Abstract
As a newly identified oncogenic long noncoding RNA (lncRNA), prostate cancer-associated transcript 6 (PCAT6) promoted cellular proliferation and colony formation of prostate cancer. However, the biological function of PCAT6 in lung cancer is still largely unknown. In this study, we found that PCAT6 is significantly increased in cancer tissues compared to normal tissues and positively correlates with metastasis of lung cancer in patients. We then examined PCAT6 expression in lung cancer cell lines and identified that PCAT6 expression was significantly elevated in lung cancer cells compared to normal human bronchial epithelial (NHBE) cells, especially in CL1-5 and H446 cells. PCAT6 knockdown significantly inhibited cellular proliferation and metastasis, as well as induced early apoptosis of lung cancer cells. Molecular analysis revealed that PCAT6 regulated the expression of two pivotal cancer-related proteins, c-Myc and p53, in lung cancer cells. However, PCAT6 was not directly combined with c-Myc and p53 as confirmed by RNA immunoprecipitation. Finally, a retrospective study further revealed that PCAT6 negatively correlates with overall survival of lung cancer patients. In conclusion, these results suggest that PCAT6 could play an oncogenic role in lung cancer progression and may serve as a biomarker for prognosis of lung cancer patients.
Key words: Long noncoding RNAs (lncRNAs), Prostate cancer-associated transcript 6 (PCAT6), Metastasis, c-Myc, p53, Lung cancer
INTRODUCTION
Long noncoding RNAs (lncRNAs) are mRNA-like transcripts ranging in length from 200 nucleotides (nt) to 100 kilobases (kb) and were initially considered as transcriptional noise due to the absence of protein-coding capacity (1). There are about 9,640 lncRNA loci in the human genome, and the number continues to grow (2). Although the functions of only a minority have been described, lncRNAs have emerged as an essential regulator in almost all aspects of biology (3–5), like differentiation (6,7), metabolism (8), embryogenesis (9), and cellular development (10). lncRNAs have been found to be involved in the regulation of gene expression through a variety of mechanisms, such as epigenetic modification, alternative splicing, nuclear import, as precursors of small RNAs, and even as regulators of mRNA modifiers or decoy elements (2,11–13). All of these have shed light on the promising future of the study of lncRNAs.
During the past decade, mounting evidence has confirmed that dysregulation of lncRNAs is involved in complex human diseases including cancer (14). Some lncRNAs have especially been found to be associated with tumor progression like invasion and metastasis and could even be used as biomarkers to predict patient prognosis (15–17). Among these lncRNAs, MALAT1 is overexpressed in several cancer types and promotes tumor progression by interacting with oncosuppressive miR-205 or enhancing Wnt signaling in renal cell carcinoma, glioblastoma, and lung cancer (18–20). lncRNA prostate cancer-associated transcript 6 (PCAT6) (also known as PCAN-R1, ncRNA-a2, and KDM5B-AS1) was first identified in keratinocytes and indirectly activates the Wnt–β catenin pathway by interacting with KLHL12 in cervical cancer cells (21). With the use of lncRNA microarray, PCAT6 was further confirmed as the most upregulated lncRNA in cancer tissues and significantly correlated with the metastasis of prostate cancer (22). Furthermore, PCAT6 enhanced cellular proliferation and colony formation of prostate cancer cells in an androgen-independent way (22). In lung cancer, PCAT6 was also found to be upregulated using Affymetrix HG-U133 Plus 2.0 Array with an lncRNA classification pipeline (23). However, no studies have been performed on the functional roles of PCAT6 in lung cancer.
As one of the most common human malignancies with an approximate 5-year survival rate less than 15% (24,25), lung cancer has become the main reason for cancer-related deaths in China, and during the past three decades, the mortality of this disease had increased by more than four times (26,27). Currently, a better understanding of the molecular pathogenesis that causes lung cancer has shown great potential for the development of new diagnostic and treatment strategies for lung cancer. Therefore, in this study, we first investigated the roles of PCAT6 on the proliferation and invasion of lung cancer cells.
We found that PCAT6 is significantly upregulated in cancer tissues compared with adjacent normal tissues and is positively correlated with the metastasis of lung cancer in patients. PCAT6 was also found to negatively correlate with overall survival of lung cancer patients with retrospective analysis. Cellular studies further confirmed that PCAT6 was overexpressed in lung cancer cell lines, especially in CL1-5 and H446 cells. PCAT6 knockdown significantly inhibited cellular proliferation and metastasis, as well as induced early apoptosis of these two lung cancer cell lines. Molecular analysis further revealed that PCAT6 regulated the expression of two pivotal cancer-related proteins, c-Myc and p53, in lung cancer cells. Thus, our data demonstrated an oncogenic role of PCAT6 in lung cancer progression, which may serve as a biomarker for prognosis evaluation of lung cancer patients.
MATERIALS AND METHODS
Participants and Tissue Samples
Fifty-eight biopsy specimens of lung cancer tissues and adjacent normal tissues were collected from the Department of Thoracic Surgery of Union Hospital (Wuhan, China). Tissue biopsy specimens were collected and immediately snap frozen in liquid nitrogen and stored at −80°C until use. All the patients with lung adenocarcinoma were proven by a clinical pathologist, and the patients’ characteristics are summarized in Table 1. The study was approved by the Ethical Review Board for Research of Union Hospital, affiliated to Tongji Medical College of Huazhong University of Science and Technology. All patients from whom these biopsy specimens were collected had signed a written informed consent before biological examination or surgery was performed.
