Abstract
Accumulating evidence has reported the significant role of miRNAs in the underlying biology of tumors, including breast cancer. The purpose for this study was to investigate the potential effects of miR-544a in breast cancer migration and invasion. The human normal breast Hs578Bst cells and the human breast cancer MCF-7 and MDA-MB-231 cells were used to analyze the expression of miR-544a by RT-PCR. The effects of miR-544a on the two kinds of breast cancer cell migration and invasion were analyzed using the Matrigel and Transwell assay, respectively. miR-544a expression on the cell metastasis-related protein expression was also analyzed using Western blotting. Compared to the normal Hs578Bst cells, miR-544a was significantly downregulated in MCF-7 cells but was upregulated in MDA-MB-231 cells (p < 0.01). The overexpressed miR-544a significantly promotes the migrated and invaded MCF-7 cells (p < 0.05), which was opposite to that in MCA-MB-231 cells (p < 0.05). Moreover, the cadherin 1 (CDH1) expression was negatively correlated to miR-544a expression in the two kinds of cells. Our study suggested that the overexpressed miR-544a may be a promoter for breast cancer migration and invasion by targeting CDH1.
Key words: Breast cancer, Cell migration, Cell invasion, miR-544a, Cadherin 1 (CDH1)
INTRODUCTION
Breast cancer remains as one of the most common malignancies among females and is becoming a leading cause for deaths worldwide (1,2). Previous papers report that a variety of factors were involved in the distant metastasis of breast cancer, which often occurs in the late stage (3). The mechanism for breast cancer metastasis is complicated, and the methods for curing breast cancer remain unsatisfactory due to the complicated mechanism (4,5). Therefore, to explore the potential mechanism for breast cancer metastasis and to develop several useful treatment methods for breast cancer is of great significance.
MicroRNAs (miRNAs) are some endogenous, highly conserved, noncoding RNAs 20 to 22 nt in length that function in a wide range of biological processes at the transcriptional or posttranscriptional level by targeting the 3′-UTR of genes (6). Various studies have demonstrated that miRNAs are involved in the progression, metastasis, and underlying biology of breast cancer via a diversity of mechanisms (7,8). For example, the upregulated miR-181a is a promoter of breast cancer metastasis (9), and miR-34a suppresses breast cancer metastasis through directly targeting Fra-1 (10,11). Recent evidence has revealed that miR-544a functions as a promoter for cancer metastasis, including lung cancer, by targeting cadherin 1 (CDH1), while miR-544a acts as an inducer for epithelial–mesenchymal transition via activating WNT signal in gastric cancer (12,13). Nevertheless, few have mentioned the potential effects of miR-544a in breast cancer metastasis.
In the current study, we analyzed the potential effects of miR-544a expression in breast cancer using MCF-7 and MDA-MB-231 cells. Comprehensive experimental methods were used to assess the effects of miR-544a expression on cell migration and invasion in the two kinds of breast cancer cells. This study aimed to investigate the possible effects of miR-544a in breast cancer metastasis and to reveal its possible mechanism.
MATERIALS AND METHODS
Cell Culture and Transfection
Human normal breast tissue Hs578Bst cells and human breast cancer MCF-7 and MDA-MBA-231 cells (obtained from American Type Culture Collection) were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 at 37°C.
The overexpressed vector for miR-544a or the silenced vector for miR-544a (Sangon Biotech, Shanghai, China) was transfected into the breast MCF-7 or MDA-MB-231 cells, respectively, using the Lipofectamine 2000 protocol (Life Technologies, USA). The scramble miR-544a was used as control.
Cell Migration Assay
For cell migration assay, Transwell assay was used as previously described (14). MCF-7 or MDA-MB-231 cells transfected with miR-544a or control plasmid (1 × 105) were seeded in the 24-well plates and grown overnight to confluence. The monolayer cells were scratched with a 20-ml pipette tip to create the wound. The floating cells were removed by washing twice with serum-free RPMI-1640 medium; after that, the medium was added to allow wound healing. The rate of wound closure was investigated using photographs 24 h later.
