Zhang F.; Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 20:1531–1541; 2014. [DOI] [PubMed] [Google Scholar]
- 38. Shen L.; Chen L.; Wang Y.; Jiang X.; Xia H.; Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neurooncol. 121(1):101–108; 2015. [DOI] [PubMed] [Google Scholar]
- 39. Miyagawa R.; Tano K.; Mizuno R.; Nakamura Y.; Ijiri K.; Rakwal R.; Shibato J.; Masuo Y.; Mayeda A.; Hirose T.; Akimitsu N. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18:738–751; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tripathi V.; Ellis J. D.; Shen Z.; Song D. Y.; Pan Q.; Watt A. T.; Freier S. M.; Bennett C. F.; Sharma A.; Bubulya P. A.; Blencowe B. J.; Prasanth S. G.; Prasanth K. V. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39:925–938; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wan Y.; Chang H. Y. HOTAIR: Flight of noncoding RNAs in cancer metastasis. Cell Cycle 9:3391–3392; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Z.; Zhou L.; Wu L. M.; Lai M. C.; Xie H. Y.; Zhang F.; Zheng S. S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 18:1243–1250; 2011. [DOI] [PubMed] [Google Scholar]
- 43. Liu X. H.; Liu Z. L.; Sun M.; Liu J.; Wang Z. X.; De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 13:464; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lv X. B.; Lian G. Y.; Wang H. R.; Song E.; Yao H.; Wang M. H. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One 8:e63516; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He X.; Bao W.; Li X.; Chen Z.; Che Q.; Wang H.; Wan X. P. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int. J. Mol. Med. 33:325–332; 2014. [DOI] [PubMed] [Google Scholar]
- 46. Huang L.; Liao L. M.; Liu A. W.; Wu J. B.; Cheng X. L.; Lin J. X.; Zheng M. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch. Gynecol. Obstet. 290:717–723; 2014. [DOI] [PubMed] [Google Scholar]
- 47. Xu Z. Y.; Yu Q. M.; Du Y. A.; Yang L. T.; Dong R. Z.; Huang L.; Yu P. F.; Cheng X. D. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 9:587–597; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu L.; Murat P.; Matak-Vinkovic D.; Murrell A.; Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry 52:9519–9527; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li L.; Liu B.; Wapinski O. L.; Tsai M. C.; Qu K.; Zhang J.; Carlson J. C.; Lin M.; Fang F.; Gupta R. A.; Helms J. A.; Chang H. Y. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 5:3–12; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brunkow M. E.; Tilghman S. M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 5:1092–1101; 1991. [DOI] [PubMed] [Google Scholar]
- 51. Bartolomei M. S.; Zemel S.; Tilghman S. M. Parental imprinting of the mouse H19 gene. Nature 351:153–155; 1991. [DOI] [PubMed] [Google Scholar]
- 52. Verhaegh G. W.; Verkleij L.; Vermeulen S. H.; den Heijer M.; Witjes J. A.; Kiemeney L. A. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur. Urol. 54:1118–1126; 2008. [DOI] [PubMed] [Google Scholar]
- 53. Medrzycki M.; Zhang Y.; Zhang W.; Cao K.; Pan C.; Lailler N.; McDonald J. F.; Bouhassira E. E.; Fan Y. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 74:6463–6473; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang E. B.; Han L.; Yin D. D.; Kong R.; De W.; Chen J. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med. Oncol. 31:914; 2014. [DOI] [PubMed] [Google Scholar]
- 55. Murphy S. K.; Huang Z.; Wen Y.; Spillman M. A.; Whitaker R. S.; Simel L. R.; Nichols T. D.; Marks J. R.; Berchuck A. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol. Cancer Res. 4:283–292; 2006. [DOI] [PubMed] [Google Scholar]
- 56. Berteaux N.; Lottin S.; Monte D.; Pinte S.; Quatannens B.; Coll J.; Hondermarck H.; Curgy J. J.; Dugimont T.; Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J. Biol. Chem. 280:29625–29636; 2005. [DOI] [PubMed] [Google Scholar]
- 57. Chen C. L.; Ip S. M.; Cheng D.; Wong L. C.; Ngan H. Y. Loss of imprinting of the IGF-II and H19 genes in epithelial ovarian cancer. Clin. Cancer Res. 6:474–479; 2000. [PubMed] [Google Scholar]
- 58. Kondo M.; Suzuki H.; Ueda R.; Osada H.; Takagi K.; Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene 10:1193–1198; 1995. [PubMed] [Google Scholar]