Table 1.
Relationship Between PCAT6 Expression and Clinicopathological Parameters of Patients With Lung Cancer
| Characteristics | Number (N = 58) | Relative PCAT6 Expression | ||
|---|---|---|---|---|
| Low | High | p-Value (Chi-Square Test) | ||
| Age (years) | 1.00 | |||
| ≤60 | 26 | 14 | 12 | |
| >60 | 32 | 17 | 15 | |
| Gender | 0.7919 | |||
| Male | 32 | 18 | 14 | |
| Female | 26 | 13 | 13 | |
| Differentiation | 0.4342 | |||
| Well/moderate | 33 | 16 | 17 | |
| Poor | 25 | 15 | 10 | |
| Tumor size | 0.0343 | |||
| ≤3 cm | 39 | 20 | 9 | |
| >3 cm | 19 | 11 | 18 | |
| Smoking history | 0.5884 | |||
| Ever | 37 | 21 | 16 | |
| Never | 21 | 10 | 11 | |
| Lymph node metastasis | <0.0001 | |||
| Positive | 36 | 11 | 25 | |
| Negative | 22 | 20 | 2 | |
| TNM stage | 0.018 | |||
| I | 25 | 18 | 7 | |
| II/III/IV | 33 | 13 | 20 | |
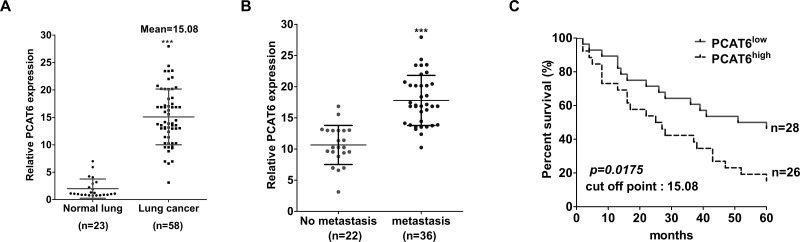
To discriminate patients with low or high PCAT6 expression, cutoff point value is 15.08.
Cell Culture
Normal human bronchial epithelial (NHBE) cells and human lung cancer cell lines H292, PC-9, CL1-5, H460, H1650, A549, H446, and H1975 were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, Shanghai Institute of Cell Biology. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, CA, USA) supplemented with 10% fetal bovine serum (Gibco), 2 mM l-glutamine (100×; Beyotime, Shanghai, China), 100 U/ml penicillin, and 100 µg/ml streptomycin (100×; Beyotime) and incubated at 37°C in humidified atmosphere containing 5% CO2.
RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from tissue specimens and cultured cells using TRIzol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. The isolated RNA was reverse transcribed to complementary DNA (cDNA) using First Strand cDNA Synthesis Kit (Toyobo, Osaka, Japan). Real-time polymerase chain reaction (PCR) was conducted with Power SYBR Green PCR Kit (Toyobo). The 2−ΔΔCt method was used to quantify the relative levels of lncRNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, and all reactions were performed in triplicate. The primer sequences were as follow: GAPDH, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); PCAT6, 5′-CAGGAACCCCCTCCTTACTC-3′ (forward) and 5′-CTAGG GATGTGTCCGAAGGA-3′ (reverse) (22).
Transfection
The small interfering RNA (siRNA) oligonucleotides were synthesized by Shanghai GenePharma Co. Ltd. (Shanghai, China), and the siRNA sequences of PCAT6 were as follows: si-NC (5′-GCGACCAA CGCCTTGATTG-3′), si-PCAT6 #1 (5′-GGTGTCTCCATCCTCATTC-3′), and si-PCAT6 #2 (5′-CTCCC AGACCTCACGTCAA-3′); si-NC was used as negative control (22). CL1-5 and H446 (4 × 105) cells were plated in six-well plates overnight and then transiently transfected with PCAT6-siRNA after cells reached 70–90% confluence using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The knockdown efficiency was determined by quantitative reverse transcription PCR (qRT-PCR) at 48 h after transfection.
Cell Proliferation Assay
CL1-5 and H446 (5 × 104) cells were transiently transfected with PCAT6-siRNA and then plated in a 24-well plate and allowed to grow for another 48 h. Cell proliferation was then determined every 24 h using CyQUANT® Cell Proliferation Assay Kit (Invitrogen) according to the manufacturer’s protocol. The fluorescence intensity was measured by using SpectraMax® i3x microplate reader (Molecular Devices, Sunnyvale, CA, USA).
RNA Immunoprecipitation
Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was used to conduct the RNA immunoprecipitation assay following the manufacturer’s instruction. The p53 and c-Myc antibodies (Santa Cruz Biotechnology, Dallas, TX, USA) used for RNA immunoprecipitation were purchased from Sixin Biotechnology Co. Ltd. (Shanghai, China). RT-PCR was used to detect the coprecipitation RNAs. Total RNAs and immunoglobulin G (IgG) isotype control were also detected to confirm that the detected signals were from RNAs that specifically bind to p53 and c-Myc.