Cell Invasion Assay
Matrigel method was used to assess the cell invasion ability as previously described (15). Briefly, after 48 h of transfection, cells were incubated with serum-free RPMI-1640 medium containing 0.01% serum albumin (BSA; Sigma-Aldrich, St. Louis, MO, USA) for another 24 h. The upper layer of Transwell was enveloped with serum-free RPMI-1640 medium supplemented with 50 mg/L Matrigel and then air dried at 4°C. After being sucked out the medium, 50 µl fresh serum-free medium containing 10 g/L BSA was added and then cultured for 30 min at 37°C. After that, Transwell was put into the 24-well plates and cultured with RPMI-1640 medium mixed with 10% FBS. Then cells in Transwell were suspended with serum-free RPMI-1640 medium. After 48 h of incubation, Transwell was washed with PBS buffer to remove the upper cells on microporous membrane, followed with fixing in ice-cold alcohol. Then the Transwell was stained with 0.1% crystal violet for 30 min, and then decolored with 33% acetic acid. The absorbance of eluents was observed at OD 570 nm using a microplate reader (Biotech, USA). Transwell in the control group was treated without Matrigel.
Real-Time PCR
Total RNA from the cells collected at 48 h was isolated using TRIzol reagent (Invitrogen) as previously described (16) and was treated with RNase-free DNase I (Promega Biotech, USA). Consequently, the concentration and purity for the isolated RNA were measured with SMA 400 UV-VIS (Merinton, Shanghai, China). Purified RNA at a density of 0.5 µg/µl with nuclease-free water was used for cDNA synthesis with the PrimerScript 1st Strand cDNA Synthesis Kit (Invitrogen, USA). Expressions of targets in OVCAR-3 cells were detected in an Eppendorf Mastercycler (Brinkman Instruments, Westbury, NY, USA) using the SYBR ExScript RT-qPCR Kit (Takara, China). Melting curve analysis of amplification products was performed at the end of each PCR to confirm that only one product was amplified and detected. Phosphoglyceraldehyde dehydrogenase (GAPDH) was chosen as the internal control. Primers used for target amplification were CDH1 sense 5′-AGCTACCCCAGGACACCCAA-3′ and antisense 5′-GCAACGCAATCAGAGTCAACG-3′; GAPDH sense 5′-GGGTGGAGCCAAACGGGTC-3′ and antisense 5′-GGAGTTGCTGTTGAAGTCGCA-3′.
Western Blotting Analysis
Cells cultured for 48 h were lysed with Radioimmunoprecipitation buffer (RIPA; Sangon Biotech, China) containing phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich), and the lysates were then centrifuged at 12,000 rpm for 10 min at 4°C. The supernatants were collected, and protein concentrations were determined using a BCA protein assay kit (Pierce, Rockford, IL, USA). The proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (17) followed by transfer onto a polyvinylidinefluoride (PVDF) membrane (Millipore). The membranes were blocked in Tris-buffered saline/Tween 20 (TBST) containing 5% nonfat milk for 1 h at room temperature and then incubated with rabbit anti-human antibodies (CDH1, 1:100 dilution; Invitrogen) overnight at 4°C. Subsequently, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rat secondary antibody (1:1,000 dilution) for 1 h at room temperature. Finally, the PVDF membranes were washed three times with 1× TBST buffer for 10 min each. The signals were detected after the membranes were incubated with a chromogenic substrate using the enhanced chemiluminescence (ECL) method. GAPDH served as the internal control.
Statistical Analysis
All experiments were conducted independently three times. All the data were expressed as the mean ± SD. Statistical analysis was performed using the SPSS 19.0 statistical software, and the significant difference for all data was calculated using a one-way analysis of variance (ANOVA). The value of p < 0.05 was considered as statistically significant.