- 59. Ma C.; Nong K.; Zhu H.; Wang W.; Huang X.; Yuan Z.; Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 35:9163–9169; 2014. [DOI] [PubMed] [Google Scholar]
- 60. Tsang W. P.; Ng E. K.; Ng S. S.; Jin H.; Yu J.; Sung J. J.; Kwok T. T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 31:350–358; 2010. [DOI] [PubMed] [Google Scholar]
- 61. Shi Y.; Wang Y.; Luan W.; Wang P.; Tao T.; Zhang J.; Qian J.; Liu N.; You Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One 9:e86295; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhu M.; Chen Q.; Liu X.; Sun Q.; Zhao X.; Deng R.; Wang Y.; Huang J.; Xu M.; Yan J.; Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 281:3766–3775; 2014. [DOI] [PubMed] [Google Scholar]
- 63. Lv J.; Ma L.; Chen X. L.; Huang X. H.; Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3beta/Cdc25A signaling pathway. J. Huazhong Univ. Sci. Technolog. Med. Sci. 34:363–369; 2014. [DOI] [PubMed] [Google Scholar]
- 64. Ariel I.; Lustig O.; Schneider T.; Pizov G.; Sappir M.; De-Groot N.; Hochberg A. The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology 45:335–338; 1995. [DOI] [PubMed] [Google Scholar]
- 65. Atala A. Re: Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. J. Urol. 190:2306; 2013. [DOI] [PubMed] [Google Scholar]
- 66. Luo M.; Li Z.; Wang W.; Zeng Y.; Liu Z.; Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333:213–221; 2013. [DOI] [PubMed] [Google Scholar]
- 67. Zhang L.; Yang F.; Yuan J. H.; Yuan S. X.; Zhou W. P.; Huo X. S.; Xu D.; Bi H. S.; Wang F.; Sun S. H. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 34:577–586; 2013. [DOI] [PubMed] [Google Scholar]
- 68. Bi H. S.; Yang X. Y.; Yuan J. H.; Yang F.; Xu D.; Guo Y. J.; Zhang L.; Zhou C. C.; Wang F.; Sun S. H. H19 inhibits RNA polymerase II-mediated transcription by disrupting the hnRNP U-actin complex. Biochim. Biophys. Acta 1830:4899–4906; 2013. [DOI] [PubMed] [Google Scholar]
- 69. Coccia E. M.; Cicala C.; Charlesworth A.; Ciccarelli C.; Rossi G. B.; Philipson L.; Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol. Cell Biol. 12:3514–3521; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nakamura Y.; Takahashi N.; Kakegawa E.; Yoshida K.; Ito Y.; Kayano H.; Niitsu N.; Jinnai I.; Bessho M. The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1; 3)(q25; q27) in a patient with B-cell lymphoma. Cancer Genet. Cytogenet. 182:144–149; 2008. [DOI] [PubMed] [Google Scholar]
- 71. Cao S.; Liu W.; Li F.; Zhao W.; Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int. J. Clin. Exp. Pathol. 7:6776–6783; 2014. [PMC free article] [PubMed] [Google Scholar]
- 72. Sun M.; Jin F. Y.; Xia R.; Kong R.; Li J. H.; Xu T. P.; Liu Y. W.; Zhang E. B.; Liu X. H.; De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 14:319; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tu Z. Q.; Li R. J.; Mei J. Z.; Li X. H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 7:4303–4309; 2014. [PMC free article] [PubMed] [Google Scholar]
- 74. Yin D.; He X.; Zhang E.; Kong R.; De W.; Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med. Oncol. 31:253; 2014. [DOI] [PubMed] [Google Scholar]
- 75. Krell J.; Frampton A. E.; Mirnezami R.; Harding V.; De Giorgio A.; Roca Alonso L.; Cohen P.; Ottaviani S.; Colombo T.; Jacob J.; Pellegrino L.; Buchanan G.; Stebbing J.; Castellano L. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS One 9:e98561; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rosa A. L.; Wu Y. Q.; Kwabi-Addo B.; Coveler K. J.; Reid Sutton V.; Shaffer L. G. Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosome Res. 13:809–818; 2005. [DOI] [PubMed] [Google Scholar]
- 77. Zhang X.; Zhou Y.; Mehta K. R.; Danila D. C.; Scolavino S.; Johnson S. R.; Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 88:5119–5126; 2003. [DOI] [PubMed] [Google Scholar]
- 78. Zhao J.; Dahle D.; Zhou Y.; Zhang X.; Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J. Clin. Endocrinol. Metab. 90:2179–2186; 2005. [DOI] [PubMed] [Google Scholar]
- 79. Benetatos L.; Vartholomatos G.; Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int. J. Cancer 129:773–779; 2011. [DOI] [PubMed] [Google Scholar]