Flow Cytometry
CL1-5 and H446 cells were transiently transfected with PCAT6-siRNA (4 × 105) and plated in six-well plates, and after a 48-h incubation the cultured cells were harvested by trypsinization. Cell apoptosis was evaluated using FITC Annexin-V Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). The cells were washed twice with cold phosphate-buffered saline (PBS) and then resuspended in 1× binding buffer at a concentration of 1.0 × 105 cells/100 µl. The cells were then stained with 5 µl of FITC Annexin-V and 5 µl of propidium iodide. Cells were gently vortexed and incubated for 15 min at 25°C in the dark. Four hundred microliters of 1× binding buffer was added to each tube and analyzed by flow cytometry.
Western Blotting
Proteins were extracted from cells with radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime) and quantified using a BCA Protein Assay Kit (Beyotime). An equivalent amount of protein was loaded on 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). The PVDF membranes were blocked with 5% nonfat milk for 1 h at 37°C and then incubated overnight at 4°C with diluted antibodies for c-Myc (1:1,000), p53 (1:1,000), Bcl-2 (1:500), Bax (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA), or GAPDH (1:5,000; Abcam, Cambridge, MA, USA). After being washed three times with TBST buffer, membranes were incubated with secondary horseradish peroxidase–goat anti-rabbit/mouse antibodies (1:10,000; Santa Cruz Biotechnology). Specific bands were detected using enhanced chemiluminescence (ECL) chromogenic substrate (Millipore) following the manufacturer’s instructions. GAPDH was the control.
Cell Invasion Assay
CL1-5 and H446 (4 × 105) cells were transiently transfected with PCAT6-siRNA and plated in six-well plates; 48 h later, cells were resuspended in serum-free media and seeded into upper Matrigel-precoated Transwell chamber (BD Biosciences). Medium containing 10% fetal bovine serum (FBS) was added in the lower chamber. After a 48-h incubation, the cells that had invaded another side of the membrane were fixed with methanol for 15 min and stained with 0.1% crystal violet for 20 min (Sigma-Aldrich). The number of invasive cells (five random fields) was captured and counted under the microscope.
Statistical Analysis
All data are presented as mean ± SD from at least three separate experiments and analyzed using the GraphPad Prism v.5.00 software (GraphPad Software, La Jolla, CA, USA). Comparison between two groups for statistical significance was performed with two-tailed Student’s t-test. For more groups, one-way analysis of variance (ANOVA) followed by Neuman–Keuls post hoc test was used. Any results with a value of p < 0.05 were considered to be statistically significant.
RESULTS
PCAT6 Is Upregulated in Lung Cancer Tissues and Related to Poor Prognosis
In order to investigate the role of PCAT6 in lung cancer progression, we first examined the expression of PCAT6 in lung cancer tissues and adjacent normal counterparts using qRT-PCR. Tissue from 58 patients was included in this study. The expression of PCAT6 was upregulated about 15 times in lung cancer tissues compared with adjacent normal tissues (p < 0.0001) (Fig. 1A). In addition, we further evaluated the correlation between PCAT6 expression and the patients’ clinicopathological characteristics (Table 1). PCAT6 expression in lung cancer was significantly correlated with tumor size (p = 0.0343), lymph node metastasis (p < 0.0001) (Fig. 1B), and TNM stage (p = 0.018). However, PCAT6 expression was not correlated with the rest of the characteristics, such as age (p = 1.00), gender (p = 0.7919), differentiation (p = 0.4342), or smoking history (p = 0.5884).
Figure 1.
Relative expression level of PCAT6 in lung cancer and adjacent normal tissues, and its relationship with clinicopathological characteristics. (A) qRT-PCR was used to examine the relative expression of PCAT6 in lung cancer tissues (n = 58) and adjacent normal tissues (n = 23) and normalized to GAPDH expression. (B) The relative expression level of PCAT6 was significantly higher in patients with lymph nodes metastasis. (C) Kaplan–Meier curve for survival analysis of lung cancer patients with high level and low level of PCAT6; four patients were excluded because they were without final information in their medical record. ***p < 0.0001.
To further validate whether PCAT6 was correlated with the patients with lung cancer, 58 patients included in this study were ranked according to the value of relative PCAT6 expression level and then divided into either the PCAT6high or PCAT6low group according to the mean value of relative PCAT6 expression (cutoff point = 15.08). A retrospective cohort study was performed by reviewing the patients’ medical record. Kaplan–Meier analysis using the log-rank test revealed that lung cancer patients with high PCAT6 expression had a significantly lower overall survival rate than those with low PCAT6 expression (15.385% vs. 46.429%, p = 0.0175) (Fig. 1C). Thus, these data indicated that PCAT6 may act as an oncogenic lncRNA in lung cancer progression and can be used as a prognostic marker for lung cancer patients.