RESULTS
miR-544a Expression in Breast Cancer Cell Lines
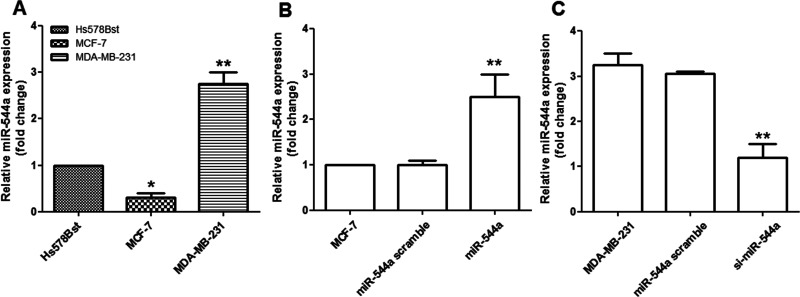
The results showed that miR-544a expression was significantly downregulated in MCF-7 cells (p < 0.05) but was significantly upregulated in MDA-MB-231 cells compared to normal Hs578Bst cells (p < 0.01) (Fig. 1A). Consequently, the overexpressed miR-544a vector or the si-miR-544a vector was transfected into the MCF-7 or MDA-MB-231 cells, respectively. The results showed that, after transfection, miR-544a expression was significantly upregulated compared to its control (p < 0.01) (Fig. 1B), while miR-544a expression was significantly downregulated by siRNA transfection in MDA-MB-231 cells (p < 0.01) (Fig. 1C).
Figure 1.
miR-544a expression on breast cancer cell lines. (A) Compared to the normal breast tissue Hs578Bst cells, miR-544a expression in MCF-7 cells was significantly decreased, while it was significantly increased in MDA-MB-231 cells. *p < 0.05 and **p < 0.01, compared to the control (Hs578Bst). (B) miR-544a expression was significantly increased by the overexpressed vector transfection in MCF-7 cells. **p < 0.01 compared to the control cells (MCF-7). (C) miR-544a expression was significantly increased by the silencing miR-544a in MDA-MB-231 cells. **p < 0.01 compared to the control cells (MDA-MB-231).
Effects of miR-544a Expression on Breast Cancer Cell Migration
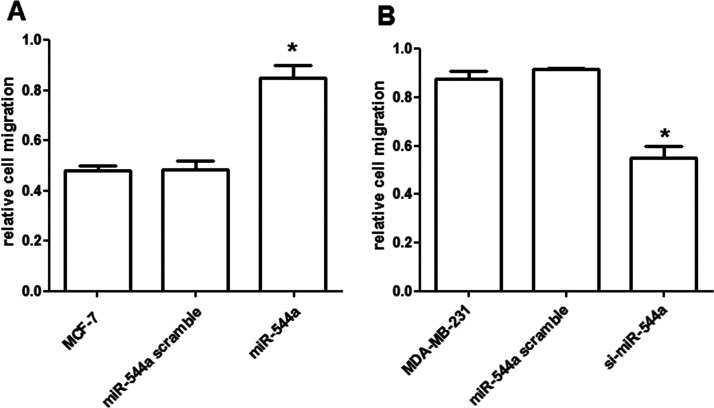
The results showed that the relative cell migration for MCF-7 cells was higher by the overexpressed miR-544a than that in its controls (p < 0.05) (Fig. 2A). On the contrary, the relative cell migration for MDA-MB-231 cells was lower by the silencing miR-544a than its controls (p < 0.05) (Fig. 2B).
Figure 2.
Effects of miR-544a expression on breast cancer cell migration. (A) The overexpressed miR-544a significantly promoted the MCF-7 cell migration. (B) The silencing of miR-544a significantly decreased the migrated MDA-MB-231 cells. *p < 0.05, compared to the control cells.
Effects of miR-544a Expression on Breast Cancer Cell Invasion
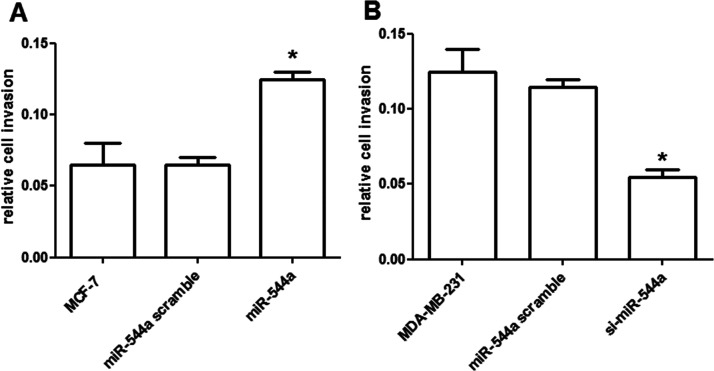
The results showed that the relative cell invasion for MCF-7 cells was higher by the overexpressed miR-544a than that in its controls (p < 0.05) (Fig. 3A). However, the relative cell invasion for MDA-MB-231 cells was lower by the silencing miR-544a than its controls (p < 0.05) (Fig. 3B).