- 80. Braconi C.; Kogure T.; Valeri N.; Huang N.; Nuovo G.; Costinean S.; Negrini M.; Miotto E.; Croce C. M.; Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30:4750–4756; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yan J.; Guo X.; Xia J.; Shan T.; Gu C.; Liang Z.; Zhao W.; Jin S. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med. Oncol. 31:879; 2014. [DOI] [PubMed] [Google Scholar]
- 82. Li Z.; Li C.; Liu C.; Yu S.; Zhang Y. Expression of the long non-coding RNAs MEG3, HOTAIR, and MALAT-1 in non-functioning pituitary adenomas and their relationship to tumor behavior. Pituitary 18(1):42–47; 2015. [DOI] [PubMed] [Google Scholar]
- 83. Jia L. F.; Wei S. B.; Gan Y. H.; Guo Y.; Gong K.; Mitchelson K.; Cheng J.; Yu G. Y. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int. J. Cancer 135:2282–2293; 2014. [DOI] [PubMed] [Google Scholar]
- 84. Lu K. H.; Li W.; Liu X. H.; Sun M.; Zhang M. L.; Wu W. Q.; Xie W. P.; Hou Y. Y. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13:461; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yin D. D.; Liu Z. J.; Zhang E.; Kong R.; Zhang Z. H.; Guo R. H. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 36(6):4851–4859; 2015. [DOI] [PubMed] [Google Scholar]
- 86. Zhou Y.; Zhong Y.; Wang Y.; Zhang X.; Batista D. L.; Gejman R.; Ansell P. J.; Zhao J.; Weng C.; Klibanski A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282:24731–24742; 2007. [DOI] [PubMed] [Google Scholar]
- 87. Panzitt K.; Tschernatsch M. M.; Guelly C.; Moustafa T.; Stradner M.; Strohmaier H. M.; Buck C. R.; Denk H.; Schroeder R.; Trauner M.; Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132:330–342; 2007. [DOI] [PubMed] [Google Scholar]
- 88. Matouk I. J.; Abbasi I.; Hochberg A.; Galun E.; Dweik H.; Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur. J. Gastroenterol. Hepatol. 21:688–692; 2009. [DOI] [PubMed] [Google Scholar]
- 89. Hammerle M.; Gutschner T.; Uckelmann H.; Ozgur S.; Fiskin E.; Gross M.; Skawran B.; Geffers R.; Longerich T.; Breuhahn K.; Schirmacher P.; Stoecklin G.; Diederichs S. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 58:1703–1712; 2013. [DOI] [PubMed] [Google Scholar]
- 90. Wang J.; Liu X.; Wu H.; Ni P.; Gu Z.; Qiao Y.; Chen N.; Sun F.; Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38:5366–5383; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhao Y.; Guo Q.; Chen J.; Hu J.; Wang S.; Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol. Rep. 31:358–364; 2014. [DOI] [PubMed] [Google Scholar]
- 92. Wang Y.; Solt L. A.; Kojetin D. J.; Burris T. P. Regulation of p53 stability and apoptosis by a ROR agonist. PLoS One 7:e34921; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang A.; Zhou N.; Huang J.; Liu Q.; Fukuda K.; Ma D.; Lu Z.; Bai C.; Watabe K.; Mo Y. Y. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 23:340–350; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang Y.; Xu Z.; Jiang J.; Xu C.; Kang J.; Xiao L.; Wu M.; Xiong J.; Guo X.; Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 25:69–80; 2013. [DOI] [PubMed] [Google Scholar]
- 95. Hou P.; Zhao Y.; Li Z.; Yao R.; Ma M.; Gao Y.; Zhao L.; Zhang Y.; Huang B.; Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 5:e1287; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Eades G.; Wolfson B.; Zhang Y.; Li Q.; Yao Y.; Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol. Cancer Res. 13(2):330–338; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yuan J. H.; Yang F.; Wang F.; Ma J. Z.; Guo Y. J.; Tao Q. F.; Liu F.; Pan W.; Wang T. T.; Zhou C. C.; Wang S. B.; Wang Y. Z.; Yang Y.; Yang N.; Zhou W. P.; Yang G. S.; Sun S. H. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25:666–681; 2014. [DOI] [PubMed] [Google Scholar]