Relative Expression of PCAT6 and its Role on Cell Growth in Lung Cancer Cell Lines
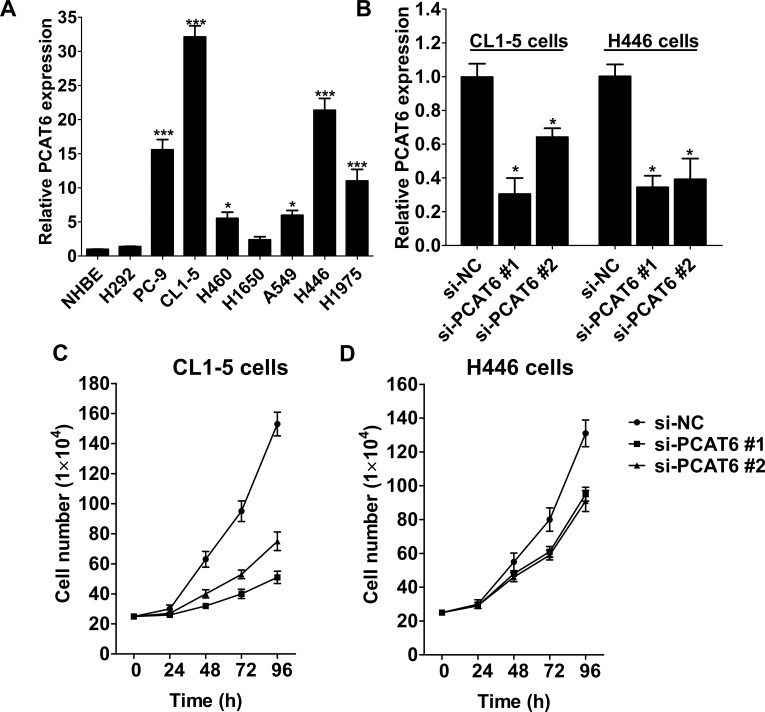
Next, to further validate the role of PCAT6 in lung cancer progression, we examined the PCAT6 expression in several lung cancer cell lines. Compared with NHBE cells, PCAT6 expression was significantly upregulated in most lung cancer cell lines, especially CL1-5 (lung adenocarcinoma) and H446 (small cell lung carcinoma) cells (Fig. 2A). We then employed two siRNAs to downregulate endogenous PCAT6 expression in lung cancer CL1-5 and H446 cells. qRT-PCR analysis revealed that si-PCAT6 #1 downregulated approximately 75% and 70% PCAT6 expression in CL1-5 and H466 cells, separately. si-PCAT6 #2 transfection decreased approximately 40% and 65% PCAT6 expression in CL1-5 and H466 cells compared with si-NC, separately (Fig. 2B). Then we evaluated the effect of PCAT6 knockdown on cellular proliferation of lung cancer cells using CyQUANT® Cell Proliferation Assay. As shown in Figure 2C and D, PCAT6 knockdown significantly suppressed the proliferation of CL1-5 and H446 cells when compared with the si-NC-transfected cells.
Figure 2.
PCAT6 expression in lung cancer cell lines and its effect in cell growth. (A) qRT-PCR was used to examine the relative expression of PCAT6 in lung cancer cell lines (H292, PC-9, CL1-5, H460, H1650, A549, H446, and H1975), and NHBE cells were used as control. (B) The expression of PCAT6 in CL1-5 and H446 cells transfected with si-NC or si-PCAT6 was measured by qRT-PCR. Growth curves of CL1-5 (C) and H446 (D) cells transfected with si-NC or si-PCAT6. Data are presented as mean ± SD, n = 3 independent experiments. *p < 0.05. ***p < 0.0001.
PCAT6 Knockdown Increased Early Apoptosis of Lung Cancer Cells
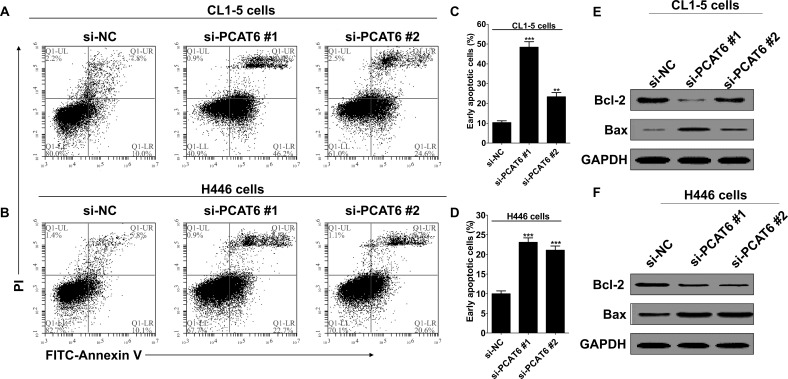
To study whether PCAT6 knockdown influence cell apoptosis, flow cytometry was used to analyze the cell apoptosis of lung cancer cells when transfected with si-PCAT6. The result indicated that PCAT6 knockdown significantly induced apoptosis of CL1-5 (Fig. 3A) and H446 cells (Fig. 3B), especially early apoptosis. Compared with the cells transfected with si-NC, early cell apoptosis increased approximately 38.1% and 13.0%, respectively, in CL1-5 cells when treated with si-PCAT6 #1 or si-PCAT6 #2 (Fig. 3C), and 13.1% and 11.1% in H446 cells (Fig. 3D). However, PCAT6 knockdown only showed a minor influence on late apoptosis, an average of 4% increase in two cell lines, and was independent of knockdown efficiency. In addition, we also noticed that cellular necrosis showed no difference with or without PCAT6 knockdown (Fig. 3A and B).