Figure 3.
Effects of miR-544a expression on breast cancer cell invasion. (A) The overexpressed miR-544a significantly promoted the MCF-7 cell invasion. (B) The silencing of miR-544a significantly decreased the invaded MDA-MB-231 cells. *p < 0.05, compared to the control cells.
Effects of miR-544a Expression on Cell Metastasis-Related Protein Expression
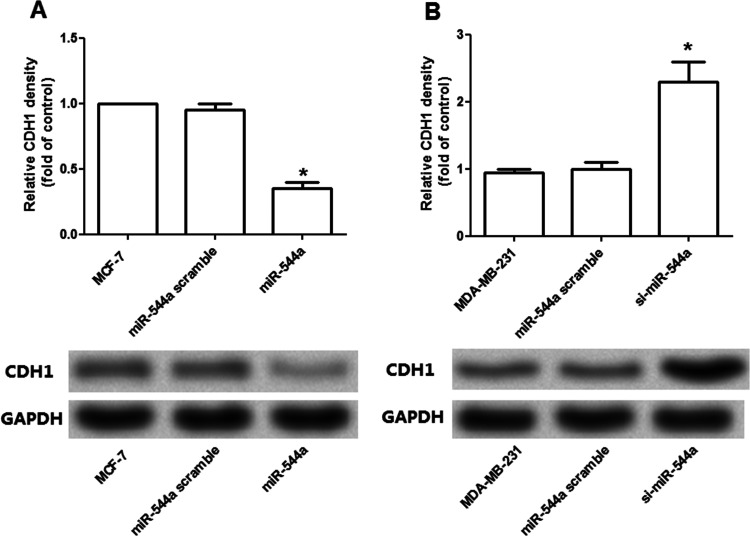
We further analyzed the potential effects of miR-544a expression on cell metastasis-related protein expression in the two kinds of breast cancer cells (Fig. 4). The results revealed that the mRNA and protein levels for CDH1 were downregulated by the overexpressed miR-544a in MCF-7 cells (p < 0.05) (Fig. 4A), but were upregulated by the silencing of miR-544a in MDA-MB-231 cells (p < 0.05) (Fig. 4B), suggesting that CDH1 expression was negatively correlated to miR-544a expression in breast cancer cells.
Figure 4.
Influence of miR-544a expression on cell metastasis-related protein expression. (A) The overexpressed miR-544a significantly decreased the mRNA and protein expression of CDH1 in MCF-7 cells. (B) The silencing of miR-544a significantly increased the mRNA and protein expression of CDH1 in MDA-MB-231 cells. *p < 0.05, compared to the control cells.
DISCUSSION
Breast cancer remains one of the most common female malignancies and is hard to cure because of its easy distant metastasis (2,11). Increasing evidence has revealed that miRNAs play pivotal roles in the biology of breast cancer migration and invasion (6,12). In this study, we investigated the potential effects of miR-544a in breast cancer metastasis and to develop its potential mechanism. The data showed that miR-544a was significantly downregulated in MCF-7 cells but was significantly overexpressed in MDA-MB-231 cells (p < 0.05). The silencing of miR-544a suppressed the MDA-MB-231 cell migration and invasion, which was opposite to that in MCF-7 cells. miR-544a expression was negatively correlated to the expression of CDH1 in the two kinds of breast cancer cells.
Previous evidence has demonstrated that MCF-7 is a low-metastatic cell, while MDA-MB-231 is a kind of high-metastatic cell (12), and Jiang et al. also proved that miR-544a was upregulated in insulinomas (18). In agreement with previous data, our results showed that miR-544a expression was highly expressed in MDA-MB-231 cells instead of in MCF-7 cells (Fig. 1), indicating that miR-544a expression was associated with a breast cancer pathogen. Therefore, we further analyzed the effects of miR-544a on breast cancer cell migration and invasion by silencing RNA transfection in MDA-MB-231 cell and overexpression in MCF-7 cells. It has been demonstrated that cell migration and invasion are the important biological processes in metastasis (19). Zhao et al. said that miR-544a was upregulated in lung cancer cell and might be a biomarker for lung cancer bone metastasis in its early diagnosis (20). In this study, the results revealed that miR-544a expression was negatively correlated with cell migration and invasion (Figs. 2 and 3), suggesting that the overexpressed miR-544a might play a role in promoting breast cancer metastasis.