- 98. Li W.; Kang Y. A new Lnc in metastasis: Long noncoding RNA mediates the prometastatic functions of TGF-beta. Cancer Cell 25:557–559; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Poliseno L.; Salmena L.; Zhang J.; Carver B.; Haveman W. J.; Pandolfi P. P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465:1033–1038; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yu G.; Yao W.; Gumireddy K.; Li A.; Wang J.; Xiao W.; Chen K.; Xiao H.; Li H.; Tang K.; Ye Z.; Huang Q.; Xu H. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear cell renal cell carcinoma progression. Mol. Cancer Ther. 13(12):3086–3097; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Poliseno L.; Haimovic A.; Christos P. J.; Vega Y Saenz de Miera E. C.; Shapiro R.; Pavlick A.; Berman R. S.; Darvishian F.; Osman I. Deletion of PTENP1 pseudogene in human melanoma. J. Invest. Dermatol. 131:2497–2500; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yu G.; Yao W.; Gumireddy K.; Li A.; Wang J.; Xiao W.; Chen K.; Xiao H.; Li H.; Tang K.; Ye Z.; Huang Q.; Xu H. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol. Cancer Ther. 13:3086–3097; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Meng J.; Li P.; Zhang Q.; Yang Z.; Fu S. A four-long non-coding RNA signature in predicting breast cancer survival. J. Exp. Clin. Cancer Res. 33:84; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Grote P.; Wittler L.; Hendrix D.; Koch F.; Wahrisch S.; Beisaw A.; Macura K.; Blass G.; Kellis M.; Werber M.; Herrmann B. G. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24:206–214; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Xu T. P.; Huang M. D.; Xia R.; Liu X. X.; Sun M.; Yin L.; Chen W. M.; Han L.; Zhang E. B.; Kong R.; De W.; Shu Y. Q. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J. Hematol. Oncol. 7:63; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. LaFlamme B. GAPLINC and gastric cancer. Nat. Genet. 46:1159; 2014. [Google Scholar]
- 107. Hu Y.; Wang J.; Qian J.; Kong X.; Tang J.; Wang Y.; Chen H.; Hong J.; Zou W.; Chen Y.; Xu J.; Fang J. Y. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 74(23):6890–6902; 2014. [DOI] [PubMed] [Google Scholar]
- 108. Sun N. X.; Ye C.; Zhao Q.; Zhang Q.; Xu C.; Wang S. B.; Jin Z. J.; Sun S. H.; Wang F.; Li W. Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer. PLoS One 9:e100340; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109. Johnson S. M.; Grosshans H.; Shingara J.; Byrom M.; Jarvis R.; Cheng A.; Labourier E.; Reinert K. L.; Brown D.; Slack F. J. RAS is regulated by the let-7 microRNA family. Cell 120:635–647; 2005. [DOI] [PubMed] [Google Scholar]
- 110. Gao Y.; Wu F.; Zhou J.; Yan L.; Jurczak M. J.; Lee H. Y.; Yang L.; Mueller M.; Zhou X. B.; Dandolo L.; Szendroedi J.; Roden M.; Flannery C.; Taylor H.; Carmichael G. G.; Shulman G. I.; Huang Y. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 42(22):13799–13811; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kallen A. N.; Zhou X. B.; Xu J.; Qiao C.; Ma J.; Yan L.; Lu L.; Liu C.; Yi J. S.; Zhang H.; Min W.; Bennett A. M.; Gregory R. I.; Ding Y.; Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 52:101–112; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shen L.; Wan Z.; Ma Y.; Wu L.; Liu F.; Zang H.; Xin S. The clinical utility of microRNA-21 as novel biomarker for diagnosing human cancers. Tumour Biol. 36(3):1993–2005; 2015. [DOI] [PubMed] [Google Scholar]
- 113. Zhang Z.; Zhu Z.; Watabe K.; Zhang X.; Bai C.; Xu M.; Wu F.; Mo Y. Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 20:1558–1568; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Leucci E.; Patella F.; Waage J.; Holmstrom K.; Lindow M.; Porse B.; Kauppinen S.; Lund A. H. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 3:2535; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Smits G.; Mungall A. J.; Griffiths-Jones S.; Smith P.; Beury D.; Matthews L.; Rogers J.; Pask A. J.; Shaw G.; VandeBerg J. L.; McCarrey J. R.; SAVOIR Consortium; Renfree M. B.; Reik W.; Dunham I. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat. Genet. 40:971–976; 2008. [DOI] [PubMed] [Google Scholar]
- 116. Keniry A.; Oxley D.; Monnier P.; Kyba M.; Dandolo L.; Smits G.; Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 14:659–665; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Li H.; Yu B.; Li J.; Su L.; Yan M.; Zhu Z.; Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5:2318–2329; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhuang M.; Gao W.; Xu J.; Wang P.; Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 448:315–322; 2014. [DOI] [PubMed] [Google Scholar]