Figure 3.
PCAT6 knockdown promoted early cell apoptosis of lung cancer cells. CL1-5 and H446 cells were transfected with si-NC or si-PCAT6, respectively. (A and B) Apoptosis was determined by flow cytometry. Q1-UL, necrotic cells; Q1-UR, late apoptotic cells; Q1-LR, early apoptotic cells. (C and D) Histogram of percentage of early apoptotic cells, according to (A) and (B). Data are presented as mean ± SD, n = 3 independent experiments. **p < 0.01. ***p < 0.0001. (E, F) Western blotting was used to detect the protein expression of Bcl-2 and Bax; GAPDH was used as control.
Next, we detected Bcl-2 and Bax expression in lung cancer cells in response to PCAT6 knockdown. Bcl-2 is known to suppress apoptosis and enhance resistance to multifarious apoptosis-inducing factors. However, Bax, a homologous gene of the Bcl-2 family, could inhibit the antiapoptotic effect of Bcl-2 by forming a heterodimer with Bcl-2 (28,29). Western blotting analysis showed that PCAT6 knockdown decreased the protein expression of Bcl-2 and, in contrast, promoted the protein expression of Bax in CL1-5 (Fig. 3E) and H446 cells (Fig. 3F).
PCAT6 Knockdown Inhibits Cell Invasion of Lung Cancer Cells
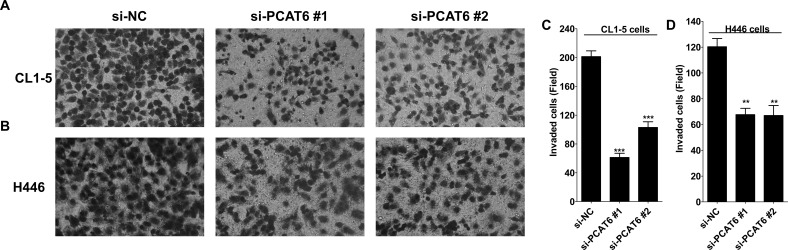
To explore whether PCAT6 was involved in regulating lung cancer cell invasion, Matrigel-based Transwell assay was performed to evaluate the influence of PCAT6 knockdown on lung cancer cell invasion. In CL1-5 cells, cell invasion was decreased about 75% and 50% with the transfection of si-PCAT6 #1 and si-PCAT6 #2, respectively (Fig. 4A and C). siPCAT6 #1 and si-PCAT6 #2 transfection suppressed a similar level of cell invasion in H446 cells, about 40% (Fig. 4B and D). Thus, these data indicate that PCAT6 promoted invasion of lung cancer cells.
Figure 4.
PCAT6 knockdown inhibited cell invasion of lung cancer cells. CL1-5 and H446 (4 × 105) cells transiently transfected with PCAT6-siRNA were plated in six-well plates; 48 h later, cells were resuspended in serum-free media and seeded into upper Matrigel-precoated Transwell chamber. Medium containing 10% FBS was added in the lower chamber. After 48 h of incubation, the cells that had invaded another side of the membrane were fixed with methanol for 15 min and stained with 0.1% crystal violet for 20 min. Five random fields were photographed (A, B), and the number of invasive cells per chamber was counted (C, D). Data are presented as mean ± SD. **p < 0.01. ***p < 0.0001.
Effect of PCAT6 on Downstream Effectors in Lung Cancer Cell Lines
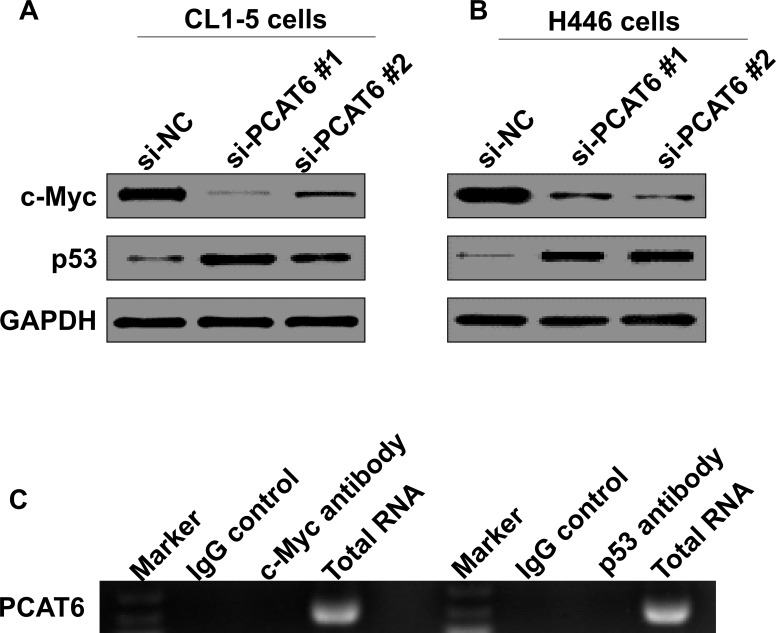
c-Myc and p53 are two pivotal proteins in regulating cancer progression, including proliferation, apoptosis, and metastasis (30–32). Furthermore, these two proteins could serve as the targets of tumor suppressive or oncogenic lncRNAs (33,34). We thus further determined whether PCAT6 regulated c-Myc and p53 expression in lung cancer cells. We noticed that PCAT6 knockdown significantly decreased c-Myc expression in both CL1-5 and H446 cells and enhanced p53 expression (Fig. 5A and B). Next, RNA immunoprecipitation was performed using antibodies against c-Myc or p53 from total cell proteins of CL1-5 cells to verify whether PCAT6 could directly bind with c-Myc or p53. However, we did not observe PCAT6 enrichment whether c-Myc or p53 antibodies were used (Fig. 5C). Thus, PCAT6 may indirectly regulate c-Myc and p53 expression in lung cancer cells.