Meanwhile, we further analyzed the possible mechanism for miR-544a in affecting breast cancer cell migration and invasion. CDH1 is a calcium-dependent cell–cell adhesion glycoprotein cadherin superfamily protein, which is involved in several biological processes in varieties of diseases including breast cancer, gastric cancer, and colorectal cancer (21–23). Loss of CDH1 function is thought to contribute to disease progression in cancer by increasing cell proliferation, invasion, and metastasis. For example, mutation in CDH1 is infrequent in women with familia lobular breast cancer (24). Similar investigations were also performed by Kuusisto et al., which revealed CDH1 mutation was observed in breast cancer (25). In this study, CDH1 expression was downregulated in breast cancer cells, which is in accordance with breast cancer metastasis. On the other side, the associate between miR-544a expression and CDH1 expression in breast cancer has not been fully mentioned. Nevertheless, miR-544a promotes lung cancer cell invasion via targeting CDH1 in vitro (12), which is similar to the report conducted by Hua et al. (26). Our results revealed that the CDH1 expression was significantly downregulated by the silencing of miR-544a, implying the negative correlation between the two factors. Therefore, we speculated that miR-544a may promote breast cancer metastasis by negatively regulating CDH1.
To sum up, the data presented in this study reveal that the overexpressed miR-544a may be a promoter in breast cancer metastasis through involvement in the migration and invasion biological processes and targeting CDH1. Our study may provide a theoretical basis for the possibility of miR-544a in illustrating the mechanism for breast cancer metastasis and may provide a basis for the potential application in the clinical treatment for breast cancer. Further experimental studies are still needed to develop the possible deep mechanism.
REFERENCES
- 1. Ly D.; Forman D.; Ferlay J.; Brinton L. A.; Cook M. B. An international comparison of male and female breast cancer incidence rates. Int. J. Cancer 132:1918–1926; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Youlden D. R.; Cramb S. M.; Cheng H. Y.; Baade P. D. Incidence and mortality of female breast cancer in the Asia-Pacific region. Cancer Biol. Med. 11:101–115; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marino N.; Woditschka S.; Reed L. T.; Nakayama J.; Mayer M.; Wetzel M.; Steeg P. S. Breast cancer metastasis. Am. J. Pathol. 183:1084–1095; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. José B.; Javier C.; Sung-Bae K.; Seock-Ah I.; Roberto H.; Young-Hyuck I.; Laslo R.; José Luiz P.; Tadeusz P.; Adam K. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med. 366:109–119; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldhirsch A.; Winer E. P.; Coates A. S.; Gelber R. D.; Piccart-Gebhart M.; Thürlimann B.; Senn H. J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 24:2206–2223; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: Therapeutic approaches based on metastasis-related miRNAs. J. Mol. Med. 89:445–457; 2011. [DOI] [PubMed] [Google Scholar]
- 7. Goh J. N.; Loo S. Y.; Datta A.; Siveen K. S.; Yap W. N.; Cai W.; Shin E. M.; Wang C.; Kim J. E.; Chan M. microRNAs in breast cancer: Regulatory roles governing the hallmarks of cancer. Biol. Rev. doi:10.1111/brv.12176; 2015. [DOI] [PubMed] [Google Scholar]
- 8. Mo Y. Y. Role of microRNAs in breast cancer. Cancer Biol. Ther. 14:201–212; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taylor M. A.; Khalid S. A.; Thompson C. L.; David D.; Schiemann W. P. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J. Clin. Invest. 123:150–163; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang S.; Li Y.; Gao J.; Zhang T.; Li S.; Luo A.; Chen H.; Ding F.; Wang X.; Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 32:4294–4303; 2013. [DOI] [PubMed] [Google Scholar]