- 119. Xi S.; Yang M.; Tao Y.; Xu H.; Shan J.; Inchauste S.; Zhang M.; Mercedes L.; Hong J. A.; Rao M.; Schrump D. S. Cigarette smoke induces C/EBP-beta-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PLoS One 5:e13764; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang J.; Tsouko E.; Jonsson P.; Bergh J.; Hartman J.; Aydogdu E.; Williams C. miR-206 inhibits cell migration through direct targeting of the actin-binding protein Coronin 1C in triple-negative breast cancer. Mol. Oncol. 8:1690–1702; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Duan F. T.; Qian F.; Fang K.; Lin K. Y.; Wang W. T.; Chen Y. Q. miR-133b, a muscle-specific microRNA, is a novel prognostic marker that participates in the progression of human colorectal cancer via regulation of CXCR4 expression. Mol. Cancer 12:164; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Liz J.; Portela A.; Soler M.; Gomez A.; Ling H.; Michlewski G.; Calin G. A.; Guil S.; Esteller M. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol. Cell 55:138–147; 2014. [DOI] [PubMed] [Google Scholar]
- 123. Cesana M.; Cacchiarelli D.; Legnini I.; Santini T.; Sthandier O.; Chinappi M.; Tramontano A.; Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tinzl M.; Marberger M.; Horvath S.; Chypre C. DD3PCA3 RNA analysis in urine--A new perspective for detecting prostate cancer. Eur. Urol. 46:182–186; discussion 187; 2004. [DOI] [PubMed] [Google Scholar]
- 125. Hessels D.; Klein Gunnewiek J. M.; van Oort I.; Karthaus H. F.; van Leenders G. J.; van Balken B.; Kiemeney L. A.; Witjes J. A.; Schalken J. A. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44:8–15; discussion 15–16; 2003. [DOI] [PubMed] [Google Scholar]
- 126. Jung M.; Xu C.; Spethmann J.; Johannsen M.; Deger S.; Stephan C.; Loening S. A.; Jung K. Re: Hessels D.; Klein Gunnewiek J. M. T.; van Oort I.; Karthaus H. F. M.; van Leenders G. J. L.; van Balken B.; Kiemeney L. A.; Witjes J. A.; Schalken J. A. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44:8–16; 2003. [DOI] [PubMed] [Google Scholar]
- 127. Costa F. F. Non-coding RNAs and new opportunities for the private sector. Drug Discov. Today 14:446–452; 2009. [DOI] [PubMed] [Google Scholar]
- 128. Hung T.; Chang H. Y. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tsai M. C.; Spitale R. C.; Chang H. Y. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 71:3–7; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Morris K. V. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides 19:299–306; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Luo M.; Li Z.; Wang W.; Zeng Y.; Liu Z.; Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 280:1709–1716; 2013. [DOI] [PubMed] [Google Scholar]
- 132. Smaldone M. C.; Davies B. J. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr. Opin. Mol. Ther. 12:607–616; 2010. [PubMed] [Google Scholar]
- 133. Gofrit O. N.; Benjamin S.; Halachmi S.; Leibovitch I.; Dotan Z.; Lamm D. L.; Ehrlich N.; Yutkin V.; Ben-Am M.; Hochberg A. DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J. Urol. 191:1697–1702; 2014. [DOI] [PubMed] [Google Scholar]
- 134. Amit D.; Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J. Transl. Med. 8:134; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Amit D.; Tamir S.; Birman T.; Gofrit O. N.; Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of IGF2-P3 and IGF2-P4 regulatory sequences. Int. J. Clin. Exp. Med. 4:91–102; 2011. [PMC free article] [PubMed] [Google Scholar]
- 136. Scaiewicz V.; Sorin V.; Fellig Y.; Birman T.; Mizrahi A.; Galula J.; Abu-Lail R.; Shneider T.; Ohana P.; Buscail L.; Hochberg A.; Czerniak A. Use of H19 gene regulatory sequences in DNA-based therapy for pancreatic cancer. J. Oncol. 2010:178174; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Mizrahi A.; Czerniak A.; Levy T.; Amiur S.; Gallula J.; Matouk I.; Abu-lail R.; Sorin V.; Birman T.; de Groot N.; Hochberg A.; Ohana P. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 7:69; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Amit D.; Hochberg A. Development of targeted therapy for a broad spectrum of cancers (pancreatic cancer, ovarian cancer, glioblastoma and HCC) mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. Int. J. Clin. Exp. Med. 5:296–305; 2012. [PMC free article] [PubMed] [Google Scholar]