Figure 5.
PCAT6 is not directly bound with c-Myc and p53. (A, B) Western blotting was used to detect c-Myc and p53 protein expression in CL1-5 and H446 cells transfected with si-NC or si-PCAT6 after 48 h. (C) RNA immunoprecipitation was performed using the p53 and c-Myc antibodies to immunoprecipitate PCAT6, and a primer was used to detect PCAT6.
DISCUSSION
With the advances of whole-genome and transcriptome-sequencing technologies, more and more lncRNAs are found to be involved in regulating cancer progression, either as oncogenes or as tumor suppressors, which further complicates the molecular pathogenesis of human cancers (35–39). Thus, deepening our understanding on how lncRNAs work is critical, not only from a mechanistic standpoint but also for the development of novel biomarkers and effective therapeutic targets for cancer patients.
As the leading cause of cancer-related deaths worldwide, lung cancer receives extensive attention, and numerous lncRNAs have been reported to participate in lung cancer progression, including HNF1A-AS1, MALAT1, H19, PCAT1 (38,40–45). In the present study, we first investigated the biological role of lncRNA PCAT6 in lung cancer progression and found that PCAT6 was significantly elevated in lung cancer tissues and was positively correlated with lung cancer metastasis. As a new identified lncRNA, PCAT6 is located in the chromosome 1 neighboring protein-coding gene KDM5B, thus named KDM5B-AS1 (22). Until now, there has been only one report found where PCAT6 expression was enhanced in prostate cancer and positively correlated with prostate cancer metastasis using Affymetrix Human Exon 1.0 ST Array and Affymetrix 3′ IVT arrays (22). In lung cancer, PCAT6 has been found to be upregulated in five published data sets from the Gene Expression Omnibus (GEO) with lncRNA expression profile analysis, namely, GSE27262, GSE19804, GSE19188, GSE30219, and GSE18842 (23), but its correlation with clinicopathological characteristics is still unclear. In this study, we found that PCAT6 was significantly associated with tumor size, TNM stage, and lymph node metastasis. Retrospective analysis further revealed that a high level of PCAT6 showed poor overall survival in patients with lung cancer. Therefore, these results suggest that PCAT6 may have the potential to be used as a prognostic marker of lung cancer patients.
In vitro studies have identified that PCAT6 promoted cellular proliferation and colony formation of prostate cancer cells (22). We then compared the expression level of PCAT6 in different lung cancer cell lines and further detected its role in regulating proliferation, apoptosis, and invasion of lung cancer cells. PCAT6 expression was relatively higher in CL1-5 (lung adenocarcinoma) and H446 (small cell lung carcinoma) cells than in other lung cancer cell lines. Two PCAT6-specific siRNAs (si-PCAT6 #1 and si-PCAT6 #2) were then employed to knock down PCAT6 expression in CL1-5 and H446 cells. Although the knockdown efficiency of si-PCAT6 #1 showed no significant difference in these two cell lines, CL1-5 cells seem to be more sensitive to si-PCAT6 treatment. PCAT6 knockdown with si-PCAT6 #1 induced a more apparent decrease of proliferation and invasion, and a more obvious increase of apoptosis in CL1-5 cells than in H446 cells. Thus, PCAT6 may function in different ways according to lung cancer subtype, and this needs to be clarified in future studies.
c-Myc and p53 are two pivotal proteins in regulating cancer progression, including proliferation, apoptosis, and metastasis (30–32). Furthermore, these two proteins could serve as the targets of tumor suppressive or oncogenic lncRNAs (33,34). In lung cancer cells, we found that PCAT6 knockdown significantly increased p53 protein expression and inhibited c-Myc in lung cancer cell lines (CL1-5 and H446), while PCAT6 was not directly bound with p53 or c-Myc protein, as identified by RNA immunoprecipitation. Thus, PCAT6 may regulate expression of these two proteins in an indirect pathway. Bcl-2 and Bax are two pivotal proteins involved in cellular apoptosis and could be regulated by p53 and c-Myc (46–48). PCAT6 knockdown significantly enhanced the expressive ratio of Bax/Bcl-2, which is usually elevated during cellular apoptosis. In the cellular apoptosis assay, we noticed that PCAT6 knockdown was more inclined to induce early apoptosis of these two lung cancer cell lines, although at different levels, but showed no significant effect on late apoptosis and necrosis. To answer why PCAT6 specifically inhibited early apoptosis of lung cancer cells, further studies are required to systematically evaluate the expression of apoptosis initiation-related genes with PCAT6 knockdown or overexpression.