- 11. Tang J.; Ahmad A.; Sarkar F. H. The role of microRNAs in breast cancer migration, invasion and metastasis. Int. J. Mol. Sci. 13:13414–13437; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mo X.; Zhang F.; Liang H.; Liu M.; Li H.; Xia H. miR-544a promotes the invasion of lung cancer cells by targeting cadherina 1 in vitro. Onco Targets Ther. 7:895–900; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yanaka Y.; Muramatsu T.; Uetake H.; Kozaki K. I.; Inazawa J. miR-544a induces epithelial–mesenchymal transition through the activation of WNT signaling pathway in gastric cancer. Carcinogenesis 36:1363–1371; 2015. [DOI] [PubMed] [Google Scholar]
- 14. Li Y.; Zhang M.; Chen H.; Dong Z.; Ganapathy V.; Thangaraju M.; Huang S. Ratio of miR-196s to HOXC8 mRNA correlates with breast cancer cell migration and metastasis. Cancer Res. 70:7894–7904; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adini A.; Fainaru O.; Udagawa T.; Connor K. M.; Folkman J.; D’Amato R. J. Matrigel cytometry: A novel method for quantifying angiogenesis in vivo. J. Immunol. Methods 342:78–81; 2009. [DOI] [PubMed] [Google Scholar]
- 16. Hummon A. B.; Lim S. R.; Difilippantonio M. J.; Ried T. Isolation and solubilization of proteins after TRIzol® extraction of RNA and DNA from patient material following prolonged storage. Biotechniques 42:467; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y.; Kuramitsu Y.; Takashima M.; Yokoyama Y.; Iizuka N.; Tamesa T.; Sakaida I.; Oka M.; Nakamura K. Identification of four isoforms of aldolase B down-regulated in hepatocellular carcinoma tissues by means of two-dimensional Western blotting. In Vivo 25:881–886; 2011. [PubMed] [Google Scholar]
- 18. Jiang X.; Shan A.; Su Y.; Cheng Y.; Gu W.; Wang W.; Ning G.; Cao Y. MiR-144/451 promote cell proliferation via targeting PTEN/AKT pathway in insulinomas. Endocrinology 156:2429–2439; 2015. [DOI] [PubMed] [Google Scholar]
- 19. Condeelis J.; Pollard J. W. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell 124:263–266; 2006. [DOI] [PubMed] [Google Scholar]
- 20. Zhao Q.; Ping L. I.; Junrong M. A.; Xijie Y. U. MicroRNAs in lung cancer and lung cancer bone metastases: Biomarkers for early diagnosis and targets for treatment. Recent Pat. Anticancer Drug Discov. 10:182–220; 2015. [DOI] [PubMed] [Google Scholar]
- 21. Pharoah P.; Guilford P.; Caldas C. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology 121:1348–1353; 2001. [DOI] [PubMed] [Google Scholar]
- 22. Haixin L.; Sjöberg-Margolin S.; Sima S.; Barbro W.; Eva J.; Kari H.; Annika L.; Igor V. CDH1 mutations are present in both ductal and lobular breast cancer, but promoter allelic variants show no detectable breast cancer risk. Int. J. Cancer 98:199–204; 2002. [DOI] [PubMed] [Google Scholar]
- 23. Zhang L.; Xiao A.; Ruggeri J.; Bacares R.; Somar J.; Melo S.; Figueiredo J.; Simões-Correia J.; Seruca R.; Shah M. A. The germline CDH1 c.48 G>C substitution contributes to cancer predisposition through generation of a pro-invasive mutation. Mutat. Res. 770:106–111; 2014. [DOI] [PubMed] [Google Scholar]
- 24. Schrader K. A.; Masciari S.; Boyd N.; Salamanca C.; Senz J.; Saunders D. N.; Yorida E.; Maines-Bandiera S.; Kaurah P.; Tung N. Germline mutations in CDH1 are infrequent in women with early-onset or familial lobular breast cancers. J. Med. Genet. 48:64–68; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuusisto K. M.; Bebel A.; Vihinen M.; Schleutker J.; Sallinen S. L. Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res. 13:R20–R20; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H.; Mo X.; Sun G. Impact of the transfection of miR-544a on the invasion and metastasis of lung cancer cells. Chin. J. Clin. Lab. Sci. 11; 2013. [Google Scholar]