In summary, our study first evaluated the biological function of PCAT6 in lung cancer progression including proliferation, apoptosis, and invasion and confirmed that PCAT6 is a novel potential oncogenic lncRNA in lung cancer. Combined with the results that high PCAT6 expression positively correlated with metastasis and showed poor overall survival in lung cancer patients, our studies suggest that PCAT6 may be a potential molecular therapeutic target or a prognostic marker for patients with lung cancer.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant No. 81000032).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Prensner J. R.; Chinnaiyan A. M. The emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang G.; Lu X.; Yuan L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta 1839:1097–1109; 2014. [DOI] [PubMed] [Google Scholar]
- 3. Spizzo R.; Almeida M. I.; Colombatti A.; Calin G. A. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene 31:4577–4587; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ponting C. P.; Oliver P. L.; Reik W. Evolution and functions of long noncoding RNAs. Cell 136:629–641; 2009. [DOI] [PubMed] [Google Scholar]
- 5. Mercer T. R.; Dinger M. E.; Mattick J. S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 10:155–159; 2009. [DOI] [PubMed] [Google Scholar]
- 6. Wang P.; Xue Y.; Han Y.; Lin L.; Wu C.; Xu S.; Jiang Z.; Xu J.; Liu Q.; Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344:310–313; 2014. [DOI] [PubMed] [Google Scholar]
- 7. Kretz M.; Siprashvili Z.; Chu C.; Webster D. E.; Zehnder A.; Qu K.; Lee C. S.; Flockhart R. J.; Groff A. F.; Chow J.; Johnston D.; Kim G. E.; Spitale R. C.; Flynn R. A.; Zheng G. X.; Aiyer S.; Raj A.; Rinn J. L.; Chang H. Y.; Khavari P. A. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493:231–235; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hung C. L.; Wang L. Y.; Yu Y. L.; Chen H. W.; Srivastava S.; Petrovics G.; Kung H. J. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA 111:18697–18702; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pauli A.; Rinn J. L.; Schier A. F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12:136–149; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guttman M.; Donaghey J.; Carey B. W.; Garber M.; Grenier J. K.; Munson G.; Young G.; Lucas A. B.; Ach R.; Bruhn L.; Yang X.; Amit I.; Meissner A.; Regev A.; Rinn J. L.; Root D. E.; Lander E. S. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477:295–300; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawaguchi T.; Tanigawa A.; Naganuma T.; Ohkawa Y.; Souquere S.; Pierron G.; Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. USA 112:4304–4309; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhuang W.; Ge X.; Yang S.; Huang M.; Zhuang W.; Chen P.; Zhang X.; Fu J.; Qu J.; Li B. Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells 33:1985–1997; 2015. [DOI] [PubMed] [Google Scholar]
- 13. Gibb E. A.; Brown C. J.; Lam W. L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 10:38; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lalevee S.; Feil R. Long noncoding RNAs in human disease: Emerging mechanisms and therapeutic strategies. Epigenomics 7:877–879; 2015. [DOI] [PubMed] [Google Scholar]
- 15. Su X.; Malouf G. G.; Chen Y.; Zhang J.; Yao H.; Valero V.; Weinstein J. N.; Spano J. P.; Meric-Bernstam F.; Khayat D.; Esteva F. J. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget 5:9864–9876; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan J. H.; Yang F.; Wang F.; Ma J. Z.; Guo Y. J.; Tao Q. F.; Liu F.; Pan W.; Wang T. T.; Zhou C. C.; Wang S. B.; Wang Y. Z.; Yang Y.; Yang N.; Zhou W. P.; Yang G. S.; Sun S. H. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25:666–681; 2014. [DOI] [PubMed] [Google Scholar]
- 17. Li H.; Yu B.; Li J.; Su L.; Yan M.; Zhu Z.; Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5:2318–2329; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirata H.; Hinoda Y.; Shahryari V.; Deng G.; Nakajima K.; Tabatabai Z. L.; Ishii N.; Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 75:1322–1331; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vassallo I.; Zinn P.; Lai M.; Rajakannu P.; Hamou M. F.; Hegi M. E. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene 35:12–21; 2016. [DOI] [PubMed] [Google Scholar]
- 20. Ji P.; Diederichs S.; Wang W.; Boing S.; Metzger R.; Schneider P. M.; Tidow N.; Brandt B.; Buerger H.; Bulk E.; Thomas M.; Berdel W. E.; Serve H.; Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22:8031–8041; 2003. [DOI] [PubMed] [Google Scholar]
- 21. Orom U. A.; Derrien T.; Beringer M.; Gumireddy K.; Gardini A.; Bussotti G.; Lai F.; Zytnicki M.; Notredame C.; Huang Q.; Guigo R.; Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Du Z.; Fei T.; Verhaak R. G.; Su Z.; Zhang Y.; Brown M.; Chen Y.; Liu X. S. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20:908–913; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang J.; Lin J.; Liu T.; Chen T.; Pan S.; Huang W.; Li S. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer 85:110–115; 2014. [DOI] [PubMed] [Google Scholar]
- 24. Siegel R. L.; Miller K. D.; Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 65:5–29; 2015. [DOI] [PubMed] [Google Scholar]
- 25. Torre L. A.; Bray F.; Siegel R. L.; Ferlay J.; Lortet-Tieulent J.; Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 65:87–108; 2015. [DOI] [PubMed] [Google Scholar]
- 26. Zhou M.; Guo M.; He D.; Wang X.; Cui Y.; Yang H.; Hao D.; Sun J. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J. Transl. Med. 13:231; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. She J.; Yang P.; Hong Q.; Bai C. Lung cancer in China: Challenges and interventions. Chest 143:1117–1126; 2013. [DOI] [PubMed] [Google Scholar]
- 28. Osorio L. M.; Jondal M.; Aguilar-Santelises M. Regulation of B-CLL apoptosis through membrane receptors and Bcl-2 family proteins. Leuk. Lymphoma 30:247–256; 1998. [DOI] [PubMed] [Google Scholar]
- 29. Podhorecka M.; Halicka D.; Klimek P.; Kowal M.; Chocholska S.; Dmoszynska A. Simvastatin and purine analogs have a synergic effect on apoptosis of chronic lymphocytic leukemia cells. Ann. Hematol. 89:1115–1124; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wade M.; Li Y. C.; Wahl G. M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13:83–96; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S. P.; Wang W. L.; Chang Y. L.; Wu C. T.; Chao Y. C.; Kao S. H.; Yuan A.; Lin C. W.; Yang S. C.; Chan W. K.; Li K. C.; Hong T. M.; Yang P. C. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat. Cell Biol. 11:694–704; 2009. [DOI] [PubMed] [Google Scholar]
- 32. Kong L. M.; Liao C. G.; Zhang Y.; Xu J.; Li Y.; Huang W.; Zhang Y.; Bian H.; Chen Z. N. A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 74:3764–3778; 2014. [DOI] [PubMed] [Google Scholar]
- 33. Huarte M.; Guttman M.; Feldser D.; Garber M.; Koziol M. J.; Kenzelmann-Broz D.; Khalil A. M.; Zuk O.; Amit I.; Rabani M.; Attardi L. D.; Regev A.; Lander E. S.; Jacks T.; Rinn J. L. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142:409–419; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang F.; Xue X.; Zheng L.; Bi J.; Zhou Y.; Zhi K.; Gu Y.; Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 281:802–813; 2014. [DOI] [PubMed] [Google Scholar]
- 35. Qiu M. T.; Hu J. W.; Yin R.; Xu L. Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol. 34:613–620; 2013. [DOI] [PubMed] [Google Scholar]
- 36. Yang F.; Bi J.; Xue X.; Zheng L.; Zhi K.; Hua J.; Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 279:3159–3165; 2012. [DOI] [PubMed] [Google Scholar]
- 37. Qiu M.; Xu Y.; Wang J.; Zhang E.; Sun M.; Zheng Y.; Li M.; Xia W.; Feng D.; Yin R.; Xu L. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 6:e1858; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Y.; Liu H.; Shi X.; Yao Y.; Yang W.; Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget 6:9160–9172; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fatima R.; Akhade V. S.; Pal D.; Rao S. M. Long noncoding RNAs in development and cancer: Potential biomarkers and therapeutic targets. Mol. Cell. Ther. 3:5; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zequn N.; Xuemei Z.; Wei L.; Zongjuan M.; Yujie Z.; Yanli H.; Yuping Z.; Xia M.; Wei W.; Wenjing D.; Na F.; Shuanying Y. The role and potential mechanisms of LncRNA-TATDN1 on metastasis and invasion of non-small cell lung cancer. Oncotarget; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loewen G.; Jayawickramarajah J.; Zhuo Y.; Shan B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 7:90; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang E.; Li W.; Yin D.; De W.; Zhu L.; Sun S.; Han L. Erratum to: c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour Biol. 37(4):5653; 2016. [DOI] [PubMed] [Google Scholar]
- 43. Zhang E. B.; Yin D. D.; Sun M.; Kong R.; Liu X. H.; You L. H.; Han L.; Xia R.; Wang K. M.; Yang J. S.; De W.; Shu Y. Q.; Wang Z. X. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 5:e1243; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nie F. Q.; Zhu Q.; Xu T. P.; Zou Y. F.; Xie M.; Sun M.; Xia R.; Lu K. H. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 35:7587–7594; 2014. [DOI] [PubMed] [Google Scholar]
- 45. Khandelwal A.; Bacolla A.; Vasquez K. M.; Jain A. Long non-coding RNA: A new paradigm for lung cancer. Mol. Carcinog. 54:1235–1251; 2015. [DOI] [PubMed] [Google Scholar]
- 46. Miyashita T.; Krajewski S.; Krajewska M.; Wang H. G.; Lin H. K.; Liebermann D. A.; Hoffman B.; Reed J. C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9:1799–1805; 1994. [PubMed] [Google Scholar]
- 47. Jin Z.; May W. S.; Gao F.; Flagg T.; Deng X. Bcl2 suppresses DNA repair by enhancing c-Myc transcriptional activity. J. Biol. Chem. 281:14446–14456; 2006. [DOI] [PubMed] [Google Scholar]
- 48. Cao X.; Bennett R. L.; May W. S. c-Myc and caspase-2 are involved in activating Bax during cytotoxic drug-induced apoptosis. J. Biol. Chem. 283:14490–14496; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]